ABSTRACT
The conference ‘The Many Faces of Epigenetics: Multidisciplinary Perspectives “over” Genetics’ was held in Oxford (6–8 December 2017) and offered a valuable window into the domain of Epigenetics and its promises. The workshop revealed that, among a wealth of discourses about Epigenetics, it is not so easy to decipher which discourses are to be trusted. Because Epigenetics is a rather old notion that has generated many debates and promises, defining precisely what has changed and where we are currently is a challenge in itself. Interestingly, the conference allowed debates beyond statements such as ‘If you don’t know the cause, you say it’s epigenetic’ (Deichmann 2016), pointing out that the lack of a precise definition of Epigenetics was no hindrance to the discussions. Finally, it highlighted the grounds of (dis)agreement among communities of natural and social scientists; but eventually the discussions showed that epigenetic tools open the path to new topics and challenges that are awaiting us.
KEYWORDS: Epigenetics, definition, explanatory tools, molecular tools, society, environment
Introduction
The conference ‘The Many Faces of Epigenetics: Multidisciplinary Perspectives “over” Genetics’ was held in Oxford (6–8 December 2017) (http://mfo.cnrs.fr/fr/epigenetique-resumes). Epigenetics is becoming increasingly relevant not only to biological and medical studies but also to Social and Political Sciences, History, Anthropology and Philosophy. Clearly, Epigenetics will contribute to the future development of social, health and environmental policies. In parallel, philosophical and social considerations may help epigeneticists to conceptually refine the epigenetic ‘paradigm’.a
Our paper aims at reporting the original ideas that we think will lead on to further discussions and research projects. Therefore, we focus on the scientific change associated with Epigenetics in the perspective of the different disciplines that were represented in Oxford. We argue that scientific change was the common thread through most debates that occurred during the conference.
After a description of the opening workshop and the questions it addressed, fundamental issues, such as the definition of Epigenetics, are listed. Finally, original topics that emerged are developed and discussed.
Description of the workshop and of its issues
The Oxford conference ‘The Many Faces of Epigenetics: Multidisciplinary Perspectives “over” Genetics’b was born from the following idea: Epigenetics is trans-disciplinary; it crosses Chemistry, Biology, Medicine, Philosophy of Science, History, Social and Political Sciences. While natural scientists have a molecular approach to Epigenetics, social sciences and humanities scholars use a different scale of analysis. Still, Epigenetics provides the opportunity to focus on the same object of study from different angles [1]. Thus, by gathering researchers from different fields, new ideas and branches of Epigenetics should emerge. Projects such as ‘Epigenetics and the embodiment of environments: which biosocial agendas for sociology and epidemiology?’, directed by Severine Louvel (UGA, France) and involving social scientists, political scientists and epidemiologists, could notably be developped.
Thanks to the Maison Française d’Oxford (MFO), the sponsorship from Sorbonne Universités, University of Oxford, French Foreign Affairs, EU COST CM1406 EpiChemBio, 75 researchers from Humanities, Social Sciences and Natural Sciences met, exchanged results and discussed ideas at the MFO in Oxford from 6th to 8 December 2017.
Since we were from different disciplinary fields, the meeting started with a ‘brainstorming’ workshop to define the vocabulary and concepts that would then be discussed during the conference. To this aim, a short survey was sent to all participants beforehand with the following questions:
How do you define Epigenetics?
Do you think that Epigenetics is transdisciplinary? If yes, how?
How do you think the other disciplines (Natural Sciences, Social Sciences and Human Sciences) see your discipline (Natural Sciences, Social Sciences and Human Sciences)?
What do you think will be the contribution of Epigenetics to your discipline in the next years?
What are the main questions/dilemmas that Epigenetics raises in your field?
What do you expect from this transdisciplinary conference?
What mattered and what did not
We expected that the definition of Epigenetics would be a controversial matter, together with heredity, but we were surprised that the discussion rapidly moved towards other questions.
Beyond definition
There is much controversy, publications and thoughts concerning the definition of Epigenetics [1–6]. The most common definition describes Epigenetics as ‘the study of heritable phenotypes that do not alter the DNA sequence’ [4,7].
By plotting the written answers (21 out of 75) that we received to question 1 of the survey, we obtained the word cloud reported in Figure 1. Clearly, it appears that the center of Epigenetics remains DNA and the genes, in particular their expression, the impact of environment and the heritability of epigenetic changes.
Figure 1.

Word cloud built from the written answers to the survey (by using www.wordclouds.com).
Unsurprisingly, it happened that the definition is not fixed, but rather broad and blurry. Importantly, when each discipline was asked to give a single word to define Epigenetics, we realised that most words were common to all (Figure 2). However, it must be noted that this analysis is biased by the disciplines and interests of the participants of the meeting; for example, no scientist from plant biology was present, and few from Developmental Biology; in addition, most of us work on humans or mammals.
Figure 2.
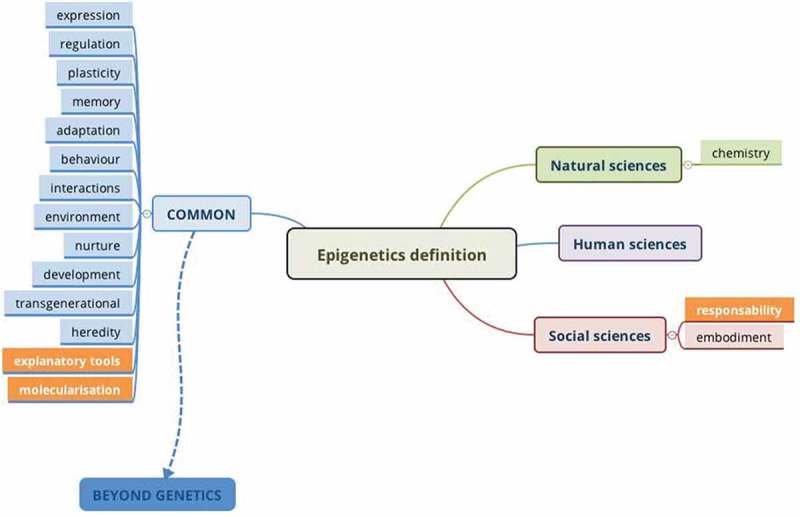
Words defining Epigenetics (built using Xmind 8). Depicted in blue are the common words to all disciplines, and in orange are those that attracted more attention.
This exercise opened the following discussions and ideas.
Few elements were only cited by some disciplines; of note is responsibility, which was discussed by the social scientists with respect to considerations coming from the field of environmental Epigenetics [8], but was hardly shared among the natural scientists.
As expected, gene expression and regulation seem to best characterise Epigenetics (Figure 2, common items in blue), together with the fact that epigenetic marks contribute to the plasticity and memory of cells, organisms and societies, and take part in adaptation. Another feature of Epigenetics shared by the different fields is its role in development (at the levels of cells, organisms and societies), together with heredity and transgenerational impact (how influences experienced by parents are transmitted to their progeny).
Most interestingly, all participants converged to state that a major contribution of Epigenetics to Science consists in offering new explanatory tools at the molecular level: shedding light on how environment (chemical, physical, social, cultural, …) impacts gene expression and behaviour. These tools also take into account the interactions between biology, nutritional input and social structures; link environment to diseases risk; and suggest how psychosocial stress and social inequality produce health disparities. This issue was developed and discussed by M. Szyf (McGill University, Ca) in a talk entitled ‘DNA methylation mediating between experience and phenotypes: implication for mental health’. The famous example of the honey bees by Kurchaski et al. was cited [9] to show that by modifying epigenetic marks (here DNA methylation) it is possible to change a phenotype (in the Dnmt3A-siRNA treated larvae, most of the adults become queens with fully developed ovaries). However, this example also illustrates that Epigenetics is only one of the factors contributing to phenotypes [10]. Finally, S. Louvel (UGA, France) showed how material and social environments change epigenetic patterns and impact health in her presentation entitled ‘About reductionism: how does environmental epigenetics conceptualize and operationalize environments across the lifespan?’. These issues and methods concerning the investigation of how social, environmental and nutritional factors influence biological mechanisms currently fall under the field of Environmental Epigenetics.
Importantly, Epigenetics provides heuristic tools allowing for the start of new research hypotheses that can be experimentally tested and improved, resulting in new ideas and increased knowledge of the complexity of living beings. It recalls what happened with the discovery of the DNA and the establishment of the Central Dogma of Molecular Biology ‘DNA → RNA → proteins’: many thought that the answers to diseases and societies could be found in genes, but the sequencing of the first human genomes [11,12] showed that the Dogma only tells part of the story. This part is undoubtedly essential, since DNA constitutes the words of the booklet of instructions of the cells, but readers and interpreters are also needed. Epigenetics reveals that a new layer is essential for improved understanding of the complexity of cells, organisms and life. This will foster the molecularisation of social and historical phenomena and will (and already does) greatly impact our knowledge [13]. Epigenetics can offer molecular understanding of the profound entanglement of social structures and human biology. One example of this is reducing social dynamics to molecular modifications of DNA, thus shedding light on medical sciences. This topic was developed by F. Panese and L. Chiapperino (University of Lausanne, CH) in ‘The biosocial economies of Epigenetics’.
In brief, the definition of Epigenetics did not monopolise the discussion, since we rapidly agreed on common terms characterising Epigenetics, and the absence of a precise definition was no limit to the discussions that followed. Epigenetics simply brings to concepts what is beyond genes.
The question of epigenetic heredity
The question of epigenetic heredity was expected to be another central point of discussion during the conference. Indeed, Epigenetics is one of the fields of investigation that challenge the gene-centered vision of heredity, which has dominated the scientific landscape for the last sixty years. It provides evidence that biological systems – notably plants and animals – do not only transmit genes across generations, revealing that trans-generational similarities cannot be exclusively explained by the replication of DNA [14–17]. More precisely, epigenetic studies show that molecular modifications of chromatin and non-coding RNAs, which can be sensitive to environmental changes, may be maintained across generations and affect both parent and offspring phenotypes. In this respect, the discipline sheds light on mechanisms by which environmentally-induced traits can be transmitted from one generation to the next [18]. This is the reason why it was initially said to rehabilitate theories regarding the heredity of acquired characters, generally qualified as ‘Lamarckian’ [3,19]. However, this simplistic analysis has been discarded since then [20].
Surprisingly, the problem of trans-generational maintenance of epigenetic marks was not at the centre of the debates. More precisely, the idea that epigenetic transmission challenges the gene-centric vision of heredity did not emerge as a key issue. Participants addressed the difference between trans-generational exposure and genuine epigenetic heredity. They agreed that whereas the former, which can be called cross-generational epigenetic maintenance, is due to the common exposure of several generations to the same environmental cue, the latter, which can be called trans-generational epigenetic maintenance, implies that a dedicated mechanism ensures the reconstruction of epigenetic marks across cell divisions (mitosis and meiosis). In his presentation ‘Transgenerational epigenetics: where are we now?’, P. Allard (UCLA, USA) shed light on elements illustrating this issue. In addition, the importance of clarifying what kind of maintenance is at stake in the famous alleged examples of epigenetic heredity was briefly discussed. This discussion was in agreement with the literature to precisely describe the various kinds of epigenetic maintenance at stake across generations [5,21].
However, no debate arose concerning the frequency or rarity of epigenetic heredity stricto sensu. More precisely, the impact of epigenetic transmission on general thinking about heredity turned out to be a non-issue for the workshop participants. This stands in striking contrast to the contributions according to which epigenetic transmission is highly challenging for the current gene-centered theory of heredity [22,23].
Hot topics, new tools and challenges
If not the definition of Epigenetics and epigenetic heredity, what then were the hot topics discussed during the conference? On what grounds did disagreement occur? It appeared that disagreements were not substantial, but mainly due to the gap between new explanatory goals and old disciplinary norms. They disappeared once the questions were redefined.
In short, what about the hopes and promises of Epigenetics?
Plasticity at the molecular level
In order to identify what the workshop brought about in terms of the current state of the art in Epigenetics, the first question to be asked was: What is different now (now that the domain of Epigenetics is rapidly developing)?
Focusing on the notion of plasticity was a useful way to assess what kind of results epigenetic studies have recently produced. Plasticity is an old notion in Life Sciences that was traditionally conceived at organismal and social levels, as testified by Maurizio Meloni (University of Sheffield, UK) in ‘A postgenomic body: Plasticity, Epigenetics, Biopolitics’. Before the discovery of DNA methylation, it was simply assumed that there probably existed mechanisms accounting for dynamics and reversibility in gene expression. These mechanisms are now extensively studied. This means that instead of being just dreamed of, the explanation of plasticity at the molecular level is on its way, to be (hopefully) followed by intervention in such processes.
Thus, the step forward has been molecular: a molecular mechanism explaining phenotypes has been discovered. For instance, plasticity has long been well-known as an important phenomenon in development. Similarly, it has been commonly assumed that something in the genome had to be plastic, because it was known that gene expression is not rigidly deterministic. However, there was no detailed explanation available of how plasticity could be brought about within cells. The lack of explanatory mechanisms that could account for the non-rigidity of gene expression was a major obstacle to understanding how genes cause phenotypes. Indeed, even though it was assumed that gene expression was somehow plastic, the precise realization of this plasticity remained unknown. Similarly, it was commonly acknowledged that Genetics, however powerful, cannot be the only explanans of development. The problem was that there was hardly any means available to harness this intuition, namely to formulate any precise hypothesis to be tested on the bench. Epigenetics has been a breakthrough in this respect.
Famous examples of plasticity related to epigenetic variations are the three following. Cubas et al. showed for the first time in 1999 that a heritable epimutation, the silencing of Lcyc gene by DNA methylation, is responsible for the naturally-occurring mutant of Linaria vulgaris (Linaria pelori), a flower plant [24]. Cavalli and Paro showed that Drosophila chromosomal elements can transmit an epigenetic state (active or repressed) to the next generations in the absence of any apparent covalent modifications of the DNA [25] and observed that the active mark set at embryonic stages is mitotically stable and inheritable even after the removal the main regulator [26]. Finally, Manning and colleagues shed light on the role of epigenetic marks in the regulatory network that controls tomatoes ripening and tomato colours [27].
Placing the genome in its biochemical context
The discoveries that have changed the course of Epigenetics bear upon mechanisms that participate in gene regulation and enrich our understanding thereof, like DNA methylation, histone modifications, nucleosome positioning, interactions between DNA methylation and chromatin, chromatin enhancers and silencers. These discoveries have had a tremendous impact on the field of Molecular Genetics, as the strict separation between genes and proteins, which was central within classical Genetics, is now fading. Similarly, close connections are now perceived between polymerase, transcription machinery and 3D structure, three items that were conceived as separated beforehand. DNA is not separated anymore from its molecular context made by chromatin and its modifications. As a result, the transcription and translation ‘machinery’ takes a new, more precise meaning, including dynamical aspects.
It is as if the lens we had for studying gene expression was coarse beforehand and has now become more powerful, so that new details are made visible. This ‘lens’ was explicated by the talks of the chemists A. Ganesan (UEA, UK): ‘Being human’ and C. Schofield (University of Oxford, UK): ‘The infinitely complicated chemistry of Epigenetics’. The details change the general picture of the genome as they muddy the waters of former oppositions (that are now seen as more confusing than enlightening) and reveal the dynamical components of the process. For instance, some of the events occurring between the DNA molecule and proteins are better known and we have now understood for sure that some are reversible. An important upshot of these discoveries is that new proteins and new problems have been identified. Epigenetics has decisively entered its new experimental phase with the ‘-omics’ scale-up and the measurements of the chemical modifications of DNA and histones.
Entering this phase means that assumptions and promises about plasticity have been replaced by testable hypotheses. Let us put forward a distinction between (i) wide-scope hypotheses that may inspire general ideas (e.g.; ‘Plasticity plays an important role in development’), (ii) hypotheses that get research done on a daily basis (e.g.; ‘Methylation is an important mechanism of how plasticity actually occurs at the molecular level’), and (iii) statements that may be popular but do not have positive effects on scientific research (e.g.; ‘Epigenetics will overcome genetic determinism’). Contrary to hypotheses in the third category, hypotheses in the second may realize the promises of the ones in the first category and enrich them. This threefold distinction enables us to better assess the status of the above presented discoveries. On one hand, they were expected, as the general hypotheses that something other than genes influences gene expression has been entertained since the very beginnings of Molecular Genetics. But on the other hand, they have been surprising, as is every discovery of an actual mechanism. As such, they are to be viewed as putting forward genuine novelty in the fields of Molecular Genetics and Biochemistry.
Relationships between genetics and epigenetics
The above leads us to dispel two related misconceptions that are common in discourses about Epigenetics. The first is to conceive Epigenetics as a scientific revolution that would discard the idea that only genes are responsible for development. The presentation on aging by W. Reik (Babraham Institut, UK) ‘Epigenetic programming in development and aging’ illustrated this point. The second is that Epigenetics is nothing more than the study of gene regulation and that accordingly, there is nothing actually new in the field. It is important to get rid of both misconceptions. First, the discovery of the molecular mechanisms that are responsible for part of the plasticity of gene expression represents a genuine progress in the field of post-Human Genome Project (HGP) research. Second, accordingly, Genetics has not been overcome by Epigenetics, and nor should it be. Far from such a misconceived view, Genetics is complemented by Epigenetics as scientists progressively realise how immensely complex gene expression is at the molecular scale. What we now know for sure is that Epigenetics is not opposed to Genetics but is an enhancement thereof. The reasoning relating one phenotypic trait to one genetic variant is translated from Genetics to Epigenetics by adding reference to epigenetic variants. Genetics is not over; on the contrary, it goes on through new meanings.
Thus, with respect to the current state of the art in Epigenetics, the workshop has allowed for a better focus on the actual dynamics of research, which is shaped by available technologies that come from the development of Genetics at the end of the twentieth century, among which HGP. This reading of the current state of the field sheds light on the frequently-heard denunciation of ‘molecularisation’. It is the other way around! Our new understanding of processes at the molecular level has opened new avenues for research that are the pre-conditions for explorations at larger, phenotypic scales.
Conclusions and perspectives
To conclude, let us emphasise that the Oxford conference was an example of a cross-disciplinary discussion fuelled by a scientific concept, Epigenetics, which has itself been at the crossroads of several disciplinary fields since it was coined in the 1940’s [28]. Moreover, the meeting provided an opportunity to launch inter-disciplinary considerations about the scientific change supposedly induced by Epigenetics and abundantly discussed in the literature [14,18,29]. It gave momentum to genuine trans-disciplinary work that could be structured in several axis of research and complement disciplinary studies that may develop in the future.
Epigenetics at the crossroads of disciplines
Discussions about Epigenetics have been favourable to cross-disciplinary exchanges since the very beginning [1]. When Conrad Waddington (1942) coined the word Epigenetics to describe the ‘mechanisms by which the genes of the genotype bring about phenotypic effects’, he already targeted the connection of embryology and genetics, two disciplines that had been long been separate. More generally, the word Epigenetics, in referring stricto sensu to whatever is beyond, around or above the genes, has a plastic meaning – many faces – thus setting the stage for pluri-disciplinary approaches.
The Oxford conference illustrated this statement. Gathering scholars from different fields of research, it was a living example of a pluri-disciplinary discussion about a still vague notion. Even if one considers that the precise definition of Epigenetics is important for scientific communication [30], and while conceptual vagueness can first appear as an obstacle to rigorous work, it can also be considered as an ingredient for scientific fruitfulness and as a facilitator of interdisciplinary exchanges [31]. Noteworthy, the vagueness of the concept of gene at first could have appeared as a source of scientific confusion but was after acknowledged as scientifically fertile [32]. Notably, Epigenetics can facilitate communication among scientists with different backgrounds like it did in Oxford, fostering rich cross-disciplinary discussions.
What kind of scientific change are we witnessing?
Epigenetics is sometimes presented as bringing revolutionary change in Biology and as providing radically better insights into the dynamical dimension of biological processes. It is said to improve our understanding of biological and medical phenomena (heredity, development, evolution, pathology, etc.). Assessing the distinguishing features, reach and significance of the ongoing change represents an exciting challenge. Scientific development is indeed a major topic in Philosophy of Science. It is generally analysed according to two main conceptions: either scientific change is a rational, cumulative process in which new theories, better fitted to facts, replace older ones, or a brutal and partially irrational process that implies a radical modification of theoretical frameworks.
The discussions during the conference showed that the description of ongoing scientific change deserves a deeper analysis than that provided by usual categories. While the natural scientists expressed a real enthusiasm about molecular mechanisms making sense of the dynamic and plastic dimension of biological phenomena, their questions and the way in which they addressed them were strikingly reminiscent of the scientific practices at work during the golden age of Molecular Genetics. More precisely, a contrast emerged between the explanatory ambitions of epigeneticists and their ways of addressing scientific problems, which extend the usual practice in Genetics of looking for (epi)genetic variant for phenotypic traits.
In this specific context, philosophers of science, sociologists and scientists are encouraged to undertake collaborative work in order to assess the extent and the nature of the scientific change induced by Epigenetics. They are urged to ask whether this change is about methodology, concepts, norms, instruments or something else and whether it can be called revolutionary or not. They are also invited to ask whether the analysis they will produce will be equally relevant in all scientific fields encompassed within the label ‘Epigenetics’.
The future of cross-disciplinary discussions
What was the nature of the Oxford conference at the end? Should it be qualified as an interdisciplinary meeting? Such a description would not fairly reflect the rich exchanges that took place between specialists and the genuine attention they paid to others’ arguments. Should it then be described as a trans-disciplinary encounter? While inter-disciplinarity implies that each scholar approaches a given topic from her specific point of view, trans-disciplinarity involves a more integrated work that is co-constructed by specialists sharing common theoretical interests. Obviously, disciplinary boundaries were tangible during the Oxford conference. True trans-disciplinarity, as such, was not reached; for example, the discussions about molecularisation and reductionism showed the limits of the dialogue across Natural and Social Sciences.
Neither inter-disciplinary nor trans-disciplinary, the meeting was a unique event in its kind. It set up the conditions for the implementation of a rich discussion that has to be fuelled within the next few years. In other words, it gave momentum to the construction of trans-disciplinary studies. One way to maintain such momentum is to organise similar events on a regular basis, and to associate them to an institutional support that could take the form of an international, inter-disciplinary network carrying out a consistent research program along various axes (e.g. ‘Epigenetic and society’, ‘Epigenetic and scientific explanation’, etc.).
Finally, the Oxford conference embodied the early stage of a challenging way of doing research, in which trans-disciplinary studies may be complementing disciplinary works. Such studies may become additional devices to address some specific issues, those that require the collaboration of specialists with different backgrounds, expertise and perspectives. The present paper, written by a chemist and two philosophers, is an example of trans-disciplinary considerations that have been facilitated by the Oxford conference. The challenge, now, is to outline the issues that could be relevantly addressed by trans-disciplinary teams. One of them regards the extent of the announced – and never seriously characterized – scientific change brought about by Epigenetics.
Funding Statement
This work was supported by the Association Sorbonne Université [NA]; European Cooperation in Science and Technology EU COST [CM1406]; Institut Pasteur [EpiChBio]; CNRS.
Notes
Meant as a discipline taking into full account the plasticity of gene expression.
The Many Faces of Epigenetics: Multidisciplinary Perspectives ‘over’ Genetics, Maison Française d’Oxford, Oxford, UK. Convenors: Paola Arimondo (CNRS), Michel Dubois (Epidapo), Ludovic Halby (MFO/CNRS), Stéphane Jettot (MFO/Paris-Sorbonne), Frédéric Thibault-Starzyk (MFO/CNRS).
Acknowledgments
The authors thank all co-organisers (Frédéric Thibault-Starzyk (MFO/CNRS), Stéphane Jettot (MFO/Sorbonne University), Ludovic Halby (MFO/CNRS) and Michel Dubois (EpiDapo), and funding agents (CNRS, Sorbonne University and COST action CM 1406 Epigenetic Chemical Biology). The authors also thank two anonymous reviewers for their enlightening comments, as well as Timothy Daly, PhD student in philosophy of science, who kindly helped us with the English, and Julia and Zoé Arimondo-Lacroix for proofreading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Nicoglou A, Merlin F.. Epigenetics: A way to bridge the gap between biological fields. Stud Hist Philos Biol Biomed Sci. 2017. December;66:73–82. PubMed PMID: 29033228. [DOI] [PubMed] [Google Scholar]
- [2].Deichmann U. Epigenetics: the origins and evolution of a fashionable topic. Dev Biol. 2016. August 1;416(1):249–254. . [DOI] [PubMed] [Google Scholar]
- [3].Bird A. Perceptions of epigenetics. Nature. 2007. May 24;447(7143):396–398. PubMed PMID: 17522671; eng. . [DOI] [PubMed] [Google Scholar]
- [4].Berger SL, Kouzarides T, Shiekhattar R, et al. An operational definition of epigenetics. Genes Dev. 2009. April 1;23(7):781–783. PubMed PMID: 19339683; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014. March 27;157(1):95–109. PubMed PMID: 24679529; PubMed Central PMCID: PMCPMC4020004. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann N Y Acad Sci. 2002. December;981:82–96. PubMed PMID: 12547675. [DOI] [PubMed] [Google Scholar]
- [7].Dupont C, Armant DR, Brenner CA. Epigenetics: definition, mechanisms and clinical perspective. Semin Reprod Med. 2009. September;27(5):351–357. . PubMed PMID: 19711245; PubMed Central PMCID: PMCPMC2791696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burris HH, Baccarelli AA. Environmental epigenetics: from novelty to scientific discipline. J Appl Toxicol. 2014. February;34(2):113–116. . PubMed PMID: 23836446; PubMed Central PMCID: PMCPMC3867531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kucharski R, Maleszka J, Foret S, et al. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008. March 28;319(5871):1827–1830. PubMed PMID: 18339900; eng. [DOI] [PubMed] [Google Scholar]
- [10].Rasmussen EM, Amdam GV. Cytosine modifications in the honey bee (Apis mellifera) worker genome. Front Genet. 2015;6:8 PubMed PMID: 25705215; PubMed Central PMCID: PMCPMC4319462. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001. February 15;409(6822):860–921. PubMed PMID: 11237011. [DOI] [PubMed] [Google Scholar]
- [12].Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001. February 16;291(5507):1304–1351. PubMed PMID: 11181995. [DOI] [PubMed] [Google Scholar]
- [13].Lock M. Comprehending the body in the era of the epigenome. Curr Anthropol. 2015;56(2):151–177. [Google Scholar]
- [14].Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009. June;84(2):131–176. PubMed PMID: 19606595. [DOI] [PubMed] [Google Scholar]
- [15].Johannes F, Porcher E, Teixeira FK, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009. June;5(6):e1000530 PubMed PMID: 19557164; PubMed Central PMCID: PMCPMC2696037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. PubMed PMID: 18767965. . [DOI] [PubMed] [Google Scholar]
- [17].Holeski LM, Jander G, Agrawal AA. Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol Evol. 2012. November;27(11):618–626. . PubMed PMID: 22940222. [DOI] [PubMed] [Google Scholar]
- [18].Richards EJ. Inherited epigenetic variation–revisiting soft inheritance. Nat Rev Genet. 2006. May;7(5):395–401. . PubMed PMID: 16534512. [DOI] [PubMed] [Google Scholar]
- [19].Jablonka E, Lamb MJ. The evolution of information in the major transitions. J Theor Biol. 2006. March 21;239(2):236–246. PubMed PMID: 16236324. . [DOI] [PubMed] [Google Scholar]
- [20].Loison L. Lamarckism and epigenetic inheritance: a clarification [journal article]. Biol Philos. 2018. August 27;33(3):29. . [Google Scholar]
- [21].Grossniklaus U, Kelly WG, Kelly B, et al. Transgenerational epigenetic inheritance: how important is it? Nat Rev Genet. 2013. March;14(3):228–235. PubMed PMID: 23416892; PubMed Central PMCID: PMCPMC4066847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jablonka E, Lamb MJ. Transgenerational epigenetic inheritance In: Pigliucci M, MüLler GB, editors. Evolution – the Extended Synthesis. Cambridge, MA: MIT Press; 2010. p. 175–208. [Google Scholar]
- [23].Bonduriansky R. Rethinking heredity, again. Trends Ecol Evol. 2012. June;27(6):330–336. . PubMed PMID: 22445060. [DOI] [PubMed] [Google Scholar]
- [24].Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999. September 9;401(6749):157–161. PubMed PMID: 10490023. . [DOI] [PubMed] [Google Scholar]
- [25].Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998. May 15;93(4):505–518. PubMed PMID: 9604927. [DOI] [PubMed] [Google Scholar]
- [26].Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999. October 29;286(5441):955–958. PubMed PMID: 10542150. [DOI] [PubMed] [Google Scholar]
- [27].Manning K, Tor M, Poole M, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006. August;38(8):948–952. PubMed PMID: 16832354. [DOI] [PubMed] [Google Scholar]
- [28].Waddington CH. The Epigenotype. Endeavour. 1942;1(1):18–20. [Google Scholar]
- [29].Meloni M, Testa G. Scrutinizing the epigenetics revolution. Biosocieties. 2014. November;9(4):431–456. PubMed PMID: 25484911; PubMed Central PMCID: PMCPMC4255066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Greally JM. A user’s guide to the ambiguous word ‘epigenetics’. Nat Rev Mol Cell Biol. 2018. April;19(4):207–208. . PubMed PMID: 29339796. [DOI] [PubMed] [Google Scholar]
- [31].Löwy I. The strength of loose concepts — boundary concepts, federative experimental strategies and disciplinary growth: the case of immunology. Hist Sci. 1992;30:371–396. [Google Scholar]
- [32].Fox Keller E. Making Sense of Life, Explaining Biological Development with Models, Metaphors and Machines, Cambridge, MA: Harvard University Press; 2002. [DOI] [PubMed] [Google Scholar]


