Abstract
Coenzyme A (CoA) regulates energy metabolism and exists in separate pools in the cytosol, peroxisomes, and mitochondria. At the whole tissue level, the concentration of CoA changes with the nutritional state by balancing synthesis and degradation; however, it is currently unclear how individual subcellular CoA pools are regulated. Liver and kidney peroxisomes contain Nudt7 and Nudt19, respectively, enzymes that catalyze CoA degradation. We report that Nudt8 is a novel CoA-degrading enzyme that resides in the mitochondria. Nudt8 has a distinctive preference for manganese ions and exhibits a broader tissue distribution than Nudt7 and Nudt19. The existence of CoA-degrading enzymes in both peroxisomes and mitochondria suggests that degradation may be a key regulatory mechanism for modulating the intracellular CoA pools.
Keywords: Coenzyme A, cell compartmentalization, mitochondria, metabolic regulation, Nudix hydrolase
Introduction
CoA is an indispensable cofactor that activates acyl groups for a multitude of metabolic reactions and the widespread posttranslational acylation of histones and other proteins (1–5). Unacylated CoA (free CoA) has also been found to directly react with cysteine residues of target proteins under conditions of metabolic stress, resulting in protein ‘CoAlation’ (6). CoA-dependent metabolism and post-translational modifications occur in multiple subcellular compartments, including the nucleus and the endoplasmic reticulum, but the cytosol, peroxisomes, and mitochondria contain the major pools of this cofactor (5,7–9). The mitochondria are estimated to contain the highest concentration of CoA (7,8,10), which is required to activate gluconeogenesis and for pathways such as ketogenesis, fatty acid oxidation, and the TCA cycle.
At the whole tissue level, the concentration of CoA changes with the metabolic state by coordinating synthesis and degradation. In the liver, activation of the biosynthetic pathway drives the increase in CoA that occurs in the transition between the fed and fasted state (11,12). Conversely, the net decrease in the concentration of hepatic CoA observed upon refeeding after a fast requires both inhibition of de novo CoA synthesis and degradation of the cofactor accumulated in the fasted state. At the subcellular level, such metabolic adaptations require modulation of the size and composition of the individual CoA pools (7,8). To date, however, our knowledge of the mechanisms regulating these subcellular CoA pools is limited.
In the peroxisomes of liver and kidneys, CoA is hydrolyzed to 3’,5’-ADP and (acyl-)phosphopantetheine by two Nudix hydrolases, Nudt7 and Nudt19, respectively (13–16). Unlike intact CoA, these smaller degradation products can exit the peroxisomes (17); thus, the activity of Nudt7 and Nudt19 likely plays an important role in the regulation of the peroxisomal CoA pool and, in turn, CoA-dependent peroxisomal metabolism (12,17,18). Consistent with such a role, the expression of Nudt7 and the activity of Nudt19 are higher in the fed state compared to the fasted state, and inversely correlate with tissue CoA levels in the liver and kidneys, respectively (13,16,19).
Nudix hydrolases hydrolyze phosphodiester bonds in a variety of substrates including nucleoside di- and triphosphates, dinucleoside polyphosphates, nucleotide sugars and cofactors like CoA and NAD(P)H (20,21). All Nudix hydrolases are characterized by the presence of the Nudix box motif, G[5X]E[7X]REXXEEXGU (where U is a hydrophobic residue and X is any residue), which coordinates a catalytically essential divalent cation (22,23). Nudix hydrolases that specifically hydrolyze CoA contain another conserved motif, (L/M)(L/F)TXR(S/A)[3X](R/K)[3X]G[3X]FPGG (PROSITE accession number PS01293), called the CoA box, which contains residues important for CoA binding and catalysis (13,24). In addition to mammals, peroxisomal Nudix hydrolases specific for CoA are present in yeast and nematodes (24,25). Plants contain cytosolic and mitochondrial isoforms (26); however, no mammalian counterparts have been identified so far.
Nudt8 is an uncharacterized Nudix hydrolase that has been annotated as a mitochondrial enzyme and is closely related to Nudt7 (21,27). In this study, we show that Nudt8 specifically hydrolyzes CoA, is indeed a mitochondrial enzyme, and is broadly expressed in mitochondria-rich organs such as the heart, kidneys, liver, and brown adipose tissue. The characterization of the first mitochondrial CoA-degrading enzyme in mammals provides a potential mechanism for the dynamic regulation of the mitochondrial CoA pool.
Materials and Methods
Materials
Reagents were purchased from the following suppliers: oligonucleotides, Lipofectamine 2000, cell culture reagents, Alexa Fluor 647- and HRP-conjugated goat anti-rabbit IgG, and MitoTracker Orange CMTMRos from Thermo Fisher Scientific; restriction enzymes from New England Biolabs; HEK293 and HEK293T cells from the American Type Culture Collection; Swift membrane stain from G-biosciences. Antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH), citrate synthase, and catalase were purchased from Cell Signaling. The Nudt7 antibody was generated as previously described (19). 3’,5’-ADP and antibodies against ATP-binding cassette, sub-family D, member 3 (PMP70) and the FLAG epitope were obtained from Sigma-Aldrich; the Dylight 488 mouse IgG was purchased from Rockland Immunochemicals. The antibody against the full-length mouse Nudt8 was raised in rabbit and purified by antigen affinity chromatography. The fluorescent CoA derivative of monobromobimane (mBB), mBB-CoA, was synthesized as previously described (13). All other reagents were of analytical grade or better and were purchased from Sigma-Aldrich or Fisher Scientific, unless otherwise stated.
Nudt8 purification and enzymatic assays
Mouse Nudt8 (accession NP_079805.1) was expressed as a C-terminal hexahistine-tagged protein from the pET21-derived plasmid pKM202, a kind gift of Suzanne Jackowski at St. Jude Children’s Research Hospital. Nudt8 was expressed in E.coli BL21 Codon Plus (DE3)-RIPL cultured in auto-inducing medium for 24 h at room temperature (28). The enzyme was purified by Ni-NTA chromatography, followed by dialysis in 20 mM Tris-HCl, pH 8.5, 1 mM EDTA, 1 mM DTT at 4 °C overnight. The combined dialyzed fractions were applied onto a POROS 50 HQ column (Thermo Fisher Scientific). Nudt8 was eluted in two peaks at 160 and 305 mM NaCl in 40 mM diethanolamine hydrochloride, pH 9.0. Only the peak eluted at 160 mM NaCl contained enzymatic activity. Active fractions containing >80% pure Nudt8 were combined and concentrated to >1 mg/ml. Glycerol was added to a final concentration of 50% before storing the protein at −20 °C. For the analysis of Nudt8 by analytical gel filtration, purified recombinant Nudt8 (50 μl) was loaded onto a Superdex 75 10/300 GL column and eluted at 0.3 ml/min with 50 mM Tris-HCl, 150 mm NaCl, 1 mM DTT, 1 mM EDTA, pH 8.5. Protein elution was monitored by measuring the absorbance at 280 nm. The activity of Nudt8 was assayed as previously described (13). The optimized standard assay mixture contained 50 ng of enzyme, 100 mM Tris-HCl pH 8.5, 0.1 mg/ml γ-globulin, 250 µM of free CoA or acyl-CoA substrates and 2 mM MnCl2, in a total volume of 40 µl. Kinetic parameters were determined by varying the substrate concentration from 0.125 to 4 mM. Data points, individually corrected for the background in reaction mixtures lacking the enzyme, were fitted to a ‘Specific Binding with Hill slope’ and ‘kcat’ models using Graphpad Prism 6 (GraphPad Software) to determine the kinetic parameters.
Animal studies
C57BL/6J male mice were purchased from the Jackson Laboratory and allowed to acclimate for at least 7 days. The mice were fed a standard chow diet (Tekland 2018S) and maintained at a room temperature of 22.2 ± 0.2 °C, room humidity of 40% ± 2%, and a 12-h light, 12-h dark cycle, with the dark cycle starting at 6:00 pm. For fasting experiments, the mice were placed on cages with grids for 24 h without food. All studies were approved by the Institutional Animal Care and Use Committees of West Virginia University.
Subcellular fractionation
All centrifugation steps were conducted at 4 °C. To obtain mitochondria-free peroxisomal fractions, we pooled the livers from two mice and adapted the procedure described by Antonenkov, V. D. et. al. (29). Briefly, the combined livers were weighed, minced, and homogenized in 4 volumes of peroxisomal homogenization medium (0.16 M sucrose, 12% PEG1500, 10 mM MOPS pH 7.4, 1 mM EDTA, 1 mM EGTA, 0.1% ethanol, 0.1 mM PMSF, 2 mM DTT) using 5–6 strokes of a motor-driven Teflon pestle at a speed of 550 rpm. The homogenate was centrifuged at 1,500 × g for 12 min, and the supernatant was applied onto a 50% Percoll cushion. After centrifuging at 23,500 × g for 1 h, the peroxisomal fraction was removed from the bottom of the cushion, diluted with 1 ml of homogenization medium, and applied onto an Optiprep density gradient. The gradient was centrifuged at 68,000 × g in a 70.1 Ti fixed angle rotor for 1 h. Fractions (1 ml) were removed starting from the bottom of the Optiprep gradient and stored at −80 °C until analyzed.
Immunoblotting, RT-PCR and immunofluorescence
For western blot analysis, subcellular fractions and tissue homogenates were fractionated by SDS-PAGE on 4–12% Bis-Tris gels and transferred onto PVDF membranes. Flash frozen tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer containing protease inhibitors and centrifuged at 10,000 × g for 10 min at 4°C. To normalize antibody signals to total protein loaded, membranes were stained with Swift membrane stain as per manufacturer’s instructions. Antibodies were used at the following dilutions: Nudt8, Nudt7 and GAPDH antibodies at 1:3,000; citrate synthase, catalase and FLAG antibodies at 1:1,000. Primary antibodies were detected by chemiluminescence with HRP-conjugated goat anti-rabbit or goat anti-mouse IgG at 1:45,000 dilution. Western blot bands were quantified using Genetools (Syngene) or ImageJ analysis software.
RNA was isolated from flash frozen tissue as previously described (13,30). Nudt8 mRNA levels were quantified in triplicate by RT-PCR using the Quantitect SYBR Green RT-PCR kit (Qiagen) and the primers 5’-CAGTTTCCCAGGCGGTAAGT-3’ and 5’-CACGTTGGCAAGTACTGGGA-3’ as the forward and reverse primers, respectively. Primers for Nudt7 and Nudt19 have been published (30). For each tissue, the relative abundance of Nudt7, Nudt8, and Nudt19 mRNA was calculated by the CT method using the averaged CT values for two calibrators, ribosomal protein L22 (Rpl22) and β2-microglobulin (B2m). Primers 5’-GTGCCTTTCTCCAAAAGGTATTT-3’ and 5’-CTCTCTTTGCTGTTGGCGAC-3’ were used as forward and reverse primers, respectively, for Rpl22. Primers 5’-ACTGACCGGCCTGTATGCTA-3’ and 5’-ATGTTCGGCTTCCCATTCTCC-3’ were used as forward and reverse primers, respectively for B2m.
To express Nudt8 in mammalian cells, Nudt8 was amplified from pKM202 using primers introducing an EcoRI restriction site and a Kozak sequence at the 5’ end and a FLAG tag and an XhoI restriction site at the 3’ end. The amplified product was subcloned into pcDNA3.1(+) (Thermo Fisher Scientific) between EcoRI and XhoI sites. HEK293 and HEK293T cells were cultured and transfected as previously described (13). The localization of mouse Nudt8 in HEK293 cells was determined by confocal microscopy, as previously reported (13). The FLAG antibody was used at a 1:1,000 dilution and incubated overnight. Dylight 488 goat anti-mouse IgG was used at a 1:5,000 dilution.
Statistical Analysis
Unless otherwise stated, all data are reported as the mean ± standard error. Statistical significance was calculated by unpaired two-tailed Student’s t test using GraphPad Prism 6 (GraphPad Software).
Results
Nudt8 specifically hydrolyzes CoA species and has a distinctive preference for manganese ions
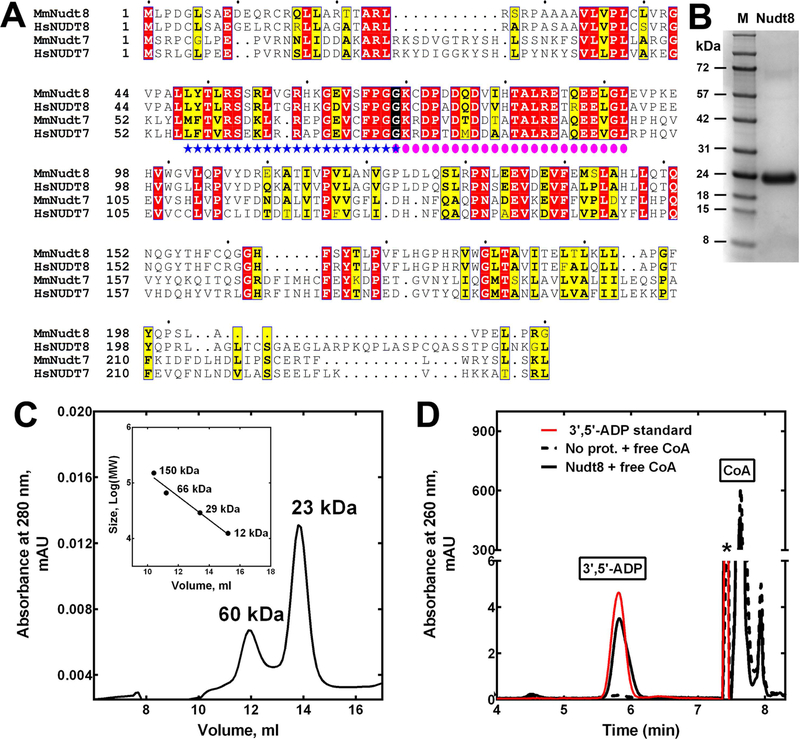
Mouse Nudt8 shares a 38% sequence identity with the CoA-degrading enzyme Nudt7 (Fig 1A). Both proteins contain the CoA and Nudix boxes and the sequence identity in these regions increases to 60%. To test the possibility that Nudt8 could hydrolyze CoA species, we expressed and purified the mouse enzyme as a 24 kDa protein with a C-terminal hexahistidine tag (Fig. 1B). Compared to recombinant Nudt7, Nudt8 was purified in significantly lower amounts. Indeed, one liter of auto-induced culture typically yielded about 15 mg of Nudt8 compared to the 70–100 mg of Nudt7 obtained per liter of standard E. coli culture (13). Interestingly, we also attempted to express human NUDT8; however, similar to a report by Carreras-Puigvert et al., we could not detect any expression (21). Analytical gel filtration analysis of recombinant mouse Nudt8 indicated that similar to Nudt7 (13), about 70% of the protein eluted as a monomer (Fig. 1C).
Fig. 1.
Expression and characterization of mouse Nudt8. (A) Alignment of the protein sequences of mouse and human Nudt8 and Nudt7. Invariant residues are highlighted in red, conserved residues in yellow, and groups of conserved residues are boxed. The Nudix box (blue stars) and the CoA box (pink circles) overlap at a glycine highlighted in black. Mm, Mus musculus; Hs, Homo sapiens; Genbank GI numbers for the aligned proteins are: MmNudt8, 13384950; HsNUDT8, 344217763; MmNudt7, 165972342; HsNUDT7, 157785656. Sequence alignments were obtained using ESPript 3 [44]. AUTHOR: Please check if Figure 1 has been reproduced from Ref. [44].. Please add the statement “Figure reproduced from Ref. [44]” to the end of figure caption if Figure 1 has been reproduced from published sources. Please also confirm if written permission has been obtained from the publisher/author if Figure 1 has been reproduced. Please ignore this query if figure parts described as being modified from other publications are sufficiently different and do not require permission statements for reproduction from the previous publisher/author. Please indicate in the query sheet as "Ignore" if this author query can be ignored(B) Coomassie blue-stained protein gel showing purified Nudt8. M, molecular weight marker. (C) Elution profile of purified, His-tagged Nudt8 from a Superdex 75 10/300 GL column. The inset shows the calibration curve used to estimate the apparent molecular weight of Nudt8. The calculated molecular weight of the recombinant protein, based on the amino acid composition, is 24.3 kDa. (D) High-performance liquid chromatography (HPLC) elution profile of the degradation product co-migrating with a 3′,5′-ADP standard formed in reaction mixtures containing Nudt8 and free CoA. * marks a solvent impurity.
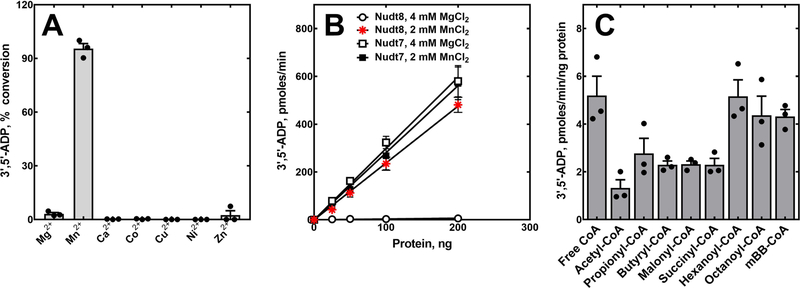
We next tested the hydrolytic activity of Nudt8 in the presence of 4 mM MgCl2 and a broad range of Nudix hydrolase substrates including ATP, ADP, NAD+, NADH, NADP+, NADPH, diadenosine triphosphate, ADP-ribose and free CoA. Product formation was detected only with free CoA as a substrate and co-elution of this product with a 3’,5’-ADP standard confirmed that Nudt8 was a CoA diphosphohydrolase (Fig. 1D). Under the assay conditions used for Nudt7 (13), the specific activity of Nudt8 was about 100-fold lower (Fig. 2B), suggesting that one or more components of the reaction mixture needed to be optimized for Nudt8. Raising the buffer pH from 8.0 to 8.5 modestly increased Nudt8 activity; however, substituting Mg2+ with Mn2+ as the catalytically essential divalent cation in the reaction mixture, increased free CoA hydrolysis from about 2% to over 90% (Fig. 2A). The optimal concentration of MnCl2 was found to be 2 mM (data not shown). Under these optimized conditions, the specific activity of Nudt8 against free CoA was comparable to that of Nudt7 with either MgCl2 or MnCl2 (Fig. 2B). Importantly, we re-screened the diphosphate-containing molecules above as potential Nudt8 substrates in the presence of 2 mM MnCl2 and confirmed that the activity of the enzyme remained specific for free CoA (data not shown).
Fig. 2.
Divalent cation requirement and substrate specificity of recombinant Nudt8. (A) The ability of different divalent cations to support the hydrolytic activity of Nudt8 was tested in reaction mixtures containing 6 μg of Nudt8, 250 μm of free CoA and 0.5 mm of each cation introduced as the chloride salt. (B) Nudt8 and Nudt7 protein curves in the presence of the indicated concentrations of MgCl2 and MnCl2. Linear regression of the curves, obtained using three independently purified Nudt8 batches in duplicate, was used to calculate the specific activity of the enzymes ± standard error. In the presence of the optimal concentration of MnCl2, the specific activity of Nudt8 (2.4 ± 0.1 pmoles·min−1·ng−1 of protein) was comparable to the specific activity of Nudt7 with either MgCl2 (3.0 ± 0.1 pmoles·min−1·ng−1 of protein) or MnCl2 (2.8 ± 0.1 pmles·min−1·ng−1 of protein). (C) CoA diphosphohydrolase activity of Nudt8 against multiple CoA substrates. Data in panels A and C are plotted as the mean (bars) of experiments conducted in duplicate using three independently purified Nudt8 batches (circles) ± SEM.
Having established that Nudt8 was a CoA diphosphohydrolase, we measured its activity against various short- and medium-chain acyl-CoAs. We could not test long-chain acyl-CoAs, such as lauroyl- and stearoyl-CoA, as these compounds precipitated even at the lowest concentration of MnCl2 required to detect Nudt8 activity. Nudt8 hydrolyzed all CoA species tested, exhibiting the highest activity toward free CoA, hexanoyl- and octanoyl-CoA and the lowest activity against acetyl-CoA (Fig. 2C). Nudt8 also readily hydrolyzed the synthetic CoA derivative mBB-CoA. Analysis of Nudt8 kinetics toward representative acyl-CoAs revealed KMs ranging from 150 µM for free CoA to 707 µM for acetyl-CoA (Table 1). The values were generally in the same range of KMs reported for Nudt7 and Nudt19 (14–16). Taking both the kcat and the KM into consideration, the specificity constants calculated for the substrates confirmed that free CoA and acetyl-CoA were the best and worst substrates for Nudt8, respectively (Table 1).
Table 1.
KM, kcat and specificity constant (kcat/KM) for Nudt8 with different CoA substrates. The kinetic parameters KM and kcat were determined, ± SEM, by non-linear regression analysis of data obtained from 3 independent experiments, each conducted in duplicate.
| Substrate | KM, µM | kcat, min−1 | kcat/KM, min−1*µM−1 |
|---|---|---|---|
| Free CoA | 150 ± 13 | 216 ± 8 | 1.44 |
| Acetyl-CoA | 707 ± 252 | 116 ± 10 | 0.16 |
| Propionyl-CoA | 318 ± 87 | 155 ± 14 | 0.49 |
| Butyryl-CoA | 224 ± 40 | 107 ± 9 | 0.48 |
| Malonyl-CoA | 233 ± 71 | 184 ± 19 | 0.79 |
| Succinyl-CoA | 329 ± 72 | 187 ± 14 | 0.57 |
| Hexanoyl-CoA | 251 ± 98 | 149 ± 11 | 0.59 |
Combined, these data indicated that Nudt8 is a novel CoA diphosphohydrolase with broad acyl-CoA substrate specificity and a distinctive preference for Mn2+, a feature not observed in the other known mammalian isoforms, Nudt7 and Nudt19 (14,15).
Nudt8 exhibits a broad tissue distribution and its expression does not change with the nutritional state
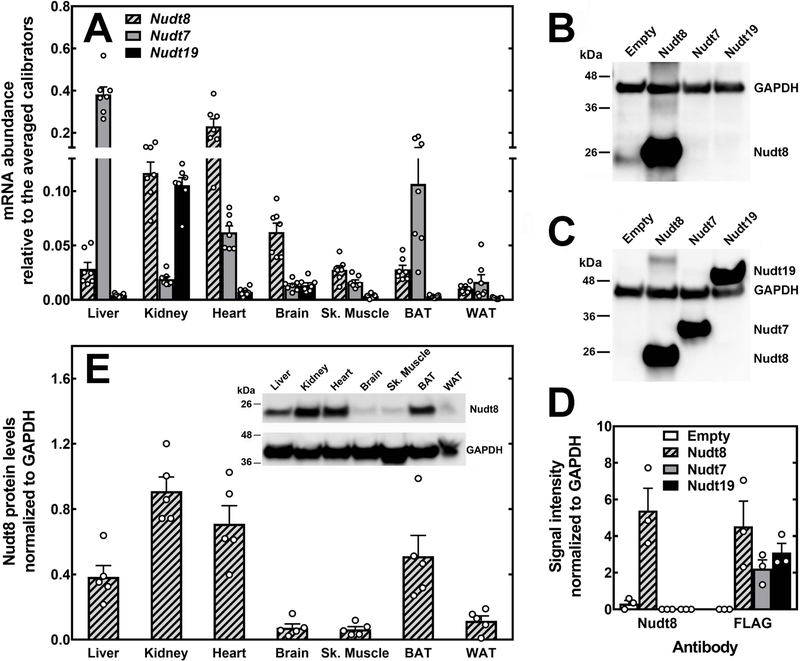
Nudt7 and Nudt19 exhibit a very narrow tissue distribution, predominating in the liver and kidneys, respectively (13,15,16). To determine the tissue distribution and relative transcript abundance of Nudt8 with respect to Nudt7 and Nudt19, we measured the mRNA levels of all three enzymes in different organs. We found that Nudt8 was the major isoform in the heart, brain, and skeletal muscle. Furthermore, Nudt8 mRNA in the kidneys was as abundant as Nudt19 mRNA (Fig. 3A). We next generated a polyclonal antibody to measure Nudt8 protein expression in each tissue. To determine the isoform specificity of the Nudt8 antibody, we analyzed lysates from HEK293T cells transfected with constructs encoding FLAG-tagged versions of Nudt8, Nudt7, and Nudt19, or the empty vector. While the FLAG antibody detected all three enzymes (Fig. 3C), the Nudt8 antibody recognized only Nudt8 (Fig. 3B), indicating isoform specificity. Western blot analysis of different tissues showed that Nudt8 was expressed at the highest levels in the kidneys, heart, brown adipose tissue, and liver (Fig. 3E). Detectable, but significantly lower levels of the enzyme were also found in the brain, skeletal muscle, and white adipose tissue. Combined, these data indicated that Nudt8 had a much broader tissue distribution than Nudt7 and Nudt19.
Fig. 3.
Relative abundance and tissue distribution of Nudt8 mRNA and protein. (A) Relative abundance of mouse Nudt8, Nudt7, and Nudt19 transcripts in selected tissues. Data are reported as the mean (bars) of measurements on individual mice (circles) ± SEM. (B–D) HEK293T cells were transfected with either an empty plasmid or plasmids carrying the coding sequences for the FLAG-tagged versions of Nudt8, Nudt7, and Nudt19. Representative western blots showing that (B) the Nudt8 antibody did not cross-react with Nudt7 or Nudt19, while (C) the FLAG antibody detected all isoforms. (D) Quantification of the Nudt8 and FLAG signals from three independent transfections (circles) ± SEM. (E) Levels of Nudt8 protein in mouse tissues as quantified by western blot analysis. Data are reported as the mean (bars) of measurements of the Nudt8 signal in tissues from individual mice (circles) ± SEM. Inset, representative western blot. GAPDH was used to normalize the Nudt8 and FLAG signals in (B) and (C) and to normalize the tissue Nudt8 signal in (E).
Nudt7 is regulated by the nutritional state and its mRNA and protein levels in the liver decrease with fasting (16,19). Conversely, the expression of Nudt19 in the kidneys remains constant between the fed and fasted states, despite the contribution of Nudt19 to the regulation of the concentration of CoA in the fed state (13). We quantified Nudt8 mRNA and protein levels in the liver, kidneys and heart of mice fed ad libitum or fasted for 24 h. Similar to Nudt19, but unlike the closely related Nudt7, Nudt8 expression did not change with the nutritional state in any of the tissue analyzed (Fig. 4), suggesting that regulation might occur through a different mechanism.
Fig. 4.
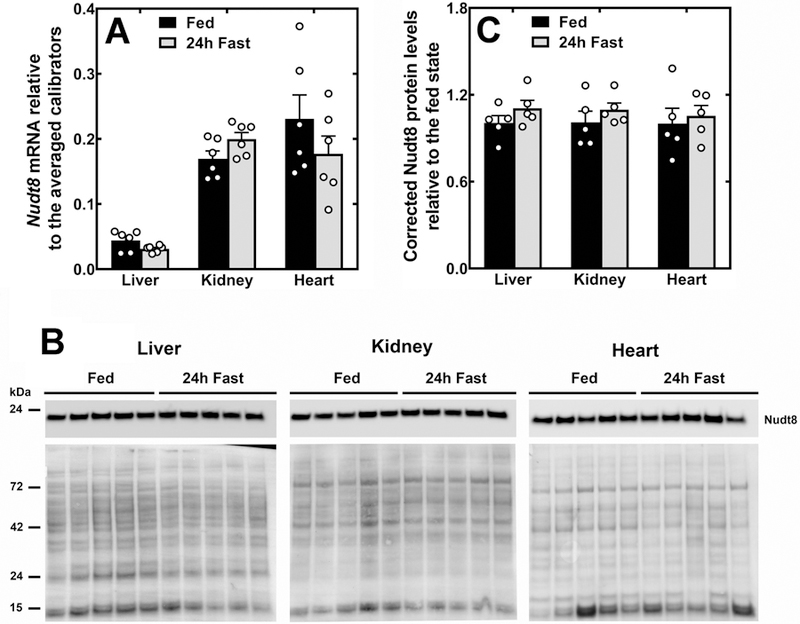
Nudt8 expression in the liver, kidneys and heart of fed and fasted mice. (A) Nudt8 mRNA in the liver, kidneys and heart of mice fed ad libitum or fasted for 24 h. (B) Western blot analysis showing that Nudt8 protein levels do not change with the nutritional state. Total protein loading is shown under each immunoblot. (C) Quantification of the western blots in (B) normalizing to total protein loading and expressing the corrected intensity relative to the fed state. Data in (A) and (C) are shown as the mean (bars) of measurements on individual mice (circles) ± SEM.
Mitochondrial localization of Nudt8
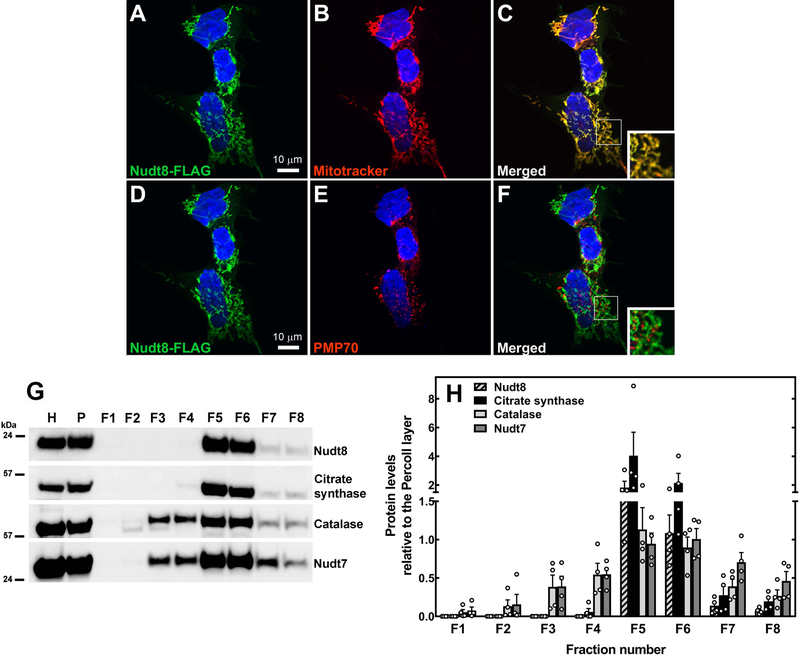
Many eukaryotic CoA diphosphohydrolases, including Nudt7 and Nudt19, localize to the peroxisomes. Nudt8 does not contain a peroxisomal targeting signal; instead, its N-terminus contains a putative mitochondrial targeting sequence within the first 24 residues, as predicted by both TargetP (31) and MitoFates (32). Furthermore, Nudt8 is annotated as a mitochondrial protein in the Mitocarta 2.0 database, an inventory of mouse and human proteins whose mitochondrial localization is supported by a combination of computational and proteomics data (27). To verify the subcellular localization of Nudt8 in intact cells, HEK293 cells were transiently transfected with a construct expressing Nudt8 with a C-terminal FLAG tag. Using the FLAG tag antibody, we found that Nudt8 co-localized with the mitochondrial marker MitoTracker Orange CMTMRos, but not with the peroxisomal marker PMP70 (Fig. 5A–F). To corroborate this conclusion in mouse tissue, we fractionated liver homogenates with a protocol that achieved some degree of separation between peroxisomes and mitochondria (Fig. 5G–H). While endogenous Nudt8 was found in mitochondria-enriched fractions (Fig. 5G–H, fractions F5-F6), exhibiting the same fractionation profile as the mitochondrial marker citrate synthase, this enzyme was not detected in fractions containing purified peroxisomes (Fig. 5G–H, fractions F2-F4), as marked by both catalase and Nudt7. Combined, these results supported the conclusion that Nudt8 localized exclusively to the mitochondria.
Fig. 5.
Mitochondrial localization of Nudt8. HEK293 cells were transfected with plasmids encoding Nudt8 with a C-terminal FLAG tag, fixed, and analyzed by confocal microscopy. (A, C, D, F) Nudt8, visualized with the FLAG antibody, is in green. (B-C) Mitochondria, visualized using Mitotracker Orange CMTMRos, are in red. (E-F) Endogenous PMP70, used as a peroxisomal marker, is in red. Cell nuclei were stained with DAPI, in blue. Insets in merged images show details at higher magnification. The results are representative of at least two independent experiments. (G-H) Subcellular fractionation of mouse livers. Nudt8 exhibited a sedimentation profile similar to the mitochondrial marker citrate synthase. Catalase and Nudt7 were used as peroxisomal markers. The results are representative of four independent experiments quantified in (H) relative to the input in the Percoll layer, P. H, liver homogenate; F1–8, fractions 1–8.AUTHOR: Figure captions are extracted from the PDF source file. Please check if this is okay
Discussion
Until now, only liver and kidney peroxisomes were known to contain enzymes capable of degrading CoA (14,15). The major finding of this work is that Nudt8 is a novel CoA diphosphohydrolase that localizes to the mitochondria and has a broad tissue distribution. Thus, our data strongly suggest that regulation of CoA levels by degradation occurs in more organs than previously thought and in both peroxisomes and mitochondria.
Recombinant mouse Nudt8 readily hydrolyzed free CoA and a variety of short-chain acyl-CoAs but exhibited a relatively low activity against acetyl-CoA (Fig. 2C and Table 1). Additionally, this enzyme showed a distinctive preference for Mn2+ ions that sets it apart from the peroxisomal isoforms Nudt7 and Nudt19, which can use Mg2+ or Mn2+ (Fig. 2B) (14,15). Given some of the properties of Mn2+, this strong metal preference could potentially play a role in the substrate specificity of Nudt8 and, in particular, in the low binding affinity that the enzyme showed towards acetyl-CoA compared to the closely related Nudt7 (16). Mn2+-ligand bonds are more flexible than the equivalent Mg2+-ligand bonds both in length and angle. Furthermore, Mn2+ binds nitrogen-containing ligands (e.g. histidines) more frequently than Mg2+ and can form bidentate complexes with aspartate and glutamate residues (33,34). In the case of Nudix hydrolases, the latter property may be particularly relevant given that the Nudix box motif G[5X]E[7X]REXXEEXGU contains three invariant glutamates (underscored) that play a key role in the coordination of the catalytically essential metal ion. While this motif is highly conserved between Nudt7 and Nudt8 (Fig. 1A), the binding of Mn2+ may impart a unique geometry to the Nudt8 active site and affect substrate binding. Crystal structures of Nudt7 and Nudt8 with substrate and preferred metal bound will be required to determine whether the presence of Mn2+ can explain how Nudt8 better discriminates between acetyl-CoA and other CoA substrates compared to Nudt7. This substrate specificity, in turn, may be relevant to the physiological function of Nudt8. Under our assay conditions, the optimal MnCl2 concentration for Nudt8 was 2 mM, which is within the physiological range of the estimated Mn2+ concentration inside the mitochondria (35,36). Interestingly, significant activation by Mn2+ has also been observed for other mitochondrial Nudix hydrolases including mouse Nudt13, an enzyme that degrades NADP(H), and AtNudt15, a CoA diphosphohydrolase in A. thaliana that, however, was not tested against acetyl-CoA (37,38). Importantly, the existence of Nudt8 and its CoA-degrading activity may explain how, even in the absence of Nudt19 and any detectable levels of Nudt7, the concentration of CoA could still be significantly decreased in the kidneys of Nudt19−/− mice during the transition from the fasted to the fed state (13).
Nudt8 was highly expressed in all the mitochondria-rich organs analyzed except the skeletal muscle (Fig. 3A). This is not unusual as, even among organs with similar mitochondrial densities, the concentration of mitochondrial proteins can vary substantially depending on the different functional emphasis of the mitochondria within each organ (39,40). Interestingly, the lower Nudt8 protein levels correlated with the lower concentration of CoA in skeletal muscle compared to the liver, heart, kidneys and brown adipose tissue (41), suggesting that Nudt8 expression in this tissue may be proportionally adjusted to the concentration of the cofactor to avoid depleting it. At the subcellular level, mitochondria contain the highest concentration of cellular CoA (7,8) and are the site of several CoA-dependent metabolic processes, including glucose and fatty acid oxidation. CoA is present in both the mitochondrial matrix and the intermembrane space. While the localization of Nudt8 places it in a position to modulate mitochondrial metabolism, the precise physiological role of this enzyme will depend on the submitochondrial CoA pool that it can actually access. Using independent spatially restricted enzyme tagging-based approaches, two studies have recently identified human NUDT8 as a resident of the inner membrane/matrix in HEK293T cells (42,43), which suggests that Nudt8 may regulate the matrix CoA pool. Such localization would also potentially place Nudt8 in the same compartment as CoA synthase (Coasy), the last enzyme in the CoA biosynthetic pathway (44–46). Coasy catalyzes the formation of dephospho-CoA from ATP and phosphopantetheine and the final phosphorylation of dephospho-CoA to CoA. Coasy has been localized to both the outer mitochondrial membrane (46) and the inner membrane/matrix (42). In the absence of regulatory mechanisms, the latter subcellular localization would allow Coasy to re-synthesize CoA from the phosphopantetheine produced by Nudt8, generating wasteful cycles of CoA synthesis and degradation. Our data indicate that in the kidneys, heart and liver, organs with high expression of Nudt8 and detectable CoA-degrading activity (13,47,48), Nudt8 levels did not change with the nutritional state. Additional studies will be required to determine whether other potential regulatory mechanisms, including posttranslational modifications or regulation by metabolites in the mitochondria, may control the activity of Nudt8 and whether Coasy and Nudt8 actually localize to the same sub-mitochondrial compartment.
In conclusion, the identification and characterization of Nudt8 as a mitochondrial CoA-degrading enzyme indicates that CoA degradation is not limited to the peroxisomes and may provide a common mechanism to regulate the subcellular pools of CoA in response to changes in the metabolic state.
Acknowledgements:
We thank Deborah Corbin, Mason Colbert, Dominique Saporito, Paolo Fagone in the Biochemistry Protein Core Facility, and the Imaging Facilities at West Virginia University for their expert technical assistance. This work was supported by West Virginia University’s School of Medicine startup foundation funding (RL), and National Institutes of Health Grants R35GM119528 (RL), F31DK118878 (EWK), and F31GM126838 (SAS). The West Virginia University Imaging Facilities were supported by the West Virginia University Cancer Institute and NIH grants P20RR016440, P30RR032138, P30GM103488 and U54GM104942.
Abbreviations:
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PMP70
ATP-binding cassette, sub-family D, member 3
- mBB
monobromobimane
References
- 1.Choudhary C, Weinert BT, Nishida Y, Verdin E, and Mann M (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nature reviews. Molecular cell biology 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 2.Hirschey MD, and Zhao Y (2015) Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Molecular & cellular proteomics : MCP 14, 2308–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resh MD (2016) Fatty acylation of proteins: The long and the short of it. Progress in lipid research 63, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniotti JL, Pedro MP, and Valdez Taubas J. (2017) The role of S-acylation in protein trafficking. Traffic [DOI] [PubMed]
- 5.Sabari BR, Zhang D, Allis CD, and Zhao Y (2017) Metabolic regulation of gene expression through histone acylations. Nature reviews. Molecular cell biology 18, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gout I (2018) Coenzyme A, protein CoAlation and redox regulation in mammalian cells. Biochemical Society transactions 46, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horie S, Ishii H, and Suga T (1981) Changes in peroxisomal fatty acid oxidation in the diabetic rat liver. Journal of biochemistry 90, 1691–1696 [DOI] [PubMed] [Google Scholar]
- 8.Van Broekhoven A, Peeters MC, Debeer LJ, and Mannaerts GP (1981) Subcellular distribution of coenzyme A: evidence for a separate coenzyme A pool in peroxisomes. Biochemical and biophysical research communications 100, 305–312 [DOI] [PubMed] [Google Scholar]
- 9.Peng Y, and Puglielli L (2016) N-lysine acetylation in the lumen of the endoplasmic reticulum: A way to regulate autophagy and maintain protein homeostasis in the secretory pathway. Autophagy 12, 1051–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Idell-Wenger JA, Grotyohann LW, and Neely JR (1978) Coenzyme A and carnitine distribution in normal and ischemic hearts. The Journal of biological chemistry 253, 4310–4318 [PubMed] [Google Scholar]
- 11.Leonardi R, Rehg JE, Rock CO, and Jackowski S (2010) Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PloS one 5, e11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shumar SA, Kerr EW, Fagone P, Infante AM, and Leonardi R (2019) Overexpression of Nudt7 decreases bile acid levels and peroxisomal fatty acid oxidation in the liver. Journal of lipid research [DOI] [PMC free article] [PubMed]
- 13.Shumar SA, Kerr EW, Geldenhuys WJ, Montgomery GE, Fagone P, Thirawatananond P, Saavedra H, Gabelli SB, and Leonardi R (2018) Nudt19 is a renal CoA diphosphohydrolase with biochemical and regulatory properties that are distinct from the hepatic Nudt7 isoform. The Journal of biological chemistry 293, 4134–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasmi L, and McLennan AG (2001) The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. The Biochemical journal 357, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ofman R, Speijer D, Leen R, and Wanders RJ (2006) Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. The Biochemical journal 393, 537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reilly SJ, Tillander V, Ofman R, Alexson SE, and Hunt MC (2008) The nudix hydrolase 7 is an Acyl-CoA diphosphatase involved in regulating peroxisomal coenzyme A homeostasis. Journal of biochemistry 144, 655–663 [DOI] [PubMed] [Google Scholar]
- 17.Antonenkov VD, and Hiltunen JK (2006) Peroxisomal membrane permeability and solute transfer. Biochimica et biophysica acta 1763, 1697–1706 [DOI] [PubMed] [Google Scholar]
- 18.Hunt MC, Tillander V, and Alexson SE (2014) Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie 98, 45–55 [DOI] [PubMed] [Google Scholar]
- 19.Leonardi R, Rock CO, and Jackowski S (2014) Pank1 deletion in leptin-deficient mice reduces hyperglycaemia and hyperinsulinaemia and modifies global metabolism without affecting insulin resistance. Diabetologia 57, 1466–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bessman MJ, Frick DN, and O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. The Journal of biological chemistry 271, 25059–25062 [DOI] [PubMed] [Google Scholar]
- 21.Carreras-Puigvert J, Zitnik M, Jemth AS, Carter M, Unterlass JE, Hallstrom B, Loseva O, Karem Z, Calderon-Montano JM, Lindskog C, Edqvist PH, Matuszewski DJ, Ait Blal H., Berntsson RPA, Haggblad M, Martens U, Studham M, Lundgren B, Wahlby C, Sonnhammer ELL, Lundberg E, Stenmark P, Zupan B, and Helleday T (2017) A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nature communications 8, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLennan AG (2006) The Nudix hydrolase superfamily. Cellular and molecular life sciences : CMLS 63, 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Pena AH, Suarez A, Duong-Ly KC, Schoeffield AJ, Pizarro-Dupuy MA, Zarr M, Pineiro SA, Amzel LM, and Gabelli SB (2015) Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus. PloS one 10, e0141716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartwright JL, Gasmi L, Spiller DG, and McLennan AG (2000) The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its derivatives. The Journal of biological chemistry 275, 32925–32930 [DOI] [PubMed] [Google Scholar]
- 25.AbdelRaheim SR, and McLennan AG (2002) The Caenorhabditis elegans Y87G2A.14 Nudix hydrolase is a peroxisomal coenzyme A diphosphatase. BMC biochemistry 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito D, Yoshimura K, Ishikawa K, Ogawa T, Maruta T, and Shigeoka S (2012) A comparative analysis of the molecular characteristics of the Arabidopsis CoA pyrophosphohydrolases AtNUDX11, 15, and 15a. Bioscience, biotechnology, and biochemistry 76, 139–147 [DOI] [PubMed] [Google Scholar]
- 27.Calvo SE, Clauser KR, and Mootha VK (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic acids research 44, D1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 29.Antonenkov VD, Sormunen RT, and Hiltunen JK (2004) The behavior of peroxisomes in vitro: mammalian peroxisomes are osmotically sensitive particles. American journal of physiology. Cell physiology 287, C1623–1635 [DOI] [PubMed] [Google Scholar]
- 30.Garcia M, Leonardi R, Zhang YM, Rehg JE, and Jackowski S (2012) Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PloS one 7, e40871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emanuelsson O, Brunak S, von Heijne G, and Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 32.Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, and Imai K (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Molecular & cellular proteomics : MCP 14, 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bock CW, Katz AK, Markham GD, and Glusker JP (1999) Manganese as a Replacement for Magnesium and Zinc: Functional Comparison of the Divalent Ions. Journal of the American Chemical Society 121, 7360–7372 [Google Scholar]
- 34.Kehres DG, and Maguire ME (2003) Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev 27, 263–290 [DOI] [PubMed] [Google Scholar]
- 35.Gunter TE, Miller LM, Gavin CE, Eliseev R, Salter J, Buntinas L, Alexandrov A, Hammond S, and Gunter KK (2004) Determination of the oxidation states of manganese in brain, liver, and heart mitochondria. Journal of neurochemistry 88, 266–280 [DOI] [PubMed] [Google Scholar]
- 36.Gunter RE, Puskin JS, and Russell PR (1975) Quantitative magnetic resonance studies of manganese uptake by mitochondria. Biophys J 15, 319–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelraheim SR, Spiller DG, and McLennan AG (2017) Mouse Nudt13 is a Mitochondrial Nudix Hydrolase with NAD(P)H Pyrophosphohydrolase Activity. The protein journal 36, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, Tanabe N, and Shigeoka S (2008) Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant physiology 148, 1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forner F, Foster LJ, Campanaro S, Valle G, and Mann M (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Molecular & cellular proteomics : MCP 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 40.Johnson DT, Harris RA, French S, Blair PV, You J, Bemis KG, Wang M, and Balaban RS (2007) Tissue heterogeneity of the mammalian mitochondrial proteome. American journal of physiology. Cell physiology 292, C689–697 [DOI] [PubMed] [Google Scholar]
- 41.Dansie LE, Reeves S, Miller K, Zano SP, Frank M, Pate C, Wang J, and Jackowski S (2014) Physiological roles of the pantothenate kinases. Biochemical Society transactions 42, 1033–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, and Ting AY (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Salokas K, Tamene F, Jiu Y, Weldatsadik RG, Ohman T, and Varjosalo M (2018) An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nature communications 9, 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aghajanian S, and Worrall DM (2002) Identification and characterization of the gene encoding the human phosphopantetheine adenylyltransferase and dephospho-CoA kinase bifunctional enzyme (CoA synthase). The Biochemical journal 365, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, de Crecy-Lagard V, and Osterman A (2002) Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. The Journal of biological chemistry 277, 21431–21439 [DOI] [PubMed] [Google Scholar]
- 46.Zhyvoloup A, Nemazanyy I, Panasyuk G, Valovka T, Fenton T, Rebholz H, Wang ML, Foxon R, Lyzogubov V, Usenko V, Kyyamova R, Gorbenko O, Matsuka G, Filonenko V, and Gout IT (2003) Subcellular localization and regulation of coenzyme A synthase. The Journal of biological chemistry 278, 50316–50321 [DOI] [PubMed] [Google Scholar]
- 47.Lopaschuk GD, and Neely JR (1987) Coenzyme A degradation in the heart: effects of diabetes and insulin. Journal of molecular and cellular cardiology 19, 281–288 [DOI] [PubMed] [Google Scholar]
- 48.Lopaschuk GD, and Neely JR (1987) Stimulation of myocardial coenzyme A degradation by fatty acids. The American journal of physiology 253, H41–46 [DOI] [PubMed] [Google Scholar]
- 49.Robert X, and Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic acids research 42, W320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]