Abstract
Historic cell culture media were designed to ensure continuous cancer cell proliferation in vitro. However, their composition does not recapitulate the tumor’s nutritional environment. Recent studies show that novel media formulations alleviate the non-physiological constraints imposed by historic media, and lead to cell culture results more relevant to tumor metabolism.
Keywords: Cancer metabolism, Physiological medium, Cancer models
A tumor is a complex, dynamic and disordered structure within an organism, composed of mixed populations of normal and cancer cells. In order to understand the role of the biological units of this biological system, more than 60 years ago researchers embraced a reductionist approach and started to culture cells in isolation. Since then, the majority of the experiments in cancer research have been performed with cell lines cultured as monolayers. This implies that a substantial advance in cancer cell biology has been achieved with cultured cells, often referred to as in vitro. It is undeniable that culturing cancer cells is informative and has advantages that overall exceed its obvious limitations. Tumors consist of different niches depending on vascularization, genetic clonality and infiltration of immune and stromal cells. Since a cell culture dish overtly differs from the growth conditions of cells in tumors, researchers continuously attempted to refine culture conditions by modulating oxygen concentration, by allowing cells to form self-contained three dimensional structures, i.e. spheroids, or by supplying extracellular matrixes with different chemical-physical properties. Recently, more complex co-culture systems have also allowed to study in vitro the interaction between different cell types.
However, most of the experiments currently ongoing are still performed in historic cell culture media, some of which were formulated at least half a century ago, and whose composition clearly differs from the nutritional environment that cells withstand in tumors. For example Eagle’s Minimal Essential Medium, MEM, and its Dulbecco’s modified version, DMEM, were designed to supply cancer cells with only those nutrients essential for their continuous proliferation. These are widely used and in 2016 more than half of the published cell culture-based studies employed one or other of these media [1]. Another frequently used cell culture medium, F12, was optimized for the clonal growth of Chinese Hamster Ovary (CHO) cells under reduced serum supplementation [2]. In general, currently available commercial cell culture media were thought to allow continuous growth of specific cell types, not to recapitulate the metabolic environment of the tissue of origin [2]. As a result they often lack metabolites normally present in human fluids, while others, such as glucose, glutamine or pyruvate, are often found in media at supra-physiological concentrations (Table 1). On the contrary compounds irrelevant for human pathophysiology, such as L-alanyl-L-glutamine dipeptide (e.g. GlutaMAX™), are commonly supplemented at millimolar concentrations, with uncharacterized, yet inevitable consequences on cell metabolism.
Table 1. Comparison between the formulations of physiological and historic media.
Formulations of Plasmax™, Human Plasma-Like Medium (HPLM), Minimal Essential Medium (MEM), Iscove's Modified Dulbecco's Medium (IMDM), Dulbecco's modified Eagles Medium (DMEM, high glucose), DMEM/F-12 nutrient mix (1:1), Ham’s F-12 Nutrient Mix (F-12), and Roswell Park Memorial Institute 1640 Medium (RPMI 1640, low glucose). The range of normal concentration values for human plasma are reported in the first column. All the concentrations are reported in μM. NA: not available. The colors represent the relative abundance of each component across the different media. White represents the level in Plasmax™, and blue and red represent lower and higher concentrations, respectively. An eightfold cutoff on color scale was applied. Grey and yellow cells are not part of the color scale.
| Human plasma | Plasmax™ | HPLM | MEM | IMDM | DMEM | DMEM/F-12 | F-12 | RPMI 1640 | |
|---|---|---|---|---|---|---|---|---|---|
| Proteinogenic Amino Acids | |||||||||
| L-Alanine | 230 - 510 [13] | 510 | 430 | NA | 281 | NA | 50 | 100 | NA |
| L-Arginine | 13 - 64 [13] | 64 | 110 | 597 | 399 | 398 | 699 | 1000 | 1149 |
| L-Asparagine | 45-130 [13] | 41 | 50 | NA | 189 | NA | 50 | 100 | 379 |
| L-Aspartic acid | 0 - 6 [13] | 6 | 20 | NA | 226 | NA | 50 | 100 | 150 |
| L-Cysteine | 23.2 - 43.8 [14] | 33 | 40 | NA | NA | NA | 100 | 200 | NA |
| L-Glutamate | 32-140 [13] | 98 | 80 | NA | 510 | NA | 50 | 100 | 136 |
| L-Glutamine | 420-720 [13] | 650 | 550 | 2000 | 4000 | 4000 | 2500 | 1000 | 2055 |
| Glycine | 170 - 330 [13] | 330 | 300 | NA | 400 | 400 | 250 | 100 | 133 |
| L-Histidine | 26 - 120 [13] | 120 | 110 | 200 | 200 | 200 | 150 | 100 | 97 |
| L-Isoleucine | 42 - 100 [13] | 140 | 70 | 397 | 802 | 802 | 416 | 31 | 382 |
| L-Leucine | 66 - 170 [13] | 170 | 160 | 397 | 802 | 802 | 451 | 100 | 382 |
| L-Lysine | 150 - 220 [13] | 220 | 200 | 399 | 798 | 798 | 499 | 199 | 219 |
| L-Methionine | 16 - 30 [13] | 30 | 30 | 101 | 201 | 201 | 116 | 30 | 101 |
| L-Phenylalanine | 41 - 68 [13] | 68 | 80 | 194 | 400 | 400 | 215 | 30 | 91 |
| L-Proline | 110-360 [13] | 360 | 200 | NA | 348 | NA | 150 | 300 | 174 |
| L-Serine | 56 - 140 [13] | 140 | 150 | NA | 400 | 400 | 250 | 100 | 286 |
| L-Threonine | 92 - 240 [13] | 240 | 140 | 403 | 798 | 798 | 449 | 100 | 168 |
| L-Tryptophan | 44.8 - 64.2 [14] | 78 | 60 | 49 | 78 | 78 | 44 | 10 | 25 |
| L-Tyrosine | 45 - 74 [13] | 74 | 80 | 199 | 462 | 399 | 214 | 30 | 111 |
| L-Valine | 150 - 310 [13] | 230 | 220 | 393 | 803 | 803 | 452 | 100 | 171 |
| Non-proteinogenic Amino Acids | |||||||||
| α-Aminobutyrate | 15 - 41 [13] | 41 | 20 | NA | NA | NA | NA | NA | NA |
| L-Citrulline | 16 - 55 [13] | 55 | 40 | NA | NA | NA | NA | NA | NA |
| L-Cystine | 30 - 65 [13] | 65 | 100 | 99 | 292 | 201.3 | 100 | NA | 207.7 |
| L-Homocysteine | 6.1 - 12.1 [15] | 9 | NA | NA | NA | NA | NA | NA | NA |
| 4-Hydroxy-L-proline | 3 - 23 [16] | 13 | 20 | NA | NA | NA | NA | NA | 152.7 |
| L-Ornitine | 27 - 80 [13] | 80 | 70 | NA | NA | NA | NA | NA | NA |
| L-Pyroglutamate | 12.2 - 15.3 [17] | 20 | NA | NA | NA | NA | NA | NA | NA |
| Amino Acids Derivatives | |||||||||
| L-Acetyl glycine | 69.7 [14] | 70 | 90 | NA | NA | NA | NA | NA | NA |
| L-Carnosine | 5.5 - 7.5 [18] | 6 | NA | NA | NA | NA | NA | NA | NA |
| Glutathione (reduced) | 32.2 - 41.8 [19] | 37 | 25 | NA | NA | NA | NA | NA | 3.3 |
| Putrescine | 0.1 - 0.3 [20] | NA | NA | NA | NA | NA | 0.5 | 1.0 | NA |
| Taurine | 45 - 130 [13] | 130 | 90 | NA | NA | NA | NA | NA | NA |
| N-Trimethylglycine (betaine) | 49.6 - 94.4 [14] | 72 | 70 | NA | NA | NA | NA | NA | NA |
| Other Components | |||||||||
| Acetate | 26.8 - 57 [14] | 42 | 60 | NA | NA | NA | NA | NA | NA |
| Acetone | 24.8 - 84 [14] | 55 | 60 | NA | NA | NA | NA | NA | NA |
| Acetyl carnitine | 2.5 - 8.6 [21] | 5 | NA | NA | NA | NA | NA | NA | NA |
| Citrate | 87.2 - 141.2 [14] | 114 | 130 | NA | NA | NA | NA | NA | NA |
| Carnitine | 34.1 - 57.3 [14] | 46 | 40 | NA | NA | NA | NA | NA | NA |
| Creatine | 8.4 - 65 [14] | 37 | 40 | NA | NA | NA | NA | NA | NA |
| Creatinine | 60.5 - 87.7 [14] | 74 | 75 | NA | NA | NA | NA | NA | NA |
| Formate | 19.5 - 46.1 [14] | 33 | 50 | NA | NA | NA | NA | NA | NA |
| Fructose | 28.0 - 34.0 [22] | NA | 40 | NA | NA | NA | NA | NA | NA |
| Galactose | 53.6 - 123 [22] | NA | 60 | NA | NA | NA | NA | NA | NA |
| D-Glucose | 4598.5 - 5344.1 [14] | 5560 | 5000 | 5556 | 25000 | 25000 | 17506 | 10011 | 11111 |
| Glycerol | 331.2 - 532 [14] | 82 | 120 | NA | NA | NA | NA | NA | NA |
| 2-Hydroxybutyrate | 23.5 - 39.1 [14] | 31 | 50 | NA | NA | NA | NA | NA | NA |
| 3-Hydroxybutyrate | 10.6 - 143.2 [14] | 77 | 50 | NA | NA | NA | NA | NA | NA |
| 3-Hydroxyisobutyrate | 19.0 - 23.0 [23] | 20 | NA | NA | NA | NA | NA | NA | NA |
| Hypoxanthine | 4.5 - 5.3 [24] | 5 | 10 | NA | NA | NA | 15 | 30 | NA |
| Lactate | 1118.2 - 1860.6 [14] | 500 | 1600 | NA | NA | NA | NA | NA | NA |
| Linoleic Acid | 45.8 - 121.8 [14] | NA | NA | NA | NA | NA | 0.15 | 0.30 | NA |
| Lipoic Acid | 0.060 - 0.094 [22] | NA | NA | NA | NA | NA | 0.5 | 1.0 | NA |
| Malonate | 12.3 - 14.7 [14] | NA | 10 | NA | NA | NA | NA | NA | NA |
| Methyl acetoacetate | for acetoacetate 4.1 - 77.1 [14] | 41 | NA | NA | NA | NA | NA | NA | NA |
| Phenol Red | NA | 25.0 | 14.0 | 26.6 | 39.9 | 39.9 | 21.5 | 3.2 | 13.3 |
| Pyruvate | 9.3 - 59.7 [14] | 100 | 50 | NA | 1000 | 1000 | 500 | 1000 | NA |
| Succinate | 23.5 [14] | 23 | 20 | NA | NA | NA | NA | NA | NA |
| Thymidine | 0.1 - 0.3 [19] | NA | NA | NA | NA | NA | 1.5 | 2.9 | NA |
| Uracil | 1.1 - 3.1 [19] | 2 | NA | NA | NA | NA | NA | NA | NA |
| Urate | 228.9 - 315.1 [19] | 270 | 250 | NA | NA | NA | NA | NA | NA |
| Urea | 3920.4 - 8228.8 [14] | 3000 | 5000 | NA | NA | NA | NA | NA | NA |
| Uridine | 1.8 - 4.4 [19] | 3 | NA | NA | NA | NA | NA | NA | NA |
| Inorganic Salts | |||||||||
| Ammonium Chloride | for NH4+ [25] for Cl- [1] | 50 | 40 | NA | NA | NA | NA | NA | NA |
| Calcium Chloride | for Ca2+ and Cl- [1] | 1800 | 2350 | 1802 | 1487 | 1802 | 1050 | 299 | NA |
| Calcium Nitrate | for Ca2+ [1] for NO3-[22] | NA | 40 | NA | NA | NA | NA | NA | 424 |
| Magnesium Chloride | for Mg2+ and Cl- [1] | NA | 480 | NA | NA | NA | 302 | 602 | NA |
| Magnesium Sulfate | for Mg2+ and SO42- [1] | 813 | 250 | 814 | 814 | 814 | 407 | NA | 407 |
| Potassium Chloride | for K+ and Cl- [1] | 5330 | 4100 | 5333 | 4400 | 5333 | 4157 | 2981 | 5333 |
| Potassium Nitrate | for K+ [1] for NO3- [22] | NA | NA | NA | 1 | NA | NA | NA | NA |
| Sodium Bicarbonate | for Na+ and HCO3- [1] | 26191 | 24000 | 26191 | 36000 | 44048 | 29024 | 14000 | 23810 |
| Sodium Chloride | for Na+ and Cl- [1] | 118706 | 105000 | 117241 | 77672 | 110345 | 120612 | 131017 | 103448 |
| Sodium Phosphate monobasic | for Na+ and PO43- [1] | 1010 | NA | 1014 | 906 | 906 | 453 | NA | NA |
| Sodium Phosphate dibasic | for Na+ and PO43- [1] | NA | 870 | NA | NA | NA | 500 | 1000 | 5634 |
| Trace Elements | |||||||||
| Ammonium Metavanadate | for NH4+ [25] for V [26] | 0.0026 | NA | NA | NA | NA | NA | NA | NA |
| Cupric Sulfate | for Cu [27] for SO42- [1] | 0.0052 | NA | NA | NA | NA | 0.0052 | 0.0100 | NA |
| Ferric Chloride | for Fe [28] for Cl- [1] | NA | NA | NA | 2 | NA | NA | NA | NA |
| Ferric Nitrate | for Fe [28] for NO3-[22] | 0.1238 | NA | NA | NA | 0.2475 | 0.1238 | NA | NA |
| Ferric Sulfate | for Fe [28] for SO42- [1] | 1.0428 | NA | NA | NA | NA | 1.5000 | 3.0000 | NA |
| Manganous Chloride | for Mn [29] for Cl- [1] | 0.0002 | NA | NA | NA | NA | NA | NA | NA |
| Sodium Selenite | for Na+ [1] for Se [29] | 0.0289 | NA | NA | 0.0980 | NA | NA | NA | NA |
| Zinc Sulfate | for Zn [29] for SO42- [1] | 1.50 | NA | NA | 0.49 | NA | 1.50 | 2.90 | NA |
| Vitamins | |||||||||
| p-Aminobenzoate | 5.0 - 32.0 [24] | NA | 7.3 | NA | NA | NA | NA | NA | 7.3 |
| Ascorbate | 57.9 - 67.3 [19] | 62 | NA | NA | NA | NA | NA | NA | NA |
| D-Biotin | 0.0006 - 0.0019 [30] | 4.100 | 0.800 | NA | 0.530 | NA | 0.014 | 0.030 | 0.820 |
| Choline | 9.2 - 19.8 [14] | 7.1 | 21.5 | 7.1 | 28.6 | 28.6 | 64.1 | 100 | 21.4 |
| Folate | 0.017 - 0.025 [31] | 2.30 | 2.30 | 2.30 | 9.10 | 9.10 | 6.00 | 2.90 | 2.27 |
| myo-Inositol | 17.1 [14] | 11.1 | 194.3 | 11.1 | 40.0 | 40.0 | 70.0 | 100.0 | 194.4 |
| Niacinamide | 0.435 - 0.445 [32] | 8.2 | 8.2 | 8.2 | 32.8 | 32.8 | 16.6 | 0.3 | 8.2 |
| D-Calcium pantothenate | 4.5 - 5.3 [22] | 2.10 | 0.52 | 2.10 | 8.40 | 8.40 | 4.70 | 1.05 | 0.52 |
| Pyridoxine | 0.007 - 0.060 [33] | 4.90 | 4.90 | 4.90 | 19.60 | 19.40 | 9.80 | 0.29 | 4.90 |
| Riboflavin | 0.0054 - 0.028 [34] | 0.30 | 0.50 | 0.27 | 1.10 | 1.10 | 0.58 | 0.01 | 0.53 |
| Thiamine | 0.078 - 0.114 [35] | 3.0 | 3.0 | 3.0 | 11.9 | 11.2 | 6.4 | 0.9 | 3.0 |
| Vitamin B12 | 0.00017 - 0.00033 [36] | 0.0050 | 0.0037 | NA | 0.0096 | NA | 0.5000 | 1.0000 | 0.0037 |
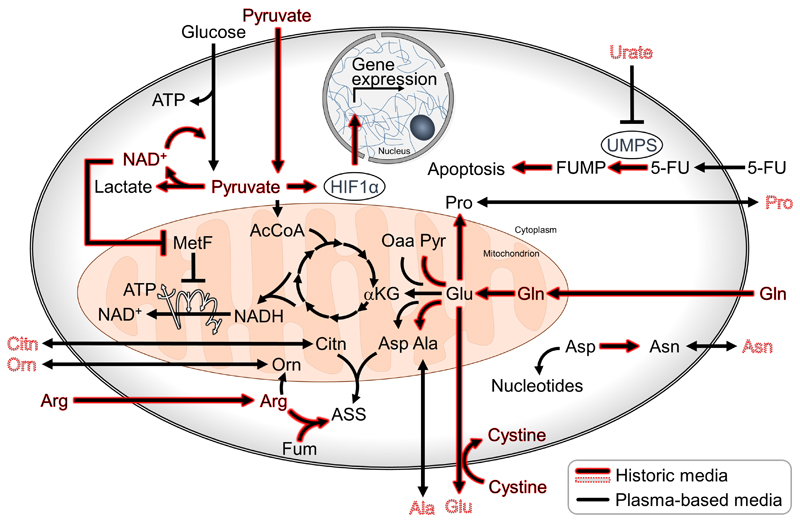
Only in recent years, has it been shown that excessive concentrations of nutrients affect the metabolism of cultured cells and lead to discrepancies in metabolic phenotypes between cultured cells and tumors. For example, the proliferation of cancer cells has been shown to depend less on mitochondrial respiration when cultured with excessive concentrations of pyruvate, as indicated by their decreased sensitivity to metformin [3] (Figure 1). Additionally, high concentrations of cystine found in historic media enhance glutamine consumption and dependency of cancer cells in culture [4]. Undoubtedly, the effects of non-physiological levels of nutrients present in culture media, are not limited to cancer cells. BrainPhys™ is an example of a medium developed to recapitulate in culture, specific functional phenotypes observed in the brain. In 2015, Bardy et al. formulated BrainPhys™ with a reduced concentration of neuroactive ions and amino acids in comparison to DMEM/F-12 and Neurobasal™ media. This specialized medium enabled researchers to study the electrical activity of neurons derived either from primary tissue, or from induced pluripotent stem cells-, as well as ex vivo brain explants, in culture [5]. The logic applied in the designing of BrainPhys™ raises questions on what is currently known about the availability of nutrients and metabolites in specialized tissues, and in the tumor environment. Are cells in tumors exposed to nutrient concentrations comparable to those of plasma? Do adjacent cells directly exchange nutrients between each other or via extracellular interstitial fluid? Answer to these broad questions remains largely speculative, however recent evidences suggest that in poorly vascularized pancreatic adenocarcinomas, the concentrations of specific nutrients in the interstitial fluid significantly deviates from the circulating levels [6].
Figure 1. Metabolic reactions observed in cancer cells cultured in historic and physiological media.
Arrows and names highlighted in red indicate reactions or metabolite levels enhanced in historic media, such as DMEM. Nutrients and metabolites with a dashed outline are absent in DMEM. 5-FU: 5-fluorouracil, AcCoA: acetyl-Coenzym A, ASS: argininosuccinate, ATP: adenosine triphosphate, Citn: citrulline, Fum: fumarate, FUMP: 5-fluorouracil monophosphate, HIF1α: hypoxia-inducible factor 1α, αKG: α-ketogluterate, MetF: metformin, NAD: nicotinamide adenine dinucleotide, Oaa: oxaloacetate, Orn: Ornithine, Pyr: pyruvate, UMPS: uridine monophosphate synthetase.
Under a reasonable assumption that the circulating levels of metabolites constitute a relevant source of nutrients for most normal and neoplastic tissues, in 2015 we formulated a medium with glucose, pyruvate and amino acids concentrations similar to human blood (Serum-like Modified Eagle’s Medium, SMEM [7, 8]). SMEM has lower concentrations of the amino acids found in DMEM, it has additional proteinogenic (e.g. alanine, glutamate) and non-proteinogenic amino acids (e.g. ornithine, citrulline), but still lacks many polar metabolites normally found in human plasma. In 2017 Cantor et al. described the effects on cancer cells of a more complex medium formulation with amino acid derivatives, ketone bodies, end products of organismal catabolism (e.g. urate) and other components at concentrations found in human plasma (HPLM, Table 1 [9]). Urate is the end product of purine catabolism, and Cantor et al. reported that it can regulate the biosynthesis of the pyrimidine nucleotides by inhibiting uracil monophosphate synthetase (Figure1). This enzyme is also responsible for the activation of the drug 5-flurouracil, therefore cancer cells cultured in HPLM have been shown to be less sensitive to this anticancer drug. These observations suggest that the formulation of the cell culture medium might have profound implications in the target identification and drug development processes, in particular when these focus on cell metabolism [10].
Plasmax™ is a more complex iteration of the afore mentioned SMEM and, similarly to HPLM, it aims to recapitulate more closely the nutrient composition of human plasma [11]. Plasmax™ formulation contains 66 organic components. Amongst these, arginine and pyruvate are ~10 fold less abundant in this medium than in historic ones, such as DMEM. In triple negative breast cancer (TNBC) cells, pyruvate stabilizes the hypoxia-inducible factor 1α (HIF1α) in a dose dependent manner, and at the concentrations supplemented in historic media (0.5-1mM), it induces a pseudo-hypoxic response even under atmospheric oxygen availability. Concomitantly, in cells cultured in media such as DMEM or RPMI, the high concentrations of arginine reverse the direction of the reaction catalyzed by the urea cycle enzyme, argininosuccinate lyase. This metabolic feature was not observed in cancer cells grown in Plasmax™, nor in mammary orthotopic xenografts. Furthermore, in only four days of culture in Plasmax™ the metabolic profile of TNBC spheroids resembled the metabolic landscape of orthotopic xenografts more closely than that obtained with historic media. This indicates that the metabolism of established cancer cell lines, isolated and cultured for many passages under the non-physiological selective pressure of historic media, can be rectified towards a more tumor-like state. In addition, these observations suggest that in vitro models could be further refined by culturing cells freshly isolated from patient-derived material directly in a more physiological medium.
In commonly employed media, essential components (e.g growth factors, non-polar nutrients, and trace elements) are largely provided by the serum. Hence, their concentration varies between different batches, thereby impairing the reproducibility of results between laboratories. In order to achieve a chemically defined medium which allows cells to be cultured without serum supplementation, essential components must be included in the formulation. Plasmax™, as well as some advanced commercial media, contain trace elements such as Fe, Se, Zn, Cu in the form of salts. Human metabolism largely depends on circulating levels of these elements bound to organic small molecules and proteins such as transferrin, selenoproteins, ceruloplasmin and albumin. Therefore, the physiological availability of these important metabolic catalysts can be achieved by supplementing relevant concentrations of the trace elements, coupled with appropriate carrier molecules. Essential components are not limited to trace elements. Vitamins, hormones, lipids and growth factors, normally contributed by serum, should also be considered in the attempt to achieve tumor-relevant media formulations.
Finally, it is common cell culture practice to incubate the cells with a fixed amount of medium for extended periods. This can lead to the exhaustion of heavily consumed nutrients (e.g. glucose) and an accumulation of metabolic products (e.g. lactate) far beyond the physiological ranges reported in humans. This consideration applies in particular to physiological media where the concentration of nutrients has not been artificially increased to overcome this problem. An option to prevent both nutrient exhaustion in spent medium and excessive concentrations of nutrients in fresh one, is offered by a bioreactor, called chemostat [12], which provides a constant flow of fresh and exhausted media. While this approach is widely employed in microbiology and biotechnology, it is far less practical for multiplex experiments used in cancer research. A gross approximation of a steady state level of nutrients and metabolic end-products in the cell supernatant, can be achieved by adjusting the ratio between cell number, volume of medium and time between medium renewals. This can be applied with only minor modifications of current cell culture practice, for example by increasing the frequency of medium renewal, or by adjusting the volume of medium proportionally to the number of cells.
In summary, for decades cell biologists have used media disconnected from physiology. In the last decade, the growing focus on cancer cell metabolism has contributed to the exacerbation of some of the artifacts observed by culturing cells in historic media. The implications of using nutritionally skewed media in cancer research will become more evident with the use of refined and more physiologically relevant culture media by a broader research community. In parallel, more efforts to understand the nutritional environment of tumors will provide us with more defined templates for designing better cellular models of cancer.
Acknowledgments
This work was supported by Cancer Research UK (C596/A17196, Award 23982).
References
- [1].Mckee TJ, Komarova SV. Is it time to reinvent basic cell culture medium? Am J Physiol Cell Physiol. 2017;312(5):C624–C626. doi: 10.1152/ajpcell.00336.2016. [DOI] [PubMed] [Google Scholar]
- [2].Yao T, Asayama Y. Animal-cell culture media: History, characteristics, and current issues. Reprod Med Biol. 2017;16(2):99–117. doi: 10.1002/rmb2.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gui DY, et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD + and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016;24(5):716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Muir A, Danai LV, Gui DY, Waingarten CY, Vander Heiden MG. Environmental cystine drives glutamine anaplerosis and sensitizes cells to glutaminase inhibition. Elife. 2017;6:e27713. doi: 10.7554/eLife.27713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bardy C, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci. 2015;112(20):E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sullivan MR, et al. Quantification of microenvironmental metabolites in murine cancer models reveals determinants of tumor nutrient availability. bioRxiv. 2018 Jan;:492652. doi: 10.7554/eLife.44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tardito S, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17(12):1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schug ZT, et al. Acetyl-CoA Synthetase 2 Promotes Acetate Utilization and Maintains Cancer Cell Growth under Metabolic Stress. Cancer Cell. 2015;27(1):57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cantor JR, et al. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 2017;169(2):258–272.e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Muir A, Vander Heiden MG. The nutrient environment affects therapy. Science. 2018;360(6392):962–963. doi: 10.1126/science.aar5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vande Voorde J, et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv. 2019;5(1):eaau7314. doi: 10.1126/sciadv.aau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Birsoy K, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508(7494):108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].American Accreditation HealthCare Commission. Plasma amino acids. [Accessed: 11-Apr-2019];2019 [Online]. Available: https://medlineplus.gov/ency/article/003361.htm.
- [14].Psychogios N, et al. The Human Serum Metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bleich S, Otto M, Zerr I, Kropp S, Kretzschmar HA, Wiltfang J. Creutzfeldt-Jakob Disease and Homocysteine Levels in Plasma and Cerebrospinal Fluid. Gerontology. 2005;51:142–144. doi: 10.1159/000082200. [DOI] [PubMed] [Google Scholar]
- [16].Cynober LA. Plasma Amino Acid Levels With a Note on Membrane Transport : Characteristics, Regulation, and Metabolic Significance. Nutrition. 2002;18:761–766. doi: 10.1016/s0899-9007(02)00780-3. [DOI] [PubMed] [Google Scholar]
- [17].Friesen RW, Novak EM, Hasman D, Innis SM. Relationship of Dimethylglycine, Choline, and Betaine with Oxoproline in Plasma of Pregnant Women and Their Newborn Infants. J Nutr. 2007;2(August):2641–2646. doi: 10.1093/jn/137.12.2641. [DOI] [PubMed] [Google Scholar]
- [18].Fonteh AN, Harrington RJ, Tsai A, Liao P, Harrington MG. Free amino acid and dipeptide changes in the body fluids from Alzheimer’s disease subjects. Amino Acids. 2007;32:213–224. doi: 10.1007/s00726-006-0409-8. [DOI] [PubMed] [Google Scholar]
- [19].Tavazzi B, et al. Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005;38:997–1008. doi: 10.1016/j.clinbiochem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- [20].Saito A, Takagi T, Chung T, Ohta K. Serum levels of polyamines in patients with chronic renal failure. Kidney Int Suppl. 1983;16:S234–237. [PubMed] [Google Scholar]
- [21].Costa CG, et al. Quantitative analysis of plasma acylcarnitines using gas chromatography chemical ionization mass fragmentography. J Lipid Res. 1997;38:173–182. [PubMed] [Google Scholar]
- [22].Diem K, Lentner C, Seldrup J. Geigy scientific tables. 8th rev. a. Basle: CIBA-GEIGY; 1981. [Google Scholar]
- [23].Avogaru A, Bier DM. Contribution of 3-hydroxyisobutyrate to the measurement of 3-hydroxybutyrate in human plasma : comparison of enzymatic and gas-liquid chromatography-mass spectrometry assays in normal and in diabetic subjects. J Lipid Res. 1989;30:1811–1817. [PubMed] [Google Scholar]
- [24].Lianiou E, Ioannou P. Simple spectrofluorometric determination of p-aminobenzoic and p-aminosalicylic acids in biological fluids by use of terbium-sensitized luminescence. Clin Chem. 1996;42(10):1659–1665. [PubMed] [Google Scholar]
- [25].Pita AM, et al. Plasma urea-cycle-related amino acids, ammonium levels, and urinary orotic acid excretion in short-bowel patients managed with an oral diet. Clin Nutr. 2003;22:93–98. doi: 10.1054/clnu.2002.0606. [DOI] [PubMed] [Google Scholar]
- [26].Sabbioni E, Kueera J, Pietra R, Vesterberg O. A critical review on normal concentrations of vanadium in human blood, serum, and urine. Sci Total Environ. 1996;188:49–58. doi: 10.1016/0048-9697(96)05164-9. [DOI] [PubMed] [Google Scholar]
- [27].Ince ATI, et al. Serum Copper, Ceruloplasmin and 24-h Urine Copper Evaluations in Celiac Patients. Dig Dis Sci. 2008;53:1564–1572. doi: 10.1007/s10620-007-0043-7. [DOI] [PubMed] [Google Scholar]
- [28].Dale JC, Burritt MF, Zinsmeister AR. Diurnal Variation of Serum Iron, Iron-Binding Capacity, Transferrin Saturation, and Ferritin Levels. Clin Chem. 2002;117(5):802–808. doi: 10.1309/2YT4-CMP3-KYW7-9RK1. [DOI] [PubMed] [Google Scholar]
- [29].Rokgauer M, Klein J, Kruse-Jarres JD. Trace Elements Reference Values for the Trace Elements Copper, Manganese, Selenium, and Zinc in the Serum / Plasma of Children, Adolescents, and Adults. J Trace Elem Med Biol. 1997;11(2):92–98. doi: 10.1016/S0946-672X(97)80032-6. [DOI] [PubMed] [Google Scholar]
- [30].Thuy LP, Sweetman L, Nyhan WL. A new immunochemical assay for biotin. Clin Chim Acta. 1991;202:191–198. doi: 10.1016/0009-8981(91)90049-i. [DOI] [PubMed] [Google Scholar]
- [31].Chiang E, Smith DE, Selhub J, Dallal G, Wang Y, Roubenoff R. Inflammation causes tissue-specific depletion of vitamin B6. Arthritis Res Ther. 2005;7(6):R1254–1262. doi: 10.1186/ar1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stratford M, Dennis M. High-performance liquid chromatographic determination of nicotinamide and its metabolites in human and murine plasma and urine. J Chromatogr. 1992;582(1–2):145–151. doi: 10.1016/0378-4347(92)80313-f. [DOI] [PubMed] [Google Scholar]
- [33].Gori AM, et al. Predictors of Vitamin B 6 and Folate Concentrations in Older Persons: The InCHIANTI Study. Clin Chem. 2006;52(7):1318–1324. doi: 10.1373/clinchem.2005.066217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hustad S, et al. Riboflavin, Flavin Mononucleotide, and Flavin Adenine Dinucleotide in Human Plasma and Erythrocytes at Baseline and after Low-Dose Riboflavin Supplementation. Clin Chem. 2002;48(7):1571–1577. [PubMed] [Google Scholar]
- [35].Hung S, Hung S, Tarng D, Yang W, Chen T, Huang T. Thiamine deficiency and unexplained encephalopathy in hemodialysis and peritoneal dialysis patients. Am J Kidney Dis. 2001;38(5):941–947. doi: 10.1053/ajkd.2001.28578. [DOI] [PubMed] [Google Scholar]
- [36].Singer DMI, Rust MMP, Wagner DGK, Elmadfa I. B-Vitamin Status and Concentrations of Homocysteine in Austrian Omnivores, Vegetarians and Vegans. Ann Nutr Metab. 2006;50:485–491. doi: 10.1159/000095828. [DOI] [PubMed] [Google Scholar]