Editors summary
Single-molecule analyses of transient protein-protein interactions are enabled using an engineered biological nanopore.Protein-protein interactions (PPIs) are essential for many cellular processes. However, the transient nature of these PPIs is difficult to quantitate due to numerous limitations of available methods. We engineered a genetically-encoded sensor for real-time sampling of transient PPIs at single-molecule resolution. Our sensor comprises a truncated outer membrane protein pore, a flexible tether, a protein receptor and a peptide adapter. When a protein ligand present in solution binds to the receptor reversible capture and release events of the receptor can be measured as current transitions between two open substates of the pore. Importantly, the binding and release of the receptor by a protein ligand can be unambiguously discriminated in a complex sample containing fetal bovine serum. Our selective nanopore sensor could be applied for single-molecule protein detection, could form the basis for a nanoproteomics platform or might be adapted to build tools for protein profiling and biomarker discovery.
Physical associations between proteins (protein-protein interactions, PPIs) underpin cell functions in normal and pathogenic conditions and are important therapeutic targets1. Existing methods detect transient PPIs at high spatial resolution using single-molecule technologies2–4 but these methods are limited by low-throughput data acquisition. Conventional methods, such as biolayer interferometry (BLI)5 and surface plasmon resonance (SPR)6 have been used in the affinity, kinetic, and thermodynamic determinations of transient PPIs in bulk phase. Both are real-time techniques, which can be used at high-throughput. SPR is limited by nonspecific protein binding on surfaces, surface heterogeneity, protein inactivation at the liquid-metal film interface, and molecular crowding effects. Isothermal titration calorimetry (ITC)7 can report binding kinetics but is not high throughput and requires large quantities of proteins. BLI, SPR, and ITC provide average kinetic or affinity parameters and cannot be used in a heterogeneous sample.
Transient PPIs can be measured using a resistive-pulse technique8 and solid-state nanopores that are large enough to permit tethering of protein receptors on their internal surface.9, 10 However, accurate identification of the anchoring site for tethered protein receptors remains difficult, which limits the sensitivity of single-molecule measurements. An alternative approach to these shortcomings is the detection of reversible PPIs using an engineered protein nanopore. Protein nanopores have the advantage of being modifiable with atomic precision. Furthermore, sensing with engineered biological nanopores is amenable to integration into nanofluidic devices and high-throughput technologies11. Biological nanopores could theoretically couple molecular precision with a parallel format12–16.
Two requirements must be fulfilled to measure binding events between two folded proteins in solution using a protein nanopore. First, reversible PPIs must occur in the aqueous phase, because the dimensions of protein-protein complex exceed the cross-sectional internal diameter of protein nanopores. Therefore, these interactions can only be detected outside the nanopore lumen17–19. Second, a transducing mechanism is required to convert the reversible physical association and dissociation of the two interacting protein partners in the aqueous phase into a high-fidelity electrical signature of the nanopore sensor.
We report an engineered protein nanopore sensor that can detect reversible PPIs in solution and at single-tethered receptor resolution. Because the detection of binding events occurs outside the ‘voltage-drop region’ of the nanopore, we can apply our sensor to detect transient PPIs in mammalian serum. A single-polypeptide chain protein was created using t-FhuA20, a monomeric β-barrel scaffold that is a truncated version of ferric hydroxamate uptake component A (FhuA)21 from Escherichia coli (Fig. 1a1). A protein receptor (110-amino acid residue RNAse barnase (Bn)22 ) was fused to t-FhuA, on the β-turns side, with a flexible (GGS)2 tether (ONLINE METHODS; Supplementary Tables 1, 2). An H102A mutant of Bn was used, because this lacks ribonuclease activity22, 23 which in turn ensures that Bn-FhuA fusion proteins are not toxic to the expression host (ONLINE METHODS). The 455-residue t-FhuA scaffold formed a transmembrane pore,20 facilitating a quiet single-channel electrical signature for long periods when added to the cis side of the chamber (Supplementary Fig. 1). Both t-FhuA termini are accessible to the β-turns′ opening of the pore (Fig. 1a1). We tested two protein pore nanostructures, in which Bn was fused to either the N-terminus (Bn(GGS)2t-FhuA) or the C-terminus (t-FhuA(GGS)2Bn) of t-FhuA. The average single-channel conductance noted with t-FhuA was 1.6 ± 0.1 nS (n = 6) at a transmembrane potential of −40 mV and in 300 mM KCl, 10 mM Tris·HCl, pH 8 (Supplementary Fig. 1). Bn(GGS)2t-FhuA and t-FhuA(GGS)2Bn showed average single-channel conductance values that were similar to that recorded with t-FhuA. Bn(GGS)2t-FhuA and t-FhuA(GGS)2Bn also exhibited stability of the open-state current for long periods. These results suggest that Bn is located outside the pore lumen, on the β-turns′ side of t-FhuA. Moreover, these findings indicate that t-FhuA tolerates large polypeptide extensions at either terminus without affecting its pore-forming ability.
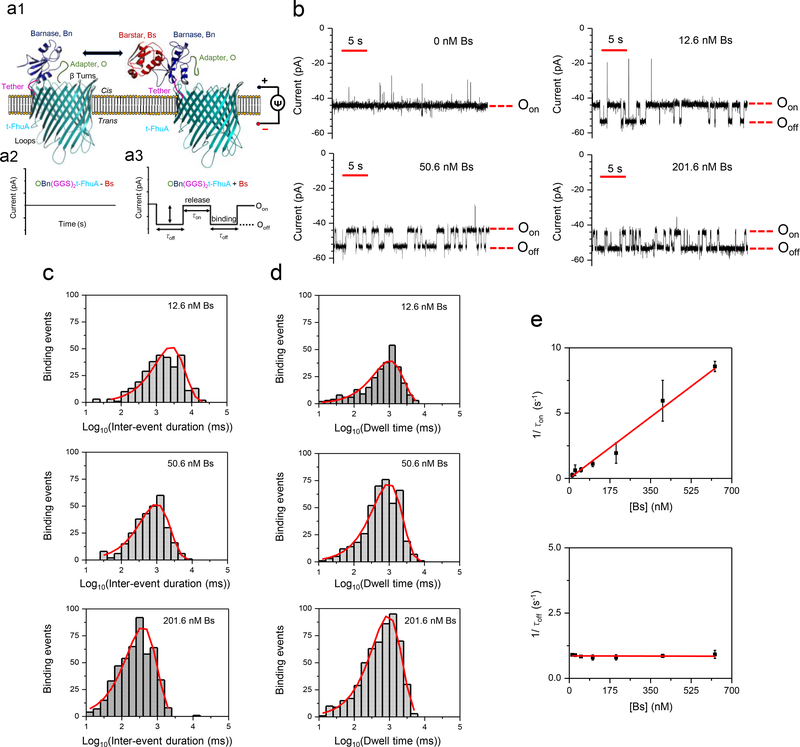
Figure 1: Measuring high-affinity PPIs using a nanopore sensor.
(a) A protein pore-based nanostructure for the real-time sampling of transient PPIs is shown in a1. The single-polypeptide chain protein comprises a t-FhuA protein pore scaffold, a flexible (GGS)2 tether, a barnase (Bn) protein receptor and an O peptide adapter. The schematic model was created in PyMol using the pdb files ID: 1BY3 (FhuA)21 and 1BRS (Bn-Bs)34. Stochastic sensing of transient PPIs using a single OBn(GGS)2t-FhuA protein is shown in a2, a3. The protein nanostructure maintains a basal open-state conductance (a2). When added to the cis side, the Bs protein ligand is expected to produce current transitions between two conductance substates (a3); (b) Reversible captures of tethered Bn by Bs were observed through well-defined current transitions. Bs was added to the cis compartment. Oon represents the Bn-released open substate, whereas Ooff represents the Bn-captured open substate. This single-channel electrical signature was replicated in three independent experiments. The applied transmembrane potential was −40 mV. Single-channel electrical traces were further low-pass 8-pole Bessel filtered at 100 Hz; (c) Representative semi-logarithmic histograms of the inter-event duration (τon) of the transient PPIs at various Bs concentrations. The τon duration (mean ± SEM) was 2338 ± 236 ms (number of events: n = 302), 1312 ± 102 ms (n = 300), and 458 ± 15 ms (n = 481) at a Bs concentration of 12.6 nM, 50.6 nM, and 201.6 nM, respectively; (d) Representative semi-logarithmic event dwell-time (τoff) histograms of the PPIs at various Bs concentrations. The τoff dwell times (mean ± SEM) determined from these histograms were 1142 ± 78 ms (number of events: n = 278), 1155 ± 72 ms (n = 424), and 1085 ± 57 ms (n = 547) at a Bs concentration of 12.6 nM, 50.6 nM, and 201.6 nM, respectively; (e) Diagrams illustrating the dependence of 1/τon and 1/τoff on the Bs concentration. The slope of the linear fit of 1/τon versus the Bs concentration, [Bs], is the association rate constant, kon, of the PPIs, because kon = 1/(τon[Bs]). The horizontal line is an average fit of the (1/τoff) data points recorded for various [Bs] values. Data points in both panels represent mean ± SD obtained from n = 3 distinct experiments.
The protein ligand we used in our initial set of measurements was the 89-residue barstar (Bs)24, which is an inhibitor of Bn ribonuclease activity. Surprisingly, the addition of 201.6 nM Bs to the cis side, either with reconstituted Bn(GGS)2t-FhuA or with t-FhuA(GGS)2Bn, failed to produce reversible alterations in the electrical signature (Supplementary Fig. 2). It is feasible that the Bn receptor adopts a conformation that blocks the accessibility of its binding site to Bs, or that Bn-Bs interactions in solution are indistinguishable (electrically silent ) in the readout. We favor the hypothesis that Bn does not obstruct ionic flux through the pore, and that transient Bn-Bs complex formation pulls Bn away from the pore opening. This hypothesis relies on single-channel measurements using t-FhuA, whose electrical signature is indistinguishable from those recorded either with Bn(GGS)2t-FhuA or t-FhuA(GGS)2Bn (Supplementary Fig. 1).
To obtain an altered single-channel electrical signature of Bn(GGS)2t-FhuA that might be sensitive to Bn-Bs specific interactions, we fused a 12-residue peptide adapter (O) to the N-terminus of Bn, resulting in OBn(GGS)2t-FhuA (Fig. 1a1; Supplementary Fig. 3a). Our peptide adapter has an unstructured and slightly negatively charged sequence25 that should span the distance between the N-terminus of Bn (its fusion point) and the pore opening. This distance is ~4 nm assuming that the flexible (GGS)2 linker is stretched-out. We hypothesized that this peptide adapter would exhibit non-specific interactions with the highly acidic entrance of the pore, resulting in a distinct and sensitive electrical signature. We found that OBn(GGS)2t-FhuA had a reduced unitary conductance of 1.23 ± 0.03 nS (n = 8) at a transmembrane potential of −40 mV (Supplementary Fig. 3b, 3c). Further, OBn(GGS)2t-FhuA exhibited a population of frequent, brief, and upwards current spikes (Supplementary Fig. 4, 5). The fusion of the peptide adapter to the N-terminus of Bn(GGS)2t-FhuA likely induces an obstruction moiety near the pore opening of t-FhuA, thereby reducing its unitary conductance.
Remarkably, when Bs was added to the cis side in the nanomolar range, we detected reversible current transitions between Oon, a lower-current amplitude open substate, and Ooff, a higher-current amplitude open substate (Fig. 1a2, 1a3, and 1b; Supplementary Table 3). We interpret these current transitions as reversible capture (Ooff) and release (Oon) events of Bn by Bs. We hypothesize that the transient capture events are accompanied by pulling the obstruction moiety away from the pore opening, increasing the open-state current to Ooff. This level is closely similar to that noted with Bn(GGS)2t-FhuA (Supplementary Fig. 3b). This interpretation is in accord with a quieter and higher-conductance open substate of the capture events (Supplementary Fig. 4, Fig. 6).
The current amplitudes of Ooff and Oon enabled an unambiguous separation of the “off” (quiet substate) and “on” (noisy substate) events, with a difference of 10 ± 2 pA (n=3). Notably, reversible current transitions were only observed when Bs was added to the cis side, but not to the trans side (Supplementary Fig. 7) which confirmed that the truncated FhuA protein pores insert into the lipid bilayer with a preferred orientation.26, 27 The pore is inserted such that the loops face the trans side and the β turns face the cis side (Fig. 1a1)20.
Next, we tested the single-molecule kinetics of the Bn-Bs interactions and showed that the frequency of Bn-Bs binding events increased with Bs concentration (Fig. 1b). The semi-logarithmic representations of the inter-event duration, τon, and dwell time, τoff, analyses are shown in Fig. 1c and Fig. 1d, respectively. Note that the center location of the peak is the logarithm of the time constant. The standard inter-event duration and dwell-time analyses of the “on” and “off” events are illustrated in Supplementary Fig. 8 and Fig. 9, respectively. We applied a logarithm likelihood ratio (LLR) test to fit models of these histograms (Fig. 1c, 1d) and this showed that “on” and “off” events had a single-exponential value distribution, suggesting a single-barrier transition of the free-energy landscape of these PPIs.
The frequency of the Bn-Bs binding events in the form of 1/τon was linearly dependent on the Bs concentration, confirming a bimolecular association process (Fig. 1e). The slope of the linear fit of event frequency was the association rate constant, kon. The reciprocal of the τoff duration, which is the dissociation rate constant, koff, was independent of the Bs concentration, confirming a unimolecular dissociation process. We obtained kon = (1.34 ± 0.04) × 107 M−1s−1 and koff = 0.86 ± 0.02 s−1, corresponding to an equilibrium dissociation constant, Kd, of 64 ± 02 nM (Supplementary Table 4). The value of this constant indicated a high-affinity PPIs of the Bn-Bs pair. This agrees well with prior kinetic measurements of Bn-Bs interactions22, 23. Furthermore, we found that the signal-to-noise ratio was not increased by an increase in Bs concentration even in the low-micromolar range (Supplementary Fig. 10). At higher Bs concentrations, the frequency of PPI events showed a nonlinear dependence on protein concentration, likely due to saturation of the Bn binding site (Supplementary Fig. 11).
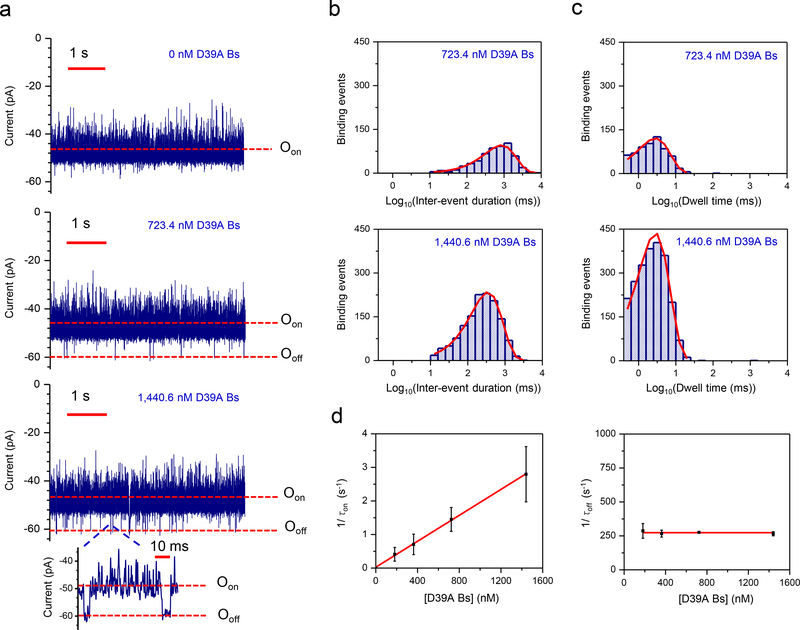
Weak PPIs, with Kd values in the micromolar or millimolar range are important in many signaling pathways28. The main difficulty in detecting weak PPIs is either high-dissociation or low-association rate constants, or both2. To test the ability of OBn(GGS)2t-FhuA to detect weaker PPIs, we carried out single-channel recordings using D39A Bs, a Bs variant with greatly reduced binding affinity to Bn. When a low-nanomolar concentration of D39A Bs was added to the cis side, reversible current transitions occurred between Oon and Ooff, but with a binding duration in the low-millisecond scale. τoff was much shorter than the value for the Bn-Bs pair (Fig. 2a). This weak interaction had inter-event duration (Fig. 2b) and dwell-time (Fig. 2c) histograms with single-exponential distributions. The standard inter-event duration and dwell-time analyses of the corresponding “on” and “off” events are reported in Supplementary Fig. 12 and Fig. 13, respectively. τon decreased when D39A Bs concentration was increased (Supplementary Table 5) whereas there was no statistically significant alteration in τoff when D39A Bs concentration was altered. The frequency of low-affinity events increased linearly with increasing the D39A Bs concentration, consistent with our results using high-affinity binder Bs (Fig. 2d). Our finding of kon = (0.193 ± 0.003) × 107 M−1s−1 and koff = 281 ± 8 s−1, to yield a Kd of 146 ± 4 μM (Supplementary Table 4) is consistent with earlier studies carried out in bulk phase23. For example, in 50 mM Tris·HCl, pH 8, transient PPIs between Bn and D39A Bs had koff and Kd values of ~17 s−1 and ~39 nM, respectively. These PPIs are ~ two orders of magnitude weaker than those recorded for Bn and Bs, which had a Kd of ~0.32 nM in the same conditions. This shows that our nanopore sensor can detect transient PPIs at protein ligand concentrations several orders of magnitude below the measured Kd. Transient PPIs cannot easily be measured at millisecond-time resolution with other biophysical methods in solution28 whereas our nanopore sensor shows promise for detecting weak PPIs with high koff values.
Figure 2: Measuring low-affinity PPIs using a nanopore sensor.
(a) Representative single-molecule captures of transient PPIs between Bn and D39A Bs using OBn(GGS)2t-FhuA. This single-channel electrical signature was replicated in three independent experiments. The applied transmembrane potential was −40 mV. The low-affinity D39A Bs was added to the cis compartment. The current traces were low-pass Bessel filtered at 1 kHz; (b) Representative semi-logarithmic histograms of the inter-event duration, τon, of the PPIs at two D39A Bs concentrations. The τon values (mean ± SEM) were 855 ± 1 (number of events: n = 560) and 378 ± 12 ms (n = 1349) at D39 Bs concentrations of 723.4 nM and 1,440.6 nM, respectively; (c) Representative semi-logarithmic event dwell-time histograms of the weak PPIs at two D39A Bs concentrations. The τoff values (mean ± SEM) determined from these histograms were 3.6 ± 0.2 ms (number of events: n = 622) and 3.6 ± 0.1 ms (n = 2240) at a D39A Bs concentration of 723.4 nM and 1,440.6 nM, respectively; (d) Diagrams illustrating the dependence of 1/τon and 1/τoff on the D39A Bs concentration, [D39A Bs]. The slope of the linear fit of 1/τon versus [D39A Bs] is the association rate constant, kon, of the PPIs. The bottom diagram shows that the τoff binding time was independent of the D39A Bs concentration. Data points in both panels represent mean ± SD obtained from either n = 3 (723.4 nM and 1440.6 nM) or n=4 (181.4 nM and 362.4 nM) distinct experiments.
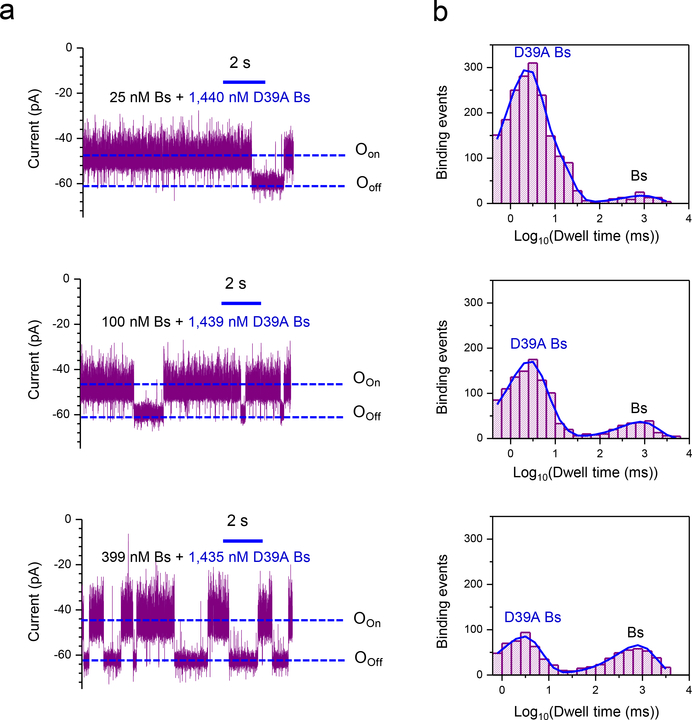
An ability to detect multiple analytes in a mixture of proteins is needed for diagnostic applications and we next tested whether our sensor could identify low- and high-affinity PPIs in a mixture. If both Bs and D39A Bs were added we detected both long-lived and brief transitions (Fig. 3a, Fig. 3b; Supplementary Tables 6, 7). Increasing the concentration of high-affinity Bs resulted in an increase in the frequency of long-lived current transitions and a reduction in the frequency of the brief binding events. These experiments revealed that our nanopore sensor can discriminate competitive interactions between two Bs variants for the same binding site.
Figure 3: Concurrent detection of weak and strong PPIs using a nanopore sensor.
(a) Representative single-channel current traces of OBn(GGS)2 t-FhuA were collected at an applied transmembrane potential of −40 mV when different mixtures of protein ligands were added to the cis compartment: (i) 1,440 nM D39A Bs and 25 nM Bs (top trace); (ii) 1,439 nM D39A Bs and 100 nM Bs (middle trace); (iii) 1,435 nM D39A Bs and 399 nM Bs (bottom trace). These single-channel electrical signatures were replicated in three independent experiments. The single-channel electrical traces were low-pass Bessel filtered at 1 kHz; (b) Representative event dwell-time histograms of the weak and strong PPIs: (i) 1,440 nM D39A Bs and 25 nM Bs (top diagram); (ii) 1,439 nM D39A Bs and 100 nM Bs (middle diagram); (iii) 1,435 nM D39A Bs and 399 nM Bs (bottom diagram). The average τoff dwell times (mean ± SEM), which corresponded to D39A Bs and Bs, respectively, were the following: (i) 4.1 ± 0.1 ms and 904 ± 1 ms (number of events: n= 1888) (top diagram); (ii) 3.4 ± 0.1 ms and 829 ± 1 ms (n = 1153) (middle diagram); (iii) 3.3 ± 0.1 ms and 827 ± 1 ms (n = 789) (bottom diagram).
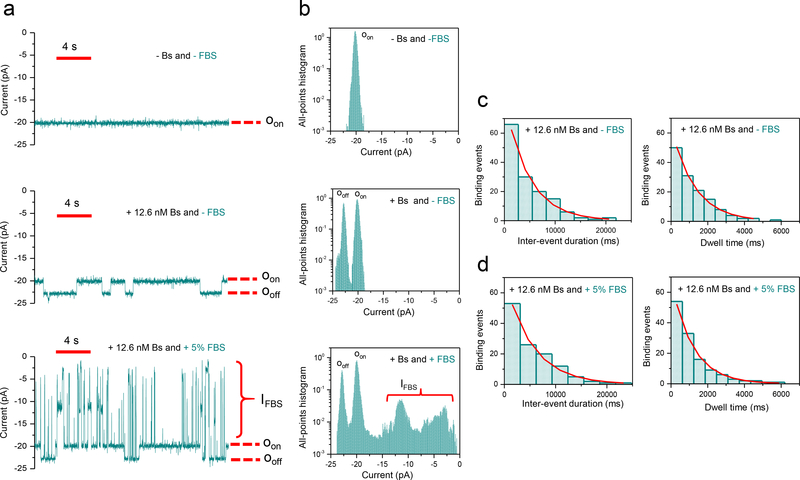
We also tested whether our sensor could detect Bs in the presence of fetal bovine serum (FBS). In the absence of Bs, OBn(GGS)2t-FhuA exhibited a uniform and quiet unitary current at a transmembrane potential of −15 mV (n=3; Fig. 4a; upper trace). Single-channel traces acquired with OBn(GGS)2t-FhuA in the presence of 12.6 nM Bs, which was added to the cis side, displayed reversible low-current amplitude transitions between Oon and Ooff (n=3; Fig. 4a; middle trace). In the presence of 5% (v/v) FBS, large-amplitude current blockades were recorded, more than likely produced by the serum constituents that partitioned into the pore lumen (n=3; IFBS; Fig. 4a; bottom trace). However, the single-channel electrical traces also showed low-amplitude, PPIs-induced current transitions that were unambiguously distinguished from large-amplitude current blockades (n=3; Fig. 4b). We chose a voltage of −15 mV because at a greater transmembrane potential of −40 mV the FBS-produced current blockades were very long (Supplementary Fig. 14). These long-lived blockades precluded a precise evaluation of the kinetic rate constants. Notably, the transient PPIs recorded at transmembrane potentials of −15 and −40 mV displayed no statistically significant distinctions in kinetic rate constants (Supplementary Tables 3, 4, and 8).
Figure 4: Single-molecule protein detection and observation of transient PPIs using a nanopore sensor in fetal bovine serum (FBS).
(a) Representative single-molecule binding events of the Bn-Bs pair using OBn(GGS)2t-FhuA in 5% (v/v) FBS at an applied transmembrane potential of −15 mV. This single-channel electrical signature was replicated in three independent experiments. Single-channel electrical traces were low-pass Bessel filtered at 40 Hz; (b) Representative all-points histograms for the duration of the displayed single-channel electrical traces in A. This single-channel electrical signature was replicated in three independent experiments; (c) Representative histograms of the inter-event durations (τon) and the event dwell times (τoff) at 12.6 nM Bs and in the absence of FBS. The τon and τoff durations (mean ± SEM) determined from these fits were 5,182 ± 200 ms (number of events: n = 140) and 1,272 ± 53 ms (n = 133), respectively; (d) Representative histograms of the inter-event durations (τon) and the event dwell times (τoff) for the Bn-Bs interactions at 12.6 nM Bs and in the presence of 5% (v/v) FBS. The τon and τoff durations (mean ± SEM) determined from these histogram fits were 5,153 ± 299 ms (number of events: n = 120) and 1,165 ± 54 ms (n = 127), respectively.
Representative standard histograms acquired in the absence and presence and of 5% (v/v) FBS, are displayed in Fig. 4c and Fig. 4d, respectively. Notably, kon and koff of the transient PPI in the absence and presence of FBS were similar (Supplementary Table 8). To test the sensitivity of our single-molecule sensor, we determined the Bs concentration in the presence of FBS using CBs=1/(τonkon), where CBs, τon, and kon are the concentration of the sampled Bs protein, inter-event duration determined in the presence of FBS, and association rate constant inferred for Bs in the absence of FBS, respectively. The average Bs concentration was 13.3 ± 5.0 nM (n=3). This value is in agreement with the actual Bs concentration added to the cis chamber (12.6 nM Bs). Therefore, our nanopore sensor can detect and quantitate a protein analyte in a complex biological fluid. Further, we were able to obtain detailed kinetic information of the transient PPIs even in these challenging conditions.
Most biophysical approaches for the quantification of kinetics of transient PPIs are carried out in bulk phase, and report average measurements of an ensemble of proteins in solution. In contrast, our method has the potential to detect and characterize subpopulations of distinct binding events in a complex biofluid environment. Furthermore, our genetically-encoded sensor shows promise for the identification of rare and brief binding events, which cannot be detected using existing technologies. We also anticipate that temperature-dependent electrical recordings might be able to illuminate enthalpic and entropic contributions to kinetic rate constants of association and dissociation of transient PPIs. Simultaneous determination of the koff values for more than one protein target, while providing the apparent kon and IC50, is quite a challenging task employing other techniques in solution. Using our single-molecule nanopore sensor, we were able to examine competitive protein interactions with the same binding site. We have shown that our nanopore sensor can probe koff in the range of 102 – 103 s-1. There is no theoretical reason to suggest that our sensor could not resolve even shorter PPIs events, which have a faster dissociation rate constant, such as extremely weak PPIs involved in the rapid responses of cell signaling2.
Because our sensor is genetically encoded, we envisage that it could form the basis of a combinatorial sensor library of different protein receptors with potential application in clinical nanoproteomics, or high-throughput screening of small-molecule drugs or peptide inhibitors. Finally, the ability to distinguish a specific PPIs in a complex biological fluid sample reveals the potential of this sensor for single-molecule protein detection29–33 in cell lysates, biopsies or blood.
ONLINE METHODS
Design and mutagenesis of the expression constructs.
All the designed genes were constructed using conventional and assembly PCR techniques, and cloned into the expression vector pPR-IBA1 using respective restriction sites. bn(ggs)2t-fhua, which encoded barnase (Bn)34 at the N-terminus of the heavily truncated t-FhuA protein20 via a flexible Gly/Ser-rich tether and BsaI site, was prepared using the bn and t-fhua genes, as well as three PCR reactions. The first PCR reaction was performed using bn as a template DNA, the forward primer (1) and reverse primer (2). The second PCR reaction was performed using t-fhua as a template DNA, as well as the forward primer (3) and reverse primer (4). The third PCR reaction was performed using PCR 1 and PCR 2 products as a template DNA, as well as the forward primer (1) and reverse primer (4).
obn[ggs]2t-fhua, which encoded the peptide adapter, O, MGDRGPEFELGT, fused at the N-terminus of Bn, a flexible Gly/Ser-rich tether, t-FhuA, as well as KpnI site at the 5’ and 3’ ends, was prepared using the bn and t-fhua genes, via three PCR reactions. The first PCR reaction was performed using bn as a template DNA, along with the forward primer (5) and reverse primer (6). For the second PCR reaction, t-fhua was used as a template DNA, along with the forward primer (7) and reverse primer (4). The third PCR reaction was performed using PCR 1 and PCR 2 products as a template DNA, along with the forward primer (5) and reverse primer (4). The bs gene encoded the barstar (Bs) protein ligand. We removed all the purification tags from the gifted plasmid (Courtesy Prof. Andreas Matouschek) using the forward primer (8) and reverse primer (9), then sub-cloned it into the pPR-IBA1 expression vector using BsaI restriction sites. The bs gene also encoded a double-alanine mutant, C40A/C82A35. The d39a bs gene, which encoded D39A Bs, was prepared by performing an inverse PCR using the forward primer (10) and reverse primer (11).
Protein expression and purification.
All the constructed genes were transformed into E. coli BL21(DE3) cells for protein expression. Bn(GGS)2t-FhuA, t-FhuA(GGS)2Bn, and OBn(GGS)2t-FhuA were expressed and purified, as previously described20. In the case of Bs and D39A Bs, transformed cells were grown in Luria-Bertani medium at 37°C until OD600 reached a value of ~0.5, after which the temperature was changed to 20°C. Bs expression was initiated by inducing the cells with IPTG when OD600 was ~0.7. After induction, the cells were further cultured for an additional period of ~18 hours at 20°C. Then, the cells were centrifugated at 3,700×g for 30 min at 4°C, followed by their resuspension in 150 mM KCl, 50 mM Tris·HCl, 5 mM EDTA, pH 8. The cell lysis was accomplished using a Model 110L microfluidizer (Microfluidics, Newton, MA). Cell lysates were centrifuged at 108,500 × g for 30 min at 4°C to separate the insoluble pellet and supernatant. The supernatant was further processed for ammonium sulfate precipitation. In the first step, ammonium sulfate was slowly dissolved into supernatant to a final concentration of 10% (w/v) at 4°C for 30 min. Later, the supernatant was centrifuged at 108,500 × g for 30 min at 4 °C to separate the precipitate and supernatant. Further, the supernatant was processed like the previous step with 40% ammonium sulfate. The collected supernatant was dialyzed extensively against 20 mM Tris·HCl, pH 8, overnight at 4°C. Dialyzed supernatant was then purified on the Q-Sepharose column (Bio-Rad, Hercules, CA) using a linear salt gradient of 0–1 M KCl, 20 mM Tris·HCl, pH 8. Pure fractions were further passed through the Superdex-75 size-exclusion column (SEC; GE Healthcare Life Sciences, Pittsburg, PA) as a refining purification step. Pure Bs variants were stored at −80°C. Protein purity was tested by the SDS-PAGE analysis.
Protein refolding.
Lyophilized Bn(GGS)2t-FhuA, t-FhuA(GGS)2Bn, and OBn(GGS)2t-FhuA were solubilized in 200 mM KCl, 8 M urea, 50 mM Tris·HCl, pH 8 to a final concentration of ~15 μM and incubated at room temperature for at least 4 hours before refolding. Later, n-dodecyl-β-D-maltopyranoside (DDM) was added to denatured samples to a final concertation of 1.5% (w/v). The protein samples were immediately dialyzed against the buffer containing 200 mM KCl, 20 mM Tris·HCl, pH 8, at 4 °C for a duration of at least 72 h. Then, the refolded protein samples were 20-fold diluted in 200 mM KCl, 20 mM Tris·HCl, pH 8, 0.5% DDM before single-channel electrical recordings. Protein concentrations were determined by their molar absorptivity at a wavelength of 280 nm.
Single-channel electrical recordings using planar lipid bilayers.
Single-molecule electrophysiology measurements employed planar lipid bilayers, as previously reported26. The proteins were added to the cis compartment, which was at ground, to a final concentration in a range between 0.3 and 1 ng/μl. The single-channel electrical currents were acquired using an Axopatch 200B patch-clamp amplifier (Axon Instruments, Foster City, CA). For all experiments, the electrolyte solution contained 300 mM KCl, 10 mM Tris·HCl, pH 8. Single-channel current transitions were collected from individual electrical traces, whose durations were within a range of 10–15 min. For both data acquisition and analysis, a pClamp 10.5 software package (Axon) was used. Figures were prepared using ClampFit 10.7 (Axon) and Origin 8.5 (OriginLab, Northampton, MA). A logarithm likelihood ratio (LLR) test was used for the single-exponential fit of the histograms.36 All single-channel electrical recordings were acquired at a temperature of 23 ± 1°C.
Detection of Bs in fetal bovine serum.
Gibco™ fetal bovine serum (FBS), Catalog no. A3160601, was obtained from Fisher Scientific (Pittsburg, PA). FBS was sterile-filtered using an 0.2-μm filter and kept at −80°C for long-time storage. A fresh aliquot of this frozen serum was thawed at 4°C and incubated at room temperature for at least 30 min before the single-channel experiment.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stewart Loh for generosity in using his FPLC instrument in the very early stage of these studies, Andreas Matouschek for his kindness in offering plasmids containing gene-encoding Bn and Bs proteins, as well as Motahareh Larimi Ghahari and Mohammad M. Mohammad for their assistance in the very early stage of this project. This work was supported by the US National Institutes of Health grants GM088403 (L.M.) and GM129429 (L.M.).
Footnotes
COMPETING INTERESTS
A.K.T. and L.M. are named inventors on two provisional patent applications (US 62/720,190 and US 62/579,982) filed by Syracuse University on this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Hayes S, Malacrida B, Kiely M & Kiely PA Studying protein-protein interactions: progress, pitfalls and solutions. Biochem. Soc. Trans. 44, 994–1004 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Yoo J, Lee TS, Choi B, Shon MJ & Yoon TY Observing Extremely Weak Protein-Protein Interactions with Conventional Single-Molecule Fluorescence Microscopy. J. Am. Chem. Soc. 138, 14238–14241 (2016). [DOI] [PubMed] [Google Scholar]
- 3.De Keersmaecker H et al. Mapping Transient Protein Interactions at the Nanoscale in Living Mammalian Cells. ACS nano (2018). [DOI] [PubMed] [Google Scholar]
- 4.Nogal B, Bowman CA & Ward AB Time-course, negative-stain electron microscopy-based analysis for investigating protein-protein interactions at the single-molecule level. J. Biol. Chem. 292, 19400–19410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez LC Protein microarrays, biosensors, and cell-based methods for secretome-wide extracellular protein-protein interaction mapping. Methods 57, 448–458 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Douzi B Protein-Protein Interactions: Surface Plasmon Resonance. Methods Mol. Biol 1615, 257–275 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Pierce MM, Raman CS & Nall BT Isothermal titration calorimetry of protein-protein interactions. Methods 19, 213–221 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Sackmann B & Neher E Single-Channel Recording, Edn. Second Edition. (Kluwer Academic/Plenum Publishers, New York; 1995). [Google Scholar]
- 9.Wei R, Gatterdam V, Wieneke R, Tampe R & Rant U Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 7, 257–263 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Ying YL, Yu RJ, Hu YX, Gao R & Long YT Single antibody-antigen interactions monitored via transient ionic current recording using nanopore sensors. Chemical communications (Cambridge, England) 53, 8620–8623 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Weichbrodt C et al. Antibiotic translocation through porins studied in planar lipid bilayers using parallel platforms. Analyst 140, 4874–4881 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Reiner JE et al. Disease detection and management via single nanopore-based sensors. Chem. Rev. 112, 6431–6451 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Deamer D, Akeson M & Branton D Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayub M & Bayley H Engineered transmembrane pores. Curr. Opin. Chem. Biol. 34, 117–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burns JR, Seifert A, Fertig N & Howorka S A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 11, 152–156 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Howorka S Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Movileanu L, Howorka S, Braha O & Bayley H Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 18, 1091–1095 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Rotem D, Jayasinghe L, Salichou M & Bayley H Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc. 134, 2781–2787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington L, Cheley S, Alexander LT, Knapp S & Bayley H Stochastic detection of Pim protein kinases reveals electrostatically enhanced association of a peptide substrate. Proc. Natl. Acad. Sci. U.S.A 110, E4417–E4426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakur AK, Larimi MG, Gooden K & Movileanu L Aberrantly large single-channel conductance of polyhistidine arm-containing protein nanopores. Biochemistry 56, 4895–4905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locher KP et al. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95, 771–778 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Schreiber G & Fersht AR Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 32, 5145–5150 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Schreiber G & Fersht AR Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol 248, 478–486 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Deyev SM, Waibel R, Lebedenko EN, Schubiger AP & Pluckthun A Design of multivalent complexes using the barnase*barstar module. Nat. Biotechnol. 21, 1486–1492 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Kudlinzki D, Schmitt A, Christian H & Ficner R Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biological chemistry 393, 1131–1140 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Mohammad MM, Howard KR & Movileanu L Redesign of a plugged beta-barrel membrane protein. J. Biol. Chem. 286, 8000–8013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammad MM et al. Engineering a rigid protein tunnel for biomolecular detection. J. Am. Chem. Soc. 134, 9521–9531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins JR, Diboun I, Dessailly BH, Lees JG & Orengo C Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Nivala J, Mulroney L, Li G, Schreiber J & Akeson M Discrimination among protein variants using an unfoldase-coupled nanopore. ACS nano 8, 12365–12375 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Kennedy E, Dong Z, Tennant C & Timp G Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol 11, 968–976 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Sze JYY, Ivanov AP, Cass AEG & Edel JB Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 8, 1552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G, Willems K, Soskine M, Wloka C & Maglia G Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 8, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restrepo-Perez L, Joo C & Dekker C Paving the way to single-molecule protein sequencing. Nat Nanotechnol 13, 786–796 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Buckle AM, Schreiber G & Fersht AR Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry 33, 8878–8889 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Guillet V, Lapthorn A, Hartley RW & Mauguen Y Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure 1, 165–176 (1993). [DOI] [PubMed] [Google Scholar]
- 36.McManus OB & Magleby KL Kinetic states and modes of single large-conductance calcium-activated potassium channels in cultured rat skeletal-muscle. J. Physiol. (Lond.) 402, 79–120 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.