Abstract
This review deals with the human adult cardiomyocyte proliferation as a potential source for heart repair after injury. The mechanism to regain the proliferative capacity of adult cardiomyocytes is a challenge. However, recent studies are promising in showing that the ‘locked’ cell cycle of adult cardiomyocytes could be released through modulation of cell cycle checkpoints. In support of this are the signaling pathways of Notch, Hippo, Wnt, Akt and Jak/Stat that facilitate or inhibit the transition at cell cycle checkpoints. Cyclins and cyclin dependant kinases (CDKs) facilitate this transition which in turn is regulated by inhibitory action of pocket protein e.g. p21, p27 and p57. Transcription factors e.g. E2F, GATA4, TBx20 up regulate Cyclin A, A2, D, E, and CDK4 as promoters of cell cycle and Meis-1 and HIF-1 alpha down regulate cyclin D and E to inhibit the cell cycle. Paracrine factors like Neuregulin-1, IGF-1 and Oncostatin M and Extracellular Matrix proteins like Agrin have been involved in cardiomyocyte proliferation and dedifferentiation processes.
A molecular switch model is proposed that transforms the post mitotic cell into an actively dividing cell. This model shows how the cell cycle is regulated through on- and off switch mechanisms through interaction of transcription factors and signaling pathways with proteins of the cell cycle checkpoints. Signals triggered by injury may activate the right combination of the various pathways that can ‘switch on’ the proliferation signals leading to myocardial regeneration.
Keywords: Cell cycle, Heart regeneration, Cardiomyocyte proliferation, Cyclins
1. Introduction
The adult heart has been viewed for decades as a post mitotic organ with typical characteristics of cardiomyocytes as terminally differentiated and being incapable of proliferation [1]. This view was supported by the fact that after birth the development and growth of the heart switches abruptly from hyperplasia to hypertrophy [2], [3]. This would mean that the cardiomyocytes exhibit non-proliferative cell cycles that increase the DNA content but without cell division. The subsequent multinucleation and polyploidization of cardiomyocytes is seen as the end of replication [4], [5], [6]. In addition, the rigid sarcomeric structure of adult cardiomyocytes makes them refractory to cytokinesis [7]. Thus, it justifies the notion that the proliferation of adult cardiomyocytes is ‘locked’. Is it possible then to ‘unlock’ the adult cardiomyocytes to regenerate the myocardium that is electrically and mechanically in sync with the surrounding heart tissue?[8]
Zebra fish model is the best studied for demonstrating adult heart regeneration. Experiments have shown that zebra fish hearts can regenerate after apical amputation of the ventricle, cryoinjury, or genetic ablation [9], [10], [11]. This regenerative capacity was also demonstrated in neonatal mice who recovered from an experimental myocardial infarction and an apical resection injury within the first day of life [12], [13]. The apical defect was replaced by cardiomyocytes as being identified by morphology and cardiac troponin T staining. Cardiomyocyte mitosis and cytokinesis were shown by colocalization of phosphohistone H3 and aurora B kinase with cardiac troponin T and sarcomere disassembly. In addition, normal systolic function was observed in the newly formed apex [14]. This remarkable regenerative potential of neonatal mice heart is, however, lost within the first week of life. The exact mechanism for this shift is not known but coincides with the developmental window when cardiomyocytes become binucleated and withdraw from the cell cycle [2].
Studies have shown that in humans the cardiomyocyte numbers increase from 1 to 4 billion from birth to an age of 20 years after which the total number of cardiomyocytes remain constant throughout life [15], [16]. The Carbon-14 birth dating method estimated that new cardiomyocytes are generated at a rate of <1% per year at 25 years decreasing to ∼0.45% at 75 years of age, so that about 45% of cardiomyocytes are exchanged during the entire life time of a person [17]. This regenerative ability, albeit small, might prove to be important owing to longer life span in humans [18]. It may be sufficient for repairing age related daily wear and tear. However, it is apparently unable to keep up with cardiomyocyte loss due to heart failure or to fill in large gaps resulting from myocardial infarction.
Regardless of its effectiveness, it is now established that human heart is capable of generating new cardiomyocytes after birth [19]. The source of these new cardiomyocytes, however, is still under debate. Are these new cardiomyocytes coming from differentiation of resident/homing stem cells in the heart or from division of pre-existing cardiomyocytes?
Studies both in zebra fish and mice have demonstrated that it is the proliferation of preexisting cardiomyocytes and not endogenous progenitor or stem cells that contribute to cardiac regeneration [20], [21], [22]. In mice, cell exhibiting specific cardiac gene markers were shown to undergo DNA replication and cytokinesis [23]. These newly regenerated cells demonstrated in turn electromechanical networking with neighboring myocardium [24]. Latest studies by He et al [25] and Li et al [26] have provided evidence that the adult mammalian heart lacks myocyte producing stem cells, so heart repair cannot be attributed to any progenitor cell and cardiomyocytes in injured myocardium are derived from preexisting cardiomyocytes [26], [27].
The discovery of endogenous adult cardiomyocytes as a source of new cardiomyocytes opens a new dimension in the field of regenerative medicine. This may enable the myocardial repair by stimulating endogenous adult cardiomyocytes proliferation [8].
The possible mechanisms underlying cardiomyocyte proliferation will be discussed.
2. Cell cycle regulators in heart regeneration
It has been shown that cardiomyocytes withdraw from cell cycle activity after birth [14]. Post-mitotic, terminally differentiated cardiomyocytes that results from this transition are associated with up-regulation of cell cycle inhibitors and down-regulation of cell cycle promoters at check points G1/S and G2/M through cyclins and cyclin-dependent kinases (CDKs) [3], [15], [28], [29]. Recent studies are investigating the mechanisms of how cardiomyocytes exit the cell cycle to identify ways to re-enter the cell cycle. Mechanism(s) to regain proliferative capacity of adult cardiomyocytes could be through modulation of cell cycle checkpoints as is shown in Fig. 1.
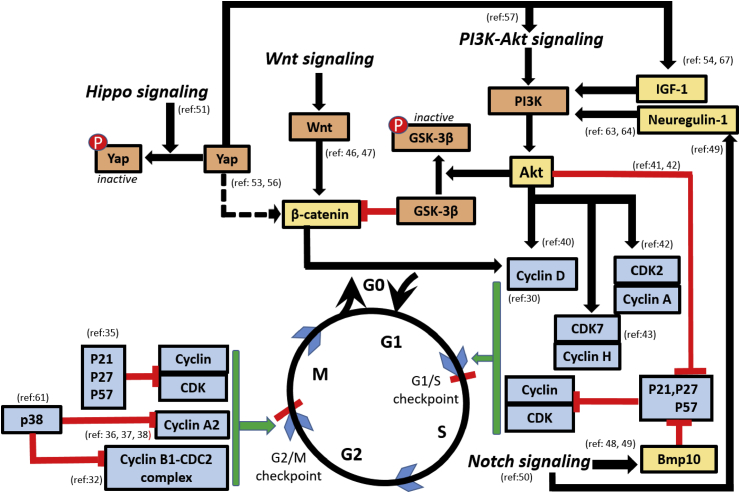
Fig. 1.
Interactions of cell cycle regulators and signaling proteins model for adult cardiomyocyte proliferation. The cell cycle after G0 is time-phased with four phases of G1, S, G2 and M with the checkpoints at the interphase of G1/S and G2/M. Signaling pathways of Notch, Hippo, Wnt and Akt facilitate or inhibit the transition at checkpoint as shown by the interacting colored arrows. Cyclins, cyclin dependent kinases (CDKs) facilitate the transition at checkpoints which in turn is regulated by inhibitory action of P21, P27, p28 and P57 on cyclin and CDKs. Akt shows a triple response of inactivating GSK-3β, releasing β-catenin to action, activates cyclin D and inhibits P27 leading to disinhibition of cyclins and CDKs. Hippo signaling, on one hand activates Yap proteins to regulate β-catenin and on the other hand, it activates insulin like growth factor-1(IGF-1) to switch on Akt signaling. Wnt signaling activates β-catenin switching on the G1 to S transition. Notch signaling together with Akt inhibits P27 and P57 proteins in order to disinhibit the cyclin-CDK facilitation of cell cycle checkpoints.
Cyclin D is a G1 phase cyclin that drives the G1/S phase transition. Cell cycle activity in cardiomyocytes expressing cyclin D2 persisted after experimental myocardial infarction in mice with resulting DNA synthesis and regression of infarct [30]. It means cyclin D2 is proliferation friendly. Contrastingly, overexpression of cyclin D1 of adult cardiomyocytes in mice resulted in DNA synthesis and multinucleation but did not proceed to proliferation [31]. Hence overexpression of cyclin D2 could be a potential target for stimulating myocardial regeneration after injury.
Cyclin B is a G2 phase cyclin. It drives the cells into and out of M phase of the cell cycle. It has been shown that the absence of cyclin B1-CDC2 complex (cell division cycle 2 kinase) mediates G2/M arrest in adult cardiomyocytes. In support of this effect, Bicknell et al has demonstrated that overexpressing this G2/M regulator protein stimulated cell division in adult rat cardiomyocytes that had previously exited the cell cycle [32].
Cyclin Dependent Kinase inhibitors (CKIs) regulate the cyclin-CDK complexes to control the cell cycle. These thus, can be manipulated to induce proliferation in cardiomyocytes. P21, p27 and p57 of the CDK interacting protein/kinase inhibitory protein family of CKIs are present in adult cardiomyocytes to switch off the cell cycle as shown in Fig. 1. Studies have shown that p27 and p57 work in synergy to control the cell cycle exit and leading to cell differentiation [33]. P21 controls cell cycle arrest and prevent re-entry into cell cycle [34]. It has been shown that knockdown of p21, p27 and p57 genes induced quiescent adult rat cardiomyocytes to actively proliferate with intact cytokinesis. This phenomenon was linked to the re-expression of neonatal genes, inhibition of adult genes and a change in adult cardiomyocyte morphology [35].
Cyclin A2, a key cell cycle regulator mediating G1-S and G2-M transition, has been shown to increase cardiomyocyte cycling and cardiac regeneration [36], [37]. In rats, delivery of a cyclin A2 expressing adenoviral vector after myocardial infarction improved cardiac function [37]. It attenuated myocardial fibrosis and caused an increase in cardiomyocyte number in the infarct zone. Delivery of the cyclin A2 gene to infarcted porcine hearts also resulted in the increase in cardiomyocyte mitosis, increase in cardiomyocyte number, and decrease in fibrosis. Time-lapse microscopy of cultured porcine cardiomyocytes demonstrated cytokinesis with preservation of sarcomeric structure [38] showing cyclin A2 as an important cell cycle regulator in adult cardiomyocytes (Fig. 1).
Cyclins and CKIs, are thus important factors that can be manipulated to re-enter the cardiomyocyte cell cycle via gene transfer or pharmacological modulation through small molecules as will be discussed further.
Recently, it was shown that overexpression of CDK1, CDK4, cyclin B1, and cyclin D1 efficiently induced cell division in post-mitotic mouse, rat, and human cardiomyocytes [39]. It was also shown through in vivo lineage tracing experiments that 15–20% of adult cardiomyocytes expressing these four factors underwent stable cell division and improved cardiac function after myocardial infarction. This demonstrates that a specific combination of genes could potentially unlock the proliferative ability in those cells that have exited the cell cycle [39].
3. The signaling pathways controlling cell cycle proteomics to stimulate adult cardiomyocyte proliferation
PI3K-Akt is an intracellular signaling pathway that is one of the drivers of cell-cycle as well as promoter of apoptosis. Studies have shown that constitutive expression of active protein kinase B or Akt can prolong the half-life of cyclin D, resulting in positive cell-cycle regulatory effects while inhibition of PI3K speeds up cyclin D1 degradation [40]. The S-phase cyclin-dependent kinase CDK2 is a target for Akt during cell cycle progression. Expression of p27, a negative regulator of cell cycle is also decreased by the PI3K-AKT pathway to facilitate G2/M phase progression [41], [42]. Studies in cardiomyocytes have shown that the number of neonatal as well as adult cardiomyocytes in the mitotic and cytokinetic phase was increased after activation of the PI3K-AKT signals [43]. Apart from its proliferation inducing potential, Akt is also shown to orchestrate fibroblast-to-cardiomyocyte reprogramming in mice [44].
Wnt/β-catenin signaling is one of the main signaling pathways to regulate the regenerative process. Wnt signaling is activated in response to cardiac injury leading to cardiac remodeling [45]. Chemical inhibition and genetic knockout of GSK3β, a signaling mediator in the Wnt/β-catenin pathway has been shown to increase cardiomyocyte cycling by inducing S phase entry [46], [47]. One of the proposed mechanisms for postnatal cell cycle arrest is the down regulation of Wnt signaling. Wnt signaling pathway is, hence, considered a potential player in stimulating cardiac regeneration.
Notch pathway of cell signaling has also been shown to modulate cardiomyocyte proliferation and trabculation in the developing heart [48], [49]. One mechanism of Notch signaling is to facilitate the activity of BMP10 (Bone Morphogenetic Protein – 10), which is essential for regulating growth and maturation of the heart [48], [49]. Experiments have shown that BMP10 knockout mice demonstrate a dramatic reduction in proliferative activity in cardiomyocytes. Another mechanism by which Notch signaling affects cardiomyocyte proliferation is by indirect control of Neuregulin expression [49]. Neuregulin induces cardiomyocyte proliferation as described in the section of extracellular and paracrine factors. Ventricular amputation injury in zebra fish activates the Notch pathway in the regenerative response specifically in the endocardium and epicardium [50]. Unexpectedly, however, it was shown that hyperactivation of Notch signaling also suppresses cardiomyocyte proliferation [50]. This would mean that cardiomyocyte proliferative renewal could be sensitive to disturbance in Notch signaling [50].
Hippo signaling pathway has generated interest in recent years as it regulates heart size by inhibiting cellular proliferation. This effect of the Hippo signaling is mediated by inhibition of a subset of Wnt target genes [51]. In adult cardiomyocyte, it was also shown that cardiomyocytes re-enter the cell cycle upon Hippo pathway disruption and proceed to cytokinesis and regeneration of the myocardium. Hippo-deficient cardiomyocytes also demonstrated reduced scar size, increased survival and proliferation after ventricular apical resection injury [52]. Further studies have shown that neonatal and adult mouse cardiomyocyte cycling was enhanced after manipulating various regulators of the Hippo pathway [52], [53], [54]. One of the mechanisms of Hippo signaling involves phosphorylation of the transcriptional co-activator YAP (yes-associated protein). A conditional deletion of YAP-1 during heart development results in impaired myocyte proliferation; hence its importance in proper fetal heart growth [55].
Overexpression of a constitutively active mutant YAP was shown to enhance myocyte proliferation and improve heart function after MI in adult mice [53]. Also, adeno-associated virus–mediated delivery of human YAP in mice post-MI produced beneficial results [56].
Hippo-YAP pathway is linked to the PI3K-AKT signaling pathway. It is also through this route that Hippo-YAP causes cardiomyocyte proliferation [57]. These results clearly show that Hippo off and on signaling is a viable target for inducing adult cardiomyocyte proliferation.
Jak/Stat signaling pathway is a developmental pathway closely linked to cardiac regeneration. Inhibition of this signaling in zebra fish has led to reduced cardiomyocyte proliferation and increased scarring after cardiac injury [58]. Experimental myocarditis model in mice demonstrated that recovery from myocarditis-induced damage was due to proliferation of pre-existing cardiomyocytes by cell cycle reentry through STAT3. This was shown by immunostaining with cell cycle markers Ki-67 and Aurora B. The cardiomyocyte nuclei were counted to determine that there was a high proportion of mononucleated cardiomyocytes that have entered the cell cycle and underwent cell division. Cell fate mapping analysis further substantiated the claim that STAT3 is not only a protective factor but also as a proliferative factor in adult mammalian cardiomyocytes [59].
p38 mitogen-activated protein kinase is a signaling molecule that induces cell cycle exit and differentiation of cardiomyocytes [60]. It is a negative regulator of mammalian cardiomyocyte division both in vitro and in vivo [61]. Expression of cyclin A2, cdc2a, and cyclin B is increased by inhibition of p38. Its inhibition and growth factor stimulation act synergistically to induce genes involved in cardiomyocyte proliferation and regeneration, including Ki67, cdc2, and cyclin A, and the cell cycle inhibitor p27. It also leads to phosphorylation of Rb, a key cell cycle regulator [61]. P38 activity, thus not only regulates cell cycle factors as shown above but also regulates neonatal and adult cardiomyocyte karyokinesis and cytokinesis [61].
4. Paracrine factors in inducing adult cardiomyocyte proliferation
Extracellular and paracrine factors can stimulate receptors on cardiomyocytes and induce proliferation through intracellular signaling pathways.
Neuregulin-1 is an agonist of ErbB1-4 which are the tyrosine kinases receptor of the epidermal growth factor receptor family [62]. This binding to the cardiomyocyte cell-surface receptor increases its kinase activity leading to stimulation of intracellular PI3-kinase signal transduction pathway [63]. Neuregulin-1- induces cell cycle proliferation proceeding to cell division in adult cardiomyocytes [63]. It induces differentiated cardiomyocytes to reenter the cell cycle from S phase and undergo both karyokinesis and cytokinesis leading to cardiomyocyte proliferation [63], [64]. This effect was also seen after myocardial infarction leading to cardiac repair and improved heart function. Thus, Neuregulin-1/ErbB4 and Neuregulin-1/ErbB2 axes can be seen as a new molecular strategy for heart regeneration. However targeting Neuregulin-Erb pathway may not be straightforward as persistent activation has been shown to ultimately lead to cardiac hyperplasia and heart failure in animal models [27], [65], [66].
Insulin like growth factor-1 (IGF-1) is a mitogen that binds to IGF-1 receptor to initiate a nexus of intracellular signaling. IGF-1 links the Wnt, Hippo and PI3K pathways for coordinating cardiac chamber morphogenesis and embryonic heart size [54], [67]. In transgenic mice, cardiac over expression of IGF-1 stimulated cardiac proliferation and subsequently caused an increase in cardiac weight. Constitutive overexpression of IGF-1 has been seen to prevent cell death after myocardial infarction and limit ventricular dilatation [68], [69].
Fibroblast growth factor-1 (FGF-1) belongs to a group of secreted signaling proteins as paracrine or endocrine signals in development, health, and disease in major organs [70]. Paracrine FGFs exert biological activities by binding to cell surface FGF receptors (FGFR) and heparan sulfate as a co-factor. The FGF-FGFR-heparan sulfate complex leads to the activation of intracellular signaling pathways including the RAS-MAPK, PI3K-AKT, phospholipase Cγ (PLCγ), and STAT pathways [71], [72], [73]. Stimulation of adult cardiomyocytes with FGF-1 together with p38 inhibition act sequentially to promote induction of DNA synthesis as well as G2/M transition in cardiomyocytes [61]. In vivo, post MI p38 inhibition facilitates FGF1-mediated cardiomyocyte mitosis within the infarct and at the border zone leading to preserved myocardial wall thickness, reduced scaring, and improved function [74]. It has also been shown that FGFR-1interacts with Fn14 (TNF receptor superfamily member fibroblast growth factor-inducible molecule 14) to enable cardiomyocyte cell cycle reentry [75].
Oncostatin M (OSM) is a pleiotropic cytokine that belongs to the Interleukin-6 family. It is involved in dedifferentiation [76], cell proliferation and inflammatory processes [77]. OSM induces cardiomyocyte dedifferentiation and activation of fetal genes, expression of progenitor cell markers and enables cell-cycle progression [78]. These effects of OSM are mediated by the Ras/Raf/MEK/Erk signaling cascade. OSM seems to be beneficial after acute myocardial infarction but prolonged dedifferentiation leads to reduction in contractile force of the heart and ultimately heart failure [78].
Agrin. Cell cycle reentry needs extracellular matrix proteins (ECM) for a coordinated proliferation-differentiation of adult cardiomyocytes [79], [80]. In vitro administration of ECM fragments from post natal day 1 (P1) but not day 7 (P7), promoted an increase in cell cycle activity in P1 and P7 isolated cardiomyocytes and co-treatment with a broad matrix metalloproteinase inhibitor resulted in reduced cardiomyocyte proliferation induced earlier [80]. It was seen that Agrin, a large extracellular hepran sulfate proteoglycan was downregulated between P1 and P7 as well as in the adult heart. Agrin dose dependently increased P1 and P7 cardiomyocyte proliferation, evidenced by immunofluorescence staining for markers of proliferation Ki67, phospho-histone H3 and aurora kinase B. Intramyocardial injection of Agrin after experimental myocardial infarction in juvenile and adult hearts also induced cardiomyocyte cell-cycle re-entry in the region adjacent to the infarcted myocardium, reduced scar size and improved cardiac function. Mechanistically, Agrin is shown to destabilize the dystrophin-glycoprotein complex, promoting myofibril disassembly with activation of downstream signaling molecules Yap and ERK [80].
Periostin, a secreted extracellular matrix protein [81] is expressed in the developing heart, but not in healthy adult ventricular myocardium [82]. It is re-expressed in adult ventricular myocardium following injury [83]. Recombinant periostin has shown to induce reentry of differentiated mononucleated cardiomyocytes into the cell cycle [84]. Periostin induced full mitotic cycle in differentiated cardiomyocytes as evidenced by an increase in cardiomyocyte DNA synthesis, aurora B kinase detection and Brd-U positive nuclei in 100% of cardiomyocyte cytokinesis [84]. Periostin-induced cardiomyocyte cell-cycle reentry was through the PI3K pathway and required various integrins. Myocardial delivery of periostin improved ventricular remodeling and function after myocardial infarction. It reduced fibrosis, infarct size and increased angiogenesis [84]. The above findings were challenged by reports of no role of periostin in cardiomyocyte proliferation and regeneration in the adult heart [85], [86]. However, recently Chen et al [87] has provided comprehensive evidence that periostin is required for cardiomyocyte proliferation, angiogenesis and regeneration of the neonatal heart post-MI. They examined the impact of genetic loss-of-function in neonatal mice following MI and showed that PI3K/Akt/GSK3b signaling mediates the pro-regenerative actions of periostin [87].
5. Transcriptional factors mediation of cardiomyocyte proliferative response
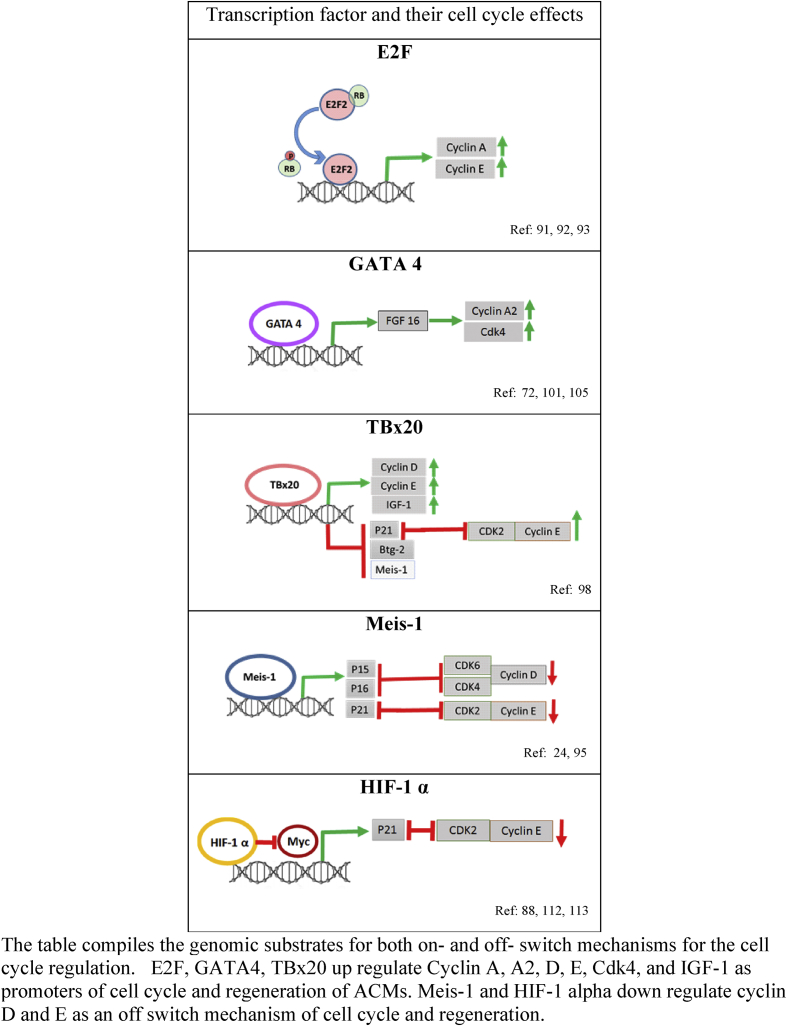
Cell cycle regulators are also controlled by various transcription factors. The role of different transcriptional factors in cell cycle mediation are summarized in Table 1.
Table 1.
Transcription factors and their role in cardiomyocyte cell cycle regulation.
E2F are a family of transcription factors that form a complex network with the pocket protein in the cell cycle to regulate cardiomyocyte proliferation and differentiation [88]. E2F-1 stimulates DNA synthesis in post mitotic rat ventricular myocytes. It leads to accumulation of myocytes in G2/M phase of the cell cycle overriding the G1/S checkpoint followed by an increase in the rate of apoptosis and mortality [89], [90] .Overexpression of E2F-2, however, is shown to stimulate cardiomyocyte cycling. Targeted expression of E2F-2 in mice showed a significant increase in the number of cardiomyocytes within S-phase, along with a significant increase in phosphorylated histon-H3 and Aurora-kinase positive cardiomyocytes demonstrating that the cardiomyocytes completed the entire cell cycle including mitosis and cytokinesis [91]. Table 1 shows the genomic response of E2F-2 on the up regulation of both cyclin A and E.
It has also been shown that knocking down the E2F regulatory pocket proteins, Rb and p107 caused an increase in the cardiomyocyte cycling [92], [93]. Since hypophosphorylated Rb binds to E2F complexes, its phosphorylation is carried out by CDK2 and CDK4, which in turn, releases the E2F complexes. E2F complexes activate transcription triggering expression of genes needed for DNA synthesis and regulation of cell cycle. Rb and p130 maintain the post mitotic state of adult cardiomyocytes. They do so by directing heterochromatin formation and silencing a subset of E2F-dependant pro-proliferation G2/M and cytokinesis genes. It has been shown that cardiomyocyte cell cycle re-entry can be achieved by reversal of this silencing mechanism [92].
Meis1 is a transcription factor and a key regulator of cell cycle arrest. Meis1 plays an important role in embryonic cardiomyocyte development [94]. It maintains high levels of the cyclin-dependent kinase inhibitors p15, p16 and p21 postnatally by directly regulating these genes [95]. Meis1-binding sequences have been found in the promoter regions of genes. Recent studies have shown that deleting Meis1 in cardiomyocytes extended the cardiomyocyte proliferative window postnatally from 7 to 14 days. It induces cell cycle reentry and induction of proliferation in adult cardiomyocytes without reducing heart function or inducing cardiomyocyte hypertrophy. Graphical representation of Meis-1in Table 1 shows its genomic effects on p15, 16 and 21, which in turn switches off the CDK6, 4 and 2 subsequently decreasing cyclin D and E. Evidence suggests therefore, that the limited postnatal cardiac regeneration in mammals is the result of a Meis1-imposed limit on the division and proliferation of cardiomyocytes [24].
Tbx20 is a transcription factor essential for cardiomyocyte proliferation during development. Its absence causes embryonic death at mid-gestation [96]. Induced knockout of Tbx20 in adult cardiomyocytes also causes severe cardiomyopathy and death [97]. This shows that Tbx20 is required for fetal as well as adult cardiomyocyte homeostasis [98]. Tbx20 binds to p21, Meis1, and Btg2 gene sequences and represses their expression [98] which in turn contributes to Tbx20-induced cardiomyocyte proliferation. This is shown by increase in the total number of cardiomyocytes and increased expression of mitotic markers with corresponding changes in cell-cycle regulatory genes. Over expression of Tbx20 causes adult cardiomyocytes to have fetal-like characteristics. These include 1. Small mononucleated cardiomyocytes; 2. Up regulation of fetal contractile protein expression of ssTNI and βMHC and 3. Induction of multiple proliferative signaling pathways like PI3K/Akt, Hippo/Yap and BMP/Smad1/5/8 signaling pathways [98]. The cardiomyocyte proliferation is seen without hypertrophy or fibrosis in normal as well as injured hearts [98].
Tbx20, therefore, possesses a dual genomic switch as shown in Table 1. On one hand, the expression of cyclin D, E and IGF-1 is switched on to drive the cardiomyocyte cycle. On the other hand it switches off the genomic expression of Meis-1, btg-2 and p21 to induce proliferation in adult cardiomyocytes.
GATA-4 is a zinc finger transcription factor that plays a key role during heart development. It regulates specific genes that mediate the development of the embryonic and neonatal heart [99], [100]. In neonatal heart, ablation of Gata4 in the cardiomyocytes decreases cardiomyocytes proliferation following apical resection and cryoinjury [101]. GATA4 is also crucial in adulthood where it regulates cardiomyocyte viability, cardiac hypertrophy, and fibrosis [102], [103]. Mechanistic studies have shown that GATA4 exerts its effects via regulation of Fgf16 (fibroblast growth factor 16) [101]. It is a cardiac-specific member of the Fibroblast Growth Factor family of signaling proteins essential for cell proliferation and tissue regeneration [72]. In the fetus, Fgf16 is required for cardiomyocyte replication during development [104]. Its expression is increased in the post natal period and throughout adulthood. It is stored in the extracellular matrix to help maintain cardiac homeostasis by upregulating the genes associated with cell proliferation and protecting the heart from injury [101], [105]. Thus GATA4/Fgf16 axis is an important player in regulation of heart regeneration by switching on cyclin A2 and CDK4 as depicted in Table 1.
Hypoxia-inducible factor (HIF)-1 α is a transcription factor complex that regulates the cellular response to hypoxia [106]. HIF is a heterodimeric protein made up of the constitutive expressed HIF-1β and the oxygen sensitive hypoxia-inducible HIF-1α [107]. Cells continuously synthesize and degrade HIF-1α protein in normoxic conditions as von Hippel–Lindau tumor suppressor protein (pVHL) interacts with the α-subunit to target for proteolysis by the ubiquitin–proteasome pathway [108], [109]. HIF-α protein escapes proteasomal degradation in hypoxic conditions, stabilizes, translocates to the nucleus and dimerizes with HIF-1β. This complex then binds to the HRE (Hyoxia responsive elements) sequences of target genes [110] and results in their transcription. Regarding the role of HIF-1 α in cardiomyocyte proliferation, studies have shown that after ventricular resection, the zebra fish heart regeneration was mediated by HIF-1 α as a crucial player in hypoxic response to injury [111]. Recently it was shown that during development, there is a distinct subset of cardiomyocytes that are hypoxic due to their location within the non-vascularized regions of the mid-gestational fetal heart. This hypoxic environment promoted fetal cardiomyocyte proliferation as part of the cell stress pathways regulated by HIF-1α [112]. Genetic studies however, have shown that HIF-1 α induce cell cycle arrest by displacing the transcription factor Myc binding from the promoter of p21 which is a CDK inhibitor and controls the cell cycle checkpoint as shown in Table 1. This action of HIF-1 α is independent of its DNA binding or transcriptional activity. HIF-1α, therefore, has at least two mechanisms for regulating gene expression: its C-terminal transactivation of genes through its HRE, and via its N-terminal counteraction of Myc activity. This indicates a divergent role for HIF-1α in cardiac proliferation and development [88], [113].
6. The ‘molecular switch’ model
Endogenous cardiac repair by stimulating adult cardiomyocytes proliferation is an exciting possibility that is showing promise in the light of animal experiments. However, there are fundamental questions that are yet to be answered before therapeutic endogenous cardiac repair becomes a reality in humans. As we know that cell cycle reentry of adult cardiomyocytes can have different results. This can be limited to the S phase, the phase of karyokinesis or it can progress all the way to cytokinesis. Recent studies suggest that only one third of the cardiomyocytes that enter the cell cycle progress through mitosis and enter cell divisions [88], [114]. This means that the proliferative potential is limited to only a subset of cardiomyocytes. What might be the characteristics of these adult cardiomyocytes that enable them to divide? Is it limited to being mononuclear and diploid myocytes or are there other morphological and functional features?
For cell division, the cardiomyocytes need to rapidly and accurately disassemble and reassemble cytoskeletal and nuclear structures in addition to cell–cell and cell matrix attachments. A recent study used time-lapse video microscopy to show myocyte proliferation in adult mouse myocytes cocultured with neonatal rat ventricular myocytes. Adult cardiomyocytes showed a continuous process of dedifferentiation with sarcomere disassembly and induction of cell cycle markers, proliferation and redifferentiation with reorganization of sarcomere and restoration of contractility [115].
Epigenetic regulation of cardiomyocyte proliferation is a new area that is being explored recently. Post-translational modification of histone proteins such as DNA methylation, de-acetylation, phosphorylation and microRNA mediated gene regulation have been found to influence expression of cell cycle factors in the adult heart [116]. Cardiac specific transcriptional factors such as GATA4, Tbx5 and cell cycle regulators Rb/p130 have been shown to form complexes with epigenetic modifying proteins and these complex lead to modifications of histones at promoter regions of cardiac and cell cycle genes which determines the adult cardiomyocyte phenotype [116].
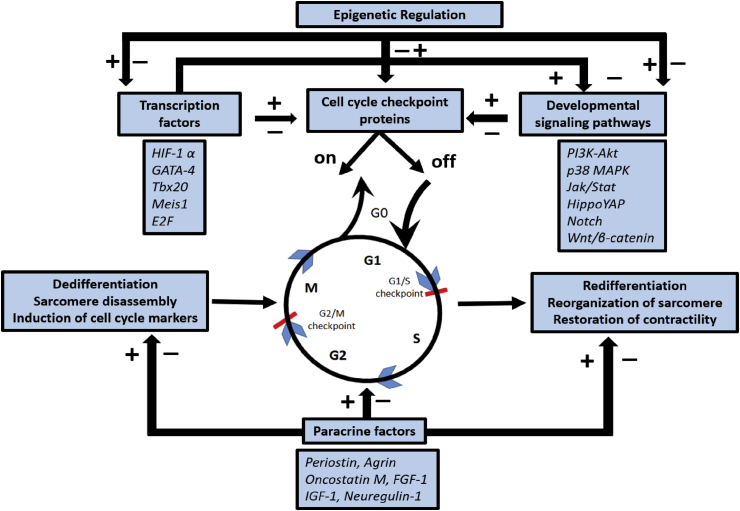
Reviewing studies of the various transcription factors, signaling proteins, along with physical interactions that play a role in adult cardiomyocyte proliferation we have modeled a molecular switch that transforms the post mitotic cell into an actively dividing cell (Fig. 2). Signals triggered by injury may activate the right combination of signaling molecules and transcription factors that can ‘switch on’ the proliferation signals leading to myocardial regeneration. What might be the right combination is yet to be defined. Another important question is how to control the “off” switch of cardiac proliferation once the “on” switch is triggered? Much needs to be done in understanding the basic mechanisms that regulate the molecular switch of adult cardiomyocyte division and proliferation before therapeutic regenerative approaches are applicable. Cardiomyocyte proliferation is an essential part of the heart regeneration story but it cannot be emphasized enough that renewal of a piece of myocardium needs contributions from cardiomyocytes, endothelial cells, smooth muscle cells, fibroblast, pacemaker cells, conducting and Purkinje cells to bring the orchestration of rhythmically contracting and relaxing heart [117].
Fig. 2.
Molecular switch model. It shows how the cell cycle is regulated through on- and off switch mechanisms through interaction of transcription factors, signaling pathways, paracrine factors and epigenetic regulators upon cell cycle checkpoint proteins as elucidated in the text.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosures
None.
Acknowledgement
Dedicated to Dr Camer Vellani, Distinguished Professor of Cardiology and former Rector of Aga Khan University, for inspiring us to pursue the scientific method of inferences.
Footnotes
Peer review under responsibility of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
References
- 1.Zak R. Cell proliferation during cardiac growth. Am J Cardiol. 1973;31(2):211–219. doi: 10.1016/0002-9149(73)91034-5. [DOI] [PubMed] [Google Scholar]
- 2.Li F., Wang X., Capasso J.M., Gerdes A.M. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28(8):1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 3.Soonpaa M.H., Kim K.K., Pajak L., Franklin M., Field L.J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271(5 Pt 2):H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 4.Olivetti G., Cigola E., Maestri R. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 1996;28(7):1463–1477. doi: 10.1006/jmcc.1996.0137. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky V., Sarkisov D.S., Arefyeva A.M., Panova N.W., Gvasava I.G. Polyploidy in cardiac myocytes of normal and hypertrophic human hearts; range of values. Virchows Arch. 1994;424(4):429–435. doi: 10.1007/BF00190566. [DOI] [PubMed] [Google Scholar]
- 6.Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel F.B., Schebesta M., Keating M.T. Anillin localization defect in cardiomyocyte binucleation. J Mol Cell Cardiol. 2006;41(4):601–612. doi: 10.1016/j.yjmcc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Senyo S.E., Lee R.T., Kuhn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014;13(3 Pt B):532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chablais F., Veit J., Rainer G., Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Panakova D., Kikuchi K. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138(16):3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haubner B.J., Adamowicz-Brice M., Khadayate S. Complete cardiac regeneration in a mouse model of myocardial infarction. Aging (Albany NY) 2012;4(12):966–977. doi: 10.18632/aging.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porrello E.R., Mahmoud A.I., Simpson E. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110(1):187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porrello E.R., Mahmoud A.I., Simpson E. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mollova M., Bersell K., Walsh S. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A. 2013;110(4):1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann O., Zdunek S., Felker A. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161(7):1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Bergmann O., Bhardwaj R.D., Bernard S. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Berlo J.H., Molkentin J.D. An emerging consensus on cardiac regeneration. Nat Med. 2014;20(12):1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltrami A.P., Urbanek K., Kajstura J. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344(23):1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 20.Jopling C., Sleep E., Raya M., Marti M., Raya A., Izpisua Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi K., Holdway J.E., Werdich A.A. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnabel K., Wu C.C., Kurth T., Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senyo S.E., Steinhauser M.L., Pizzimenti C.L. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali S.R., Hippenmeyer S., Saadat L.V., Luo L., Weissman I.L., Ardehali R. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci U S A. 2014;111(24):8850–8855. doi: 10.1073/pnas.1408233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L., Li Y., Pu W. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med. 2017;23(12):1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., He L., Huang X. Genetic Lineage Tracing of Non-myocyte Population by Dual Recombinases. Circulation. 2018 Aug 21;138(8):793–805. doi: 10.1161/CIRCULATIONAHA.118.034250. [DOI] [PubMed] [Google Scholar]
- 27.Vagnozzi R.J., Molkentin J.D., Houser S.R. New myocyte formation in the adult heart: endogenous sources and therapeutic implications. Circ Res. 2018;123(2):159–176. doi: 10.1161/CIRCRESAHA.118.311208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks G., Poolman R.A., Li J.M. Arresting developments in the cardiac myocyte cell cycle: role of cyclin-dependent kinase inhibitors. Cardiovasc Res. 1998;39(2):301–311. doi: 10.1016/s0008-6363(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 29.Poolman R.A., Brooks G. Expressions and activities of cell cycle regulatory molecules during the transition from myocyte hyperplasia to hypertrophy. J Mol Cell Cardiol. 1998;30(10):2121–2135. doi: 10.1006/jmcc.1998.0808. [DOI] [PubMed] [Google Scholar]
- 30.Pasumarthi K.B., Nakajima H., Nakajima H.O., Soonpaa M.H., Field L.J. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96(1):110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 31.Soonpaa M.H., Koh G.Y., Pajak L. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J Clin Invest. 1997;99(11):2644–2654. doi: 10.1172/JCI119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bicknell K.A., Coxon C.H., Brooks G. Forced expression of the cyclin B1-CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem J. 2004;382(Pt 2):411–416. doi: 10.1042/BJ20031481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P., Wong C., DePinho R.A., Harper J.W., Elledge S.J. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12(20):3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poolman R.A., Gilchrist R., Brooks G. Cell cycle profiles and expressions of p21CIP1 AND P27KIP1 during myocyte development. Int J Cardiol. 1998;67(2):133–142. doi: 10.1016/s0167-5273(98)00320-9. [DOI] [PubMed] [Google Scholar]
- 35.Di Stefano V., Giacca M., Capogrossi M.C., Crescenzi M., Martelli F. Knockdown of cyclin-dependent kinase inhibitors induces cardiomyocyte re-entry in the cell cycle. J Biol Chem. 2011;286(10):8644–8654. doi: 10.1074/jbc.M110.184549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhry H.W., Dashoush N.H., Tang H. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J Biol Chem. 2004;279(34):35858–35866. doi: 10.1074/jbc.M404975200. [DOI] [PubMed] [Google Scholar]
- 37.Woo Y.J., Panlilio C.M., Cheng R.K. Therapeutic delivery of cyclin A2 induces myocardial regeneration and enhances cardiac function in ischemic heart failure. Circulation. 2006;114(1 Suppl):I206–I213. doi: 10.1161/CIRCULATIONAHA.105.000455. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro S.D., Ranjan A.K., Kawase Y. Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci Transl Med. 2014;6(224):224ra227. doi: 10.1126/scitranslmed.3007668. [DOI] [PubMed] [Google Scholar]
- 39.Mohamed T.M.A., Ang Y.S., Radzinsky E. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173(1):104–116 e112. doi: 10.1016/j.cell.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parekh P., Motiwale L., Naik N., Rao K.V. Downregulation of cyclin D1 is associated with decreased levels of p38 MAP kinases, Akt/PKB and Pak1 during chemopreventive effects of resveratrol in liver cancer cells. Exp Toxicol Pathol. 2011;63(1–2):167–173. doi: 10.1016/j.etp.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Brazil D.P., Yang Z.Z., Hemmings B.A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29(5):233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Maddika S., Ande S.R., Wiechec E., Hansen L.L., Wesselborg S., Los M. Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. J Cell Sci. 2008;121(Pt 7):979–988. doi: 10.1242/jcs.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beigi F., Schmeckpeper J., Pow-Anpongkul P. C3orf58, a novel paracrine protein, stimulates cardiomyocyte cell-cycle progression through the PI3K-AKT-CDK7 pathway. Circ Res. 2013;113(4):372–380. doi: 10.1161/CIRCRESAHA.113.301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H., Dickson M.E., Kim M.S., Bassel-Duby R., Olson E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112(38):11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergmann M.W. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res. 2010;107(10):1198–1208. doi: 10.1161/CIRCRESAHA.110.223768. [DOI] [PubMed] [Google Scholar]
- 46.Tseng A.S., Engel F.B., Keating M.T. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem Biol. 2006;13(9):957–963. doi: 10.1016/j.chembiol.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Kerkela R., Kockeritz L., Macaulay K. Deletion of GSK-3beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J Clin Invest. 2008;118(11):3609–3618. doi: 10.1172/JCI36245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H., Shi S., Acosta L. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131(9):2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grego-Bessa J., Luna-Zurita L., del Monte G. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L., Borikova A.L., Ben-Yair R. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci U S A. 2014;111(4):1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heallen T., Zhang M., Wang J. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heallen T., Morikawa Y., Leach J. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xin M., Kim Y., Sutherland L.B. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110(34):13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin M., Kim Y., Sutherland L.B. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4(196):ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Gise A., Lin Z., Schlegelmilch K. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109(7):2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Z., von Gise A., Zhou P. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115(3):354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Z., Zhou P., von Gise A. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ Res. 2015;116(1):35–45. doi: 10.1161/CIRCRESAHA.115.304457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang Y., Gupta V., Karra R., Holdway J.E., Kikuchi K., Poss K.D. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci U S A. 2013;110(33):13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyawaki A., Obana M., Mitsuhara Y. Adult murine cardiomyocytes exhibit regenerative activity with cell cycle reentry through STAT3 in the healing process of myocarditis. Sci Rep. 2017;7(1):1407. doi: 10.1038/s41598-017-01426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eriksson M., Leppa S. Mitogen-activated protein kinases and activator protein 1 are required for proliferation and cardiomyocyte differentiation of P19 embryonal carcinoma cells. J Biol Chem. 2002;277(18):15992–16001. doi: 10.1074/jbc.M107340200. [DOI] [PubMed] [Google Scholar]
- 61.Engel F.B., Schebesta M., Duong M.T. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19(10):1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuller S.J., Sivarajah K., Sugden P.H. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J Mol Cell Cardiol. 2008;44(5):831–854. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- 63.Bersell K., Arab S., Haring B., Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 64.Wadugu B., Kuhn B. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol. 2012;302(11):H2139–H2147. doi: 10.1152/ajpheart.00063.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yutzey K.E. Regenerative biology: neuregulin 1 makes heart muscle. Nature. 2015;520(7548):445–446. doi: 10.1038/520445a. [DOI] [PubMed] [Google Scholar]
- 66.Sysa-Shah P., Xu Y., Guo X. Cardiac-specific over-expression of epidermal growth factor receptor 2 (ErbB2) induces pro-survival pathways and hypertrophic cardiomyopathy in mice. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hertig C.M., Kubalak S.W., Wang Y., Chien K.R. Synergistic roles of neuregulin-1 and insulin-like growth factor-I in activation of the phosphatidylinositol 3-kinase pathway and cardiac chamber morphogenesis. J Biol Chem. 1999;274(52):37362–37369. doi: 10.1074/jbc.274.52.37362. [DOI] [PubMed] [Google Scholar]
- 68.Reiss K., Cheng W., Ferber A. Overexpression of insulin-like growth factor-1 in the heart is coupled with myocyte proliferation in transgenic mice. Proc Natl Acad Sci U S A. 1996;93(16):8630–8635. doi: 10.1073/pnas.93.16.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kajstura J., Cheng W., Reiss K., Anversa P. The IGF-1-IGF-1 receptor system modulates myocyte proliferation but not myocyte cellular hypertrophy in vitro. Exp Cell Res. 1994;215(2):273–283. doi: 10.1006/excr.1994.1343. [DOI] [PubMed] [Google Scholar]
- 70.Itoh N., Ohta H., Nakayama Y., Konishi M. Roles of FGF signals in heart development, health, and disease. Front Cell Dev Biol. 2016;4:110. doi: 10.3389/fcell.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carter E.P., Fearon A.E., Grose R.P. Careless talk costs lives: fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol. 2015;25(4):221–233. doi: 10.1016/j.tcb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brewer J.R., Mazot P., Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30(7):751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engel F.B., Hsieh P.C., Lee R.T., Keating M.T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A. 2006;103(42):15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Novoyatleva T., Sajjad A., Pogoryelov D., Patra C., Schermuly R.T., Engel F.B. FGF1-mediated cardiomyocyte cell cycle reentry depends on the interaction of FGFR-1 and Fn14. FASEB J. 2014;28(6):2492–2503. doi: 10.1096/fj.13-243576. [DOI] [PubMed] [Google Scholar]
- 76.Song H.Y., Kim M.R., Lee M.J. Oncostatin M decreases adiponectin expression and induces dedifferentiation of adipocytes by JAK3- and MEK-dependent pathways. Int J Biochem Cell Biol. 2007;39(2):439–449. doi: 10.1016/j.biocel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 77.Hohensinner P.J., Kaun C., Rychli K. The inflammatory mediator oncostatin M induces stromal derived factor-1 in human adult cardiac cells. FASEB J. 2009;23(3):774–782. doi: 10.1096/fj.08-108035. [DOI] [PubMed] [Google Scholar]
- 78.Kubin T., Poling J., Kostin S. Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell. 2011;9(5):420–432. doi: 10.1016/j.stem.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 79.Morikawa Y., Heallen T., Leach J., Xiao Y., Martin J.F. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547(7662):227–231. doi: 10.1038/nature22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bassat E., Mutlak Y.E., Genzelinakh A. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547(7662):179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takeshita S., Kikuno R., Tezuka K., Amann E. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 1993;294(Pt 1):271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snider P., Hinton R.B., Moreno-Rodriguez R.A. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102(7):752–760. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimazaki M., Nakamura K., Kii I. Periostin is essential for cardiac healing after acute myocardial infarction. J Exp Med. 2008;205(2):295–303. doi: 10.1084/jem.20071297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhn B., del Monte F., Hajjar R.J. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13(8):962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 85.Lorts A., Schwanekamp J.A., Elrod J.W., Sargent M.A., Molkentin J.D. Genetic manipulation of periostin expression in the heart does not affect myocyte content, cell cycle activity, or cardiac repair. Circ Res. 2009;104(1):e1–e7. doi: 10.1161/CIRCRESAHA.108.188649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taniyama Y., Katsuragi N., Sanada F. Selective blockade of periostin Exon 17 preserves cardiac performance in acute myocardial infarction. Hypertension. 2016;67(2):356–361. doi: 10.1161/HYPERTENSIONAHA.115.06265. [DOI] [PubMed] [Google Scholar]
- 87.Chen Z., Xie J., Hao H. Ablation of periostin inhibits post-infarction myocardial regeneration in neonatal mice mediated by the phosphatidylinositol 3 kinase/glycogen synthase kinase 3beta/cyclin D1 signalling pathway. Cardiovasc Res. 2017;113(6):620–632. doi: 10.1093/cvr/cvx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahuja P., Sdek P., MacLellan W.R. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87(2):521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agah R., Kirshenbaum L.A., Abdellatif M. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J Clin Invest. 1997;100(11):2722–2728. doi: 10.1172/JCI119817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yurkova N., Shaw J., Blackie K. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res. 2008;102(4):472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- 91.Ebelt H., Zhang Y., Kampke A. E2F2 expression induces proliferation of terminally differentiated cardiomyocytes in vivo. Cardiovasc Res. 2008;80(2):219–226. doi: 10.1093/cvr/cvn194. [DOI] [PubMed] [Google Scholar]
- 92.Sdek P., Zhao P., Wang Y. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J Cell Biol. 2011;194(3):407–423. doi: 10.1083/jcb.201012049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.MacLellan W.R., Garcia A., Oh H. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol. 2005;25(6):2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wamstad J.A., Alexander J.M., Truty R.M. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mahmoud A.I., Kocabas F., Muralidhar S.A. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497(7448):249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greulich F., Rudat C., Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91(2):212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 97.Shen T., Aneas I., Sakabe N. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiang F.L., Guo M., Yutzey K.E. Overexpression of Tbx20 in adult cardiomyocytes promotes proliferation and improves cardiac function after myocardial infarction. Circulation. 2016;133(11):1081–1092. doi: 10.1161/CIRCULATIONAHA.115.019357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Monzen K., Shiojima I., Hiroi Y. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19(10):7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Molkentin J.D., Lin Q., Duncan S.A., Olson E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 101.Yu W., Huang X., Tian X. GATA4 regulates Fgf16 to promote heart repair after injury. Development. 2016;143(6):936–949. doi: 10.1242/dev.130971. [DOI] [PubMed] [Google Scholar]
- 102.Bisping E., Ikeda S., Kong S.W. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci U S A. 2006;103(39):14471–14476. doi: 10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oka T., Maillet M., Watt A.J. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 104.Lu S.Y., Sontag D.P., Detillieux K.A., Cattini P.A. FGF-16 is released from neonatal cardiac myocytes and alters growth-related signaling: a possible role in postnatal development. Am J Physiol Cell Physiol. 2008;294(5):C1242–C1249. doi: 10.1152/ajpcell.00529.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hotta Y., Sasaki S., Konishi M. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev Dynam. 2008;237(10):2947–2954. doi: 10.1002/dvdy.21726. [DOI] [PubMed] [Google Scholar]
- 106.Semenza G.L. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 1985;88(4):1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 107.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maxwell P.H., Wiesener M.S., Chang G.W. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 109.Ohh M., Park C.W., Ivan M. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2(7):423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 110.Weidemann A., Johnson R.S. Biology of HIF-1alpha. Cell Death Differ. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 111.Jopling C., Sune G., Faucherre A., Fabregat C., Izpisua Belmonte J.C. Hypoxia induces myocardial regeneration in zebrafish. Circulation. 2012;126(25):3017–3027. doi: 10.1161/CIRCULATIONAHA.112.107888. [DOI] [PubMed] [Google Scholar]
- 112.Guimaraes-Camboa N., Stowe J., Aneas I. HIF1alpha represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev Cell. 2015;33(5):507–521. doi: 10.1016/j.devcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koshiji M., Kageyama Y., Pete E.A., Horikawa I., Barrett J.C., Huang L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23(9):1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bettencourt-Dias M., Mittnacht S., Brockes J.P. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J Cell Sci. 2003;116(Pt 19):4001–4009. doi: 10.1242/jcs.00698. [DOI] [PubMed] [Google Scholar]
- 115.Wang W.E., Li L., Xia X. Dedifferentiation, proliferation, and redifferentiation of adult mammalian cardiomyocytes after ischemic injury. Circulation. 2017;136(9):834–848. doi: 10.1161/CIRCULATIONAHA.116.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oyama K., El-Nachef D., Zhang Y., Sdek P., MacLellan W.R. Epigenetic regulation of cardiac myocyte differentiation. Front Genet. 2014;5:375. doi: 10.3389/fgene.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ahmad H.R., Hashmi S. Is biological repair of heart on the horizon? Pak J Med Sci. 2017;33(4):1042–1046. doi: 10.12669/pjms.334.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]