Abstract
Young children are infected by a diverse range of enteric pathogens in high disease burden settings, suggesting pathogen contamination of the environment is equally diverse. This study aimed to characterize across- and within-neighborhood diversity in enteric pathogen contamination of public domains in urban informal settlements of Kisumu, Kenya, and to assess the relationship between pathogen detection patterns and human and domestic animal sanitation conditions. Microbial contamination of soil and surface water from 166 public sites in three Kisumu neighborhoods was measured by enterococcal assays and quantitative reverse transcription polymerase chain reaction (qRT-PCR) for 19 enteric pathogens. Regression was used to assess the association between observed sanitary indicators of contamination with enterococci and pathogen presence and concentration, and pathogen diversity. Seventeen types of pathogens were detected in Kisumu public domains. Enteric pathogens were codetected in 33% of soil and 65% of surface water samples. Greater pathogen diversity was associated with the presence of domestic animal feces but not with human open defecation, deteriorating latrines, flies, or disposal of human feces. Sanitary conditions were not associated with enterococcal bacteria, specific pathogen concentrations, or “any pathogen”. Young children played at 40% of observed sites. Managing domestic animal feces may be required to reduce enteric pathogen environmental contamination in high-burden settings.
Graphical Abstract

INTRODUCTION
Recent multicountry epidemiological studies of diarrheal disease etiology in high-burden settings have shown a wide diversity of enteric pathogen infections within and between individual children, and between populations and population subgroups (i.e., by age).1,2 The diversity in pathogens causing childhood enteric infections shows that children under the age of five years (<5yr) in these settings are chronically exposed to a range of environmentally transmitted enteric pathogens. Relatively little is known about the extent of environmental enteric pathogen contamination in these settings and the patterns with which enteric pathogen contamination occurs over space and time. A handful of studies have reported high frequencies of detection of different types of enteric pathogens in Indian and Tanzanian household environments, confirming that household exposure pathways pose a risk to children.3–5 However, recent multimillion-dollar randomized, controlled trials of household water, sanitation, and hygiene (WASH) interventions found inconsistent impact of household WASH on pediatric diarrhea.6–8 This suggests that children may be infected by non-household exposure pathways, such as through play in public areas near their household. In low-income settings where sanitation coverage is low, neighborhood public areas are often used for open defecation and disposal of untreated human waste by many households, as well as for domestic animal husbandry.9 Thus, levels of pathogen contamination in public areas may far exceed contamination levels in privately owned domestic areas, and public areas could pose a disproportionally high risk of infection by enteric pathogens for exposed children.
The overarching goal of the Social Microbes Study is to examine enteric pathogen transmission patterns in high disease burden settings, from sources of fecal contamination through the environment to children, using pathogen distribution and diversity as indicators of transmission pathways. This is the first Social Microbes report to be released, which focuses on testing the hypothesis that public areas within low-income neighborhoods with high disease burden are contaminated by a diverse set of enteric pathogens, and testing whether observed indicators of human and animal feces contamination would be associated with increased presence and total diversity in pathogens. We also measured how frequently <5yr children are observed in public areas and whether we saw unsafe exposure behaviors to assess whether public domain exposures are viable pathways for <5yr enteric infection.
Environmental fecal contamination is usually measured by general fecal indicator bacteria, fecal source tracking markers, or focused detection of specific pathogens of interest as indicators of exposure risk. Fecal Escherichia coli and enterococcal bacteria are popular, low-cost indicators but typically correlate poorly with other infectious pathogens in sewage, ecological soils, surfaces, and waters.10–13 They may be particularly unsuitable indicators for measuring risks from feces in ecological systems where gut bacteria become a naturalized part of soil, water, and surface microbial communities.14 Host-specific fecal source tracking markers have improved capacity to distinguish human versus animal fecal contamination of the environment, but the reliability of fecal source tracking markers for predicting infectious pathogens is currently unclear.10,15 Selection of one or a few specific types of enteric pathogens is often used to understand transmission patterns of specific types of pathogens.16,17 However, dozens of enteric viruses, bacteria, protozoans, and helminthic species circulate at different times of the year in endemic settings, so reliance on any one to predict risks from all pathogens transmitted by fecal–oral exposure pathways is risky.1,2 In this study, environmental samples were tested for a wide range of common enteric viruses, bacteria, protozoans, and helminths involved in fecal–oral disease transmission in high-burden settings to reduce the likelihood of exposure misclassification (classifying samples as uncontaminated on the basis of one indicator) or inaccurate concentration estimates for environmental exposure pathways.
Pathogen diversity was an important systems-level evaluation metric in this study that was adopted on the basis of the theory that diversity in enteric pathogen contamination would occur not just across high disease burden neighborhoods but also at fine spatial scales. If environmental microbial contamination by human and animal feces is pervasive in high disease burden settings, with multiple pathogens circulating at any given time, then there is a strong chance that multiple enteric pathogens co-occur in some environments (e.g., open defecation sites) at the same time. Exposure of children to these uniquely hazardous environments could increase their risk of infection by multiple pathogen types. The risk of exposure to any given pathogen from soil or surface water is influenced by the recent presence of a fecal source but also by the pathogen’s capacity to persist in specific environments, like soil or surface water, and overall infection rate in a host population. The evolutionary traits that govern persistence of microbes may be unique to members of a phylogenetic group (e.g., nonobligate bacterial replication in the environment) as well as reflecting species-specific ecological adaptations. Examining both the pathogen-specific and higher-order taxonomic differences in pathogens in the environment could provide insight as to the types of pathogens potentially transmitted via different child behaviors, for example, ingesting soil versus water. From a programming standpoint, capacity to identify high-risk areas or exposure pathways, in a milieu of existing elevated background contamination, could improve how well investments reduce <5yr enteric disease in high-burden settings. Pathogen diversity could improve the identification of high- versus low-risk areas or exposure pathways and the fecal sources contributing to environmental contamination.
This paper describes enteric pathogen detection frequencies and diversity patterns in soil and surface water from public areas of three low-income, urban neighborhoods of Kisumu, Kenya, with low sanitation coverage. Then we examine the relationship between human and domestic animal fecal sources and pathogen detection and diversity patterns.
MATERIALS AND METHODS
Study Population.
Kisumu is the third largest city in Kenya and has a population of approximately 409 928 inhabitants.18 Up to 60% of the city’s population resides in peri-urban informal settlements, which have emerged due to economic migration and a lack of affordable housing.19 Kisumu County has a high prevalence of diarrhea (18% two-week period prevalence) with most cases of diarrhea occurring in children less than three years of age.20 The child mortality rate is 105 deaths per 1000 live births, and the prevalence of severe childhood stunting (>–2 standard deviations below the reference norm) is approximately 25%.20 This study took place in three established informal settlements, Nyalenda A [population density (pd) = 8953 persons per square kilometer] Nyalenda B (pd = 6886/km2), and Obunga (pd = 1913 km2). Cohabitation with domestic animals is common, and most residents rely upon open defecation or sanitation facilities shared by eight or more households.21,22
Pilot of Environmental Sampling and Microbiology Methods.
Prior to the primary study in Kenya, a pilot sampling project was conducted in Iowa in June 2015 (similar climate conditions) to understand how often fecal indicators and enteric pathogens would be detected in a watershed impacted by agriculture, free-range animal management (cows), and concentrated animal feeding operations (pigs). This information is presented in Supporting Information Sections G and H as a comparative baseline for understanding the importance of fecal indicator and pathogen contamination in Kisumu, Kenya.
Sample Site Selection.
A cross-sectional observational survey with environmental sampling was performed in Kenya in July 2015. The study was designed to ensure sampling sites were randomly distributed across special areas of interest to optimize measurement of variability in pathogen diversity and co-occurrence patterns and to prevent introduction of observer bias in selection of environmental sampling locations. Neighborhood boundary parameters were visualized utilizing Batchgeo (Google), and these spaces were defined geographically by the northernmost, westernmost, easternmost, and southernmost latitude or longitude values in rectangular form. For each neighborhood, 60 latitude and longitude pairs that fell within these specified ranges were randomly generated by utilizing the Web site geomidpoint.com, prior to field-based data collection (Figure S1). Global Positioning System (GPS) coordinates were entered into a Waytracker mobile phone app, and daily routes were identified to navigate between ~15 coordinates per day. Observers navigated to the coordinates (±3 m), which was considered the center of a site, defined as all areas falling within a 25 m radius around the central coordinates. If the coordinates fell within a private household yard or business, the nearest set of coordinates outside that private space but within 25 m of the random coordinates were identified. Sites were visited at various times during the day (morning and afternoon), with site visits lasting approximately 20 min.
Public Site Observation.
Study teams systematically documented sanitary conditions at each site using an observational survey implemented via mobile phone app (FieldLogs, Trekea Mobile Inc.). Observers documented landscape features such as surface waters, grasslands, and altitude. Development of infrastructure was noted, including roads, drains, dams, industry, housing, public water sources, and the presence, physical condition, and hygiene of public or communal latrines. Indicators of human open defecation or unsafe disposal of excreta included observed “flying toilets” (plastic bags containing excreta), used diapers, piles of human feces, emptying of latrines into drains or land around the latrine, septage emptied from a latrine next to the latrine, and visual confirmation of an adult or child actively defecating in the open. Presence and type of domestic animals and their feces were recorded. Observers recorded whether, during that ~10–15 min spot observation, children approximately <5 yrs were observed in the public area, the number present, and any behaviors that would result in hand or mouth contact with environmental fomites (touching soil, surface water, animals, or objects on the ground, swimming, eating food, eating dirt, mouthing hands). Additionally, enumerators documented whether any children were defecating in the open at the time of observation. Ecological conditions that could influence pathogen presence and persistence, specifically daily temperature and relative humidity, were extracted from National Oceanic and Atmospheric Administration (NOAA) data collected at the Kisumu Airport. Observers did not include areas within private housing or businesses adjoining or overlapping with the site area to avoid potential bias in conditions observed from privately held property. However, private conditions that impacted public space, such as drains or sewage leaching from the household or animals roaming between public and private areas, were recorded.
Sample Collection.
Standard operating protocols were implemented to ensure standardized hygienic sampling and processing of environmental materials. Approximately 5 g of soil was collected at every site by inserting an alcohol-sterilized scoop into the ground at a 45° angle to a depth of 5 cm (half the length of the scoop) and transferring the soil into a sterile WhirlPak bag (Sigma–Aldrich, St. Louis, MO). Surface water was collected, if present, by skimming water into a WhirlPak bag. Collection bags were stored on ice packs in a cooler and transported to the laboratory within 6 h of collection. In Kenya, four samples were collected at seven randomized sites in each neighborhood to account for anticipated variance in pathogen distributions at public sites caused by the final spatial scale of sites per neighborhood.
Indicator Analysis.
One gram of soil was measured into 10 mL of phosphate-buffered saline (PBS), and the mixture was vortexed for 30 s and then mixed on a rotator for 20 min. Solid matter was allowed to settle for 5 min and elute was removed for enterococcal assays. Enterococci are recommended by the U.S. Environmental Protection Agency for identifying fecal material in fresh and marine recreational waters,23 although both enterococci and E. coli are considered inadequate indicators in tropical settings like Hawaii.15,24 There is no global recommendation on appropriate indicators for tropical waters, so enterococci were chosen based upon the EPA policy. Enterococci were enumerated by vacuum filtration of three serial dilutions of surface water (10, 1, and 0.1 mL) or soil (1, 0.1, and 0.01 mL) rinsing through a white gridded 0.45 μm mixed cellulose esters filter (product GSWP04700, Millipore, Billerica, MA), and culturing filters for 18–24 h at 37 °C on mEI agar (EPA Method 1600). Colony-forming units (cfu) of enterococci were counted according to the manufacturer’s recommendations.
DNA and RNA Extraction.
DNA and RNA was extracted from 0.5 g of soil by use of the FastDNA and FastRNA SPIN kits for soil (MP Biomedicals, Solon, OH), including a bead beating step. A 10 mL volume of surface water was processed by adjusting water to 2.5 mM MgCl2 and pH 10.0 and then vacuum-filtering through a 0.45 μm mixed cellulose esters filter. This volume was chosen to ensure we could systematically sample a wide range of surface waters (floodwater puddles, drains, rivers) representing potential exposure hazards in Kisumu. This method was chosen because it more efficiently removes inhibitors than ultrafiltration and is practical for laboratories with limited capacity and for isolation of many different types of pathogens.25,26 Filters were frozen at –20 °C for 1–4 weeks and transported to the University of Iowa, where they were stored at –80 °C until extraction (1 week). Filters were cut in half and processed by use of the FastDNA and FastRNA for soil kits (equivalent to 5 mL of water).
Quantitative PCR on Environmental Samples.
The concentration and quality of DNA and RNA was measured on a Nanodrop UV–vis spectrophotometer (Thermo Scientific). Duplicate 6 and 0.6 μL volumes of nucleic acid extract were tested for inhibition by the QuantiFast pathogen internal control kit (Qiagen, Germantown, MD). These methods effectively removed most amplification inhibitors (Table S1). Samples that were not inhibited were analyzed by combining 20 μL of DNA and RNA each (total 40 μL) with AgPath polymerase and then performing quantitative reverse transcription polymerase chain reaction (qRT-PCR) using a 19-pathogen microfluidic TaqMan array card (TAC) on a QuantiStudio 12K Flex real-time PCR system with array card block (ThermoFisher, Chicago, IL).27 Inhibited samples were diluted 1:10 with deionized (di) H2O before TAC analysis. The card format included five types of viruses (adenovirus 40/41, astrovirus, sapovirus, norovirus GII, and rotavirus), nine types of bacteria [enteroaggregative E. coli (EAEC), enter-opathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), shiga toxin-expressing E. coli (STEC), Campylobacter jejuni, Shigella, Vibrio cholerae, Salmonella enterica, and Clostridium difficile], three protozoans (Cryptosporidium spp., Giardia lamblia, and Entamoeba histolytica), and two helminths (Ascaris and Trichuris). Exponential curves and multicomponent plots were visually examined to validate positive amplification. Gene targets with real amplification in one well were reanalyzed and considered positive if amplification was detected again. Gene targets with cycle threshold (CT) values over 35 were cross-validated by performing a 14-cycle preamplification reaction with pathogen-specific primers (0.2 μM each), to increase the starting concentration of pathogen DNA/cDNA in a sample,28 and then reanalyzed by qRT-PCR. Samples were classified as positive for a specific gene target if amplification was verified at a lower cycle threshold and negative if unverified. This resulted in presence and concentration data for 24 pathogen gene targets representing 19 types of pathogen taxa per sample (detection of either gene target for ETEC, EPEC, EAEC, and STEC was positive at a taxa level).
The concentration of each pathogen per reaction volume was estimated by comparing the CT for a pathogen gene target in a sample to a standard curve generated by qRT-PCR of a 6–7-fold serial dilution of a positive control of known concentration for each of the 19 qRT-PCR targets (Table S2). For samples that were repeated by use of the preamplification step, the initial concentration was estimated by comparison to a standard processed by preamplification. If samples determined to be positive still reflected signs of significant inhibition, concentrations were excluded from analysis to avoid biasing statistical estimates of mean and standard deviation. Otherwise, final concentrations of gene copy per gram of soil or per milliliter of surface water were generated by multiplying the concentration of each gene target by the dilution factors introduced by processing. Concentrations for samples with no detectable amplification were not transformed to avoid inflating statistics caused by relatively high methodological lower limit of detection (LLOD) for some pathogens.
Statistical Analysis.
Data were analyzed by use of SAS version 9.4 (SAS Institute, Cary, NC). Descriptive statistics of variables were reported as proportions or mean and standard deviation.
Microbial contamination outcomes of interest included the following: (l) presence of enterococcal fecal indicator bacteria, (2) log10 number of enterococcal cfu, (3) presence of any pathogen gene targets, (4) pathogen diversity, defined by the sum count of all unique types of enteric pathogens detected in an individual soil or water sample (either target for pathogenic bacteria defined as one group), and (5) log10 gene copy of pathogen gene targets.
Exposure variables representing potential sources of fecal contamination were any human or animal feces observed versus none observed; indicators of open defecation versus none observed; presence of any type of domestic animal present versus none observed; flies present versus none observed; and a categorical variable for latrines present in good condition, latrines present in bad condition, or no latrines present. Frequently latrines in both good and bad condition were present at a site. In this case the site was classified as latrines in bad condition, based upon the default assumption that just one deteriorating latrine could introduce contamination into soil and water. Potential confounders included type of sample (soil versus water), altitude in meters, relative humidity, temperature in Celsius, and landscape use (housing area versus undeveloped).
Generalized linear mixed models (glmm) with binary log link, robust standard errors, and random intercept for site ID and neighborhood with exchangeable correlation structure to account for spatial clustering at each level were used to test whether sites with any of the exposure variables were more likely to have at least one type of enteric pathogen detected. Identical glmm models with Poisson log link were used to test whether sites with any of the same potential fecal sources were more likely to have increased sum counts in pathogen diversity. Censored regression models were used to assess whether exposure variables were associated with increased concentration of enteric pathogen gene copy number to account for left-censoring of data. Each modeling process involved analyzing the effect of each exposure variable independently on the pathogen outcome, while adjusting for landscape type, daily temperature, relative humidity, altitude, and type of sample (soil vs water). Full models included all variables simultaneously to measure individual effects, accounting for co-occurrence of exposure variables at a site. On the basis of full model results, a subanalysis of association between specific types of domestic animals and pathogen diversity was performed by replacing the binary domestic animals variable with presence/absence variables for specific categories of animals. A one-sided Mantel test, using the geographic distances between sites, was used to test for spatial clustering of sites with compositional similarity between pairs of sites in terms of the number of pathogens detected at both sites.
RESULTS
Characteristics of Study Sites.
A total of 166 public sites were inspected for this study. Ecological conditions and the proportion of sites developed with housing were relatively similar in the three surveyed neighborhoods, with the exception of higher mean elevation in Obunga, more sites with indicators of open defecation or open feces disposal in Nyalenda B, fewer sites with shared latrines in Obunga, and more latrines in good structural condition in Nyalenda B (Table 1).
Table 1.
Ecological and Sanitation Characteristics of Public Domains of Three Kisumu Neighborhoodsa
| observed sanitary indicators | Nya-A, N = 55 | Nya-B, N = 53 | Obu, N = 58 | total, N = 166 |
|---|---|---|---|---|
| Ecological Factors | ||||
| landscape developed with housing, n (%) | 40 (72.7) | 39 (73.6) | 45 (77.6) | 124 (74.7) |
| landscape with open lots/fields, n (%) | 15 (27.3) | 14 (26.4) | 13 (22.4) | 42 (75.3) |
| relative humidity, % (SD) | 61.6 (3.1) | 60.1 (0.4) | 62.8 (1.1) | 61.6 (2.2) |
| altitude, m (SD) | 1147.3 (3.9) | 1145.8 (4.7) | 1162.1 (8.8) | 1152 (9.7) |
| temperature, °C (SD) | 24.4 (1.7) | 23.4 (1.1) | 22.1 (0.99) | 23.3 (1.4) |
| surface water present, n (%) | 15 (27.3) | 11 (20.8) | 14 (26.4) | 40 (24.1) |
| Potential Fecal Sources | ||||
| human/animal feces on ground, n (%) | 37 (67.3) | 36 (67.9) | 44 (75.9) | 117 (70.5) |
| human open defecation indicators, n (%) | 13 (23.6) | 19 (35.8) | 15 (25.9) | 47 (28.3) |
| domestic animals present, n (%) | 41 (75.6) | 42 (79.3) | 45 (77.6) | 128 (77.1) |
| pig | 2 (3.6) | 2 (3.8) | 2 (3.5) | 6 (3.6) |
| chicken | 22 (39.3) | 23 (43.4) | 34 (58.6) | 79 (47.3) |
| cattle | 8 (14.3) | 11 (20.8) | 12 (20.7) | 31 (18.6) |
| dog | 9 (16.1) | 13 (24.5) | 6 (10.3) | 28 (16.8) |
| sheep/goat | 5 (8.9) | 12 (22.6) | 13 (22.4) | 30 (17.9) |
| flies present, n (%) | 51 (96.2)b | 50 (94.3) | 56 (96.6) | 157 (95.7)b |
| shared or public latrine on-site, n (%) | 45 (81.8) | 44 (83) | 39 (67.2) | 130 (78.3) |
| functional | 0 (0) | 9 (17.0) | 0 (0) | 9 (5.4) |
| dilapidated | 44 (100) | 36 (67.9) | 39 (100) | 121 (72.9) |
| children under 5 present, n (%) | 24 (43.6) | 17 (32.1) | 31 (53.5) | 72 (43.4) |
Total of 145 single sample sites and 21 multisample (more than one soil sample) sites. Nya-A, Nyalenda A neighborhood; Nya-B, Nyalenda Bneighborhood; Obu, Obunga neighborhood.
Denominator = 163.
Children less than five years of age were observed at 40% (66 of 166) sites (median two children per site, range 1–10), including infants (defined as unable to stand and walk) at 9% (15 of 166) of sites. At least one <5yr child was observed crawling on the ground or sitting on the ground playing in dirt or mud at 24 sites, with a third of these observations (n = 8) recording infants crawling or sitting in the dirt. Children were observed playing with objects in the dirt at four sites. One infant and one child ~ 12–24 months were observed playing in a water puddle or drain water at two sites. Children were seen mouthing their hands at three sites (multiple ages), and one infant was eating soil at one site. Open defecation was not observed.
Microbial Detection, Concentration, and Neighborhood-Level Diversity in Kenya.
A total of 185 public site soil samples and 51 water samples were collected and tested for enterococci and enteric pathogens. Enterococcal colonies were isolated from 100% of surface water samples and 74.6% of soil samples (Figure 1). At least one type of enteric pathogen was detected in 92% of water samples and 82% of soil samples. This included 20 gene markers representing 16 types of enteric pathogens in water and 20 gene markers representing 17 types of pathogens in soil. The most common pathogens in soil were Cryptosporidium (67.0%), Giardia (17.8%), ETEC (13.5%), EAEC (10.8%), and EPEC (8.6%). The most common pathogens in water were Cryptosporidium (70.0%), EAEC (58.0%), ETEC (56.0%), EPEC (46%), and human adenovirus (36.0%). The relative abundance of different biological phyla and types of pathogens differed between pooled soil and water samples. Protozoan organisms accounted for 61.2% of microbial organisms in soil, followed by 30.6% pathogenic bacteria, 5.8% helminths, and 2.3% viruses (Figure 2). In contrast, pathogenic bacteria were more abundant in water (57.5%), followed by 26.9% protozoans, 5.8% viruses, and 2.3% helminths.
Figure 1.
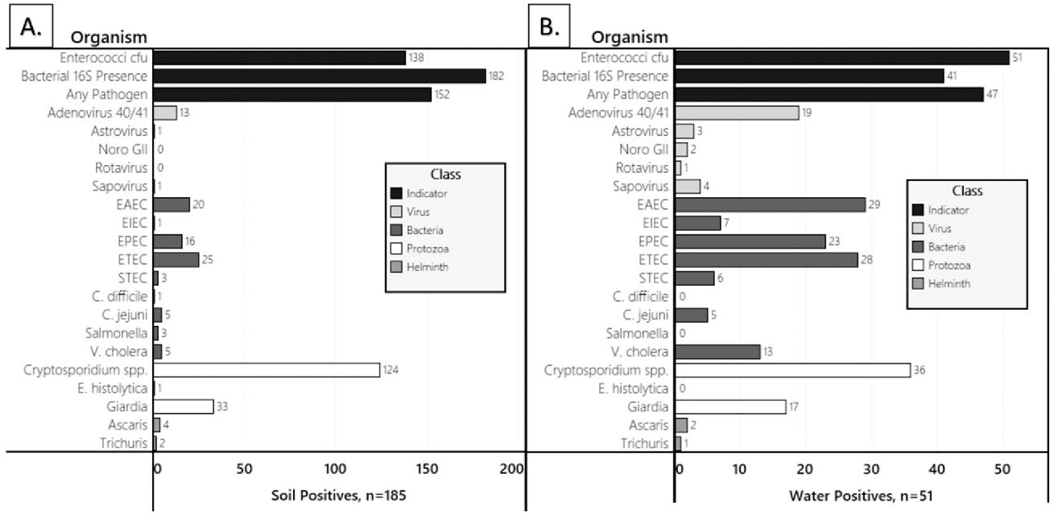
Detection frequency for enterococcal bacteria, bacterial 16S DNA, and enteric virus, bacteria, protozoan, or helminth pathogens in (A) soil and (B) surface water from Kisumu neighborhoods.
Figure 2.
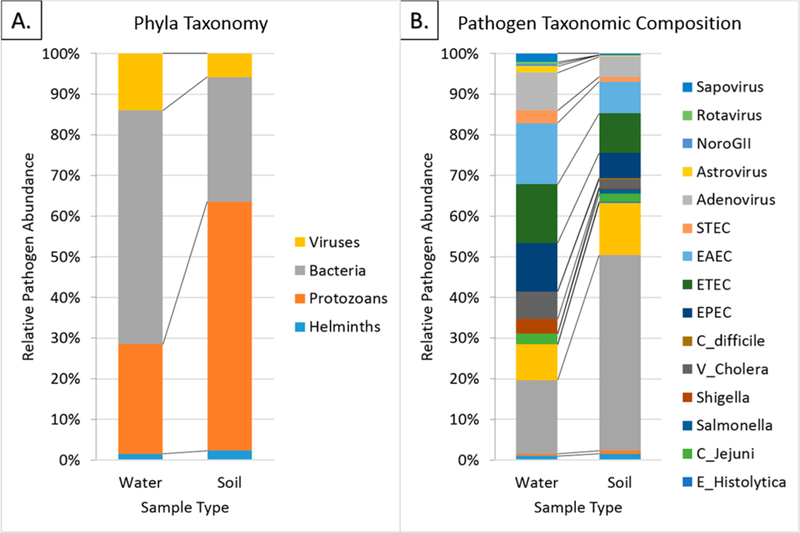
Relative abundance of pathogen gene copies for (A) phylogenetic groups and (B) categories of enteric pathogens in soil and surface water from Kisumu neighborhoods.
Enterococcal concentrations were about 3 log units higher in concentration in surface water than in soil (Table S3). Mean pathogen concentrations in positive samples were 2–5 log units higher than the limit of detection, although wide standard deviations for all pathogens reflected variability in concentration ranges (Table S3). Concentrations of pathogens in 1 g of soil were consistently 1–2 log units higher in concentration than those in 1 mL of surface water. In soil, the adenovirus 40/41 hexon, ETEC eltB, Salmonella invA, and STEC stx2 genes were all detected at >7.0 log10 units/g, whereas C. difficile tcdB was the lowest concentration at 4.2 log10 units/g. In surface water, adenovirus 40/41 hexon was the highest at 6.6 log10 units/mL, followed by EAEC aaiC, ETEC eltB, and STEC stx1 and stx2 genes at 5.6 log10 units/mL, and astrovirus capsid, norovirus GI/GII ORF1–2, and Campylobacter cadF at <3 log10 units/mL.
Within-Site Enteric Pathogen Diversity.
Two or more enteric pathogens were codetected in 35% (64/185) of soil samples (median 1; range 0–7) and in 69% (35/51) of surface water samples (median 4; range 0–10) (Figure 3). Patterns of pathogen codetection were heterogeneous, with only two patterns (Cryptosporidium spp. + EPEC + ETEC + EAEC and Cryptosporidium spp. + Giardia spp. + ETEC + EAEC) reoccurring twice at a total of two sites each among the 35 water samples with ≥2 pathogens detected. Of the 64 soil samples with ≥2 pathogens codetected, Cryptosporidium spp. + ETEC + EAEC occurred three times, Cryptosporidium spp. + Giardia spp. + and ETEC occurred twice, and the remainder were various combinations of Cryptosporidium spp. with other types of pathogens. The Cryptosporidium spp. assays might have detected species that do not typically infect humans.29 If Cryptosporidium spp. results are excluded from the analysis, then 44% (82/185) of soil samples and 80% (41/51) of water samples contained at least one pathogen, while two or more enteric pathogens were codetected in 18% (33/185) of soil samples (median 0; range 0–6), and 65% (33/51) of surface water samples (median 3; range 0–9).
Figure 3.
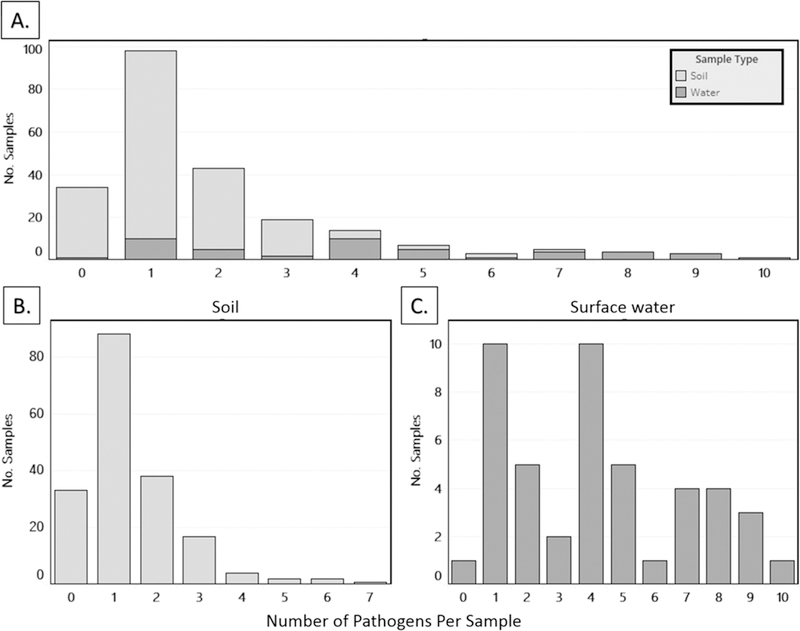
Histogram of diversity in enteric pathogens detected in (A) soil and surface water combined, (B) soil, and (C) surface water samples from Kisumu neighborhoods.
Association between Sanitary Conditions and Enterococcal Contamination.
Observation of feces, indicators of human open defecation, domestic animals, presence and poor condition of latrines, and flies were not associated with the presence (Table 2A) or increased concentration (Table 2B) of enterococcal indicators of bacterial fecal contamination in water and soil, after adjustment for development of the terrain, relative humidity, altitude, ambient temperature, and type of sample.
Table 2.
Association between Human Sanitation and Animal Feces Sources and the Presence and Concentration of Enterococci in Soil from Kisumu Neighborhood Public Domainsa
| (A) Presence of Enterococci | (B) Log10 | Concentration of Enterococci | |||||
|---|---|---|---|---|---|---|---|
| exposure | enterococci present, % (n positive/N) |
unadj OR (95% CI) |
adj OR (95% CI) |
exposure | mean log10 concn (SD) |
unadj RR (95% CI) |
adj RR (95% CI) |
| observed feces on ground | 80.7 (125/155) | 2.67 (0.41, 17.44) | 2.42 (0.62, 9.45) | observed feces on ground | 2.5 (2.0) | 1.65 (0.92, 2.96) | 1.25 (0.66, 2.37) |
| no feces on ground | 76.1 (54/71) | ref | ref | no feces on ground | 2.4 (2.1) | ref | ref |
| observed open defecation indicators | 71.0 (44/62) | 1.42 (0.31, 6.55)b | 0.86 (0.22, 3.28) | observed open defecation indicators | 2.1 (2.0) | 1.63 (0.87, 3.05)b | 1.18 (0.60, 2.33) |
| no open defecation indicators | 82.3 (135/164) | ref | ref | no open defecation indicators | 2.6 (2.0) | ref | ref |
| domestic animals | 83.0 (146/176) | 2.88 (0.35, 24.03) | 3.42 (0.69, 17.03) | domestic animals | 2.6 (2.0) | 1.82 (0.86, 3.85) | 1.50 (0.63, 3.56) |
| no domestic animals | 66.0 (33/50) | ref | ref | no domestic animals | 2.0 (2.1) | ref | ref |
| flies | 79.9 (17½14) | 1.73 (0.11, 27.31) | 0.72 (0.05, 11.39) | flies | 2.5 (2.0) | 3.02 (1.39, 6.56) | 1.88 (0.74, 4.76) |
| no flies | 57.1 (4/7) | ref | ref | no flies | 0.6 (0.9) | ref | ref |
| latrine condition: none | 71.1 (32/45) | ref | ref | latrine condition: none | 1.9 (2.0) | ref | ref |
| latrine condition: good | 66.7 (8/12) | 0.78 (0.10, 5.85) | 0.59 (0.05, 7.40) | latrine condition: good | 1.2 (1.6) | 0.67 (0.23, 1.94) | 0.67 (0.22, 2.09) |
| latrine condition: bad | 82.5 (139/169) | 1.07 (0.19, 6.03) | 1.03 (0.19, 5.56) | latrine condition: bad | 2.7 (2.0) | 1.50 (0.70, 3.22) | 1.31 (0.57, 3.01) |
All models are adjusted for landscape features, relative humidity, altitude, and ambient temperature. Adjusted models also include all five exposure variables at the same time. SD, standard deviation; OR, odds ratio; RR, risk ratio; CI, confidence interval; ref, reference value. Surface water is not included because all samples were positive. Statistical significance (P < 0.05) is indicated by boldface type.
The unadjusted OR and RR for this model reflect significant confounding by an ecological variable. Bivariate association for open defecation indicators and presence of enterococci is unadj OR = 0.50 (95% CI 0.19, 1.34), and the bivariate model for open defecation indicators and log10 concentration of enterococci is unadj RR = 0.92 (95% CI 0.48, 1.76).
Association between Sanitation Conditions and Pathogen Diversity.
Observation of feces, indicators of human open defecation, domestic animals, presence and poor condition of latrines, and flies were not associated with the detection of any type of enteric pathogen, after adjustment for development of the terrain, relative humidity, altitude, ambient temperature, and type of sample (Table 3A). However, the presence of domestic animals was significantly associated with increased diversity in enteric pathogens in fully adjusted models, although observation of feces, indicators of human open defecation, presence and poor condition of latrines, and flies were not associated with increased diversity (Table 3B). Subanalysis of animal types in the fully adjusted model found that chickens (adjOR = 1.64; 95% CI 1.22, 2.22), cattle (adjOR = 1.36; 95% CI 1.08, 1.72), and goats and sheep (adjOR = 1.39; 95% CI 1.00, 1.94) were associated with increased pathogen diversity, while pigs (adjOR = 0.80; 95% CI 0.36, 1.77) and dogs (adjOR = 1.20; 95% CI 0.87, 1.65) were not significantly associated. No association was observed between different fecal sources and increased concentration of individual types of pathogens (data not shown).
Table 3.
Association between Human Sanitation and Animal Feces Sources and Presence and Diversity in Enteric Pathogens from Kisumu Neighborhood Public Domainsa
| (A) Presence of Any Pathogen | (B) Pathogen Diversity | ||||||
|---|---|---|---|---|---|---|---|
| exposure | any pathogen, % (n positive/N) |
inadj OR (95% CI) |
adj OR (95% CI) |
exposure | mean diversity (SD) |
unadj RR (95% CI) |
adj RR (95% CI) |
| observed feces on ground | 83.9 (130/155) | 1.41 (0.41, 4.87) | 1.01 (0.25, 4.04) | observed feces on ground | 1.9 (1.9) | 1.04 (0.80, 1.34) | 1.0 (0.75, 1.33) |
| no feces on ground | 83.1 (59/71) | ref | 1.02 ref | no feces on ground | 1.8 (2.0) | ref | 2.0 ref |
| observed open defecation indicators | 82.3 (51/62) | 1.46 (0.41, 5.19) | 0.98 (0.22, 4.52) | observed open defecation indicators | 1.4 (1.5) | 0.95 (0.70, 1.29) | 0.88 (0.63, 1.22) |
| no open defecation indicators | 84.2 (138/164) | ref | ref | no open defecation indicators | 2.0 (2.1) | ref | ref |
| domestic animals | 85.3 (151/177) | 1.60 (0.43, 5.91) | 1.53 (0.28, 8.54) | domestic animals | 2.0 (2.1) | 1.39 (1.03, 1.89) | 1.44 (1.01, 2.06) |
| no domestic animals | 77.6% (38/49) | ref | ref | no domestic animals | 1.3 (1.2) | ref | ref |
| flies | 84.7 (182/215) | 17.1 (0.70, 418.0) | 5.15 (0.20, 130.1) | flies | 1.9 (2.0) | 2.35 (1.00, 5.56) | 2.00 (0.84, 4.79) |
| no flies | 42.9 (3/7) | ref | ref | no flies | 0.6 (0.8) | ref | ref |
| latrine condition: none | 84.4 (38/45) | ref | ref | latrine condition: none | 1.5 (1.7) | ref | ref |
| latrine condition: good | 66.7 (8/12) | 0.16 (0.01, 2.72) | 0.70 (0.10, 4.99) | latrine condition: good | 0.8 (0.8) | 0.77 (0.39, 1.51) | 0.78 (0.40, 1.53) |
| latrine condition: bad | 84.6 (143/169) | 0.78 (0.17, 3.54) | 0.98 (0.21, 4.52) | latrine condition: bad | 2.0 (2.0) | 1.12 (0.79, 1.58) | 1.08 (0.74, 1.57) |
Adjusted for landscape features, relative humidity, altitude, and ambient temperature. SD, standard deviation; OR, odds ratio; RR, risk ratio; CI,confidence interval; ref, reference value. Statistical significance (P < 0.05) is indicated by boldface type.
Spatial Clustering of Sites with High Pathogen Diversity.
High pathogen diversity was not statistically associated with lower distance between pairs of sites (p-value = 0.22).
DISCUSSION
This study confirmed that neighborhood landscapes in Kisumu, Kenya, are contaminated by many enteric viral, bacterial, protozoan, and helminthic enteric pathogen species, at both neighborhood and localized (within 25 m radius) levels of spatial scale. At least one type of pathogen was detected at 80% of public sites, with a third of soil samples and three-quarters of surface water samples co-contaminated by multiple taxa of enteric pathogens. Even if Cryptosporidium detections are excluded, in the event that the species detected were not human-infective, multiple pathogens were still detected in a fifth of soil sampels and two-thirds of water samples. This evidence addresses major knowledge gaps about enteric pathogen co-occurrence patterns in the environment, especially for public areas that are “ground zero” for contamination from open defecation and animal feces. The most abundant types of pathogen taxa varied for soil and surface water, with protozoans being most abundant in soil and pathogenic bacteria being most abundant in surface water. Domestic animals (specifically chickens, cows, and goats/sheep)—rather than human sources of fecal waste—were associated with increased pathogen diversity, whereas enterococcal bacteria, presence of any pathogen type, and concentrations of individual enteric pathogen taxa were not significantly associated with human and animal sources of fecal contamination. Finally, this study validated that <5yr old children play in these contaminated public settings, confirming that child exposure to and infection by pathogens in public areas is plausible. Therefore, interventions that prevent neighborhood-level animal fecal contamination may be necessary for reducing enteric disease burden in <5yr children in Kisumu. These interventions could include creation of protected child play spaces with finished, cleanable floors, promoting the safe disposal of animal waste, or animal penning designs that safely collect animal waste away from children.
These findings confirmed our hypothesis that pathogen diversity in Kisumu neighborhoods would mirror the leading etiologic causes of symptomatic and asymptomatic childhood infections in low-income, high-burden endemic countries in Asia and Africa.1,2,27 Many of the most common causes of moderate to severe diarrhea in children in Kenya (Cryptosporidium spp., ETEC ST toxin-expressing E. coli, adenovirus 40/41, C. jejuni)1,30 were the most frequently detected in the environment in Kisumu, although rotavirus and Shigella/EIEC were rare. This suggests that widespread contamination by those pathogens in the environment is responsible for causing enteric infection in children. Our findings also confirmed our hypothesis that multipathogen contamination of soil and water would be detected at locations where children play. This further suggests that children could ingest multiple pathogens during play in just one area outside the household. Pathogen diversity is a novel approach for characterizing environmental exposure risks in low-income countries and for identifying potential fecal sources associated with environmental contamination. However, microbial community characterization has been used for decades to understand ecological health and to monitor the impact of natural or man-made actions on system function.31–33 In public health, microbiome community characterization has become a platform for understanding human susceptibility to enteric infection, symptomology and severity of disease, and vaccine failure.34,35 A few studies have used the microbial community approach to understand the relationships between environmental microbiota, fecal indicator bacteria, and pathogenic bacteria in surface water and soil, although they did not examine infrastructure conditions driving this contamination.36,37 Rather than sequencing, we used customized multipathogen qPCR tools for disease-targeted detection of the viral, bacterial, protozoan, and helminthic causes of pediatric diarrhea.27 Additionally, we accounted for potential interrelated conditions underlying pathogen co-occurrence in our analysis rather than treating pathogen occurrence as an isolated, independent event.
The inclusion of pathogen diversity as an indicator of interrelated contamination conditions was important for understanding the relative intensity and determinants of contamination of public areas across Kisumu, as well as between Kenya and a reference site in Iowa. Consistent with prior studies, enterococci were omnipresent in Kenya and Iowa soil and surface water, even though evidence of human fecal contamination was absent in Iowa and animal feces were less common.15 Evidence on fecal microbes in soil is limited, but our detection rates were similar to at least one other study.38 The lack of association between human and animal fecal source risk factors with enterococci in Kenya reinforces that enterococcal indicators are not optimal tools for predicting or quantifying exposure to enteric pathogens or for fecal source tracking. An “any pathogen” indicator was also not statistically associated with fecal sources. However, the stark differences in overall pathogen detection frequencies between Kenya and an Iowa watershed impacted by farm and agriculture helped calibrate our expectations as to what contamination patterns should look like in a setting where open defecation is absent, and they highlighted how alarming the pathogen detection levels in Kenya are by comparison. Sampling at different times, for example, different seasons, or sampling from agriculturally impacted areas in a tropical state (e.g., Florida), may have produced different results.
Neighborhood-level pathogen contamination in Kenya corresponded with the frequent observation of human and animal feces in public areas. These neighborhoods have low levels of household sanitation coverage, and domestic animal ownership is common in Kisumu.21,22 We expected open defecation, dilapidated latrines, and domestic animals to be associated with pathogen diversity, given the evidence of their role in diarrhea in children.39,40 In spite of widespread human feces in Kisumu, only domestic animals were associated with increased diversity in pathogens in the environment. Domestic animal reservoirs may contribute to pediatric disease burden by facilitating a rapid turnover in the types of pathogen species that children are exposed to, a situation where exposure-based immunity provides little benefit. We are unaware of evidence of this hypothesis, but theoretically domestic animals may be more important disease vectors than humans. First, animals (dogs, chickens, pigs, ducks) are more likely to be coprophagous (eat feces than humans) and therefore have higher risks of infection.41 Second, animals are less likely to be treated for diarrheal symptoms of enteric infection than children42 and therefore shed pathogens for longer times due to persistent infection.Third, dozens of animals may be kept by a household versus one or a few children,21 therefore density of potential animal disease vectors is higher than child vectors. The lack of association between human sanitation and pathogen presence or diversity might reflect lower levels of pathogen infection rates in humans relative to animals. We think that the lack of association with all sanitary conditions and pathogen concentration is due to ecological conditions (humidity, UV, properties of soil) playing a strong mediating role on pathogen persistence and fate in the environment.
This study has several limitations. It is cross-sectional and cannot establish the direction of causality. Pathogen contamination could be a proxy for sites with more abundant animal food sources (feces, grassland), which thus attract domestic animals or flies to those sites. Even if that is the case, this is likely a circular relationship that involves animals defecating at the feeding site. Our observational criteria may not have distinguished well between unsafe latrines (those that are deteriorated and/or sometimes unused) and safe latrines (those that effectively contain excreta and prevent release into the environment). Without an in-depth inspection of the underground integrity of the latrine pits, the degree to which excreta are effectively contained cannot be assessed. Our sample size may have not provided sufficient power to detect important associations between fecal sources and microbial outcomes. There was no prior source of information to predict frequencies of enteric pathogen co-occurrence, especially for 19 enteric pathogens.
Inherent heterogeneity in pathogen distributions in the environment and insufficient spacing scale between sites (sites too far apart) may explain why we did not find many repeat patterns of pathogen co-occurrence or spatial hot zones. Such patterns would provide even more precise metrics for linking widespread contamination to specific fecal sources, compared to the relatively simple approach of simply summing the number of pathogen types detected. In the handful of repeat patterns noted, the pathogens detected were the most common overall, suggesting presence is more a function of probability and sample size than impacts from types of fecal sources. In particular, the most common pathogen gene detected was Cryptosporidium spp. 18S, which could indicate contamination by species rarely detected in humans (Cryptosporidium meleagridis, Cryptosporidium canis, Cryptosporidium felis, Cryptosporidium muris, or Cryptosporidium suis) rather than the species (Cryptosporidium hominus and Cryptosporidium parvum) commonly linked with human infection.29 This may overestimate the abundance of clinically relevant pathogens in public areas of Kisumu, although even after exclusion of Cryptosporidium-positive counts, pathogen detection and codetection was still very high. Research involving larger numbers of environmental samples is needed to understand whether co-occurrence patterns of specific groups of pathogens is common. Either way, the substantial amount of heterogeneity in pathogen detection patterns highlights the risk for misclassification of environmental exposures if the presence of an individual pathogen type is used as a proxy for the presence of any fecal pathogens or for fecal contamination overall. While some associations with trends toward significance may surpass significance cutoff thresholds with larger sample sizes, we are confident that it would not change the overall conclusion that domestic animals are important contributors to multipathogen contamination in public domains of Kisumu.
Finally, detection of pathogen DNA or RNA in soil and water by PCR does not confirm viability or infectivity of pathogens. PCR methods may detect extracellular DNA or intact but noninfectious cells. By filtering sample eluate through a 0.45 μm pore size filter, our sampling methods may have removed some extracellular material, although nonviable cellular DNA or RNA could still be trapped on the filter. PCR was chosen as the most systematic way to detect viral, bacterial, protozoan, and helminthic classes of fecal pathogens, many of which have no alternative detection methods. Culture-based assays could have been used for some bacteria but may have been equally inaccurate due to the presence of viable but nonrecoverable bacteria (e.g., V. cholerae, Shigella dysenteriae, C. jejuni, and enterotoxigenic E. coli).17,43–45 Since we did not adopt a pre-enrichment step, it is possible that our enterococcal concentration data underestimate actual live concentrations of enterococci in soil or water. This also would introduce intermethod variability in detection methods across viral, bacterial, protozoan, and helminthic taxa. Adjusting concentrations for pathogen decay under these ecological conditions may improve the accuracy of estimated PCR concentrations. However, there is no such information for many enteric pathogens included in our assay, so such an approach could not be systematic. Also problematic is that feces contamination by animals and humans in Kisumu is ongoing. Failing to counteradjust pathogen concentration estimates for periodic reintroduction of pathogens in the environment would underestimate final concentration estimates. While some pathogen DNA detected in Kisumu may be nonviable, it is more likely to be relatively recent contamination because free and cellular DNA/RNA is typically degraded by native soil or water microbiota within a matter of days or weeks.46–51
In conclusion, this study addresses two neglected realms in the WASH sector. First, we demonstrate that public domains are highly contaminated and may pose a substantial risk of enteric pathogen exposure for children. Young children will continue to play in soil and surface water in the public domain because they live in crowded conditions and at a certain age are drawn to engage in social play with their peers. In light of this, WASH interventions should invest in improving public sanitary conditions to prevent child exposure to enteric pathogens and possibly to reduce the childhood enteric disease burden that persists in many low- and middle-income countries. Second, even if high levels of household WASH coverage are achieved and human open defecation is eliminated, a high baseline level of enteric disease caused by contact with animal feces will persist. Domestic animal management must be included in the WASH agenda to reduce pathogen contamination at the neighborhood level, and potentially within households as well. Forthcoming research by this group is examining the generalizability of these findings in other geographical settings using improved assessment methods. Yet, the conditions in Kisumu—low-income urban settlements with high population density and limited public health infrastructure—are common in low-income settlements around the world, suggesting pervasive pathogen contamination of the environment in such settings is a universal problem.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by start-up funding from the University of Iowa to K.K.B., a career development award from the University of Iowa Environmental Health Sciences Research Center (NIH P30 ES005605), and a pilot grant from the University of Iowa Water Sustainability Initiative. We appreciate the support of Lucy DesJardin and Nancy Hall at the Iowa State Hygiene Laboratory, who allowed our group access to a ViiA7 thermocyler. We thank Julius Ochieno, Lily Lukorito, Horace Phoxydee, Mandy Larson, Lena Swander, Gocale Nicoue, and Kevin Tsai, who assisted with the collection of data, development of assays, or data management. Craig Ellermeier at the University of Iowa generously donated several strains of C. difficile for development of standard controls.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b01528.
Additional text describing RT-PCR inhibition and standard curves, back calculations of final concentrations, and microbial detection and diversity in Iowa watershed; two figures showing location of public domain sampling sites in Kisumu, Kenya, and detection of pathogens from an agricultural Iowa watershed; three tables listing soil and surface water samples from Kenya with evidence of inhibition of the QuantiFast Internal Control, source of reference DNA or RNA used for standard curves, and concentrations of enterococcal indicator bacteria and enteric pathogen DNA and RNA gene copies (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Kotloff KL; Nataro JP; Blackwelder WC; Nasrin D; Farag TH; Panchalingam S; Wu Y; Sow SO; Sur D; Breiman RF; Faruque AS; Zaidi AK; Saha D; Alonso PL; Tamboura B; Sanogo D; Onwuchekwa U; Manna B; Ramamurthy T; Kanungo S; Ochieng JB; Omore R; Oundo JO; Hossain A; Das SK; Ahmed S; Qureshi S; Quadri F; Adegbola RA; Antonio M; Hossain MJ; Akinsola A; Mandomando I; Nhampossa T; Acacio S; Biswas K; O’Reilly CE; Mintz ED; Berkeley LY; Muhsen K; Sommerfelt H; Robins-Browne RM; Levine MM Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013, 382 (9888), 209–22. [DOI] [PubMed] [Google Scholar]
- (2).Platts-Mills JA; Babji S; Bodhidatta L; Gratz J; Haque R; Havt A; McCormick BJ; McGrath M; Olortegui MP; Samie A; Shakoor S; Mondal D; Lima IF; Hariraju D; Rayamajhi BB; Qureshi S; Kabir F; Yori PP; Mufamadi B; Amour C; Carreon JD; Richard SA; Lang D; Bessong P; Mduma E; Ahmed T; Lima AA; Mason CJ; Zaidi AK; Bhutta ZA; Kosek M; Guerrant RL; Gottlieb M; Miller M; Kang G; Houpt ER; The MAL-ED Network Investigators.. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. Health 2015, 3 (9), e564–e575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pickering AJ; Julian TR; Marks SJ; Mattioli MC; Boehm AB; Schwab KJ; Davis J Fecal contamination and diarrheal pathogens on surfaces and in soils among Tanzanian households with and without improved sanitation. Environ. Sci. Technol. 2012, 46 (11), 5736–43. [DOI] [PubMed] [Google Scholar]
- (4).Mattioli MC; Pickering AJ; Gilsdorf RJ; Davis J; Boehm AB Hands and water as vectors of diarrheal pathogens in Bagamoyo, Tanzania. Environ. Sci. Technol. 2013, 47 (1), 355–63. [DOI] [PubMed] [Google Scholar]
- (5).Odagiri M; Schriewer A; Daniels ME; Wuertz S; Smith WA; Clasen T; Schmidt WP; Jin Y; Torondel B; Misra PR; Panigrahi P; Jenkins MW Human fecal and pathogen exposure pathways in rural Indian villages and the effect of increased latrine coverage. Water Res. 2016, 100, 232–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Humphrey JH; Prendergast AJ; Ntozini R; Gladstone M; Colford J The Sanitation Hygiene Infant Nutrition Efficacy (SHINE) Trial; American Society for Tropical Medicine and Hygiene Annual Meeting, Baltimore, MD, November 7, 2017. [Google Scholar]
- (7).Null C; Stewart CP; Pickering AJ; Dentz HN; Arnold BF; Arnold CD; Benjamin-Chung J; Clasen T; Dewey KG; Fernald LCH; Hubbard AE; Kariger P; Lin A; Luby SP; Mertens A; Njenga SM; Nyambane G; Ram PK; Colford JM Jr. Effects of water quality, sanitation, handwashing, and nutritional interventions on diarrhoea and child growth in rural Kenya: a cluster-randomised controlled trial. Lancet Glob Health 2018, 6 (3), e316–e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Luby SP; Rahman M; Arnold BF; Unicomb L; Ashraf S; Winch PJ; Stewart CP; Begum F; Hussain F; Benjamin-Chung J; Leontsini E; Naser AM; Parvez SM; Hubbard AE; Lin A; Nizame FA; Jannat K; Ercumen A; Ram PK; Das KK; Abedin J; Clasen TF; Dewey KG; Fernald LC; Null C; Ahmed T; Colford JM Jr. Effects of water quality, sanitation, handwashing, and nutritional interventions on diarrhoea and child growth in rural Bangladesh: a cluster randomised controlled trial. Lancet Glob Health 2018, 6 (3), e302–e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Medgyesi DN; Brogan JM; Sewell DK; Creve-Coeur JP; Kwong LH; Baker KK Where Children Play: Young child exposure to environmental hazards during play in public areas in a transitioning internally displaced persons community in Haiti. Int. J. Environ. Res. Public Health 2018, 15 (8), 1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Harwood VJ; Levine AD; Scott TM; Chivukula V; Lukasik J; Farrah SR; Rose JB Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 2005, 71 (6), 3163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ferguson AS; Layton AC; Mailloux BJ; Culligan PJ; Williams DE; Smartt AE; Sayler GS; Feighery J; McKay LD; Knappett PS; Alexandrova E; Arbit T; Emch M; Escamilla V; Ahmed KM; Alam MJ; Streatfield PK; Yunus M; van Geen A Comparison of fecal indicators with pathogenic bacteria and rotavirus in groundwater. Sci. Total Environ. 2012, 431, 314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Haramoto E; Katayama H; Oguma K; Yamashita H; Tajima A; Nakajima H; Ohgaki S Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci. Technol 2006, 54 (11–12), 301–8. [DOI] [PubMed] [Google Scholar]
- (13).Verbyla ME; Iriarte MM; Mercado Guzman A; Coronado O; Almanza M; Mihelcic JR Pathogens and fecal indicators in waste stabilization pond systems with direct reuse for irrigation: Fate and transport in water, soil and crops. Sci. Total Environ. 2016, 551–552, 429–37. [DOI] [PubMed] [Google Scholar]
- (14).Desmarais TR; Solo-Gabriele HM; Palmer CJ Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 2002, 68 (3), 1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Byappanahalli MN; Nevers MB; Korajkic A; Staley ZR; Harwood VJ Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76 (4), 685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Islam MS; Talukder KA; Khan NH; Mahmud ZH; Rahman MZ; Nair GB; Siddique AK; Yunus M; Sack DA; Sack RB; Huq A; Colwell RR Variation of toxigenic Vibrio cholerae O1 in the aquatic environment of Bangladesh and its correlation with the clinical strains. Microbiol. Immunol. 2004, 48 (10), 773–7. [DOI] [PubMed] [Google Scholar]
- (17).Alam M; Sultana M; Nair GB; Siddique AK; Hasan NA; Sack RB; Sack DA; Ahmed KU; Sadique A; Watanabe H; Grim CJ; Huq A; Colwell RR Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (45), 17801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).The 2009 Kenya Population and Housing Census; Kenyan National Bureau of Statistics, 2010; https://www.knbs.or.ke/publications/.
- (19).Situation analysis of informal settlements in Kisumu; UNHABITAT publication 783/05E; Cities without Slums Sub-Regional Programme for Eastern and Southern Africa, Kenya Slum Upgrading Programme, 2005; https://unhabitat.org/books/situation-analysis-of-informal-settlements-in-kisumu/.
- (20).Kenya - Multiple Indicator Cluster Survey 2011, Nyanza Province; Kenya National Bureau of Statistics, 2013; http://microdata.worldbank.org/index.php/catalog/2660.
- (21).Barnes AN; Mumma J; Cumming O Role, ownership and presence of domestic animals in peri-urban households of Kisumu, Kenya. Zoonoses Public Health 2018, 65 (1), 202–214. [DOI] [PubMed] [Google Scholar]
- (22).Simiyu S; Swilling M; Cairncross S; Rheingans R Determinants of quality of shared sanitation facilities in informal settlements: case study of Kisumu, Kenya. BMC Public Health 2017, 17 (1), 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).U.S. Recreational Water Quality Criteria; U.S. Environmental Protection Agency; http://water.epa.gov/scitech/swguidance/standards/criteria/health/recreation/index.cfm (accessed January 26, 2018).
- (24).Griffith JF; Cao Y; McGee CD; Weisberg SB Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res. 2009, 43 (19), 4900–7. [DOI] [PubMed] [Google Scholar]
- (25).Ahmed W; Harwood VJ; Gyawali P; Sidhu JP; Toze S Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015, 81 (6), 2042–−9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Haramoto E; Katayama H; Asami M; Akiba M Development of a novel method for simultaneous concentration of viruses and protozoa from a single water sample. J. Virol. Methods 2012, 182 (1–2), 62–9. [DOI] [PubMed] [Google Scholar]
- (27).Liu J; Gratz J; Amour C; Kibiki G; Becker S; Janaki L; Verweij JJ; Taniuchi M; Sobuz SU; Haque R; Haverstick DM; Houpt ER A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J. Clin Microbiol 2013, 51 (2), 472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ishii S; Kitamura G; Segawa T; Kobayashi A; Miura T; Sano D; Okabe S Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl. Environ. Microbiol. 2014, 80 (24), 7505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Stroup SE; Roy S; McHele J; Maro V; Ntabaguzi S; Siddique A; Kang G; Guerrant RL; Kirkpatrick BD; Fayer R; Herbein J; Ward H; Haque R; Houpt ER Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J. Med. Microbiol. 2006, 55 (9), 1217–1222. [DOI] [PubMed] [Google Scholar]
- (30).Liu J; Platts-Mills JA; Juma J; Kabir F; Nkeze J; Okoi C; Operario DJ; Uddin J; Ahmed S; Alonso PL; Antonio M; Becker SM; Blackwelder WC; Breiman RF; Faruque AS; Fields B; Gratz J; Haque R; Hossain A; Hossain MJ; Jarju S; Qamar F; Iqbal NT; Kwambana B; Mandomando I; McMurry TL; Ochieng C; Ochieng JB; Ochieng M; Onyango C; Panchalingam S; Kalam A; Aziz F; Qureshi S; Ramamurthy T; Roberts JH; Saha D; Sow SO; Stroup SE; Sur D; Tamboura B; Taniuchi M; Tennant SM; Toema D; Wu Y; Zaidi A; Nataro JP; Kotloff KL; Levine MM; Houpt ER Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016, 388 (10051), 1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Balint M; Bahram M; Eren AM; Faust K; Fuhrman JA; Lindahl B; O’Hara RB; Opik M; Sogin ML; Unterseher M; Tedersoo L Millions of reads, thousands of taxa: microbial community structure and associations analyzed via marker genes. FEMS Microbiol Rev. 2016, 40 (5), 686–700. [DOI] [PubMed] [Google Scholar]
- (32).Xiao XY; Wang MW; Zhu HW; Guo ZH; Han XQ; Zeng P Response of soil microbial activities and microbial community structure to vanadium stress. Ecotoxicol. Environ. Saf. 2017, 142, 200–206. [DOI] [PubMed] [Google Scholar]
- (33).Brandt KK; Amezquita A; Backhaus T; Boxall A; Coors A; Heberer T; Lawrence JR; Lazorchak J; Schonfeld J; Snape JR; Zhu YG; Topp E Ecotoxicological assessment of antibiotics: A call for improved consideration of microorganisms. Environ. Int. 2015, 85, 189–205. [DOI] [PubMed] [Google Scholar]
- (34).Harris VC; Armah G; Fuentes S; Korpela KE; Parashar U; Victor JC; Tate J; de Weerth C; Giaquinto C; Wiersinga WJ; Lewis KD; de Vos WM Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J. Infect. Dis. 2017, 215(1), 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).David LA; Weil A; Ryan ET; Calderwood SB; Harris JB; Chowdhury F; Begum Y; Qadri F; LaRocque RC; Turnbaugh PJ Gut microbial succession follows acute secretory diarrhea in humans. mBio 2015, 6 (3), No. e00381–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mwaikono KS; Maina S; Sebastian A; Schilling M; Kapur V; Gwakisa P High-throughput sequencing of 16S rRNA Gene Reveals Substantial Bacterial Diversity on the Municipal Dumpsite. BMC Microbiol. 2016, 16 (1), 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sun H; He X; Ye L; Zhang XX; Wu B; Ren H Diversity, abundance, and possible sources of fecal bacteria in the Yangtze River. Appl. Microbiol. Biotechnol 2017, 101 (5), 2143–2152. [DOI] [PubMed] [Google Scholar]
- (38).Ben Said L; Klibi N; Dziri R; Borgo F; Boudabous A; Ben Slama K; Torres C Prevalence, antimicrobial resistance and genetic lineages of Enterococcus spp. from vegetable food, soil and irrigation water in farm environments in Tunisia. J. Sci. Food Agric. 2016, 96 (5), 1627–33. [DOI] [PubMed] [Google Scholar]
- (39).Penakalapati G; Swarthout J; Delahoy MJ; McAliley L; Wodnik B; Levy K; Freeman MC Exposure to Animal Feces and Human Health: A Systematic Review and Proposed Research Priorities. Environ. Sci. Technol. 2017, 51 (20), 11537–11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Freeman MC; Garn JV; Sclar GD; Boisson S; Medlicott K; Alexander KT; Penakalapati G; Anderson D; Mahtani AG; Grimes JET; Rehfuess EA; Clasen TF The impact of sanitation on infectious disease and nutritional status: A systematic review and meta-analysis. Int. J. Hyg. Environ. Health 2017, 220 (6), 928–949. [DOI] [PubMed] [Google Scholar]
- (41).Dock G; Bass CC Hookworm disease; etiology, pathology, diagnosis, prognosis, prophylaxis, and treatment. C.V. Mosby Co.: St. Louis, 1910; https://archive.org/details/hookwormdiseasee00dock.
- (42).Kagira JM; Kanyari PW Questionnaire survey on urban and peri-urban livestock farming practices and disease control in Kisumu municipality, Kenya. J. S. Afr. Vet. Assoc. 2010, 81 (2), 82–86. [DOI] [PubMed] [Google Scholar]
- (43).Rollins DM; Colwell RR Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 1986, 52 (3), 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Islam MS; Hossain MA; Khan SI; Khan MN; Sack RB; Albert MJ; Huq A; Colwell RR Survival of Shigella dysenteriae type 1 on fomites. J. Health Popul Nutr. 2001, 19 (3), 177–182. [PubMed] [Google Scholar]
- (45).Lothigius A; Sjoling A; Svennerholm AM; Bolin I Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 2010, 108 (4), 1441–9. [DOI] [PubMed] [Google Scholar]
- (46).Romanowski G; Lorenz MG; Sayler G; Wackernagel W Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl. Environ. Microbiol. 1992, 58 (9), 3012–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Dejean T; Valentini A; Duparc A; Pellier-Cuit S; Pompanon F; Taberlet P; Miaud C Persistence of environmental DNA in freshwater ecosystems. PLoS One 2011, 6 (8), e23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bae S; Wuertz S Decay of host-associated Bacteroidales cells and DNA in continuous-flow freshwater and seawater microcosms of identical experimental design and temperature as measured by PMA-qPCR and qPCR Water Res. 2015, 70, 205–13. [DOI] [PubMed] [Google Scholar]
- (49).Eichmiller JJ; Borchert AJ; Sadowsky MJ; Hicks RE Decay of genetic markers for fecal bacterial indicators and pathogens in sand from Lake Superior. Water Res. 2014, 59, 99–111. [DOI] [PubMed] [Google Scholar]
- (50).Kim M; Wuertz S Survival and persistence of host-associated Bacteroidales cells and DNA in comparison with Escherichia coli and Enterococcus in freshwater sediments as quantified by PMA-qPCR and qPCR. Water Res. 2015, 87, 182–92. [DOI] [PubMed] [Google Scholar]
- (51).Rogers SW; Donnelly M; Peed L; Kelty CA; Mondal S; Zhong Z; Shanks OC Decay of bacterial pathogens, fecal indicators, and real-time quantitative PCR genetic markers in manure-amended soils. Appl. Environ. Microbiol. 2011, 77 (14), 4839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


