ABSTRACT
Extracellular vesicles (EVs) are central to intercellular communication and play an important role in cancer progression and development. Osteosarcoma (OS) is an aggressive bone tumour, characterized by the presence of malignant mesenchymal cells. The specific tumour-driving genetic alterations that are associated with OS development are currently poorly understood. Mesenchymal stem cells (MSCs) of osteogenic lineage have been postulated as likely candidates as the cells of origin for OS, thus indicating that MSCs and OS stroma cells may be related cell types. Therefore, this study set out to examine the EV-mediated intercellular crosstalk of MSCs and OS. MSCs and pre-osteoblasts were treated with OS-EVs at different time points, and the epigenetic signature of OS-EVs was assessed by methylation analysis of LINE-1 (long interspersed element) and tumour suppressor genes. In addition, surface markers and expression of specific genes were also evaluated. Our data indicated that OS-EVs mediated LINE-1 hypomethylation in MSCs, whereas an opposite effect was seen in pre-osteoblasts, indicating that MSCs but not pre-osteoblasts were susceptible to epigenetic transformation. Thus, OS-EVs modulated the fate of MSCs by modulating the epigenetic status, and also influenced the expression of genes related to bone microenvironment remodelling. Overall, this study provided evidence that epigenetic regulation appears to be an early event in the transformation of MSCs during the development of OS. Elucidating the mechanisms of EV-mediated communication may lead to new avenues for therapeutic exploitation.
KEYWORDS: Osteosarcoma, mesenchymal stem cells, osteogenesis, extracellular vesicles, LINE-1 methylation, DNA methylation, flow cytometry
Introduction
Osteosarcoma (OS) is the most common primary heterogeneous malignant tumour of long bones affecting children and adolescents. Though relatively rare, it is still one of the highest causes of cancer-related deaths in paediatric patients [1,2]. It is characterized by increased osteoid production, a strong tendency for recurrence and an extremely high metastatic potential [3,4]. For the last three decades, 5-year survival rates for metastatic and recurrent OS is below 20% and has remained unchanged [5]. Therefore, novel treatment approaches are urgently needed. Identification of tumour driving genetic alterations associated with OS development is hindered due to the lack of precursor lesions, presence of complex karyotypes, high genetic instability, and lack of recurrent genetic alterations in OS [6]. The cells of origin for OS also remain unknown. Recent data have suggested a hypothesis that under the influence of certain environmental and epigenetic signals, an MSC-derived osteogenic progenitor or an undifferentiated MSC may be the cell of origin for OS [7–10]. Thus, understanding the cellular origin of OS will have direct implications on improving treatment approaches through identification of new therapeutic targets.
Extracellular vesicles (EVs) are membrane-bound biologically active nano-vesicles found in all bodily fluids. They are secreted by most cell types that are involved in both physiological and pathological conditions [11,12]. The complex and specific cargo of EVs (lipids, proteins, mRNA, microRNA and other non-coding RNA) as well as their composition differ depending on their cellular origin [13,14]. EVs mediate intercellular communication through the bidirectional exchange of their cargo with neighbouring cells. They have been shown to induce epigenetic changes by affecting the DNA methylation status and change the phenotype of several recipient cell types [15–17]. Further, it has been suggested that EV-associated mRNA and proteins are involved in epigenetic regulation, thus affecting the expression of tumour promoting or suppressing genes [15].
Tumour-derived EVs are a driving force in the transformation of MSCs [18–20]. However, the influence of OS-derived EVs (OS-EVs) on the epigenetic reprograming of MSCs and pre-osteoblasts, specifically the global LINE-1 (long interspersed element) methylation status, is still largely unknown, and the mechanisms underlying the conversion of normal MSCs into tumour-favourable MSCs remain unclear [21]. Global LINE-1 hypomethylation is viewed as an early event in normal cell transformation in a wide range of cancers and benign neoplasms [22]. However, conflicting data also exists [23,24]. Addressing these questions may yield new insights as to the cell of origin of OS, with the prospect of finding and exploiting molecular drivers underlying OS.
Thus, we examined the consequences of OS-EV treatment on the epigenetic reprograming of MSCs and pre-osteoblasts by analyzing their global LINE-1 status and methylation analysis of selected tumour suppressor genes. Taken together, our data showed that OS-EVs mediated an epigenetic response in MSCs, but not in pre-osteoblasts, thus indicating that MSCs but not pre-osteoblast were more susceptible to epigenetic transformation. There were no changes seen in the methylation status of tumour suppressor genes in MSCs. As early indicators of transformation, OS-EV treated MSCs and pre-osteoblasts showed higher expression of genes related to bone microenvironment remodelling and also a significant upregulation of intercellular adhesion molecule (ICAM1/CD54). Thus, OS-EVs dictated the fate of MSCs by modulating their epigenetic status. Overall, this study provided evidence that epigenetic regulation appears to occur early in the transformation of MSCs.
Materials and methods
The workflow is described in Figure 1.
Figure 1.
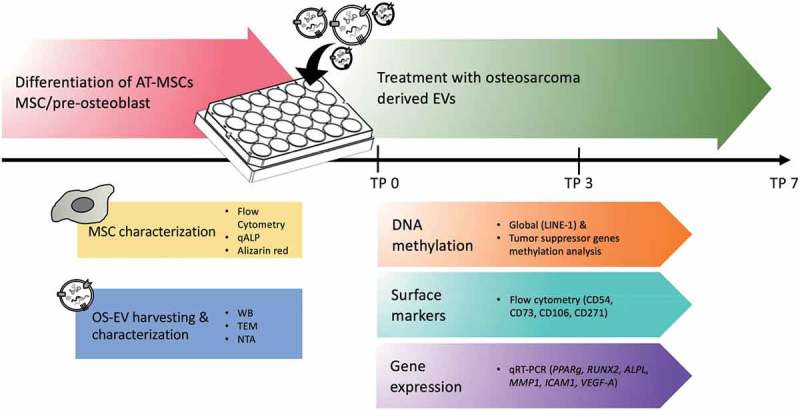
Flowchart of the study design. MSCs/pre-osteoblasts were treated with OS-EVs at different time points (TP 0, 3 and 7 days). MSCs and OS-EVs were characterized by various methods. OS-EV treated MSCs/pre-osteoblasts samples were further analyzed for DNA methylation, surface marker and gene expression.
Cell lines and cell culture
Human osteosarcoma cell line 143B (HOS143b; ATCC® CRL-8303™) was purchased from ATCC and cultured according to the manufacturer’s instructions. Briefly, cells were cultured in RPMI Medium 1640 + GlutaMAX (Gibco Life Technologies, ref. 61870–010 lot. 1896169), supplemented with 1% antibiotics (100 U/ml penicillin, 0.1 mg/ml streptomycin; Lonza ref. DE 17–602 E, lot. 5MB 068) and 10% foetal bovine serum (FBS; Gibco Life Technologies, South American ref. 10270106, lot. 42G8468K). Human osteosarcoma line U2OS (ATCC® HTB-96™) cells were cultured in maintenance media (MM) consisting of Dulbecco’s modified Eagle’s medium/Ham’s Nutrient Mixture F-12 with 1% L-alanyl-L-glutamine (DMEM/F-12 1:1 GlutaMAX; Gibco ref. 31331–028, lot. 1765999), 1% antibiotics and 10% FBS (Foetal Bovine Serum, South American ref. 10270–106, lot. 42G8468K).
Adipose tissue was provided by the Department of Plastic Surgery (Laser Tilkka Ltd, Finland). All tissue donors (n = 6) were female, age range 32–55 years, average 41 years; BMI range 22–29, average 25. The study was carried out under approval of the ethical committee of Helsinki and Uusimaa Hospital District and with informed consent from the donors (Ethical approval DNro: 217/13/03/02/2015). Human adipose tissue mesenchymal stromal/stem cells (AT-MSCs) were obtained from water-assisted lipotransfer liposuction aspirates [25,] using mechanical and enzymatic isolation as described previously [25]. Once AT-MSCs had adhered to the culture flask surface, non-adherent populations were washed away with PBS, and fresh culture media was added. Cells were cultured in MM. Human bone marrow MSCs (BM-MSCs) were received as a kind gift from Assoc. Prof Susanna Miettinen (University of Tampere). The BM-MSCs were expanded in MM similarly to AT-MSCs.
AT-MSC characterization
AT-MSCs (n = 4) were characterized using BD Accuri C6 flow cytometer (Becton Dickinson, Franklin Lakes) to confirm the mesenchymal origin of the cells. Allophycocyanin (APC)-conjugated monoclonal antibodies against CD14 (clone: M5E2), CD19 (clone: HIB19), CD34 (clone: 581), CD45RO (clone: UCHL1), CD54 (clone: HA58), CD73 (clone: AD2), CD90 (clone: 5E10), CD105 (clone: 266) and HLA-DR (clone: G46-6) (BD Pharmingen, Becton Dickinson) were used. Analysis was performed on 100,000 cells per sample and the positive expression was defined as the level of fluorescence greater than 99% of the corresponding unstained cell sample [26].
Osteogenic differentiation assay
Osteogenically differentiated MSCs were termed pre-osteoblasts. For pre-osteoblast differentiation, MSCs were cultured for 14 days in osteogenic media (OM). Briefly, cells were left to adhere for 24 h in MM and osteogenic differentiation was induced using osteogenic media (MM) supplemented with 50 µM L-ascorbic acid 2-phosphate, 10 mM β-glycerophosphate disodium salt hydrate and 5 nM dexamethasone (all Sigma-Aldrich). Early osteogenic response was assessed by quantitative alkaline phosphatase activity (qALP) assays and mineralization was assessed using Alizarin Red staining (ARS) as described previously [26] after 3 weeks of osteogenic induction [27].
EV isolation and treatments
EV isolation
EV-depleted FBS was prepared by 19 h of ultracentrifugation at 100,000g using an SW28 rotor, k-factor 246 (Beckmann-Coulter) +4°C. Only the light-coloured top layers of the supernatant were used in the subsequent analyses. Cell lines (HOS and MSCs) were maintained in culture medium containing FBS, but prior to EV harvesting, the cells were switched into media containing EV-depleted FBS for up to 72 h. Every 24 h up to 72 h, conditioned media (CM) was collected and stored at −80°C. Fresh EV-depleted media was added every time after collection of the cell cultures. EVs collected from conditioned OS media (OS-EVs) were extracted using ultracentrifugation at 100,000g for 2 h at +4°C to collect the EV pellet, filtered in PBS (0.1µm filter) and then ultracentrifugation was repeated. Isolated pellets were suspended in filtered PBS and stored in low protein binding microcentrifuge tubes (Protein LoBind Tube, Eppendorf) at −80°C.
Nanoparticle tracking analysis (NTA)
The EVs were analyzed by NTA to determine vesicle concentration and size distribution using (0.1 μm) DPBS to obtain the optimal detection concentration of 106–109 particles/ml, and three 60 s videos were recorded using camera level 13. The data was analyzed using NTA software 3.0 with the detection threshold 5 [27].
Transmission electron microscopy (TEM)
EV samples were prepared for EM and imaged as described previously [27,28]. Briefly, samples from 2–3 biological replicates were viewed with transmission EM using Tecnai 12 (FEI Company) operating at 80 kV. Images were taken with Gatan Orius SC 1000B CCD-camera (Gatan Inc.) with 4008 × 2672 px image size and no binning.
Western blotting
Western blotting was performed as described previously [27] using antibodies against Hsp70 (70 kDa heat shock protein, BD Biosciences, cat#554243, 1:100) CD63 (Lysosomal-Associated Membrane Protein 3, BD Biosciences, cat#556019, 1:1000), calnexin (Cell Signaling Technology, cat#2679, 1:800), TSG101 (ESCRT-I Complex Subunit TSG101, Sigma-Aldrich, cat# SAB2702167, 1:500). OS-EV samples and as a control, protein from AT-MSCs lysates, were denatured at 95°C for 5 min in reducing Laemmli sample buffer, separated using Mini-PROTEAN® TGX™ 4–20% gradient SDS-PAGE gel (Bio-Rad) with page ruler prestained protein ladder (Thermo Scientific) as a standard and blotted on nitrocellulose membranes, 0.2 µm (#162–0112, Bio-Rad). Blocking and antibody incubations were performed in Odyssey blocking buffer (LI-COR) with 0.1% Tween-20. Membranes were probed with IRDye® 800CW Goat anti-Mouse (LI-COR) at 1:15,000 for 2 hours at room temperature (RT). After incubation, membranes were washed three times in TBS-T for 10 minutes at RT and imaged on an Odyssey FC Imager (LI-COR).
EV treatments
MSCs and pre-osteoblast cells were plated in 24 well plates at a cell density of 2.5 −7.5x104 cells per well, then treated with OS-EVs and analyzed at time points 3, 7 and 14. Untreated MSCs and pre-osteoblasts were used as controls. EVs were added at day 1 (24h after cell adhesion), day 3 (48h after first dose) and day 5 (96h after first dose) (Figure 1). On day 7, CM was collected and cells were analyzed with methylation assays, flow cytometry and quantitative real-time PCR (qRT-PCR). MSCs treated with the same amount of autologous EVs were used as an experimental control. The experiments were repeated 4 times.
LINE-1 methylation analysis
DNA extraction was done using NucleoSpin® Tissue XS (Macherey-Nagel, Ref 740,901). Custom-made LINE-1 kit as described previously [29] was used. Briefly, three areas inside the LINE-1 promoter sequence and seven control probes were included in the kit. 70ng of DNA was used to perform MS-MLPA reaction using the SALSA MS-MLPA Reagents kit P300-A1 Human DNA Reference-2 (MRC-Holland), following a MS-MLPA standard reaction protocol as described in MRC-Holland instructions (http://www.mrc-holland.com). Methylation dosage ratio was calculated as described previously [29]. Three controls were used; commercially available colon cancer cell line RKO (ATCC® CRL-2577), pooled blood sample from healthy individual, and one unmethylated sample generated using GenomePlex complete whole genome amplification (WGA) kit [29,30]. LINE-1 probes and controls used in the analysis were received as kind gift from Prof. Päivi Peltomäki (University of Helsinki).
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)
The methylation status of tumour suppressor genes were assessed using commercially available MS-MLPA kit (ME001-C2 and ME002-C1 Tumour suppressor, from MRC Holland) according to the manufacturer’s instructions (http://www.mrc-holland.com) using 150ng of DNA for the analysis. The methylation dosage ratios (Dm) were calculated as described previously [31]. A cut-off value of Dm ≥0.20 was considered to indicate promoter methylation.
qRT-PCR
Total RNA was extracted from MSCs treated with and without OS-EVs (n = 3) using miRCURY RNA isolation kit (Cell and plant, Exiqon cat#300110). RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). All PCR reactions were conducted in triplicates in QuantStudio 5 (Applied Biosystems), using TaqMan assays (Thermo Fisher Scientific). The expression of the following genes was quantified: intercellular adhesion molecule 1 (ICAM1, assay ID Hs00164932_m1) matrix metallopeptidase 1 (MMP1, assay ID Hs00899658_m1), runt-related transcription factor 2 (RUNX2, assay ID Hs01047973_m1), alkaline phosphatase, liver/bone/kidney (ALPL, assay ID Hs01029144_m1), vascular endothelial growth factor A, (VEGF-A, Hs00900055_m1), peroxisome proliferator activated receptor gamma, (PPARγ, Hs00234592_m1). Results were normalized with the housekeeping gene, ribosomal protein lateral stalk subunit P0 (RPLP0, assay ID Hs99999902_m1) and calculated using the ddCt-method [32].
Flow cytometry analysis
MSCs receiving EV treatment (OS-EVs and MSC-EVs) were analyzed with flow cytometry. Briefly, MSCs and pre-osteoblasts with or without OS-EV treatment, n = 3 with 3 repeats, as well as MSCs receiving autologous EVs were analysed using BD Accuri C6 flow cytometer (Becton Dickinson, Franklin Lakes) using monoclonal antibodies conjugated with APC against CD54, CD73, CD106 (VCAM-1) (Clone 51-10C9) and CD271 (Clone C40-1457). As controls, HOS-143b (n = 2), U2OS (n = 1), BM-MSCs (n = 3) were analysed using APC-conjugated monoclonal antibodies against CD14, CD19, CD34, CD45RO, CD54, CD73, CD90, CD105, CD106, CD271 and HLA-DR, all purchased from BD PharmingenTM (Becton Dickinson). Analysis was performed on 1 × 105 events per sample and positive expression was defined as the level of fluorescence > 99% of the corresponding unstained cell sample [26].
Statistical analysis
Statistical analyses were performed with GraphPad Prism 6 (GraphPad Software Inc.) to analyse the effects of OS-EVs on MSC. Cultures were analysed using ANOVA two- way analysis of variance. Bonferroni post-hoc tests were used to determine individual significant differences. The results were considered significant when the Bonferroni corrected p-value was below 0.05.
Results
MSC displayed typical stem cell features and osteogenic differentiation potential
In concordance with the guidelines defined by the International Society for Cellular Therapy (ISCT), the MSCs displayed characteristic morphology of MSCs (Figure 2(a,b)) and also displayed the immunophenotypical profile of MSCs (Figure 2(e)). MSCs demonstrated a surface marker profile of CD73/CD90/CD105 positive and CD14/CD19/CD45/HLA-DR negative expression, also moderate expression of CD34 was detected. Further, MSCs also expressed moderate levels of CD54 (ICAM-1) (Figure 2(e)). The osteogenic differentiation potential of MSCs was also evaluated by qALP and ARS. The results showed that chemically induced osteogenesis enhanced the osteogenic differentiation and mineralization process as compared to control conditions (Figure 2(c,d,f)).
Figure 2.
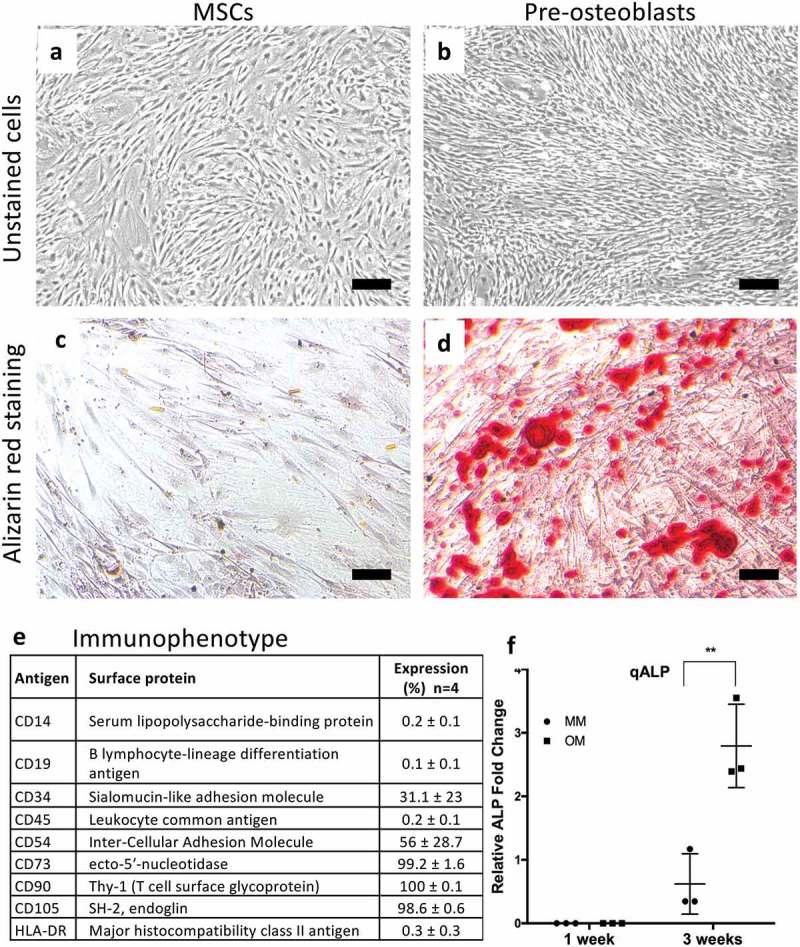
Morphological characterization of adipose-tissue derived mesenchymal stem cells (AT-MSCs) by light microscopy after culturing in the maintenance media (MSCs; a) and osteogenic media (pre-osteoblasts; b) for 14 days. Scale bars are 100 μm. The level of mineralization was assessed by Alizarin red staining in both non‐treated AT-MSCs (MSC) (c) and osteogenically treated AT-MSCs (pre-osteoblast) (d) after 14 days. (e) Surface marker expression (%) of undifferentiated AT-MSCs analyzed by flow cytometric analysis. (f) Early osteogenic response measured by alkaline phosphatase (ALP) activity assay. Values are reported as mean ±SD and the assay was performed at 7 and 21 days (**p < 0,05; n = 3) .
Characterization of OS-EVs
EVs were characterized using WB, TEM and NTA. TEM revealed that the OS-EVs were intact and that EVs of various shapes and sizes are secreted by OS (Figure 3(a)). Western blotting results confirmed the presence of EVs as all the EV markers (CD63, Hsp70, and TSG101) showed the positive band, whereas there was no band in calnexin, indicating the purity of the EV samples (Figure 3(b)). According to the NTA results, the majority of the particles were in the size range of 50–200 nm (Figure 3(c)) with a size distribution range of approx. 50–500 nm (Figure 3(d)). Particles between 50 and 400 nm in diameter were termed as EVs. Distribution of particle fractions is shown in Figure 3(e).
Figure 3.

OS-EVs characterization. (a) Confirmation of EVs by TEM. Scale bar 500 nm (b) Western blotting showed presence of EV markers (TSG101, Hsp70 and CD63). Absence of calnexin protein indicate purity of the samples. (c–d) EV concentration and size distribution analyzed by nanoparticle tracking analysis (NTA), y-axis depict particle concentration (particles 106/ml). PBS was used as a negative control. (e) Table showing distribution of the particles, with D10, D50 and D90 representing the midpoint (D50) and range (D10, D90) of the particle sizes of a given sample.
Treatment of MSCs with OS-EVs induced global LINE-1 hypomethylation
Both MSCs and pre-osteoblasts were treated with OS-EVs (+EVs) and analyzed at 0, 3, 7 and 14 to examine the epigenetic effects induced by the treatments. Untreated (-EVs) MSCs and pre-osteoblasts were used as control, and time point 0 served as a baseline with no treatment. As shown in Figure 4(a), MSCs treated with OS-EVs exhibited decreased LINE-1 expression at time point 3 which returned to baseline at time point 7, and was maintained until time point 14 (time point 14 data not shown). For pre-osteoblasts (Figure 4(b)), LINE-1 hypermethylation was seen at time points 3 and 7 and was also seen at time point 7 for the control group (MSCs receiving autologous EVs).
Figure 4.
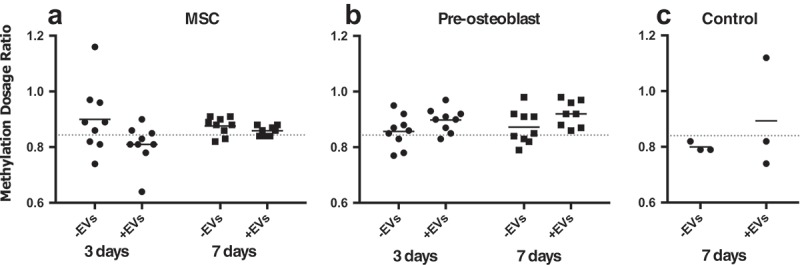
Methylation dosage ratio for LINE-1 in MSC and pre-osteoblast untreated and treated with OS-EVs at time point 3 and 7. AT-MSC were treated with autologous MSC-EVs were used as an experimental control. Dotted line represents baseline measurement at time point 0.
Tumour suppressor genes methylation status was not affected in MSCs treated with OS-EVs
To further investigate the epigenetic effects mediated by OS-EVs treatment, methylation analysis of 29 tumour suppressor genes was performed on both +EVs and -EVs MSCs and pre-osteoblasts. The analyzed methylation status of the genes investigated was not affected in MSCs and pre-osteoblasts with +EV (data not shown).
Expression of MMP1, ICAM1, VEGF-A and RUNX1 was affected in MSCs treated with OS-EVs
Effect of OS-EV treatments was also evaluated on gene expression level on both MSCs and pre-osteoblasts (Figure 5). OS-EV treated MSCs and pre-osteoblasts showed significantly elevated expression of MMP1, ICAM1, and VEGF-A as a response to OS-EV treatment (*p > 0.05). However, EV treatment had a more pronounced effect on MMP1 expression in MSCs when compared to pre-osteoblasts. As expected, adipose marker PPARγ expression was reduced after OS-EV treatment (**p > 0.001). Due to donor variation ALPL and RUNX2 showed no heterogeneous gene expression.
Figure 5.
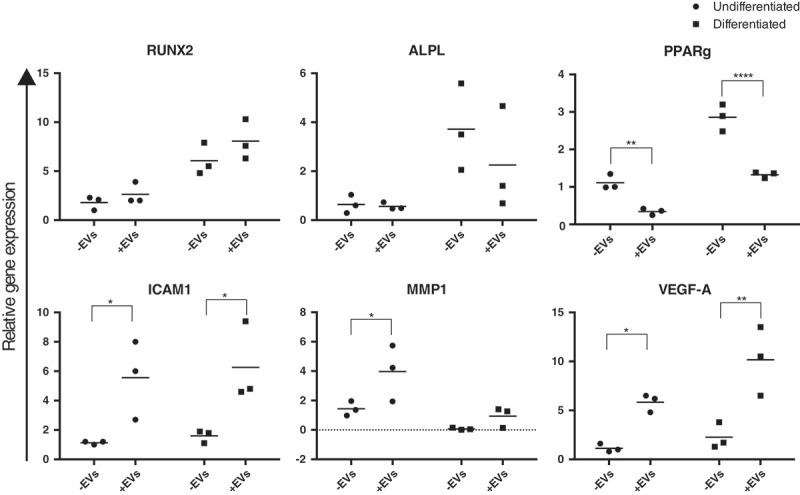
Real-time quantitative PCR analysis of RUNX2, ALPL, PPARγ, ICAM1, MMP1 and VEGF-A expression in MSCs and pre-osteoblasts untreated (-EVs) and treated (+EVs) with OS-EVs at time point 3. Biological replicates are represented by dots, and means by bars. Asterisks indicate statistical significance, * p = 0.05; ** p = 0.005; *** p = 0.0005; **** p < 0.0001.
Surface marker expression analysis
Surface marker expression was also assessed in MSCs and pre-osteoblast +EVs and -EVs (Table 1). AT-MSCs treated with autologous MSCs (+autoEVs), osteosarcoma cell lines (HOS143b and U2OS) as well as BM-MSCs were added as controls for marker expression. The flow cytometric analysis showed (Figure 6(a)) that the only marker affected by the treatment was CD54 (ICAM1). A relative dose specific response was detected, as no change in CD54 expression level was seen at lowest OS-EV concentration (5×104), and at an intermediate concentration (4×105), higher expression was seen at highest dose (1×107particles) (Figure 6 (a,b)). In addition CD106 (Vascular cell adhesion protein 1, VCAM-1) was analyzed, which is differentially expressed in AT-MSCs (negative), and BM-MSCs (positive) and reportedly present in rat osteosarcoma cell line COS1NR [33,34]. The results showed that VCAM-1 was expressed at moderate levels only in BM-MSCs, with no expression detected in AT-MSCs. Next, CD271 (low-affinity nerve growth factor receptor, LNGFR) was analyzed which is reportedly present in osteosarcoma stem cells [35,36]. The data showed that CD271 was present only in U2OS control cells, with moderate expression present in BM-MSCs and no expression detected in AT-MSCs.
Table 1.
Surface marker expression of MSCs and pre-osteoblast untreated or treated with OS-EVs (-EVs/+EVs), n = 3 repeated 3 times. As controls, MSCs treated with autologous EVs, as well as cell lines (HOS143b, U2O and BM-MSCs) were used. CD, cluster of differentiation.
| OS-EV treatment group |
Controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| MSC -EVs |
MSC +EVs | Pre-osteo -EVs |
Pre-osteo +EVs |
MSC +autoMSC EVs (n = 3) | HOS143b (n = 2) |
U2OS (n = 1) |
BM-MSC (n = 3) |
|
| CD14 | 0.3 ± 0.1 | 0.2 | 0.3. ± 0.1 | |||||
| CD19 | 0.2 ± 0.1 | 0.1 | 0.1. ± 0.1 | |||||
| CD34 | 2.2 ± 2.8 | 0.2 | 1.1. ± 1.7 | |||||
| CD45 | 0.4 ± 0.2 | 0.3 | 0.3 ± 0.1 | |||||
| CD54 | 27.6 ± 19.1 | 46.7 ± 38.4 | 50.0 ± 8.8 | 79.4 ± 6.1 | 7.2 ± 0.45 | 99.7 ± 0.7 | 5.9 | 12.6 ± 2.3 |
| CD73 | 98.1 ± 2.6 | 98.8 ± 1.9 | 99.5 ± 0.2 | 99.7 ± 0.1 | 98.0 ± 0.7 | 99 ± 1.2 | 0.4 | 99.4 ± 0.4 |
| CD90 | 4 ± 4.1 | 99 | 97.6 ± 0.0 | |||||
| CD105 | 97.7 ± 0.9 | 37.1 | 96.6 ± 1.2 | |||||
| HLA-DR | 0.9 ± 0.3 | 0.5 | 1.3 ± 0.4 | |||||
| CD106 | 0.1 ± 0.1 | 0.6 ± 0.7 | 0.3 ± 0.2 | 0.3 ± 0.2 | 0.1 ± 0.7 | 0 | 0.2 | 41.4 |
| CD271 | 0.0 ± 0.1 | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | 1.9 | 86.9 | 24.5 |
Figure 6.
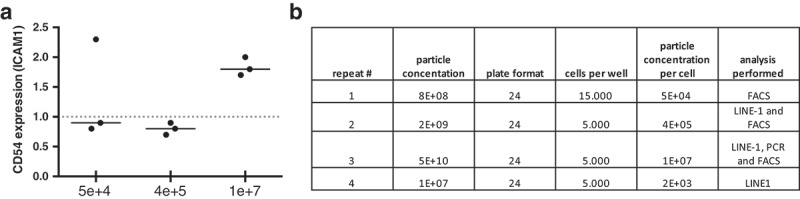
(a) Surface marker ICAM/CD54 expression measured by flow cytometry for MSCs untreated and treated with OS-EVs. X-axis depict OS-EVs concentrations. Dotted line represents baseline measurement (MSCs untreated with OS-EVs). (b) Table showing repeats and amount of OS-EVs applied per MSC.
Discussion
It is well known that the tumour microenvironment is correlated with tumorigenesis and cancer progression. MSCs have been reported to be isolated at high frequency from OS samples demonstrating their importance in the OS microenvironment [37]. In recent years, EVs have shown to be powerful mediators of intercellular communication with the ability to manipulate the local and systemic tumour microenvironment [17,38,39]. Having a bimodal role in tumorigenesis, EVs modulate either immunostimulatory or inhibitory functions or both [40]. The balance between these two responses is therefore likely to be the determining factor for the outcome of disease. EVs are therefore an excellent tool for studying the interaction between the tumour and its surrounding cells.
Cross-talk between MSCs and OS has demonstrated the oncogenic potential of OS-EVs in recipient cells [37,41,42]. For example, in an in vivo mouse study, OS-EVs carrying functional TGFβ (transforming growth factor β) were internalized by MSCs and altered the MSCs phenotype towards a pro-tumorigenic and pro-metastatic phenotype by activating oncogenic IL&-STAT3 signalling pathways, thereby promoting tumour growth and metastasis [21]. In recent years, the composition of OS-EVs cargo has been investigated, indicating the presence of proteins related to tumour progression and metastasis [43,44], and the presence of repetitive DNA, including satellites and transposable elements [45]. However, the impact of OS-EVs on the epigenome of MSCs is still unclear and the consequences of these interactions and their impact on tumour behaviour need to be addressed. Based on the reported evidence at genetic level, OS-EVs alter the MSC physiology, thus, this study addressed the implications of OS-EVs in the epigenetic reprogramming of MSCs.
To date, there is no consensus about EV dose required in cell culture and animal model experiments. EV concentrations reported in literature are not clearly stated and there is also considerable variation with regards to the experimental setup and the cell line studied. Since the concentrations for EV treatments are not standardized, we tested several EV concentrations to determine dose-dependent effect of OS-EVs. Methylation and flow cytometric analysis were performed with different EV concentrations to set up a dose-response pattern, however due to lack of material gene expression was repeated only with one EV concentration. LINE-1 analysis was not affected by different EV doses, however CD54 expression by flow cytometry showed a dose-dependent response.
The complex bioactive cargo of EVs, including molecules with epigenetic reprogramming capability, has been reported to induce an epigenetic response in recipient cells by affecting their methylation status, thereby promoting genomic instability [46]. Moreover, since subtle changes in the microenvironment can stimulate the transformation of cells, we applied OS-EVs to introduce the first hit of transformation for MSCs in to order to investigate whether OS-EVs affect the global LINE-1 methylation status of MSCs. Secondly, the study was set out to analyze whether MSCs or pre-osteoblasts are susceptible to epigenetic reprogramming, potentially highlighting the cell type directing sarcomagenesis.
Our data indicated that a significant decline in LINE-1 methylation occurred in MSCs after 3 days of induction with OS-EVs which was maintained until day 7, but returned to normal levels after extended exposure to OS-EVs (time point 14; data not shown), concluding that epigenetic regulation appears to play a role in transformation of MSCs. On the contrary, LINE-1 hypomethylation was not observed in pre-osteoblasts treated with OS-EVs (Figure 4), indicating that pre-osteoblasts are less susceptible to global LINE-1 hypomethylation. In addition, our data indicated that hypomethylation occurs early during the transformation of MSCs and subsequently, as the methylation levels return to normal level after extended exposure to OS-EVs, several oncogenic hits may be required for maintenance of hypomethylation and full transformation of MSCs. Transformation of normal cells is a multistep process, which is linked to accumulation of genetic and epigenetic alterations. To address the timing of genome-wide methylation and to determine whether it is causative for the transformation of MSCs, Wild and co-authors [47] developed a model for producing fully transformed MSCs by introducing five consecutive oncogenic hits, concluding that in vitro transformation of MSCs induces gradual hypomethylation and it is not causative for transformation. The discrepancy between our results and those of Wild et al may be explained by several factors, such as different methods used for MSC transformation, the fact that LINE-1 methylation was studied only at late stage of MSC transformation by Wild et al in contrast to our study. However, it should be noted that the actual epigenetic changes occurring during the in vivo transformation may not recapitulate in a simplified cell model.
The cell of origin for OS is presumed to be an intermediate precursor between MSCs and osteoblasts [48]. In our study, under the influence of environmental signals, i.e., OS-EV treatment, epigenetic modifications underpin the transformation of MSCs but not of pre-osteoblasts, however, conclusions regarding the cell of origin is difficult to draw with our limited data.
As global hypomethylation is known to affect genome-wide methylation levels [46,49], our study further evaluated whether OS-EVs may regulate the promotor hypermethylation status of tumour suppressor genes. The methylation status of the 29 tumour suppressor genes was not affected by the treatment with OS-EVs, indicating that short term exposure of the MSCs to OS-EVs was not sufficient to induce promotor hypermethylation of the selected genes and that this event occurs at a later stage of transformation.
Global LINE-1 hypomethylation contribute to cancer development by inducing genetic instability, and predisposing the cells to chromosomal defects, transcription disruption, insertion mutations, and thus influencing the overall gene expression [50–52]. OS-EV treated cells have been shown to acquire tumour-phenotype characteristics such as increased adhesion, proliferation, migration rate, and anchorage-independent growth [42]. Subsequently, the effects of OS-EVs on recipient cells were investigated by focusing on analyzing the expression of genes (MMP1, VEGF-A, ICAM1) related only to bone microenvironment remodelling. Both MSCs and pre-osteoblasts showed higher expression of these genes after OS-EV treatment (Figure 6). Matrix metalloproteinase-1 (MMP1) is involved in extracellular matrix (ECM) remodelling, regulation of cell signalling in bone microenvironment, and in facilitating invasion and metastasis [53]. Its high expression is related to poor prognosis in OS patients [54]. Binding of vascular endothelial growth factor (VEGF-A) to its receptor results in the expression of MMPs, which subsequently allows for ECM degradation and formation of new vessels. Thus, overexpression of VEGF-A in OS is associated with lung metastasis and poor overall survival [55]. Intercellular adhesion molecule 1 (ICAM1/CD54) is an adhesion molecule that regulates inflammation, immune response, and intercellular signalling [56,57]. It is expressed in many cancers and facilitates cancer cell invasion through the ECM. Increased expression of ICAM1 is also reported in OS cells co-cultured with MSCs [37]. This result is further supported by protein level data analyzed by flow cytometry. Taken together, these results indicate, that MSCs may have transformed towards a cancer associated fibroblast phenotype by the OS-EVs treatment.
Numerous in vitro investigations have demonstrated that fat-induction factors such as PPARγ inhibit osteogenesis, and conversely, bone-induction factors such as RUNX2 deter adipogenesis [58]. The expression of osteogenesis (RUNX2 and ALPL) and adipogenesis (PPARγ) related genes was assessed in our study. The expression of all genes was elevated by osteogenic conditions when compared with the control, but OS-EV treatment provided a unique signature to the regulation of the genes. OS-EVs upregulated the expression of RUNX2, conversely ALPL and PPARγ were downregulated in response to OS-EV treatment (Figure 6). Specifically, the TGFβ/BMPs signalling pathway is known to have dual roles in regulating adipogenic and osteogenic differentiation of MSCs, as expression of RUNX2 and PPARγ can be regulated by either the Smad or the p38 MAPK pathway, depending on the composition and concentration of signalling cues in the microenvironment [58]. Therefore, as our results show the upregulation of RUNX2 in MSCs promoted osteogenesis, while inhibiting adipogenesis, shown by the expression of PPARγ. OS cells are known to share many features with osteoprogenitors, such as expression of osteogenic markers (RUNX2, ALPL). Furthermore, the more aggressive OS phenotypes often resemble early osteoprogenitors, while less aggressive tumours appear to share similarities with osteogenic MSCs that have progressed further along the differentiation cascade. For instance, ALPL shows lower expression in OS tumour cells compared to committed osteoblastic cell line [59], a trend also seen in our ALPL expression results. However, our results may be inadequate to prove the full commitment of MSCs into a transformed phenotype, but point towards MSCs being primed by the cargo present in the OS-EV vesicle fraction.
There is yet very little data on the concentration of EVs needed for a cell response or a therapeutic effect, so we studied the concentration of OS-EV vesicles given based on surface marker expression, and we found that ICAM1/CD54 is expressed in a dose dependent manner. The response is likely primarily an immune response, however, it is not excluded that it is associated with MSCs presenting a transformed phenotype.
Conclusions
Our study investigated the effects induced by OS-EVs on MSCs and revealed a number of significant findings. Firstly, we showed for the first time that OS-EVs are implicated in epigenetic reprograming of MSC and that the transformation of MSCs, but not pre-osteoblasts, is epigenetically regulated via global LINE-1 hypomethylation; secondly, the study demonstrates the oncogenic power of OS-EVs on MSCs gene expression related to bone microenvironment remodelling, and lastly, we saw a dose-dependent effect of OS-EVs on adhesion molecule expression ICAM1/CD54 in MSCs. Additional studies are needed to investigate the elements responsible for global LINE-1 hypomethylation in OS-EVs, and for identifying the mechanisms by which LINE-1 hypomethylation affects MSC behaviour.
Funding Statement
This research was supported by University of Helsinki project funding [WBS490302, WBS73714112], Helsinki University Hospital State funding for university-level health research [Y1014SUL05, TYH2016130], the Finnish Dental Society Apollonia, Egyptian Ministry of Higher Education (MoHE), Finnish-Norwegian medical foundation, and Selma and Maja-Lisa Selander Foundation.
Acknowledgments
The authors thank Flow Cytometry Core Facility, EV Core Facility and Electron Microscopy Unit of the Institute of Biotechnology (all University of Helsinki). Assoc. prof Susanna Miettinen (University of Tampere) is thanked for providing bone marrow stem cells and plastic surgeon Hilkka Peltoniemi is thanked for providing lipoaspirate samples. Laboratory technicians Heidi Husu and Anne Kivimäki are thanked for excellent technical assistance, and Ph.D Satu Valo, Ph.D Walter Pavicic and Prof Päivi Peltomäki are thanked for scientific support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Longhi A, Errani C, De Paolis M, et al. Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treat Rev. 2006;32:423–436. [DOI] [PubMed] [Google Scholar]
- [2].Ta HT, Dass CR, Choong PF, et al. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–263. [DOI] [PubMed] [Google Scholar]
- [3].Abarrategi A, Tornin J, Martinez-Cruzado L, et al. Osteosarcoma: cells-of-origin, cancer stem cells, and targeted therapies. Stem Cells Int. 2016;2016:3631764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Geller DS, Gorlick R.. Osteosarcoma: A review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- [5].Allison DC, Carney SC, Ahlmann ER, et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martin JW, Squire JA, Zielenska M.. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. [DOI] [PubMed] [Google Scholar]
- [8].Xiao W, Mohseny AB, Hogendoorn PC, et al. Mesenchymal stem cell transformation and sarcoma genesis. Clin Sarcoma Res. 2013;3:10–3329-3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sottnik JL, Campbell B, Mehra R, et al. Osteocytes serve as a progenitor cell of osteosarcoma. J Cell Biochem. 2014;115:1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lye KL, Nordin N, Vidyadaran S, et al. Mesenchymal stem cells: from stem cells to sarcomas. Cell Biol Int. 2016;40:610–618. [DOI] [PubMed] [Google Scholar]
- [11].Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. [DOI] [PubMed] [Google Scholar]
- [12].Lee TH, D’Asti E, Magnus N, et al. Microvesicles as mediators of intercellular communication in cancer–the emerging science of cellular ‘debris’. Semin Immunopathol. 2011;33:455–467. [DOI] [PubMed] [Google Scholar]
- [13].Andaloussi SEL, Mager I, Breakefield XO, et al. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. [DOI] [PubMed] [Google Scholar]
- [14].Penfornis P, Vallabhaneni KC, Whitt J, et al. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer. 2016;138:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qian Z, Shen Q, Yang X, et al. The role of extracellular vesicles: an epigenetic view of the cancer microenvironment. Biomed Res Int. 2015;2015:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [17].Vallabhaneni KC, Penfornis P, Dhule S, et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget. 2015;6:4953–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Wang S, Zhu R, et al. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFkappaB-TLR signaling pathway. J Hematol Oncol. 2016;9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chowdhury R, Webber JP, Gurney M, et al. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget. 2015;6:715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haga H, Yan IK, Takahashi K, et al. Tumour cell-derived extracellular vesicles interact with mesenchymal stem cells to modulate the microenvironment and enhance cholangiocarcinoma growth. J Extracell Vesicles. 2015;4:24900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baglio SR, Lagerweij T, Perez-Lanzon M, et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin Cancer Res. 2017;23:3721–3733. [DOI] [PubMed] [Google Scholar]
- [22].Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. [DOI] [PubMed] [Google Scholar]
- [23].Yegnasubramanian S, Haffner MC, Zhang Y, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Florl AR, Steinhoff C, Muller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Peltoniemi HH, Salmi A, Miettinen S, et al. Stem cell enrichment does not warrant a higher graft survival in lipofilling of the breast: A prospective comparative study. J Plast Reconstr Aesthet Surg. 2013;66:1494–1503. [DOI] [PubMed] [Google Scholar]
- [26].Lindroos B, Boucher S, Chase L, et al. Serum-free, xeno-free culture media maintain the proliferation rate and multipotentiality of adipose stem cells in vitro. Cytotherapy. 2009;11:958–972. [DOI] [PubMed] [Google Scholar]
- [27].Kornilov R, Puhka M, Mannerstrom B, et al. Efficient ultrafiltration-based protocol to deplete extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2018;7:1422674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kaur S, Abu-Shahba AG, Paananen RO, et al. Small non-coding RNA landscape of extracellular vesicles from human stem cells. Sci Rep. 2018;8:15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pavicic W, Joensuu EI, Nieminen T, et al. LINE-1 hypomethylation in familial and sporadic cancer. J Mol Med (Berl). 2012;90:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gylling AH, Nieminen TT, Abdel-Rahman WM, et al. Differential cancer predisposition in lynch syndrome: insights from molecular analysis of brain and urinary tract tumors. Carcinogenesis. 2008;29:1351–1359. [DOI] [PubMed] [Google Scholar]
- [32].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [33].Lindroos B, Suuronen R, Miettinen S. The potential of adipose stem cells in regenerative medicine. Stem Cell Rev. 2011;7:269–291. [DOI] [PubMed] [Google Scholar]
- [34].Tsukamoto S, Honoki K, Fujii H, et al. Mesenchymal stem cells promote tumor engraftment and metastatic colonization in rat osteosarcoma model. Int J Oncol. 2012;40:163–169. [DOI] [PubMed] [Google Scholar]
- [35].Otoukesh B, Boddouhi B, Moghtadaei M, et al. Novel molecular insights and new therapeutic strategies in osteosarcoma. Cancer Cell Int. 2018;18:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tian J, Li X, Si M, et al. CD271+ osteosarcoma cells display stem-like properties. PLoS One. 2014;9:e98549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cortini M, Avnet S, Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. [DOI] [PubMed] [Google Scholar]
- [38].Deregibus MC, Cantaluppi V, Calogero R, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. [DOI] [PubMed] [Google Scholar]
- [39].Deregibus MC, Tetta C, Camussi G. The dynamic stem cell microenvironment is orchestrated by microvesicle-mediated transfer of genetic information. Histol Histopathol. 2010;25:397–404. [DOI] [PubMed] [Google Scholar]
- [40].Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vallabhaneni KC, Hassler MY, Abraham A, et al. Mesenchymal stem/stromal cells under stress increase osteosarcoma migration and apoptosis resistance via extracellular vesicle mediated communication. PLoS One. 2016;11:e0166027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Urciuoli E, Giorda E, Scarsella M, et al. Osteosarcoma-derived extracellular vesicles induce a tumor-like phenotype in normal recipient cells. J Cell Physiol. 2018;233:6158–6172. [DOI] [PubMed] [Google Scholar]
- [43].Jerez S, Araya H, Thaler R, et al. Proteomic analysis of exosomes and exosome-free conditioned media from human osteosarcoma cell lines reveals secretion of proteins related to tumor progression. J Cell Biochem. 2017;118:351–360. [DOI] [PubMed] [Google Scholar]
- [44].Garimella R, Washington L, Isaacson J, et al. Extracellular membrane vesicles derived from 143B osteosarcoma cells contain pro-osteoclastogenic cargo: A novel communication mechanism in osteosarcoma bone microenvironment. Transl Oncol. 2014;7:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bronkhorst AJ, Wentzel JF, Ungerer V, et al. Sequence analysis of cell-free DNA derived from cultured human bone osteosarcoma (143B) cells. Tumour Biol. 2018;40:1010428318801190. [DOI] [PubMed] [Google Scholar]
- [46].Zhu X, You Y, Li Q, et al. BCR-ABL1-positive microvesicles transform normal hematopoietic transplants through genomic instability: implications for donor cell leukemia. Leukemia. 2014;28:1666–1675. [DOI] [PubMed] [Google Scholar]
- [47].Wild L, Funes JM, Boshoff C, et al. In vitro transformation of mesenchymal stem cells induces gradual genomic hypomethylation. Carcinogenesis. 2010;31:1854–1862. [DOI] [PubMed] [Google Scholar]
- [48].Yang Y, Yang R, Roth M, et al. Genetically transforming human osteoblasts to sarcoma: development of an osteosarcoma model. Genes Cancer. 2017;8:484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ehrlich M, Jiang G, Fiala E, et al. Hypomethylation and hypermethylation of DNA in wilms tumors. Oncogene. 2002;21:6694–6702. [DOI] [PubMed] [Google Scholar]
- [50].Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. [DOI] [PubMed] [Google Scholar]
- [51].Iskow RC, McCabe MT, Mills RE, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Phokaew C, Kowudtitham S, Subbalekha K, et al. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36:5704–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bjornland K, Flatmark K, Pettersen S, et al. Matrix metalloproteinases participate in osteosarcoma invasion. J Surg Res. 2005;127:151–156. [DOI] [PubMed] [Google Scholar]
- [54].Uchibori M, Nishida Y, Nagasaka T, et al. Increased expression of membrane-type matrix metalloproteinase-1 is correlated with poor prognosis in patients with osteosarcoma. Int J Oncol. 2006;28:33–42. [PubMed] [Google Scholar]
- [55].Chen D, Zhang YJ, Zhu KW, et al. A systematic review of vascular endothelial growth factor expression as a biomarker of prognosis in patients with osteosarcoma. Tumour Biol. 2013;34:1895–1899. [DOI] [PubMed] [Google Scholar]
- [56].Makrilia N, Kollias A, Manolopoulos L, et al. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest. 2009;27:1023–1037. [DOI] [PubMed] [Google Scholar]
- [57].Gahmberg CG, Tolvanen M, Kotovuori P. Leukocyte adhesion–structure and function of human leukocyte beta2-integrins and their cellular ligands. Eur J Biochem. 1997;245:215–232. [DOI] [PubMed] [Google Scholar]
- [58].Chen Q, Shou P, Zheng C, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wagner ER, Luther G, Zhu G, et al. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma. 2011;2011:325238. [DOI] [PMC free article] [PubMed] [Google Scholar]


