Abstract
GPR124 is involved in embryonic development and remains expressed by select organs. The importance of GPR124 during development suggests that its aberrant expression might participate in tumor growth. Here we show that both increases and decreases in GPR124 expression in glioblastoma cells reduce cell proliferation by differentially altering the duration mitotic progression. Using mass spectrometry-based proteomics, we discovered that GPR124 interacts with ch-TOG, a known regulator of both microtubule (MT)-plus-end assembly and mitotic progression. Accordingly, changes in GPR124 expression and ch-TOG similarly affect MT assembly measured by real-time microscopy in cells. Our study describes a novel molecular interaction involving GPR124 and ch-TOG at the plasma membrane that controls glioblastoma cell proliferation by modifying MT assembly rates and controlling the progression of distinct phases of mitosis.
Keywords: cell proliferation, G protein-coupled receptors and microtubules, glioblastomas
1 |. INTRODUCTION
G protein-coupled receptors (GPCRs) control many facets of cell biology, including cell migration, proliferation, differentiation, and survival. Accordingly, time-dependent changes in the expression of GPCRs play critical roles during embryogenesis and tissue development, and disruption of GPCR activities may be involved in pathological processes, including cancer (Bar-Shavit et al., 2016; Cherry & Stella, 2014). Canonical signaling through GPCRs occurs through G proteins and β-arrestins, whereas noncanonical signaling through GPCRs occurs through dimerization with tyrosine kinases and scaffolding with intracellular enzymes (Ellisdon & Halls, 2016; Kamato et al., 2015; Kenakin, 2017). Genomewide analyses suggest that ~750 genes encode for GPCRs, and more than half of these receptors do not have an identified endogenous ligand and remain essentially uncharacterized with respect to their coupling mechanisms (Fredriksson & Schioth, 2005; Tang, Wang, Li, Luo, & Liu, 2012; Wacker, Stevens, & Roth, 2017). Thus, the involvement of many GPCRs in pathological processes such as cancer represents a cardinal area of research (Bar-Shavit et al., 2016; Dorsam & Gutkind, 2007; Liu et al., 2016).
GPR124 (also known as Tumor Endothelial Marker 5) is an orphan adhesion GPCR that was originally identified in endothelial cells that form the neovasculature that invades colorectal tumors (St Croix et al., 2000). Shortly after its discovery, GPR124 was shown to play an essential role in embryonic brain angiogenesis and blood-brain barrier formation (Anderson et al., 2011; Cullen et al., 2011; Kuhnert et al., 2010). Accordingly, this receptor is expressed at high levels in endothelial cells that form the vasculature of the developing brain and plays a role in endothelial cell migration and differentiation without affecting cell proliferation (Kuhnert et al., 2010). Furthermore, genetic deletion of GPR124 in endothelial cells leads to impaired angiogenic sprouting and results in leaky blood vessels (Cullen et al., 2011; Wang, Cho, Wu, Siwko, & Liu, 2014). It is also known that GPR124 remains expressed at low levels in select adult tissues, and that its signaling partners include Wnt7A/B and disk-large tumor suppressor gene that regulates the activity of the tumor suppressors adenomatous polyposis coli and protein phosphatase and tensin homologue proteins (Noda, Vallon, & Kuo, 2016; Yamamoto et al., 2004; Zhou & Nathans, 2014). Thus, a role for GPR124 has been established in endothelial cell migration and differentiation involved in neovascularization of both the developing brain and select solid tumors, such as colorectal cancer.
A role of GPR124 in tumor growth when expressed by cancer cells has not been tested. One report indicates that changes in GPR124 expression affect the proliferation of HUVEC cells, suggesting a role for GPR124 in controlling the proliferation of immortalized cells and possibly cancer cells (Vallon, Rohde, Janssen, & Essler, 2010). Furthermore, several reports indicate that GPR124 expression is increased in some cancers, including urothelial carcinoma and lung adenocarcinoma (Gao et al., 2014; Williams et al., 2010), and decreased in other cancers, such as prostate cancer (Hunter et al., 2016). One study showed that GPR124 expression can be either greatly increased or greatly decreased depending on the patient-derived (PD)-glioblastoma sphere and glioblastoma cell line (Fève et al., 2014). Based on this premise, we studied how increases and decreases in GPR124 expression affects glioblastoma cell proliferation and used stable isotope labeling of amino acids in cell culture (SILAC)/mass spectrometry analysis and fluorescence microscopy to determine the molecular and cellular mechanisms of this response.
2 |. MATERIALS AND METHODS
2.1 |. Cell culture
All cells were cultured at 37°C in a 5% CO2 humidified atmosphere unless otherwise noted. The T98G, U87MG, HeLa, and HCT116 cell lines (ATCC, Manassas, VA) were cultured with DMEM supplemented with 10% FBS, 2 mM L-glutamine, 5 mM NaHCO3, 5 mM HEPES, 100 U/mL penicillin, and 100 μg/mL streptomycin or RPMI supplemented with 10% FBS, 100 U/mL penicillin, and 100 μL/mL streptomycin.
2.2 |. mRNA from noncancerous human brain and PD-GBM spheres grown in culture
The ten noncancerous human brain mRNA samples (PGCs 42, 71, 76, 80, 82, 81, 84, 86, 87, and 88) were harvested from human postmortem brains (age range 26–52 years old, 5 males/5 females) and their microarray data and cases studies deposited at the GEO database (accession # GSE11882; Berchtold et al., 2008, 2014). The seven PD-GBM spheres (SNs 276, 272, 243, 264, 227, 260, and 217) and MGG8 sphere were grown and mRNA harvested as described (Cherry et al., 2016).
2.3 |. Vectors and generation of stable and transient transgenic cell lines
Stable GPR124 knockdown cells were generated in T98G cells using MISSION® pLKO.1-puro lentiviral constructs purchased from Sigma- Aldrich (GPR124 targeting sequence: 5′—ATGTGGAAGCACAAGTTCAGC—3′). The pLKO.1-puro nontarget shRNA plasmid (Sigma-Aldrich) was used as a control plasmid. Stable cell lines were grown as single clones generated under puromycin selection (2 μg/mL). Clones were grown and individually assessed for GPR124 expression using qPCR. Stable U87MG cells with GPR124-myc overexpression were generated by transfecting pcDNA3.1-GPR124-myc (kindly provided by Brad St Croix, Tumor Angiogenesis Section, Mouse Cancer Genetics Program, National Cancer Institute, National Institutes of Health, Frederick, MD) into U87MG cells using the Lipofectamine® 2000 reagent (Thermo). An empty pcDNA3.1™ (Invitrogen) vector was used as a control. Stable lines were grown as single clones generated under G418 selection (1 mg/mL) and individually assessed for the presence of GPR124-myc by qPCR and immunofluorescence staining. Transient overexpression of GPR124-myc was conducted using lentiviral transduction. GPR124-myc was cloned into a lentiviral construct (referred to as pBS-4821) containing GFP under an IRES promoter. An empty pBS-4821 vector was used as a control. Transient overexpression of GPR124-mCherry was conducted using the Amaxa Nucleofector™ system according to the manufacturer’s recommendations (Lonza, Basel, Switzerland). GPR124-mCherry was generated by cloning mCherry into the pcDNA3.1-GPR124-myc vector. An empty mCherry vector used as a control was created by cloning mCherry into the pEGFP-C1 (Clontech) vector. Transient knockdown of GPR124 in HeLa cells was conducted using siRNA (targeting sequence: 5′—CCGACUAAACAUAUCUGGA—3′, Sigma). The Universal Control #1 siRNA (UNI, Sigma) or Silencer® Negative control #1 siRNA (Thermo) were used as controls. SiRNA was transfected using Lipofectamine® RNAiMAX reagent (Thermo Fisher) according to the manufacturer’s protocol. Genomic EGFP-FKBP fusion to the chTOG gene was constructed using CRISPR Cas9 cleavage and homology-directed repair (HDR). Chromosomal sequences were analyzed with the program CRISPRdirect (http://crispr.dbcls.jp/) to identify target sites. Custom oligonucleotides encoding CRISPR target sites (IDT, Skokie IL) were ligated to pX459 version 2 (AddGene, Boston MA) to create the CRISPR targeting vector. Synthetic HDR template DNA (BioBasic, Montreal, QC) was made containing the last exon (exon 44) of the chTOG gene fused to fluorescent protein and FKBP sequences, flanked by approximately 600 bp of genomic sequence from each side of the exon. HCT116 cells were co-transfected with linearized HDR template and a CRISPR targeting plasmid, then treated from 24 to 72 hr. Posttransfection with 0.4 μg/mL puromycin to transiently select cells containing the targeting vector. Surviving cells containing fluorescent fusion proteins were selected, grown to homogeneous populations, and analyzed by PCR of genomic DNA and Western blots to identify populations in which both alleles of the locus were recombinant.
2.4 |. Cell viability and proliferation
Cell viability was assessed with the WST-1 reagent (Roche, Pleasanton, CA) according to the manufacturer’s protocol. All WST-1 experiments were performed in phenol red-free DMEM media. To measure cell proliferation, [3H]thymidine (5 μCi/mL, Perkin-Elmer, Waltham, MA) was added to cells and was measured at the indicated times by adding 1 M NaOH for 10 min on ice and quantifying radioactivity.
2.5 |. Real time qPCR
RNA was extracted from cells using the PerfectPure RNA kit (5-Prime, Gaithersburg, MD), and qPCR was performed with the Roche LightCycler® 480 Probes Master kit and Universal Probe Library (UPL) system (Roche, Indianapolis, IN) according to the manufacturer’s instructions on a Stratagene MX3000P qPCR machine (Stratagene, San Diego, CA). β-Actin, HPRT, and YWHAZ were used to generate a geometric mean for normalization. Intron-spanning primers for GPR124 (Forward: 5′—GGCTCCTTCCTGGGACTG—3′; Reverse: 5′—GCACTGTGCTGATGATGTTGTT-3′, UPL probe #67), p-Actin (Forward: 5′—ATTGG CAATGAGCGGTTC—3′; Reverse: 5′—CGTGGATGCCACAGGACT—3′, UPL probe #11), HPRT (Forward: 5′—TGATAGATCCATTCCTATGACTGTAGA—3′; Reverse: 5′—CAAGACATTCTTTCCAGTTAAAGTTG—3′ UPL probe # 22), and YWHAZ (Forward: 5′—GCAATTACTGAGAG ACAACTTGACA—3′; Reverse: 5′—TGGAAGGCCGGTTAATTTT—3′ UPL probe # 2) were designed using the Roche UPL Assay Design Center.
2.6 |. Time-lapse microscopy
HeLa cells stably expressing H2B-GFP were used to visualize nuclei during live cell imaging. H2B-GFP HeLa were transfected with siRNA targeting GPR124 or control siRNA (see above) or transduced with virus containing GPR124-myc or a control vector (see above) 30 hr prior to imaging. Cells were plated in 35 mm HiQ4 4 chamber glass bottom imaging dishes at a density of 10,000 cells per chamber. Immediately before imaging, media was replaced with CO2-independent media with 10% FBS. Images were acquired on a Nikon Biostation IM-Q (Nikon) as stacks of 4 × 3-μm Z-steps every 5–10 min for 24–30 hr. Time-lapse movies were analyzed by hand using Fiji.
2.7 |. Live cell imaging of MT polymerization
HeLa cells were transfected with EB3-GFP to label assembling MTs (Wordeman, Wagenbach, & von Dassow, 2007). Cells were transfected with GPR124-mCherry or CMV-mCherry empty vector for GPR124 overexpression studies. For GPR124 knockdown studies, HeLa cells were transfected with siRNA targeting GPR124 or scrambled siRNA, as above. For ch-TOG overexpression studies, HCT-116 cells were transfected with ch-TOG-GFP or GFP empty vector. Interphase cells were imaged at a single Z-plane at 500-ms intervals at on a Deltavision microscope system (Applied Precision, Issaquah, WA) using a ×60 1.42 NA lens (Olympus, Tokyo, Japan). Mitotic cells were imaged at 37°C using 3 × 1.0-μm sections at 2-s intervals and deconvolved using SoftWorx 5.0 (Applied Precision). MT assembly rates were scored in interphase cells using Fiji TrackMate (Jaqaman et al., 2008). For mitotic cells, the CDK1 inhibitor RO3306 (10 μM, Sigma) was applied overnight and washed with CO2-independent media for 30 min to increase mitotic index. Cells were imaged at 3 × 0.5-μm sections at 1 s intervals for 45 seconds as above. Spindle and astral MT assembly was measured with Fiji by generating kymographs of the EB3-GFP channel. The Fiji Directionality plugin was used to calculate the angles of the EB3-GFP tracks. Speed was calculated using the equation velocity = , where μm=distance per pixel, t=frame time, θ=angle of the EB3-GFP comet in the kymograph (Figure S5). Data are expressed as the average of the spindle or astral MT assembly rates for each cell.
2.8 |. Fluorescent immunocytochemistry
Cells fixed in 4% paraformaldehyde in PBS at 37°C for 20 min and permeabilized in 0.5% Triton X-100 for 5 min at room temperature. The anti-ch-TOG antibody (1:250, BioLegend, or 1:50, Abcam) was applied overnight at room temperature. Actin was labeled using either fluorescein-phalloidin (1:40, Molecular Probes) or ActinGreen-488 (2 drops/mL, Molecular Probes) applied for 1 hr at room temperature. Tubulin and centromeres were labeled using the anti-tubulin monoclonal DM1-alpha (1:250, Sigma) or anti-centromere (Antibodies Inc., Davis, CA) applied for 1 hr at room temperature. Coverslips were mounted using ProLong® Gold containing DAPI (Molecular Probes, Eugene, OR). Fluorescent images were collected as 0.5-μm Z-stacks on a Deltavision microscope system (Applied Precision/GEHealthcare, Issaquah, WA) using a ×60 1.42 NA lens (Olympus, Tokyo, Japan). Selected images were deconvolved using SoftWorx 5.0 (Applied Precision/GEHealthcare), and representative images are presented as a flat Z-projection.
For quantitative analysis of ch-TOG in mitotic cells, the CDK1 inhibitor RO3306 (10 μM, Sigma) was applied overnight and washed with CO2-independent media for 30 min. Regions containing the entire cell or the mitotic spindle were selected on summed slices Z-projections. The pixel intensity was quantified and normalized to the area of each region. The background was subtracted by quantifying the pixel intensity in a region outside of the cell and subtracting from the pixel intensity measured inside the cell.
For assessment of spindle profiles, T98G and U87MG cells were stained as above to visualize tubulin and centromeres. Mitotic stages were scored by hand using a Nikon FX-A microscope with a ×60 1.4 NA lens (Tokyo, Japan).
2.9 |. TIRF-FRAP
Crispr modified Hct116 cells expressing ch-TOG-FKBP-GFP homozygously from the endogenous promotor (described above) were transfected with GPR124-Cherry and cultured on poly-D-lysine coated MatTeK dishes. Cells were switched to CO2-independent media (Gibco) and imaged on a Deltavision microscope (GEHealthcare) outfitted with 4-laser TIRF capabilities, Olympus 60×, 1.49 NA TIRF objective and Ultimate focus (GEHealthcare) at 37°C. Ch-TOG-FKBP-GFP was photobleached within 100 nm of the cell membrane using a single 488 laser pulse. Recovery of GFP fluorescence was monitored using adaptive intervals ranging from 0.2–1.5 s intervals. Images were corrected for photobleaching using an unbleached region of the cell membrane and arbitrary fluorescence units normalized using ImageJ ver. 2.0 (2018). Nonlinear fit of photo recovery curve was fitted using PRISM. The R2 for the curves was 0.59 and 0.74 for plus and minus GPCR-cherry, respectively. The proportion of fluorescence recovery was 0.51 +/− 0.03 (N = 13 cells) and 0.76 +/− 0.04 (N = 10 cells) for plus and minus GRCR-cherry, respectively.
2.10 |. Membrane solubilization and immunoprecipitation
All steps were performed on ice or at 4°C. Cells expressing GPR124-myc were harvested by scraping into TME buffer (25 mM Tris-HCl pH 7.4, 5 mM MgCl2, 4 mM EDTA) containing a cocktail of protease inhibitors (Sigma), lysed with 40 strokes in a dounce homogenizer, and centrifuged at 100 × g in a tabletop centrifuge for 10 min to pellet nuclei. The supernatant was collected and diluted with TME containing digitonin to a final concentration of 50 mg/mL (Sigma). Lysates were placed on a rotator at 4°C for 2 hr and then centrifuged at 15,700 × g. Supernatants were collected, and protein concentration was determined using the DC™ Protein Assay (Bio-Rad, Hercules, CA). For immunoprecipitation, at least 1 mg of protein was incubated with anti-myc antibody (1:100, Cell Signaling Technologies) at 4°C for 2 hr. For competition with myc peptide, myc antibody and myc peptide (5 μg/mL, Sigma) were preincubated for 30 min at room temperature. Mouse IgA-conjugated agarose beads were spiked into lysates and incubated for 1 hr at 4°C. Beads were washed ×5 with TME buffer. For subsequent western blotting, proteins were eluted by heating to 70°C in ×4 LDS sample buffer (Invitrogen) containing 10% β-mercaptoethanol. For subsequent analysis by mass spectrometry, proteins were reduced with 10 mM DTT and alkylated with 600 mM chloracetamide (Sigma). Proteins were eluted by heating to 70°C in ×4 LDS sample buffer containing 20 mM DTT. Beads were pelleted by centrifugation, and supernatant was loaded into a 4–20% Bis-Tris polyacrylamide gel for western blotting or mass spectrometry.
2.11 |. Western blotting
Lysates were prepared as above. Proteins were separated on 4–20% polyacrylamide Mini-PROTEAN® gels (BioRad) and transferred onto PVDF membranes. Membranes were probed with myc (1:1,000; Cell Signaling Technologies, Danvers, MA) or ch-TOG (1:1,000, BioLegend, San Diego, CA) primary antibodies overnight at 4°C and then visualized using goat-anti-rabbit HRP-linked secondary antibodies (1:2,000, Invitrogen) or mouse TrueBlot® (1:1,000, Rockland Antibodies & Assays, Limerick, PA).
2.12 |. Stable isotope labeling of amino acids in cell culture
Metabolic labeling of amino acids using SILAC was completed as described previously (Lau, Suh, Golkowski, & Ong, 2014; Ong, 2010; Ong & Mann, 2006) with SILAC DMEM media supplemented with 10% dialyzed FBS (Sigma) and either light (L-lysine and L-arginine [Fisher]) or heavy ([13C6, 15N2] L-lysine [Sigma-Isotec, St Louis, MO] and [13C6,15N4] L-arginine [Cambridge Isotope Laboratories, Andover, MA]) isotope-enriched amino acids. Cells were split into two groups regarded as “heavy” and “light.” SILAC media was applied to cells for at least 5 cell doublings to ensure complete labeling of the proteome, which was verified by mass spectrometry. Membranes were solubilized as above and immunoprecipitation was performed in preparation of mass spectrometry. Each SILAC labeling experiment consisted of two parts completed in parallel: (a) the forward experiment in which a competing myc peptide (5 μg/mL, Sigma) was applied to the heavy condition and (b) the reverse experiment in which the myc peptide was applied to the light condition. Full competition of the GPR124 complex by the myc peptide was verified by western blot analysis (data not shown).
2.13 |. LC-MS analysis of SILAC reactions
Proteins were separated on a 4–20% polyacrylamide gel and stained with SimplyBlue™ SafeStain (Invitrogen). Lanes were cut into five pieces by protein molecular weight. Proteins were digested with trypsin, and peptides were extracted and desalted on C18 StageTips (Ong, 2010). Peptides were analyzed on an Orbitrap Elite (Thermo, Bremen Germany) using 90 min gradients of 3–35% acetonitrile at 200 nL/min (Thermo Dionex RSLCnano, Sunnyvale, CA) as described previously (Lau et al., 2014). Proteins were identified using MaxQuant (version 1.3.0.5; Cox et al., 2011; Cox & Mann, 2008). Protein “hits” were identified as described previously (Ong & Mann, 2006). Statistical significance was determined using one sample Student’s t-tests of the absolute value of the normalized heavy: light peptide ratios of the forward and reverse experiments. A protein was considered statistically relevant if the normalized ratios of each experiment were significantly different from 0.
2.14 |. Statistical analysis
The GraphPad Prism software (v5.01, La Jolla, CA) was used for statistical analysis. Data are presented as the mean ± SEM, and statistical significance was determined using Student’s t-test, a one-way ANOVA followed by a Dunnet’s or Tukey posthoc test, or a two-way ANOVA followed by a Bonferroni posthoc test.
3 |. RESULTS
3.1 |. Glioblastoma cells express low and high levels of GPR124 and changes in its expression inhibits cell proliferation
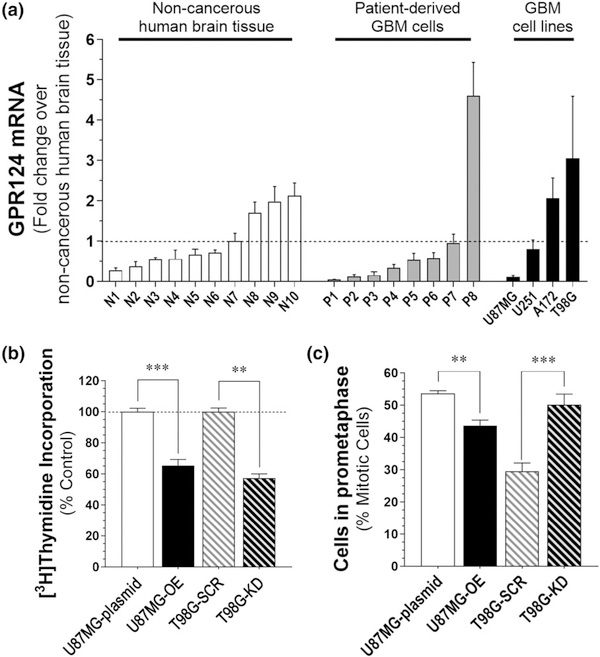
We measured GPR124 mRNA levels (gene name: ADGRA2) by qPCR in ten samples of noncancerous human brain (N1-N10; average = reference control), eight samples of PD-glioblastoma spheres grown in culture and four samples of human glioblastoma cell lines grown in culture (Table S1 provides sample nomenclature and information). Compared with reference control, GPR124 expression was significantly increased in one PD-glioblastoma sphere (P8) and two glioblastoma lines (T98G and A172) and was significantly decreased in three PD-glioblastoma spheres (P1, P2, and P3) and one glioblastoma line (U87MG; Figure 1a). Combined with the study by Fève et al. (2014), these results show that GPR124 expression may be aberrantly expressed in a subset of glioblastoma tumors. To determine if changes in GPR124 expression affect glioblastoma cell proliferation, we selected a cell line with low GPR124 expression, U87MG cells, and generated stable cell lines that overexpresses GPR124 (U87MG-OE). We also selected a cell line with high GPR124 expression, T98G cells, and generated stable cell lines with decreased GPR124 expression (T98G-KD). [3H]Thymidine incorporation assays of these genetically-altered glioblastoma cell lines showed that both increase and decrease in GPR124 expression significantly reduces cell proliferation (Figure 1b). We investigated the possibility that reduced cell proliferation results from altered MT dynamics and, accordingly, mitotic progression (Rao, Yamada, Yao, & Dai, 2009). Thus, we scored the proportion of cells in each stage of mitosis to determine if altered GPR124 expression in U87MG and T98G cells affects the proportion of cells in prometaphase, an index of timely progression through mitosis (Luo et al., 2009). We found that the U87MG-OE cell population contained 22% less cells in prometaphase than the control U87MG cells population, and the T98G-KD cell population contained 70% more cells in prometaphase than the control T98G cell population (Figure 1c). These results suggest that changes in GPR124 expression in either direction might affect mitotic progression.
FIGURE 1.
Glioblastoma cells express low and high levels of GPR124, and changes in its expression differentially regulate cell proliferation and mitotic progression. (a) GPR124 mRNA (ADGRA2) expression measured by qPCR in noncancerous human brain tissue, patient-derived glioblastoma spheres and glioblastoma cell lines. Specifics about human samples are in Table S1. Data are the mean ± SEM of three experiments expressed as a fold-change over the average ADGRA2 expression in all noncancerous brain tissues. (b) Cell proliferation measured by [3H] thymidine incorporation in U87MG cells expressing a control pcDNA vector (U87MG-plasmid) compared with U87MG cells with increased GPR124 expression (U87MG-OE), and of T98G cells expressing a scrambled shRNA control (T98G-SCR) compared with T98G cells with reduced GPR124 expression (T98G-KD). Data are the mean ± SEM of at least three experiments. ***p < .001, **p < .01, Student’s t-test compared to corresponding control. (c) Proportion of cell in prometaphase determined by fluorescent microscopy for U87MG-plasmid cells compared with U87MG-OE, and of T98G-CTR cells compared with T98G-KD cells. Data are mean ± SEM of at least three experiments. **p < .01 compared with corresponding control, Student’s t-test
3.2 |. Increase and decrease in GPR124 expression affect distinct phases of mitosis
To determine if and how changes in GPR124 expression influence mitosis and extend our results obtained with U87MG and T98G cells, we studied mitotic progression in HeLa cells, a commonly used cell line with well-known mitotic progression (Meraldi, Draviam, & Sorger, 2004; Puck & Steffen, 1963). qPCR analysis of GPR124 mRNA showed that HeLa cells express average levels compared to U87MG and T98G cells allowing to study this response in one model system (Figure S1). Thus, we used lentivirus infection to either increase or decrease GPR124 expression in HeLa cells, and measured the cell cycle timing by time-lapse microscopy (Kim et al., 2016). In agreement with the results gathered in U87MG cells, the time spent in prometaphase was reduced by 40% in HeLa cells with overexpressed GPR124 (HeLa-OE) compared with HeLa cells expressing a control virus (HeLa-vector; Figure 2a). Furthermore, the times spent in metaphase and anaphase/telophase were increased by 32% and 8%, respectively, resulting in no change in overall mitotic timing in HeLa-OE cells compared to HeLa-vector (Figure 2a). In agreement with the results gathered with T98G cells, a reduction of GPR124 expression in HeLa cells (HeLa-KD) increased the time spent in prometaphase by 118% compared to HeLa cells expressing a control scramble siRNA (HeLa-SCR; Figure 2a). Here, the times spent in metaphase was increased by 46% and the time spent in anaphase/telophase was reduced by 20%, resulting in an increase in the total time spent in mitosis by 46% when comparing HeLa-KD and HeLa-SCR (Figure 2a). These data show that increases and decreases in GPR124 expression differentially influence mitotic progression by changing the timing of distinct phases of mitosis.
FIGURE 2.
Changes in GPR124 expression differentially modulate select phases of mitosis and incidence of lagging chromosomes. (a) Timelapse microscopy of HeLa cells with overexpressed (HeLa-OE) and knocked down (HeLa-KD) GPR124 compared with controls expressing either an empty expression vector (HeLa-vector) or a scrambled siRNA (HeLa-SCR). Data are the mean ± SEM of at least three independent experiments. *p < .1, **p < .01, ***p < .01 compared with corresponding control, Student’s t-test. (b) Representative images of time-lapse microscopy for HeLa-vector and HeLa-OE cells. Scale, 10 μm, arrows depict lagging chromosomes. (c) Prevalence of lagging chromosomes in HeLa-vector, HeLa-OE, HeLa-SCR, and HeLa-KD cells. Data are the mean ± SEM of at least three experiments. **p < .01 compared with corresponding control, Student’s t-test
The accurate and timely establishment of kinetochore-MT attachments during prometaphase and metaphase is required for correct chromosome segregation during mitosis, and the progression to anaphase is paused by the spindle assembly checkpoint (SAC) until the chromosomes have formed proper bipolar attachments to spindle MTs (Ertych et al., 2014). Erroneous kinetochore-MT attachments occurring during metaphase that evade the SAC can result in lagging chromosomes during anaphase and their quantification provides an index of chromosome instability (CIN; Cimini et al., 2001a; Stolz et al., 2010). Thus, HeLa cells overexpressing GPR124 spend less time in prometaphase (Figure 2b) but also manifest more mitotic errors resulting in a 78% increase in the prevalence of lagging chromosomes (Figure 2c). This result suggests that increased GPR124 expression promotes accelerated prometaphase that prevents cells from correcting erroneous kinetochore-MT attachments. By contrast, decreased GPR124 expression in HeLa cells did not change the prevalence of lagging chromosomes (Figure 2d). Because lagging chromosomes arise from aberrant MT attachments to kinetochores (Cimini et al., 2001a), these results raise the possibility that GPR124 might impact chromosome segregation by interfering with MT dynamics or behavior within the spindle.
3.3 |. GPR124 interacts with ch-TOG
To determine whether GPR124 interacts with proteins involved in glioblastoma cell proliferation, we sought to identify constituents of the GPR124 protein complex in U87MG cells using SILAC/mass spectrometry (Lau et al., 2014; Ong et al., 2002). Analysis of the proteins that were pulled down with GPR124-myc stably expressed in U87MG cells suggested that cytoskeleton-associated protein-5 (CKAP5, commonly referred to as ch-TOG) associates with GPR124. (SILAC ratios for CKAP5 were greater than two-fold enriched in GPR124-myc pulldown experiments; p = 0.001, one-way Student’s t-test comparing with a theoretical value of 1). Additional proteins that were associated with GPR124 are reported in Table S2. Ch-TOG is a MT polymerase that regulates plus-end MT assembly and mitotic progression, and is indispensable for the proper formation of bipolar spindles (Brouhard et al., 2008; Charrasse et al., 1998; Gard & Kirschner, 1987; Nithianantham et al., 2018). It is well-known that partial depletion of ch-TOG reduces MT assembly rates during prometaphase and metaphase (Cassimeris & Morabito, 2004a; Sironi et al., 2011).
We validated the interaction between GPR124 and ch-TOG using three independent approaches. First, ch-TOG co-immunoprecipitated with GPR124-myc expressed in U87MG cells (Figure 3a). Co-immunoprecipitation controls are in Figure S2. Second, endogenous ch-TOG and overexpressed mCherry-tagged GPR124 co-localized on the plasma membrane in HeLa cells undergoing mitosis and in interphase (Figure 3b,c). Ch-TOG and GPR124 also co-localized in filopodial extensions in interphase HeLa cells (Figure 3c, arrows) and it localizes more visibly to the plasma membrane in cells expressing GPR124 than control HeLa cells that are not overexpressing GPR124 (Figure 3d). Third, we investigated the behavior of endogenous ch- TOG-GFP in Hct116 CRISPR cells both in the presence and absence of exogenous GPR124 expression. We hypothesized that MT tipped with ch-TOG interact only transiently with the plasma and that membrane-associated GPR124 may alter the time course of this interaction. To test this, we used live total internal reflection fluorescence (TIRF) microscopy to measure the turnover of ch-TOG-GFP at MTs tips next to the plasma membrane. We observed that ch-TOG-GFP associated MTs that were actively assembling along the membrane did not generally co-localize with GPR124-cherry. In contrast, MT tips labeled with ch-TOG that exhibited co-localization with GPR124- cherry were often transiently stationary (Figure 4a-c, solid circles vs dotted circles). To test the hypothesis that ch-TOG was stabilized at membranes by GPR124, we photobleached the ch-TOG-GFP and recorded its recovery in cells expressing exogenous GPR124 versus those that do not. Using fluorescence recovery after photobleaching (FRAP), we found that ch-TOG fluorescence recovery is remarkably slower in cells expressing highly levels of GPR124 (Figure 4d,e), a result that is consistent with a greater stable proportion of ch-TOG-GFP in the vicinity of plasma membranes. Together, these results suggest that ch-TOG can interact with GPR124 expressed at the plasma membrane.
FIGURE 3.
GPR124 interacts with ch-TOG and these proteins co-localize at the plasma membrane. (a) Ch-TOG co-immunoprecipitates with GPR124-myc. Representative blots from at least three experiments performed with U87MG cells stably expressing GPR124-myc. (b) Tubulin and ch-Tog co-localize in mitotic control and GPR124-KD HeLa cells (HeLa-KD), and GPR124 and ch-Tog co-localize in mitotic HeLa cells overexpressing GPR124 (HeLa-OE). Scale = 10 μm. (c) GPR124 and ch-Tog co-localize in interphase HeLa cells overexpressing GPR124. Scale = 10 μm. Co-localized GPR124 and ch-Tog in filopodial extensions is shown in c’ and c” (Scale = 2 μm). (d) Tubulin and ch-Tog co-localize in interphase control and GPR124-KD (HeLa -KD) cells. Scale = 10 μm. Representative immunocytochemical images of ch-TOG (green), tubulin (red), and GPR124-mCherry (red) are shown. DAPI is shown in blue
FIGURE 4.
Dynamic interactions between GPR124 and ch-TOG. (a) TIRF image of ch-TOG-GFP (green) and GPR124-mCherry (red) at time = 0. Some MT ends appear to co-localize with GPR124 (dotted circles 1 and 2) and some do not (solid circles 3 and 4). (b) A trails overlay of the same cell after 60 s of imaging. MT ends continue assembling when not associated with GPR124 (solid circles, arrow) whereas MT ends that associate with GPR124 remain stationary (dotted circles). (c) Close-up of MT ends in the numbered circles in (a). (d) Example of photobleached and recovered ch-TOG in an area of cell membrane. (e) Comparison of ch-TOG recovery time course and extent in cells expressing (red) or not expressing (green) GPR124. (f) Microtubule plus end tips (green) colocalize with GPR124-mCherry (red) in discreet areas on the plasma membrane (marked by arrows). Scale = 2 μm. (g,h) Representative western blots of GPR124-myc and ch-TOG co-immunprecipitation of U87MG cells stably expressing GPR124-myc treated with either vehicle control (0.1% DMSO, CTR) and vinblastine (10 μM). Whole lysate (Lys) and immuno-precipitated (IP; H). Ch-TOG protein amount was normalized to the amount of GPR124 in each immunoprecipitation and quantified in 3 experiments. *p < .05, Student’s t-test compared to CTR
Analysis of fixed interphase HeLa cells overexpressing GPR124, revealed that GPR124 overlaps with MTs in discreet locations where a MT plus-end contacted the plasma membrane (Figure 4f). To test whether this interaction might involve functional MTs, we treated U87MG-OE cells with the MT-destabilizing drug vinblastine and measured the ch-TOG that co-immunoprecipitated with GPR124-myc. Vinblastine treatment resulted in a 70% reduction in the amount of ch-TOG that co-immunoprecipitated with GPR124-myc, suggesting that this interaction depends on the presence of assembling MTs close the plasma membrane (Figure 4g,h). Collectively, these results suggest the existence of a transient molecular interactions between plasma membrane-associated GPR124, ch-TOG, and proximal MTs.
3.4 |. Changes in GPR124 expression influence MT assembly rates and ch-TOG expression
To test the functional significance of a GPR124-ch-TOG complex on MT function, we first tested whether changes in GPR124 expression affects MT plus-end assembly rates in the mitotic spindle of HeLa cells undergoing mitosis by tracking MT with GFP-tagged EB3 and time lapse microscopy, which was developed based on our previous work (Stumpff, Wagenbach, Franck, Asbury, & Wordeman, 2012; Wordeman, Decarreau, Vicente, & Wagenbach, 2016; Figure S3). Both increased and decreased GPR124 expression significantly enhanced spindle MT assembly rates by 9% (p = 0.01) and 18% (p < 0.001), respectively, over corresponding controls (Figure 5a,b). Next, we tested if changes GPR124 expression affect ch-TOG expression in subcellular compartments using quantitative immunofluorescence analysis. Increased GPR124 expression in HeLa cells resulted in increased ch-TOG expression by 61% in whole mitotic HeLa cells and by 49% in the spindle compartment (Figure 5c,e). By contrast, decreased GPR124 expression did not affect ch-TOG expression in HeLa cells (Figure 5d,f). It is possible that increased GPR124 expression enhances spindle MT assembly by increasing ch-TOG expression. However, the enhanced spindle MT assembly observed when GPR124 expression is decreased is likely to be independent of a change in ch-TOG expression. Indeed, decreased ch-TOG expression in cells is known to reduce MT assembly rates in mitotic spindles (Ertych et al., 2014). There are other pathways that can increase spindle MT assembly rates in addition to the ch-TOG pathway (Stolz, Ertych, & Bastians, 2015) and we are presently investigating which alternate pathway is impacted upon GPR124 depletion. In accordance with GPR124 overexpression, simply increasing ch-TOG expression in HCT116 cells, which have been extensively characterized with regard to MT assembly and express moderate levels of GPR124 (Figure S1; Ertych et al., 2014), enhanced spindle MT assembly by 22% (p = 0.001; Figure S4). Thus, increased GPR124 expression results in increasing ch-TOG expression in the spindle and ensuing increased ch-TOG expression might underlie the enhanced spindle MT assembly rates.
FIGURE 5.
Changes in GPR124 expression differentially affect MT assembly and ch-TOG expression. (a,b) Real-time microscopy measurements of MT assembly rates in the mitotic spindle of HeLa cells overexpressing GPR124 (HeLa-OE) compared with control cells expressing an empty vector (HeLa-vector; a), and of HeLa cells with knocked down GPR124 (HeLa-KD) compared with control cells expressing a scrambled siRNA (HeLa-SCR; b). (c,d) Immunocytochemistry measurements of whole-cell ch-TOG expression in HeLa cells with overexpressed (c) and knocked down (d) GPR124 compared with controls. (e,f) Immunocytochemistry measurements of spindle ch-TOG expression in HeLa cells with overexpressed (e) and knocked down (f) GPR124 compared with controls. All data were collected over three experiments and represented as individual cells. Bars are the mean ± SEM. **p < .01, ***p < .001, Student’s t-test
4 |. DISCUSSION
Our study identified a novel molecular interaction involving GPR124 and ch-TOG residing at the plasma membrane that regulates MT plusend assembly and mitotic progression in cancer cells, including glioblastoma cells. We show that both increases and decreases in GPR124 expression significantly reduce glioblastoma cell proliferation through distinct molecular mechanisms that differentially disrupts mitotic progression and chromosome segregation in these cells. Changes in the expression of several GPCRs have been involved in tumor growth. Two examples include G2A, a GPCR discovered by a cDNA expression library screen for oncogenes (Weng et al., 1998), and GPR171 (also known as H963), which was identified through GENT microarray database analysis (Wittenberger, Schaller, & Hellebrand, 2001). Changes in the expression of either G2A or GPR171 altered cell proliferation and tumor growth (Weng et al., 1998; Wittenberger et al., 2001); however, the molecular mechanism that links changes in the expression of these GPCRs to the modifications of cell proliferation remains unknown. In glioblastomas, the upregulation of GPR56 is thought to contribute to their migration and invasiveness of healthy brain tissue (Shashidhar et al., 2005). Our study provides an additional example whereby changes in GPCR expression, here in both directions by the adhesion receptor GPR124, regulates cell proliferation of glioblastomas and other cancer model systems.
Several reports have suggested the existence of a molecular interaction between plasma membrane-associated GPCRs and MTs that control cell proliferation (Roychowdhury & Rasenick, 2008; Schappi, Krbanjevic, & Rasenick, 2014). For example, activation of sphingosine-1-phosphate receptors 5 (S1PR5) results in accumulation of S1PR5 at centrosomes where it modifies centrosomal function (Gillies et al., 2009). Other examples include metabotropic glutamate receptors and melatonin receptors that directly bind tubulin through their intracellular loops (Ciruela & Mcllhinney, 2001; Ciruela, Robbins, Willis, & Mcllhinney, 1999; Jarzynka et al., 2006; Saugstad, Yang, Pohl, Hall, & Conn, 2002). The molecular interaction that we discovered here involving GPR124 and ch-TOG adds an additional branch to our understanding of molecular network by which GPCRs located at the plasma membrane might affect fundamental MT-dependent functions such as cell proliferation.
4.1 |. GPR124 in the control of MT dynamics, mitosis, and chromosome segregation
A delay in mitotic timing occurs when incorrect MT-kinetochores attachments accumulate and must be resolved during prometaphase and metaphase (Kops & Shah, 2012; McEwen, Heagle, Cassels, Buttle, & Rieder, 1997). Attachments should be strong enough to collect and orient chromosomes properly within the spindle while maintaining an optimal level of MT turnover to allow for error correction (Lampson & Grishchuk, 2017). Too many erroneous attachments or excessively strong attachments can overwhelm the error correction machinery, leading to lagging chromosomes and chromosome instability (Gregan, Polakova, Zhang, Tolić-Nørrelykke, & Cimini, 2011). Accordingly, increased MT assembly rates are also strongly correlated with enhanced CIN (Ertych et al., 2014; Lampson & Grishchuk, 2017). Consistent with these published observations, increased GPR124 expression shortened prometaphase and prolonged metaphase. Moreover, MT assembly rates and the prevalence of lagging chromosomes were increased upon increased GPR124 expression. These findings suggest that changes in GPR124 expression affect glioblastoma cell proliferation by modifying MT dynamics and disturbing mitosis to favor CIN. Our results should also be considered in the context of the anti-tumor activity of many cancer drugs that rely on their ability to tip the balance of cell division from low to high levels of CIN and ensuing cell death. This is particularly relevant considering how intermediate levels of CIN are thought to allow tumorigenesis in the absence of cell death (Birkbak et al., 2011). How the anti-tumor activity of different drugs, including MTA, is affected by changes in GPR124 expression remains to be studied.
4.2 |. GPR124 interacts with ch-TOG, a partner that regulates MT dynamics
Our current understanding of ch-TOG pertains to its role in centrosomal MT nucleation and dynamics (Cassimeris, Becker, & Carney, 2009; Cassimeris & Morabito, 2004a; Gergely, Draviam, & Raff, 2003) and little is known about the function of this enzyme during interphase beyond that it binds to MT plus ends and promotes increased MT dynamics (Brouhard et al., 2008; Charrasse et al., 1998; Gard & Kirschner, 1987). Our study adds a novel facet to our understanding of ch-TOG activity. First, we show that ch-TOG may be localized to the plasma membrane through its interaction with GPR124, as ch-TOG is absent from the cell cortex of HeLa cells except in instances where GPR124 is overexpressed. From these data, we can propose a mechanism in which increased expression of GPR124, a binding partner of ch-TOG, triggers a compensatory increase in ch-TOG expression that enhances MT assembly rates throughout the cell. In agreement with this model, we found increased MT assembly rates are promoted by an increase in GPR124 expression.
GPR124 is associated with ch-TOG at the plasma membrane during interphase and mitosis, suggesting that this recruitment to the plasma membrane may affect the distribution and function of ch-TOG during mitosis. In line with this notion, our finding that treatment with vinblastine leads to a substantial decrease in ch-TOG association with GPR124 suggests that this interaction requires dynamic MTs. This mechanism would support the hypothesis that GPR124 is homeostatic with respect to ch-TOG expression in the cell and regulates its level accordingly to prevent over- or under-expression of this critical mitotic protein in different functional contexts. The expression of mitotic protein regulators is tightly controlled and exogenous disruption of this balance by either enhanced or reduced expression of these regulators has the potential to influence mitosis and promote aberrant proliferation (Bastians, 2015). Because ch-TOG has been implicated in strengthening kinetochore-MT attachment and opposing kinetochore fiber MT turnover necessary for error correction (Cheeseman, Harry, McAinsh, Prior, & Royle, 2013; Miller, Asbury, & Biggins, 2016), this results also provides a likely explanation for the increase in lagging chromosomes during anaphase upon increasing GPR124 expression. We also measured increased ch-TOG protein levels within the spindle and the appearance of lagging chromosomes during anaphase. These molecular responses are all features that have been correlated with hyper-stable but erroneous kinetochore-MT attachments leading to chromosome instability (Ertych et al., 2014; Lampson & Grishchuk, 2017).
Our results suggest that GPR124 interacts with ch-TOG and that changes in the amount of GPR124-ch-TOG complex at the plasma membrane is functionally relevant by affecting ch-TOG levels and ensuing activity in the whole-cell and the spindle. Accordingly, given that GPR124 is a membrane-associated protein that increases MT polymerization rates, an intracellular MT polymerizing enzyme would be expected to facilitate this response. These results are consistent with a model whereby ch-TOG mediates the cellular effects of enhanced GPR124 expression.
4.3 |. GPR124 is involved in proliferation through multiple, competing mechanisms
Our finding that both increased and decreased GPR124 expression result in decreased proliferation of glioblastomas suggests the presence of multiple competing pathways linked to GPR124 that might regulate proliferation. Increasing and decreasing GPR124 expression in glioblastoma cells reduced proliferation, suggesting both anti- and pro-proliferative signals can be mediated by this protein. As no changes in ch-TOG levels occurred upon GPR124 knockdown, it is possible that GPR124 mediates pro-proliferative signaling through a ch-TOG-independent mechanism. The recent finding that GPR124 is an essential cofactor for transduction of Wnt7a signaling could provide an additional mechanism by which GPR124 contributes to proliferative signaling (Noda et al., 2016). The role of WNT7A in the regulation of cancer cell proliferation is still unclear. In cervical cancer and nonsmall cell lung cancer, WNT7A has been found to function as a tumor suppressor (Calvo et al., 2000; Ramos-Solano et al., 2015; Winn et al., 2005). However, high levels of WNT7A have been found to contribute significantly to the progression of ovarian cancer (Yoshioka et al., 2012) and overexpression of WNT7A was significantly associated with poorer survival outcomes in patients with colorectal cancer (Wang et al., 2015). Thus, the activation of WNT7A signaling by GPR124 could affect proliferation in a manner that is highly dependent on cancer cell type. In our study, increased GPR124 expression reduced proliferation of glioblastoma cells in a ch-TOG-dependent mechanism and decreased GPR124 expression reduced proliferation of glioblastoma cells in a mechanism that could depend on its ability to activate WNT7A signaling. The presence of these competing mechanisms adds significant complexity to GPR124 signaling, and the balance between these systems requires future studies.
In summary, we have identified a novel molecular complex involving GPR124 and the MT polymerizing enzyme ch-TOG that affects glioblastoma cell proliferation by modulating mitotic progression and chromosome segregation. These mitotic modifications may underlie fundamental mechanisms of tumor growth such as CIN. Our findings add to our current understanding of the molecular interactions that exist between GPCRs and MT, and how it links changes in the expression of select GPCRs to the control of tumor growth.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Bryce Sopher (Department of Neurosurgery, University of Washington, Seattle, WA) for generating lentiviral constructs and Chizuru Kinoshita (Department of Neurosurgery, University of Washington, Seattle, WA) for generating lentiviral particles for transient overexpression of GPR124; Brian Haas and Yi Hsing Lin (Department of Pharmacology, University of Washington, Seattle, WA) for technical help and generating stable cell lines, Brad St. Croix (Tumor Angiogenesis Section, Mouse Cancer Genetics Program, National Cancer Institute, National Institutes of Health, Frederick, MD) for providing the pcDNA3.1-GPR124-myc expression vector, Greg Foltz and Parvinder Hothi (Swedish Hospital, Seattle, WA) for providing PD-GBM mRNA, and Nicole Berthold and Carl Cotman (Department of Neurobiology and Behavior, University of California Irvine, Irvine, CA) for noncancerous human brain mRNA. Funding sources: This work was supported by National Institutes of Health grants: DA0144861 and NS106924 (N. Stella), GM069429 (L. Wordeman), AR065459 (S.E. Ong), AG000538 and AG051807 (N. Berthold and C. Cotman), NS084217 and NS099929 (R. S. Morrison).
Funding information
National Institute of General Medical Sciences, Grant/Award Number: GM069429; National Institute of Neurological Disorders and Stroke, Grant/Award Number: NS106924; National Institute on Drug Abuse, Grant/Award Number: DA0144861; National Institutes of Health, Grant/Award Numbers: NS099929, NS084217, AG051807, AG000538, AR065459, GM069429, NS106924, DA0144861
Footnotes
DECLARATION OF INTERESTS
A.E.C., J.J.V., K.S., C.X., R.S.M., S.O., N.B., and L.W. have no conflict of interests to disclose. NS is part-time employed by Stella Consulting LLC. and the terms of this arrangement have been reviewed and approved by the University of Washington in accordance its policies governing outside work and financial conflicts of interest in research.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Anderson KD, Pan L, Yang XM, Hughes VC, Walls JR, Dominguez MG, … Wei Y (2011). Angiogenic sprouting into neural tissue requires Gpr124, an orphan G protein-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America, 108, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Shavit R, Maoz M, Kancharla A, Nag JK, Agranovich D, Grisaru-Granovsky S, & Uziely B (2016). G protein-coupled receptors in cancer. International Journal of Molecular Sciences, 17, 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastians H (2015). Causes of chromosomal instability. Recent Results in Cancer Research, 200, 95–113. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, … Trojanowski JQ (2008). Gene expression changes in the course of normal brain aging are sexually dimorphic. Proceedings of the National Academy of Sciences, 105, 15605–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Sabbagh MN, Beach TG, Kim RC, Cribbs DH, & Cotman CW (2014). Brain gene expression patterns differentiate mild cognitive impairment from normal aged and Alzheimer’s disease. Neurobiology of Aging, 35, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, … Swanton C (2011). Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Research, 71, 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, … Hyman AA (2008). XMAP215 is a processive microtubule polymerase. Cell, 132, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, … Brambilla E (2000). Altered HOX and WNT7A expression in human lung cancer. Proceedings of the National Academy of Sciences of the United States of America, 97, 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Becker B, & Carney B (2009). TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motility and the Cytoskeleton, 66, 535–545. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, & Morabito J (2004a). TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Molecular Biology of the Cell, 15, 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, & Larroque C (1998). The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. Journal of Cell Science, 111(Pt 10), 1371–1383. [DOI] [PubMed] [Google Scholar]
- Cheeseman LP, Harry EF, McAinsh AD, Prior IA, & Royle SJ (2013). Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. Journal of Cell Science, 126, 2102–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AE, Haas BR, Naydenov AV, Fung S, Xu C, Swinney K, … Coy J (2016). ST-11: A new brain-penetrant microtubule-destabilizing agent with therapeutic potential for glioblastoma multiforme. Molecular Cancer Therapeutics, 15, 2018–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AE, & Stella N (2014). G protein-coupled receptors as oncogenic signals in glioma: Emerging therapeutic avenues. Neuroscience, 278, 222–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, & Salmon E (2001a). Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. The Journal of Cell Biology, 153, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruela F, & McIlhinney RA (2001). Metabotropic glutamate receptor type 1alpha and tubulin assemble into dynamic interacting complexes. Journal of Neurochemistry, 76, 750–757. [DOI] [PubMed] [Google Scholar]
- Ciruela F, Robbins MJ, Willis AC, & McIlhinney RA (1999). Interactions of the C terminus of metabotropic glutamate receptor type 1alpha with rat brain proteins: Evidence for a direct interaction with tubulin. Journal of Neurochemistry, 72, 346–354. [PubMed] [Google Scholar]
- Cox J, & Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, & Mann M (2011). Andromeda: A peptide search engine integrated into the MaxQuant environment. Journal of Proteome Research, 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Cullen M, Elzarrad MK, Seaman S, Zudaire E, Stevens J, Yang MY, … Hilton MB (2011). GPR124, an orphan G protein-coupled receptor, is required for CNS-specific vascularization and es tablishment of the blood-brain barrier. Proceedings of the National Academy of Sciences of the United States of America, 108, 5759–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsam RT, & Gutkind JS (2007). G-protein-coupled receptors and cancer. Nature Reviews Cancer, 7, 79–94. [DOI] [PubMed] [Google Scholar]
- Ellisdon AM, & Halls ML (2016). Compartmentalization of GPCR signalling controls unique cellular responses. Biochemical Society Transactions, 44, 562–567. [DOI] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuss S, Burfeind P, … Bastians H (2014). Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nature Cell Biology, 16, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fève M, Saliou J-M, Zeniou M, Lennon S, Carapito C, Dong J, … Cianférani S (2014). Comparative expression study of the Endo-G protein coupled receptor (GPCR) repertoire in human Glioblastoma cancer stem-like cells, U87-MG cells and non malignant cells of neural origin unveils new potential therapeutic targets. PLoS One, 9, e91519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, & Schioth HB (2005). The repertoire of G-protein-coupled receptors in fully sequenced genomes. Molecular Pharmacology, 67, 1414–1425. [DOI] [PubMed] [Google Scholar]
- Gao Y, Fan X, Li W, Ping W, Deng Y, & Fu X (2014). miR-138–5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochemical and Biophysical Research Communications, 446, 179–186. [DOI] [PubMed] [Google Scholar]
- Gard DL, & Kirschner MW (1987). A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plusend. The Journal of Cell Biology, 105, 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, & Raff JW (2003). The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes & Development, 17, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies L, Lee SC, Long JS, Ktistakis N, Pyne NJ, & Pyne S (2009). The sphingosine 1-phosphate receptor 5 and sphingosine kinases 1 and 2 are localised in centrosomes: Possible role in regulating cell division. Cellular Signalling, 21, 675–684. [DOI] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang L, Tolić-Nørrelykke IM, & Cimini D (2011). Merotelic kinetochore attachment: Causes and effects. Trends in Cell Biology, 21, 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SM, Rowley SM, Clouston D, Li J, Lupat R, Krishnananthan N, … Campbell IG (2016). Searching for candidate genes in familial BRCAX mutation carriers with prostate cancer. Urologic Oncology: Seminars and Original Investigations, 34, 120.e9–120.e16. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, & Danuser G (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods, 5, 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzynka MJ, Passey DK, Ignatius PF, Melan MA, Radio NM, Jockers R, … Witt-Enderby PA (2006). Modulation of melatonin receptors and G-protein function by microtubules. Journal of Pineal Research, 41, 324–336. [DOI] [PubMed] [Google Scholar]
- Kamato D, Rostam MA, Bernard R, Piva TJ, Mantri N, Guidone D, … Little PJ (2015). The expansion of GPCR transactivation-dependent signalling to include serine/threonine kinase receptors represents a new cell signalling frontier. Cellular and Molecular Life Sciences, 72, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T (2017). A scale of Agonism and allosteric modulation for assessment of selectivity, bias, and receptor mutation. Molecular Pharmacology, 92, 414–424. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Lee H-S, Lee NC, Goncharov NV, Kumeiko V, Masumoto H, … Larionov V (2016). Development of a novel HAC-based “gain of signal” quantitative assay for measuring chromosome instability (CIN) in cancer cells. Oncotarget, 7, 14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, & Shah JV (2012). Connecting up and clearing out: How kinetochore attachment silences the spindle assembly checkpoint. Chromosoma, 121, 509–525. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Mancuso MR, Shamloo A, Wang HT, Choksi V, Florek M, … Heilshorn SC (2010). Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science, 330, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, & Grishchuk EL (2017). Mechanisms to avoid and correct erroneous kinetochore-microtubule attachments. Biology (Basel), 6, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HT, Suh HW, Golkowski M, & Ong SE (2014). Comparing SILAC- and stable isotope dimethyl-labeling approaches for quantitative proteomics. Journal of Proteome Research, 13, 4164–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, An S, Ward R, Yang Y, Guo X-X, Li W, & Xu T-R (2016). G protein-coupled receptors as promising cancer targets. Cancer Letters, 376, 226–239. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, … Elledge SJ (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell, 137, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, & Rieder CL (1997). Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. The Journal of Cell Biology, 137, 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, & Sorger PK (2004). Timing and checkpoints in the regulation of mitotic progression. Developmental Cell, 7, 45–60. [DOI] [PubMed] [Google Scholar]
- Miller MP, Asbury CL, & Biggins S (2016). A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell, 165, 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantham S, Cook BD, Beans M, Guo F, Chang F, & Al-Bassam J (2018). Structural basis of tubulin recruitment and assembly by microtubule polymerases with tumor overexpressed gene (TOG) domain arrays. eLife, 7, e38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Vallon M, & Kuo CJ (2016). The Wnt7’s tale: A story of an orphan who finds her tie to a famous family. Cancer Science, 107, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE (2010). Unbiased identification of protein-bait interactions using biochemical enrichment and quantitative proteomics. Cold Spring Harbor Protocols, 2010, pdb.prot5400. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, & Mann M (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- Ong SE, & Mann M (2006). A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nature Protocols, 1, 2650–2660. [DOI] [PubMed] [Google Scholar]
- Puck TT, & Steffen J (1963). Life cycle analysis of mammalian cells. I. a method for localizing metabolic events within the life cycle, and its application to the action of Colcemide and sublethal doses of X-irradiation. Biophysical Journal, 3, 379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Solano M, Meza-Canales ID, Torres-Reyes LA, Alvarez-Zavala M, Alvarado-Ruiz L, Rincon-Orozco B, … Rosl F (2015). Expression of WNT genes in cervical cancer-derived cells: Implication of WNT7A in cell proliferation and migration. Experimental Cell Research, 335, 39–50. [DOI] [PubMed] [Google Scholar]
- Rao CV, Yamada HY, Yao Y, & Dai W (2009). Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: A perspective from genetic studies in mice. Carcinogenesis, 30, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, & Rasenick MM (2008). Submembraneous microtubule cytoskeleton: Regulation of microtubule assembly by heterotrimeric Gproteins. The FEBS Journal, 275, 4654–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugstad JA, Yang S, Pohl J, Hall RA, & Conn PJ (2002). Interaction between metabotropic glutamate receptor 7 and alpha tubulin. Journal of Neurochemistry, 80, 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schappi JM, Krbanjevic A, & Rasenick MM (2014). Tubulin, actin and heterotrimeric G proteins: Coordination of signaling and structure. Biochimica et Biophysica Acta, 1838, 674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidhar S, Lorente G, Nagavarapu U, Nelson A, Kuo J, Cummins J, … Foehr ED (2005). GPR56 is a GPCR that is over-expressed in gliomas and functions in tumor cell adhesion. Oncogene, 24, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Sironi L, Solon J, Conrad C, Mayer TU, Brunner D, & Ellenberg J (2011). Automatic quantification of microtubule dynamics enables RNAi-screening of new mitotic spindle regulators. Cytoskeleton, 68, 266–278. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, … Vogelstein B (2000). Genes expressed in human tumor endothelium. Science, 289, 1197–1202. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ertych N, & Bastians H (2015). A phenotypic screen identifies microtubule plus end assembly regulators that can function in mitotic spindle orientation. Cell Cycle, 14, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B, … Petersen I (2010). The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nature Cell Biology, 12, 492–499. [DOI] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, & Wordeman L (2012). Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Developmental Cell, 22, 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XL, Wang Y, Li DL, Luo J, & Liu MY (2012). Orphan G protein-coupled receptors (GPCRs): Biological functions and potential drug targets. Acta Pharmacologica Sinica, 33, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon M, Rohde F, Janssen KP, & Essler M (2010). Tumor endothelial marker 5 expression in endothelial cells during capillary morphogenesis is induced by the small GTPase Rac and mediates contact inhibition of cell proliferation. Experimental Cell Research, 316, 412–421. [DOI] [PubMed] [Google Scholar]
- Wacker D, Stevens RC, & Roth BL (2017). How ligands illuminate GPCR molecular pharmacology. Cell, 170, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cho S-G, Wu X, Siwko S, & Liu M (2014). G-protein coupled receptor 124 (GPR124) in endothelial cells regulates vascular endothelial growth factor (VEGF)-induced tumor angiogenesis. Current Molecular Medicine, 14, 543–554. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wei J, Zhang S, Li G, Zhang T, Yu X, … Liu M (2015). Over-expression of Wnt7alpha protein predicts poor survival in patients with colorectal carcinoma. Tumour Biology, 36, 8781–8787. [DOI] [PubMed] [Google Scholar]
- Weng Z, Fluckiger A-C, Nisitani S, Wahl MI, Le LQ, Hunter CA, … Witte ON (1998). A DNA damage and stress inducible G proteincoupled receptor blocks cells in G2/M. Proceedings of the National Academy of Sciences, 95, 12334–12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SV, Platt FM, Hurst CD, Aveyard JS, Taylor CF, Pole J, … Knowles MA (2010). High-resolution analysis of genomic alteration on chromosome arm 8p in urothelial carcinoma. Genes, Chromosomes and Cancer, 49, 642–659. [DOI] [PubMed] [Google Scholar]
- Winn RA, Marek L, Han SY, Rodriguez K, Rodriguez N, Hammond M, … Barry N (2005). Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. The Journal of Biological Chemistry, 280, 19625–19634. [DOI] [PubMed] [Google Scholar]
- Wittenberger T, Schaller HC, & Hellebrand S (2001). An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. Journal of Molecular Biology, 307, 799–813. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Decarreau J, Vicente JJ, & Wagenbach M (2016). Divergent microtubule assembly rates after short-versus long-term loss of end-modulating kinesins. Molecular Biology of the Cell, 27, 1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, & von Dassow G (2007). MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. The Journal of Cell Biology, 179, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Irie K, Asada M, Mino A, Mandai K, & Takai Y (2004). Direct binding of the human homologue of the drosophila disc large tumor suppressor gene to seven-pass transmembrane proteins, tumor endothelial marker 5 (TEM5), and a novel TEM5-like protein. Oncogene, 23, 3889–3897. [DOI] [PubMed] [Google Scholar]
- Yoshioka S, King ML, Ran S, Okuda H, MacLean JA 2nd, McAsey ME, … Hayashi K (2012). WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/beta-catenin pathway. Molecular Cancer Research, 10, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, & Nathans J (2014). Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Developmental Cell, 31, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.