Abstract
Astrocytes are critical for the development and function of the central nervous system. In developing brains, immature astrocytes undergo morphological, molecular, cellular, and functional changes as they mature. Although the mechanisms that regulate the maturation of other major cell types in the central nervous system such as neurons and oligodendrocytes have been extensively studied, little is known about the cellular and molecular mechanisms that control astrocyte maturation. Here, we identified molecular markers of astrocyte maturation and established an in vitro assay for studying the mechanisms of astrocyte maturation. Maturing astrocytes in vitro exhibit similar molecular changes and represent multiple molecular subtypes of astrocytes found in vivo. Using this system, we found that astrocyte‐to‐astrocyte contact strongly promotes astrocyte maturation. In addition, secreted signals from microglia, oligodendrocyte precursor cells, or endothelial cells affect a small subset of astrocyte genes but do not consistently change astrocyte maturation. To identify molecular mechanisms underlying astrocyte maturation, we treated maturing astrocytes with molecules that affect the function of tumor‐associated genes. We found that a positive feedback loop of heparin‐binding epidermal growth factor‐like growth factor (HBEGF) and epidermal growth factor receptor (EGFR) signaling regulates astrocytes maturation. Furthermore, HBEGF, EGFR, and tumor protein 53 (TP53) affect the expression of genes important for cilium development, the circadian clock, and synapse function. These results revealed cellular and molecular mechanisms underlying astrocytes maturation with implications for the understanding of glioblastoma.
Keywords: astrocyte, maturation, cell–cell interaction, growth factor
1. INTRODUCTION
Astrocytes constitute about 40% of all cells in the human brain. Long thought to be passive support cells, astrocytes have been recently shown to be critical for the development and function of the central nervous system. Astrocytes promote neuron survival (Banker, 1980), stimulate synapse formation and function (Allen, 2014; Allen et al., 2012; Allen & Eroglu, 2017; Chung, Allen, & Eroglu, 2015; Eroglu et al., 2009; Farhy‐Tselnicker et al., 2017; Khakh & McCarthy, 2015; Kucukdereli et al., 2011; Molofsky et al., 2014; Pfrieger & Barres, 1997; Singh et al., 2016; Stogsdill et al., 2017; Ullian, Sapperstein, Christopherson, & Barres, 2001), and engulf extra synapses formed during development (Chung et al., 2013; Vainchtein et al., 2018). Furthermore, astrocytes are important for neural transmitter recycling (Maragakis & Rothstein, 2004; Rothstein et al., 1994), ion and water homeostasis, synaptic transmission, synaptic plasticity (Haydon & Nedergaard, 2015; Nedergaard, 1994), the integrity of the blood brain barrier, and the regulation of blood flow (Takano et al., 2006). Astrocytes are involved in a range of neurological disorders, including neurodevelopmental disorders (Ballas, Lioy, Grunseich, & Mandel, 2009; Cheng, Lau, & Doering, 2016; Jacobs, Nathwani, & Doering, 2010; Lioy et al., 2011), epilepsy (Carmignoto & Haydon, 2012; Coulter & Steinhauser, 2015; Ding et al., 2007; Tian et al., 2005), neurodegeneration (Benraiss et al., 2016; Itagaki, McGeer, Akiyama, Zhu, & Selkoe, 1989; Tong et al., 2014; Yamanaka et al., 2008), traumatic brain injury (Anderson et al., 2016; Bush et al., 1999; Herrmann et al., 2008), and stroke (Gallo & Deneen, 2014; Molofsky et al., 2012). Astrocyte dysfunction contributes to disease progression in some diseases (Liddelow et al., 2017; Yamanaka et al., 2008) whereas astrocytes facilitate neural protection and repair in others (Anderson et al., 2016; Bush et al., 1999; Herrmann et al., 2008). Astrocytes are a key cell type in understanding the health and disease of the central nervous system.
Similar to two other major cell types of the central nervous system, neurons and oligodendrocytes, astrocytes are derived from embryonic radial glial cells (Molofsky & Deneen, 2015; Schmechel & Rakic, 1979). After an early neurogenic phase (around embryonic day nine to shortly after birth in the mouse cerebral cortex), radial glial cells undergo a “gliogenic switch” and begin to generate immature astrocytes (shortly before birth until 2 weeks after birth in the mouse cerebral cortex) (Deneen et al., 2006; Freeman, 2010; Molofsky & Deneen, 2015). Immature astrocytes migrate away from ventricular zones to populate cortical layers where they undergo additional rounds of proliferation before exiting the cell cycle (Ge, Miyawaki, Gage, Jan, & Jan, 2012; Tien et al., 2012). Immature and mature astrocytes differ in their morphology (Bushong, Martone, & Ellisman, 2004), gene expression (Chaboub et al., 2016; Molofsky et al., 2013; Zhang et al., 2016), function (Geisert & Stewart, 1991; Rudge & Silver, 1990; Smith, Rutishauser, Silver, & Miller, 1990), and injury responses (Smith, Miller, & Silver, 1986). Migrating immature astrocytes typically have one or two short processes and appear unipolar or bipolar. Mature astrocytes, in contrast, have hundreds of peripheral processes contacting synapses and blood vessels, and exhibit bushy morphology (Bushong et al., 2002, 2004). Our group and others recently showed that as astrocytes mature, they undergo transcriptome‐wide changes in their gene expression (Chaboub et al., 2016; Molofsky et al., 2013; Zhang, Sloan, et al., 2016). Maturing astrocytes also undergo corresponding changes in function. Immature astrocytes strongly promote neurite outgrowth whereas mature astrocytes have limited capacity in supporting neurite outgrowth (Geisert & Stewart, 1991; Rudge & Silver, 1990; Smith et al., 1990). In response to traumatic injury, mature astrocytes can deposit laminin and form glial scars that seal off injury sites, whereas immature astrocytes have limited scar‐forming propensities (Smith et al., 1986). The broad range of morphological, cellular, molecular, and functional transformations mentioned above occur in a relatively short period of time. In mice, astrocyte maturation occurs during the first 3–4 postnatal weeks (Bushong et al., 2004; Ge et al., 2012; Nixdorf‐Bergweiler, Albrecht, & Heinemann, 1994) and in humans the first postnatal year (Zhang, Sloan, et al., 2016). Previous work found roles for thyroid hormone (Manzano, Bernal, & Morte, 2007), bone morphogenetic proteins (Scholze, Foo, Mulinyawe, & Barres, 2014), neuroligin‐2 (Stogsdill et al., 2017), and neuronal activity (Morel, Higashimori, Tolman, & Yang, 2014) in regulating astrocyte maturation. Compared to the wealth of information on neuron and oligodendrocyte maturation, the signals and mechanisms that control the timing of astrocyte maturation remain poorly understood. Studying the mechanisms that regulate astrocyte maturation is important not only for understanding brain development, but also for the investigation of neurological disorders. For example, glioblastoma is the most common type of primary malignant brain tumor in adults. Glioblastoma cells share similar molecular markers with immature astrocytes (Brennan et al., 2013; Verhaak et al., 2010; Zhang, Sloan, et al., 2016). Glioblastoma cells and immature astrocytes, but not mature astrocytes, are proliferative and migratory. Therefore, understanding the mechanisms that control astrocyte maturation may shed light on the treatment of glioblastoma.
The mechanisms underlying the maturation of neurons and oligodendrocytes have been extensively investigated. Studies have identified transcriptional regulation, hormonal signals, and intrinsic clock mechanisms that control the maturation of neurons and oligodendrocytes (Barres, Lazar, & Raff, 1994; Batista‐Brito & Fishell, 2009; Dugas, Ibrahim, & Barres, 2007; Emery et al., 2009). Studies of astrocyte development have mostly focused on the gliogenic switch and the specification of astrocyte cell fate. For example, Notch signaling, LIF, and CNTF family secreted factors, the transcription factors STAT3, Sox9, NFIA/B, and SCL are all important in the specification of astrocytes (Bonni et al., 1997; Deneen et al., 2006; Freeman, 2010; Glasgow et al., 2017; Kang et al., 2012; Muroyama, Fujiwara, Orkin, & Rowitch, 2005; Tiwari et al., 2018; Zhang et al., 2016). What signal(s) triggers the next step of astrocyte development, that is, the transition from lineage‐committed immature astrocytes to mature astrocytes, remains elusive. Are the signals that induce maturation derived from astrocytes themselves or other cell types? Do cell autonomous, noncell autonomous mechanisms, or both regulate astrocyte maturation?
To study astrocyte maturation, a robust and sensitive in vitro maturation assay is useful. The most commonly used astrocyte culture method involves the selection and expansion of astrocytes in a serum‐containing medium (McCarthy & de Vellis, 1980). In healthy brains, however, serum components do not contact astrocytes because of the presence of the blood brain barrier. It is only during barrier breakdown in injury and disease do serum components contact astrocytes and induce reactive astrogliosis, which includes a spectrum of cellular, molecular and functional changes (Sofroniew, 2014). Indeed, transcriptome profiling studies showed that serum‐selected astrocytes exhibited characteristics of reactive astrocytes found in injured and diseased brains (Foo et al., 2011; Zamanian et al., 2012). Reactive astrocytes in adult brains can assume properties of immature astrocytes and re‐enter the cell cycle (Sofroniew, 2014). Serum‐selected astrocytes remain proliferative for weeks over multiple passages, resembling immature astrocytes (McCarthy & de Vellis, 1980). Furthermore, serum‐selected astrocytes exhibit polygonal fibroblast‐like morphology, unlike the process‐bearing morphology of astrocytes in vivo. Moreover, serum is not chemically defined and its composition varies between vendors and across batches.
To overcome these challenges, we first established a serum‐free astrocyte culture system that allows in vitro maturation. Using this system, we identified molecular markers for astrocyte maturation. Although glia fibrillary acidic protein (GFAP) has been a very useful astrocyte marker in previous astrocyte development studies, using GFAP alone is not sufficient to characterize astrocyte maturation as all astrocytes do not express GFAP and the reactivity states of astrocytes strongly affect GFAP expression. We discovered a group of molecular markers for astrocyte maturation and established an in vitro assay of astrocyte maturation using serum‐free cultures of immunopanning‐purified murine astrocytes. We found that astrocyte‐to‐astrocyte contact powerfully stimulated astrocyte maturation, whereas signals from other glia and vascular cell types did not have similar effects. Furthermore, we found that HBEGF and EGFR inhibit, and TP53 promotes astrocyte maturation. We showed that astrocytes exhibit a positive feedback loop in HBEGF‐EGFR signaling and that HEBGF, EGFR, and TP53 regulate genes involved in cilium formation, circadian clock, and the regulation of synapse formation and function. These results show that both cell–cell interactions and cell autonomous signals control astrocyte maturation.
2. MATERIALS AND METHODS
2.1. Vertebrate animals
All animal experimental procedures were approved by the Chancellor's Animal Research Committee at the University of California, Los Angeles and conducted in compliance with national and state laws and policies. We used rat cells for in vitro maturation characterization, astrocyte‐microglia, astrocyte‐oligodendrocyte precursor cell (OPC) and astrocyte‐endothelial cell coculture experiments because of the higher yield and consistency of rat astrocyte, microglia, OPC, and endothelial cell cultures compared to corresponding mouse cell cultures. We used mouse cells for clustered regularly interspaced short palindromic repeats (CRISPR) genome editing and fluorescence‐activated cell sorting (FACS) experiment because of the availability of CRISPR single guide RNA (sgRNA) design information for the mouse genes. We used mouse cells for all RNA‐sequencing (RNA‐seq) experiments.
2.2. Purification and culturing of rat astrocytes
We purified immature rat astrocytes according to a previously published immunopanning protocol with modifications (Foo et al., 2011). Briefly, we coated three 150 mm diameter petri dishes first with species‐specific secondary antibodies (Zhang, Sloan, et al., 2016) and then with an antibody against CD45 (BD 550539), a hybridoma supernatant against the O4 antigen (Zhang, Sloan, et al., 2016), and an antibody against Itgb5 (eBioscience 14‐0497‐82), respectively. We dissected cerebral cortices of postnatal day two rat pups and dissociated cortices into a single cell suspension by papain digestion. We then depleted microglia/macrophages and OPCs from the single cell suspension by incubating the suspension sequentially on the CD45 and O4‐coated petri dishes. We then incubated the single cell suspension on the Itgb5‐coated petri dish. After washing away nonadherent cells, we lifted astrocytes bound to the Itgb5‐coated dish using trypsin and plated them on poly‐d‐lysine coated plastic coverslips in a serum‐free medium containing Dulbecco's modified Eagle's medium (DMEM) (LifeTechnologies 11960069), Neurobasal (LifeTechnologies 21103049), sodium pyruvate (LifeTechnologies 11360070), SATO (Foo et al., 2011), glutamine (LifeTechnologies 25030081), N‐acetyl cysteine (Sigma A8199) and HBEGF (Sigma E4643) on 24‐well culture plates. We replaced half of the medium with fresh medium every 2–3 days. To test whether the astrocyte‐derived signal(s) that promotes astrocyte maturation is contact‐dependent or secreted, we plated 20 × 103 astrocytes on coverslips in each well as described above and an additional 180 × 103 astrocytes on a 1 μm‐diameter pore size insert (Corning, 08‐771‐9).
2.3. Purification and culture of mouse astrocytes
To purify immature mouse astrocytes, we coated three petri dishes first with species‐specific secondary antibodies (Zhang, Sloan, et al., 2016) and then with an antibody against CD45, a hybridoma supernatant against the O4 antigen, and an antibody against HepaCAM (R&D Systems, MAB4108), respectively. We dissected cerebral cortices of postnatal Day 2 mouse pups and performed immunopanning purification as described above, except for using the HepaCAM‐coated dishes for selecting mouse astrocytes.
2.4. Collection of astrocyte‐conditioned medium
We purified astrocytes from P8 rat pups and cultured them as described above. We collected medium from the 3–5 days in vitro (DIV) astrocyte cultures, spun them at 500 g × 3 min to discard cell debris, and concentrated the conditioned medium to ~10 mg protein per ml using the Pierce™ Protein Concentrator (Thermos Scientific, 88525). To test the effect of astrocyte‐conditioned medium (ACM) on astrocyte maturation, we added 75 μg/mL of conditioned medium to cultured rat astrocytes and determined maturation marker gene expression by qPCR after 7 days of culture. We chose this concentration of conditioned medium because previous studies showed that reactive ACM at as low as 50 μg/mL had strong neurotoxic effects (Liddelow et al., 2017).
2.5. Immunocytochemistry
We fixed and permeablized astrocytes with 4% paraformaldehyde and 0.2% Triton‐X100 in phosphate buffered saline (PBS). After blocking in 10% donkey serum, we stained astrocytes with the following primary antibodies: anti‐GFAP (1:1,500. Biolegend, 829401), Ki67 (1:200. Invitrogen MA5‐14520), enhanced green fluorescent protein (EGFP) (1:1,000. Aves Labs GFP‐1020), mCherry (1:600. Clontech 632543), Sox9 (1:2,000. Millipore AB5535), Bromodesoxyuridine (BrdU) (1:500, Novusbio NB500‐169), and fluorescent secondary antibodies (Invitrogen). For BrdU staining, cells were pretreated by 2N hydrochloric acid for 20 min before blocking. After three washes in PBS, we mounted the coverslips with VectorShield with DAPI (Vector Labs H1200) and imaged them using an Evos FL Auto 2 inverted fluorescence microscope (Invitrogen) with ×10, and ×20 lenses and the Imager M2 upright fluorescence microscope (Zeiss) with ×10, ×20, and ×40 lenses. We cropped the images with the FIJI and Photoshop softwares.
2.6. Identification of astrocyte maturation markers by RNA‐seq
We previously purified human astrocytes, neurons, microglia, oligodendrocytes, and endothelial cells and performed RNA‐seq transcriptome profiling (Zhang et al., 2014; Zhang, Sloan, et al., 2016). Our human astrocyte samples included immature astrocytes harvested from gestation week 18 fetal brains and mature astrocytes from patients eight to 63 years of age. For each gene, we calculated the ratio of its expression at the mature stage (averaged from all 12 patients eight to 63 years of age) and the immature stage (averaged from six patients at gestation week 18 fetal stage). We chose genes with the highest ratios as mature astrocyte markers and the lowest ratios as immature astrocyte markers.
2.7. Purification of microglia and co‐culturing of microglia and astrocytes
We used an immunopanning purification method for rat microglia based on a published protocol that minimized microglia activation (Bohlen et al., 2017; Bohlen, Bennett, & Bennett, 2018). Briefly, we first anesthetized postnatal Day 6 rat pups with ketamine and xylazine. We then perfused them with Dulbecco's PBS to flush out the blood to minimize leukocyte contamination. The purification procedures were performed at 4°C as quickly as possible to minimize microglia activation. We used whole brain without olfactory bulb and cerebellum and dissociated the tissue using a dounce homogenizer (Fisher Scientific 06‐435A). After centrifugation and resuspension, we incubated cells on an anti‐CD11b antibody (Biolegend 2018‐01)‐coated petri dish, washed away nonadherent cells, and lifted bound microglia by TrypLE digestion (ThermoFisher 12605010). We then plated the microglia and the astrocytes purified by immunopanning described above separated by a one‐micrometer pore size porous insert in a serum‐free coculture medium containing the astrocyte growth medium described above supplemented with sodium selenite (100 ng/mL. Sigma), insulin (5 μg/mL. Sigma), cholesterol (1.5 μg/mL. Avanti Polar Lipids), CSF‐1 (10 ng/mL. Peprotech) and TGF‐β2 (2 ng/mL. Peprotech), which kept both cell types healthy in culture for at least a week. For the BrdU incorporation assay, we added 15 μg/mL BrdU in the medium 6 days after the start of coculturing and fixed cells 24 hr afterward.
2.8. Purification of OPCs and co‐culturing of OPCs and astrocytes
We purified OPCs from cerebral cortices of postnatal Day 6 rats by immunopanning. We coated four petri dishes first with secondary antibodies (Zhang, Sloan, et al., 2016) and then with the following primary antibodies: an antibody against CD45 to deplete microglia and macrophages, an antibody against HepaCAM to deplete astrocytes (Zhang, Sloan, et al., 2016), and a hybridoma supernatant against the O4 antigen to bind OPCs. We dissociated cortices by papain digestion (Foo et al., 2011) and we sequentially incubated the single cell suspension on the CD45 and HepaCAM‐coated petri dishes to deplete contaminating cell types, and then incubated the suspension on the O4‐coated petri dishes to bind OPCs. After washing off nonadherent cells, we lifted the OPCs by trypsin digestion and plated the OPCs and the astrocytes separated by a one‐micrometer pore size porous insert in a serum‐free coculture medium containing the astrocyte growth medium described above supplemented with d‐biotin (10 ng/mL. Sigma), Trace element B (×1. Cellgro 99‐175‐CI), insulin (5 μg/mL), Cilliary neurotrophic factor (10 ng/mL. Peprotech), Platelet‐derived growth factor (20 ng/mL. Peprotech), neurotrophin 3 (1 ng/mL. Peprotech), and Forskolin (4.2 ng/mL. Sigma), which kept both cell types healthy in culture for at least a week.
2.9. Purification of endothelial cells and co‐culturing of endothelial cells and astrocytes
We purified endothelial cells from the cerebral cortices of postnatal day 14 rats based on a published protocol (Zhou, Sohet, & Daneman, 2014). Briefly, we digested cortices with papain, depleted microglia and macrophages by immunopanning using anti‐CD45 antibody‐coated petri dishes, and selected endothelial cells by immunopanning using anti‐CD31 antibody (BD 550300) coated petri dishes. We plated the endothelial cells and the astrocytes separated by a one‐micrometer pore size porous insert in a serum‐free coculture medium containing DMEM, 20% fetal calf serum (Invitrogen), fibroblast growth factor (10 ng/mL. Peprotech) and puromycin (3 μg/mL. Sigma), which kept both cell types healthy in culture for at least a week.
2.10. Quantitative real time PCR (qRT‐PCR)
We purified mRNA using the PureLink RNA mini kit (Invitrogen 12183018A) using a protocol provided by the manufacturer. We then generated cDNA using the SuperScript III First‐Strand Synthesis SuperMix (Invitrogen 18080400). We designed primers spanning exon‐exon junctions to avoid amplification of genomic DNA. We used the PowerUp SYBR Green Master Mix (Applied Biosystems A25742) and ran qRT‐PCR reactions on a Lightcycler 96 thermocycler (Roche). We determined the CTs of each gene's by qRT‐PCR in duplicates or triplicates. When determining fold changes in gene expression, the CTs of each gene were normalized to the CTs of the housekeeping gene Gapdh from the same sample.
2.11. Generation of lentiviral constructs and lentivirus packaging
We cloned the human GFAP promotor into a third generation lentivirus backbone. We inserted the CRISPR‐associated protein 9 (Cas9) coding sequence and EGFP coding sequence connected in frame by the T2A peptides downstream of the human GFAP promotor. In a second construct, we inserted sgRNAs targeting GFAP, Sox9, EGFR, and TP53 genes, the P2A peptide, and the coding sequence for mCherry downstream of the human GFAP promotor. To package lentiviruses, we transfected low passage number (<11) human embryonic kidney 293 cells (ATCC CRL3216) with the third generation lentivirus packaging mix containing pVSV‐G, pMDL, pRSV, and the DNA constructs described above using polyethylenimine (Polysciences 23966‐1). We harvested the supernatant over 72 hr after transfection and then concentrated lentiviruses solutions 100 times using the LentiX concentrator (Clontech 631232).
2.12. CRISPR genome editing in cultured mouse astrocytes
We added 1–20 μL of 100× concentrated lentiviruses encoding cas9‐EGFP and sgRNA‐mCherry to each well of mouse astrocytes at 2 div. We changed the medium 72 hr after infection. We analyzed cells infected with both the cas9‐EGFP and sgRNA‐mCherry viruses 7–21 days after infection.
2.13. FACS
We analyzed cultured mouse astrocytes by FACS at 7, 14, and 21 days after infection. We lifted astrocytes by trypsin digestion and stopped trypsin digestion with an ovomucoid solution (Zhang, Sloan, et al., 2016). We then spun down astrocytes and resuspended them in a solution containing 50% neurobasal, 50% DMEM, 0.5% glucose, and 5 mM EDTA. We analyzed endogenous fluorescence of Cas9‐EGFP and sgRNA‐mCherry lentiviruses infected astrocytes with a BD LSRII analyzer. We analyzed noninfected samples as negative controls. We also analyzed samples infected by a single virus (Cas9‐EGFP or sgRNA‐mCherry) to calculate the compensation for spectral overlap. We analyzed the FACS data with the Flowjo software.
2.14. RNA‐seq
We harvested astrocytes purified from P2 mouse cerebral cortex and cultured in serum‐free conditions for 2, 7, and 14 days for RNA‐seq. To inhibit EGFR signaling, we added 0.05 μM of the EGFR inhibitor PD168393 at 2 div and harvested cells at 3 div. To inhibit P53, we added 5 μM of the P53 inhibitor Pifithrin‐α at 2 div and harvested cells at 4 div. We used 2–3 biological replicates per condition. We purified total RNA using the miRNeasy Mini kit (Qiagen Cat# 217004) and analyzed RNA concentration and integrity with TapeStation (Agilent) and Qubit. All samples have RNA integrity numbers higher than 7. We then generated cDNA using the Nugen Ovation V2 kit (Nugen), fragmented cDNAs using the Covaris sonicator, and generated sequencing libraries using the Next Ultra RNA Library Prep kit (New England Biolabs) with 10 cycles of PCR amplification. We sequenced the libraries with the Illumina HiSeq 4,000 sequencer and obtained 12.9 ± 2.8 million (mean ± standard deviation [SD]) 50 bp single‐end reads per sample.
2.15. RNA‐seq data analysis
We mapped sequencing reads to mouse genome MM10 using the STAR package (Dobin et al., 2013) and HTSEQ (Anders, Pyl, & Huber, 2015) to obtain raw counts. We then used the EdgeR‐Limma‐Voom packages in R to obtain Reads per Kilobase per Million Mapped Reads (RPKM) values and calculated differential gene expression. To perform Gene Ontology analysis, we uploaded gene lists from each condition to the DAVID bioinformatics Resource (Huang, Sherman, & Lempicki, 2009) and used the GOTERM_BP_DIRECT option.
2.16. Data deposition
We deposited all RNA sequencing data (2019) to the Gene Expression Omnibus under accession number GSE125610. To review the dataset, go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125610.
2.17. Comparison of astrocyte maturation marker gene expression in vivo and in vitro
To compare astrocyte maturation marker gene expression in vivo and in vitro, we downloaded E13.5 and E17.5 astrocyte gene expression data as determined by microarray from (Molofsky et al., 2013). We performed background subtraction and quantile normalization using the RMA method with the Affy package in R. We also download normalized E14.5 and P4 astrocyte gene expression data from (Chaboub et al., 2016). We determined differential gene expression using the Limma package in R. We compared 2 div versus 14 div in vitro astrocyte gene expression from our study using the Limma package in R.
2.18. Heterochronic cocultures
We cultured astrocytes under three conditions: (a) younger astrocytes alone: we purified astrocytes from P2 rat cerebral cortex and plated them at 30 thousand per well density; (b) younger + younger astrocytes: we purified astrocytes from P2 rat cerebral cortices and plated them at 100,000 per well density; and (c) younger + older astrocytes. We first purified astrocytes from P7 rat cerebral cortex and plated them in multiple wells at varying densities. We then infected them with lentiviruses encoding EGFP described above at 2 div. We titrated the amount of lentiviruses and used the volume that resulted in >98% infection rate as determined by fluorescent imaging at 6 div. We then plated younger astrocytes purified from P2 rats 7 days after culturing P7 astrocytes. After 7 days of coculture, we fixed astrocytes and stained them with antibodies against Ki67 and EGFP. We counted total cell density based on DAPI labeling. We only used wells with similar total cell densities under conditions b and c for final analyses because we found that astrocyte density has a robust effect on astrocyte maturation.
2.19. Determination of astrocyte, OPC, and microglia densities in vivo
We use in situ hybridization images of the astrocyte marker Glul, the OPC marker Pdgfra, and the microglia marker Tmem119 from sagittal sections of P14 mouse from the Allen Brain Atlas (Lein et al., 2007). Three images from matching medial‐lateral positions were used for each gene. We selected cerebral cortex as regions of interest, used ImageJ to set the same threshold for all images, and used the Analyze Particle function with minimum particle size of 10 pixels to count cell numbers.
2.20. Analysis of astrocyte subpopulation markers
We used the list of subpopulation A–E specific marker genes (False discovery rate < 0.1) from (John Lin et al., 2017) and plotted the average expression of all specific marker genes for each subpopulation in each of our samples.
2.21. Comparison of the transcriptome profiles of astrocytes in vitro with glioblastoma subtypes
We downloaded the expression data of the core 840 genes that distinguished between proneural, neural, classical, and mesenchymal subtypes of glioblastoma from 173 glioblastoma samples from the TCGA dataset (Verhaak et al., 2010). We centered the data before comparing the TCGA dataset with our data since the transcriptome profiles were obtained from different platforms. To center the TCGA data, we calculated median and SD across all TCGA samples for each gene. Then we centered the expression of each gene in each sample using the following formula: centered data = (raw expression – medium)/SD. We performed the same procedures for our own data. We then calculated the Pearson correlation between every of our samples and every of the TCGA samples.
3. RESULTS
3.1. Establishment of an in vitro assay for astrocyte maturation
To establish an in vitro assay for astrocyte maturation, we avoided serum‐selection, which keeps astrocytes immature and proliferative, and modified an immunopanning purification method (Foo et al., 2011) to purify immature astrocytes and culture them in a chemically‐defined serum‐free medium. We purified neonatal rat astrocytes and observed that immunopanned rat astrocytes in serum‐free media exhibited process‐bearing morphology and gradually increased branch complexity and arbor size over a couple of weeks in vitro (Figure 1a). Although initially proliferative, rat astrocytes in serum‐free culture gradually reduced proliferation rates over a 2‐week period (Figure 1a,b). The timeline parallels the observation that astrocyte proliferation slows down and eventually stops over the first 2 weeks after birth in vivo (Ge et al., 2012; Nixdorf‐Bergweiler et al., 1994).
Figure 1.
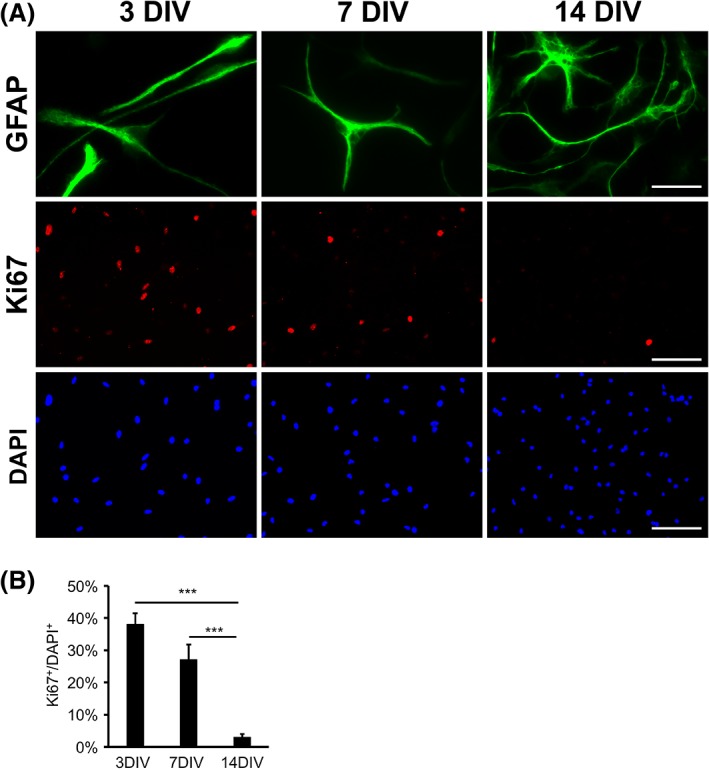
Process outgrowth and proliferation of immunopanned rat astrocytes in serum‐free culture. (a) Morphological maturation and the decrease of proliferation of immunopanned rat astrocytes in serum‐free culture. Scale bars: 25 μm in the GFAP images and 100 μm in the Ki67 and DAPI images. (b) Quantification of the percentage of astrocytes positive for the proliferation marker Ki67. Error bars represent the standard error of the means in all figures. *p < .05. **p < .01. ***p < .001. N.S., not significant in all figures. Two‐tailed t‐test was used in all figures except for the RNA‐seq data analyses [Color figure can be viewed at wileyonlinelibrary.com]
To characterize the molecular changes of astrocyte maturation, we analyzed an RNA‐seq dataset that we previously generated (Zhang, Sloan, et al., 2016). We constructed the dataset by the acute purification of human astrocytes from patients at a variety of ages, ranging from 18 week gestation to 63 years. We compared the gene expression of fetal and adult human astrocytes and identified markers of immature and mature astrocytes, respectively (Figure 2a). To examine the expression of immature and mature astrocyte markers by rat astrocytes in serum‐free culture, we performed qRT‐PCR and found that the expression of immature markers decreased and mature markers increased in cultured rat astrocytes in serum‐free media (Figure 2b).
Figure 2.
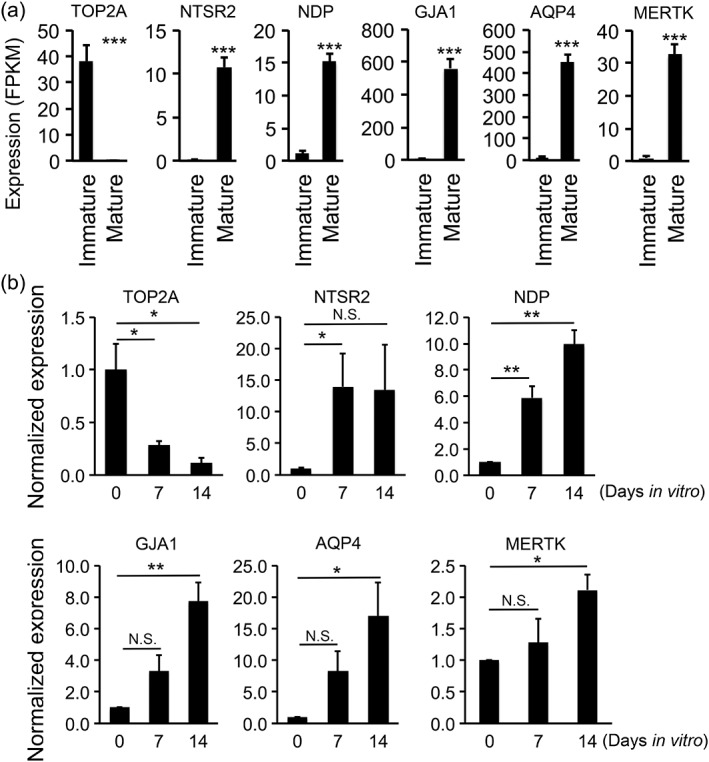
Immunopanned rat astrocytes gradually mature in serum‐free media. (a) Identification of astrocyte maturation markers by RNA‐seq. We performed RNA‐seq of acutely purified human astrocytes at the immature (18‐week gestation) and mature (8–63 years old) stages (Zhang, Sloan, et al., 2016) and identified six of the top differentially expressed genes at the immature and mature stages. FPKM, fragment per kilobase of transcripts per million mapped reads. (b) Immunopanned rat astrocytes gradually matured in serum‐free media. We performed qRT‐PCR to determine the expression of maturation marker genes by rat astrocytes grown in serum‐free media at 0, 7, and 14 div. The expression level was first normalized to the house‐keeping gene Gapdh and then normalized to the expression at 0 div
To systematically characterize the molecular changes of astrocyte maturation in vitro at the transcriptome level, we performed RNA‐seq of mouse astrocytes at 2, 7, and 14 div. We found that gene expression changes as astrocytes mature in vitro mirrors those observed during astrocyte maturation in vivo based on human and mouse astrocyte transcriptomes we recently characterized (Zhang et al., 2014; Zhang, Sloan, et al., 2016; Tables 1 and 2). In addition, we compared our data with astrocyte maturation datasets published by other groups (Chaboub et al., 2016; Molofsky et al., 2013). These spinal cord developing astrocyte microarray datasets identified mature and immature astrocyte marker genes. We compared the in vivo datasets and our in vitro data and found that the majority of immature and mature astrocyte marker genes exhibit similar temporal changes in our in vitro astrocyte maturation dataset and in vivo datasets published by others (Figure 3). It is not surprising that all genes did not show consistent temporal profiles because these studies used spinal cord tissue and the microarray method whereas our study used cerebral cortex tissue and the RNA‐seq method.
Table 1.
Differentially expressed genes at 7 div compared to 2 diva
| Gene | Log fold change | FDR |
|---|---|---|
| Up‐regulated genes | ||
| Timp3 | 2.28 | 0.00012175 |
| 6330403K07Rik | 2.47 | 0.00013227 |
| Gpm6a | 1.63 | 0.00013312 |
| Ttyh1 | 2.28 | 0.00014983 |
| Slc6a11 | 1.69 | 0.00015643 |
| Apoe | 3.41 | 0.00015643 |
| Tril | 1.89 | 0.00015643 |
| Draxin | 2.06 | 0.00015643 |
| Peli2 | 1.43 | 0.00015643 |
| Asrgl1 | 1.56 | 0.00015643 |
| Sez6 | 2.45 | 0.00015643 |
| Bmp3 | 3.91 | 0.00020206 |
| Sox8 | 1.30 | 0.00025483 |
| Spon1 | 1.98 | 0.00027613 |
| Plekhb1 | 1.85 | 0.00027613 |
| Smoc1 | 1.92 | 0.00030392 |
| Adcyap1r1 | 1.55 | 0.00031074 |
| Pcdh11x | 2.06 | 0.00031074 |
| Lrrc4c | 2.36 | 0.00031074 |
| Itgb8 | 1.26 | 0.00031784 |
| Down‐regulated genes | ||
| Dgkk | −2.37 | 0.00012175 |
| Tbc1d1 | −1.58 | 0.00015643 |
| Slc6a6 | −2.63 | 0.00015643 |
| Lrrc8b | −1.36 | 0.00024285 |
| F2rl2 | −3.32 | 0.0002579 |
| Arhgap18 | −1.32 | 0.00027613 |
| Ptgfrn | −1.62 | 0.00027613 |
| Fam135b | −4.41 | 0.00031784 |
| Fbn2 | −1.31 | 0.00032356 |
| Sh3pxd2b | −1.43 | 0.00032356 |
| Vav3 | −1.19 | 0.0003566 |
| Nhsl2 | −2.98 | 0.00040225 |
| Ltbp1 | −2.97 | 0.00049772 |
| Tubb6 | −1.80 | 0.00053476 |
| Mest | −1.12 | 0.00053476 |
| Ntng1 | −1.78 | 0.00053476 |
| Fam167a | −2.68 | 0.00053476 |
| Sfrp1 | −1.56 | 0.00053476 |
| Cacng5 | −2.02 | 0.00053476 |
| Angpt2 | −1.79 | 0.00060389 |
We ranked genes by false discovery rate (FDR) and listed top 20 up‐ and down‐regulated genes. Genes with counts per million (CPM) less than three in at least three out of all six samples were not listed in all tables. Fold change is shown as the logarithm to the base two in all tables.
Table 2.
Differentially expressed genes at 14 div compared to 2 diva
| Gene | Log fold change | FDR |
|---|---|---|
| Up‐regulated genes | ||
| Tril | 2.56 | 3.71E−06 |
| Cp | 3.12 | 1.32E−05 |
| Wnt5a | 3.06 | 1.34E−05 |
| Spon1 | 2.76 | 2.00E−05 |
| Apoe | 4.58 | 2.48E−05 |
| Mlc1 | 2.06 | 2.72E−05 |
| 6330403K07Rik | 3.23 | 2.72E−05 |
| Timp3 | 2.68 | 3.22E−05 |
| Cacna2d3 | 4.01 | 3.35E−05 |
| Cryab | 3.59 | 3.60E−05 |
| Galnt10 | 2.88 | 3.99E−05 |
| Tmem47 | 2.00 | 4.13E−05 |
| Gpm6a | 1.74 | 4.22E−05 |
| Svep1 | 3.91 | 4.36E−05 |
| Pcdh11x | 2.19 | 4.36E−05 |
| Plcd4 | 2.14 | 4.61E−05 |
| Serinc5 | 1.79 | 4.69E−05 |
| Wls | 2.47 | 4.91E−05 |
| Ryr3 | 3.75 | 5.29E−05 |
| Itih5 | 3.03 | 5.47E−05 |
| Down‐regulated genes | ||
| Slc6a6 | −3.15 | 3.71E−06 |
| Fam135b | −5.34 | 3.71E−06 |
| Map1b | −3.29 | 4.88E−06 |
| Dgkk | −2.84 | 2.30E−05 |
| Sfrp1 | −2.16 | 2.48E−05 |
| Pfkp | −2.23 | 2.72E−05 |
| Sh3pxd2b | −1.64 | 3.22E−05 |
| Ltbp1 | −3.67 | 3.35E−05 |
| Ntng1 | −2.82 | 3.99E−05 |
| Tagln2 | −2.59 | 4.36E−05 |
| Coro2b | −1.66 | 4.69E−05 |
| Uaca | −1.73 | 4.69E−05 |
| Angpt2 | −1.98 | 4.91E−05 |
| Cadps | −2.05 | 5.29E−05 |
| Vcan | −1.54 | 5.72E−05 |
| Cav1 | −2.79 | 6.66E−05 |
| Gng3 | −2.60 | 6.66E−05 |
| Efnb2 | −2.31 | 6.66E−05 |
| Nhsl2 | −2.99 | 8.30E−05 |
| Fam167a | −2.69 | 8.30E−05 |
We ranked genes by false discovery rate (FDR) and listed top 20 up‐ and down‐regulated genes.
Figure 3.
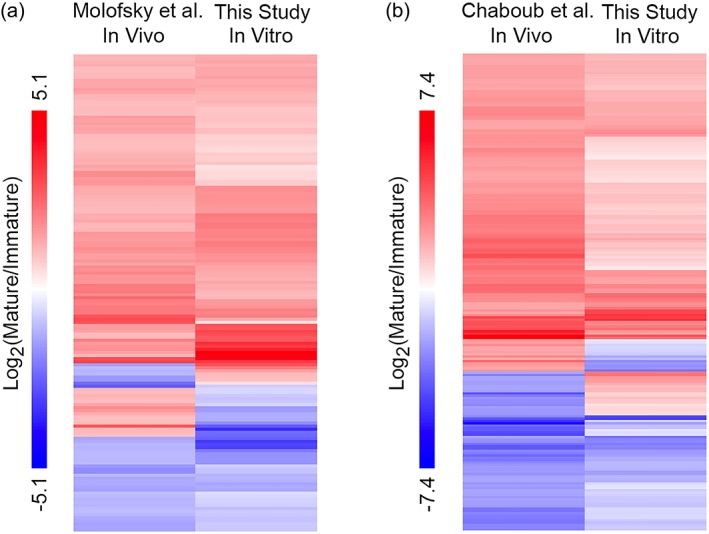
Comparison of astrocyte maturation marker gene expression in vivo and in vitro. (a) Differentially expressed genes between immature and mature astrocytes in vivo (Molofsky et al., 2013) and in vitro (this study). Genes with two‐fold or higher differences with FDR <0.05 were included. (b) Differentially expressed genes between immature and mature astrocytes in vivo (Chaboub et al., 2016) and in vitro (this study). Genes with four fold or higher differences with FDR <0.05 were included [Color figure can be viewed at wileyonlinelibrary.com]
All together, these comparisons suggest that immunopanned astrocytes in serum‐free media exhibited spontaneous maturation in vitro that by and large parallels the time scale and molecular changes of astrocyte maturation in vivo. Thus, serum‐free cultures of immunopanned murine astrocytes can serve as a model for investigating the mechanism of astrocyte maturation.
3.2. An astrocyte‐derived signal(s) promotes astrocyte maturation
The observation that purified astrocytes exhibited spontaneous maturation without input from other cell types suggests that either cell autonomous mechanisms or astrocyte‐ astrocyte interactions can trigger maturation. To test whether astrocyte‐astrocyte interactions affect maturation, we plated rat astrocytes at low (20 × 103 cells per well on a 24‐well plate), medium (50 × 103 cells per well), and high (200 × 103 cells per well) densities. We found that rat astrocytes at higher densities down‐regulated immature markers and upregulated mature markers faster, and were less proliferative than those at lower densities (Figure 4(a,c)), suggesting that an astrocyte‐derived signal(s) promoted astrocyte maturation. Furthermore, to test whether this mechanism is conserved in evolution, we performed similar experiments using mouse astrocytes and found similar density‐dependent maturation changes (Figure 4d) demonstrating that mouse and rat astrocytes share the regulation of maturation by astrocyte‐astrocyte interactions.
Figure 4.
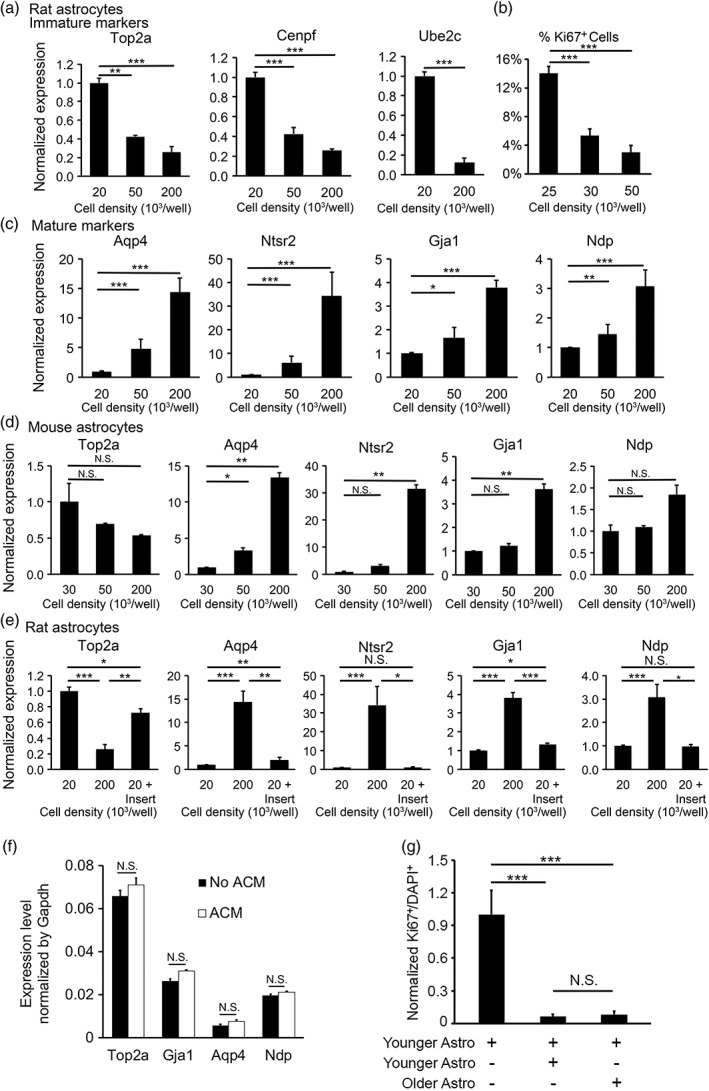
Astrocyte‐to‐astrocyte contact promotes astrocyte maturation. (a–c) Rat astrocytes matured faster at high cell density. We determined the expression of immature (a) and mature (c) astrocyte markers by qRT‐PCR. The expression for each gene was normalized to the house‐keeping gene Gapdh. The expression at each cell density was normalized to the level at 20 × 103 cells per well. (b) We determined the percentage of proliferating cells by quantifying Ki67 positive cells at 14 div. (d) Similar to rat astrocytes, mouse astrocytes matured faster at high cell densities. (e) We grew rat astrocytes at three conditions: Low density (20 × 103 cells per well), high density (200 × 103 cells per well), and low density with inserts (20 × 103 cells per well with additional 180 × 103 cells per well on porous inserts). Secreted signals freely diffused across the porous inserts whereas cells separated by the inserts do not make direct contacts. (f) Expression of astrocyte maturation markers as determined by qPCR in the presence and absence of 75 μg/mL ACM. (g) Younger and older astrocytes similarly promote astrocyte maturation. Younger astrocytes alone: Astrocytes purified from P2 rats plated at low density (30,000 per well). Younger + younger astrocytes: Astrocytes purified from P2 rats plated at high density (100,000 per well). Younger + older astrocytes: We first purified astrocytes from P7 rats and cultured them for 7 days before adding younger astrocytes purified from P2 rats. We labeled older astrocytes with lentiviruses encoding EGFP at >98% efficiency. We plated cells at varying densities and only used wells with the same final density in the latter two conditions for the final analyses. We quantified the percentage of proliferating cells by counting Ki67+ cells out of all DAPI stained cells. In the heterochronic condition, we only counted younger astrocytes (not labeled by EGFP). Percentage of Ki67+ cells was normalized to the younger astrocyte alone condition
3.3. Astrocyte‐to‐astrocyte contact promotes astrocyte maturation
To test whether the astrocyte derived signal(s) that promotes astrocyte maturation is secreted or contact‐dependent, we grew astrocytes in two layers separated by porous inserts that prevented cell contact but allowed the diffusion of secreted molecules. We found that astrocytes separated by inserts did not affect the expression of some maturation markers (Ntsr2 and Ndp) and exhibited much weaker effects than astrocytes in contact on the expression of other maturation markers (Top2a, Aqp4, and Gja1) (Figure 4e). To further examine the effect of astrocyte‐secreted signals on astrocyte maturation, we collected ACM and treated immature astrocytes. We found that ACM did not significantly change the expression of astrocyte maturation marker genes (Figure 4f). Therefore, most of the effect of astrocyte‐astrocyte interactions on astrocyte maturation is mediated by direct contact.
3.4. Younger and older astrocytes similarly promote astrocyte maturation
In developing mouse brains, the majority of astrocytes are generated between P0 and P14 (Ge et al., 2012). As astrocytes mature, each cell may contact younger, older, or same‐aged neighbor astrocytes (up to about 2‐week difference in age). To determine whether younger astrocytes, older astrocytes, or both can promote the maturation of astrocytes, we devised a “heterochronic” coculture experiment. We cultured astrocytes in the following three configurations: (a) younger astrocytes: astrocytes purified from P2 rats at low density (30,000 per well); (b) younger astrocytes + younger astrocytes: astrocytes purified from P2 rats at high density (100,000 per well); and (c) younger astrocytes + older astrocytes: younger astrocytes purified from P2 rats at low density (30,000 per well) and older astrocytes purified from P7 rats and cultured for an additional 7 days for in vitro maturation before the coculture experiment. To distinguish between younger and older astrocytes in heterochronic cocultures (Condition 3), we infected older astrocytes with lentiviruses encoding EGFP (Figure 8a) and then washed out the virus‐containing supernatant before using these cells for coculture experiments. We titrated virus dosage and only used cultures with >98% infection rate based on EGFP fluorescence. Younger astrocytes were not labeled. Therefore, there was no ambiguity in the age of the astrocytes in the heterochronic coculture. Since astrocyte maturation is affected by density (Figure 4a,e), we plated younger and older astrocytes at a variety of densities, counted cell numbers based on DAPI stain, and only used younger + younger and younger + older cocultures with similar total cell density for the final analyses. We stained astrocytes with proliferative cell marker Ki67 and found that younger and older astrocytes similarly inhibited astrocyte proliferation and promoted maturation (Figure 4g). The older astrocytes we tested are 12 days older than the younger astrocytes, representing the typical age difference in a population of maturing astrocytes in vivo. We did not test whether adult or aging adult astrocytes could promote the maturation of young astrocytes because adult or aging astrocytes exhibit poor survival in serum‐free conditions.
Figure 8.
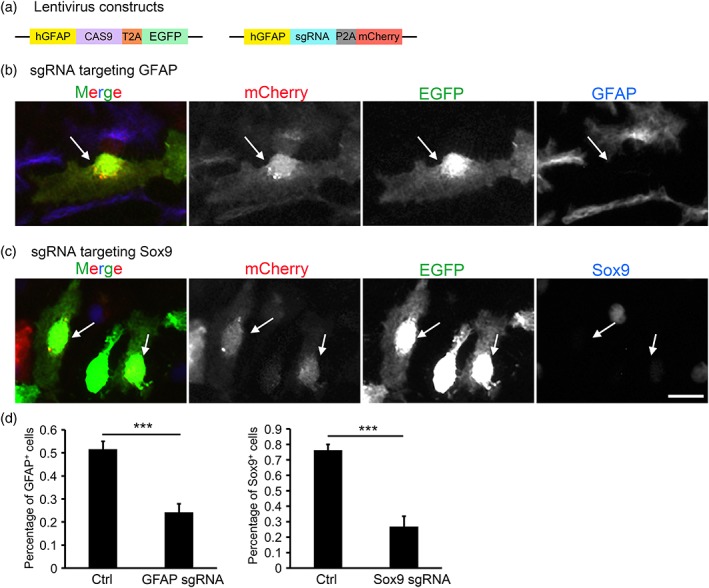
CRISPR genome editing in cultured mouse astrocytes. (a) We generated two lentivirus constructs containing the human GFAP promotor, Cas9, sgRNA, and fluorescent reporters EGFP and mCherry. (b,c) We infected cultured mouse astrocytes with lentiviruses encoding CAS9‐EGFP and sgRNAs targeting GFAP (b) and Sox9 (c) with the mCherry reporter. Images were taken 7 days after infection. Arrows point to cells doubly infected with lentiviruses encoding CAS9 and sgRNA. Doubly infected cells had reduced GFAP (b) or Sox9 (c) protein expression (d). Quantification of percentage of GFAP and Sox9 positive cells with or without CRISPR genome editing [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Secreted signals from microglia, OPCs, or endothelial cells change the expression of some but not all astrocyte maturation marker genes
Is astrocyte maturation triggered by a specific signal(s) from neighboring astrocytes or nonspecific signals from any other cell types? To test this, we purified microglia, OPCs, and endothelial cells from rat brains and co‐cultured each cell type with astrocytes separated by porous inserts. We determined the expression of astrocyte maturation markers by qRT‐PCR. We found that both microglia and OPC significantly reduced the expression of Top2a, a gene associated with cell proliferation and transcription but had no effect on other astrocyte maturation markers (Figure 5a,b). Therefore, microglia and OPC may affect astrocyte gene expression, either by producing a signal(s) that induce gene expression changes or by depleting nutrients in the media that affect astrocyte gene expression. Interestingly, endothelial cells induced a 3.5 fold increase in the expression of the gene encoding the water channel Aqp4 by astrocytes but had no effect on other astrocyte maturation markers (Figure 5c). The Aqp4 protein is important for water homeostasis and transport between the blood and the brain (Papadopoulos & Verkman, 2013) and it is the target of auto‐antibodies in an auto‐immune neurological disorder, neuromyelitis optica (Lennon, Kryzer, Pittock, Verkman, & Hinson, 2005). Aqp4 is located at astrocyte endfeet contacting blood vessels. Previous studies showed that vascular signals are important for the endfeet localization of Aqp4 (Armulik et al., 2010). Our results identified a novel mechanism by which vascular cells regulate astrocyte Aqp4 levels and added to the accumulating evidence that vascular cells and astrocytes regulate each other's development and function (Lee, Martinez‐Lozada, Krizman, & Robinson, 2017; Mi, Haeberle, & Barres, 2001). Although signaling between astrocytes and microglia, OPCs, and endothelial cells changed the expression of a small subset of genes, these interactions did not consistently induce changes of the expression of all astrocyte maturation markers, as did astrocyte‐to‐astrocyte contact. Therefore, astrocytes recognize specific signals from neighboring astrocytes that promote maturation.
Figure 5.
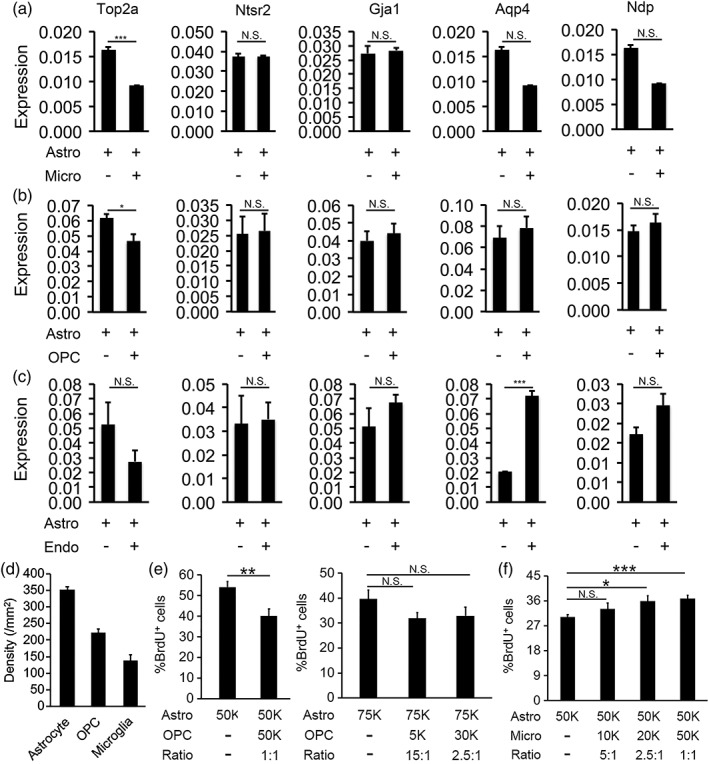
Microglia, OPCs, or endothelial cells change the expression of a subset of astrocyte maturation markers. Comparison of astrocyte maturation marker expression in pure astrocyte cultures versus (a) astrocyte‐microglia cocultures, (b) astrocyte‐OPC cocultures, and (c) astrocyte‐endothelial cell cocultures. Gene expression was determined by qRT‐PCR and normalized to the expression of the housekeeping gene, Gapdh. (d) Densities of astrocytes, OPCs, and microglia in developing mouse cerebral cortex in vivo. Cell number was counted based on Glul, Pdgfra, and Tmem119 in situ hybridization signal from P14 images from the Allen brain atlas. (e,f) Percentage of proliferating BrdU+ astrocytes in astrocyte‐OPC and astrocyte‐microglia cocultures with varying cell ratios. 50 k = 50,000 cells per well on a 24‐well plate
To examine whether the coculture conditions of different cell types we used in vitro is relevant for the developing brain in vivo, we assessed relative densities of different cell types in vivo and in vitro. To determine the ratio of astrocytes, OPCs, and microglia in vivo during astrocyte development, we counted cells labeled by cell type‐specific markers Glul, PDGFRa, and Tmem119 in the cerebral cortex at P14, a time point in the middle of astrocyte maturation in mouse (P0–P30) based on in situ hybridization images from the Allen Brain Atlas (Lein et al., 2007). We found that the ratio of astrocytes to OPCs is 1.6:1 and astrocytes to microglia 2.5:1 at P14 in vivo (Figure 5d). We then plated astrocytes, OPCs, and microglia at varying densities (Figure 5e,f). We examined the effect of OPCs and microglia on astrocyte proliferation using BrdU labeling of proliferating astrocytes. We found that OPCs inhibits astrocyte proliferation when OPCs and astrocytes are plated at 1:1 ratio but had no effect when the astrocyte‐to‐OPC ratio is 2.5:1 (Figure 5e). Since the astrocyte‐to‐OPC ratio in vivo is 1.6:1 (Figure 5d), between the two ratios we tested in vitro, whether there are enough OPCs in vivo to affect astrocyte proliferation remains uncertain. In addition, we found that microglia affected astrocyte proliferation at the astrocyte‐to‐microglia ratios of 1:1, and 2.5:1, but not 5:1 (Figure 5f). Since the astrocyte‐to‐microglia ration in vivo is 2.5:1 during astrocyte maturation, the effect we found in microglia‐astrocyte interaction in vitro is likely relevant in vivo.
What are the molecules secreted by OPC and microglia that may affect astrocyte development? We recently purified astrocyte, OPC, microglia, endothelial cells, and neurons from developing mouse brains and performed RNA‐seq (Zhang et al., 2014). We found that dozens of genes encoding secreted proteins are expressed specifically or enriched in OPC and microglia (Figure 6). OPC secreted proteins include Matn4, Lum, and Dcn, which have been previously reported to inhibit cell proliferation (Uckelmann et al., 2016; Vij, Roberts, Joyce, & Chakravarti, 2004; Xaus, Comalada, Cardó, Valledor, & Celada, 2001). Microglia secreted proteins include Ccl3, Ccl4, and Gdf15, with previously reported roles in promoting cell proliferation (Li, Ma, Zheng, & Zhang, 2018; Tsai et al., 2013). These proteins are among the candidate signals that mediate the effect of OPC and microglia on astrocyte development.
Figure 6.
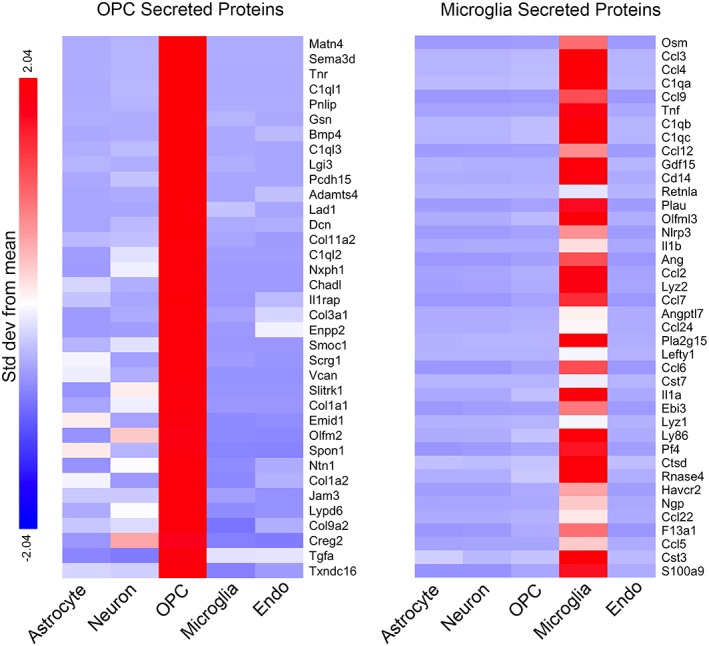
OPC and microglia secreted proteins. We purified astrocytes, neurons, OPCs, microglia, and endothelial (endo) cells from P7–P17 mouse cerebral cortex by immunopanning and FACS and performed RNA‐seq (Zhang et al., 2014). The top 40 genes most highly enriched in OPC and microglia encoding secreted proteins are shown. All the genes listed have high expression (FPKM >10). Secreted protein annotations are obtained from Uniprot. Color represents SD from means for each gene [Color figure can be viewed at wileyonlinelibrary.com]
3.6. HBEGF inhibits astrocyte maturation
Growth factor signaling is important for the development of multiple cell types. To explore whether growth factor signaling regulates astrocyte maturation, we examined the expression of growth factor receptors by astrocytes. Astrocytes highly express EGFR (Cahoy et al., 2008; Zhang et al., 2014; Zhang, Sloan, et al., 2016). Therefore, we tested the effect of an EGFR ligand, HBEGF, on astrocyte maturation. We compared the expression of astrocyte maturation markers by qRT‐PCR in the presence and absence of HBEGF. We found that HBEGF‐treated astrocytes expressed maturation markers at lower levels than control astrocytes (Figure 7a), suggesting that HBEGF inhibited astrocyte maturation.
Figure 7.
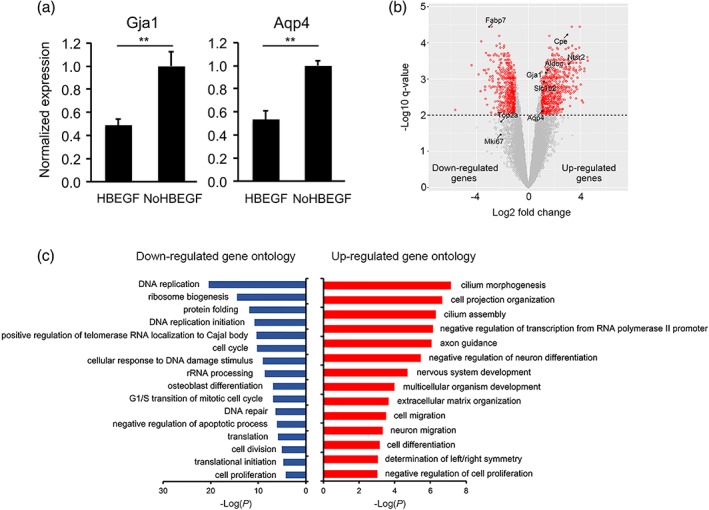
HBEGF inhibits astrocyte maturation. (a) Astrocytes were treated with or without 5 ng/mL HBEGF. The expression of Gja1 and Aqp4 were determined by qRT‐PCR and normalized to the expression of the housekeeping gene, Gapdh. (b) Volcano plot showing differential gene expression under HBEGF withdrawal conditions. Q‐value is the p value adjusted for multiple comparisons using the EdgeR‐Limma‐Voom package in R. (c) Top gene ontology terms enriched in up‐ or down‐regulated genes under HBEGF withdrawal conditions [Color figure can be viewed at wileyonlinelibrary.com]
3.7. CRISPR genome editing in cultured astrocytes
In addition to identifying the roles of cell–cell interaction and growth factor signaling in astrocyte maturation, we also wanted to test what cell intrinsic mechanism(s) regulates astrocyte maturation. To identify cell autonomous signals that control astrocyte maturation, we employed CRISPR genome editing in cultured mouse astrocytes. We first tested previously generated lentiviruses in which the expression of Cas9 and sgRNAs was driven by generic EF1a and SSFV promotors. Neither of these promoters induced sufficient gene expression in mouse astrocytes in vitro (data not shown). We therefore cloned the human GFAP promotor into the lentivirus vector backbone. We inserted sequences encoding Cas9 and EGFP downstream of the human GFAP promotor in one construct and sequences encoding sgRNA and mCherry in another construct (Figure 8a). We found that these lentiviruses efficiently infected mouse astrocytes and drove expression of fluorescent markers (Figure 8b,c).
To test the efficiency of CRISPR genome editing in cultured mouse astrocytes, we designed sgRNAs targeting the astrocyte marker genes GFAP and Sox9. We chose these genes because high quality antibodies against GFAP and Sox9 are available, thus allowing us to evaluate the extent and time course of protein elimination after CRISPR genome editing. We infected mouse astrocytes in vitro with lentiviruses encoding Cas9 and sgRNAs against GFAP (Figure 8b) and Sox9 (Figure 8c). We found substantial reduction of GFAP and Sox9 proteins at 7 days after infection (Figure 8b–d). Therefore, CRISPR genome editing is an efficient approach to knocking out candidate genes in mouse astrocytes in vitro.
3.8. The tumor‐associated genes EGFR and TP53 promote and inhibit the proliferation and/or survival of immature astrocytes, respectively
As astrocytes mature, they slow down and eventually stop proliferating in vivo (Ge et al., 2012; Nixdorf‐Bergweiler et al., 1994) and in vitro (Figure 1). Tumor‐associated genes that affect the cell cycle may therefore affect astrocyte maturation. Mouse, rat, and human astrocytes all highly express the tumor‐associated genes EGFR and TP53 (Foo et al., 2011; Zhang et al., 2014; Zhang, Sloan, et al., 2016). We therefore tested the roles of EGFR and TP53 in the proliferation of immature astrocytes.
We infected cultured mouse astrocytes with lentiviruses encoding Cas9‐EGFP and sgRNA‐mCherry targeting EGFR and TP53. EGFP and mCherry double positive cells were knockout cells and other cells were wild type cells. The changes of the percentage of knockout cells in the entire population over time reflect the proliferation and/or survival of knockout cells compared to wild type cells. We analyzed the percentage of knockout cells at 7, 14, and 21 days after infection. We found that EGFR sgRNA/Cas9 double positive knockout cells decreased over time whereas TP53 sgRNA/Cas9 double positive knockout cells increased over time in the astrocyte population (Figure 9). Therefore, EGFR promotes and TP53 inhibits the proliferation and/or survival of immature astrocytes, respectively.
Figure 9.
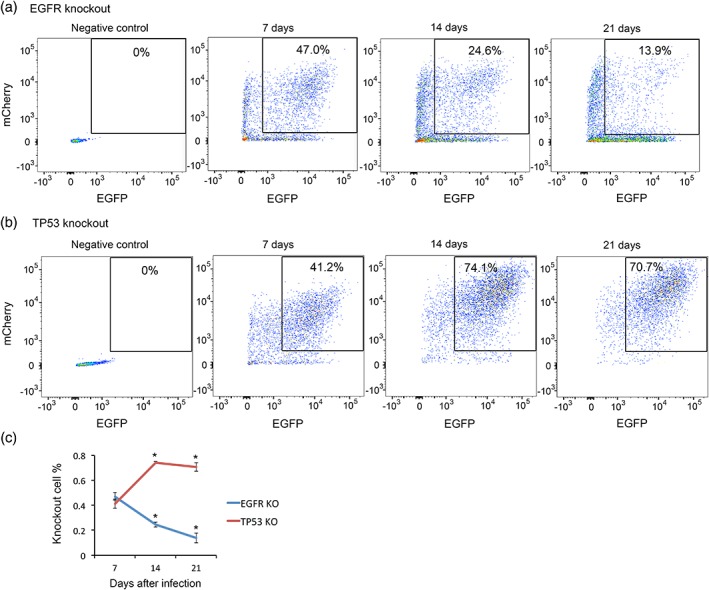
EGFR and TP53 regulate astrocyte progenitor proliferation/survival. We infected mouse astrocyte progenitors with lentiviruses encoding Cas9‐EGFP and sgRNA‐mCherry. The sgRNAs targeted EGFR (a) and TP53 (b). FACS plots showed that the percentage of doubly infected EGFR knockout cells (a) gradually decreased whereas TP53 knockout cells increased (b). (c) Percentage of knockout cells at 7, 14, and 21 days after infection [Color figure can be viewed at wileyonlinelibrary.com]
3.9. Molecular mechanisms underlying the effect of HBEGF, EGFR, and P53 on astrocyte maturation
To identify molecular mechanisms underlying the effect of HBEGF, EGFR, and P53 on astrocyte maturation, we performed RNA‐seq of maturating astrocytes treated with HBEGF, an EGFR inhibitor PD168393, and a P53 inhibitor Pifithrin‐α (Figures 7, 10, 11 and Tables 3, 4, 5). We first examined the expression of astrocyte maturation markers under these conditions and confirmed our finding that HBEGF and EGFR inhibits astrocyte maturation and P53 promotes astrocyte maturation (Figures 7, 10 and Tables 3, 4, 5).
Figure 10.
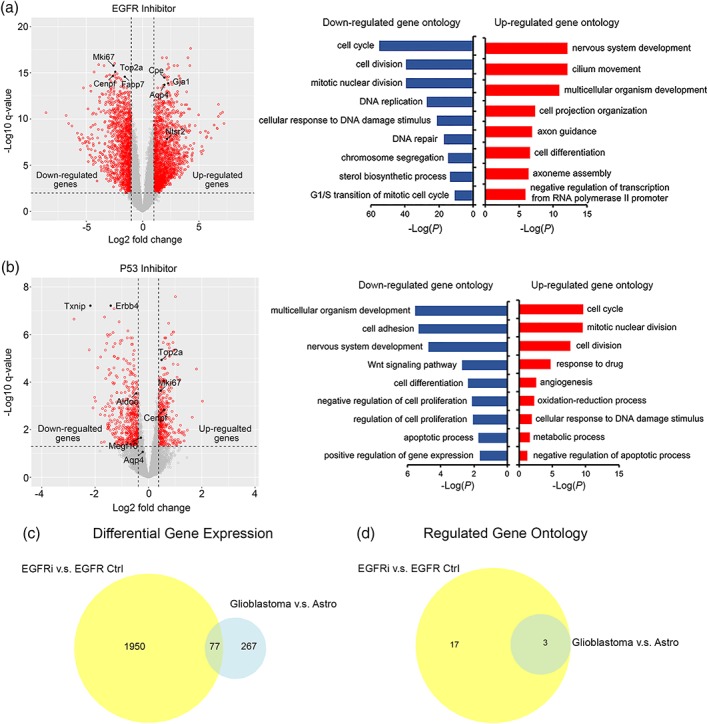
Genes differentially expressed by astrocytes with EGFR inhibition and P53 inhibition. (a,b) Volcano plots showing differentially expressed gene with EGFR and P53 inhibition. Gene ontology terms enriched in genes up‐ and down‐regulated with EGFR and P53 inhibition. (c) Comparison of differentially expressed genes by EGFR inhibition, P53 inhibition, and glioblastoma. (d) Comparison of gene ontology terms differentially affected by EGFR inhibition, P53 inhibition, and glioblastoma [Color figure can be viewed at wileyonlinelibrary.com]
Figure 11.
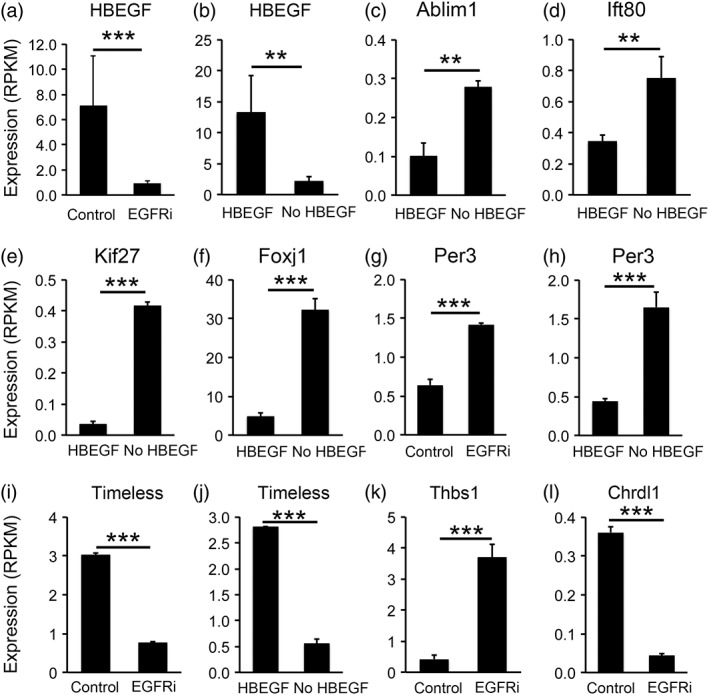
Examples of genes differentially expressed under the HBEGF withdrawal and EGFR inhibition conditions. Expression is shown as RPKM determined by RNA‐seq. (a,b) HBEGF. (c–f) Genes associated with cilium formation. (g–j) circadian clock genes. (k,l) genes that regulation synapse formation (Thbs1) and maturation (Chrdl1)
Table 3.
Differentially expressed genes with EGFR inhibitiona
| Gene | Log fold change | FDR |
|---|---|---|
| Up‐regulated genes | ||
| Gfap | 4.27 | 2.31E−18 |
| Zbtb20 | 1.74 | 2.33E−17 |
| Ntrk2 | 2.13 | 2.33E−17 |
| Miat | 4.03 | 2.33E−17 |
| Ndrg2 | 2.27 | 1.17E−16 |
| Tnr | 3.10 | 1.66E−16 |
| Igfbp5 | 2.80 | 1.66E−16 |
| Ahnak | 4.20 | 1.66E−16 |
| Apoe | 2.34 | 2.68E−16 |
| Gnao1 | 2.17 | 4.06E−16 |
| Dtna | 1.92 | 1.63E−15 |
| Clu | 3.23 | 2.15E−15 |
| Nrep | 2.30 | 2.64E−15 |
| Tmem132b | 2.10 | 2.64E−15 |
| 3110039I08Rik | 2.07 | 3.24E−15 |
| Cst3 | 1.92 | 3.24E−15 |
| Cpe | 1.93 | 3.24E−15 |
| Csmd1 | 3.34 | 4.06E−15 |
| Pxdn | 2.21 | 5.77E−15 |
| 6330403K07Rik | 2.53 | 1.03E−14 |
| Down‐regulated genes | ||
| Txn1 | −2.48 | 9.63E−17 |
| Fbln2 | −3.89 | 1.32E−16 |
| Mki67 | −2.59 | 1.66E−16 |
| Inka2 | −3.42 | 1.68E−16 |
| Top2a | −2.42 | 7.78E−16 |
| Adam12 | −4.31 | 8.07E−16 |
| Hip1 | −2.26 | 1.29E−15 |
| Ldha | −2.49 | 1.63E−15 |
| Hmgcr | −2.92 | 1.69E−15 |
| Rhoq | −3.17 | 2.15E−15 |
| Cenpf | −2.65 | 2.23E−15 |
| Tagln2 | −2.45 | 2.53E−15 |
| Cyp51 | −3.23 | 2.53E−15 |
| Fabp7 | −1.58 | 2.64E−15 |
| Nes | −2.13 | 3.24E−15 |
| Kcnc1 | −2.24 | 3.54E−15 |
| Sulf2 | −2.33 | 4.13E−15 |
| Serpine2 | −1.96 | 4.45E−15 |
| Hmgcs1 | −2.88 | 6.25E−15 |
| Nucks1 | −1.88 | 6.43E−15 |
We ranked genes by false discovery rate (FDR) and listed top 20 up‐ and down‐regulated genes.
Table 4.
Differentially expressed genes with P53 inhibitiona
| Gene | Log fold change | FDR |
|---|---|---|
| Up‐regulated genes | ||
| Sparcl1 | 1.01 | 2.57E−08 |
| Dusp4 | 1.11 | 1.84E−05 |
| Diras2 | 1.80 | 2.82E−05 |
| Inpp4b | 1.07 | 6.24E−05 |
| Gm2115 | 1.17 | 7.74E−05 |
| Npnt | 1.16 | 8.58E−05 |
| Rnase1 | 2.03 | 0.00063234 |
| Gcnt4 | 1.05 | 0.00250861 |
| Gpc5 | 1.64 | 0.00300998 |
| Tuba4a | 1.20 | 0.00651057 |
| Down‐regulated genes | ||
| Tnr | −2.34 | 3.00E−11 |
| Igsf9b | −1.64 | 4.46E−09 |
| Miat | −1.80 | 7.38E−09 |
| Erbb4 | −1.41 | 6.15E−08 |
| Txnip | −2.18 | 6.15E−08 |
| Pdgfra | −1.29 | 8.23E−08 |
| Mfsd2a | −1.64 | 1.84E−07 |
| Gpr17 | −2.79 | 2.25E−07 |
| Myt1 | −1.10 | 2.92E−07 |
| Cp | −1.33 | 8.24E−07 |
| Pcdh15 | −1.43 | 1.00E−06 |
| Sox10 | −1.54 | 1.00E−06 |
| Dcc | −1.51 | 1.17E−06 |
| Sh3bp4 | −1.05 | 1.82E−06 |
| Arrdc4 | −1.56 | 2.35E−06 |
| CT010467.1 | −1.25 | 2.39E−06 |
| Enpp2 | −1.06 | 2.39E−06 |
| Bmp3 | −1.32 | 3.30E−06 |
| Zfp488 | −2.22 | 4.42E−06 |
| Prkcq | −1.61 | 4.53E−06 |
We ranked genes by false discovery rate (FDR) and listed top 10–20 up‐ and down‐regulated genes.
Table 5.
Differentially expressed genes with HBEGF withdrawala
| Gene | Log fold change | FDR |
|---|---|---|
| Up‐regulated genes | ||
| Gfap | 3.97 | 3.59E−05 |
| Draxin | 3.36 | 3.59E−05 |
| Cpe | 3.01 | 5.96E−05 |
| Mt‐Rnr1 | 1.62 | 6.40E−05 |
| Gabrb1 | 2.76 | 8.63E−05 |
| Igfbp5 | 3.38 | 0.00012041 |
| Tspan18 | 3.11 | 0.00013543 |
| Ntrk2 | 1.72 | 0.00013543 |
| Cyp1b1 | 3.93 | 0.00013543 |
| Sfrp1 | 2.21 | 0.00013706 |
| Cfap54 | 3.09 | 0.00013706 |
| Zbtb20 | 1.60 | 0.00013706 |
| Gramd1b | 2.28 | 0.00014896 |
| Cfap44 | 3.66 | 0.00014896 |
| Foxj1 | 2.88 | 0.00014896 |
| Tmem132b | 1.85 | 0.00015487 |
| Ggta1 | 3.43 | 0.00019063 |
| Dapp1 | 2.22 | 0.00019921 |
| Hspa2 | 3.36 | 0.00020204 |
| Igsf11 | 1.85 | 0.00020326 |
| Down‐regulated genes | ||
| Fabp7 | −3.02 | 3.59E−05 |
| Kcnc1 | −2.83 | 3.59E−05 |
| Slc4a4 | −2.24 | 6.40E−05 |
| Inka2 | −3.63 | 9.16E−05 |
| Olig1 | −2.20 | 0.0001065 |
| Ccnd1 | −2.45 | 0.00012041 |
| Slc5a3 | −1.86 | 0.00012041 |
| Egr1 | −2.16 | 0.00013543 |
| Olig2 | −1.91 | 0.00013543 |
| Dusp6 | −3.27 | 0.00019063 |
| Ncl | −1.56 | 0.00019063 |
| Ipo5 | −1.88 | 0.00019921 |
| Npm1 | −1.81 | 0.00019921 |
| Slc25a5 | −2.07 | 0.00020326 |
| Akap12 | −2.58 | 0.00020326 |
| Ccnd2 | −1.61 | 0.00020326 |
| Tubb6 | −2.44 | 0.00022065 |
| Hspd1 | −1.88 | 0.00022065 |
| Igfbp3 | −2.67 | 0.00022065 |
| Serbp1 | −1.47 | 0.00022065 |
We ranked genes by false discovery rate (FDR) and listed top 20 up‐ and down‐regulated genes.
To gain mechanistic understanding of the regulation of astrocyte maturation, we examined differentially expressed genes in astrocytes with or without HBEGF, the EGFR inhibitor, and the P53 inhibitor. HBEGF is a growth factor secreted by astrocytes, microglia, and endothelial cells in the brain (Zhang et al., 2014). HBEGF binds receptors including EGFR, Erbb2, and Erbb4 that initiate intracellular signaling (Roskoski, 2014). Astrocytes secrete HBEGF and express its receptors (Cahoy et al., 2008; Foo et al., 2011; Zhang et al., 2014). Therefore, HBEGF likely has autocrine function in astrocytes. Interestingly, the HBEGF gene itself is one of the most significantly changed genes in astrocytes treated with HBEGF and the EGFR inhibitor (Figure 11a,b). We found that withdrawal of HBEGF from astrocytes decreased HBEGF expression by astrocytes. Similarly, inhibiting EGFR decreased HBEGF expression. Therefore, a positive feedback loop operates in developing astrocytes. This loop may amplify a small decrease in HBEGF levels by further decreasing HBEGF production, which leads to astrocyte maturation. The positive feedback loop amplifies signals and may therefore ensure the nonreversible maturation of astrocytes during normal brain development. Consistent with this hypothesis, we observed a gradual decrease of HBEGF expression as astrocytes mature. Conversely, either increasing HBEGF or enhancing EGFR signaling leads to further increases in HBEGF levels and therefore promotes the immature astrocyte state. In brain injury and diseases, astrocytes may revert to a reactive state with properties of immature astrocytes (Foo et al., 2011; Sofroniew, 2014; Zamanian et al., 2012). In glioblastoma, tumor cells have molecular and phenotypic resemblance to immature astrocytes (Verhaak et al., 2010; Zhang, Sloan, et al., 2016). Future investigation of the involvement of the HBEGF‐EGFR positive feedback loop in the context of reactive astrogliosis and glioma genesis may reveal key regulators of these processes. The effect of HBEGF withdrawal and EGFR inhibition on the reduction of growth factor gene expression is specific to HBEGF, as the expression of some other growth factors are increased (HGF, FGF11) or not changed (EGF), ruling out the possibility that HBEGF withdrawal or EGFR inhibition results in compromised cell health and reduced production of all growth factors.
The RNA‐seq analyses revealed that the top Gene Ontology terms downregulated for EGFR inhibition and upregulated for P53 inhibition is cell cycle/cell division (Figure 10 and Table 8). This finding reiterates the well‐known effects of P53 on suppression of proliferation and EGFR on promoting proliferation, and the links between cell cycle regulation and astrocyte maturation. The gene expression data confirmed our earlier results of the effect of EGFR and TP53 CRISPR knockout on astrocyte population composition as determined by FACS (Figure 9).
Table 8.
Gene ontology (GO) terms enriched in genes affected by EGFR inhibition and glioblastomaa
| Term ID | Term name | FDR |
|---|---|---|
| GO terms shared by genes affected by EGFR inhibition and glioblastoma | ||
| GO:0007049 | Cell cycle | 3.03E−10 |
| GO:0007067 | Mitotic nuclear division | 2.99E−06 |
| GO:0051301 | Cell division | 3.63E−05 |
| GO terms only enriched in genes affected by EGFR inhibition | ||
| GO:0006260 | DNA replication | 9.85E−09 |
| GO:0007059 | Chromosome segregation | 1.83E−06 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 6.01E−05 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 8.51E−05 |
| GO:0000070 | Mitotic sister chromatid segregation | 8.56E−05 |
| GO:0006974 | Cellular response to DNA damage stimulus | 1.21E−04 |
| GO:0007399 | Nervous system development | 2.81E−04 |
| GO:0016126 | Sterol biosynthetic process | 8.09E−04 |
| GO:1904874 | Positive regulation of telomerase RNA localization to Cajal body | 8.98E−04 |
| GO:0007018 | Microtubule‐based movement | 0.001232 |
| GO:0006270 | DNA replication initiation | 0.001693 |
| GO:0006730 | One‐carbon metabolic process | 0.004834 |
| GO:0007411 | Axon guidance | 0.005193 |
| GO:0008203 | Cholesterol metabolic process | 0.012354 |
| GO:0043407 | Negative regulation of MAP kinase activity | 0.042922 |
| GO:0006694 | Steroid biosynthetic process | 0.046516 |
| GO:0006695 | Cholesterol biosynthetic process | 0.047637 |
GO terms are ranked by FDR.
We performed gene ontology analyses of differentially expressed genes in astrocytes treated with the above‐mentioned conditions. We found that some of the most highly enriched gene ontology terms associated with HBEGF withdrawal and EGFR inhibition are cilium assembly and morphogenesis (Figures 7c and 10a). Indeed, many cilium‐associated genes exhibit increased expression by astrocytes under HBEGF withdrawal or EGFR inhibition conditions (Figure 11c–f). Primary cilia are slender microtubule‐based organelles that project from the cell body (Álvarez‐Buylla & Ihrie, 2014). Cilia are important for Sonic Hedgehog function. Mutations in cilium genes are associated with a variety of diseases collectively called ciliopathies. Our results showed for the first time that HBEGF‐EGFR signaling inhibits cilium assembly in developing astrocytes, to the best of our knowledge.
Period and Timeless are the molecular gears of the circadian clock that controls rhythmic behaviors in cells and organisms (Dubowy & Sehgal, 2017). Their mRNA and protein levels exhibit daily rhythmic oscillations. We collected treated and control samples at the same time of the day and found that HBEGF withdrawal and EGFR inhibition increased Period 3 and decreased Timeless gene expression (Figure 11g–j). Furthermore, HBEGF withdrawal decreased and P53 inhibition increased the expression of the gene encoding the Timeless interacting protein Tipin. These results suggest that signals that regulate astrocyte maturation have strong influences on the operation of the circadian clock in developing astrocytes.
The time window of astrocyte maturation overlaps with the period of synapse formation. Astrocytes secrete important protein signals that induce synapse formation (Allen et al., 2012; Blanco‐Suarez, Liu, Kopelevich, & Allen, 2018; Christopherson et al., 2005; Eroglu et al., 2009; Farhy‐Tselnicker et al., 2017; Kucukdereli et al., 2011; Pfrieger & Barres, 1997; Stogsdill et al., 2017; Ullian et al., 2001). To test whether HBEGF, EGFR, and P53 affect astrocytic interaction with synapses, we examined the expression of genes synapse‐inducing proteins. We found that EGFR inhibition in astrocytes increased the expression of genes inducing synapse formation (Thbs1, Thbs3, Sparcl1, Nlgn2, Figure 11k) but decreased the expression of a gene that drives synapse maturation and limits plasticity (Chrdl1 Figure 11l). Therefore, EGFR regulation of astrocyte maturation may contribute to the precise temporal control of synapse formation, maturation, and the time window of circuit plasticity. Furthermore, we found that HBEGF withdrawal induced some similar changes as EGFR inhibition (Thbs1 and Thbs3 up‐regulation) whereas P53 inhibition induced some opposite changes as EGFR inhibition (Thbs3 down‐regulation).
3.10. Molecular pathways affected by EGFR and P53 inhibition partially overlap with those affected by glioblastoma
Glioblastoma cells share molecular and phenotypic similarities with immature astrocytes (Verhaak et al., 2010; Zhang, Sloan, et al., 2016). We showed that tumor‐associated genes EGFR and TP53 affect astrocyte maturation including changing the expression of molecular pathways mentioned above. Are the same pathways affected by glioblastoma? To address this question, we compared RNAseq data generated in this study to a dataset we recently generated by comparing healthy adult astrocytes and glioblastoma cells from human patients (Zhang, Sloan, et al., 2016) (Figure 10c,d, Tables 6, 7, 8). We found that a substantial portion of genes and pathways affected by EGFR inhibition in developing astrocytes are also affected by glioblastoma. For example, both EGFR inhibition and glioblastoma affect genes involved in cell cycle and cell division (Table 8). In addition, there are genes and pathways specifically affected by EGFR inhibition during astrocyte development but not by glioblastoma, such as the gene ontology terms, nervous system development, and axon guidance (Table 8).
Table 6.
Differentially expressed genes shared by EGFR inhibition versus control and glioblastoma versus astrocytesa
| Genes | EGFRi LogFC | EGFRi FDR | GBM LogFC | GBM FDR |
|---|---|---|---|---|
| Cadm3 | 3.43 | 0.0002394 | −3.29 | 0.00806944 |
| Bub1 | −2.87 | 0.00038194 | 5.23 | 0.00042527 |
| Cse1l | −1.40 | 0.00049666 | 2.05 | 0.00884872 |
| Mdc1 | −1.69 | 0.00065999 | 1.00 | 1.25E−90 |
| Serpine2 | −1.97 | 0.00065999 | −3.07 | 0.00202009 |
| Ctnna2 | 2.34 | 0.00066317 | −3.14 | 0.00569524 |
| Pxdn | 2.17 | 0.00069189 | 5.46 | 0.00427848 |
| Mtbp | −1.59 | 0.00075641 | 2.10 | 0.00320239 |
| Cdca5 | −2.01 | 0.0010244 | 3.74 | 0.00859505 |
| Nek2 | −2.75 | 0.00154249 | 4.03 | 0.00177904 |
| Serbp1 | −1.26 | 0.00160475 | 1.42 | 0.00585271 |
| Ccna2 | −2.54 | 0.00172862 | 4.04 | 0.00537184 |
| Mcm4 | −1.71 | 0.00188994 | 1.84 | 0.00047873 |
| Cdh20 | −1.48 | 0.00191899 | −4.38 | 0.00766177 |
| Cdca8 | −2.77 | 0.00191899 | 2.74 | 0.00040685 |
| Pgap1 | −1.11 | 0.00199076 | 1.55 | 0.00537184 |
| Slco1c1 | 1.41 | 0.00199808 | −4.25 | 0.00727681 |
| Foxm1 | −2.16 | 0.00202998 | 4.41 | 0.00119993 |
| Tpx2 | −2.80 | 0.00217451 | 6.30 | 0.00320239 |
| Ube2c | −2.75 | 0.00218558 | 5.51 | 0.005064 |
We listed top 20 genes ranked by FDR in EGFR inhibition (EGFRi) with at least two fold difference and FDR < 0.01 in both EGFRi versus control and glioblastoma versus astrocyte comparisons.
Table 7.
Genes differentially expressed with EGFR inhibition but not differentially expressed in the glioblastoma versus astrocyte comparisona
| Genes | EGFRi LogFC | EGFRi FDR |
|---|---|---|
| Gm37694 | 2.64 | 8.85E−05 |
| Rhoq | −3.20 | 0.00017575 |
| Rrm1 | −1.63 | 0.00017575 |
| Mad2l1 | −2.26 | 0.00019026 |
| Mmp15 | −2.84 | 0.00019898 |
| Ncapg2 | −2.25 | 0.000263 |
| Miat | 3.99 | 0.00028567 |
| Ndrg2 | 2.25 | 0.00028567 |
| Hspa4 | −1.42 | 0.00028567 |
| Txn1 | −2.50 | 0.0002858 |
| Sfpq | −1.12 | 0.0002858 |
| Spred3 | −1.62 | 0.0002858 |
| Selenop | 1.50 | 0.00035539 |
| Mthfd2 | −4.40 | 0.00036567 |
| H2afx | −1.40 | 0.00036567 |
| Hip1 | −2.28 | 0.00036567 |
| Usp1 | −2.01 | 0.00037416 |
| Timeless | −1.99 | 0.00037709 |
| Lamp2 | 1.07 | 0.00037709 |
| Pole | −2.04 | 0.00041609 |
We used the following criteria: fold change >2 (Log fold change >1), FDR <0.01, and RPKM >1. We identified genes meeting all three criteria in EGFR inhibition versus control but not in the glioblastoma versus astrocyte comparison. We ranked genes by FDR and listed the top 20 genes.
3.11. Maturing astrocytes in vitro represent multiple molecular subtypes of astrocytes and exhibit maturation‐associated subpopulation composition changes
Emerging evidence suggests the heterogeneity of astrocyte populations (Chai et al., 2017; Farmer et al., 2016; Hochstim, Deneen, Lukaszewicz, Zhou, & Anderson, 2008; John Lin et al., 2017; Tsai et al., 2012; Zhang & Barres, 2010). For example, a recently study identified five subtypes of astrocytes with differing molecular and functional profiles (John Lin et al., 2017). Moreover, these distinct subpopulations of astrocytes are represented in human glioblastoma. To determine whether our in vitro astrocyte maturation system represents all or a subset of astrocyte subpopulations, we examined the expression of subpopulation‐specific marker genes by astrocytes in vitro. We found that cultured astrocytes express markers of all five subpopulations (Figure 12a–d) suggesting that our in vitro culture model is likely a comprehensive representation of all major subpopulations of astrocytes found in the cerebral cortex in vivo. Interestingly, we found that treatment that alters astrocyte maturation differentially affect subpopulation composition (Figure 12a–d). For example, subpopulation B has been associated with tumor aggressiveness and tumor‐associated epileptic seizures. We found that EGFR inhibition increased and P53 inhibition decreased the expression of subpopulation B‐specific marker gene expression (Figure 12b,c). These results suggest that the serum‐free in vitro astrocyte maturation system we developed may be suitable for molecular studies that gain insight into the regulation of astrocyte development relevant for glioblastoma.
Figure 12.
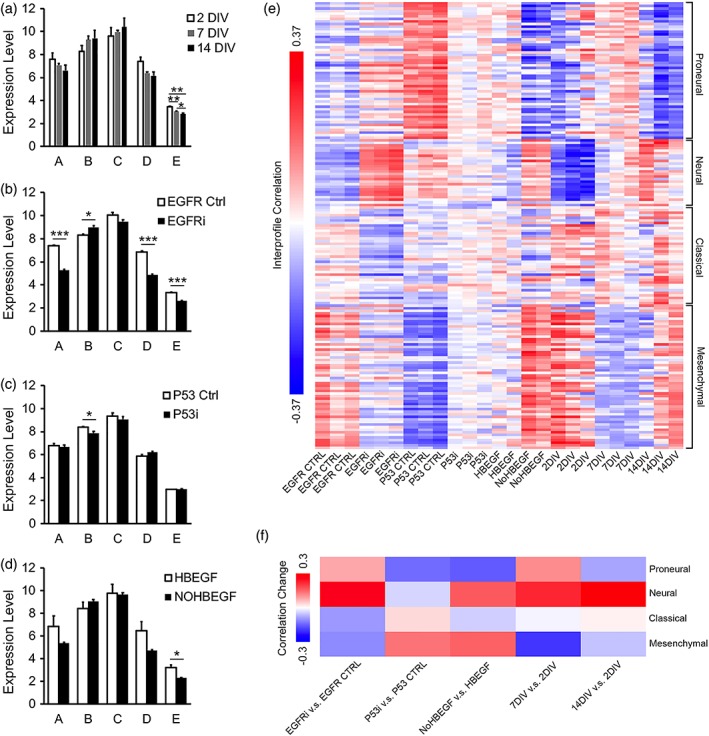
Comparison of astrocytes in vitro with subpopulations of astrocytes in vivo and glioblastoma subtypes. Expression of subpopulation specific genes by astrocytes in vitro at different maturation stages (a), with EGFR inhibition (b), P53 inhibition (c), and HBEGF withdrawal (d). Expression is shown as RPKM. (e) Interprofile Pearson correlations of the transcriptome profiles of astrocytes in vitro and glioblastoma subtypes. (f) Changes of the correlation between transcriptome profiles of astrocytes in vitro and glioblastoma subtypes with EGFR inhibition, P53 inhibition, HBEGF withdrawal, and maturation state changes [Color figure can be viewed at wileyonlinelibrary.com]
3.12. Comparison of astrocyte maturation with subtypes of glioblastoma
Glioblastoma in human patients can be molecularly defined as four subtypes: proneural, neural, classical, and mesenchymal (Verhaak et al., 2010). To further explore the molecular parallel between maturing astrocytes and glioblastoma, we compared the similarities of the transcriptome profiles of maturing astrocytes treated with HBEGF, EGFR inhibitor, and P53 inhibitor with 173 samples of human glioblastoma including all four subtypes from the TCGA dataset (Verhaak et al., 2010). We calculated interprofile correlations and found that HBEGF, EGFR inhibitor, and P53 inhibitor differentially changed the similarities of developing astrocytes with subtypes of glioblastoma (Figure 12e,f). For example, EGFR inhibition and HBEGF withdrawal both increased the correlation of developing astrocytes to the Neural subtype of glioblastoma, whereas P53 inhibition decreased correlation to the Proneural subtype. These results expanded our understanding of the molecular similarities between developing astrocytes and glioblastoma.
In summary, we identified molecular markers for astrocyte maturation, established an in vitro model of astrocyte maturation, and found that astrocyte‐to‐astrocyte contact and a positive feedback loop of HBEGF‐EGFR signaling promoted maturation. In addition OPCs, microglia, and endothelial cells affect astrocyte gene expression. Furthermore, we found that tumor associated genes HBEGF, EGFR, and P53 affected genes involved in cilium formation, the circadian clock, and synaptic function in developing astrocytes. These findings uncovered cellular and molecular mechanisms underlying astrocyte maturation.
4. DISCUSSION
After the initial astrocyte cell fate specification regulated by the Notch, Jak/STAT, Sox9, NFIA/B, CNTF/LIF pathways, immature astrocytes undergo morphological, cell biological, transcriptional, and functional changes before they become mature (Freeman, 2010; Molofsky & Deneen, 2015). In mouse, astrocyte maturation occurs within the first several weeks after birth (Bushong et al., 2004; Ge et al., 2012; Nixdorf‐Bergweiler et al., 1994) and in humans, the first year of infant life (Zhang, Sloan, et al., 2016). Astrocyte maturation occurs in parallel with synapse formation and refinement. For example, synapse density gradually increases in human cerebral cortex until it reaches the peak at 6 month to 1 year of age (Huttenlocher, 1979; Huttenlocher, de Courten, Garey, & Van der Loos, 1982), when human astrocytes begin to express high levels of the phagocytic receptor, Mertk, important for synapse elimination (Chung et al., 2013; Zhang, Sloan, et al., 2016). Afterward, synapse density gradually decreases (Huttenlocher, 1979; Huttenlocher et al., 1982). The previously reported roles of astrocytes in synapse formation and elimination (Chung et al., 2013; Pfrieger & Barres, 1997; Ullian et al., 2001), and the precise temporal correlation between astrocyte maturation and synapse density raise the hypothesis that astrocyte maturation may trigger neural circuit maturation. The first year of infancy is a vulnerable stage for neurodevelopmental disorders such as autism spectrum disorders (Wan et al., 2013). Understanding the mechanisms that regulate astrocyte maturation is an important step in the investigation of the contribution of astrocyte to neural circuit development and neurodevelopmental disorders. Interestingly, we recently found that 74 autism‐associated genes are enriched in astrocytes compared to neurons, microglia, oligodendrocytes, and endothelial cells in human brains (Zhang, Sloan, et al., 2016), supporting the hypothesis that astrocyte development defects may contribute to autism.
In this study, we made the following discoveries on the mechanisms that regulate astrocyte maturation: (a) astrocyte‐to‐astrocyte contact promotes astrocyte maturation; (b) a positive feedback loop of HBEGF‐EGFR signaling regulates astrocyte maturation; (c) the tumor‐associated genes TP53 promotes astrocyte maturation; and (d) microglia, OPCs, and endothelial cells affect the expression of a subset of genes during astrocyte development. Therefore, a range of cell autonomous and nonautonomous mechanisms cooperate in regulating the timing of astrocyte maturation.
Astrocytes occupy nonoverlapping territories in the central nervous system and exhibit the “tiling” phenomenon (Bushong et al., 2002). Astrocytes are distributed throughout every brain region. Such tiling phenomenon is conserved in evolution from fruit flies to mammals (Bushong et al., 2002; Oberheim et al., 2009; Stork, Sheehan, Tasdemir‐Yilmaz, & Freeman, 2014). How do astrocytes detect the presence of neighboring astrocytes and how do they determine their locations in developing brains? Immature astrocytes gradually decrease their proliferation rates as they mature (Ge et al., 2012; Nixdorf‐Bergweiler et al., 1994). We found that astrocyte‐to‐astrocyte contact inhibits proliferation and promotes maturation. Therefore, once proliferating immature astrocytes reach the density that allows neighboring astrocytes to make contacts, they would stop proliferation and ensure that neighboring astrocytes occupy nonoverlapping territories. The molecule(s) that mediates astrocyte‐to‐astrocyte contact induced maturation remains unknown. Interestingly, astrocytes express some of the molecules implicated in dendritic tiling (Stk38, the mammalian homolog of Trc in Drosophila melanogaster; Emoto, Parrish, Jan, & Jan, 2006) and the mosaic spacing of retinal amacrine cells and horizontal cells (Megf10) (Cahoy et al., 2008; Chung et al., 2013; Kay, Chu, & Sanes, 2012; Zhang et al., 2014; Zhang, Sloan, et al., 2016). Future studies testing the involvement of these molecules in astrocyte tiling may be fruitful. Although glioblastoma cells share many molecular markers with immature astrocytes, these tumor cells do not stop proliferation upon contact with neighboring cells. Identification of the molecules involved in astrocyte tiling and how glioblastoma cells override these molecular signals may contribute to the understanding and treatment of glioblastoma.
Developing astrocytes are in close contact with other cell types, such as neurons, microglia, OPCs, and endothelial cells. How communications between different cell types affect neural development is not fully understood. Previously, studies showed that neurons and neural activities induce morphological maturation and gene expression of astrocytes (Duan, Anderson, Stein, & Swanson, 1999; Koulakoff, Ezan, & Giaume, 2008; Morel et al., 2014; Schlag et al., 1998; Swanson et al., 1997; Yang et al., 2009). In this study, we tested the effect of microglia, OPCs, and endothelial cells on astrocyte maturation. Interestingly, we found that microglia and OPC affect astrocyte proliferation, whereas endothelial cells strongly promote the expression of the water channel gene Aqp4 in immature astrocytes. These results support the hypothesis that communications between cell types affect neural development. In the future, it will be important to systematically investigate the effect of microglia, OPC, and endothelial cell derived signals on astrocyte development in healthy and diseased brains. For example, concussion to developing brains may induce microglia activation. How would activated versus resting microglia differ in their effect on astrocyte development? Our discoveries of the effect of microglia, OPCs, and endothelial cells on astrocyte gene expression are some of the first steps in understanding the roles of signaling across cell types in astrocyte development. The regulation of astrocyte maturation in health and disease is a complex process. Clearly, other factors and pathways that regulate astrocyte maturation are yet to be defined.
In summary, we discovered novel cell autonomous and nonautonomous mechanisms that regulate astrocyte maturation. These findings add to our understanding of brain development in health and disease.
ACKNOWLEDGMENTS
The authors thank the UCLA Janis V. Giorgi Flow Cytometry Core Laboratory, Integrated Molecular Technology Core, and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, University of California Los Angeles BioSequencing Core Facility for their services, Mahnaz Akhavan and Suhua Feng for their technical support. This work was supported by the NIH/NIMH T32MH073526 and Achievement Rewards for College Scientists Foundation Los Angeles Founder Chapter to M.C.K. The NIH/NINDS R00NS089780, R01NS109025, National Center for Advancing Translational Sciences UCLA CTSI Grant UL1TR001881, UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Innovation Award, and the Friends of the Semel Institute for Neuroscience & Human Behavior Friends Scholar Award to Y.Z.
Li J, Khankan RR, Caneda C, et al. Astrocyte‐to‐astrocyte contact and a positive feedback loop of growth factor signaling regulate astrocyte maturation. Glia. 2019;67:1571–1597. 10.1002/glia.23630
Jiwen Li and Rana R. Khankan authors contributed equally to this article.
Funding information Achievement Rewards for College Scientists Foundation Los Angeles Founder Chapter; National Center for Advancing Translational Sciences UCLA CTSI Grant, Grant/Award Number: UL1TR001881; National Institute of Mental Health, Grant/Award Number: T32MH073526; National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: R00NS089780, R01NS109025; the Friends of the Semel Institute for Neuroscience & Human Behavior Friends Scholar Award; UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Innovation Award
REFERENCES
- Allen, N. J. (2014). Astrocyte regulation of synaptic behavior. Annual Review of Cell and Developmental Biology, 30, 439–463 Available from http://www.ncbi.nlm.nih.gov/pubmed/25288116 [DOI] [PubMed] [Google Scholar]
- Allen, N. J. , Bennett, M. L. , Foo, L. C. , Wang, G. X. , Chakraborty, C. , Smith, S. J. , & Barres, B. A. (2012). Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature, 486, 410–414 Available from http://www.nature.com/doifinder/10.1038/nature11059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, N. J. , & Eroglu, C. (2017). Cell biology of astrocyte‐synapse interactions. Neuron, 96, 697–708 Available from http://www.ncbi.nlm.nih.gov/pubmed/29096081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez‐Buylla, A. , & Ihrie, R. A. (2014). Sonic hedgehog signaling in the postnatal brain. Seminars in Cell & Developmental Biology, 33, 105–111 Available from http://www.ncbi.nlm.nih.gov/pubmed/24862855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq––A python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169 Available from https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. A. , Burda, J. E. , Ren, Y. , Ao, Y. , O'Shea, T. M. , Kawaguchi, R. , … Sofroniew, M. V. (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature, 532, 195–200 Available from http://www.ncbi.nlm.nih.gov/pubmed/27027288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik, A. , Genové, G. , Mäe, M. , Nisancioglu, M. H. , Wallgard, E. , Niaudet, C. , … Betsholtz, C. (2010). Pericytes regulate the blood–brain barrier. Nature, 468, 557–561 Available from http://www.nature.com/doifinder/10.1038/nature09522 [DOI] [PubMed] [Google Scholar]
- Ballas, N. , Lioy, D. T. , Grunseich, C. , & Mandel, G. (2009). Non‐cell autonomous influence of MeCP2‐deficient glia on neuronal dendritic morphology. Nature Neuroscience, 12, 311–317 Available from http://www.ncbi.nlm.nih.gov/pubmed/19234456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker, G. A. (1980). Trophic interactions between astroglial cells and hippocampal neurons in culture. Science, 209, 809–810 Available from http://www.ncbi.nlm.nih.gov/pubmed/7403847 [DOI] [PubMed] [Google Scholar]
- Barres, B. A. , Lazar, M. A. , & Raff, M. C. (1994). A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development, 120, 1097–1108 Available from http://www.ncbi.nlm.nih.gov/pubmed/8026323 [DOI] [PubMed] [Google Scholar]
- Batista‐Brito, R. , & Fishell, G. (2009). Chapter 3 the developmental integration of cortical interneurons into a functional network. Current Topics in Developmental Biology, 87, 81–118 Available from http://www.ncbi.nlm.nih.gov/pubmed/19427517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benraiss, A. , Wang, S. , Herrlinger, S. , Li, X. , Chandler‐Militello, D. , Mauceri, J. , … Goldman, S. A. (2016). Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nature Communications, 7, 11758 Available from http://www.nature.com/doifinder/10.1038/ncomms11758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco‐Suarez, E. , Liu, T.‐F. , Kopelevich, A. , & Allen, N. J. (2018). Astrocyte‐secreted Chordin‐like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron, 100, 1116–1132.e13 Available from http://www.ncbi.nlm.nih.gov/pubmed/30344043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen, C. J. , Bennett, F. C. , & Bennett, M. L. (2018). Isolation and culture of microglia. Current Protocols in Immunology, e70 Available from http://doi.wiley.com/10.1002/cpim.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen, C. J. , Bennett, F. C. , Tucker, A. F. , Collins, H. Y. , Mulinyawe, S. B. , & Barres, B. A. (2017). Diverse requirements for microglial survival, specification, and function revealed by defined‐medium cultures. Neuron, 94, 759–773.e8 Available from http://www.ncbi.nlm.nih.gov/pubmed/28521131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni, A. , Sun, Y. , Nadal‐Vicens, M. , Bhatt, A. , Frank, D. A. , Rozovsky, I. , … Greenberg, M. E. (1997). Regulation of gliogenesis in the central nervous system by the JAK‐STAT signaling pathway. Science, 278, 477–483 Available from http://www.ncbi.nlm.nih.gov/pubmed/9334309 [DOI] [PubMed] [Google Scholar]
- Brennan, C. W. , Verhaak, R. G. W. , McKenna, A. , Campos, B. , Noushmehr, H. , Salama, S. R. , et al. (2013). The somatic genomic landscape of Glioblastoma. Cell, 155, 462–477 Available from http://www.ncbi.nlm.nih.gov/pubmed/24120142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, T. G. , Puvanachandra, N. , Horner, C. H. , Polito, A. , Ostenfeld, T. , Svendsen, C. N. , … Sofroniew, M. V. (1999). Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar‐forming, reactive astrocytes in adult transgenic mice. Neuron, 23, 297–308 Available from http://www.ncbi.nlm.nih.gov/pubmed/10399936 [DOI] [PubMed] [Google Scholar]
- Bushong, E. A. , Martone, M. E. , & Ellisman, M. H. (2004). Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. International Journal of Developmental Neuroscience, 22, 73–86 Available from http://www.ncbi.nlm.nih.gov/pubmed/15036382 [DOI] [PubMed] [Google Scholar]
- Bushong, E. A. , Martone, M. E. , Jones, Y. Z. , Ellisman, M. H. , Lin, J. H. C. , Wang, F. , … Nedergaard, M. (2002). Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. The Journal of Neuroscience, 22, 183–192 Available from http://www.ncbi.nlm.nih.gov/pubmed/11756501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy, J. D. , Emery, B. , Kaushal, A. , Foo, L. C. , Zamanian, J. L. , Christopherson, K. S. , … Barres, B. A. (2008). A Transcriptome database for astrocytes, neurons, and Oligodendrocytes: A new resource for understanding brain development and function. The Journal of Neuroscience, 28, 264–278 Available from http://www.ncbi.nlm.nih.gov/pubmed/18171944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmignoto, G. , & Haydon, P. G. (2012). Astrocyte calcium signaling and epilepsy. Glia, 60, 1227–1233 Available from http://www.ncbi.nlm.nih.gov/pubmed/22389222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub, L. S. , Manalo, J. M. , Lee, H. K. , Glasgow, S. M. , Chen, F. , Kawasaki, Y. , … Deneen, B. (2016). Temporal profiling of astrocyte precursors reveals parallel roles for Asef during development and after injury. The Journal of Neuroscience, 36, 11904–11917 Available from http://www.ncbi.nlm.nih.gov/pubmed/27881777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, H. , Diaz‐Castro, B. , Shigetomi, E. , Monte, E. , Octeau, J. C. , Yu, X. , … Khakh, B. S. (2017). Neural circuit‐specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron, 95, 531–549.e9 Available from http://www.ncbi.nlm.nih.gov/pubmed/28712653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. , Lau, S. K. M. , & Doering, L. C. (2016). Astrocyte‐secreted thrombospondin‐1 modulates synapse and spine defects in the fragile X mouse model. Molecular Brain, 9, 74 Available from http://www.ncbi.nlm.nih.gov/pubmed/27485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson, K. S. , Ullian, E. M. , Stokes, C. C. A. , Mullowney, C. E. , Hell, J. W. , Agah, A. , … Barres, B. A. (2005). Thrombospondins are astrocyte‐secreted proteins that promote CNS synaptogenesis. Cell, 120, 421–433 Available from http://www.ncbi.nlm.nih.gov/pubmed/15707899 [DOI] [PubMed] [Google Scholar]
- Chung, W.‐S. , Allen, N. J. , & Eroglu, C. (2015). Astrocytes control synapse formation, function, and elimination. Cold Spring Harbor Perspectives in Biology, 7, a020370 Available from http://www.ncbi.nlm.nih.gov/pubmed/25663667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W.‐S. , Clarke, L. E. , Wang, G. X. , Stafford, B. K. , Sher, A. , Chakraborty, C. , … Barres, B. A. (2013). Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature, 504, 394–400 Available from http://www.ncbi.nlm.nih.gov/pubmed/24270812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter, D. A. , & Steinhauser, C. (2015). Role of astrocytes in epilepsy. Cold Spring Harbor Perspectives in Biology, 5, a022434–a022434 Available from http://www.ncbi.nlm.nih.gov/pubmed/25732035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen, B. , Ho, R. , Lukaszewicz, A. , Hochstim, C. J. , Gronostajski, R. M. , & Anderson, D. J. (2006). The transcription factor NFIA controls the onset of Gliogenesis in the developing spinal cord. Neuron, 52, 953–968 Available from http://www.ncbi.nlm.nih.gov/pubmed/17178400 [DOI] [PubMed] [Google Scholar]
- Ding, S. , Fellin, T. , Zhu, Y. , Lee, S.‐Y. , Auberson, Y. P. , Meaney, D. F. , … Haydon, P. G. (2007). Enhanced Astrocytic Ca2+ signals contribute to neuronal Excitotoxicity after status Epilepticus. The Journal of Neuroscience, 27, 10674–10684 Available from http://www.ncbi.nlm.nih.gov/pubmed/17913901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , … Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21 Available from https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, S. , Anderson, C. M. , Stein, B. A. , & Swanson, R. A. (1999). Glutamate induces rapid upregulation of astrocyte glutamate transport and cell‐surface expression of GLAST. The Journal of Neuroscience, 19, 10193–10200 Available from http://www.ncbi.nlm.nih.gov/pubmed/10575016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy, C. , & Sehgal, A. (2017). Circadian rhythms and sleep in Drosophila melanogaster . Genetics, 205, 1373–1397 Available from http://www.ncbi.nlm.nih.gov/pubmed/28360128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas, J. C. , Ibrahim, A. , & Barres, B. A. (2007). A crucial role for p57Kip2 in the intracellular timer that controls Oligodendrocyte differentiation. The Journal of Neuroscience, 27, 6185–6196 Available from http://www.ncbi.nlm.nih.gov/pubmed/17553990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, B. , Agalliu, D. , Cahoy, J. D. , Watkins, T. A. , Dugas, J. C. , Mulinyawe, S. B. , … Barres, B. A. (2009). Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell, 138, 172–185 Available from http://www.ncbi.nlm.nih.gov/pubmed/19596243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, K. , Parrish, J. Z. , Jan, L. Y. , & Jan, Y.‐N. (2006). The tumour suppressor hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature, 443, 210–213 Available from http://www.ncbi.nlm.nih.gov/pubmed/16906135 [DOI] [PubMed] [Google Scholar]
- Eroglu, Ç. , Allen, N. J. , Susman, M. W. , O'Rourke, N. A. , Park, C. Y. , Özkan, E. , … Barres, B. A. (2009). Gabapentin receptor α2δ‐1 is a neuronal Thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell, 139, 380–392 Available from http://www.ncbi.nlm.nih.gov/pubmed/19818485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy‐Tselnicker, I. , van Casteren, A. C. M. , Lee, A. , Chang, V. T. , Aricescu, A. R. , & Allen, N. J. (2017). Astrocyte‐secreted Glypican 4 regulates release of neuronal Pentraxin 1 from axons to induce functional synapse formation. Neuron, 96, 428–445.e13 Available from http://www.ncbi.nlm.nih.gov/pubmed/29024665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, W. T. , Abrahamsson, T. , Chierzi, S. , Lui, C. , Zaelzer, C. , Jones, E. V. , … Murai, K. K. (2016). Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science (80‐), 351, 849–854 Available from http://www.ncbi.nlm.nih.gov/pubmed/26912893 [DOI] [PubMed] [Google Scholar]
- Foo, L. C. , Allen, N. J. , Bushong, E. A. , Ventura, P. B. , Chung, W.‐S. , Zhou, L. , … Barres, B. A. (2011). Development of a method for the purification and culture of rodent astrocytes. Neuron, 71, 799–811 Available from http://linkinghub.elsevier.com/retrieve/pii/S0896627311006490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, M. R. (2010). Specification and morphogenesis of astrocytes. Science, 330, 774–778 Available from http://www.ncbi.nlm.nih.gov/pubmed/21051628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo, V. , & Deneen, B. (2014). Glial development: The crossroads of regeneration and repair in the CNS. Neuron, 83, 283–308 Available from http://www.ncbi.nlm.nih.gov/pubmed/25033178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, W.‐P. , Miyawaki, A. , Gage, F. H. , Jan, Y. N. , & Jan, L. Y. (2012). Local generation of glia is a major astrocyte source in postnatal cortex. Nature, 484, 376–380 Available from http://www.ncbi.nlm.nih.gov/pubmed/22456708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisert, E. E. , & Stewart, A. M. (1991). Changing interactions between astrocytes and neurons during CNS maturation. Developmental Biology, 143, 335–345 Available from http://www.ncbi.nlm.nih.gov/pubmed/1991556 [DOI] [PubMed] [Google Scholar]
- Glasgow, S. M. , Carlson, J. C. , Zhu, W. , Chaboub, L. S. , Kang, P. , Lee, H. K. , … Deneen, B. (2017). Glia‐specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nature Neuroscience, 20, 1520–1528 Available from http://www.ncbi.nlm.nih.gov/pubmed/28892058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon, P. G. , & Nedergaard, M. (2015). How do astrocytes participate in neural plasticity? Cold Spring Harbor Perspectives in Biology, 7, a020438 Available from http://www.ncbi.nlm.nih.gov/pubmed/25502516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, J. E. , Imura, T. , Song, B. , Qi, J. , Ao, Y. , Nguyen, T. K. , … Sofroniew, M. V. (2008). STAT3 is a critical regulator of Astrogliosis and scar formation after spinal cord injury. The Journal of Neuroscience, 28, 7231–7243 Available from http://www.ncbi.nlm.nih.gov/pubmed/18614693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstim, C. , Deneen, B. , Lukaszewicz, A. , Zhou, Q. , & Anderson, D. J. (2008). Identification of positionally distinct astrocyte subtypes whose identities are specified by a Homeodomain code. Cell, 133, 510–522 Available from http://www.ncbi.nlm.nih.gov/pubmed/18455991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4, 44–57 Available from http://www.nature.com/articles/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huttenlocher, P. R. (1979). Synaptic density in human frontal cortex—Developmental changes and effects of aging. Brain Research, 163, 195–205 Available from http://www.ncbi.nlm.nih.gov/pubmed/427544 [DOI] [PubMed] [Google Scholar]
- Huttenlocher, P. R. , de Courten, C. , Garey, L. J. , & Van der Loos, H. (1982). Synaptogenesis in human visual cortex‐‐evidence for synapse elimination during normal development. Neuroscience Letters, 33, 247–252 Available from http://www.ncbi.nlm.nih.gov/pubmed/7162689 [DOI] [PubMed] [Google Scholar]
- Itagaki, S. , McGeer, P. L. , Akiyama, H. , Zhu, S. , & Selkoe, D. (1989). Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. Journal of Neuroimmunology, 24, 173–182 Available from http://www.ncbi.nlm.nih.gov/pubmed/2808689 [DOI] [PubMed] [Google Scholar]
- Jacobs, S. , Nathwani, M. , & Doering, L. C. (2010). Fragile X astrocytes induce developmental delays in dendrite maturation and synaptic protein expression. BMC Neuroscience, 11, 132 Available from http://www.ncbi.nlm.nih.gov/pubmed/20955577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin, C.‐C. , Yu, K. , Hatcher, A. , Huang, T.‐W. , Lee, H. K. , Carlson, J. , … Deneen, B. (2017). Identification of diverse astrocyte populations and their malignant analogs. Nature Neuroscience, 20, 396–405 Available from http://www.ncbi.nlm.nih.gov/pubmed/28166219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P. , Lee, H. K. , Glasgow, S. M. , Finley, M. , Donti, T. , Gaber, Z. B. , … Deneen, B. (2012). Sox9 and NFIA coordinate a transcriptional regulatory Cascade during the initiation of Gliogenesis. Neuron, 74, 79–94 Available from http://www.ncbi.nlm.nih.gov/pubmed/22500632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, J. N. , Chu, M. W. , & Sanes, J. R. (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature, 483, 465–469 Available from http://www.nature.com/articles/nature10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh, B. S. , & McCarthy, K. D. (2015). Astrocyte calcium signaling: From observations to functions and the challenges therein. Cold Spring Harbor Perspectives in Biology, 7, a020404 Available from http://www.ncbi.nlm.nih.gov/pubmed/25605709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulakoff, A. , Ezan, P. , & Giaume, C. (2008). Neurons control the expression of connexin 30 and connexin 43 in mouse cortical astrocytes. Glia, 56, 1299–1311 Available from http://www.ncbi.nlm.nih.gov/pubmed/18512249 [DOI] [PubMed] [Google Scholar]
- Kucukdereli, H. , Allen, N. J. , Lee, A. T. , Feng, A. , Ozlu, M. I. , Conatser, L. M. , … Eroglu, C. (2011). Control of excitatory CNS synaptogenesis by astrocyte‐secreted proteins Hevin and SPARC. Proceedings of the National Academy of Sciences, 108, E440–E449 Available from http://www.ncbi.nlm.nih.gov/pubmed/21788491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. L. , Martinez‐Lozada, Z. , Krizman, E. N. , & Robinson, M. B. (2017). Brain endothelial cells induce astrocytic expression of the glutamate transporter GLT‐1 by a notch‐dependent mechanism. Journal of Neurochemistry, 143, 489–506 Available from http://www.ncbi.nlm.nih.gov/pubmed/28771710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein, E. S. , Hawrylycz, M. J. , Ao, N. , Ayres, M. , Bensinger, A. , Bernard, A. , et al. (2007). Genome‐wide atlas of gene expression in the adult mouse brain. Nature, 445, 168–176 Available from http://www.nature.com/articles/nature05453 [DOI] [PubMed] [Google Scholar]
- Lennon, V. A. , Kryzer, T. J. , Pittock, S. J. , Verkman, A. S. , & Hinson, S. R. (2005). IgG marker of optic‐spinal multiple sclerosis binds to the aquaporin‐4 water channel. The Journal of Experimental Medicine, 202, 473–477 Available from http://www.jem.org/lookup/doi/10.1084/jem.20050304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Ma, Y.‐M. , Zheng, P.‐S. , & Zhang, P. (2018). GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. Journal of Experimental & Clinical Cancer Research, 37, 80 Available from http://www.ncbi.nlm.nih.gov/pubmed/29636108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow, S. A. , Guttenplan, K. A. , Clarke, L. E. , Bennett, F. C. , Bohlen, C. J. , Schirmer, L. , … Barres, B. A. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541, 481–487 Available from http://www.nature.com/articles/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy, D. T. , Garg, S. K. , Monaghan, C. E. , Raber, J. , Foust, K. D. , Kaspar, B. K. , … Mandel, G. (2011). A role for glia in the progression of Rett's syndrome. Nature, 475, 497–500 Available from http://www.ncbi.nlm.nih.gov/pubmed/21716289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano, J. , Bernal, J. , & Morte, B. (2007). Influence of thyroid hormones on maturation of rat cerebellar astrocytes. International Journal of Developmental Neuroscience, 25, 171–179 Available from http://linkinghub.elsevier.com/retrieve/pii/S0736574807000196 [DOI] [PubMed] [Google Scholar]
- Maragakis, N. J. , & Rothstein, J. D. (2004). Glutamate transporters: Animal models to neurologic disease. Neurobiology of Disease, 15, 461–473 Available from http://www.ncbi.nlm.nih.gov/pubmed/15056453 [DOI] [PubMed] [Google Scholar]
- McCarthy, K. D. , & de Vellis, J. (1980). Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. The Journal of Cell Biology, 85, 890–902 Available from http://www.ncbi.nlm.nih.gov/pubmed/6248568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Haeberle, H. , & Barres, B. A. (2001). Induction of astrocyte differentiation by endothelial cells. The Journal of Neuroscience, 21, 1538–1547 Available from http://www.ncbi.nlm.nih.gov/pubmed/11222644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A. V. , & Deneen, B. (2015). Astrocyte development: A guide for the perplexed. Glia, 63, 1320–1329 Available from http://www.ncbi.nlm.nih.gov/pubmed/25963996 [DOI] [PubMed] [Google Scholar]
- Molofsky, A. V. , Glasgow, S. M. , Chaboub, L. S. , Tsai, H.‐H. , Murnen, A. T. , Kelley, K. W. , … Oldham, M. C. (2013). Expression profiling of Aldh1l1‐precursors in the developing spinal cord reveals glial lineage‐specific genes and direct Sox9‐Nfe2l1 interactions. Glia, 61, 1518–1532 Available from http://www.ncbi.nlm.nih.gov/pubmed/23840004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A. V. , Kelley, K. W. , Tsai, H.‐H. , Redmond, S. A. , Chang, S. M. , Madireddy, L. , … Rowitch, D. H. (2014). Astrocyte‐encoded positional cues maintain sensorimotor circuit integrity. Nature, 509, 189–194 Available from http://www.ncbi.nlm.nih.gov/pubmed/24776795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A. V. , Krencik, R. , Krenick, R. , Ullian, E. M. , Ullian, E. , Tsai, H. , … Rowitch, D. H. (2012). Astrocytes and disease: A neurodevelopmental perspective. Genes & Development, 26, 891–907 Available from http://genesdev.cshlp.org/cgi/doi/10.1101/gad.188326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, L. , Higashimori, H. , Tolman, M. , & Yang, Y. (2014). VGluT1+ neuronal Glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic Astroglia. The Journal of Neuroscience, 34, 10950–10962 Available from http://www.ncbi.nlm.nih.gov/pubmed/25122895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama, Y. , Fujiwara, Y. , Orkin, S. H. , & Rowitch, D. H. (2005). Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature, 438, 360–363 Available from http://www.ncbi.nlm.nih.gov/pubmed/16292311 [DOI] [PubMed] [Google Scholar]
- Nedergaard, M. (1994). Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science, 263, 1768–1771 Available from http://www.ncbi.nlm.nih.gov/pubmed/8134839 [DOI] [PubMed] [Google Scholar]
- Nixdorf‐Bergweiler, B. E. , Albrecht, D. , & Heinemann, U. (1994). Developmental changes in the number, size, and orientation of GFAP‐positive cells in the CA1 region of rat hippocampus. Glia, 12, 180–195 Available from http://doi.wiley.com/10.1002/glia.440120304 [DOI] [PubMed] [Google Scholar]
- Oberheim, N. A. , Takano, T. , Han, X. , He, W. , Lin, J. H. C. , Wang, F. , … Nedergaard, M. (2009). Uniquely hominid features of adult human astrocytes. The Journal of Neuroscience, 29, 3276–3287 Available from http://www.ncbi.nlm.nih.gov/pubmed/19279265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos, M. C. , & Verkman, A. S. (2013). Aquaporin water channels in the nervous system. Nature Reviews Neuroscience, 14, 265–277 Available from http://www.ncbi.nlm.nih.gov/pubmed/23481483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger, F. W. , & Barres, B. A. (1997). Synaptic efficacy enhanced by glial cells in vitro. Science, 277, 1684–1687 Available from http://www.ncbi.nlm.nih.gov/pubmed/9287225 [DOI] [PubMed] [Google Scholar]
- RNA sequencing data . RNA sequencing data is uploaded to Gene Expression Omnibus under accession GSE125610. 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125610.
- Roskoski, R. (2014). The ErbB/HER family of protein‐tyrosine kinases and cancer. Pharmacological Research, 79, 34–74 Available from http://www.ncbi.nlm.nih.gov/pubmed/24269963 [DOI] [PubMed] [Google Scholar]
- Rothstein, J. D. , Martin, L. , Levey, A. I. , Dykes‐Hoberg, M. , Jin, L. , Wu, D. , … Kuncl, R. W. (1994). Localization of neuronal and glial glutamate transporters. Neuron, 13, 713–725 Available from http://www.ncbi.nlm.nih.gov/pubmed/7917301 [DOI] [PubMed] [Google Scholar]
- Rudge, J. S. , & Silver, J. (1990). Inhibition of neurite outgrowth on astroglial scars in vitro. The Journal of Neuroscience, 10, 3594–3603 Available from http://www.ncbi.nlm.nih.gov/pubmed/2230948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag, B. D. , Vondrasek, J. R. , Munir, M. , Kalandadze, A. , Zelenaia, O. A. , Rothstein, J. D. , & Robinson, M. B. (1998). Regulation of the glial Na+−dependent glutamate transporters by cyclic AMP analogs and neurons. Molecular Pharmacology, 53, 355–369 Available from http://www.ncbi.nlm.nih.gov/pubmed/9495799 [DOI] [PubMed] [Google Scholar]
- Schmechel, D. E. , & Rakic, P. (1979). A golgi study of radial glial cells in developing monkey telencephalon: Morphogenesis and transformation into astrocytes. Anatomy and Embryology, 156, 115–152 Available from http://link.springer.com/10.1007/BF00300010 [DOI] [PubMed] [Google Scholar]
- Scholze, A. R. , Foo, L. C. , Mulinyawe, S. , & Barres, B. A. (2014). BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One, 9, e110668 Available from http://dx.plos.org/10.1371/journal.pone.0110668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. K. , Stogsdill, J. A. , Pulimood, N. S. , Dingsdale, H. , Kim, Y. H. , Pilaz, L.‐J. , … Eroglu, C. (2016). Astrocytes assemble Thalamocortical synapses by bridging NRX1α and NL1 via Hevin. Cell, 164, 183–196 Available from http://www.ncbi.nlm.nih.gov/pubmed/26771491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. M. , Miller, R. H. , & Silver, J. (1986). Changing role of forebrain astrocytes during development, regenerative failure, and induced regeneration upon transplantation. The Journal of Comparative Neurology, 251, 23–43 Available from http://doi.wiley.com/10.1002/cne.902510103 [DOI] [PubMed] [Google Scholar]
- Smith, G. M. , Rutishauser, U. , Silver, J. , & Miller, R. H. (1990). Maturation of astrocytes in vitro alters the extent and molecular basis of neurite outgrowth. Developmental Biology, 138, 377–390 Available from http://www.ncbi.nlm.nih.gov/pubmed/2318341 [DOI] [PubMed] [Google Scholar]
- Sofroniew, M. V. (2014). Astrogliosis. Cold Spring Harbor Perspectives in Biology, 7, a020420 Available from http://www.ncbi.nlm.nih.gov/pubmed/25380660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill, J. A. , Ramirez, J. , Liu, D. , Kim, Y. H. , Baldwin, K. T. , Enustun, E. , … Eroglu, C. (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature, 551, 192–197 Available from http://www.ncbi.nlm.nih.gov/pubmed/29120426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork, T. , Sheehan, A. , Tasdemir‐Yilmaz, O. E. , & Freeman, M. R. (2014). Neuron‐glia interactions through the heartless FGF receptor signaling pathway mediate morphogenesis of drosophila astrocytes. Neuron, 83, 388–403 Available from http://www.ncbi.nlm.nih.gov/pubmed/25033182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R. A. , Liu, J. , Miller, J. W. , Rothstein, J. D. , Farrell, K. , Stein, B. A. , & Longuemare, M. C. (1997). Neuronal regulation of glutamate transporter subtype expression in astrocytes. The Journal of Neuroscience, 17, 932–940 Available from http://www.ncbi.nlm.nih.gov/pubmed/8994048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, T. , Tian, G.‐F. , Peng, W. , Lou, N. , Libionka, W. , Han, X. , & Nedergaard, M. (2006). Astrocyte‐mediated control of cerebral blood flow. Nature Neuroscience, 9, 260–267 Available from http://www.ncbi.nlm.nih.gov/pubmed/16388306 [DOI] [PubMed] [Google Scholar]
- Tian, G.‐F. , Azmi, H. , Takano, T. , Xu, Q. , Peng, W. , Lin, J. , … Nedergaard, M. (2005). An astrocytic basis of epilepsy. Nature Medicine, 11, 973–981 Available from http://www.ncbi.nlm.nih.gov/pubmed/16116433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, A.‐C. , Tsai, H.‐H. , Molofsky, A. V. , McMahon, M. , Foo, L. C. , Kaul, A. , … Rowitch, D. H. (2012). Regulated temporal‐spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development, 139, 2477–2487 Available from http://www.ncbi.nlm.nih.gov/pubmed/22675209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, N. , Pataskar, A. , Péron, S. , Thakurela, S. , Sahu, S. K. , Figueres‐Oñate, M. , … Berninger, B. (2018). Stage‐specific transcription factors drive Astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell, 23, 557–571.e8 Available from http://www.ncbi.nlm.nih.gov/pubmed/30290178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, X. , Ao, Y. , Faas, G. C. , Nwaobi, S. E. , Xu, J. , Haustein, M. D. , … Khakh, B. S. (2014). Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington's disease model mice. Nature Neuroscience, 17, 694–703 Available from http://www.ncbi.nlm.nih.gov/pubmed/24686787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.‐H. , Li, H. , Fuentealba, L. C. , Molofsky, A. V. , Taveira‐Marques, R. , Zhuang, H. , … Rowitch, D. H. (2012). Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science, 337, 358–362 Available from http://www.ncbi.nlm.nih.gov/pubmed/22745251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S.‐C. , Lin, S.‐J. , Lin, C.‐J. , Chou, Y.‐C. , Lin, J.‐H. , Yeh, T.‐H. , … Tsai, C.‐H. (2013). Autocrine CCL3 and CCL4 induced by the oncoprotein LMP1 promote Epstein‐Barr virus‐triggered B cell proliferation. Journal of Virology, 87, 9041–9052 Available from http://www.ncbi.nlm.nih.gov/pubmed/23760235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann, H. , Blaszkiewicz, S. , Nicolae, C. , Haas, S. , Schnell, A. , Wurzer, S. , … Essers, M. A. G. (2016). Extracellular matrix protein Matrilin‐4 regulates stress‐induced HSC proliferation via CXCR4. The Journal of Experimental Medicine, 213, 1961–1971 Available from http://www.jem.org/lookup/doi/10.1084/jem.20151713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian, E. M. , Sapperstein, S. K. , Christopherson, K. S. , & Barres, B. A. (2001). Control of synapse number by glia. Science, 291, 657–661 Available from http://www.ncbi.nlm.nih.gov/pubmed/11158678 [DOI] [PubMed] [Google Scholar]
- Vainchtein, I. D. , Chin, G. , Cho, F. S. , Kelley, K. W. , Miller, J. G. , Chien, E. C. , … Molofsky, A. V. (2018). Astrocyte‐derived interleukin‐33 promotes microglial synapse engulfment and neural circuit development. Science, 359, 1269–1273 Available from http://www.ncbi.nlm.nih.gov/pubmed/29420261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak, R. G. W. , Hoadley, K. A. , Purdom, E. , Wang, V. , Qi, Y. , Wilkerson, M. D. , … Hayes, D. N. (2010). Integrated genomic analysis identifies clinically relevant subtypes of Glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 17, 98–110 Available from https://www.sciencedirect.com/science/article/pii/S1535610809004322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij, N. , Roberts, L. , Joyce, S. , & Chakravarti, S. (2004). Lumican suppresses cell proliferation and aids Fas‐Fas ligand mediated apoptosis: Implications in the cornea. Experimental Eye Research, 78, 957–971 Available from http://linkinghub.elsevier.com/retrieve/pii/S0014483504000028 [DOI] [PubMed] [Google Scholar]
- Wan, M. W. , Green, J. , Elsabbagh, M. , Johnson, M. , Charman, T. , & Plummer, F. (2013). Quality of interaction between at‐risk infants and caregiver at 12‐15 months is associated with 3‐year autism outcome. Journal of Child Psychology and Psychiatry, 54, 763–771 Available from http://doi.wiley.com/10.1111/jcpp.12032 [DOI] [PubMed] [Google Scholar]
- Xaus, J. , Comalada, M. , Cardó, M. , Valledor, A. F. , & Celada, A. (2001). Decorin inhibits macrophage colony‐stimulating factor proliferation of macrophages and enhances cell survival through induction of p27(Kip1) and p21(Waf1). Blood, 98, 2124–2133 Available from http://www.ncbi.nlm.nih.gov/pubmed/11567999 [DOI] [PubMed] [Google Scholar]
- Yamanaka, K. , Chun, S. J. , Boillee, S. , Fujimori‐Tonou, N. , Yamashita, H. , Gutmann, D. H. , … Cleveland, D. W. (2008). Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nature Neuroscience, 11, 251–253 Available from http://www.ncbi.nlm.nih.gov/pubmed/18246065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Gozen, O. , Watkins, A. , Lorenzini, I. , Lepore, A. , Gao, Y. , … Rothstein, J. D. (2009). Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron, 61, 880–894 Available from http://www.ncbi.nlm.nih.gov/pubmed/19323997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian, J. L. , Xu, L. , Foo, L. C. , Nouri, N. , Zhou, L. , Giffard, R. G. , & Barres, B. A. (2012). Genomic analysis of reactive astrogliosis. The Journal of Neuroscience, 32, 6391–6410 Available from http://www.ncbi.nlm.nih.gov/pubmed/22553043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , He, X. , Liu, L. , Jiang, M. , Zhao, C. , Wang, H. , … Lu, Q. R. (2016). Hdac3 interaction with p300 histone Acetyltransferase regulates the Oligodendrocyte and astrocyte lineage fate switch. Developmental Cell, 36, 316–330 Available from http://www.ncbi.nlm.nih.gov/pubmed/26859354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , & Barres, B. A. (2010). Astrocyte heterogeneity: An underappreciated topic in neurobiology. Current Opinion in Neurobiology, 20, 588–594 Available from https://linkinghub.elsevier.com/retrieve/pii/S0959438810001029 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Chen, K. , Sloan, S. A. , Bennett, M. L. , Scholze, A. R. , O'Keeffe, S. , … Wu, J. Q. (2014). An RNA‐sequencing Transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of Neuroscience, 34, 11929–11947 Available from http://www.ncbi.nlm.nih.gov/pubmed/25186741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Sloan, S. A. , Clarke, L. E. , Caneda, C. , Plaza, C. A. , Blumenthal, P. D. , … Barres, B. A. (2016). Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron, 89, 37–53 Available from http://www.ncbi.nlm.nih.gov/pubmed/26687838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Sohet, F. , & Daneman, R. (2014). Purification of endothelial cells from rodent brain by Immunopanning. Cold Spring Harbor Protocols, 2014, 65–77 pdb.prot074963. Available from http://www.ncbi.nlm.nih.gov/pubmed/24371317 [DOI] [PubMed] [Google Scholar]


