Abstract
The highly reproducible inheritance of chromosomes during mitosis in mammalian cells involves nuclear envelope breakdown, increased chromatin compaction, loss of long-range intrachromosomal interactions, loss of enhancer–promoter proximity, displacement of many transcription regulators from the chromatin and a marked decrease in RNA synthesis. Despite these dramatic changes in the mother cell, daughter cells are able to faithfully re-establish the parental chromatin and gene expression features characteristic of the cell type. Pioneering studies of mitotic chromatin signatures showed that despite global repression of transcription, the Hsp70 gene promoter retains an open chromatin conformation, which was proposed to allow the reactivation of the Hsp70 gene upon completion of mitosis — a phenomenon termed mitotic bookmarking. It was later shown that various cell-type-specific transcription factors, such as GATA-binding factor 1 (GATA1) in erythroblasts and forkhead box protein A1 (FOXA1) in hepatocytes, remain bound at a subset of their interphase binding sites in mitosis. Such bookmarking transcription factors remain on chromosomes in mitosis and have been shown to enable a subset of genes to be reactivated in a timely fashion upon mitotic exit. In addition, sensitive new methods to measure transcription revealed that mitotic cells retain residual transcription at a large number of genes. Furthermore, genes recover their interphase level of transcription in distinct waves. Thus, gene expression is precisely regulated as cells pass through mitosis to ensure faithful propagation of cell identity and function through cellular generations.
Early studies of gene expression in living cells indicated that when cells divide, RNA polymerase II (Pol II) is less abundant in mitotic chromosomes and that production of nascent transcripts is greatly reduced or halts altogether. This dramatic reduction in global RNA synthesis during mitosis is concordant with major changes in chromosome architecture, including breakdown of the nuclear envelope, chromosome condensation and loss of the long-range interactions that facilitate interactions between distal enhancers and promoters, which are generally important for transcription1,2. Furthermore, many transcription factors are undetectable in mitotic chromosomes3 and phosphorylation events inhibit transcription machinery, including Pol II (through phosphorylation of its subunit Gdown1) and zinc-finger transcription factors4,5. Hence, mitotic cells have been typically considered to be transcriptionally silent. Nevertheless, dividing cells possess mechanisms that ensure transcriptional memory propagation from mother to daughter cells.
Transcriptional memory is essential for multicellular organisms as it ensures maintenance of cellular identity across cellular generations. Two distinct models were proposed with regard to how a transcriptome of a cell, defined as the pattern and amplitude of gene expression in a particular cell type, is properly reconstituted during exit from mitosis. The neutral model held that during mitotic exit, transcription factors — guided by residual histone modifications — would re-bind their target genes en masse, leading to globally synchronized induction of gene expression. A more deterministic model held that the subset of transcription factors exhibit mitotic chromosome binding and act as ‘bookmarking’ factors during mitosis, enabling the proper activation of genes during mitotic exit. However, to date, functional data on bookmarking factors can explain only a fraction of the reconstitution of the transcriptome in daughter cells. New, highly sensitive techniques have recently yielded a third model, which holds that many if not most genes are not transcriptionally silent during mitosis but instead are transcribed at a low level that may be primarily maintained by local control of promoter architecture. Furthermore, the new techniques show that during mitotic exit, genes are expressed in distinguishable temporal waves, thereby excluding the model in which genes would be synchronously reactivated en masse and providing strong evidence for a form of epigenetic control of gene expression.
In this Review, we focus on the latest findings regarding the inheritance of gene expression patterns through mitosis. We start with a brief overview of mitotic bookmarking by transcription factors and histone modifications. We refer the reader to other recent reviews on mitotic bookmarking6-10 and here focus primarily on Pol II targeting to mitotic chromatin and the evidence of nascent transcription in mitotic cells. These latest data change our understanding of transcriptional memory maintenance in dividing cells from a simple off–on model, with transcription factor binding and unbinding kinetics as a switch, to a model focusing on precise modulation of gene regulatory regions and interactions between them in mitotic cells, which fine-tunes the kinetics of gene reactivation after mitotic exit.
Mitotic bookmarking
Early studies of transcription factor binding in mitosis led to a conclusion that a subset of factors are retained in mitotic chromatin3 and that genes that are active in interphase can also maintain features of transcriptionally active chromatin in mitosis11. These findings gave rise to the concept of bookmarking as a mitotic epigenetic mark that enables the proper re-expression of active genes postmitosis (FIG. 1). The two mechanisms thought to be involved in bookmarking, from these early observations, were transcription factors and histone post-translational modifications.
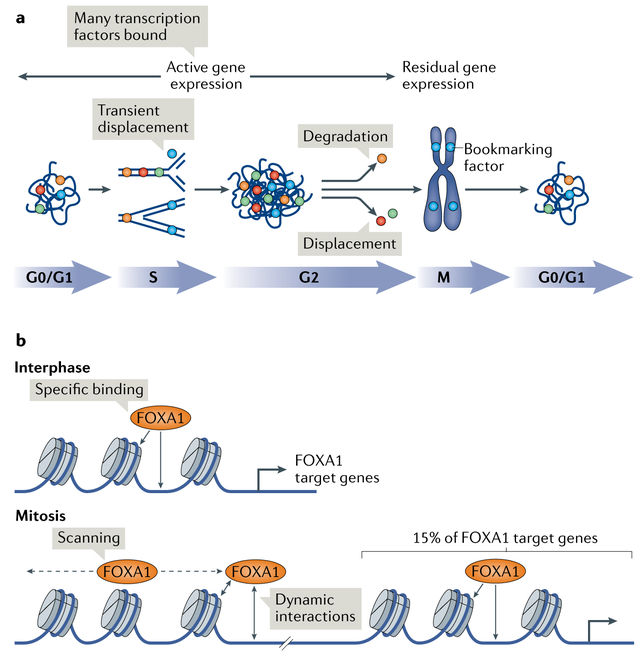
Fig. 1 ∣. Mitotic bookmarking by transcription factors.
a ∣ In interphase (G0/G1, S and G2), transcription factors robustly bind to chromatin to promote high levels of gene expression. In the mitotic phase of the cell cycle (M), a fraction of transcription factors, termed bookmarking transcription factors (blue circles), remain associated with the chromosomes, which exhibit low, residual gene expression, whereas non-bookmarking factors are excluded either by displacement (red and green circles) or through selective degradation (orange circles). Upon mitotic exit, when cells enter G1 phase or G0 (quiescence phase), factors that were displaced in mitosis re-bind as the chromosomes decondense. At the same time, factors that were degraded can be re-expressed and associate with chromatin. All transcription factors are transiently displaced from the DNA during replication in S phase to allow the progression of replication forks. In G2 phase, the active chromatin is temporarily accessible to all factors until another round of mitosis. b ∣ In interphase, bookmarking transcription factors, such as forkhead box protein A1 (FOXA1), display specific binding to chromatin to promote the expression of their target genes. By contrast, in mitosis, these factors associate with the chromatin more loosely and dynamically and scan the chromatin. This scanning may promote tight association of these factors to specific gene regulatory regions upon mitotic exit. In addition, a proportion of bookmarking transcription factors can bind specifically to chromatin also in mitosis.
Mitotic bookmarking by transcription factors.
The first class of mitotic bookmarking factors to be identified was CCAAT/enhancer-binding protein (C/EBP), guanylate-binding protein (GBP) and heat shock factor protein 1 (HSF1), which, by in vivo footprinting, exhibited conserved DNA binding patterns during mitosis at the heat shock protein 70 (Hsp70) promoter3. Later, the mitotic binding of HSF2 was demonstrated to be necessary for the proper postmitotic expression of the Hsp70 gene12,13. Subsequently, general transcription factors, such as transcription factor IID (TFIID), transcription initiation factor IIB (TFIIB) and TATA-box binding protein (TBP), were shown to remain associated with the mitotic chromatin in different cell types13-16. Notably, TFIID, TFIIB and TBP are all promoter transcription factors.
An increasing number of tissue-specific and architectural transcription factors have been identified as potential bookmarking factors on the basis of their retention in mitotic chromatin (FIG. 1), including Runt-related transcription factor 2 (RUNX2), GATA-binding factor 1 (GATA1), forkhead box protein A1 (FOXA1), hepatocyte nuclear factor 1β (HNF1β), oestrogen-related receptor-β (ESRRβ), octamer-binding protein 4 (OCT4; also known as POU5F1), MYC proto-oncogene, BHLH transcription factors, SRY box 2 (SOX2), CCCTC-binding factor (CTCF), p300, bromodomain-containing protein 4 (BRD4) and myeloid/lymphoid or mixed-lineage leukaemia protein 1 (MLL; also known as KMT2A)17-30 (TABLE 1). Of note, various studies indicate that fixation artefacts associated with cell visualization by immunofluorescence could potentially be responsible for the underestimation of the number of transcription factors associated with mitotic chromatin21,23,31. Thus, the dogma stating that tissue-specific transcription factors are typically excluded from mitotic chromatin has to be re-explored. The use of systematic alternative fixation protocols and live imaging of fusion proteins with GFP or HALO-tag are helping to clarify this aspect of mitotic bookmarking21,32.
Table 1 ∣.
Examples of mitotic bookmarking factors
| Bookmarking factor | System | Role (specific function) | Refs |
|---|---|---|---|
| TFIID, TFIIB and TBP | HeLa (human, cervical cancer) | General transcription factors | 13-16 |
| CTCF | HeLa (human, cervical cancer) | Insulator | 28 |
| FOXI1 | PAC2 (zebrafish, fibroblasts) | Forkhead transcription factor (development of cochlea and vestibulum) | 79 |
| RUNX2 | Saos-2 (human, osteosarcoma) | Runt transcription factor (osteoblastic differentiation and skeletal morphogenesis) | 80 |
| BRD4 | NIH3T3 (mouse, fibroblasts) | Chromatin reader | 26 |
| MLL | HeLa (human, cervical cancer) | Transcription co-activator | 24 |
| GATA1 | G1E (mouse, erythroid precursors) | Transcription factor (erythroid development) | 18 |
| FOXA1 | HUH7 (human, hepatocarcinoma) | Forkhead transcription factor (liver development) | 19 |
| RBPJ | F9 (mouse, testis) | Transcription regulator (regulation of Notch signalling pathway) | 81 |
| PARP1 | HEK293 (mouse, kidney) | Poly(ADP-ribosyl)transferase | 82-84 |
| ORC1 | U2OS (human, osteocarcinoma) | Origin of replication complex protein | 85 |
| RING1 and BMI1 | HeLa (human, cervical cancer) | Transcription repressors of the Polycomb group complex 1 | 86 |
| HNF1β | IMCD3 (mouse, kidney), MDCK (dog, kidney) and mouse kidney | POU-homeobox transcription factor (kidney and pancreas development) | 20,21 |
| ESRRβ, SOX2, OCT4 and KLF4 | Embryonic stem cells | Pluripotency transcription factors | 2,23,27,37 |
BMI1, Polycomb repressive complex; BRD4, bromodomain-containing protein 4; CTCF, CCCTC-binding factor; ESRRβ, oestrogen-related receptor-β; FOXA1, forkhead box protein A1; GATA1, GATA-binding protein 1; HNF1β, hepatocyte nuclear factor 1β; KLF4, Kruppel-like factor 4; MLL, myeloid/lymphoid or mixed-lineage leukaemia protein 1; OCT4, octamer-binding protein 4; ORC1, origin recognition complex subunit 1; PARP1, poly(ADP-ribose) polymerase 1; RBPJ, recombining binding protein suppressor of hairless; RING1, RING finger protein 1; RUNX2, Runt-related transcription factor 2; SOX2, SRY box 2; TBP, TATA-box binding protein; TFIID, transcription factor IID.
How bookmarking factors, and not other transcription factors, remain associated with mitotic chromatin remains unclear. As expected, DNA binding is necessary for bookmarking factors to remain bound to mitotic chromatin19,21-23. Fluorescence recovery after photo-bleaching (FRAP) and single-particle tracking studies indicate that various bookmarking transcription factors are dynamically interacting with and scanning chromatin during mitosis, whereas the general transcription factor TBP is more stably bound to promoters of genes that are active in interphase19,22,23,27,32. Point mutations that affect specific DNA binding by the bookmarking factor FOXA1, but allow nonspecific DNA binding, still permitted efficient retention of the factor on mitotic chromosomes in living cells. However, mutations of nonspecific DNA binding markedly reduced mitotic chromosome binding19. This nonspecific DNA binding has been postulated to ‘store’ the factor in the vicinity of chromatin and to give the factor a priority over other factors in scanning the genome during mitotic exit to efficiently activate gene expression, analogous to the role of FOXA1 as a pioneer factor in embryonic development10. It will be interesting to determine whether the nonspecific DNA binding mode on mitotic chromatin is a more general property of bookmarking transcription factors.
Despite the global decrease of gene expression during mitosis, mitotic chromatin remains accessible to nucleases and thus does not exhibit the extreme compaction that it was previously thought to have23,33-35. It is possible that the dynamic movement of bookmarking transcription factors in mitosis has roles beyond transcription and may contribute to the structural properties of mitotic chromosomes, which are globally condensed yet lack local long-range interactions, such as topologically associated domains (TADs), and remain accessible to nucleases.
Which genetic loci are bound by gene-specific bookmarking factors and which are not? A general finding from genome-wide mapping studies (for example, for GATA1 in erythroid cells18, FOXA1 in liver cells19 and ESRRβ in embryonic stem cells22) shows that only a subset of sites bound in interphase remain bound by bookmarking transcription factors during mitosis. Thus, for FOXA1, the factor transitions from a low nuclear mobility state, with highly specific binding in interphase, to a high-mobility, low-specificity state in mitosis. Nevertheless, it is important to note that 15% of interphase targets of FOXA1 are still specifically occupied by this transcription factor in mitosis19 (FIG. 1b). ESRRβ, a transcription factor regulating pluripotency in embryonic stem cells, binds preferentially in mitosis near subsets of genes expressed in early G1, nicely demonstrating an example of a bookmarking factor helping to reactivate genes during mitotic exit22.
The functional relevance of the mitotically retained transcription factors has been studied by genetic impairment in mitosis, with observed defects in the levels20 or timing18,19,22,32 of the postmitotic re-expression of the bookmarked genes. However, as noted by the authors of these studies, it is difficult to discern whether the effects are due to the impairment of bookmarking in mitosis or transcription factor re-binding during early G1.
Preservation of histone modifications through mitosis.
Another level of bookmarking may lie in the retention of histone post-translational modifications in mitosis, including both activating and repressive histone marks, which are believed to instruct the daughter cell as to which genes were expressed or silenced in the parental cell and thus which should be reactivated or remain repressed after completion of cell division (FIG. 2).
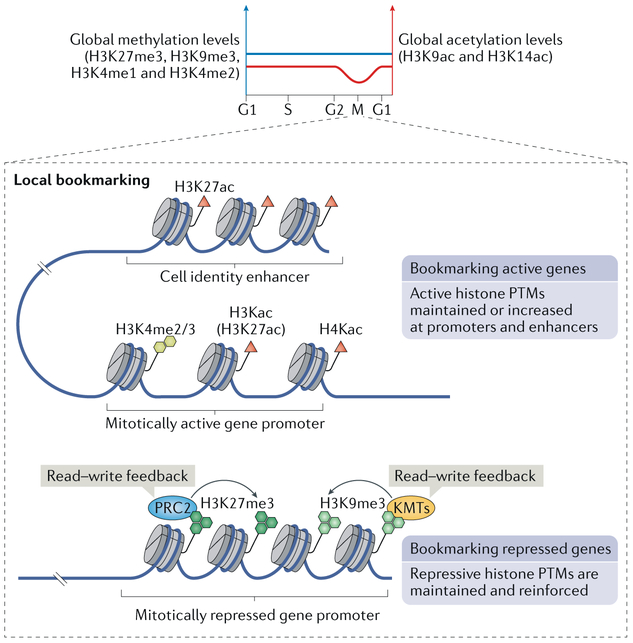
Fig. 2 ∣. Histone modifications in mitosis.
In mitosis, global levels of histone methylation are maintained, whereas histone acetylation is globally reduced. On a local scale, histone post-translational modifications (PTMs) can serve as bookmarks, allowing propagation of transcription memory through mitosis. It has been found that active histone PTMs, including histone H3 dimethylation and trimethylation at Lys4 (H3K4me2 and H3K4me3, respectively), as well as histone H3 and H4 Lys acetylation (H3Kac and H4Kac, respectively), are high at promoters of genes that are active in a given cell type. A notable PTM in this context is Lys27 acetylation of histone H3 (H3K27ac), which is globally stable or even increased in mitosis and marks both promoters of active genes, such as housekeeping genes, and cell identity enhancers. In contrast to this bookmarking of active genes, it has also been proposed that cells bookmark transcriptionally silent genes with repressive histone PTMs. In this context, trimethylation of histone H3 at Lys27 and Lys9 (H3K27me3 and H3K9me3) is deposited by machineries (Polycomb repressive complex 2 (PRC2) and lysine methyltransferases (KMTs) SUV39H1/SUV39H2 and SETDB1/ESET, respectively) that possess a read–write feedback mechanism of histone modification (they recognize existing marks and can increase their density), which allows them to maintain and reinforce gene silencing. M, mitotic phase of the cell cycle.
Globally, levels of histone methylation are retained and levels of histone acetylation decrease in mitosis36-39. More specifically, trimethylation of histone H3 at Lys27 (H3K27me3) and Lys9 (H3K9me3) as well as mono-methylation and dimethylation at Lys4 (H3K4me1 and H3K4me2) are largely retained in mitotic chromatin, whereas levels of histone H3 acetylation at Lys9 (H3K9ac) and Lys14 (H3K14ac) are reduced32,36,37,40 (FIG. 2). Interestingly, however, at the local level, promoter regions retain stable levels of the active chromatin marks, including H3K4 dimethylation and trimethylation36 but also H3 and H4 acetylation41 (FIG. 2). The most interesting histone post-translational modification as a candidate for gene bookmarking is H3K27ac, levels of which have been described either as globally stable2,29 or as increased42 during mitosis. H3K27ac is a modification typical of active enhancers. In mitosis, it localizes to promoters of housekeeping genes as well as to enhancers of early postmitotic re-expressed genes, prominently including cell identity genes2,37 (FIG. 2). In one estimate, around 50% of enhancers and 90% of super-enhancers appear to retain H3K27ac during mitosis37. However, these studies are largely correlative, and it needs to be more explicitly shown whether H3K27ac indeed bookmarks early G1 genes for efficient re-expression after mitosis. It will also be interesting to determine how the H3K27ac bookmark is affixed to particular loci during mitosis to elicit a potential bookmarking function and to identify the partner factors specifying H3K27ac location on the mitotic genome.
More recently, it has been argued that repressive histone modifications, including H3K27me3 and H3K9me3, rather than the activating modifications, are the key epigenetic marks responsible for mitotic bookmarking. Machineries depositing these modifications possess feedforward, read–write mechanisms (they recognize these modifications and reinforce them), which allow propagation of these marks without the need for their re-initiation in the progeny43. By this view, it would be more important for the cell to ‘remember’ which genes to keep off than which to keep on (FIG. 2).
Mitotic transcription
Mitotic chromosomes have long been assumed to be transcriptionally inactive owing to their compact structure and to the lack of ability to detect Pol II activity on mitotic chromatin, yet this dogma has been challenged by the recent development of highly sensitive assays showing that mitotic chromosomes are in fact transcriptionally active.
Transcription is maintained at basal levels of during mitosis.
The most widely cited evidence of transcriptional silence in mitosis comes from a study published in 1962 that exposed an asynchronous population of cells to a 5-minute radiolabelling of nascent RNAs, which yielded no positive signal in mitotic cells after a 12-day exposure period44. Interestingly, a similar, more recent study that employed radiolabelling of individual Pol-II-dependent transcripts detected low levels of nascent transcripts, even after salt and detergent washes, indicating that Pol II is active in mitosis45. However, they were unable to crosslink Pol II to mitotic chromatin with formaldehyde and thus concluded that transcription is silenced in mitosis. This study was followed over the next decade by several others aiming at crosslinking transcription machinery to mitotic chromatin, all of which concluded that Pol II and various other components of the transcription machinery are evicted from mitotic chromatin. In addition, nuclear envelope breakdown in mitosis could contribute to a decrease in Pol II local concentration.
Interestingly, a slightly different picture of Pol II–chromatin association in mitosis was obtained from studies in which inhibitors of transcription elongation, including α-amanitin46 and flavopiridol47, were used. The latter study employed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP–seq) to investigate Pol II–chromatin interactions in interphase and mitotic HeLa cells. It was demonstrated that Pol II binding to chromatin was not detected in the mitotic population under normal conditions, but the addition of flavopiridol resulted in detectable Pol II accumulation at promoters in mitotic cells47. This observation suggests that antibody-based assays are not sensitive enough to demonstrate mitotic transcription events under normal conditions (BOX 1). A more recent study using Pol II ChIP–seq, coupled to the modelling of interphase contamination of their sample, confirmed that there are indeed low levels of Pol II bound to promoters in mitosis39. A knock-in of a functional HALO–Pol II further confirmed recruitment of Pol II to mitotic chromatin, although at much lower levels than in interphase32.
Box 1∣. Old and new technologies for detecting mitotic transcription.
Unlike mapping steady-state mRNAs, which provides a static readout of a cell’s transcriptome at the time of cell lysis, the mapping of primary transcripts allows for quantification of transcriptional activity. The most common methods developed for mapping primary transcripts involve the analysis of labelled nascent transcripts in isolated nuclei because the plasma membrane is not permeable to most RNA precursors. However, the nuclear envelope breaks down in mitosis, precluding the ability to isolate nuclei for direct labelling of transcripts76. Thus, genome-wide studies of transcriptional activity during mitosis and mitotic exit have, until recently, relied on assessing the association of RNA polymerase II (Pol II) with chromatin2,47,77. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP–seq) of Pol II in murine erythroblasts arrested in mitosis by nocodazole treatment revealed a spike-like increase in Pol II binding to chromatin 90 minutes after the nocodazole was removed2. Similarly, an earlier study detected bulk recruitment of Pol II to chromatin, around the same time after release from mitotic block, by immunofluorescence19. However, the dynamic range of antibody-based methods is much less than from direct measurements of nascent transcription by sequencing as they rely on antibody–epitope interactions76. Furthermore, the cell fixation necessary for antibody-based methods has been shown to artefactually cause protein exclusion from mitotic chromatin — although transcription factors could not be visualized in mitotic chromatin by immunofluorescence, fluorescently labelled versions of the same proteins colocalized with mitotic chromatin in live cells21,23,31.
EU-RNA–seq has been recently developed to directly measure nascent transcripts while circumventing the issue of nuclear envelope breakdown during mitosis. EU-RNA–seq involves pulse-labelling nascent transcripts in vivo in intact cells by adding the cell-permeable uridine analogue 5-ethynyluridine (EU). Unlike bromouridine (BrU), the most commonly used uridine analogue for nascent RNA labelling and sequencing, EU added to the growth medium will enter the cell, make its way to the nucleus and be incorporated into nascent RNAs within a matter of minutes19,78. After isolation of RNA at the desired pulse time point, a click reaction is then used to conjugate the EU-incorporated RNAs to biotin. Streptavidin-coated beads can then be used to isolate EU-RNAs from the bulk RNA. The first strand synthesis of cDNA can occur with the EU-RNA bound to the streptavidin beads. The cDNA products can then be used to generate libraries of all the cDNAs and subjected to high-throughput DNA sequencing. By these means, detailed and quantitative measurements of the levels and dynamics of active transcription in mitosis and mitotic exit can be obtained53. Furthermore, synthetic EU-RNAs can be ‘spiked’ (that is, added at known levels) into the cellular EU-RNA preparations, before library preparation, allowing a quantitative comparison of transcription levels between different samples for the different mitotic exit time points53.
Notably, various studies have convincingly described Pol II-dependent transcription at the centromere48,49. In mammalian cells, centromeric transcription is required for proper chromosome segregation (Pol II recruits shugoshin 1 (SGO1), which in turn protects the centromeres from premature separation during sister chromatid resolution)48. These studies establish that mitotic chromatin is not refractory to transcription. In line with this, early autoradiography assays that focused on the transcription of individual protein coding regions could detect RNA synthesis in mitotic cells, which corresponded to between 5% and 40% of interphase levels, depending on the gene assayed50-52. Far more sensitive techniques used in modern studies have confirmed the prevalence of low-level transcription in mitosis (see next subsection).
New model for transcriptional memory propagation during mitosis.
A highly sensitive technique, called EU-RNA–seq (BOX 1), was recently developed in conjunction with custom spike-in controls to globally quantify mitotic transcription53. Of the approximately 28,000 transcripts expressed in an asynchronous cell population, up to 8,000 transcripts displayed measurable expression in the mitotic population, and the mean reduction in expression of these genes from the asynchronous population to the mitotic population was approximately fivefold. Similarly, using fluorescein isothiocyanate-labelled uridine triphosphate (UTP) incorporation along chromosome arms as a readout of transcription also showed a fivefold reduction in transcription levels in mitotic cells as compared with asynchronous cells53. The levels of transcription largely differed (reaching a hundredfold range) among the genes expressed in mitosis, reflecting the variability seen in individual mitotic transcript levels seen previously50-52. This wide range of mitotic RNA synthesis suggests that mitosis is not strictly a period of transcriptional silence but could be associated with altered gene regulation, whereby globally transcription is largely reduced (to subdetection levels) but individual genes are kept transcriptionally active.
The detection of low transcription of thousands of genes in mitosis, along with the aforementioned bookmarking by the basal promoter factors TFIID, TFIIB and TBP, leads to a simple new model for the mitotic inheritance of cell-specific transcriptome patterns: basal promoter activity appears to be permissive in mitotic chromatin, and the maintenance of open chromatin at promoters13 together with active transcription of the gene, albeit at a low level (FIG. 3), allows robust re-expression of the genes at mitotic exit, thereby functioning as an epigenetic mechanism that guarantees transcriptional memory propagation through mitosis. In this way, continued transcription not only preserves the local chromatin state but also provides a pool of RNAs for translation in early G1. By extension of this model, it could be that the tissue-specific bookmarking factors are responsible for the re-establishment of enhancer–promoter loops during mitotic exit to reinforce expression of cell-type-specific genes (see next section).
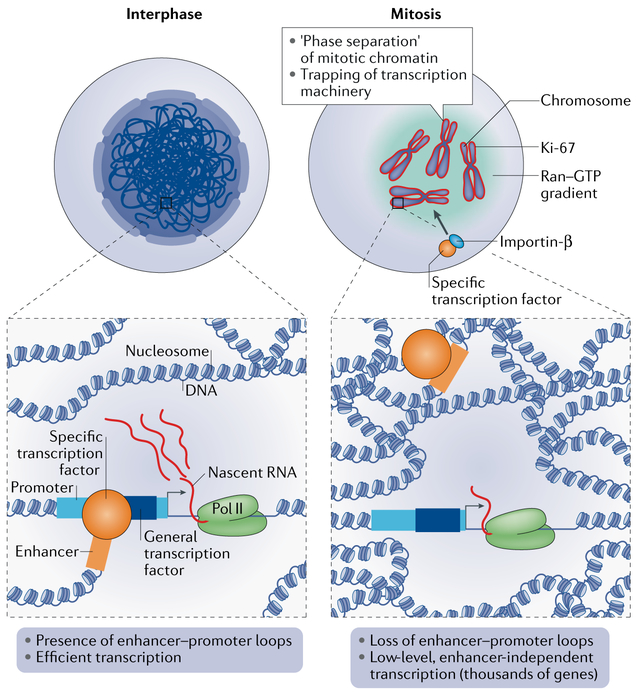
Fig. 3 ∣. Mitotic cells maintain low levels of transcription.
In interphase, enhancer–promoter loops in the nucleus are formed through interactions between general transcription factors at promoters and specific transcription factors at distal enhancers, along with architectural proteins in chromatin.The enhancer–promoter interactions allow for the robust expression of target genes. In mitosis, enhancer–promoter loops are disassembled. However, thousands of genes continue to be expressed at low levels42, presumably owing to the maintenance of general transcription factors at promoters32. At the same time, various specific transcription factors remain bound to chromatin. These observations argue against a complete shut-down of transcription in mitosis. Localization of transcription factors to mitotic chromatin may be facilitated through various means. One mechanism may involve coating of the chromosomes with proliferation marker protein Ki-67, which acts as a ‘surfactant’ that phase-separates mitotic chromatin from the surrounding cytoplasm56,57, which may contribute to trapping of transcription machinery within chromatin. In addition, the importin-β–Ran–GTP system may provide an active mechanism that drives localization of bookmarking factors to mitotic chromatin21.
Mitotic chromosomes and transcription.
It is unclear how mitotic transcription has been overlooked in the past few decades, but it may be due to the assumption, which prevailed until very recently, that mitotic chromatin is simply too compact for transcription machinery to access it. This view has recently changed with the detection of bookmarking transcription factors and open chromatin-associated histone marks in mitotic cells. Further evidence that mitotic chromatin is not as inaccessible as previously thought is provided by chromatin accessibility assays. Early studies used dimethyl sulfate (DMS) footprinting and potassium permanganate assays to show that accessibility is maintained in mitosis at the promoters of genes that were active before mitosis began3,11. More recently, a genome-wide comparison of DNase I hypersensitivity in asynchronous and mitotic erythroblasts indicated that chromatin accessibility is globally maintained, especially at promoters, although accessibility was diminished at enhancers33 (FIG. 3). Another recent study used assay for transposase-accessible chromatin with sequencing (ATAC–seq) to show that asynchronous and mitotic cells feature similar global chromatin accessibility patterns23. These studies clearly demonstrate that the pronounced compaction of chromatin during the transition from interphase to mitosis1 does not markedly affect its local accessibility and thus, possibly, its potential for basal transcription (FIG. 3).
Despite the breakdown and detachment of the nuclear envelope from mitotic chromosomes, recent studies have shown that much of a mitotic chromosome is not made up of chromatin, indicating that several nuclear components remain associated with the chromatin54,55. In fact, as much as 30–50% of the total mass of mitotic chromosomes is non-histone protein, with a considerable portion of these proteins forming a layer that coats each mitotic chromosome54,55. One of these proteins is proliferation marker protein Ki-67, which is located in the nucleolus in interphase and relocates to the chromosomal surface in late prophase56,57. Ki-67 is believed to be a ‘biological surfactant’ assisting chromosome individualization, as knockdown of Ki-67 causes the individual mitotic chromosomes to clump together54,56. Furthermore, Ki-67 is required for the compaction and packing of mitotic chromosomes54. It is intriguing to consider that global mitotic transcription may contribute to the recruitment and/or function of the non-histone proteins to mitotic chromatin. Conversely, the mitotic chromosome surface may serve to protect mitotic transcription. Ki-67 is an amphiphilic protein, with the carboxy-terminal domain interacting with chromatin and the amino-terminal, largely unstructured domain being exposed to the cytoplasm. Thus, Ki-67 may drive phase separation of the mitotic chromatin from the surrounding cytoplasm to protect the chromatin in the absence of the nuclear envelope. This separation could serve to trap transcription machinery within the mitotic chromatin mass to allow for the low-level transcription observed in mitosis50-53 (FIG. 3). Another example of a protein shown to be associated with mitotic chromatin is yeast protein Mif2 (orthologue of mammalian centromere protein C (CENPC)), which binds to centromeric chromatin throughout the cell cycle by establishing interactions with centromeric histone variant CENPA58.
Although they have not been detected in mitotic chromatin preparations, the importin proteins, which, together with a small GTPase Ran, mediate nucleo-cytosolic transport in interphase, may also support ongoing transcription in mitosis by regulating mitotic protein localization21. As noted above, HNF1β is one of the bookmarking transcription factors that binds to mitotic chromatin. Specific mutations yield HNF1β that is sensitive to temperature with respect to mitotic chromosome binding, whereby the binding is abolished at restrictive temperatures. However, when mitotic cells were shifted to the permissive temperature, HNF1β was shown to relocalize to mitotic chromatin, and this relocalization was very rapid, suggesting that an active mechanism driving relocalization is at play. The addition of importazole, which specifically inhibits importin-β59, resulted in the diminished and delayed relocalization of temperature-sensitive HNF1β to chromatin21. As the perinuclear Ran–GTP gradient is maintained in mitosis to support mitotic spindle assembly60, a Ran–importin-β-based system may also function in localization of bookmarking factors to mitotic chromatin (FIG. 3).
Taken together, diverse chromosomal proteins are retained or bind to mitotic chromosomes in addition to transcription factors and histones, and further work is needed to determine how the structural and gene regulatory proteins may functionally interact in the context of mitotic transcription.
Gene reactivation during mitotic exit
The time course of ChIP–seq analysis for Pol II in murine erythroblasts that were exiting a nocodazole-induced mitotic arrest revealed a large, transient spike in RNA Pol II binding to promoters, which occurred approximately 90 minutes into the 360-minute release period2. Similarly, single-molecule RNA fluorescent in situ hybridization (FISH) for nascent transcripts in murine erythroblasts indicated a temporary increase in nascent transcript levels approximately 90 minutes into mitotic exit. The authors thus concluded that transcription is globally reactivated in a large burst, during which transcript levels temporarily exceed that in interphase before decreasing back to interphase levels. This discovery of a ‘spike’ in transcription amplitude during mitotic exit was also found in three additional cell types by single-molecule RNA FISH61. Again, after the initial burst, the transcript levels subsided shortly after, before cytokinesis. Both studies attributed the spike in transcript levels to the decondensation of chromatin.
Direct labelling of nascent transcripts is a much more sensitive approach to detection than chromatin–protein crosslinking-based methods, such as ChIP–seq. Interestingly, EU-RNA–seq of human hepatoma cells exiting a nocodazole-induced mitotic arrest detected waves of reactivation preceding and following a main burst53 (FIG. 4). That is, some genes were found to be activated earlier than the main burst, whereas others were activated later. Of those that did spike approximately 80 minutes into the 300-minute time course, approximately 50% retained their transcription level for the duration of the exit, with the remaining genes either increased or decreased after the initial spike53. Notably, the first increase in transcription was of genes involved in basic cell functions such as translation and organelle expansion, whereas genes involved in cell-type-specific functions were lowly expressed early in mitotic exit and increased as the cells progressed to G1 (REF.53). Thus, reactivation of genes involved in growth and rebuilding of daughter cells seems to be prioritized over reactivation of genes for more specific cell functionalities. These findings indicate that the dynamics of gene reactivation during mitotic exit are far more complex than previously assumed, presumably reflecting changes in regulatory networks over time, shifting from predominately housekeeping networks to those of the more specialized cell type.
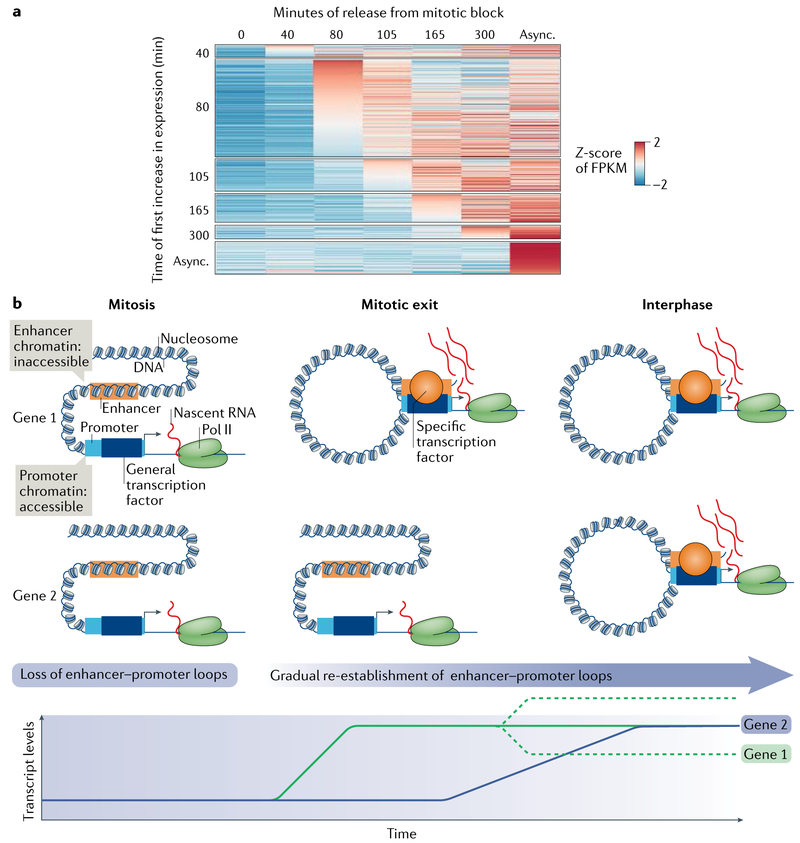
Fig. 4 ∣. Transcription reactivation during mitotic exit.
a ∣ Heat map depicting the timing of induction of transcription of genes at different times during release from a mitotic block (that is, during mitotic exit). Each line in the heat map represents the FPKM (fragments per kilobase of transcript model per million reads mapped (a normalized read density that reflects transcript expression level)) Z-score (standard deviation (or distance from) the mean) of a different gene at consecutive time points; blue colour indicates low expression and red colour indicates high expression. The genes were grouped by their common induction times (columns). The y-axis represents the time point at which the genes first increase ≥1.5-fold over levels at time 0. The x-axis represents the FPKM at the indicated time point. b ∣ Many genes are continuously transcribed from their promoter during mitosis in an enhancer-independent fashion. During mitotic exit, the enhancer–promoter loops that existed in the parental cell are re-formed, thus determining the timing of increase in the transcription of target genes. This reactivation of gene expression differs for different genes, exhibiting a wave pattern, with genes required for cell growth and other housekeeping functions reactivated first followed by the activation of cell-type-specific genes (which also follow a sequential order of expression). After the initial burst of expression, the levels of transcripts can decrease, stabilize or further increase. By early G1, interphase chromatin structure and gene expression patterns have been fully re-established, resulting in a cell-type-specific transcriptome. Async., asynchronous; Pol II, RNA poLymerase II.
Three recent studies have investigated the role of regulatory chromatin regions in mitotic transcription and postmitotic gene reactivation. In the first, chromatin accessibility was measured by treatment with DNase I followed by high-throughput sequencing33. The authors found that although the accessibility of promoters is retained in mitosis, it is largely diminished at enhancers. The same team later used chromatin conformation capture (3C) to show that the loops between enhancers and promoters, which indicate a functional enhancer, are lost in mitosis23. In addition to chromatin accessibility, the production of RNAs from enhancers leading to the generation of enhancer RNAs (eRNAs) has also been known to be a sign of active enhancers. As with the chromatin accessibility studies, assessment by EU-RNA–seq also found eRNAs to be greatly diminished or absent in mitosis53. Thus, it is likely that the basal levels of transcription in mitosis function largely in an enhancer-independent manner (FIGS 3,4b), and enhancer activation during mitotic exit, likely by the binding of specific transcription factors, promotes different times of gene reactivation (FIG. 4b).
An interesting point in the regulation of mitotic transcription and gene reactivation at mitotic exit is the control of protein levels. This protein control is particularly important for the regulation of cell-type-specific genes, which need to be expressed in a specific order to ensure the maintenance of cell identity62. Studies in embryonic stem cells showed that the presence of bookmarking transcription factors SOX2 (REFS27,63) and OCT4 (REF.37) is required to maintain pluripotency through cell division. Because mitosis is short in comparison with the remainder of the cell cycle, most of the mRNAs and proteins produced in interphase can persist through cell division and are sufficient for various functions in early G1. Interestingly however, as reported in HUH7 hepatoma cells, cell-specific transcription factors can exhibit variable stability in mitosis: the transcription factors that have the most prominent role in liver cell identity remained most stable in mitosis, whereas those that are involved later in liver cell development were more often displaced from the chromatin or selectively degraded in mitosis19. Together, these studies suggest a model in which certain transcription factors are degraded in mitosis to restrict their unscheduled activity during mitosis or early in mitotic exit, whereas others continue to be produced during mitosis to maintain their stable levels. This regulation could contribute to the control over the timing at which particular genes are expressed in daughter cells, thereby allowing the establishment of a cell-type-specific transcriptome. Therefore, it will be of great interest to investigate the regulation of protein stability in mitosis and how it may or may not affect the transcriptome as cells transition from mitosis to interphase. Such a model could also explain why housekeeping genes, which are regulated by fewer64 and stronger65 enhancers, are those that are the first to increase during mitotic exit53.
Conclusion and perspective
The recent discovery that low levels of transcription continue through mitosis is in contrast to the long-standing model of mitotic transcriptional silence. It was thought that transcription was reactivated, genome-wide, in a large burst as transcription factors and other transcription machinery components were again able to access the chromatin in early G1. However, now it is clear that the pattern of gene activation is more complex, with genes being reactivated at different time points. These findings shift our understanding of transcriptional memory through mitosis from an off–on model to one of more nuanced regulation at the level of individual genes. Nevertheless, several questions about the molecular mechanisms and regulation of gene reactivation following mitosis need to be answered in order to understand how transcription networks are properly re-established after cell division.
In the immediate future, it will be of great interest to assess the role of global transcription in mitosis and whether it serves to maintain the transcriptional signature of chromatin, recruit architectural proteins or provide RNAs for translation in early G1. However, when designing strategies to interfere with mitotic transcription, it will be necessary to employ clever engineering approaches to circumvent the inhibition of transcription at the centromere, which is required for proper chromosome segregation and hence directly affects cell fate42.
Given the relationship between transcription and chromatin structure, it will be of interest to link mitotic transcription and chromosomal structural features together on a global scale. Circularized chromosome conformation capture sequencing (4C–seq) studies, which enable the unbiased detection of all genomic loci that physically contact a genomic locus of interest, performed during mitotic exit showed that TADs are lost in mitosis and re-form at the same time as replication domain boundaries, the timing of which is cell-type-specific66. This finding indicates that TADs, which are specified independently of the cell type, serve as boundaries for replication domains, which are cell-type-specific67-72. It would be interesting to see whether the timing of TAD or replication domain boundary establishment is related to transcription reactivation waves during mitotic exit and whether the timing of gene reactivation is cell-type-specific, mirroring replication timing. Another question to address is how the timing of gene activation within each TAD relates to their interphase expression level.
Interactions between chromatin and the nuclear envelope are known to regulate transcription and to impart cell identity73-75. Despite this important role in the regulation of gene expression, it is currently not clear which components establish these contacts. Given that the nuclear envelope is disassembled in mitosis, it is also unclear how the chromatin is organized in the daughter nucleus such that the proper contacts are made with the newly formed nuclear envelope. Curiously, mitotic chromosome condensation begins at sites where chromatin contacts the inner nuclear membrane54, and it is likely that nuclear envelope components, such as the lamina and nucleoporins, stay associated with the chromatin during mitosis. If so, this mechanism may provide another form of mitotic bookmarking, which may facilitate the reassembly of the envelope around the chromatin in the daughter nuclei mirroring the parental genome organization. If transcription has a role in envelope reassembly, then there may be a correlation between regions of envelope reassembly and the activation of the genes in those regions during mitotic exit. Clearly, diverse cell biological and gene regulatory questions remain in order to understand how transcription networks are properly re-established after cell division.
Epigenetic control of gene expression.
Mechanisms independent of DNA alterations through which the gene expression programme particular to a cell type is recapitulated in daughter cells after cell division.
Footprinting.
A method of detecting where proteins are bound to DNA on the basis of their accessibility to enzymes such as DNase I or chemicals such as DMS or potassium permanganate.
General transcription factors.
Proteins that act at promoters to enable transcription of many classes of genes.
Promoter transcription factors.
General transcription factors that localize to promoters.
Architectural transcription factors.
Transcription factors, such as CTCF (CCCTC-binding factor), that bring regions of the genome together to form higher-order chromatin structures, which have important roles in gene expression regulation.
HALO-tag.
A modified enzyme that binds to a synthetic ligand target and can be fused to a protein of interest for visualization.
Pioneer factor.
Transcription factor that can directly bind nucleosomal DNA in chromatin.
Topologically associated domains.
(TADs). Boundary-insulated chromosomal segments within which sequences preferentially contact each other.
Housekeeping genes.
Genes that are responsible for the general functions of a cell, such as growth and metabolism, independent of the cell’s specialized function.
Click reaction.
The copper-catalysed conjugation of an azide with an alkyne to yield a five-part heteroatom ring.
Interphase contamination.
Transcriptional signal in a population of mitotic cells that results from contaminating interphase cells in that population. Importantly for studies of mitotic transcription, the high transcriptional activity of the contaminating interphase cells, relative to the low level that occurs in mitotic cells, can lead to overestimation of transcription during mitosis.
Spike-in controls.
Short, known sequences of DNA or RNA that are added to a sample at a known quantity at the beginning of a high-throughput sequencing assay to help normalize for synthesis and recovery when comparing different samples.
DNase I hypersensitivity.
The ability of a segment of DNA in chromatin to be digested by DNase I owing to the accessibility of this segment (lack of local compaction).
Assay for transposase-accessible chromatin with sequencing.
(ATAC–seq). A method that uses the Tn5 transposase to probe the genome for accessibility.
Amphiphilic protein.
A protein that is hydrophobic on one end and hydrophilic on the other end.
Histone variant.
A substitute for one of the canonical histones in a nucleosome.
Nocodazole.
A compound that prevents microtubule polymerization, thereby blocking the formation of the metaphase plate.
Enhancer RNAs.
(eRNAs). RNAs that are generated from the site of enhancer sequences, presumably as a by-product of Pol II activity.
Acknowledgements
The authors thank P. Navarro (Institut Pasteur) for comments on the manuscript and M. Song for help in its preparation. Research on mitotic transcription has been supported by US National Institutes of Health (NIH) grant T32GM00812 to K.C.P., by Fondation pour la Recherche Medicale grant 40334 to J.L. and by NIH grant GM36477 to K.S.Z.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Molecular Cell Biology thanks D. Suter, P. Navarro and other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Naumova N et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsiung CC et al. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 30, 1423–1439 (2016).This study, using ChIP–seq and Capture-C in murine erythroblasts in mitosis and during mitotic exit, shows a large spike in Pol II binding and a re-formation of enhancer–promoter loops approximately 90 minutes into mitotic exit.
- 3.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K & Wu C Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83, 29–38 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Guo J, Turek ME & Price DH Regulation of RNA polymerase II termination by phosphorylation of Gdown1. J. Biol. Chem. 289, 12657–12665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dovat S et al. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 16, 2985–2990 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidi SK et al. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat. Rev. Genet 11, 583–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egli D, Birkhoff G & Eggan K Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol 9, 505–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadauke S & Blobel GA Mitotic bookmarking by transcription factors. Epigenet. Chromatin 6, 6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Festuccia N, Gonzalez I, Owens N & Navarro P Mitotic bookmarking in development and stem cells. Development 144, 3633–3645 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zaret KS Genome reactivation after the silence in mitosis: recapitulating mechanisms of development? Dev. Cell 29, 132–134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michelotti EF, Sanford S & Levens D Marking of active genes on mitotic chromosomes. Nature 388, 895–899 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Xing H et al. Mechanism of hsp70i gene bookmarking. Science 307, 421–423 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Xing H, Vanderford NL & Sarge KD The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol 10, 1318–1323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segil N, Guermah M, Hoffmann A, Roeder RG & Heintz N Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10, 2389–2400 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Hinkley CS, Henry RW & Huang S TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13, 276–284 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christova R & Oelgeschlager T Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol 4, 79–82 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Young DW et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445, 442–446 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Kadauke S et al. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caravaca JM et al. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 (2013).This study shows that FOXA1 quantitatively remains on mitotic chromosomes via primarily nonspecific DNA and nucleosomal interactions while also retaining specific binding at a subset of sites.
- 20.Verdeguer F et al. A mitotic transcriptional switch in polycystic kidney disease. Nat. Med 16, 106–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerner J et al. Human mutations affect the epigenetic/bookmarking function of HNF1B. Nucleic Acids Res. 44, 8097–8111 (2016).This study shows that formaldehyde causes artefactual cytoplasmic localization of the bookmarking factor HNF1β and that inhibition of importin delays the mitotic relocalization of a temperature-sensitive HNF1β mutant to chromatin.
- 22.Festuccia N et al. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol 18, 1139–1148 (2016).In this study, an ESRRβ–GFP fusion is stably integrated into embryonic stem cells and live imaging showed that it is highly dynamic in mitosis and associates with both specific and nonspecific binding sites.
- 23.Teves SS et al. A dynamic mode of mitotic bookmarking by transcription factors. eLife 5, e22280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blobel GA et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36, 970–983 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Sung E, Donlin-Asp PG & Corces VG A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat. Commun 4, 1464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey A, Nishiyama A, Karpova T, McNally J & Ozato K Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 20, 4899–4909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deluz C et al. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 30, 2538–2550 (2016).This study performs mitosis-specific knockdown of SOX2, showing that mitotic bookmarking by SOX2 contributes to pluripotency maintenance and is necessary for neuroectodermal differentiation but is dispensable for reprogramming towards induced pluripotency.
- 28.Burke LJ et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291–3300 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong MM et al. Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLOS ONE 9, e99989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao R, Nakamura T, Fu Y, Lazar Z & Spector DL Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol 13, 1295–1304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallier C et al. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell 14, 3414–3426 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teves SS et al. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 (2018).This study focuses on the promoter binding factor TBP in mitotic bookmarking, emphasizing the role of the promoter region in mitotic memory.
- 33.Hsiung CC et al. Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 25, 213–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol 168, 41–54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou HD et al. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 (2017).This study establishes an electron microscopy tomography of chromatin (ChromEMT) to show that overall, the structure of chromosome fibres is unaltered in mitotic chromosomes and that chromatin chains bend at various lengths to perform the high level of compaction reached in mitotic chromosomes.
- 36.Valls E, Sanchez-Molina S & Martinez-Balbas MA Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem 280, 42592–42600 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Liu Y et al. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 19, 1283–1293 (2017).This study shows that mitotic-specific knockdown of OCT4 impairs pluripotency maintenance and reveals that H3K27ac in mitosis marks promoters of housekeeping genes and enhancers of pluripotency genes.
- 38.Wilkins BJ et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science 343, 77–80 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Javasky E et al. Study of the mitotic chromatin shows involvement of histone modifications in bookmarking and reveals nucleosome deposition patterns. Preprint at 10.1101/233056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F & Higgins JM Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 23, 175–184 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Kouskouti A & Talianidis I Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24, 347–357 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y et al. Transcriptional landscape of the human cell cycle. Proc. Natl Acad. Sci. USA 114, 3473–3478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinberg D & Vales LD Chromatin domains rich in inheritance. Science 361, 33–34 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Prescott DM & Bender MA Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res 26, 260–268 (1962). [DOI] [PubMed] [Google Scholar]
- 45.Parsons GG & Spencer CA Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol 17, 5791–5802 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsui SI, Weinfeld H & Sandberg AA Quantitative conservation of chromatin-bound RNA polymerases I and II in mitosis. Implications for chromosome structure. J. Cell Biol 80, 451–464 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang K et al. Mitotic transcriptional activation: clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol. Cell 60, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H et al. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell 59, 426–436 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Chen CC et al. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev. Cell 34, 73–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gariglio P, Buss J & Green MH Sarkosyl activation of RNA polymerase activity in mitotic mouse cells. FEBS Lett. 44, 330–333 (1974). [DOI] [PubMed] [Google Scholar]
- 51.Johnson TC & Holland JJ Ribonucleic acid and protein synthesis in mitotic HeLa cells. J. Cell Biol 27, 565–574 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konrad CG Protein synthesis and rna synthesis during mitosis in animal cells. J. Cell Biol 19, 267–277 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palozola KC et al. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122 (2017).This study uses a new method to detect low-level transcription occurring globally during mitosis and reveals that enhancer usage correlates with dynamic gene reactivation during mitotic exit.
- 54.Booth DG et al. 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol. Cell 64, 790–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohta S et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cuylen S et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535, 308–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautier T, Robert-Nicoud M, Guilly MN & Hernandez-Verdun D Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J. Cell Sci 102, 729–737 (1992). [DOI] [PubMed] [Google Scholar]
- 58.Xiao H et al. Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres. Genes Dev. 31, 1958–1972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderholm JF et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol 6, 700–708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clarke PR & Zhang C Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol 9, 464–477 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Vankova Hausnerova V & Lanctot C Transcriptional output transiently spikes upon mitotic exit. Sci. Rep 7, 12607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwasaki H et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 20, 3010–3021 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deluz C, Strebinger D, Friman ET & Suter DM The elusive role of mitotic bookmarking in transcriptional regulation: insights from Sox2. Cell Cycle 16, 601–606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osterwalder M et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arnold PL A reducing role for boron. Nature 502, 458 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Dileep V et al. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 25, 1104–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pope BD et al. Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen RS et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiratani I et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 20, 155–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pope BD et al. Replication-timing boundaries facilitate cell-type and species-specific regulation of a rearranged human chromosome in mouse. Hum. Mol. Genet 21, 4162–4170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rivera-Mulia JC et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 25, 1091–1103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryba T et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20, 761–770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacinto FV, Benner C & Hetzer MW The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 29, 1224–1238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ibarra A, Benner C, Tyagi S, Cool J & Hetzer MW Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toda T et al. Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21,618–634.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Core LJ, Waterfall JJ & Lis JT Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prasanth KV, Sacco-Bubulya PA, Prasanth SG & Spector DL Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 14, 1043–1057 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jao CY & Salic A Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA 105, 15779–15784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan J, Xu L, Crawford G, Wang Z & Burgess SM The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol. Cell. Biol 26, 155–168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young DW et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl Acad. Sci. USA 104, 3189–3194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lake RL, Tsai RF, Choi I, Won KJ & Fan HY Specific mitotic chromatin association of the major notch effector RBPJ and its implication for transcriptional memory. NCBI https://www.ncbi.nlm.nih.gov/bioproject/PRJNA196580 (2013). [Google Scholar]
- 82.Lodhi N, Ji Y & Tulin A Mitotic bookmarking: maintaining post-mitotic reprogramming of transcription reactivation. Curr. Mol. Biol. Rep 2, 10–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calderon MR et al. Ligand-dependent corepressor contributes to transcriptional repression by C2H2 zinc-finger transcription factor ZBRK1 through association with KRAB-associated protein-1. Nucleic Acids Res. 42, 7012–7027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lodhi N, Kossenkov AV & Tulin AV Bookmarking promoters in mitotic chromatin: poly(ADP-ribose) polymerase-1 as an epigenetic mark. Nucleic Acids Res. 42, 7028–7038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kara N, Hossain M, Prasanth SG & Stillman B Orc1 binding to mitotic chromosomes precedes spatial patterning during G1 phase and assembly of the origin recognition complex in human cells. J. Biol. Chem 290, 12355–12369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arora M, Packard CZ, Banerjee T & Parvin JD RING1A and BMI1 bookmark active genes via ubiquitination of chromatin-associated proteins. Nucleic Acids Res. 44, 2136–2144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]