Abstract
We designed an epi-illumination SPIM system which utilizes a single objective and has an identical sample interface as an inverted fluorescence microscope with no additional reflection elements. It achieves subcellular resolution and single-molecule sensitivity and is compatible with common biological sample holders, including multi-well plates. We demonstrated multicolor fast volumetric imaging, single-molecule localization microscopy, parallel imaging of sixteen cell lines and parallel recording of cellular responses to perturbations.
For over a decade, selective-plane illumination microscopy (SPIM), or light-sheet microscopy, has been successfully used for 3D imaging applications in developmental and cell biology, anatomical science, biophysics and neuroscience1. Almost all light-sheet microscopes need at least two objectives close together, hence restricting the sample mounting format. In a horizontal SPIM configuration2,3 where the optical pathways are parallel to the optical table, small tubes or cylinders of agarose gel hold the sample in the space surrounded by objectives. To accommodate traditional mounting protocols such as samples prepared on glass coverslips, “dipping” configurations4–6 were developed, with perpendicular optical pathways and the objectives pointing downwards. However, the necessity to prepare samples on tiny coverslips or pedestals complicates cell culture practices. An “open-top” configuration with the objectives pointing upwards7–11 potentially allows a SPIM system to be operated like an inverted fluorescence microscope and accepts conventional biological sample formats. This configuration seamlessly integrates with common cell biology procedures such as drug treatment and parallel imaging using multi-well plates12. Among such methods, Oblique Plane Microscopy7 and Swept Confocally Aligned Planar-Excitation microscopy (SCAPE)10 use a single objective lens for illumination and detection without additional reflecting elements13,14 that mechanically limits sample scanning. The sample is illuminated obliquely, resulting in a tilted illumination plane. This tilting is corrected by a remote imaging module in the detection path so that all light arrives on the camera in focus. However, the remote imaging module leads to a loss of numerical aperture (NA), such that both systems have a NA < 0.77,10, whereas a high NA is essential to achieving the resolution for subcellular imaging and light collection efficiency for single-molecule detection.
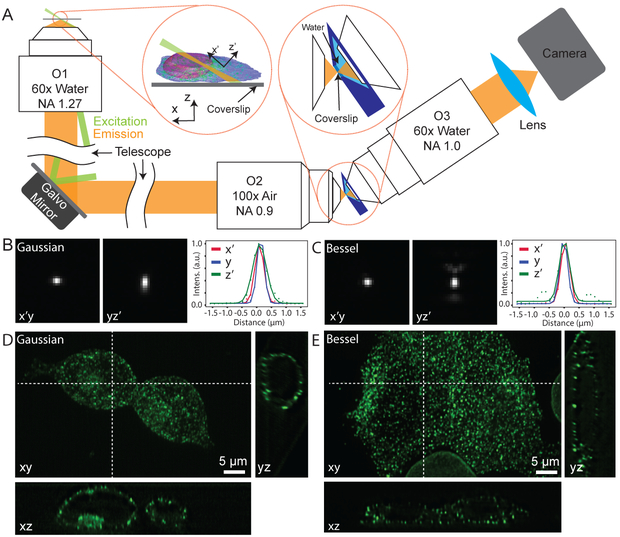
Here, we designed a single-objective oblique epi-illumination SPIM (eSPIM) system to solve the problem of limited resolution and sensitivity (Fig. 1A, Supplementary Figs. S1, S2 and S3). In this design, a high NA water-immersion objective (O1) is used for both illumination and fluorescence collection. The remote imaging module contains two objectives (O2 and O3) arranged at the same angle as the illumination light sheet tilting angle. While this angled arrangement creates an in-focus image of the illuminated plane on the camera, it is exactly the cause of the NA loss7,10 because it shifts part of the light cone generated by O2 outside of the collectable range of O3. To solve this problem, we used a mismatched pair of objectives for the remote imaging module: an air objective for O2 and a water-immersion objective for O3. A 3D-printed water container separates the focal space of the two objectives by a piece of coverglass at the intermediate image plane, with one side being air, the other side being water, and the z’ position of the coverglass adjusted to minimize spherical aberration. The refractive index difference between the working media of O2 and O3 compresses the angle of the O2 light cone, thus minimizing NA loss (Supplementary Fig. S4). Alternatively, O3 can be seen as equivalent to an air objective with NA = 1 where the collection solid angle is 2π. All light transmitted by O2 can then be collected by O3, resulting a high collection efficiency. The effective detection NA of our system is estimated to be ~ 1.20 along the y axis and ~ 1.06 along the x’-axis. The resulting collection efficiency is ~28.2%, equivalent to that of a water-immersion objective of NA ≈ 1.17. We used a Galvo mirror to scan the excitation light sheet and de-scan the image in order to obtain volumetric data. Optical simulation further confirmed performance consistency within a volume of 70 μm × 70 μm × 20 μm (Supplementary Fig. S5). The overall transmission of the remote imaging module is measured to be 62% to 73% depending on the wavelength (see Methods).
Figure 1.
Optical setup and spatial resolution of the system. (A) Schematic of the experimental setup. O1 is used for both illumination (shown in green) and fluorescence (shown in yellow) detection. A remote imaging module composed of two objective lenses O2 and O3 is used to image the oblique plane. A water container (shown in dark blue) in the focal space of O2 and O3 separates them by a glass coverslip, with one side being air medium and the other side water. Left inset shows the definition of the coordinate system. Right inset shows the detailed design of the remote imaging module. (B-C) PSF of the eSPIM with Gaussian and Bessel excitations, measured with 45 nm green fluorescent beads. Representative cross-sections of the PSF in the x’y-plane, in the yz’- plane and intensity plots along the three axes of a bead are shown. The FWHMs are 362.4 nm, 285.1 nm and 533.8 nm along the x’-, y- and z’- axes with Gaussian excitation, and are 346.3 nm, 309.5 nm, 408.9 nm along the three axes with Bessel excitation. (D-E) eSPIM images with Gaussian and Bessel excitations of a mixture of HEK 293T cells with endogenous clathrin A or lamin A/C labeled by mNG211 knock-in. Maximum intensity projection of the 3D dataset in the xy-plane is shown together with representative cross-sections in the xz-plane and in the yz-plane. B and C are raw data without deconvolution. D and E are data after deconvolution using the Richardson-Lucy algorithm with 10 iterations. At least five experiments were repeated independently with similar results for B-E.
In our system, the illumination light sheet can be either a Gaussian or a Bessel15 beam, where the Bessel beam has longer propagation distance and narrower width but some side lobes (see Methods and Supplementary Fig. S6). Measured by imaging fluorescent beads (Fig. 1B–C), the x’- and y-resolutions of our system were 339 ± 18 nm and 316 ± 8 nm (see Methods, full-width at half maximum, or FWHM, mean ± standard deviation), respectively. The z’-resolution was 443 ± 29 nm with the Bessel beam and 596 ± 32 nm with the Gaussian beam (FWHM, mean ± standard deviation). Within an imaging volume of about 70 μm in y, 20 μm in z and Galvo scan range of ~ 100 μm in x, the aberration was minor and our microscope has a performance close to diffraction limit with a Strehl ratio of 0.90 (Supplementary Fig. S7 and S8). This imaging volume is similar to that reported by Lattice Light Sheet Microcopy6 and can be further increased by scanning the sample stage if necessary.
To demonstrate the performance of our system in live cell microscopy, we imaged a variety of subcellular structures in cell lines grown in 8-well coverglass-bottom chambers, including microtubules and mitochondria in HeLa cells (transient over-expression of EGFP-α-tubulin and mRuby2-TOM20, respectively) (Supplementary Figs. S9 and Movies 1–2), endogenously labeled clathrin structures in HEK 293T cells (mNeonGreen211 knock-in for CLTA gene) (Fig. 1D–E and Supplementary Movie 3–4), two-color imaging of nuclear lamina and lysosomes in HEK 293T cells (mNeonGreen211 knock-in for LMNA gene and LysoTracker Deep Red staining, respectively) (Supplementary Figs. S10 and Movie 5–6) and B16F1 mouse cells stably expressing membrane marker CAAX-EGFP (Supplementary Movie 7). The puncta of clathrin structures showed similar resolution-limited dimensions (~ 300 nm laterally and ~ 600 nm axially) as measured earlier with fluorescent beads, demonstrating the practical spatial resolution of our system and enabling tracking of the movement of clathrin structures across the entire volume of cells. Even for endogenously labeled proteins, which typically exhibit much lower fluorescence signal than over-expressed ones, we were able to acquire live cell movies at 0.5 to 2 volumes per second continuously for more than 10 minutes. We verified the greatly reduced photobleaching of our system compared to a spinning disk confocal microscope when recording a similar level of fluorescence signal from the same sample (Supplementary Fig. S11). Our system is also suitable for fast volumetric imaging because the Galvo mirror is the only moving mechanical element in the system; thus, the scanning speed can be pushed to the limit of camera readout. To demonstrate this fast imaging capacity, we imaged Drosophila S2 cells with lysosomes labeled with LysoTracker Deep Red at 14.7 volumes per second (see Supplementary Fig. 12 and Movies 8–10).
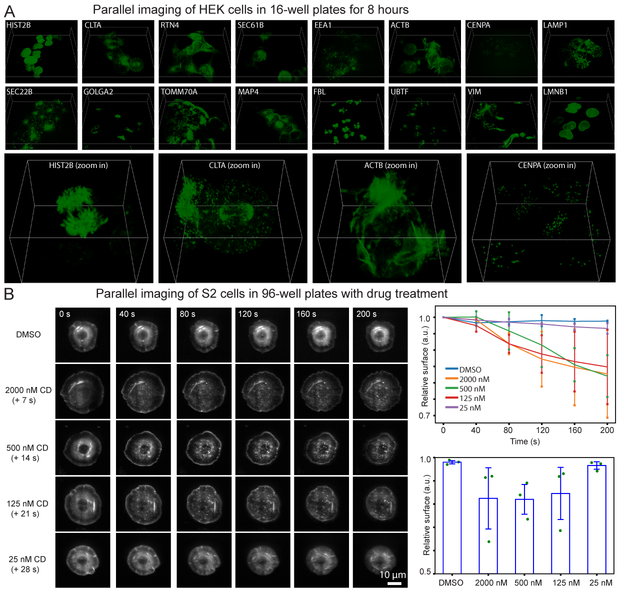
The inverted configuration of our system makes it compatible with open multi-well plates, enabling parallel imaging of many samples as well as chemical perturbation of the samples during image acquisition. To demonstrate these capabilities, we first performed long-term parallel imaging of 16 gene-edited HEK 293T cell lines plated in a 16-well plate, each cell line expressing a protein endogenously labelled by split mNeonGreen2 (Fig. 2A and Supplementary Movies 11–12). The cells were imaged continuously for 8.4 hours, equivalent to a total imaging time of 134.4 hours if imaged in serial. To stabilize the focus drift over such a long period of time, we built an eSPIM system on a commercial inverted microscope that is equipped with a focus stabilization system. The low phototoxicity was evident from the capture of normal mitosis events, including the visualization of clathrin associated with spindles during mitosis. Next, we recorded Drosophila S2 cells stably expressing mRFP-actin in response to the actin drug, Cytochalasin D (CD) in a 96-well plate. 3D images were acquired over time for each well in parallel. Different concentrations of CD were added to the wells after the first round of image acquisition. CD-treated cells clearly show the retraction of the actin-filled leading edge (Fig. 2B and Supplementary Movie 13), with the full dose-response curve acquired in one imaging session. These experiments illustrate the potential of our system in high-content and high-throughput volumetric imaging.
Figure 2.
Parallel volumetric live imaging of cells in multiwell plates. (A) HEK 293T cells were endogenously labeled with split mNeonGreen2. 3D views of a representative time point for each well are shown at the top. Four zoomed-in 3D views are shown at the bottom. The box sizes are 93.1 μm × 93.1 μm × 16.0 μm for the sixteen full field of view images, and 26.6 μm × 26.6 μm × 16.0 μm for the four zoomed-in images. Three experiments were repeated independently with similar results. (B) Drosophila S2 cells stably expressing mRFP-actin were treated with DMSO or Cytochalasin D, an inhibitor of actin polymerization. Representative max intensity projection images of the cells with different treatments and at different time points are shown. Right-top panel shows the normalized surface areas of the cells as a function of time. Right-bottom graph shows the surface areas change of the cells after treatment relative to before treatment. Values are averaged from three experiments, with error bars indicating the standard deviation. Green dots show all the data points.
The high light collection efficiency associated with the high detection NA of our system enables single-molecule imaging. As a demonstration, we performed single molecule localization microscopy of S2 cells stained for microtubules over multiple focal planes throughout the volume of a cell (Supplementary Fig. S13). The ~ 50 nm width of microtubules was revealed with an average of > 2400 photons detected for each photoswitching event for the Alexa Fluor 647 dye. This photon number is ~ 30% of that detected by a 1.40 NA oil immersion objective on a wide-field microscope using near-total-internal-reflection illumination, which is consistent with our measurement of the remote imaging module transmission (when considering the collection efficiency difference between oil and water immersion objectives for near-surface signal. See Supplementary Fig. S13). The oblique light sheet restricts excitation to a selective plane, reducing out-of-focus bleaching and out-of-focus background. Therefore, our system can be particularly useful for the imaging of densely labeled cells with point-accumulation-for-imaging-in-nanoscale-topography (PAINT)16 and for high-content screening17.
In summary, our microscope design provides a versatile platform to image live samples at high spatial-temporal resolution. We have shown that it can accommodate multi-well plates where drug treatment and parallel imaging can be performed effortlessly. We have also shown that it can be built as an add-on unit to existing inverted fluorescent microscopes (in a similar manner as a confocal spinning disk unit), converting a conventional epifluorescence microscope into a SPIM system. Our design is also inherently compatible with numerous methods to improve the performance of light-sheet microscopy, including digitally scanned light sheet (to obtain more uniform illumination), parallel imaging to increase the speed18 (Supplementary Note 1) and the use of adaptive optics19,20 to reduce optical aberrations and to improve image quality.
Methods
Optical setup
A water-immersion objective (O1, Nikon CFI Plan Apo IR 60XWI) of NA 1.27 was used for both illumination and fluorescence collection. The illumination light came from three lasers (Vortran Stradus 488nm and 642nm, Coherent Sapphire 561nm). The beams were combined by two dichroic mirrors, collimated by a telescope composed of two achromatic lens, expanded by two cylindrical lenses (Thorlabs CL 50mm and CL 200mm) to form an elongated shape, and then clipped by a mechanical slit conjugated with the pupil plane of O1. In this configuration, the oblique light sheet is a Gaussian beam. To generate a Bessel light sheet, we apply the same strategy as reported by Zhao et al.15. The slit was replaced with a photomask (HTA Photomask, Supplementary Fig. S14). The photomask can generate the desired light distribution at the pupil plane of O1 by amplitude modulation. The full-width at half-maximum (FWHM) of the Gaussian and Bessel beams are respectively 730 nm and 554 nm along the z’-axis, 12.8 um and 18.6 μm along the x’-axis (theoretical estimate, see Supplementary Fig. S6).
The illumination beam was then reflected by a dichroic mirror (DM, Chroma ZT405/488/561/640rpc) and intersected the back aperture of O1 off-center to generate an oblique light sheet at the focal space of O1. The offset was adjusted so that the illumination light sheet has an angle of 30° with respect to the actual focal plane of O1. We limited the effective NA of the illumination light sheet to ~ 0.3 so that it is not clipped by the edge of O1. The remote imaging module consisted of two objective lenses (O2, Nikon CFI LU Plan Fluor EPI P 100× NA 0.9 and O3, Nikon CFI Fluor 60XW NA 1.0). Imaging an oblique plane requires obtaining a perfect (i.e. aberration-free) intermediate image of a volume. Optical systems simultaneously satisfying both the sine and Herschel conditions fulfill this requirement21. To achieve these conditions, the pupil planes of O1 and O2 are conjugated by two 4f-systems (L1–L4) and the lateral magnification from the sample space to the intermediate image is set to be 1.33, which is the ratio between the refraction indexes of the working media of O1 and O2. Under this condition, the axial magnification is also 1.33.
The optical axis of O3 was 30° relative to that of O2 so that O3 could re-image the intermediate image in focus. A 3D-printed water container separated the focal space of the two objectives by a glass coverslip, with one side being air and the other side water. The water container was mounted on a translation stage (Thorlabs DT12) so that it could be translated along the optical axis of O3. The objective lens O1 was mounted on a manual translation stage (Thorlabs CT1) for focus adjustment. The objective lens O3 was mounted on a piezo stage (Thorlabs DRV517) so that its focus could be finely tuned. By mounting all components of the remote imaging module on the same translation stage, we found that their alignment is robust and stable, and routine realignment is unnecessary. The fluorescence was filtered by either individual band-pass filters (Chroma ET525/50m, ET605/70m and ET705/72m) or a quad-band filter (Chroma ZET405/488/561/640x) and then detected by a scientific CMOS (sCMOS) camera. Three sCMOS cameras were used: PCO Edge and Hamamatsu ORCA-Flash 4.0 for conventional fluorescence imaging and Tuscen 400BSI for single-molecule imaging. The pixel size of the cameras at the sample space was 133 nm for conventional fluorescence imaging and 167 nm for single molecule imaging.
A Galvo mirror (Thorlabs, GVS011) was conjugated to both the pupil planes of O1 and O2. Rotating the Galvo mirror scans the oblique light sheet across the sample (along the x-axis), with the incident angle kept at 60°. The Galvo mirror also descans the intermediate image at the focal space of O2 so that the intermediate image is always projected at the focal plane of O3. Neither the microscope stage nor the objective O1 need to move, ensuring mechanical stability. The scanning frequency of the Galvo mirror can be as fast as a few hundred Hz. Hence, the imaging speed was mostly limited by the readout time of the camera and the power of the excitation laser. The frame rate of the camera is as fast as 800 frames per second for a region of interest of 640 × 256 pixels. The Galvo mirror scans the light sheet faithfully across ~ 100 μm. The imaging volume is ~ 100 μm * 70 μm * 20 μm and can be further increased through tiling22. Outside of this range, either the illumination or the fluorescence light starts to be cropped. It is also possible to scan the sample with the microscope stage (PI, PILine M-687.UN) for longer ranges, albeit at slower speed. The entire system is controlled by the open-source, freely available software Micro-Manager23.
A list of materials used to construct the eSPIM microscope can be found in Supplementary Note 2.
Transmission efficiency of the remote imaging module
To measure the transmission efficiency of the remote imaging module (consisting of O2, O3 and the water chamber), we temporally removed CL1-CL3 and the slit and placed a mirror at the focal plane of O1 so that a collimated laser light is reflected backwards. The reflected beam can pass through DM1 and is attenuated. The laser beam then enters O2 perpendicular to its aperture plane and the size of the laser beams is ~ 4 mm in diameter. We measured the transmission efficiency by calculating the ratio of the light power that exits and enters the remote imaging module (i.e. before O2 and after O3). The transmission efficiency were respectively 73% at 488 nm, 67% at 561 nm and 62% at 641 nm.
Optical alignment
The optical alignment (see Supplementary Note 3 for a detailed alignment procedure) stays stable for months without the need to readjust. We typically check and adjust the position of the glass coverslip on the water chamber about every three months so ensure optimal performance of the microscope. The alignment of the other optical elements is more robust and an annual check should be enough.
eSPIM microscope based on a commercial microscope stand
Apart from the prototype system described above, we also built a second eSPIM system using a Nikon Ti-E microscope. The Ti-E stand has a tube lens already installed inside. The tube lens has the same focal length as the lens L1 (Supplementary Figure 1) and replaces L1. The rest of the eSPIM optics was identical and was built outside the left side-port of the Ti-E microscope. All other components and functionalities of the Ti-E stand were intact, including a second camera directly attached to the right side-port to acquire wide-field epi-fluorescence and bright-field transmitted light images, as well as additional objectives on the nose piece for the acquisition of wide-field images at different magnifications. The Ti-E microscope has a Perfect Focus System which greatly facilitate long-term recording. It also offers many ready-to-use functionalities. The parallel long-term imaging of HEK 293T cells was performed on this setup. All other experiments were performed on the prototype system.
Characterization of resolution with fluorescent beads
45 nm fluorescent beads were imaged to characterize the resolution of the system. The beads were embedded in 2% agarose gel and then sandwiched between a glass coverslip and a glass slide. The sample was then placed on the microscope stage with the coverslip side facing the objective. The excitation wavelength was 488 nm and the corresponding emission filter is a bandpass 525/50 filter. The raw images were slices across the sample in the x’-y plane, 30° to the x-y plane. Figures 1B and 1C show the cross-sections of bead images in the x’-y and y-z’ planes. The FWHMs of the bead images were 339 ± 18 nm and 316 ± 8 nm (mean ± standard deviation for all values in this section) along the x’- and y-axes. It was 596 ± 32 nm along the z’-axis at the waist of the light sheet with the Gaussian excitation beam. At 20 μm away from the waist (along the x’-axis), this values goes up to 1227 nm ± 287 μm, both due to the divergence of the light sheet and increased aberrations. When using the Bessel beam, the FWHM of the PSF along the z’-axis is 443 ± 29 nm at the beam waist and 667 ± 300 nm at 20 μm away from the waist (along the x’-axis), clearly improving the axial resolution. These values were obtained by averaging 5 to 7 beads.
Evaluation of chromatic aberrations
200 nm multicolor fluorescent beads were embedded in 2% agarose gel and sandwiched between a glass coverslip and a glass slide. We then sequentially acquired volumetric images of the beads in multiple channels (i.e. with respectively 488 nm, 561 nm and 642 nm laser excitation). We then performed cross-correlation of the images in pair to find out the shifts between the different channels. The shifts between the 488 and 561 channels are 0.1 nm, –1.7 nm, and –29.9 nm along the x’-, y- and z’-axes; the shifts between the 561 and 642 channels are –37.1 nm, 13.0 nm, and −104.2 nm along the x’-, y- and z’-axes. These values are much smaller than the resolution of the microscope, the chromatic aberrations of our system is therefore negligible.
Data acquisition, processing and viewing
Micro-Manager23 was used for device control and multi-dimensional data acquisition. The Galvo scanner was set up as a DA (digital-analogue)-z stage, controlled by one Analog Output channel of the NI PCIe 6323 DAQ card. The lasers emission states were controlled via an Arduino Uno board. The Galvo scanner and the lasers were hardware-synchronized23 through the TTL output of the camera.
The raw SPIM data were obtained in the x’-y-z’ space (Figure 1A). The data were then de-skewed, and deconvolved using the measured PSF and rotated to the x-y-z space. This process was performed with a free-online package (https://www.flintbox.com/public/project/31374/) produced by Janelia Farm Research Campus. Deconvolution was applied to the cell images shown in Figures 1D, 1E and Figure 2 (same as the previously used image processing procedure for Lattice Light Sheet microscopy6), but not to the bead image for resolution measurement in Figures 1C and 1D.
The free software ChimeraX24 by UCSF was used to view and demonstrate the volumetric data in 4D.
Global Exposures with Rolling Shutter
The rolling shutter mode of our sCMOS cameras provides readout time as short as 1 ms for a region of interest of 200 rows, facilitating fast imaging. Although this mode is fast, the readout of each row is no longer simultaneous. When the frame rate is close to the maximum readout speed of the camera, this asynchronous readout causes PSF distortion (Supplementary Fig. S15). Although global shutter mode can solve this problem, it increases the readout noise and slows down the readout by a factor of 2. Therefore, we implemented global exposure with rolling shutter by triggering pulsed laser illumination only during the time when all rows were exposing (Supplementary Fig. S15). The effective imaging time for each frame was thus the exposure time plus the readout time. Global exposure was realized through the NI PCIe 6323 DAQ card using LabVIEW programming.
Cell culture and transfection
The conditions for cell culture, transfection of HEK 293T and HeLa cells were described in our previous publication25. The knock-in cell lines, including the sequences for the guide RNAs and donor DNAs, were previously generated in the same publication25. To prepare sample for microscopy, knock-in cells were grown on an 8-well glass bottom chamber (Thermo Fisher Scientific). In order to achieve better cell attachment, 8-well chamber was coated with fibronectin (Sigma-Aldrich) for one hour before seeding cells.
B16F1 (ATCC-CRL-6323) mouse melanoma cells were cultured in Dulbecco’s modified Eagle medium with 4.5 g/mL glucose and supplemented with 10% fetal bovine serum and penicillin/streptomycin. A stable cell line expressing CAAX-EGFP was generated using standard lentiviral infection protocols. Cells were maintained at 37C and 5% CO2. Glass chambers were coated with 50ug/ml mouse laminin diluted in PBS and incubated overnight at 4C. The day of imaging, B16F1 cells were trypsinized and plated on laminin-coated glass and began to spread within 30 minutes.
Drosophila S2 cells were cultured in Schneider’s Drosophila Medium (Gibco) supplemented with 10% heat inactivated fetal bovine serum and penicillin/streptomycin (50 μg/ml). The cells were plated into 8-well plates coated with 0.5 mg/mL solution of Concanavalin A and then incubated for 1 day. The next day 225 μL 50 nM LysoTracker Deep Red dye solution was added to each well for 30 minutes prior to imaging.
Generation of HEK 293T cell lines for parallel imaging
To endogenously label proteins in HEK 293T cells, we used the split mNeonGreen2 (mNG2) system as previously described25. Briefly, HEK 293T stably expressing mNG21–10 fragment were first generated by lentiviral transduction. Subsequently, endogenous labeling was carried out by CRISPR-mediated homologous recombination using the single-guide RNA (sgRNA)/Cas9 ribonucleoprotein complexes (RNPs) electroporation method26. The RNPs were assembled in vitro from purified S. pyogenes Cas9-NLS protein (Macrolab, UC Berkeley) and synthetic sgRNA (Alt-R reagents, Integrated DNA Technologies). RNPs were co-delivered into nocodazole-treated cells together with single-stranded oligonucleotide donors (ssODNs, Ultramer reagents, Integrated DNA Technologies) by electroporation (Amaxa shuttle system, Lonza; program CM-130; 50 pmol RNP and 100 pmol ssODN per 200,000 cells). Finally, fluorescent cells were isolated 5–7 days post electroporation by fluorescence-activated cell sorting (SH800S system, SONY Biotechnology) and subsequently plated on fibronectin-treated glass chambers for live cell imaging. sgRNA spacer and ssODN sequences used in this study are available in Supplementary Note 4.
Prior to image acquisition, the 16 cell lines were seeded in 16-well chambered coverglass (Grace Biolabs) and allowed to attach to the fibronectin-coated coverglass surface overnight. During imaging, the temperature and CO2 concentration were maintained with a stage-top incubator (Tokai Hit) and the Perfect Focusing System of the Ti-E microscope was engaged to ensure the consistency of focal plane position when moving from well to well and during the image recording time period. 3D volumes were acquired sequentially through the 16 wells and the time-lapse acquisition was performed at a 9 min interval.
Parallel imaging of S2 cells with drug treatment and the leading edge intensity measurement
For actin drug treatment experiments, we imaged live S2 cells stably expressing mRFP-actin in the presence of Cytochalasin D or DMSO. We marked the coordinates of cells to be imaged in adjacent wells of the 96-well plate, acquired a baseline image of each cell, and then simultaneously added Cytochalasin D and DMSO directly on the microscope stage. The drug was prepared at 2x concentration in imaging media before addition to cells to achieve the targeted concentration. The experiments were repeated three times on cells plated in different wells of the same 96-well plate.
The leading edge intensity of the Drosophila S2 cells treated with DMSO and Cytochalasin D is measured from the max intensity projection images. The contour of the leading edge in the images is first recognized manually in each frame. The surface area of the leading edge is then determined in units of the pixel number by counting the number of pixels within the contour. The surface areas are normalized to that of the first frame of a time lapse data.
STORM imaging
For STORM imaging, wild-type S2 cells were plated onto Lab-Tek II 8-well chamber. Before cell seeding, each chamber was coated with 0.5 mg/ml concanavalin A for 1 h and followed by three times phosphate buffered saline (PBS) wash. Then cells were seeded into the chambers for 45 mins, washed one time with PBS, fixed with 3% paraformaldehyde and 0.1% glutaraldehyde in PBS for 10 min, treated with 0.1% NaBH4 for 7 min, then permeabilized and blocked with 0.5% Triton-X 100 and 3% bovine serum albumin for 10 min. Staining of tubulin was done with mouse monoclonal anti–β-tubulin antibody (1:200, 1 h incubation; T8328 Sigma) and secondary antibody (1:200, 1h incubation, Donkey anti Mouse, Jackson Immuno Research, labeled with Alexa Fluor 647, the ratio between antibody and dye is about 1:0.7).The sample was post-fixed (3% PFA and 0.1% gluteraldehyde in PBS, 10 minutes)and stored at 4 °C before imaging.
The sample was illuminated by the 642 laser and was imaged at 50 Hz. A total of 10 thousand frames were obtained. Single molecule localization analysis was performed with home-written C++ software described previously27. The data was drift corrected using direct cross-correlation method28.
Life sciences reporting summary
Further details on experimental design are available in the Life Science Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Code availability
Custom-written code is available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We acknowledge Tom Goddard and Thomas Ferrin from UCSF for their help in using ChimeraX. We thank Edaeni Hamid from Nikon for her help in providing the technical information of the objective lenses. We thank Tuscen Photonics Co. LTD. for generously loaning the 400BSI sCMOS camera to us. This project is supported by National Institutes of Health (R33EB019784, R21EB022798 and R01GM124334 to B.H., R35GM118119 to R.D.M.), UCSF Program for Breakthroughs in Biomedical Research and Byers Award in Basic Science (to B.H.). V.P. was supported by a predoctoral fellowship from the American Heart Association. X.C was supported by an international exchange fellowship from the Chinese Scholar Council. B.H. is a Chan Zuckerberg Biohub Investigator. R.D.M is supported by the Howard Hughes Medical Institute. M.D.L. is supported by the Chan Zuckerberg Biohub.
Footnotes
Conflict of interests
A patent application has been filed covering the reported microscope design.
Reference
- 1.Power RM & Huisken J Nat. Methods 14, 360–373 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Huisken J, Swoger J, Bene FD, Wittbrodt J & Stelzer EHK Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Tomer R, Khairy K, Amat F & Keller PJ Nat. Methods 9, 755 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Wu Y et al. Proc. Natl. Acad. Sci. U. S. A 108, 17708–17713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holekamp TF, Turaga D & Holy TE Neuron 57, 661–672 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Chen B-C et al. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunsby C Opt. Express 16, 20306–20316 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Li T et al. Sci. Rep 4, 7253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGorty R et al. Opt. Express 23, 16142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouchard MB et al. Nat. Photonics 9, 113–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strnad P et al. Nat. Methods 13, 139–142 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Maioli V et al. Sci. Rep 6, 37777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galland R et al. Nat. Methods 12, 641–644 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Meddens MBM et al. Biomed. Opt. Express 7, 2219–2236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao T et al. Sci. Rep 6, 26159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharonov A & Hochstrasser RM Proc. Natl. Acad. Sci 103, 18911–18916 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beghin A et al. Nat. Methods 14, 1184–1190 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Dean KM et al. Optica 4, 263–271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Royer LA et al. Nat. Biotechnol 34, 1267–1278 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Mcgorty R, Xie D & Huang B Opt. Express 25, 17798–17810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botcherby EJ, Juškaitis R, Booth MJ & Wilson T Opt. Commun 281, 880–887 (2008). [Google Scholar]
- 22.Kumar M & Kozorovitskiy Y Opt. Lett 44, 1706–1709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein AD et al. J. Biol. Methods 1, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goddard TD et al. Protein Sci. Publ. Protein Soc (2017). doi: 10.1002/pro.3235 [DOI] [Google Scholar]
- 25.Feng S et al. Nat. Commun 8, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonetti MD, Sekine S, Kamiyama D, Weissman JS & Huang B Proc. Natl. Acad. Sci 113, E3501–E3508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang B, Wang W, Bates M & Zhuang X Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y et al. Opt. Express 22, 15982–15991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.