Abstract
Retrograde monosynaptic tracing with EnvA-pseudotyped rabies virus has been employed to identify the afferent and efferent connectivity of transplanted human embryonic stem (hES) cell-derived neurons in animal models. Due to the protracted development of transplanted human neurons in host animals, it is important that those transplanted cells express avian leukosis and sarcoma virus subgroup A receptor (TVA) and rabies glycoprotein G (Rgp) for a period of up to several months to enable identification of the synaptic inputs from host neurons to grafted neurons through this rabies virus-based method. Here, we report the generation of an engineered hES cell line through CRISPR/Cas9-mediated targeting to the AAVS1 locus of an EnvA-pseudotyped rabies virus-based tool for retrograde monosynaptic tracing. This engineered hES cell line, named H1-CAG-GTRgp, expresses GFP, TVA and Rgp. Upon transplantation of H1-CAG-GTRgp-derived neural progenitor cells (NPCs) into the rat brain after traumatic injury, the grafted neurons derived from H1-CAG-GTRgp cells expressed GFP, TVA, and Rgp stably for up to 6 months post-transplantation and received robust synaptic inputs from host neurons in the target regions of the orthotopic neural circuitry. The retrograde monosynaptic tracing hES cell line provides an efficient approach to analyze transplant connectivity for the comprehensive assessment of host-donor cell innervation.
Keywords: Neural progenitors, AAVS1 locus, EnvA-pseudotyped rabies virus, Retrograde monosynaptic tracing, Transplantation
1. Introduction
Preclinical studies have suggested that transplantation of neural stem cells (NSCs), or their progenies, provides a promising therapeutic approach for many devastating neurological disorders.1, 2, 3 This strategy is based on the idea that grafted neural cells are capable of replacing lost neurons after nervous system injury and reconstructing vital aspects of neuronal connectivity. Grafted neurons can incorporate into the neuronal circuitry and form synaptic connections with host neurons.4, 5, 6, 7, 8 However, the extent to which grafted neurons can integrate into existing circuits is not clear. A key question is whether grafted neurons will receive orthotopically afferent inputs from host neurons and function properly.
Rabies virus can infect neurons through axon terminals and spread between two coupled synapses in a retrograde direction.9 EnvA-pseudotyped rabies virus is a recombinant virus in which the genes coding for rabies glycoprotein and envelope are replaced by genes coding for fluorescent protein and EnvA, respectively. Therefore, the virus selectively infects TVA-transfected neurons.10, 11, 12, 13 As an exquisite tool for neural circuit tracing, the recombinant rabies virus plays an important role in mapping direct synaptic connections and afferent synaptic inputs in the central nervous system.14, 15, 16, 17 Recently, EnvA-pseudotyped rabies viruses was employed to identify the synaptic inputs from host neurons to grafted neurons in different experimental animal models. This powerful monosynaptic tracing method has explicitly shown that grafted neurons receive highly orthotopic afferent inputs.18, 19, 20, 21
Transplanted cells expressing TVA, Rgp and fluorescent protein are usually generated by lentivirus infection, which enables infection by EnvA-pseudotyped rabies viruses. Human brain development is a protracted process that begins in the third gestational week with the differentiation of neural progenitor cells and extends at least through late adolescence, arguably throughout the lifespan.22 hES cell-derived cortical cells engraft robustly and mature following a species-specific timeline in the mouse newborn cortex.23 Given the protracted timeframe for the functional maturation of human cells, reconstruction of neuronal circuitry may reach its maximum at a very late time point. Therefore, for the use of rabies viruses-based trans-synaptic tracing at a late time point after transplantation, grafted cells must maintain the expression of TVA, Rgp and fluorescent protein. Transgenic human pluripotent stem cells (hPSCs) established through random transgene insertion generally cannot sustain transgene expression upon differentiation, especially in differentiation toward functional neurons.24 However, targeted insertion of transgenes into a safe harbor site offers an opportunity for stable transgene expression.24, 25
In order to maintain the expression levels of GFP, TVA, and Rgp in transgenic hES cells for rabies virus-based neuronal tracing, we generated a H1-CAG-GTRgp knock-in cell line using CRISPR/Cas9-mediated targeting to the AAVS1 locus. This genetic manipulation did not affect pluripotency and the neural differentiation capacity of the hES cells. Our results demonstrate that grafted neurons can stably expressed GFP, TVA, and Rgp for up to 6 months post-transplantation (mpt). Synaptic inputs from host neurons received by grafted cells were detected at 3 mpt and 6 mpt. Thus, this engineered hES cell line provides an efficient approach to analyze transplant connectivity for the comprehensive assessment of host-donor cell innervation.
2. Materials and methods
2.1. Antibodies and drugs
The following antibodies and drugs were used in this study: anti-OCT4 (Santa Cruz Biotechnology, sc-5279), anti-SSEA4 (Invitrogen, 414000), anti-PAX6 (Covance Research, PRB-278P-100), anti-PAX6 (Abcam, ab78545), anti-Ki67 (Abcam, ab15580), anti-SOX1 (Millipore, AB15766), anti-SOX2 (RD, MAB2018), anti-NESTIN (Millipore, ABD69), anti-NESTIN (BD, 611658), anti-DCX (Cell Signaling, 4604), anti-NKX2.1 (Millipore, MAB5460), anti-FOXG1 (Abcam, ab18259), anti-COUP-TF1 (Millipore, ABE1425), anti-HNA (Millipore, MAB1281), anti-STEM121 (Takara, Y40410), anti-NEUN (Millipore, ABN78), anti-GFAP (Dako, Z0334), anti-GFAP (Millipore, MAB360), anti-SATB2 (Abcam, ab51502), anti-CTIP2 (Abcam, ab18465), anti-TBR1 (Abcam, ab31940), anti-MAP2 (Abcam, ab32454), anti-MAP2 (Millipore, MAB3418), anti-synapsin 1 (Calbiochem, 574777), anti-PSD95 (Abcam, ab13552), Y-27632 (Sigma, S1049), G418 (Sigma, A1720), thiazovivin (Selleck, S1459), SB431542 (Selleck, S1067), dorsomorphin Selleck, S7840), insulin (Gibco. I9278), and heparin (Sigma, H3149).
2.2. Cell culture
The human ES cell lines H1 (WA01, WiCell Research Institute, Inc.) and CRISPR/Cas9 gene knock-in cell lines were cultured in 6-well tissue culture plates (Greiner Bio-One) coated with Matrigel (BD Bioscience, 354277) in defined mTeSR1 medium (STEMCELL Technologies, #05850). The medium was changed daily. Cells were passaged at a 1:4 ratio every 3–4 days after reaching approximately 80% confluence using 0.5 mM EDTA for 5 min at 37 °C. All cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air.
2.3. Production of knock-in hES cell lines through CRISPR/Cas9
The Cas9 plasmid pX330 was purchased from Addgene. Guide RNA (gRNA) targeting the AAVS1 locus was designed on the CRISPR design platform (crispr.mit.edu) and inserted into the pX330 plasmid. Donor DNA was composed of left/right homology arms to the AAVS1 locus (DNA fragments of approximately 1–1.5 kb in length at genomic AAVS1 locus), CAG promoter-GFP-TVA-glycoprotein G and PGK-neomycin cassette. The pX330 plasmid and donor DNA were electroporated into H1 cells (Nucleofector® II Device, Lonza, AAB-1001, B-16 program). The transfected cells were plated onto Matrigel-coated 6-well tissue culture plates with 10 μM Y-27632 for 1 day. Positive clones were selected using 100 μg/mL G418 in mTeSR1 medium. Individual clones were manually picked, expanded in 24-well tissue culture plates, and then divided into two portions for passage and genomic DNA extraction. Genotyping was performed by genomic PCR. The sequences of sgRNA and primers are listed in Table S1.
2.4. Knock-in hES cell line genotyping
Site specific targeted transgenes insertion at the AAVS1 locus was confirmed by genomic DNA PCR and sequencing of correlated PCR products. Genomic DNAs of selected positive clones were extracted with the TIANamp Genomic DNA Kit (Tiangen, DP348). For genomic PCR analysis, Phanta® Max Super-Fidelity DNA polymerase (Vazyme, P505-3) was used in all PCR reactions. The sequences of Primer-F and Primer-R are listed in Table S1.
2.5. Flow cytometry analysis
hES cells were digested with Accutase (Sigma, A6964) for 10–15 min at 37 °C and fixed with 4% paraformaldehyde for approximately 15 min at room temperature. Cells were washed twice with PBS and permeabilized with 90% methanol for 30 min at 4 °C. After the cells were washed, they were incubated with primary antibodies and isotype control antibodies for 30 min at 37 °C. The cells were then washed and incubated with secondary antibodies for 30 min at 37 °C. The cells were resuspended in 200 μL PBS followed by two washes with PBS, and then analyzed with an Accuri C6 (BD Biosciences).
2.6. Quantitative real-time PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen, TR118) and reverse-transcribed to cDNAs using oligo dT primers (Takara, 3806) and ReverTra Ace ® qPCR RT Kit (Toyobo, 6151001). qRT-PCR was performed using primers against SOX2, OCT4, and NANOG on a CFX96 real-time PCR detection system (BIO-RAD). GAPDH was used as an internal control to normalize the gene expression data. All qRT-PCR reactions were performed in triplicate. qRT-PCR primers are listed in Table S1.
2.7. Teratoma formation
ES cells cultured on Matrigel-coated 6-well tissue culture plates were digested by Accutase and suspended in 30% Matrigel in DMEM/F12 (HyClone, SH30023.01). A total of 100 μL of the cell suspension containing 1 × 106 cells was implanted subcutaneously into the ventral flank of NOD-SCID mice. Teratomas were surgically dissected between 6 weeks and 8 weeks after engraftment and fixed in 4% paraformaldehyde followed by embedding in paraffin. Teratomas were sectioned and processed for hematoxylin and eosin staining.
2.8. Karyotype analysis
The detailed method for karyotype analysis has been described in our previous study (Lihui Wang, et al. Natuer Methods, 2013, 10, p84–89). In brief, cells were grown in 6-well plates and demecolcine (Dahui Biotech) was added to a final concentration of 50 μg/mL for 130 min. Cells were then trypsinized, pelleted by centrifugation at 2000×g for 5 min, resuspended in 8 mL of 0.075 M KCl and incubated for 20 min at 37 °C. A fixative solution composed of 1 part of acetic acid and 3 parts methanol was added to a final volume of 10 mL, mixed gently, and incubated for 10 min at 37 °C. After further centrifugation, the supernatant was removed, and 10 mL of ice-cold fixative solution was added. Cells were then dropped on a cold slide and incubated at 75 °C for 3 h. Belts were treated with trypsin and colorant, and metaphases were analyzed on an Olympus BX51 microscope.
2.9. Neural stem cell induction and maintenance
Neural induction of hES cells using monolayer culture was begun by differentiating hES cells to neuroepithelia cells in N2B27 medium (DMEM/F12: Neurobasal 1:1, 0.5% N2, 1% B27, 2 mM Glutamax, 1 × NEAA, 5 μg/mL insulin, 2 μg/mL heparin) containing 5 μM SB431542 and 5 μM dorsomorphin for 8 days. After passaging, the cells were cultured in N2B27 medium for another 8 days. A total of 20 ng/mL bFGF was added around Day 12 to Day 13 based on the appearance of rosette-like structures. Then, neural stem cells were obtained by manually picking rosette-like structures on plated cells and continuously propgating these cells on Matrigel-coated plates in N2B27 medium.
2.10. Visual cortex lesion and cell transplantation
The cerebral cortex traumatic injury model, in which the lesion area mainly included the visual cortex, was used in this study as the grafted cells tend to have a posterior cortical identity. All of the animal experiments were carried out in full compliance with the IACUC at GIBH (NO. 2012017). Adult male SD rats were anesthetized via an intraperitoneal injection of 1.0–1.5% pentobarbital (40–60 mg/kg body weight). After brain exposure using a scalpel and skull bore, a piece of neocortex 4 mm2 in area (AP: 4–6 mm posterior to the Bregma, LM: 3–5 mm to the midline, which is mainly located in visual area.) and 2 mm in depth, was sucked away unilaterally. One week after injury, the neural stem cells were digested into single cells with Accutase, suspended in DMEM/F12 (1 × 105/μL), and microinjected in 2 sites immediately around the lesion area (2 μL per site). All of the experimental animals received cyclosporin A (10 mg/kg bodyweight, S.C.) injection daily beginning 2 days before transplantation.
2.11. Immunohistochemistry
Animals were deeply anesthetized and sacrificed by perfusion with chilled saline followed by 4% paraformaldehyde. Then, the whole brain was removed, post-fixed overnight in 4% paraformaldehyde, dehydrated at 30% sucrose for 2–3 days, and embedded in OCT compound. The brains were sliced on coronal planes at 40 μm thickness using a Leica CM3050S Cryostat. For immunostaining, the slices were washed 3 times in PBS, permeabilized with 1% Triton X-100 in PBS for 30 min, and incubated in blocking buffer (PBS containing 10% serum and 0.25% Triton X-100) for 60–120 min. Brain sections were then incubated in primary antibodies diluted in blocking buffer at 4 °C overnight. The next day, sections were washed 3 times in PBS and incubated with secondary antibodies and DAPI for 60 min. After the sections were washed with PBS, the slides were mounted with Dako fluorescence mounting medium.
3. Results
3.1. The H1-CAG-GTRgp knock-in cell line maintained pluripotency and differentiated into neural progenitor cells in vitro
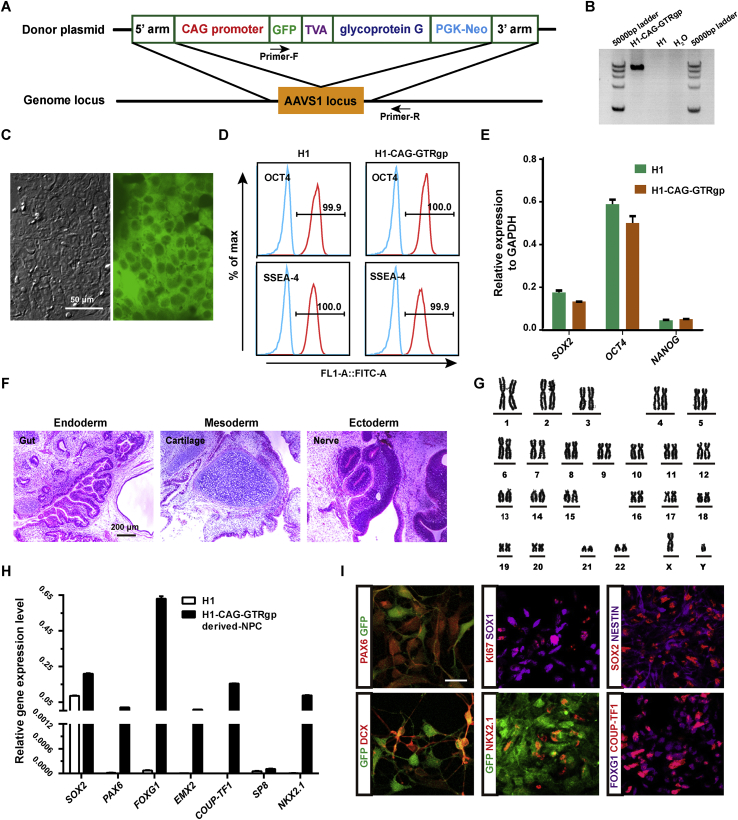
The AAVS1 safe harbor site is a preferred target for gene knock-in due to its consistent and robust transgene expression.26 CRISPR/Cas9-mediated gene targeting was applied to generate a retrograde monosynaptic tracing hES cell line, H1-CAG-GTRgp, which showed constitutive expression of GFP, TVA and Rgp. Donor DNA with homology arms to AAVS1 genomic regions and the PGK-neomycin cassette (Fig. 1A) was electroporated into hES cell line H1 together with the PX330 plasmid. Neomycin resistant clones were screened by genomic DNA PCR using a pair of primers, Primer-R and Primer-F (Fig. 1A and B). Correct insertion of the construct in the AAVS1 locus was identified by PCR product sequencing (data not shown). Two positive clones were cultured and passaged continuously. GFP fluorescence could be detected in both clones during long-term culture (Fig. 1C). The results from qRT-PCR and FACS analysis showed that the H1-CAG-GTRgp hES cells maintained pluripotency and characteristics of pluripotent stem cells (Fig. 1D and E). Moreover, the cells had a normal karyotype (Fig. 1G), and were able to form the teratomas containing cells of all three germ layers (Fig. 1F).
Fig. 1.
H1-CAG-GTRgp cells differentiated into neural progenitor cells with stable expression of GFP in vitro. A. Schematic diagram of the gene knock-in construct targeting the AAVS1 Locus. Donor plasmid containing homology arms to AAVS1 genomic regions (5′ and 3′ arms), CAG promoter, GFP, TVA, Rgp and PGK-neomycin cassette. B. Genotyping of the H1-CAG-GTRgp cell clones using the Primer-F and Primer-R primers, as depicted in (A). C. Representative GFP expression of the positive clones. Scale bar = 50 μm. D. OCT4 and SSEA4 expression levels in H1 and H1-CAG-GTRgp hES cells determined by FACS. E. SOX2, OCT4 and NANOG expression levels in H1 and H1-CAG-GTRgp hES cells determined by qRT-PCR. F. Hematoxylin and eosin staining of the teratomas formed by H1-CAG-GTRgp hES cells. Scale bar = 50 μm. G. Karyotype of the H1-CAG-GTRgp hES cells. H. qRT-PCR analysis of different NPC markers in H1 hES cells and induced NPCs derived from H1-CAG-GTRgp hES cells. I. Representative immunofluorescent images of induced neuroepithelial cells, immature neuronal cells and neural progenitor cells derived from H1-CAG-GTRgp hES cells. Scale bar = 20 μm.
H1-CAG-GTRgp knock-in cells could be induced into primitive neuroepithelial cells in a monolayer culture system using the dual-SMAD inhibition protocol. qRT-PCR analysis results showed that the expression levels of forebrain NPC markers FOXG1, PAX6, and EMX2 increased dramatically compared to H1 HES cells (Fig. 1H), while the expression levels of the midbrain NPC marker EN1 or hindbrain NPC markers GBX2 and HOXB2 were lower (Fig. S1A). The expression of some NPC markers PAX6, SOX1, NESTIN, SOX2, and FOXG1, together with immature neuron marker DCX was further confirmed by immunostaining (Fig. 1I). Unexpectedly, the ventral forebrain marker NKX2.1 was also up-regulated (Fig. 1H and I), which explained the incidence of GABAergic neurons upon further differentiation in vitro27 and in vivo (Fig. S1B). Moreover, a COUP-TF1HIGH/SP8LOW expression pattern indicated that these forebrain neural progenitors tended to have a posterior parietal-sensory and occipital-visual cortical fate rather than a frontal-motor fate28, 29 (Fig. 1H and I). Notably, H1-CAG-GTRgp cells (Fig. 1C) and their derivatives (Fig. 1I), like neural epithelial (NE) cells, NPCs and immature neuronal cells, maintained detectable expression level of GFP.
3.2. H1-CAG-GTRgp-derived NPCs differentiated into cortical pyramidal projection neuron-like cells and formed synapses in a rat model of traumatic brain injury
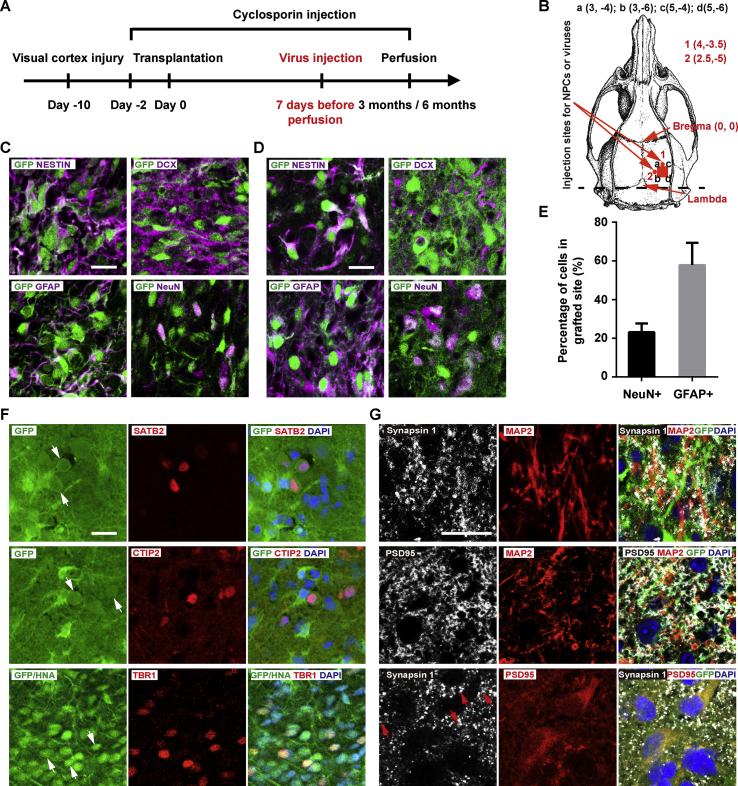
To test whether H1-CAG-GTRgp could be used as a universal tool for mapping direct synaptic connections and analyzing neuronal circuits between grafted cells and host neurons, we then studied the differentiation of H1-CAG-GTRgp-derived NPCs in a rat model of traumatic brain injury. As described above, we differentiated H1-CAG-GTRgp cells into forebrain NPCs and transplanted them into the damaged visual cortex of adult rats 46 days after in vitro differentiation (Fig. 2A and B). The cell fates of the grafted cells were studied at 3 mpt and 6 mpt.
Fig. 2.
Grafted NPCs differentiated into cortical projection neuron-like cells with stable expression of GFP and formed synapses in the transplanted site. A. Experimental scheme. EnvA-pseudotyped rabies viruses was injected 7 days before the animals were sacrificed. B. Schematic diagram of intracortical transplantation in the rat visual cortex. The red square represents the site of injury in the visual cortex. The Position of the red square was defined by X and Y axes of a, b, c and d. Red dot 1 and red dot 2 represent injection sites for NPCs or virus. The Bregma and Lambda points are indicated by arrows. C. The differentiation of grafted cells at 3 mpt. Many NESTIN+/GFP+ and DCX+/GFP+ human cells, but few GFAP+/GFP+ cells and NeuN+/GFP+ neurons were found. Scale bar = 20 μm. D. The differentiation of grafted cells at 6 mpt. More GFAP+/GFP+ cells and NeuN+/GFP+ neurons were found. Scale bar = 20 μm. E. Bar graphs summarized from bottom panel in D. At 6 mpt, the percentage was 57.76% for GFAP+ cells and 23.16% for NeuN+ cells. F. Immunostaining at 6 mpt revealed that human cells expressed markers of cortical pyramidal projection neurons, including deeper layer markers TBR1 and CTIP2 and upper layer marker SATB2. Some of the positive cells are indicated with white arrows. Scale bar = 20 μm. G. Representative images of synapsin 1 and PSD95 staining at 6 mpt. Some clusters containing synapsin 1 and PSD95 are indicated with red arrows. Scale bar = 20 μm.
Immunohistochemical analysis at 3 months and 6 months after intracortical transplantation showed that the majority of the grafted cells distributed around the implantation site and expressed GFP stably. Compared to the differentiation of grafts at 3 mpt (Fig. 2C), the grafts at 6 mpt exhibited fewer NESTIN+/GFP+ and DCX+/GFP+ human cells and more GFAP+/GFP+ cells and NeuN+/GFP+ neurons (Fig. 2D). GFAP+/GFP+ cells became the major population at 6 mpt (57.76% of GFP positive cells), whereas NeuN+/GFP+ cells increased to 23.16% (Fig. 2D and E). Furthermore, the fluorescence intensity of NeuN staining increased with time.
Some differentiated human neurons expressed markers of cortical pyramidal projection neurons, including upper layer marker SATB2 and deeper layer markers TBR1 and CTIP2 at both 3 mpt (data not shown) and 6 mpt (Fig. 2F). The TBR1+, CTIP2+ and SATB2+ cells tended to group separately. No obvious layer distribution for these cortical projection neuron-like cells was identified. Among the differentiated human neurons, we also observed a very small population of GAD67+ cells together with few TH+ cells and CHAT+ cells (Fig. S1B).
Immunohistochemical staining of presynaptic marker synapsin 1 and postsynaptic marker PSD95 in brain tissue sections at 6 mpt showed that many synapses were formed around dendrites of differentiated neurons (Fig. 2G), suggesting that synaptic connections could be formed between human and host neurons. All these data indicated that robust engraftment and neuronal/glial differentiation occurred in vivo, and that GFP expression was sustained in more mature stages of neural cells.
3.3. Intracortically grafted NPCs were infected by EnvA-pseudotyped rabies virus specifically and received synaptic inputs from host neurons
H1-CAG-GTRgp cells could differentiate into cortical pyramidal projection neuron-like cells and form synaptic connections with human or host neurons in vivo. We then analyzed the synaptic inputs that were sent from neurons in different host brain areas to grafted neurons using monosynaptic retrograde tracing at 3 mpt and 6 mpt. After human NPCs were transplanted to the rat model of traumatic injury to the visual cortex, EnvA-pseudotyped rabies viruses was injected at the location of the graft 7 days before 3 mpt or 6 mpt, and animals were perfused 7 days thereafter (Fig. 2A and B).
The EnvA-pseudotyped rabies virus, in which Rgp was replaced with mCherry, was used in this study. The titer of the virus was approximately 1.0 × 10 7 IU/mL. Because of the interaction between EnvA viral glycoprotein and its cognate receptor, TVA, pseudotyped rabies virus could selectively infect transplanted cells expressing TVA. EnvA-pseudotyped rabies virus was then complemented with Rgp at transplanted cells, labeling first-order presynaptic neurons with mCherry. We infected the H1 and H1-CAG-GTRgp hES cells with EnvA-pseudotyped rabies viruses to verify the specificity of the pseudotype rabies virus. We did not observe any mCherry fluorescence signal in H1 hES cells (Fig. S1C).
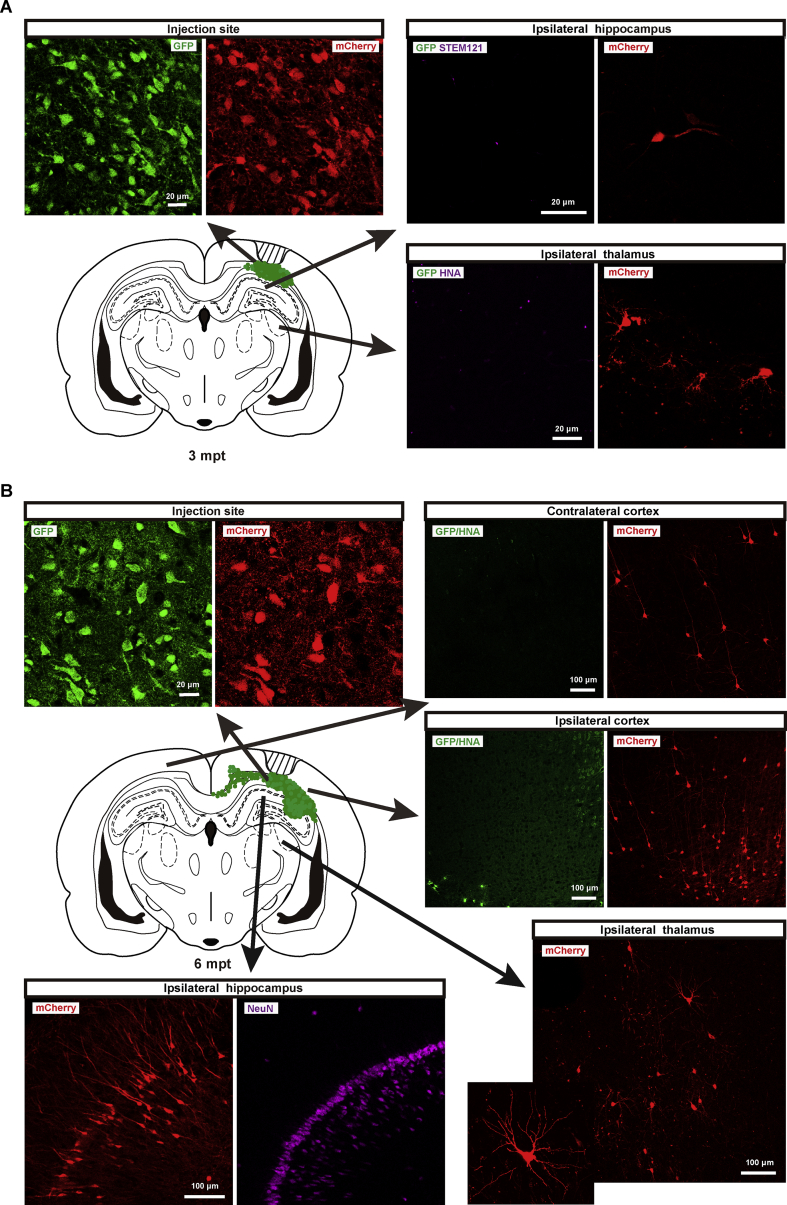
The brain slice imaging results showed that a large number of grafted cells were GFP+/mCherry+, which represented the successful infection of the injected EnvA-pseudotyped rabies viruses (Fig. 3A and B). At 6 mpt, GFP+ cells were distributed broadly in the brain areas around the lesion site, mainly in the visual cortex and corpus callosum, and some of them were in the CA1 to CA3 areas of hippocampus. Meanwhile, GFP-/mCherry+ host neurons were detected in different sites around the whole brain, including ipsilateral and contralateral visual cortex of the traumatic lesion site, CA1 and subiculum areas of the ipsilateral hippocampus, and dorsal lateral geniculate nucleus (DLG) of the ipsilateral thalamus. The absence of human nuclei antibody (HNA) signal and GFP fluorescence suggested that these mCherry+ cells were not grafted human cells (Fig. 3 B). Only a few GFP-/mCherry+ host neurons were observed in the DLG of the ipsilateral thalamus and ipsilateral hippocampus at 3 mpt (Fig. 3A), which might be due to a smaller proportion of NeuN+ cells among grafted cells (Fig. 2C and D).
Fig. 3.
Intracortically grafted NPCs received synaptic inputs from host neurons at 3 mpt and 6 mpt. A. Representative images of brain slices at 3 mpt. GFP+/mCherry+ cells represent grafted cells infected by viruses, while GFP-/mCherry+ cells represent host neurons. The STEM121 antibody, HNA antibody and GFP fluorescence was used to confirm the location of grafted cells. Green dots represent grafted cells. B. Fluorescence photomicrographs showed more GFP-/mCherry+ host cells in the corpus callosum, hippocampus and thalamus at 6 mpt than at 3 mpt. Immunostaining of NeuN indicated that GFP-/mCherry+ host cells were neurons. Green dots represent grafted cells.
Therefore, our data revealed that the grafted cells received orthotopically synaptic inputs from host neurons at 3 mpt and 6 mpt (Fig. 3A and B). The discrimination capability of H1-CAG-GTRgp cell-derived progenies for EnvA-pseudotyped rabies virus could last for a long time in vivo.
4. Discussion
Transgene expression via random integration into the genome is subjected to position effects and silencing. In addition, random gene insertion might interrupt or activate neighboring genes. Most commonly, transgenes are introduced into human ES cells via methods such as viral transduction or plasmid transfection. While expression of transgenes can be regulated in stem cells and their differentiated progenies at an early stage, such transgenes are often downregulated upon stem cell differentiation and even no longer expressed in mature cells, especially in functional neurons.24, 30, 31, 32, 33, 34 Meanwhile, random insertion comes with another disadvantage, which is that uneven expression among cell populations could cause variation among experiments.30, 31, 34, 35 Genomic safe harbor sites are transcriptionally active, thereby allowing robust and stable gene expression. Furthermore, a transgene insertion at genomic safe harbors have no adverse effect on the host cell genome. Although no genomic site has been fully validated as a genomic “safe harbor”, some candidates from the human genome have been proposed, including the adeno-associated virus site 1 (AAVS1) locus, the chemokine (CC motif) receptor 5 (CCR5) locus, and the human orthologue of the mouse ROSA26 locus.36 Among these candidates in human cells, AAVS1 has been accepted as having a high gene expression, and being a safe genomic location.37 Cells derived from the H1-CAG-GTRgp knock-in cell line could stably express GFP, TVA and Rgp in different neural cells stably and be infected specifically by EnvA-pseudotyped rabies virus in vivo at 3 mpt and 6 mpt.
Our data showed that grafted neurons could receive synaptic inputs from host neurons, which was consistent with previous reports.18, 21 The connection between grafted human cells and host cells indicated the potential ability to rebuild injured neural circuits by the transplantation of human neural progenitors. This engineered hES cell line will substantially help to analyze transplant connectivity for the comprehensive assessment of host-donor cell innervation. Such retrograde rabies mono-trans-synaptic tracers provide a useful tool to monitor the functional integration of grafted NPCs and host neural networks. In the future, the CAG promoter can be replaced with neuron-specific synapsin promoter, thus allowing the specific infection of mature neurons so that the synaptic inputs from host neurons to mature grafted neurons can be mapped.
Author contributions
Xiaofen Zhong, Yiping Guo and Guangjin Pan conceived the project. Xiaofen Zhong, Yiping Guo and Qi Xing composed the manuscript. Qi Xing generated and characterized the H1-CAG-GTRgp cell line. Zhenghui Su and Wenhao Huang generated the neural progenitor cells derived from H1-CAG-GTRgp cell line. Qi Xing, Chunhua Liu and Yiping Guo generated the rat models of traumatic brain injury and transplanted neural progenitor cells. Aiping Lin carried out all the brain sectioning and immunochemical fluorescence staining. Aiping Lin and Wenjing Guo performed brain slice imaging.
Conflict of interest
The authors declare that there is no conflicts of interest.
Acknowledgments
EnvA-pseudotyped rabies virus was a gift from Dr. Weixiang Guo. We would like to thank our lab members for their support. This work was supported by the National Key Research and Development Program of China, Stem Cell and Translational Research (2017YFA0102600), the National Natural Science Foundation of China (81571238, 81771356), the Frontier and Key Technology Innovation Special Grant from the Department of Science and Technology of Guangdong Province (2014B020225006, 2014B020225002, 2015B020228003, 2014B050504008); the Natural Science Foundation of Guangdong Province, China (2016A030313167), the Science and Technology Planning Project of Guangdong Province, China (2017B030314056), the Bureau of Science and Technology of Guangzhou Municipality, China (201803040016), and the “Hundred Talents Program” of the Chinese Academy of Science (to Dr. Zhong).
Footnotes
Peer review under responsibility of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cr.2019.01.002.
Contributor Information
Yiping Guo, Email: Guo_yiping@gibh.ac.cn.
Xiaofen Zhong, Email: zhong_xiaofen@gibh.ac.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Steinbeck J.A., Studer L. Moving stem cells to the clinic: potential and limitations for brain repair. Neuron. 2015;86:187–206. doi: 10.1016/j.neuron.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weston N.M., Sun D. The potential of stem cells in treatment of traumatic brain injury. Curr Neurol Neurosci Rep. 2018;18:1. doi: 10.1007/s11910-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson L.H., Bjorklund A. Reconstruction of brain circuitry by neural transplants generated from pluripotent stem cells. Neurobiol Dis. 2015;79:28–40. doi: 10.1016/j.nbd.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Bonner J.F., Connors T.M., Silverman W.F., Kowalski D.P., Lemay M.A., Fischer I. Grafted neural progenitors integrate and restore synaptic connectivity across the injured spinal cord. J Neurosci. 2011;31:4675–4686. doi: 10.1523/JNEUROSCI.4130-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espuny-Camacho I., Michelsen K.A., Linaro D. Human pluripotent stem-cell-derived cortical neurons integrate functionally into the lesioned adult murine visual cortex in an area-specific way. Cell Rep. 2018;23:2732–2743. doi: 10.1016/j.celrep.2018.04.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muneton-Gomez V.C., Doncel-Perez E., Fernandez A.P. Neural differentiation of transplanted neural stem cells in a rat model of striatal lacunar infarction: light and electron microscopic observations. Front Cell Neurosci. 2012;6:30. doi: 10.3389/fncel.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tornero D., Wattananit S., Gronning Madsen M. Human induced pluripotent stem cell-derived cortical neurons integrate in stroke-injured cortex and improve functional recovery. Brain. 2013;136:3561–3577. doi: 10.1093/brain/awt278. [DOI] [PubMed] [Google Scholar]
- 8.Tsupykov O., Kyryk V., Smozhanik E. Long-term fate of grafted hippocampal neural progenitor cells following ischemic injury. J Neurosci Res. 2014;92:964–974. doi: 10.1002/jnr.23386. [DOI] [PubMed] [Google Scholar]
- 9.Dum R.P., Strick P.L. Transneuronal tracing with neurotropic viruses reveals network macroarchitecture. Curr Opin Neurobiol. 2013;23:245–249. doi: 10.1016/j.conb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocas L., Fernandez G. Monosynaptic tracing in developing circuits using modified rabies virus. Methods Mol Biol. 2017;1538:353–366. doi: 10.1007/978-1-4939-6688-2_23. [DOI] [PubMed] [Google Scholar]
- 11.Wickersham I.R., Finke S., Conzelmann K.K., Callaway E.M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickersham I.R., Lyon D.C., Barnard R.J. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickersham I.R., Sullivan H.A., Seung H.S. Axonal and subcellular labelling using modified rabies viral vectors. Nat Commun. 2013;4 doi: 10.1038/ncomms3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande A., Bergami M., Ghanem A. Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proc Natl Acad Sci USA. 2013;110:E1152–E1161. doi: 10.1073/pnas.1218991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamichi K., Amat F., Moussavi F. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vivar C., Potter M.C., Choi J. Monosynaptic inputs to new neurons in the dentate gyrus. Nat Commun. 2012;3:1107. doi: 10.1038/ncomms2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watabe-Uchida M., Zhu L., Ogawa S.K., Vamanrao A., Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Doerr J., Schwarz M.K., Wiedermann D. Whole-brain 3D mapping of human neural transplant innervation. Nat Commun. 2017;8:14162. doi: 10.1038/ncomms14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkner S., Grade S., Dimou L. Transplanted embryonic neurons integrate into adult neocortical circuits. Nature. 2016;539:248. doi: 10.1038/nature20113. [DOI] [PubMed] [Google Scholar]
- 20.Grealish S., Heuer A., Cardoso T. Monosynaptic tracing using modified rabies virus reveals early and extensive circuit integration of human embryonic stem cell-derived neurons. Stem Cell Rep. 2015;4:975–983. doi: 10.1016/j.stemcr.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tornero D., Tsupykov O., Granmo M. Synaptic inputs from stroke-injured brain to grafted human stem cell-derived neurons activated by sensory stimuli. Brain. 2017;140:692–706. doi: 10.1093/brain/aww347. [DOI] [PubMed] [Google Scholar]
- 22.Stiles J., Jernigan T.L. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espuny-Camacho I., Michelsen K.A., Gall D. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Qian K., Huang C.L., Chen H. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cell. 2014;32:1230–1238. doi: 10.1002/stem.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockemeyer D., Wang H.Y., Kiani S. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ordovas L., Boon R., Pistoni M. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Hu L., Liu C. 5-HT2 receptors mediate functional modulation of GABAa receptors and inhibitory synaptic transmissions in human iPS-derived neurons. Sci Rep. 2016;6:20033. doi: 10.1038/srep20033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C., Tsai S.Y., Tsai M.J. COUP-TFI: an intrinsic factor for early regionalization of the neocortex. Genes Dev. 2001;15:2054–2059. doi: 10.1101/gad.913601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armentano M., Chou S.J., Tomassy G.S., Leingaertner A., O'Leary D.D.M., Studer M. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci. 2007;10:1277–1286. doi: 10.1038/nn1958. [DOI] [PubMed] [Google Scholar]
- 30.Cherry S.R., Biniszkiewicz D., van Parijs L., Baltimore D., Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 32.Liew C.G., Draper J.S., Walsh J., Moore H., Andrews P.W. Transient and stable transgene expression in human embryonic stem cells. Stem Cell. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 33.Smith J.R., Vallier L., Lupo G., Alexander M., Harris W.A., Pedersen R.A. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313:107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Yao S.Y., Sukonnik T., Kean T., Bharadwaj R.R., Pasceri P., Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Laker C., Meyer J., Schopen A. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72:339–348. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat Rev Canc. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 37.van Rensburg R., Beyer I., Yao X.Y. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.