Abstract
Archaic admixture is increasingly recognized as an important source of diversity in modern humans, and Neanderthal haplotypes cover 1%–3% of the genome of present-day Eurasians. Recent work has shown that archaic introgression has contributed to human phenotypic diversity, mostly through the regulation of gene expression. Yet the mechanisms through which archaic variants alter gene expression and the forces driving the introgression landscape at regulatory regions remain elusive. Here, we explored the impact of archaic introgression on transcriptional and post-transcriptional regulation. We focused on promoters and enhancers across 127 different tissues as well as on microRNA (miRNA)-mediated regulation. Although miRNAs themselves harbor few archaic variants, we found that some of these variants may have a strong impact on miRNA-mediated gene regulation. Enhancers were by far the regulatory elements most affected by archaic introgression: up to one-third of the tissues we tested presented significant enrichments. Specifically, we found strong enrichments of archaic variants in adipose-related tissues and primary T cells, even after accounting for various genomic and evolutionary confounders such as recombination rate and background selection. Interestingly, we identified signatures of adaptive introgression at enhancers of some key regulators of adipogenesis, raising the interesting hypothesis of a possible adaptation of early Eurasians to colder climates. Collectively, this study sheds new light on the mechanisms through which archaic admixture has impacted gene regulation in Eurasians and, more generally, increases our understanding of the contribution of Neanderthals to the regulation of acquired immunity and adipose homeostasis in modern humans.
Keywords: Neanderthal, archaic introgression, gene regulation, promoters, enhancers, miRNAs, T-cells, immunity, adipose tissue, adaptation
Main Text
The sequencing of the genomes of extinct human forms, such as Neanderthals or Denisovans, has enabled the mapping of archaic variants in the genomes of modern humans.1, 2, 3, 4, 5, 6, 7 This archaic introgression has functional consequences today, as introgressed variants have been reported to alter a variety of phenotypes ranging from skin pigmentation to sleeping patterns and mood disorders.8, 9 Furthermore, several studies have shown that Neanderthal haplotypes are enriched in regulatory variants, with respect to non-archaic haplotypes,10, 11 suggesting that archaic introgression might impact complex, organismal phenotypes through changes in gene expression. Indeed, up to one-quarter of Neanderthal-introgressed haplotypes have been estimated to present cis-regulatory effects across tissues and there is a bias toward downregulation of Neanderthal alleles in brain and testes.12 Furthermore, genes involved in innate immunity and interactions with RNA viruses have been reported to be enriched in Neanderthal ancestry.13, 14 Archaic variants affect, in particular, transcriptional responses to viral challenges.11, 15 A depletion of Neanderthal introgression has recently been documented in conserved coding regions and, surprisingly, in promoters,16 suggesting that archaic introgression could affect gene expression through promoter-independent mechanisms. One such example is found in post-transcriptional regulation by miRNAs; such regulation has been reported to contribute to phenotypic differences between archaic and modern humans.17, 18 Thus, the relative contributions of transcriptional and post-transcriptional mechanisms to the effects of archaic variants on gene expression remain to be determined.
Our understanding of the selective forces that shaped the landscape of archaic introgression is also rapidly growing. In most cases, archaic variants were selected against, and regions of higher selective constraint, in particular those that are X-linked or contain testis-expressed and meiotic-related genes, were depleted in archaic ancestry.1, 2, 19 Some studies have also suggested that Neanderthals had a reduced effective population size6, 7 because of a prolonged bottleneck or a deeply structured population.6, 20, 21 Natural selection in Neanderthals would thus have been less efficient in purging deleterious mutations,22, 23 a large proportion of which were removed from the genome of modern humans after their admixture with Neanderthals.16 However, archaic variants have also contributed, in some cases, to human adaptation15, 24, 25, 26, 27, 28 shortly after their introduction into modern humans or after an initial period of genetic drift.29, 30 Given the rapid evolution of regulatory regions and their potential adaptive nature,31, 32 the evolutionary dynamics of Neanderthal introgression at regulatory elements needs to be explored in further detail.
In this study, we aimed to increase knowledge about the impact archaic introgression has had on transcriptional and post-transcriptional mechanisms; we focused on promoter-, enhancer-, and microRNA (miRNA)-mediated regulation.33, 34 To this end, we first characterized the set of variants of putative Neanderthal origin — archaic SNPs (aSNPs) — as those for which one allele is both present in the Neanderthal Altai genome6 and absent in the Yoruba African population of the 1000 Genomes Project35 (see Supplemental Data). We further required aSNPs to be located in genomic regions where Neanderthal introgression has already been detected in Europe or Asia.1 We then investigated deviations in the presence or absence of aSNPs among specific classes of functional elements by measuring the density of aSNPs with respect to that of non-aSNPs in the European (CEU) and Asian (CHB) populations of the 1000 Genomes Project.35 We then compared the relative density of aSNPs at specific functional regions to that in the rest of the genome. Genomic regions were considered as enriched or depleted in aSNPs if the resulting odds ratio (OR) was significantly different from 1.
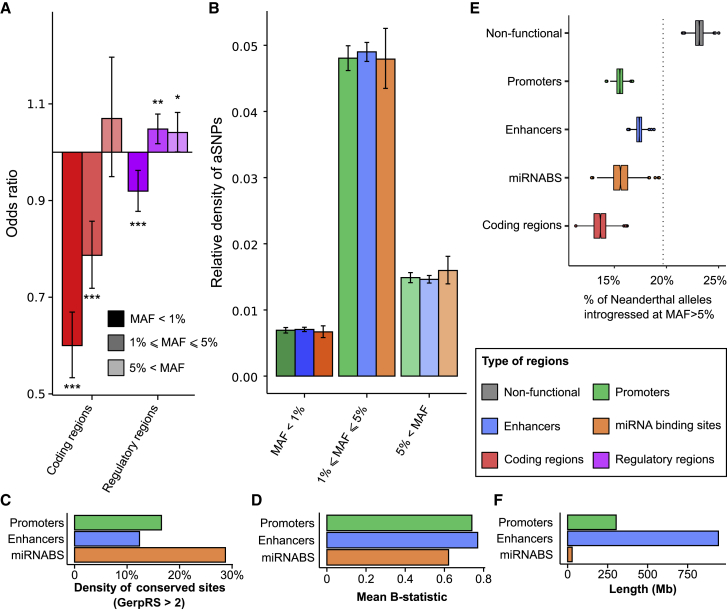
Overall, we observed a strong depletion of aSNPs in coding regions (OR = 0.71, p value < 10−4) and similar levels of introgression at regulatory regions compared to those of non-functional elements. We then divided genetic variants according to the frequency of their minor allele, which corresponds to the Neanderthal allele in 99.8% of all aSNPs. Minor-allele frequency (MAF) was computed in Eurasian populations combined, and variants were split into three bins (rare – MAF < 1%, low frequency – 1% ≤ MAF < 5%, and common – MAF ≥ 5%). In doing so, we found that the depletion in coding regions was driven by rare and low-frequency variants (OR < 0.79, p value < 2 × 10−5, Figure 1A), whereas regulatory regions were weakly enriched in low-frequency and common aSNPs (OR > 1.04, p value < 0.05). We then used the ancestral or derived state of each aSNP to distinguish between derived alleles that originated in the Neanderthal lineage (i.e., derived aSNPs, 91% of all aSNPs) and ancestral alleles that were re-introduced by Neanderthals after the fixation of the derived allele in the human lineage (i.e., ancestral aSNPs, 9% of all aSNPs, Figure S1). When comparing derived aSNPs to non-archaic variants of similar derived allele frequency (DAF), we observed a depletion of both coding and regulatory regions in rare archaic variants (DAF < 1%, OR < 0.91, p < 2 × 10−4), and there was no significant enrichment of common and low-frequency aSNPs in regulatory regions. Interestingly, when focusing on variants presenting a DAF ≥ 95% (i.e., ancestral aSNPs at ≤ 5% frequency), we observed an enrichment of archaic variants in regulatory regions (OR > 1.25, p < 2 × 10−5), highlighting the contribution that ancestral alleles re-introgressed by Neanderthals make to gene regulation in humans.
Figure 1.
Enrichment of Neanderthal Variants in Regulatory Regions
(A) Odds ratio depicting the excess or depletion of Neanderthal variants in coding regions and regulatory elements (promoters, enhancers, and miRNA binding sites) compared to the remainder of the genome. Enrichments are shown for three bins of minor-allele frequencies (MAFs), together with 95% bootstrap confidence intervals: ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗p value < 0.001.
(B) Relative density of aSNPs in promoters, enhancers, and miRNA binding sites in different MAF bins, with 95% bootstrap confidence intervals.
(C and D) Comparison of density of conserved sites (GerpRS > 2) and mean B statistic of promoters, enhancers, and miRNA binding sites.
(E) Percentage of alleles that are fixed in Neanderthal, absent from the African Yoruba from Nigeria (YRI), and introgressed at a MAF > 5% in Eurasians. For each type of region, box plots show the variability of the estimates based on 1,000 bootstrap resamples of 100 kb genomic windows. The dashed vertical line indicates the genome-wide average.
(F) Total length of promoters, enhancers, and miRNA binding sites.
To understand how introgression has impacted genetic diversity across various types of regulatory elements, we then investigated the relative density of aSNPs across promoters, enhancers, and miRNA binding sites. Although this metric differed markedly across frequency bins, it did not differ across categories of regulatory elements, despite their important differences in strength of negative or background selection (Figure 1B–1D). However, when measuring the rate at which Neanderthal alleles were introgressed in Europe or Asia, we found that they were less likely to reach high frequency (MAF > 5%) in coding or regulatory regions with respect to non-functional regions (p < 10−10, Figures 1E and S2). This effect was less marked among enhancers, and that, together with the larger size of enhancers (Figure 1F), suggests that Neanderthal variants are quantitatively more likely to affect gene regulation via modification of enhancer activity than though changes of promoter or miRNA binding sites.
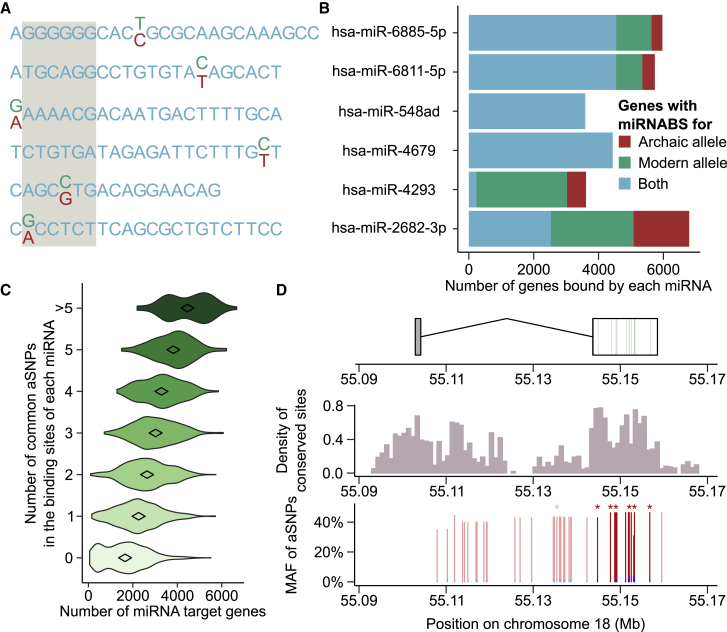
Given the low fraction of the genome that is covered by miRNAs and miRNA-binding sites (miRNABS) (Figure 1F), they are expected to be, quantitatively, the least affected by archaic introgression. Indeed, we only found six aSNPs that overlap the sequence of mature miRNAs, two of which alter the seed region (Figure 2A): rs74904371 in miR-2682-3p (MAFCHB = 0, MAFCEU = 3%) and rs12220909 in miR-4293 (MAFCHB = 17%, MAFCEU = 0). The presence of aSNPs in four of these miRNAs, particularly those located in seed regions, affected the set of genes they bind (Figure 2B and Table S1). We also detected 2,909 aSNPs in miRNABS, 29% of which were common (Table S2). We found a direct linear relationship between the number of genes bound by a miRNA and the number of aSNPs in its binding sites (r = 0.56, p value < 10−10, Figure 2C), suggesting that introgression affected miRNABS independently of their cognate miRNAs. As a pertinent example, the ONECUT2 locus (MIM: 604894) presents the highest number of aSNPs that alter conserved miRNABS (Figure 2D) and has been previously reported to be a likely target of adaptive introgression.24 This gene, which encodes a member of the onecut family of transcription factors, contains 13 aSNPs that alter miRNABS, six of which are highly conserved (GerpRS > 2). Interestingly, these aSNPs fall within the 0.4% most-differentiated aSNPs between Europeans and Asians at the genome-wide level (FST > 0.38). We also detected aSNPs, mostly population specific, that alter conserved miRNABS at several key immune genes, including CXCR5 (MIM: 601613; MAFCHB = 16%, MAFCEU = 1%), TLR6 (MIM: 605403; MAFCHB = 8%, MAFCEU = 0), IL7R (MIM: 146661; MAFCHB = 8%, MAFCEU = 0), and IL21 (MIM: 605384; MAFCHB = 0, MAFCEU = 8%).
Figure 2.
Effects of Archaic Introgression on miRNA-Mediated Regulation
(A) Representation of the archaic (red) and modern (green) human alleles for the six miRNAs presenting a Neanderthal-introgressed variant in their mature sequence. The seed region of the miRNAs is shaded in gray.
(B) Total number of genes bound by the archaic and/or modern human allele of each of the six miRNAs harboring a Neanderthal variant in their mature sequence.
(C) Relationship between the number of targets of each miRNA and the number of common aSNPs in the corresponding miRNA binding sites.
(D) Introgression of aSNPs altering the miRNABS at the ONECUT2 locus (MIM: 604894). Gene structure is shown in the upper panel, and miRNA binding sites that are altered by archaic introgression are highlighted in green. The middle panel represents the density of conserved sites (GerpRS > 2) in 1,000 bp windows, and the bottom panel represents the repartition and frequency of archaic alleles at the locus (blue for CEU, red for CHB). aSNPs that overlap miRNABS are represented with a darker shade, and aSNPs that disrupt a conserved site are marked with stars.
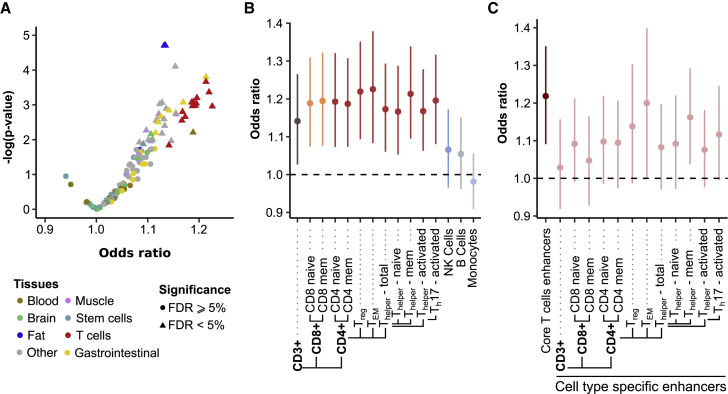
Next, we focused on how archaic introgression has affected promoters and enhancers. Given the tissue-specific impact of archaic introgression on gene regulation,10, 12 we searched for enrichments in Neanderthal ancestry across regulatory elements in 127 different tissues.33 The impact of archaic introgression in promoters was similar to that in the remainder of the genome in all tissues and frequency bins (Table S3). Conversely, we found that enhancers are enriched in common aSNPs in 42 tissues (FDR < 5%, Figure 3A and Table S4), and we detected similar patterns in CEU and CHB populations (r = 0.62, Figure S3). Among the 42 tissues presenting significant enrichments, adipose-derived mesenchymal stem cells (AdMSCs) and mesenchymal stem-cell-derived adipocytes were the most enriched (OR > 1.13, p value < 3 × 10−5), followed by fetal heart (OR = 1.15, p value = 8 × 10−5), small intestine (OR = 1.21, p value = 2 × 10−4), and different T cell tissues (OR > 1.14, p value < 1.5 × 10−2). When restricting our analyses to derived aSNPs (and using SNPs with DAF < 50% as background set), we replicated the enrichments at enhancers for 27 tissues (FDR < 5%, Tables S3 and S4), indicating that the impact of archaic introgression for these tissues is driven by Neanderthal-derived variants.
Figure 3.
Effects of Archaic Introgression at Enhancers
(A) Volcano plot illustrating the enrichment of common aSNPs in the enhancers of 127 different tissues from the Epigenomic Roadmap Consortium. Tissues with FDR < 5% (triangles) are significantly enriched.
(B) Enrichments of common aSNPs in the enhancers of different immune tissues. Vertical bars indicate 95% confidence intervals computed by bootstrap analysis.
(C) Enrichment of common aSNPs in the enhancers that are active in more than half of the investigated T cell subtypes (dark red, referred to as “core T cells”) and in enhancers that are active in each T cell subtype and are not part of core T cell enhancers (light red, referred to as “cell-type-specific enhancers”). Vertical bars indicate 95% confidence intervals computed by bootstrap.
(B and C) Note that CD4+ T cells are separated on the basis of CD25 so that Treg (CD25+), TEM (CD25Iow), and Thelper (CD25−) are distinguished from one another.
Focusing on circulating immune cell types (Figure 3B), we found enrichments among enhancers of various types of primary T cells, the most significant being CD4+/CD25− memory T cells (OR = 1.21, p value = 2.2 × 10−4), whereas enhancers of B cells, monocytes, and natural killer cells exhibited a density of common aSNPs similar to genome-wide expectations. We also observed that shared enhancers across different T cell subtypes (i.e., active in more than half of T cell subtypes, “core T cell enhancers”) display an enrichment in aSNPs (OR = 1.22, p value = 5 × 10−4, Figure 3C) with respect to more specialized enhancers that are only active in a small fraction of T cell subtypes.
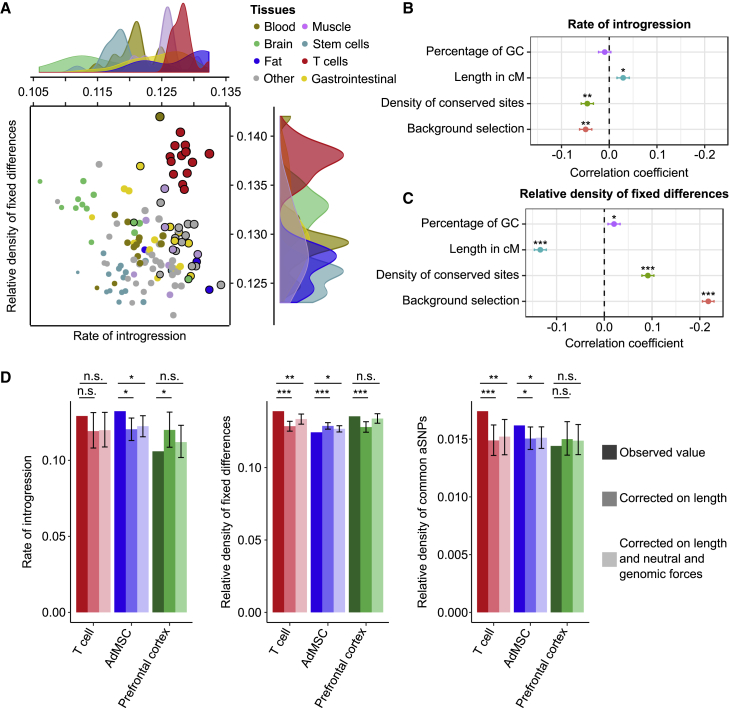
We sought to assess whether the enrichment in aSNPs detected in enhancers resulted from an excessive divergence of these elements in the Neanderthal lineage or from a higher rate of archaic introgression at enhancers. We quantified the number of fixed human-Neanderthal differences at enhancers across the 127 tissues and focused on sites where both the Altai and Vindija Neanderthal genomes6, 7 differ from the ancestral sequence. We uncovered large tissue variability; we found that enhancers active in induced pluripotent stem cells presented the highest divergence (290 differences/Mb) and that those active in pancreas cells showed the lowest (220 differences/Mb). However, given that the number of fixed differences strongly correlates with genetic diversity (i.e., density of common variants, r = 0.71, p value < 10−20), we measured the ratio of the number of fixed differences between humans and Neanderthals differences to that of common, segregating SNPs in the region. Using this metric, we found that enhancers of T cells displayed the strongest divergence (7% increase compared to the mean across tissues, Wilcoxon p value < 2 × 10−8), whereas stem cells showed the lowest (4% decrease, Wilcoxon p value < 7 × 10−6) (Figure 4A). Focusing on the rate of introgression, which is defined as the proportion of Neanderthal-descended alleles that are present in the human genome at a MAF > 5%, we found that enhancers of T cells showed the highest percentage (5% increase, Wilcoxon p value < 2 × 10−5), whereas brain cells showed the lowest percentage (7% decrease, Wilcoxon p value < 4 × 10−5) (Figure 4A).
Figure 4.
Factors Shaping Human-Neanderthal Divergence and Archaic Introgression at Enhancers
(A) Comparison of the relative density of fixed human-Neanderthal differences and rate of introgression in the enhancers of the 127 tissues studied. The size of the circles is proportional to the relative density of common aSNPs in the enhancers of the corresponding tissue; a black circle is added when the relative density of common aSNPs is significantly higher in these enhancers (FDR < 5%) than in the rest of the genome. The density of each tissue category along the two axes is also presented.
(B and C) Genome-wide correlations, using 100 kb windows, between either the rate of Neanderthal introgression (B) or the relative density of fixed human-Neanderthal differences (C) and neutral and selective forces. ∗p value < 10−2, ∗∗p value < 10−10, and ∗∗∗p value < 10−20. For each correlation, horizontal lines indicate 95% confidence interval.
(D) Observed values of rate of introgression and relative density of fixed differences and common aSNPs at the enhancers of core T cells, AdMSCs, and prefrontal cortex, with respect to expectations based on 100 kb windows matched for length of enhancers alone or for length of enhancers, percentage of GC, recombination rate, density of conserved sites, and mean B statistic of their enhancers (see Supplemental Data). n.s. = not significant; ∗p value < 0.05, ∗∗p value < 0.01, and ∗∗∗p value < 10−3. Errors bars indicate 95% confidence intervals of the expected values obtained by resampling.
We then explored the factors that might drive, at the genome-wide level, the detected variation in Neanderthal divergence and archaic introgression. Using 100 kb windows, we correlated divergence and introgression with metrics that capture local variation in neutral (mutation and recombination) and selected (negative and background selection) diversity. Specifically, we measured the percentage of guanine-cytosine (GC) to account for their higher mutability, genetic size as measure of recombination rate, density of conserved sites (GerpRS > 2) as a measure of negative selection, and background selection derived as (1 − B), where B is the mean B statistic in the window.36 We found that background selection correlated with a lower rate of archaic introgression (r = −0.049, p value < 10−15, Figure 4B), consistent with previous findings,1, 2 but also correlated with increased local divergence (r = 0.22, p value < 10−20, Figure 4C) and reduced density of both common variants and fixed differences (r = −0.46 and −0.05, respectively, p value < 5 × 10−20, Figure S4). We also found that negative selection and recombination rate correlated with both divergence and introgression, even after we adjusted for background selection (Figure S5).
To understand further how these factors could account for the variation in divergence and introgression detected at enhancers (Figure S6), we focused on three model tissues: T cells (enhancers with high divergence and introgression), AdMSCs (enhancers with low divergence and high introgression), and prefrontal cortex (enhancers with high divergence and low introgression) (Figure 4D). When correcting for the various neutral and selective factors, we found that introgression at T cell enhancers did not exceed that of other tissues (p value > 0.11), but the high divergence and relative density of aSNPs remained significant (p value < 8 × 10−3). For AdMSCs, introgression remained higher than expected (p value = 4 × 10−3), leading to an excess of aSNPs despite their depletion in divergence (p value = 3.8 × 10−2). For enhancers active at the prefrontal cortex, all variables were within expected bounds. Collectively, these analyses indicate that variation of several neutral and selective factors is not sufficient to explain the excess of Neanderthal introgression detected at enhancers. Some enhancers might have undergone past adaptation in the Neanderthal lineage or adaptive introgression in modern humans, as illustrated by T cells and AdMSCs, respectively.
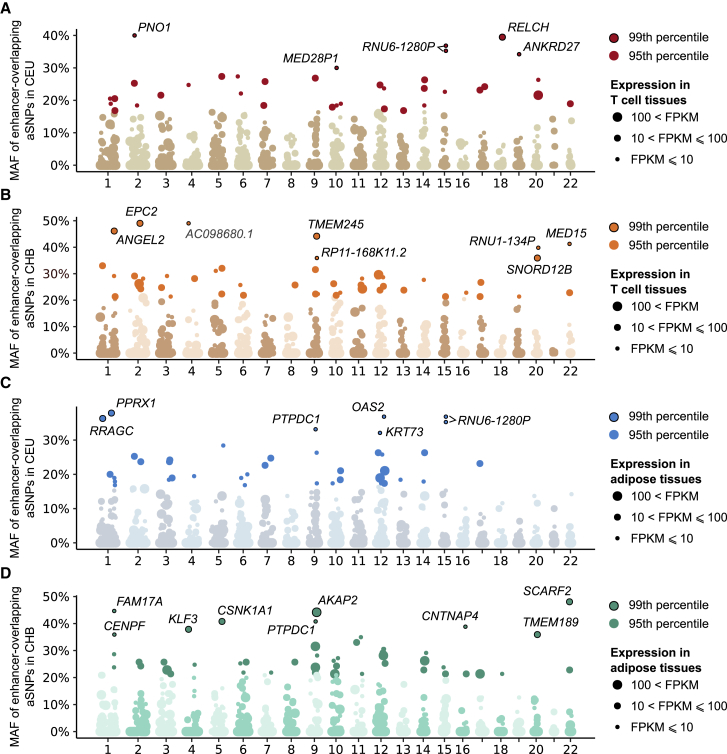
Finally, we explored the impact of archaic introgression at enhancers on gene expression. To identify genes whose expression is altered by Neanderthal introgression at enhancers, we focused on tissues where data from promoter capture Hi-C were available37, 38 and assigned each enhancer located in a promoter-interacting region to the corresponding gene(s). Archaic variants at enhancers predicted to interact with a gene were strongly enriched in eQTLs (OR = 2.6, p value < 10−3, Supplemental Note 1 and Figure S7), further supporting the regulatory potential of aSNPs. Genes interacting with T cell enhancers that harbor common aSNPs (n = 1,629, Table S5) were not enriched in any specific biological function. However, 285 of these genes are highly expressed in T cells (fragments per kilobase of transcript per million mapped reads [FPKM] > 100) and include known regulators of the immune response (e.g., CXCR4 [MIM: 162643], IL7R [MIM: 146661], IL10RA [MIM: 146933], NFKBIA [MIM: 164008], and PTPRC [MIM: 151460]). We found 14 loci presenting signatures of adaptive introgression; i.e., these were genes that interact with enhancers harboring very-high-frequency aSNPs (99th percentile of MAF: MAFCEU > 0.29 or MAFCHB > 0.35; Figures 5A and 5B). Among these, we found ANKRD27, which is associated with eosinophilic esophagitis (MIM: 610247),39 and MED15 (MIM: 607372), which is involved in several cancers.40, 41, 42 With respect to adipose-related tissues, we identified 690 genes — 43 of which were highly expressed (FPKM > 100) in the adipose tissue — interacting with AdMSC enhancers that contain common aSNPs (Table S6). These genes were enriched in functions related to the regulation of cell motility (GO: 2000145, p value < 2.0 × 10−8) and insulin-like growth factor binding protein complex (GO: 0016942, p value < 2.7 × 10−5) (Table S7). We detected 16 aSNPs at AdMSC enhancers that present strong signatures of adaptive introgression (Figures 5C and 5D).
Figure 5.
Manhattan Plots of Genes Interacting with Enhancers That Contain Archaic Variants
(A and B) Genome-wide distribution of MAFs in CEU or CHB at aSNPs that overlap enhancers active in T Cells (core T cell enhancers). For each window of 1 Mb along the genome, only the aSNP with the highest MAF is shown. Point sizes reflect FPKM of the most expressed genes (max FPKM across T lymphocytes from Blueprint database58) among genes interacting with the enhancer in T cells.37
(C and D) Similar plots for enhancers active in AdMSCs. Point sizes reflect the FPKM of the most expressed gene (max FPKM in GTEx tissues Adipose—Subcutaneous and Adipose—Visceral [Omentum]59) among genes interacting with the enhancer in adipose tissue.38
This study reconstructs the history of how Neanderthal introgression has affected various types of regulatory elements as well as the mechanistic bases through which archaic variants have altered gene regulation. Previous studies have shown that archaic variants are more likely to correlate with gene expression than non-archaic variants segregating at the same frequency.10, 11 Our approach differs from these studies in that it excludes indirect effects from non-archaic variants segregating on introgressed haplotypes and that it focuses on the direct regulatory potential of archaic alleles. In doing so, we find little evidence for an enrichment of common archaic variants in regulatory regions taken as a whole; these results might seem at odds with previous studies. Yet one should note that the functional impact of the archaic material we measure can be decomposed in two separate components: (1) the frequency at which Neanderthal haplotypes are introgressed into the human lineage, which corresponds to the rate of introgression measured by the f4-ratio statistics,16 and (2) the degree of human-Neanderthal divergence at regulatory elements, which determines the probability that introgressed haplotypes carry a functional variant. Indeed, when focusing on the rate of introgression, we find that Neanderthal alleles were introgressed at a lower rate in regulatory regions, consistent with recent findings for promoter regions.16 This lower introgression rate is nevertheless compensated by a higher human-Neanderthal divergence at regulatory regions (Figure S8), which is consistent with an increased probability that Neanderthal haplotypes are associated with gene expression.10, 11
We also explored how Neanderthal introgression has impacted miRNA-mediated regulation, and we showed that although miRNAs harbor few archaic variants per se, some of them might impact strongly miRNA-mediated gene regulation and disease risk. For example, the archaic allele at miR-4293 (rs12220909) is responsible for the loss of 95% of its targets and has been associated with diminished cancer susceptibility.43, 44 Archaic introgression has also affected miRNA binding sites, as illustrated by ONECUT2 (MIM: 604894), where an archaic haplotype that is present at a high frequency in Asia (MAFCHB = 0.49) alters multiple conserved miRNA binding sites. ONECUT2 is involved in liver, pancreas, and nervous-system development45 and has recently been proposed as a regulator of tumor growth in ovarian cancer (MIM: 167000).46
Finally, our study reveals that archaic introgression has impacted enhancers in a tissue-specific manner, reflecting either high human-Neanderthal differentiation, as observed in T cell enhancers, or increased archaic introgression, as detected in AdMSCs. Interestingly, the AdMSC enhancers impacted by archaic introgression interact preferentially with genes involved in the regulation of adipocyte differentiation and adipogenesis. These include receptors such as PDGFRB (MIM: 173410) and TGFBR2 (MIM: 190182), the insulin growth factor IGF1 (MIM: 147440) and its binding partners IGFBP2 (MIM: 146731) and IGFBP3 (MIM: 146732), and the CXCR4 chemokine (MIM: 162643).47, 48, 49, 50, 51, 52, 53 Furthermore, two of the enhancers harboring archaic variants at the highest frequencies interact with key adipocyte-differentiation regulators, such as KLF3 (MIM: 609392) and PRRX1 (MIM: 167420),54, 55 suggesting that introgression at AdMSC might have been adaptive in humans. In support of this notion, Dannemann and colleagues have found that more than half of aSNPs associated with gene expression in subcutaneous adipose tissue had increased in frequency over the last 10,000 years, whereas the majority of aSNPs had decreased in frequency over the same period of time.10 Given the proposed adaptation of Neanderthals to cold environments,56 it is tempting to speculate that archaic alleles at enhancers of AdMSCs provided a selective advantage to early modern humans during their migration out of Africa. This hypothesis becomes particularly interesting in light of previously reported cases of adaptive introgression at the LEPR (MIM: 601007) locus and at the locus of the WARS2 (MIM: 604733) and TBX15 (MIM: 604127) genes, both loci being involved in the regulation of adipose tissue differentiation and body-fat distribution.25, 57 Further studies aiming to functionally characterize the regulatory effects of Neanderthal variants on adipocyte differentiation and fat distribution are now required, as these archaic variants might have contributed to the adaptation of early Eurasians to colder climates.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), and the Agence Nationale de la Recherche (ANR) grants: “IEIHSEER” ANR-14-CE14-0008-02 and “TBPATHGEN” ANR-14-CE14-0007-02. The laboratory of L.Q.M. has received funding from the French government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10- LABX-62-IBEID). M.S. was funded by the Ecole Doctorale “Complexité du vivant,” Sorbonne Université (contract n°2532/2016).
Published: May 30, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.016.
Contributor Information
Lluis Quintana-Murci, Email: quintana@pasteur.fr.
Maxime Rotival, Email: maxime.rotival@pasteur.fr.
Web Resources
1000 Genomes Project, http://www.internationalgenome.org/
Epigenomic Roadmaps, https://egg2.wustl.edu/roadmap/web_portal/
Genotype-Tissue Expression (GTEx) project, https://gtexportal.org/home/
miRanda Software, http://www.microrna.org/microrna/
Neanderthal Genomes, http://cdna.eva.mpg.de/neandertal/
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sankararaman, S., Mallick, S., Dannemann, M., Prufer, K., Kelso, J., Paabo, S., Patterson, N., and Reich, D. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354-357. [DOI] [PMC free article] [PubMed]
- 2.Sankararaman S., Mallick S., Patterson N., Reich D. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sankararaman, S., Mallick, S., Patterson, N., and Reich, D. (2016). The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26, 1241-1247. [DOI] [PMC free article] [PubMed]
- 3.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]; Vernot, B., and Akey, J.M. (2014). Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017-1021. [DOI] [PubMed]
- 4.Vernot B., Tucci S., Kelso J., Schraiber J.G., Wolf A.B., Gittelman R.M., Dannemann M., Grote S., McCoy R.C., Norton H. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vernot, B., Tucci, S., Kelso, J., Schraiber, J.G., Wolf, A.B., Gittelman, R.M., Dannemann, M., Grote, S., McCoy, R.C., Norton, H., et al. (2016). Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235-239. [DOI] [PMC free article] [PubMed]
- 5.Browning S.R., Browning B.L., Zhou Y., Tucci S., Akey J.M. Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell. 2018;173:53–61.e9. doi: 10.1016/j.cell.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Browning, S.R., Browning, B.L., Zhou, Y., Tucci, S., and Akey, J.M. (2018). Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell 173, 53-61.e9. [DOI] [PMC free article] [PubMed]
- 6.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prufer, K., Racimo, F., Patterson, N., Jay, F., Sankararaman, S., Sawyer, S., Heinze, A., Renaud, G., Sudmant, P.H., de Filippo, C., et al. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43-49. [DOI] [PMC free article] [PubMed]
- 7.Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P., Hajdinjak M., Vernot B., Skov L., Hsieh P., Peyrégne S. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prufer, K., de Filippo, C., Grote, S., Mafessoni, F., Korlević, P., Hajdinjak, M., Vernot, B., Skov, L., Hsieh, P., Peyregne, S., et al. (2017). A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655-658. [DOI] [PMC free article] [PubMed]
- 8.Dannemann M., Kelso J. The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet. 2017;101:578–589. doi: 10.1016/j.ajhg.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dannemann, M., and Kelso, J. (2017). The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet. 101, 578-589. [DOI] [PMC free article] [PubMed]
- 9.Simonti C.N., Vernot B., Bastarache L., Bottinger E., Carrell D.S., Chisholm R.L., Crosslin D.R., Hebbring S.J., Jarvik G.P., Kullo I.J. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351:737–741. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simonti, C.N., Vernot, B., Bastarache, L., Bottinger, E., Carrell, D.S., Chisholm, R.L., Crosslin, D.R., Hebbring, S.J., Jarvik, G.P., Kullo, I.J., et al. (2016). The phenotypic legacy of admixture between modern humans and Neandertals. Science 351, 737-741. [DOI] [PMC free article] [PubMed]
- 10.Dannemann M., Prüfer K., Kelso J. Functional implications of Neandertal introgression in modern humans. Genome Biol. 2017;18:61. doi: 10.1186/s13059-017-1181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dannemann, M., Prufer, K., and Kelso, J. (2017). Functional implications of Neandertal introgression in modern humans. Genome Biol. 18, 61. [DOI] [PMC free article] [PubMed]
- 11.Quach H., Rotival M., Pothlichet J., Loh Y.E., Dannemann M., Zidane N., Laval G., Patin E., Harmant C., Lopez M. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quach, H., Rotival, M., Pothlichet, J., Loh, Y.E., Dannemann, M., Zidane, N., Laval, G., Patin, E., Harmant, C., Lopez, M., et al. (2016). Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167, 643-656.e17. [DOI] [PMC free article] [PubMed]
- 12.McCoy R.C., Wakefield J., Akey J.M. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168:916–927.e12. doi: 10.1016/j.cell.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; McCoy, R.C., Wakefield, J., and Akey, J.M. (2017). Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell 168, 916-927.e12. [DOI] [PMC free article] [PubMed]
- 13.Deschamps M., Laval G., Fagny M., Itan Y., Abel L., Casanova J.L., Patin E., Quintana-Murci L. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 2016;98:5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deschamps, M., Laval, G., Fagny, M., Itan, Y., Abel, L., Casanova, J.L., Patin, E., and Quintana-Murci, L. (2016). Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98, 5-21. [DOI] [PMC free article] [PubMed]
- 14.Enard D., Petrov D.A. Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell. 2018;175:360–371.e13. doi: 10.1016/j.cell.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Enard, D., and Petrov, D.A. (2018). Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell 175, 360-371.e13. [DOI] [PMC free article] [PubMed]
- 15.Sams A.J., Dumaine A., Nédélec Y., Yotova V., Alfieri C., Tanner J.E., Messer P.W., Barreiro L.B. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 2016;17:246. doi: 10.1186/s13059-016-1098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sams, A.J., Dumaine, A., Nedelec, Y., Yotova, V., Alfieri, C., Tanner, J.E., Messer, P.W., and Barreiro, L.B. (2016). Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 17, 246. [DOI] [PMC free article] [PubMed]
- 16.Petr M., Pääbo S., Kelso J., Vernot B. Limits of long-term selection against Neandertal introgression. Proc. Natl. Acad. Sci. USA. 2019;116:1639–1644. doi: 10.1073/pnas.1814338116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petr, M., Paabo, S., Kelso, J., and Vernot, B. (2019). Limits of long-term selection against Neandertal introgression. Proc. Natl. Acad. Sci. USA 116, 1639-1644. [DOI] [PMC free article] [PubMed]
- 17.Lopez-Valenzuela M., Ramírez O., Rosas A., García-Vargas S., de la Rasilla M., Lalueza-Fox C., Espinosa-Parrilla Y. An ancestral miR-1304 allele present in Neanderthals regulates genes involved in enamel formation and could explain dental differences with modern humans. Mol. Biol. Evol. 2012;29:1797–1806. doi: 10.1093/molbev/mss023. [DOI] [PubMed] [Google Scholar]; Lopez-Valenzuela, M., Ramirez, O., Rosas, A., Garcia-Vargas, S., de la Rasilla, M., Lalueza-Fox, C., and Espinosa-Parrilla, Y. (2012). An ancestral miR-1304 allele present in Neanderthals regulates genes involved in enamel formation and could explain dental differences with modern humans. Mol. Biol. Evol. 29, 1797-1806. [DOI] [PubMed]
- 18.Gunbin K.V., Afonnikov D.A., Kolchanov N.A., Derevianko A.P., Rogaev E.I. The evolution of Homo sapiens denisova and Homo sapiens neanderthalensis miRNA targeting genes in the prenatal and postnatal brain. BMC Genomics. 2015;16(Suppl 13):S4. doi: 10.1186/1471-2164-16-S13-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gunbin, K.V., Afonnikov, D.A., Kolchanov, N.A., Derevianko, A.P., and Rogaev, E.I. (2015). The evolution of Homo sapiens denisova and Homo sapiens neanderthalensis miRNA targeting genes in the prenatal and postnatal brain. BMC Genomics 16 (Suppl 13), S4. [DOI] [PMC free article] [PubMed]
- 19.Jégou B., Sankararaman S., Rolland A.D., Reich D., Chalmel F. Meiotic genes are enriched in regions of reduced archaic ancestry. Mol. Biol. Evol. 2017;34:1974–1980. doi: 10.1093/molbev/msx141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jegou, B., Sankararaman, S., Rolland, A.D., Reich, D., and Chalmel, F. (2017). Meiotic genes are enriched in regions of reduced archaic ancestry. Mol. Biol. Evol. 34, 1974-1980. [DOI] [PMC free article] [PubMed]
- 20.Kuhlwilm M., Gronau I., Hubisz M.J., de Filippo C., Prado-Martinez J., Kircher M., Fu Q., Burbano H.A., Lalueza-Fox C., de la Rasilla M. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kuhlwilm, M., Gronau, I., Hubisz, M.J., de Filippo, C., Prado-Martinez, J., Kircher, M., Fu, Q., Burbano, H.A., Lalueza-Fox, C., de la Rasilla, M., et al. (2016). Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429-433. [DOI] [PMC free article] [PubMed]
- 21.Rogers A.R., Bohlender R.J., Huff C.D. Early history of Neanderthals and Denisovans. Proc. Natl. Acad. Sci. USA. 2017;114:9859–9863. doi: 10.1073/pnas.1706426114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rogers, A.R., Bohlender, R.J., and Huff, C.D. (2017). Early history of Neanderthals and Denisovans. Proc. Natl. Acad. Sci. USA 114, 9859-9863. [DOI] [PMC free article] [PubMed]
- 22.Harris K., Nielsen R. The genetic cost of Neanderthal Introgression. Genetics. 2016;203:881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harris, K., and Nielsen, R. (2016). The genetic cost of Neanderthal Introgression. Genetics 203, 881-891. [DOI] [PMC free article] [PubMed]
- 23.Juric I., Aeschbacher S., Coop G. The strength of selection against Neanderthal introgression. PLoS Genet. 2016;12:e1006340. doi: 10.1371/journal.pgen.1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Juric, I., Aeschbacher, S., and Coop, G. (2016). The strength of selection against Neanderthal introgression. PLoS Genet. 12, e1006340. [DOI] [PMC free article] [PubMed]
- 24.Racimo F., Marnetto D., Huerta-Sánchez E. Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol. 2017;34:296–317. doi: 10.1093/molbev/msw216. [DOI] [PMC free article] [PubMed] [Google Scholar]; Racimo, F., Marnetto, D., and Huerta-Sanchez, E. (2017). Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol. 34, 296-317. [DOI] [PMC free article] [PubMed]
- 25.Racimo F., Gokhman D., Fumagalli M., Ko A., Hansen T., Moltke I., Albrechtsen A., Carmel L., Huerta-Sánchez E., Nielsen R. Archaic adaptive introgression in TBX15/WARS2. Mol. Biol. Evol. 2017;34:509–524. doi: 10.1093/molbev/msw283. [DOI] [PMC free article] [PubMed] [Google Scholar]; Racimo, F., Gokhman, D., Fumagalli, M., Ko, A., Hansen, T., Moltke, I., Albrechtsen, A., Carmel, L., Huerta-Sanchez, E., and Nielsen, R. (2017). Archaic adaptive introgression in TBX15/WARS2. Mol. Biol. Evol. 34, 509-524. [DOI] [PMC free article] [PubMed]
- 26.Gittelman R.M., Schraiber J.G., Vernot B., Mikacenic C., Wurfel M.M., Akey J.M. Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr. Biol. 2016;26:3375–3382. doi: 10.1016/j.cub.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gittelman, R.M., Schraiber, J.G., Vernot, B., Mikacenic, C., Wurfel, M.M., and Akey, J.M. (2016). Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr. Biol. 26, 3375-3382. [DOI] [PMC free article] [PubMed]
- 27.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]; Racimo, F., Sankararaman, S., Nielsen, R., and Huerta-Sanchez, E. (2015). Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359-371. [DOI] [PMC free article] [PubMed]
- 28.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huerta-Sanchez, E., Jin, X., Asan, Bianba, Z., Peter, B.M., Vinckenbosch, N., Liang, Y., Yi, X., He, M., Somel, M., et al. (2014). Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194-197. [DOI] [PMC free article] [PubMed]
- 29.Jagoda E., Lawson D.J., Wall J.D., Lambert D., Muller C., Westaway M., Leavesley M., Capellini T.D., Mirazón Lahr M., Gerbault P. Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol. Biol. Evol. 2017;35:623–630. doi: 10.1093/molbev/msx314. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jagoda, E., Lawson, D.J., Wall, J.D., Lambert, D., Muller, C., Westaway, M., Leavesley, M., Capellini, T.D., Mirazon Lahr, M., Gerbault, P., et al. (2017). Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol. Biol. Evol. 35, 623-630. [DOI] [PMC free article] [PubMed]
- 30.Dannemann M., Racimo F. Something old, something borrowed: admixture and adaptation in human evolution. Curr. Opin. Genet. Dev. 2018;53:1–8. doi: 10.1016/j.gde.2018.05.009. [DOI] [PubMed] [Google Scholar]; Dannemann, M., and Racimo, F. (2018). Something old, something borrowed: admixture and adaptation in human evolution. Curr. Opin. Genet. Dev. 53, 1-8. [DOI] [PubMed]
- 31.Kudaravalli S., Veyrieras J.B., Stranger B.E., Dermitzakis E.T., Pritchard J.K. Gene expression levels are a target of recent natural selection in the human genome. Mol. Biol. Evol. 2009;26:649–658. doi: 10.1093/molbev/msn289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kudaravalli, S., Veyrieras, J.B., Stranger, B.E., Dermitzakis, E.T., and Pritchard, J.K. (2009). Gene expression levels are a target of recent natural selection in the human genome. Mol. Biol. Evol. 26, 649-658. [DOI] [PMC free article] [PubMed]
- 32.Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M., Park T.J., Deaville R., Erichsen J.T., Jasinska A.J. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Villar, D., Berthelot, C., Aldridge, S., Rayner, T.F., Lukk, M., Pignatelli, M., Park, T.J., Deaville, R., Erichsen, J.T., Jasinska, A.J., et al. (2015). Enhancer evolution across 20 mammalian species. Cell 160, 554-566. [DOI] [PMC free article] [PubMed]
- 33.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., Kheradpour, P., Zhang, Z., Wang, J., Ziller, M.J., et al.; Roadmap Epigenomics Consortium (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. [DOI] [PMC free article] [PubMed]
- 34.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed]
- 35.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]; 35. 1000 Genomes Project Consortium, Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Korbel, J.O., Marchini, J.L., McCarthy, S., McVean, G.A., Abecasis, G.R. (2015). A global reference for human genetic variation. Nature 526, 68-74. [DOI] [PMC free article] [PubMed]
- 36.McVicker G., Gordon D., Davis C., Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]; McVicker, G., Gordon, D., Davis, C., and Green, P. (2009). Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5, e1000471. [DOI] [PMC free article] [PubMed]
- 37.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., BLUEPRINT Consortium Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; Javierre, B.M., Burren, O.S., Wilder, S.P., Kreuzhuber, R., Hill, S.M., Sewitz, S., Cairns, J., Wingett, S.W., Varnai, C., Thiecke, M.J., et al.; BLUEPRINT Consortium (2016). Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369-1384.e19. [DOI] [PMC free article] [PubMed]
- 38.Pan D.Z., Garske K.M., Alvarez M., Bhagat Y.V., Boocock J., Nikkola E., Miao Z., Raulerson C.K., Cantor R.M., Civelek M. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 2018;9:1512. doi: 10.1038/s41467-018-03554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pan, D.Z., Garske, K.M., Alvarez, M., Bhagat, Y.V., Boocock, J., Nikkola, E., Miao, Z., Raulerson, C.K., Cantor, R.M., Civelek, M., et al. (2018). Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 9, 1512. [DOI] [PMC free article] [PubMed]
- 39.Sleiman P.M., Wang M.L., Cianferoni A., Aceves S., Gonsalves N., Nadeau K., Bredenoord A.J., Furuta G.T., Spergel J.M., Hakonarson H. GWAS identifies four novel eosinophilic esophagitis loci. Nat. Commun. 2014;5:5593. doi: 10.1038/ncomms6593. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sleiman, P.M., Wang, M.L., Cianferoni, A., Aceves, S., Gonsalves, N., Nadeau, K., Bredenoord, A.J., Furuta, G.T., Spergel, J.M., and Hakonarson, H. (2014). GWAS identifies four novel eosinophilic esophagitis loci. Nat. Commun. 5, 5593. [DOI] [PMC free article] [PubMed]
- 40.Syring I., Weiten R., Müller T., Schmidt D., Steiner S., Kristiansen G., Müller S.C., Ellinger J. The knockdown of the Mediator complex subunit MED15 restrains urothelial bladder cancer cells’ malignancy. Oncol. Lett. 2018;16:3013–3021. doi: 10.3892/ol.2018.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Syring, I., Weiten, R., Muller, T., Schmidt, D., Steiner, S., Kristiansen, G., Muller, S.C., and Ellinger, J. (2018). The knockdown of the Mediator complex subunit MED15 restrains urothelial bladder cancer cells’ malignancy. Oncol. Lett. 16, 3013-3021. [DOI] [PMC free article] [PubMed]
- 41.Weiten R., Müller T., Schmidt D., Steiner S., Kristiansen G., Müller S.C., Ellinger J., Syring I. The Mediator complex subunit MED15, a promoter of tumour progression and metastatic spread in renal cell carcinoma. Cancer Biomark. 2018;21:839–847. doi: 10.3233/CBM-170757. [DOI] [PubMed] [Google Scholar]; Weiten, R., Muller, T., Schmidt, D., Steiner, S., Kristiansen, G., Muller, S.C., Ellinger, J., and Syring, I. (2018). The Mediator complex subunit MED15, a promoter of tumour progression and metastatic spread in renal cell carcinoma. Cancer Biomark. 21, 839-847. [DOI] [PubMed]
- 42.Shaikhibrahim Z., Offermann A., Halbach R., Vogel W., Braun M., Kristiansen G., Bootz F., Wenzel J., Mikut R., Lengerke C. Clinical and molecular implications of MED15 in head and neck squamous cell carcinoma. Am. J. Pathol. 2015;185:1114–1122. doi: 10.1016/j.ajpath.2014.12.010. [DOI] [PubMed] [Google Scholar]; Shaikhibrahim, Z., Offermann, A., Halbach, R., Vogel, W., Braun, M., Kristiansen, G., Bootz, F., Wenzel, J., Mikut, R., Lengerke, C., et al. (2015). Clinical and molecular implications of MED15 in head and neck squamous cell carcinoma. Am. J. Pathol. 185, 1114-1122. [DOI] [PubMed]
- 43.Fan L., Chen L., Ni X., Guo S., Zhou Y., Wang C., Zheng Y., Shen F., Kolluri V.K., Muktiali M. Genetic variant of miR-4293 rs12220909 is associated with susceptibility to non-small cell lung cancer in a Chinese Han population. PLoS ONE. 2017;12:e0175666. doi: 10.1371/journal.pone.0175666. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fan, L., Chen, L., Ni, X., Guo, S., Zhou, Y., Wang, C., Zheng, Y., Shen, F., Kolluri, V.K., Muktiali, M., et al. (2017). Genetic variant of miR-4293 rs12220909 is associated with susceptibility to non-small cell lung cancer in a Chinese Han population. PLoS ONE 12, e0175666. [DOI] [PMC free article] [PubMed]
- 44.Zhang P., Wang J., Lu T., Wang X., Zheng Y., Guo S., Yang Y., Wang M., Kolluri V.K., Qiu L. miR-449b rs10061133 and miR-4293 rs12220909 polymorphisms are associated with decreased esophageal squamous cell carcinoma in a Chinese population. Tumour Biol. 2015;36:8789–8795. doi: 10.1007/s13277-015-3422-2. [DOI] [PubMed] [Google Scholar]; Zhang, P., Wang, J., Lu, T., Wang, X., Zheng, Y., Guo, S., Yang, Y., Wang, M., Kolluri, V.K., Qiu, L., et al. (2015). miR-449b rs10061133 and miR-4293 rs12220909 polymorphisms are associated with decreased esophageal squamous cell carcinoma in a Chinese population. Tumour Biol. 36, 8789-8795. [DOI] [PubMed]
- 45.Kropp P.A., Gannon M. Onecut transcription factors in development and disease. Trends Dev. Biol. 2016;9:43–57. [PMC free article] [PubMed] [Google Scholar]; Kropp, P.A., and Gannon, M. (2016). Onecut transcription factors in development and disease. Trends Dev. Biol. 9, 43-57. [PMC free article] [PubMed]
- 46.Lu T., Wu B., Yu Y., Zhu W., Zhang S., Zhang Y., Guo J., Deng N. Blockade of ONECUT2 expression in ovarian cancer inhibited tumor cell proliferation, migration, invasion and angiogenesis. Cancer Sci. 2018;109:2221–2234. doi: 10.1111/cas.13633. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; Lu, T., Wu, B., Yu, Y., Zhu, W., Zhang, S., Zhang, Y., Guo, J., and Deng, N. (2018). Blockade of ONECUT2 expression in ovarian cancer inhibited tumor cell proliferation, migration, invasion and angiogenesis. Cancer Sci. 109, 2221-2234. [DOI] [PMC free article] [PubMed] [Retracted]
- 47.Gao Z., Daquinag A.C., Su F., Snyder B., Kolonin M.G. PDGFRα/PDGFRβ signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development. 2018;145:dev155861. doi: 10.1242/dev.155861. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao, Z., Daquinag, A.C., Su, F., Snyder, B., and Kolonin, M.G. (2018). PDGFRα/PDGFRβ signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development 145, dev155861. [DOI] [PMC free article] [PubMed]
- 48.Onogi Y., Wada T., Kamiya C., Inata K., Matsuzawa T., Inaba Y., Kimura K., Inoue H., Yamamoto S., Ishii Y. PDGFRβ regulates adipose tissue expansion and glucose metabolism via vascular remodeling in diet-induced obesity. Diabetes. 2017;66:1008–1021. doi: 10.2337/db16-0881. [DOI] [PubMed] [Google Scholar]; Onogi, Y., Wada, T., Kamiya, C., Inata, K., Matsuzawa, T., Inaba, Y., Kimura, K., Inoue, H., Yamamoto, S., Ishii, Y., et al. (2017). PDGFRβ regulates adipose tissue expansion and glucose metabolism via vascular remodeling in diet-induced obesity. Diabetes 66, 1008-1021. [DOI] [PubMed]
- 49.Kim Y.J., Hwang S.J., Bae Y.C., Jung J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]; Kim, Y.J., Hwang, S.J., Bae, Y.C., and Jung, J.S. (2009). MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 27, 3093-3102. [DOI] [PubMed]
- 50.Chang H.R., Kim H.J., Xu X., Ferrante A.W., Jr. Macrophage and adipocyte IGF1 maintain adipose tissue homeostasis during metabolic stresses. Obesity (Silver Spring) 2016;24:172–183. doi: 10.1002/oby.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang, H.R., Kim, H.J., Xu, X., and Ferrante, A.W., Jr. (2016). Macrophage and adipocyte IGF1 maintain adipose tissue homeostasis during metabolic stresses. Obesity (Silver Spring) 24, 172-183. [DOI] [PMC free article] [PubMed]
- 51.Yau S.W., Russo V.C., Clarke I.J., Dunshea F.R., Werther G.A., Sabin M.A. IGFBP-2 inhibits adipogenesis and lipogenesis in human visceral, but not subcutaneous, adipocytes. Int. J. Obes. 2015;39:770–781. doi: 10.1038/ijo.2014.192. [DOI] [PubMed] [Google Scholar]; Yau, S.W., Russo, V.C., Clarke, I.J., Dunshea, F.R., Werther, G.A., and Sabin, M.A. (2015). IGFBP-2 inhibits adipogenesis and lipogenesis in human visceral, but not subcutaneous, adipocytes. Int. J. Obes. 39, 770-781. [DOI] [PubMed]
- 52.Chan S.S., Schedlich L.J., Twigg S.M., Baxter R.C. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am. J. Physiol. Endocrinol. Metab. 2009;296:E654–E663. doi: 10.1152/ajpendo.90846.2008. [DOI] [PubMed] [Google Scholar]; Chan, S.S., Schedlich, L.J., Twigg, S.M., and Baxter, R.C. (2009). Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am. J. Physiol. Endocrinol. Metab. 296, E654-E663. [DOI] [PubMed]
- 53.Yao L., Heuser-Baker J., Herlea-Pana O., Zhang N., Szweda L.I., Griffin T.M., Barlic-Dicen J. Deficiency in adipocyte chemokine receptor CXCR4 exacerbates obesity and compromises thermoregulatory responses of brown adipose tissue in a mouse model of diet-induced obesity. FASEB J. 2014;28:4534–4550. doi: 10.1096/fj.14-249797. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yao, L., Heuser-Baker, J., Herlea-Pana, O., Zhang, N., Szweda, L.I., Griffin, T.M., and Barlic-Dicen, J. (2014). Deficiency in adipocyte chemokine receptor CXCR4 exacerbates obesity and compromises thermoregulatory responses of brown adipose tissue in a mouse model of diet-induced obesity. FASEB J. 28, 4534-4550. [DOI] [PMC free article] [PubMed]
- 54.Sue N., Jack B.H., Eaton S.A., Pearson R.C., Funnell A.P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J.C. Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sue, N., Jack, B.H., Eaton, S.A., Pearson, R.C., Funnell, A.P., Turner, J., Czolij, R., Denyer, G., Bao, S., Molero-Navajas, J.C., et al. (2008). Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28, 3967-3978. [DOI] [PMC free article] [PubMed]
- 55.Du B., Cawthorn W.P., Su A., Doucette C.R., Yao Y., Hemati N., Kampert S., McCoin C., Broome D.T., Rosen C.J. The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-β (TGFβ) signaling. J. Biol. Chem. 2013;288:3036–3047. doi: 10.1074/jbc.M112.440370. [DOI] [PMC free article] [PubMed] [Google Scholar]; Du, B., Cawthorn, W.P., Su, A., Doucette, C.R., Yao, Y., Hemati, N., Kampert, S., McCoin, C., Broome, D.T., Rosen, C.J., et al. (2013). The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-β (TGFβ) signaling. J. Biol. Chem. 288, 3036-3047. [DOI] [PMC free article] [PubMed]
- 56.Steegmann A.T., Jr., Cerny F.J., Holliday T.W. Neandertal cold adaptation: Physiological and energetic factors. Am. J. Hum. Biol. 2002;14:566–583. doi: 10.1002/ajhb.10070. [DOI] [PubMed] [Google Scholar]; Steegmann, A.T., Jr., Cerny, F.J., and Holliday, T.W. (2002). Neandertal cold adaptation: Physiological and energetic factors. Am. J. Hum. Biol. 14, 566-583. [DOI] [PubMed]
- 57.Sazzini M., Schiavo G., De Fanti S., Martelli P.L., Casadio R., Luiselli D. Searching for signatures of cold adaptations in modern and archaic humans: hints from the brown adipose tissue genes. Heredity (Edinb) 2014;113:259–267. doi: 10.1038/hdy.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sazzini, M., Schiavo, G., De Fanti, S., Martelli, P.L., Casadio, R., and Luiselli, D. (2014). Searching for signatures of cold adaptations in modern and archaic humans: hints from the brown adipose tissue genes. Heredity (Edinb) 113, 259-267. [DOI] [PMC free article] [PubMed]
- 58.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen, L., Ge, B., Casale, F.P., Vasquez, L., Kwan, T., Garrido-Martin, D., Watt, S., Yan, Y., Kundu, K., Ecker, S., et al. (2016). Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398-1414.e24. [DOI] [PMC free article] [PubMed]
- 59.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]; GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580-585. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.