Abstract
Background
Multiple Sclerosis (MS) results from genetic predisposition and environmental variables, including elevated Body Mass Index (BMI) in early life. This study addresses the effect of BMI on the epigenome of monocytes and disease course in MS.
Methods
Fifty-four therapy-naive Relapsing Remitting (RR) MS patients with high and normal BMI received clinical and MRI evaluation. Blood samples were immunophenotyped, and processed for unbiased plasma lipidomic profiling and genome-wide DNA methylation analysis of circulating monocytes. The main findings at baseline were validated in an independent cohort of 91 therapy-naïve RRMS patients. Disease course was evaluated by a two-year longitudinal follow up and mechanistic hypotheses tested in human cell cultures and in animal models of MS.
Findings
Higher monocytic counts and plasma ceramides, and hypermethylation of genes involved in negative regulation of cell proliferation were detected in the high BMI group of MS patients compared to normal BMI. Ceramide treatment of monocytic cell cultures increased proliferation in a dose-dependent manner and was prevented by DNA methylation inhibitors. The high BMI group of MS patients showed a negative correlation between monocytic counts and brain volume. Those subjects at a two-year follow-up showed increased T1 lesion load, increased disease activity, and worsened clinical disability. Lastly, the relationship between body weight, monocytic infiltration, DNA methylation and disease course was validated in mouse models of MS.
Interpretation
High BMI negatively impacts disease course in Multiple Sclerosis by modulating monocyte cell number through ceramide-induced DNA methylation of anti-proliferative genes.
Fund
This work was supported by funds from the Friedman Brain Institute, NIH, and Multiple Sclerosis Society.
Keywords: Obesity, Neurodegeneration, Lipids, Epigenetics, Immunity
Research In Context.
Evidence before this study
Multiple Sclerosis (MS) defines a neurological condition characterized by inflammatory demyelination and axonal/neuronal degeneration, which is thought to result from the integration of genetic predisposition and environmental variables, and clinically progresses towards permanent disability. Previous studies on high Body Mass Index (BMI) in childhood as a risk factor for the disease suggest that BMI can induce long term effects, but the underlying mechanisms and impact on disease course remain elusive.
Added value of this study
This study investigates the effect of BMI on the epigenome of monocytes sorted from the blood of recently diagnosed therapy naïve MS patients. By conducting blinded immunophenotyping, and genome-wide DNA methylation analysis, we identified DNA hypermethylation of genes negatively regulating proliferation as part of the mechanism responsible for the higher numbers of circulating monocytes in MS patients with high BMI. Unbiased lipidomic profiling of plasma samples revealed the presence of more abundant ceramide species in MS patients with high BMI, which likely reflected the synergy between de novo synthesis from dietary fats and recycling of damaged lipids due to the disease process. Cell-based approaches in cultured monocytes further revealed the nuclear localization of uptaken ceramides and validated their role as inducers of DNA hypermethylation on anti-proliferative genes, resulting in increased cell counts. Finally, we report an association between increased monocyte numbers, reduced brain volume and worsened clinical disability at the two-year follow-up in MS patients with high BMI; a finding that was reproduced in animal models of MS. Overall, these data identify the BMI-dependent increase of specific ceramide species as an important determinant of disease course in MS, acting, at least in part, by modulating the epigenome of monocytes.
Implications of all the available evidence
The reported DNA methylation changes caused by ceramides in monocytes and previous work on neurotoxic ceramides highlight the concept that the long-lasting deleterious effect of BMI on MS disease course could be hampered by modification of lifestyle variables, such as diet. It is suggested that plasma ceramide levels in MS patients are derived not only from the degradation of myelin lipids but also from the dietary intake of saturated fats. Future studies are needed to evaluate dietary guidelines limiting specific saturated fatty acids that can be used for ceramide synthesis and possibly define DNA methylation inhibitors as potential supplementary therapeutic strategies.
Alt-text: Unlabelled Box
1. Introduction
It is becoming increasingly evident that the growing “epidemic” of fast food and processed food consumption in the United States and its association with increased Body Mass Index (BMI) is impacting the overall health status of the population [[1], [2], [3]]. Alarming statistics have recently demonstrated a link between emergency room visits and BMI, not just for obese patients, but also for overweight individuals [4,5]. While much of the discussion on the exponentially increasing health care costs associated with obesity is focused on the prevention of cardiovascular disorders and metabolic syndromes [6], a growing body of literature suggests an impact of BMI on a wide variety of other health conditions, including neurological disorders, such as Alzheimer's disease [[7], [8], [9]], Parkinson's disease [10,11] and Multiple Sclerosis (MS) [[12], [13], [14]].
MS is a debilitating neurological disorder characterized by inflammatory demyelination and axonal/neuronal degeneration, which variably progresses towards permanent disability [15]. Epidemiological studies have carefully assessed the BMI as risk factor for MS [12,13,[16], [17], [18], [19], [20]] and many studies have supported the concept of long-term effects of the BMI on the disease, as obesity or high body mass in childhood or adolescence will induce disease onset many years later [12,13,19,20]. Finally, recent studies have highlighted a relationship between BMI and disease severity [[21], [22], [23], [24], [25]] and to some extent, progression [26,27], thereby further supporting the need to investigate the molecular mechanisms responsible for these long-term effects.
Increased lipid synthesis in multiple tissues, including adipose tissue, liver, skeletal muscle and plasma has been associated with high BMI [[28], [29], [30], [31]]. This is of high interest as lipids are increasingly recognized as important signaling molecules regulating a multitude of cellular processes, including proliferation, metabolism and migration [32]. Most relevant to MS is the role of sphingolipids [33] and more specifically ceramides, whose altered levels have been reported both in the brains [34,35], cerebrospinal fluid [36] and plasma [37] of MS patients. Ceramides are gaining substantial interest as biomarkers for a wide variety of metabolic, cardiovascular and neurological disorders [38,39] however their precise mechanism of action is not fully understood and has often been described as cell-specific [40]. In this study we detected intriguing parallels between elevated plasma levels of ceramide species, hypermethylation of anti-proliferative genes in monocytes, increased monocytic counts and worsened disease course in MS patients with high BMI compared to those with normal BMI. Given the strong epidemiological data on the long-term effects of BMI, we further hypothesized that ceramides act as nuclear signaling lipids and epigenomic modulators with the ability to impact DNA methylation and affect specific cellular functions, such as monocyte proliferation.
2. Materials and methods
2.1. Human studies
2.1.1. Primary cohort
All subjects were recruited at the Corinne Goldsmith Dickinson Center for Multiple Sclerosis at Mount Sinai. The protocol was approved by Mount Sinai's Institutional Review Board and all subjects provided written informed consent. Subjects were enrolled from February 2014 through December 2015. Eligible subjects aged 18–60 met criteria for Relapsing Remitting MS [41] or Clinically Isolated Syndrome along with a brain MRI consistent with MS as determined by the treating physician, and were treatment-naïve with respect to MS disease-modifying therapy. Exclusion criteria included the diagnosis of another autoimmune disease, treatment with immunosuppressants, diagnosis of diabetes, recent high dose corticosteroids (<30 days), or recent antibiotics (<90 days). Details of each subject's demographic and clinical history and weight were recorded (See Table S1). Expanded Disability Status Scale (EDSS) [42] was performed by a study co-investigator or in certain cases abstracted from a recent clinical visit.
Follow up clinical information was obtained through review of the medical records of all subjects in the spring of 2017. The most recent clinical visit was chosen as the follow up time point and EDSS was abstracted from this visit. All chart notes from the time of enrollment to the follow up time point were reviewed to assess for disease activity, which was defined as any new clinical MS relapse or MRI activity. An MS relapse was defined as the appearance of a new neurological symptom lasting >24 h in the absence of fever or infection [41] and was determined by the treating physician (all MS specialists) for clinical care purposes independently from the study. MRI activity was defined as the appearance of a new T2 lesion or new gadolinium-enhancing lesion on clinical MRI scan, again determined by the treating MS expert independently from the study. The frequency of disease activity in normal and high BMI groups was analyzed using chi-squared test on GraphPad Prism v7.0. The distribution of therapies in the two groups was also evaluated using chi-squared test on GraphPad Prism v7.0. Evaluation of associations between BMI at baseline and EDSS changes from follow-up to baseline were assessed by ordinal logistic regression on R. Multivariate analysis was conducted for BMI, age, gender, and race and partial regressions for BMI were calculated, which assessed the independent association between BMI and EDSS change and corrected for the other variables. P values <.05 were considered significant.
2.1.2. Validation cohort for monocyte counts and brain volume
The validation cohort was obtained from a prospectively-acquired natural history cohort collected under NIH protocol 09-I-0032: Comprehensive multimodal analysis of neuroimmunological diseases of the CNS (clinicaltrials.gov identifier: NCT00794352). All patients signed the informed consent and the study was approved by NIH Institutional Review Board (CNS IRB). All patients were assigned alphanumeric codes and the personnel generating immunophenotyping or volumetric MRI data were blinded to the diagnostic conclusion, clinical data or BMI status. Only treatment-naïve subjects fulfilling MS diagnostic criteria [41] and classified as RRMS by treating clinician were included (See Table S2 for demographic and clinical information).
2.1.3. Healthy control cohort for monocyte counts and brain volume
Healthy subjects were prospectively recruited between June 2013 and May 2017 as part of a Natural History protocol “Comprehensive Multimodal Analysis of Neuroimmunological Diseases of the Central Nervous System” (ClinicalTrials.gov identifier: NCT00794352) at the National Institutes of Health. The inclusion criteria were age 18 years or older and vital signs within normal range at the time of screening visit. The exclusion criteria included systemic disorder or central nervous system disease of any kind, history of alcohol or substance abuse, MRI contraindications and history of auditory disorder (See Table S3 for demographic information).
2.1.4. Healthy control cohort for lipidomic analysis
Forty plasma samples for lipidomic analysis of healthy controls (see Table S4 for demographic information). were purchased from BioIVT. They included 20 samples from healthy controls with normal BMI (10 males and 10 females with a BMI range of 21–24) and 20 from high BMI subjects (10 males and 10 females with a BMI range of 26–40).
2.1.5. Immunophenotyping
For the primary RRMS cohort, samples were analyzed blinded. Fresh whole blood was stained in FACS staining buffer for monocytes (CD45+CD3−CD14+ cells), CD8 T cells (CD45+CD3+CD8+ cells), and CD4 T cells (CD45+CD3+CD4+ cells) and CD4 T cell subsets, naïve/Th1 (CD3+CD4+CCR4−CCR6− cells), Th17 (CD3+CD4+CCR4+CCR6+ cells), and Treg (CD3+CD4+CD127−CD25+ cells). To minimize variability, staining and acquisition were performed on the same day of blood draw within 3 h using the optimized antibody cocktail. Data were acquired on a Becton Dickinson LSR Fortessa (BD) equipped with five lasers (355 nm, 407 nm, 488 nm, 532 nm, and 633 nm wavelengths) with 22 PMT detectors at the Human Immune Monitoring Center at Icahn School of Medicine. Data were acquired using DIVA 6.1.2 software (BD). A minimum of 500,000 cells was recorded from corresponding tubes in order to accurately assess minor cell populations. Compensation was performed with unstained cells and BD compensation beads particle sets. FlowJo 9.4 software (Treestar Inc.) was used for post-acquisition analysis. Debris and doublets were excluded using light scatter measurements and major cell populations were identified based on their forward and side scatter properties. Subsequently, cells were gated using CD45 for leukocytes. Each cell population was represented as percentages of the parent population (i.e. CD4+T cells as % of CD3+ cells, Treg as % of total CD4+ T cells etc.). Absolute cell counts were calculated by multiplying the percentage of each cell type compared to the total population by the white blood cell count.
For the validation RRMS cohort and healthy control cohort, whole blood immunophenotyping was performed as previously described [43]. All samples were labeled with a prospectively assigned alpha-numeric code and personnel performing the studies were blinded to the diagnosis of the subject. Specimen collection, handling and processing was performed according to written standard operating procedures (SOPs). Immunophenotyping of peripheral blood cells was performed on anticoagulated blood within 60 min of ex vivo collection after osmotic lysis of erythrocytes. A minimum of 106 blood cells were stained according to a previously established protocol [44], which included blocking of Fc receptors by 2% intravenous immunoglobulin (IVIg). Monocytes were identified based on forward/side scatter and gating on cell lineage markers (CD45+CD3−CD14+ cells). Cells were immediately acquired on a BD LSR II with High Throughput Sampler (HTS) delivery system and analyzed with FACSDiva 6.1 software (all BD Biosciences). Gating was based on isotype controls. Sample acquisition, gating and sample exclusion (based on the review of quality of the staining and of absolute numbers of acquired events to assess reliability of data) was done on coded samples.
Statistical differences in immune cell counts between normal and high BMI categories were adjusted for covariates, age and gender, by One-way MANCOVA on SPSS, followed by pair-wise comparisons between the two groups with Bonferroni correction to test for statistical significance. P values <.05 were considered significant.
2.1.6. Lipidomic Analysis
All lipid standards were purchased from Avanti Polar Lipids, Nu-Chek Prep, Sigma-Aldrich, Cambridge Isotope Laboratory, Cayman Chemical Company, Avanti Polar Lipids, or Santa Cruz Biotechnology, Inc. All solvents are of HPLC or LC/MS grade and were acquired from Sigma-Aldrich, Fisher Scientific, or VWR International.
Blinded lipidomic analyses of plasma samples randomized as to the BMI group, were performed at BERG, LLC. A cocktail of deuterium-labeled and odd chain phospholipid standards from diverse lipid classes was added to 25 μl plasma. Standards were chosen so that they represented each lipid class and were at designated concentrations chosen to provide the most accurate quantitation and dynamic range for each lipid species. 4 ml chloroform:methanol (1:1, v/v) was added to each sample and the lipid extraction were performed as previously described [45,46]. Lipid extraction was automated using a customized sequence on a Hamilton Robotics STARlet system (Hamilton, Reno, NV) to meet the high-throughput requirements. Lipid extracts were dried under nitrogen and reconstituted in 68 μl chloroform: methanol (1:1, v/v). Samples were flushed with nitrogen and stored at −20 °C. Samples were diluted 50 times in isopropanol: methanol: acetonitrile: water (3:3:3:1, by vol.) with 2 mM ammonium acetate in order to optimize ionization efficiency in positive and negative modes. Electrospray ionization-MS was performed on a TripleTOF® 5600+ (SCIEX, Framingham, MA), coupled to a customized direct injection loop on an Ekspert microLC200 system (SCIEX). 50 μl of sample was injected at a flow-rate of 6 μl/min. Lipids were analyzed using a customized data independent analysis strategy on the TripleTOF® 5600+ allowing for MS/MSALL high resolution and high mass accuracy analysis as previously described [47]. Quantification was performed using an in-house library on MultiQuant™ software (SCIEX).
Abundance of lipids was quantified as relative to a lipid standard with known concentration and ionization efficiency. Lipid standards represented each lipid class so that abundance of lipids within one class could be compared. Data was filtered and normalized prior to statistical analysis. Lipids that had missing values in over 50% samples because they were below the detection limit were excluded. For remaining lipids, missing values were imputed by sampling a random uniform distribution ranging from the lowest value detected to one half the minimum value detected. Lipids were normalized using a variance stabilization procedure to reduce the effect of lipid abundance on sample variation for each species per lipid class and this was followed by median centering and log transformation. Because the MS patients and healthy control samples were obtained at distinct time points, they were run in separate batches and therefore independently analyzed for intra-group differences between low and high BMI. To determine statistically significant differences in the abundance of lipids between high and normal BMI groups within each lipid class, we used multiple t-tests with 5% FDR correction on Graphpad Prism v7.0 and q values less than p < .05 were considered significant.
The relationship between differentially abundant ceramides (only 18:1 backbone) and brain volume was analyzed by Pearson's correlation. Only patients who had lipidomic and MRI information were included in the analysis and p values <.05 were considered significant. Graphpad Prism v7.0 was used to perform the statistical analysis and create the heat map.
2.1.7. DNA methylation analysis
CD14 cells were positively isolated using magnetic beads (Miltenyi Biotec) and DNA was extracted with QIAamp DNA Blood Mini Kit (Qiagen). 500 ng of genomic DNA from each sample were bisulfite-treated with Methylamp One-Step DNA Modification Kit (EpiGentek) and methylation levels were measured at ~450,000 CpG sites by using the Illumina 450 K Array at the New York Genome Center in two different batches, which were randomized and blinded to the personnel running the samples. Methylation values for CpG sites were measured as beta values, or the ratio of signal intensity between the methylated probe and the sum of intensities of both methylated and unmethylated probes, and as M values, or the log2 ratio of the signal intensity of the methylated probe versus that of the unmethylated one. Because beta values have more intuitive value for biological interpretation, while M values are more valid for obtaining statistical significance [48], beta value differences were used to methylation difference values and M values were used to determine statistical significance. Probes with a detection p value of >0.01 and bead count <3 in >5% of the samples were filtered out. Non-CG probes, those close to SNPs (as defined by Zhou et al. [49]) or aligned to multiple locations (as defined by Nordlund et al. [50]), or on sex chromosomes, were removed. Beta values and M values were normalized with BMIQ v1.6 to correct for Type-I and Type-II probe bias. Batch effects were removed by using the combat algorithm implemented in the ChAMP package [51]. PCA analysis of the normalized data did not reveal any outliers in the samples. As we have previously described [52,53], our analysis was focused on genomic regions rather than individual CpG sites as regulatory DNA modifications generally involve multiple consecutive CpGs. To identify differentially methylated regions between overweight/obese MS patients (BMI ≥ 25) and normal BMI MS patients (BMI < 25), we utilized LIMMA package [54] and used a multilinear regression model, which identified significant correlations (p < .05) between M values at each individual CpG site and BMI (i.e. partial regression) after controlling for age, sex, race, EDSS, disease duration, and batch effect. We then used a 1 kb sliding window as implemented in the package DMRcate [55] to define genomic regions with closely located CpG sites that were either differentially methylated or not. Overlapping regions were removed. Based on these regions that were spanning the whole genome, we combined each CpG-specific p value from our linear regression model within a single region with Stouffer's method [56]. This was followed by 1% FDR correction for multiple hypothesis testing. Differentially methylated regions (DMRs) were defined as regions with an FDR-adjusted combined p value of <0.01. Data analysis was performed in R Studio by utilizing the R packages ChAMP [51] for data preprocessing and normalization, LIMMA for linear regression [54], and DMRCATE for the 1 kb sliding window function and the p value combination with the Stouffer method [55] and q value [57] for FDR correction.
Pathway enrichment analysis was performed on hypermethylated CpGs using the Kyoto Encylopedia of Genes and Genomes (KEGG) database on Enrichr [58,59]. Genes containing individual CpGs, which correlated with BMI (p < .05) after adjusting for clinical and demographic variables, were used to broaden the gene base. Enrichment scores for KEGG terms were calculated as the –log (adjusted p value). Genes with the largest methylation differences were determined by averaging the beta value differences of each CpG within the DMR.
2.1.8. MRI Measurements
In a primary cohort of MS patients, brain volumetric analysis was conducted on available T1-weighted sequences (TE = 9.5–24.0 ms, TR = 359–612 ms) and normalized brain volume (NBV) was measured on baseline T1-weighted images using SIENAX [60]. Personnel were blinded to the BMI status of the patients. Due to the nature of the data, acquired for clinical purposes, further segmentation in gray and white matter tissues was not performed. Quantification of T1-weighted lesion load was also conducted on available T1-weighted sequences (TE = 9.5–24.0 ms, TR = 359–612 ms) at baseline and the two-year follow-up. T1 lesions were identified and outlined, for each patient, using a semi-automated technique based on user-supervised local thresholding (Jim version 7; Xinapse Systems, http://www.xinapse.com) at each time point. Changes in lesion load were computed as the difference between lesion volume measured on follow-up scan and baseline lesion volume. Changes in NBV could not be calculated since the available sequences from baseline and follow-up were not comparable.
In a validation cohort of MS patients and healthy controls, the volume of brain structures were calculated using LesionTOADS [61] tissue segmentation with T1- and T2-weighted MRI images. Total brain volume was defined as the sum of white and gray matter and brain parenchymal fraction was calculated as the total brain percentage of the sum of total brain and total CSF volume.
Correlations between monocyte counts and brain volume were performed using linear regression. Evaluation of changes in T1 lesion volume were performed using chi-squared test. All analyses were performed on Graphpad Prism v7.0 and p values <.05 were considered significant.
2.2. Cell culture experiments
2.2.1. Monocytic cell line culture
THP1 cells (TIB-202, ATCC) were derived from a one-year old male infant with acute monocytic leukemia. Cells were cultured using complete RPMI (RPMI 1640 Medium (Sigma), 10% FBS, 1% Penicillin-Streptomycin, 1% L-Glutamine, 1% HEPES, 1% Non-essential amino acids, 1% MEM Sodium Pyruvate, 0.05 mM 2-Mercaptoethanol). For visualization of NBD-labeled Ceramide C16, cells were treated with 5 uM NBD-Ceramide C16 for 24 h. To assess the effects of a single ceramide, cells were treated with 100 nM Ceramide C16:0 (Avanti Polar Lipids) for 1, 12, or 24 h, or untreated, and harvested simultaneously. Cells were treated with a cocktail of ceramides at different concentrations – 1. Low: 100 nM Ceramide C16:0 (Avanti Polar Lipids), 300 nM Ceramide C22:0 (Avanti Polar Lipids), 1 uM Ceramide C24:1 (Cayman Chemicals) and 2. High: 120 nM Ceramide C16:0 (Avanti Polar Lipids), 420 nM Ceramide C22:0 (Avanti Polar Lipids), 1.3 uM Ceramide C24:1 (Cayman Chemicals), with or without 1 uM 5-aza-2′-deoxycytidine (Sigma) for 1 h followed by a 23 h wash period or for 24 h. For visualization of NBD-labeled Ceramide C16, cells were treated with 5 uM NBD-Ceramide C16 for 24 h.
2.2.2. Immunostaining
THP1 cells were grown on LabTekII chamber slides during the ceramide treatment and then fixed with 4% PFA for 15 min. DNA methylation levels were assessed by anti-5-methylcytosine staining as previously described [62]. Briefly, cells were permeabilized with 0.4% Triton X-100 for 15 min followed by denaturation with 2 N HCl for 15 min and neutralization with 100 mM TrisHCl for 10 min. Cells were blocked with 10% goat serum, 3% bovine serum albumin (BSA) in PBS containing 0.1% Triton X-100 for 1 h. Cells were incubated in anti-5-methylCytosine (Eurogentec, 1:500) and anti-Ki67 (Abcam, 1:1000) to measure proliferation at 4C overnight. Cells were stained with anti-mouse AlexaFluor 546 (Invitrogen, 1:500) and anti-rabbit AlexaFluor 488 (Invitrogen, 1:500) the next day for 2 h followed by washing and mounting with Fluoromount-G containing DAPI. For visualization of NBD-Ceramide C16, cells were blocked with 10% goat serum, and 0.1% Triton X-100 in PGBA (0.1 M phosphate buffer, 0.1% gelatin, 1% BSA, 0.002% sodium azide) for 1 h followed by overnight incubation with anti-4-fluoro-7-nitrobenzofurazan (NBD) (BioRad, 1:100) at 4C. Cells were stained with anti-rabbit AlexaFluor 488 (Invitrogen, 1:500) the next day for 1 h and mounted with Fluoromount-G containing DAPI.
2.2.3. Imaging
Images were acquired at 20× on the Zeiss LSM800 and 3 images per well were taken. For each experiment, 3–4 wells were used per condition, and a total of 3 independent experiments were performed. Mean intensity was analyzed on ImageJ. Briefly, DAPI+ nuclei were used to create Regions of Interest (ROIs) using the Analyze Particles function and added to the ROI manager. Mean gray values of each ROI were calculated for 5-methylcytosine and Ki67 stainings. An arbitrary threshold for each staining was set to determine 5-methylcytosine and Ki67 positive cells that was validated by manual counting. Percentage of positive cells over total cells was calculated for each image. The percentages for all images within one well were averaged followed by averaging of percentages in wells for each experiment. For statistical analysis, Student's t-test was performed between two groups, and Dunnett's multiple comparisons test was used among three groups and was calculated on Graphpad Prism v7.0. P values <.05 were considered significant. 3D reconstruction of cells treated with NBD-ceramide C16 was performed on Imaris.
2.2.4. MTT
The MTT assay was performed as previously described [63]. THP1 cells were treated with different doses of 5-aza-2′-deoxycytidine (0, 0.5, 1, 2, 5 uM). After 24 h of treatment, 5 mg/ml MTT was added to the cell media to a final concentration of 0.05 mg/ml and allowed to incubate for 3 h to allow MTT reduction. The formazan precipitate formed by viable cells was solubilized in DMSO, and read spectrophotometrically at 540 nm with background subtraction at 655 nm.
2.2.5. Sequenom mass array EpiTyper
Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). DNA purity was assessed by measuring the A260/A280 ratio using NanoDrop. 200 ng genomic DNA was sodium bisulfite–treated using the Methylamp One-step Modification Kit (Epigentek). Primers were designed using EpiDesigner software: NRXN1 intron forward: GTGTTATTGAGGAGTTAATGGTTGA, NRXN1 intron reverse: ATTTCCCCTCTATATTTAACAATCACA, FZD7 promoter forward: TTATTGAGAAGTGATTTGGAAGTGA, FZD7 promoter reverse: ACCTCACAAAAATCTAAACAAACCA, TP63 promoter forward: GTTTTAAATTGTTAATAATAAGTGTGGTT, TP63 promoter reverse: AAAATTTAAACACTTCTCTCCTTCA, TP63 exon forward: TTTTTTTGTAAATATGTATGAAGGAGAGA, TP63 exon reverse: AAAAAATTATATCACATTCAAACTCACC. Forward primers were designed with a 10-mer tag (AGGAAGAGAG) and reverse primers designed with a T7-promoter tag (CAGTAATACGACTCACTATAGGGAGAAGGCT), as per the manufacturer's guidelines. PCR products were then processed as previously described to determine methylation levels. CpGs overlapping silent peaks, outside the mass spectrometry analytical window (low or high mass), or lacking sufficient coverage (methylation level determined for <90% of samples) were filtered out before subsequent analysis. Statistical analysis for the overall region (taking all individual CpGs into account) was performed by Two-way ANOVA on Graphpad Prism v7.0 and p values <.05 were considered significant.
2.2.6. Quantitative real time PCR
RNA was extracted from THP1 cells using Trizol (Invitrogen) extraction and isopropanol precipitation. RNA samples were resuspended in water and further purified with RNeasy columns with on-column DNase treatment (Qiagen). RNA purity was assessed by measuring the A260/A280 ratio using NanoDrop. For qRT-PCR, RNA was reverse-transcribed with qScript cDNA Supermix (Quanta) and performed using PerfeCTa SYBR Green FastMix, ROX (Quanta) at the Epigenetics Core at the Advanced Science Research Center at CUNY. Primer sequences are as follows: NRXN1 forward: TAAGTGGCCTCCTAATGACCG, NRXN1 reverse: TCGCACCAATACGGCTTCTTT, FZD7 forward: GTGCCAACGGCCTGATGTA. FZD7 reverse: AGGTGAGAACGGTAAAGAGCG TP63 forward: TCCTCAGGGAGCTGTTATCC, TP63 reverse: ATTCACGGCTCAGCTCATGG, RPLP0 forward: GCGACCTGGAAGTCCAAC, RPLP0 reverse: GTCTGCTCCCACAATGAAAC. 3 technical triplicates per biological replicate, 3 biological replicates per experiment, and 3 independent experiments were performed. After normalization to the housekeeping gene, RPLP0, CT values of technical triplicates were averaged for each biological replicate, followed by averaging of biological replicates for each independent experiment. Values were normalized to the average value of control conditions (i.e. “Untreated” or “Low”). Statistical analysis was performed on Graphpad Prism v7.0: Student's t-test was performed between two groups, and Dunnett's multiple comparisons test was used among three groups. P values <.05 were considered significant.
2.2.7. Generation of NBD-Ceramide C16
NBD Sphingosine was purchased from Avanti® Polar Lipids, Inc. (Cat. Nr. 810205). All other reagents and solvents were purchased from commercial sources (Sigma-Aldrich, Fisher Scientific, etc.) and were used without further purification. Reaction progress was monitored by LC-MS analysis. HPLC (1260 infinity II, Agilent) was used for purification of NBD-Ceramide C16:0.
Stock solutions of palmitic acid (0·05 M), TBTU (0·05 M), DIPEA (0·05 M), and NBD sphingosine (0·01 M) in EtOAc were prepared. Palmitic acid (3·00 ul, 0·15 umol), TBTU (3·00 ul, 0·15 umol), and DIPEA (6·00 ul, 0·30 umol) were added to a microcentrifuge tube (1·5 ml) and incubated for 1 h under mild orbital shaking. NBD sphingosine (10·0 ul, 0·10 umol) was added and the reaction mixture was incubated for 16 h under mild orbital shaking (starting material completely consumed by LC-MS). MeOH was added and the reaction mixture was transferred into a glass vial. The crude mixture was purified by preparative HPLC (H2O & 1% Formic Acid (10%) in CH3CN to CH3CN) to yield NBD-Ceramide C16:0 as a yellow solid with approximately 80% recovery rate (yield estimate used for concentration calculations: 57·3 μg, 0·08 umol). HRMS (EI+): m/z calcd. For [C40H70N5O6] +: 716·5321, found: 716·5326 ([M + H]+).
2.3. Animal experiments
2.3.1. Animals
All experiments were performed according to IACUC-approved protocols and mice were maintained in a temperature- and humidity-controlled facility on a 12-h light-dark cycle with food and water ad libitum. Female C57/bl6 mice (The Jackson Laboratory) were fed a low-fat diet (2914, Envigo) or high fat diet (TD.96132, Envigo) starting at 3–4 weeks of age. Experimental autoimmune encephalitis (EAE) was induced after 5 weeks of diet as previously described [64]. Briefly, mice were injected subcutaneously with 200 μg of MOG35–55 thoroughly emulsified in Complete Freund's Adjuvant (CFA) with 500 μg Mycobacterium tuberculosis dissolved in 200 μl of a solution consisting of equal amounts of 1.5 mg/ml MOG peptide, 2.5 mg/ml Mycobacterium tuberculosis and 50% CFA. 500 ng of pertussis toxin dissolved in 200 μl of PBS were administered intraperitoneally at the time of the MOG35–55 injection and 2 days thereafter. Disease progression was monitored daily using a standard EAE scoring system (modified from the Hooke Lab recommendations) as following: 0 = no visible symptoms, 1 = flaccid tail, 1·5 = limp tail and hind leg inhibition, 2 = limp tail and weak hind legs, 2·5 = limp tail and dragging of hind legs, 2·75 = limp tail and paralysis of one hindlimb, 3 = limp tail and complete hindlimb paralysis, 3·5 = limp tail, complete hindlimb paralysis, and weakness of forelimbs, 4 = quadriplegia, and 5 = death.
For analysis of infiltrating monocytes in the CNS, Cx3cr1-Gfp mice (B6.129P-Cx3cr1tm1Litt/J) and Ccr2-Rfp mice (B6.129(Cg)-Ccr2tm2.1Ifc/J) were purchased from The Jackson Laboratory and crossed to generate Cx3cr1Gfp/+;Ccr2Rfp/+ mice. Heterozygous female mice were used for the experiments and fed a low-fat diet (2914, Envigo) or high fat diet (TD.96132, Envigo) starting at 4 weeks of age for a period of 5 weeks.
2.3.2. Analysis of disease course
Changes in weight were calculated after 5 weeks of diet as a measure of weight gain and were assessed by Student's t-test in low fat diet and high fat diet groups. Blood glucose test strips (Contour) were used to measure blood glucose levels as an indication of pre-diabetes and differences were assessed by Student's t-test. EAE scores were recorded to evaluate disease course. The difference in weight from baseline weight (weight at immunization) was calculated for each time point throughout disease course. Differences in weight change for individual time points were assessed by Sidak's multiple comparisons test. Differences in clinical severity on individual time points were assessed by Mann-Whitney U test. Area under the curve and maximum EAE score were also assessed for each mouse and compared in low fat diet and high fat diet fed mice by Mann-Whitney U test. Calculations and statistical analysis were performed on Graphpad Prism v7.0 and p values <.05 were considered significant.
2.3.3. Immunostaining
Mice were sacrificed at the peak of EAE for histological analysis. Mice were anaesthetized and perfused with 4% (w/v) paraformaldehyde. Spinal cords were removed, post-fixed, and cryopreserved in 30% (w/v) sucrose, embedded in OCT and sectioned coronally (12 μm). Sections were blocked with 10% goat serum, and 0.1% Triton X-100 in PGBA (0.1 M phosphate buffer, 0.1% gelatin, 1% BSA, 0.002% sodium azide) for 1 h followed by overnight incubation with primary antibodies – NFH (Millipore, 1:400), CD45 (BD Biosciences, 1:100), Iba1 (Wako, 1:500), and 5-methylCytosine (Zymo Research, 1:100) at 4C overnight. For 5-methylCytosine staining, antigen retrieval was performed using sodium citrate buffer at 95C for 15 min prior to blocking. Sections were washed and incubated with secondary antibodies, anti-mouse AlexaFluor 546 (Invitrogen, 1:500), anti-rabbit AlexaFluor 488 (Invitrogen, 1:500) and anti-rat AlexaFluor 647 (Invitrogen, 1:500), followed by counterstaining with DAPI (1:20,000) and mounting with Immunomount (Electron Microscopy Sciences). Fluoromyelin Green (Invitrogen, 1:300) was applied to sections for 20 min at room temperature after incubation with primary antibodies.
2.3.4. Imaging
To assess demyelination, axonal loss, and monocyte infiltration, images were acquired at 20× on the Zeiss LSM800. At least three images per section were acquired in two sections per animal and at least three animals were analyzed for each condition. Images were taken as a Z-stack and the maximum projection was acquired using the Zeiss Zen software. Areas were calculated on ImageJ. Fluoromyelin and NFH+ areas were delineated manually. To calculate CD45+ area, images were thresholded and the area calculated using the “analyze particles” function. Regions of interest (ROIs) were defined as CD45+ areas, and Iba1+ area was calculated within CD45+ areas using a threshold. Total Iba1+CD45+ area for each image was calculated as the sum of Iba1+ area for all CD45+ areas or ROIs in each image. ROIs were limited to the lesion area. To assess DNA methylation levels, images were acquired at 63× on the Zeiss LSM800. 3 images per section were taken and 2 sections per animal and 3 animals per condition were assessed. Intensity was defined as mean gray value and calculated on Image J. 5 representative nuclei of Iba1+ cells were selected manually in each image and 5mc and DAPI intensity were calculated. For statistical analysis, Student's t-test was performed on Graphpad Prism v7.0 and p values <.05 were considered significant.
2.3.5. Flow cytometry
Brains and spinal cords were removed and homogenized. Mononuclear cells were separated with a 30%/70% Percoll (GE Healthcare) gradient as previously reported [65]. Single-cell suspensions were analyzed on a FACSAria II Cell Sorter (BD) equipped with five lasers (355 nm, 407 nm, 488 nm, 532 nm, and 633 nm wavelengths) running DIVA 6.1.2 software (BD). Data were analyzed with FlowJo v10.0.7 software (Tree Star). Briefly, mononuclear cells were identified by size (forward and side scatter) and doublets excluded. Infiltrating monocytes were identified as CCR2-RFP+ and microglia as CX3CR1-GFP+. Percentages of CCR2-RFP+ and CX3CR1-GFP+ cells of total singlets were calculated for 5 mice per group. For statistical analysis, Student's t-test was performed on Graphpad Prism v7.0 and p values <.05 were considered significant.
2.4. Quantification and statistical analysis
Analysis of immunophenotyping was performed on SPSS. DNA methylation analysis and ordinal logistic regression of BMI and EDSS change were analyzed using R. All other statistical analyses were done using GraphPad Prism. For all graphs, error bars are mean ± SEM. All statistical details for each graph can be found in the figure legends.
2.5. Data and software availability
Sequencing data deposited in NCBI's Gene Expression Omnibus Series accession number GSE103929.
3. Results
3.1. Immune profiling reveals increased monocytic counts in MS patients with high BMI
The discovery cohort consisted of fifty-four patients who had been recently diagnosed with Relapsing-Remitting MS (RRMS) and had been recruited to participate in the study prior to the initiation of disease-modifying therapy (“therapy-naïve”). Patients were classified by BMI as normal (average BMI = 21·8 ± 1·9, n = 27) or high (average BMI = 30·2 ± 5·0, n = 27). At baseline, the clinical disability score, as defined by the Kurtzke Expanded Disability Status Scale (EDSS), did not differ by BMI (MS with normal BMI: median ± SD = 2·0 ± 1.1; MS with high BMI: median ± SD = 1·0 ± 1·0), as patients were recruited at the early stages of disease (Table S1).
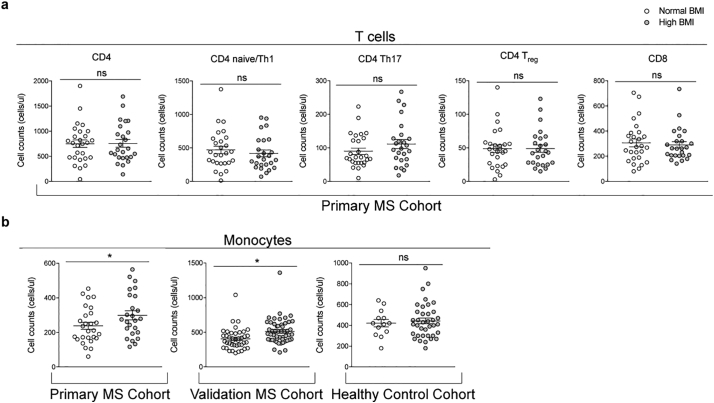
Since MS is an immune-mediated demyelinating disease, we sought to define whether the peripheral immune profiles differed in patients with different BMIs. Immunophenotyping was conducted using flow cytometry for the detection of surface markers. The total number of circulating CD4 T cells and related subsets, CD8 T cells and monocytes (CD14+) were calculated (Fig. S1). CD8 T cells and total CD4 T cell counts were not significantly different among MS patients with differing BMI, nor were CD4 naïve/Th1 (CCR4− and CCR6−), Th17 (CCR4+ and CCR6+) and Treg (CD25hi and CD127−) subsets (Fig. 1A). However, the number of CD14+ monocytes was significantly higher in MS patients within the high BMI group, after adjusting for age and sex statistically, a finding that was validated in a second independent validation cohort of 91 therapy-naïve RRMS patients (normal BMI average = 21·4 ± 1·9, n = 42; high BMI average = 29·6 ± 4·4, n = 51) and was not detected in 50 healthy controls (normal BMI average = 21·5 ± 2·3, n = 13; high BMI average = 30·2 ± 4·5, n = 37) (Fig. 1B). This suggested that in out cohorts, the number of circulating monocytes was uniquely affected by the interplay between high BMI and MS disease state.
Fig. 1.
Immune profiling reveals a BMI-dependent increase in blood monocyte counts in MS patients but not healthy individuals. Immune cell populations from fresh whole blood were quantified using flow cytometry. (a) Differences in T cell counts due to BMI were assessed in a primary cohort of MS patients (n = 52). (b) Differences in monocyte counts due to BMI were assessed in a primary cohort of MS patients (n = 52), a validation cohort of MS patients (n = 91), and a cohort of healthy individuals (n = 50). One-way MANCOVA was used to adjust for age and sex, followed by pair-wise comparisons with Bonferroni correction to determine statistically significant differences in the two BMI groups (normal BMI = white dots, high BMI = gray dots) (*p < .05).
3.2. Plasma lipidomic analysis reveals increased abundance of ceramides in MS patients with high BMI
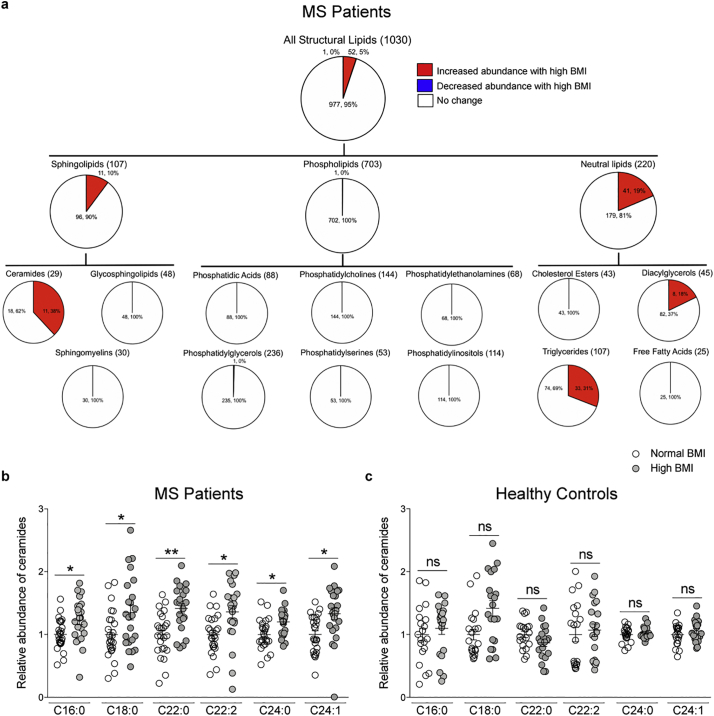
Because differences in BMI have been associated with lipid changes in several tissues [28,29,31], we asked whether MS patients with normal or high BMI also differed in circulating lipids. Plasma samples were analyzed using an MS/MSALL platform which identified over 2000 lipids (within 13 lipid classes), and the relative abundance for each lipid species was calculated in reference to reliable standards with known abundance and ionization efficiency for each lipid class. Lipids were excluded if values were below the detection limit in over 50% of samples, yielding 1030 lipid species that were considered suitable for further analysis. Differentially abundant lipids were identified by multiple t-tests with 5% FDR correction for each lipid class, revealing sphingolipids and neutral lipids as more abundant in the high BMI group than in normal BMI MS patients (Fig. S2A). The largest percentage difference in abundance was detected for the ceramides (38%), a subclass of sphingolipids, followed by triglycerides (30%) and diacylglycerols (18%) within the neutral lipids class (Fig. 2A). To further determine whether the ceramide abundance in the high BMI group of MS patients was specific to the disease state, or simply reflected the effect of high body mass, we conducted a similar analysis on a cohort of 40 plasma samples from healthy controls with either a normal (average = 22·6.5 ± 1·1, n = 20) or high BMI (average = 29·4 ± 3·4, n = 20) (Fig. 2B, Fig. S2B). Interestingly, the ceramide species with differential abundance in the plasma of MS patients with high BMI, were not similarly elevated in healthy individuals with high BMI, thereby suggesting that, at least within the limited size of the cohort included in our study, the specific ceramide species detected in patients were the likely result of the process of myelin destruction, characteristic of the disease state (Fig. 2C). This concept was further supported by an inverse correlation between specific ceramides and brain volume (Fig. S3). Overall these data suggested that the increased ceramide species detected in MS patients with high BMI, were the likely result of the combined de novo synthesis of ceramides from dietary fats and their recycling from myelin, damaged by the disease process.
Fig. 2.
Lipidomic profiling reveals increased abundance of ceramide species in MS patients with high BMI, but not in healthy individuals. Unbiased lipidomic analysis was performed on plasma samples from MS patients (n = 48) and healthy individuals (n = 40) using an MS/MSALL platform. Lipids that differed in abundance due to BMI were assessed using multiple t-tests with 5% FDR correction, q < 0.05 were considered significant (*q < 0.05, **q < 0.01). (a) Pie chart representation of numbers and percentage of lipids in MS patients by lipid family and class. Red = increased abundance, blue = decreased abundance, white = no change in patients with high BMI. (b) Relative abundance of ceramide species with 18:1 backbone that were significantly different in abundance in MS patients with high BMI (gray dots) compared to those with normal BMI (white). (c) The most highly significantly abundant ceramide species in MS patients were not statistically different in healthy individuals with high BMI (gray dots) compared to those with normal BMI (white). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Ceramide treatment increases DNA methylation and enhances proliferation in cultured monocytes
To begin defining a potential relationship between elevated ceramide levels and increased monocyte counts in high BMI MS patients, we used an in vitro approach and treated monocytic cell lines with different ceramides.
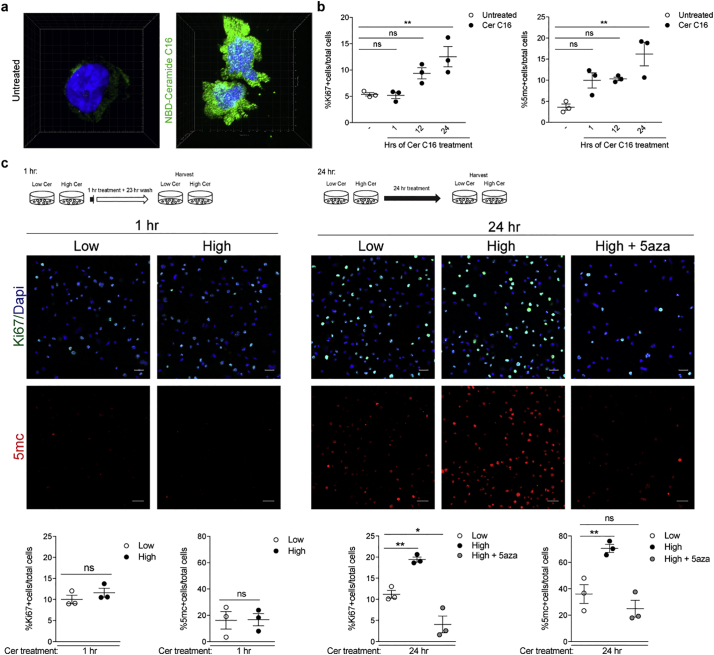
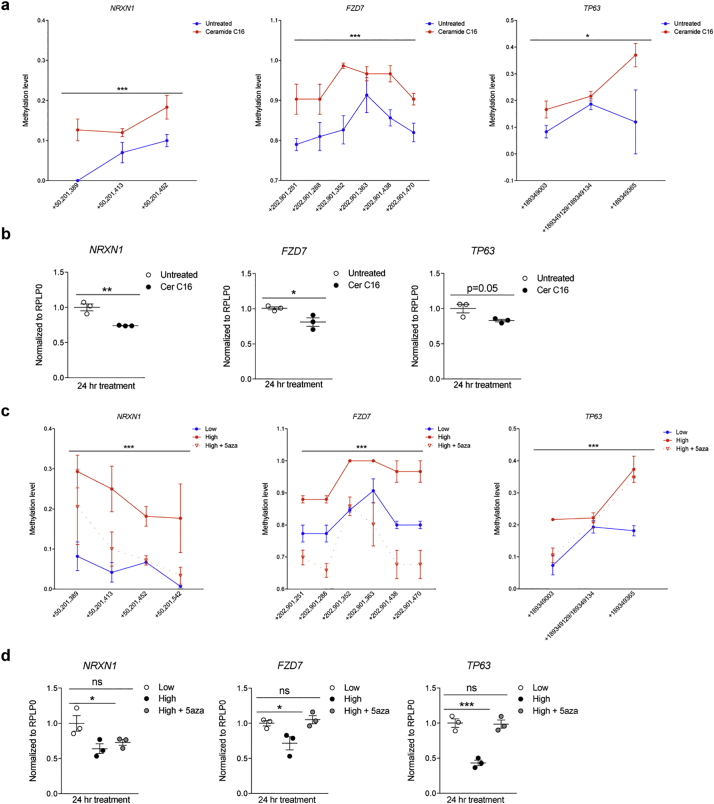
To define whether plasma ceramides could be uptaken by monocytes, we exposed cultured cells to fluorescently-labeled ceramide C16 (i.e. synthetically conjugated to the small fluorophore nitrobenzoxadiazole to form NBD-ceramide C16) for 24 h, and collected several confocal images. A 3D reconstruction of those images revealed both nuclear and cytoplasmic localization of the labeled ceramide within monocytic cells (Fig. 3A). Next, we performed a time course of ceramide C16 on monocytes, which revealed increased proliferation after 12 or 24 h of treatment, compared to baseline (Fig. 3B). Intriguingly, in the ceramide C16 treated cells we also detected increased DNA methylation which occurred as early as 1 h after exposure and preceded the effects on proliferation. This finding was of great interest as previous studies had reported an effect of fatty acids on DNA methylation in monocytes [[66], [67], [68], [69], [70]]. To further validate these results, we conducted a similar experiment but exposed the cells to the cocktail of ceramide species which we identified as differentially abundant in the normal and high BMI groups of MS patients (Ceramide C16:0, C22:0, and C24:1). Each cocktail was prepared based on the physiological ceramide concentrations detected in human plasma [37,71,72] and reflected the relative high or low abundance of ceramide species detected in our lipidomic analysis. Monocytes exposed for 24 h to the cocktail corresponding to the ceramide species levels detected in the plasma samples from MS patients with high BMI showed increased proliferation and DNA methylation compared to cells exposed to the lower concentrations of ceramides, equivalent to those detected in the normal BMI MS group (Fig. 3C). Importantly, the effect of ceramide on proliferation was prevented by co-treatment with the DNA methylation inhibitor, 5-aza-2′-deoxycytidine (5-aza) at doses of 5aza that were well tolerated by the cells (Fig. S4). The effect on proliferation and DNA methylation was not detected with treatment of shorter duration (Fig. 3C). Together, these results support a relationship between ceramide-induced effects on DNA methylation and increased monocyte counts, as detected in the high-BMI MS patients compared to those with normal BMI.
Fig. 3.
Exposure to ceramides results in dose-dependent increase in proliferation and DNA methylation. (a) Cultured monocytes (i.e. THP1 cells) were treated with fluorescently labeled Ceramide C16 (NBD-Ceramide C16) or untreated for 24 h. 3D reconstruction of the cells was conducted on Imaris to visualize localization of NBD-Ceramide (green). (b) Cultures either untreated or treated with Ceramide C16 (Cer C16) for 1, 12, or 24 h were fixed at the same time. After immunocytochemistry for Ki67 to detect proliferating cells or5-methylcytosine as marker for global DNA methylation the percentage of cells relative to the total DAPI+ nuclei was calculated and plotted in the graphs. Statistical differences in the percentage of Ki67+ cells and percentage of 5mc + cells relative to total DAPI+ nuclei were assessed by Student's t-test. (c) Cultures were treated for 1 (left panels) or 24 (right panels) hours with concentrations of a mixture of ceramides C16, C22, and C24:1, mimicking those detected in the plasma of MS patients with normal (Low) or high BMI (High) in the absence or presence of the DNA methylation inhibitor, 5-aza-2′-deoxycytidine (5-aza). After fixation, cells were stained for Ki67 (green) as proliferation marker, 5-methylcytosine (5mC, red), as marker for global DNA methylation and DAPI (blue), to identify nuclei. Confocal images were acquired at 20× magnification using the Zeiss LSM800 confocal (scale bar = 25um). High and High+5-aza groups were compared to Low Cer at 24 h using Dunnett's multiple comparisons test (*p < .05, **p < .01) (n = 3 independent biological replicates, each conducted in triplicate wells and analyzing 3 images/well). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Differential DNA methylation in monocytes sorted from MS patients with high or normal BMI
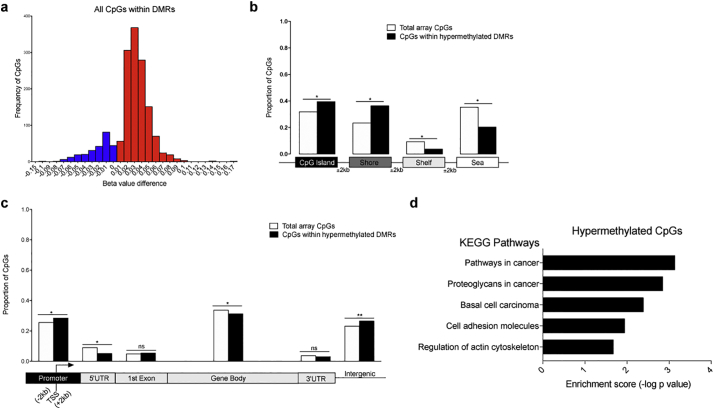
The previously mentioned epidemiological evidence on the long-term consequence of high BMI in childhood and adolescence on the risk to develop MS [17,19,20], the recognition of the overall importance of epigenetic changes as the interface between genetic predisposition and environmental variables and the documented responsiveness of monocytic DNA methylation to lipids [[66], [67], [68], [69], [70]], collectively informed our decision to conduct an unbiased genome-wide analysis of DNA methylation in monocytes sorted from MS patients differing by BMI. We used the Illumina 450 K Array, which accounts for approximately 485,000 CpG sites and covers 99% of all RefSeq genes and 98% of gene-associated CpG islands in the human genome, thereby providing an extensive and reliable coverage of the gene-rich regions. Individual CpGs were evaluated using a multilinear regression model to identify the contribution of BMI on DNA methylation changes after controlling for age, race, sex, EDSS and disease duration. We adopted a sliding window of 1 kb to interrogate the genome [55] and used well-accepted criteria of analysis [73] to define genomic regions with similarly regulated CpGs, which we termed Differentially Methylated Regions (DMRs). The overall DNA methylation effect of each clustered region was then computed using Stouffer's method for combined p values and followed by FDR correction for multiple hypothesis testing. This analysis identified 160 DMRs as hypermethylated and 18 DMRs as hypomethylated in the monocytes sorted from the MS patients with high BMI compared to those with normal BMI (Fig. S5). Therefore, we observed skewing of monocyte-derived DNA towards hypermethylation and focused on hypermethylated DMRs for further analysis (Fig. 4A).
Fig. 4.
Genome-wide distribution of DNA methylation marks in monocytes of high BMI MS patients. DNA methylation analysis was conducted on DNA from monocytes of MS patients using the Infinium HumanMethylation450 BeadChip. DNA samples from the monocytes of MS patients with high BMI (n = 23) were compared to those from patients with normal BMI (n = 25). Differentially methylated regions (DMRs) were identified using a 1 kb sliding window and significantly different CpGs were determined by evaluating the combined p values of CpG sites within each region, using the Stouffer's method with a 1% FDR correction. (a) Methylation values of individual CpGs within DMRs were measured as beta values, or the signal intensity from the methylated probe in relation to the sum of intensities from both methylated and unmethylated probes. Beta values of CpGs in MS samples with high BMI were subtracted from those in with normal BMI and a frequency histogram of the beta value differences was plotted to visualize the frequency of beta value differences in a given range. Positive beta value differences, indicating DNA hypermethylation in high BMI MS samples, are plotted in red, whereas negative differences, indicating DNA hypomethylation, are in blue. (b) Genomic distribution of the hypermethylated DMR in patients with high BMI (black bars), compared to the distribution of all the CpGs in the array (white bar). The relative proportions of CpGs in islands, shores (2 kb flanking the islands), shelves (2 kb flanking the shores) and sea (regions outside the previous three categories) are indicated. (c) Distribution of the hypermethylated CpGs distributed within in MS patients with high BMI (black bars) within distinct gene features, compared to the distribution of all the CpGs in the array (white bar). The relative proportion of differentially methylated CpGs in high BMI MS patients is represented relative to RefSeq gene promoters (2 kb from TSS), 5’UTR, 1st exon, gene bodies, 3’UTR and intergenic regions. Note that the CpGs hypermethylated in MS patients with high BMI were enriched at promoters and intergenic region and depleted at gene bodies, consistent with a repressive pattern of DNA methylation. Enrichment defined as statistically significantly different from the proportion of total array CpGs in a given location, which was calculated by chi- squared test, *p < .05, **p < .01. (d) Pathway enrichment analysis was performed on hypermethylated CpGs using the KEGG database on Enrichr. Enrichment score was calculated as the -log (adjusted p value). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When the CpG distribution of the hypermethylated DMRs was compared to the CpG distribution of all tested probes in the Illumina 450 K array, we detected enrichment in CpG islands and shores (±2 kb from islands) and concomitant reductions in the shelves (±2 kb from shores) and open sea (the remainder of genomic regions) (Fig. 4B). Further evaluation of the DMRs by gene features revealed enrichment of DMRs at promoters and intergenic regions and depletion at the 5’UTR and within gene bodies (Fig. 4C). Such a distribution is suggestive of transcriptional repression. To begin defining the potential functional relevance of these differences in DNA methylation, we then performed a pathway enrichment analysis of the hypermethylated genes in the monocytes of high BMI patients, which revealed cancer-related pathways, enriched for genes regulating cell proliferation. Enrichment in the categories of cell adhesion molecule (CAM) and actin cytoskeleton were also indicative of biological processes related to cell proliferation (Fig. 4D).
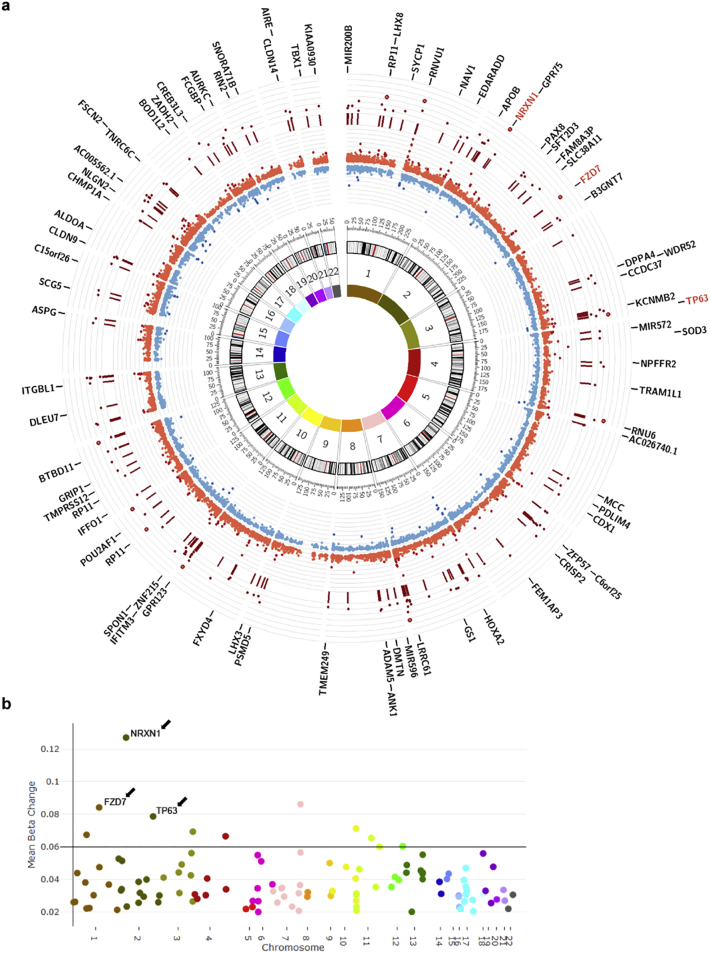
The chromosomal distribution of DMRs visualized by Circos plot did not reveal any preferential chromosomal distribution (Fig. 5A), however visualization of the genes with the highest average methylation using a Manhattan plot (Fig. 5B) identified genes, whose function had been linked to negative regulation of cell proliferation. These genes included: NRXN1 (Neurexin 1alpha), a membrane protein with the ability to negatively regulate hematopoiesis [74]; TP63 (Tumor Protein P63), a member of the p53 family of transcription factors, which negatively regulates proliferation in an isoform-specific manner [75]; and FZD7 (Frizzled class receptor 7), a receptor for several WNT ligands, which regulates proliferation in a P63-dependent manner [76]. Collectively, these data identify DNA hypermethylation of genes negatively regulating cell proliferation as part of the mechanism underlying the higher monocyte counts detected in high-BMI MS patients.
Fig. 5.
Chromosomal distribution and identification of genes with most significantly hypermethylated regions in MS patients with high BMI identifies negative regulators of cell proliferation. (a) Circos plot representing the chromosomal distribution of DMRs and CpGs within DMRs. Chromosomal numbers are indicated in the innermost circle. The inner red and blue circle depict the beta value differences for each CpG within a DMR. The more hypermethylated the CpG (bright red) the farther it is from the center of the circos plot, and the more hypomethylated the CpG (blue) the closer it is to the center of the plot. Dark red lines in the outer circle denote the genomic location of the DMRs and dark red dots indicate the mean beta value difference for each DMR. Genes overlapping DMRs are indicated in the outermost circle and genes with the greatest methylation differences are highlighted in red. (b) Manhattan plot highlighting the overall methylation difference, or mean beta change, for each DMR on each chromosome. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Hypermethylation and decreased transcript levels of regulators of proliferation in ceramide-treated monocytes
To functionally relate these findings to the results of the unbiased lipidomic analysis, we asked whether treatment of cultured human monocytic lines with ceramides would induce DNA hypermethylation at the same genomic locations identified in the monocytes directly sorted from the patients' blood. DNA methylation at target genes (i.e. NRXN1, FZD7 and TP63) was measured using a mass-spectrometry-based approach (i.e. Sequenom EpiTyper), which validated the occurrence of statistically significant hypermethylation induced by ceramide treatment at the previously identified genomic locations (Fig. 6A). The ceramide-induced hypermethylation was associated with decreased transcript levels of NRXN1, FZD7 and TP63, compared to untreated monocytes (Fig. 6B). The experiment was then repeated by treating cultures with a cocktail of ceramides resembling those found in MS patients with high or normal BMI. DNA hypermethylation (Fig. 6C) and reduced transcript levels (Fig. 6D) were only detected in cells exposed to ceramide concentrations equivalent to those found in the high BMI group of MS patients, and the effect was prevented by the presence of pharmacological inhibitors of DNA methylation. Of note, despite the occurrence of a sizeable ceramide-dependent DNA hypermethylation of NRXN1 at CpG +50,201,389, its transcript levels were not fully rescued by co-treatment with 5aza. This could be in part explained by a more complex regulation of this gene transcript possibly relying on additional epigenomic repressive marks. Overall these results further strengthen the link between the elevated plasma ceramide levels detected in MS patients with high BMI and DNA hypermethylation of genes negatively regulating cell proliferation in monocytes.
Fig. 6.
Exposure to ceramides results in increased DNA methylation and reduced transcription of negative regulators of cell proliferation. (a) Cultured human monocytes (i.e. THP1 cells) were either untreated (blue) or treated with Ceramide C16 (red) for 24 h. DNA was harvested and processed for Sequenom Mass Array EpiTyper, in order to quantify differential methylation of CpGs (Y axis) at the indicated genomic locations (X axis). The gene name is shown on top of each graph. (b) samples from the same cells described in (a) were used for RNA isolation and quantification of the corresponding gene transcripts by using real time-PCR and normalized to reference gene transcripts. (c) Cultures were treated for 24 h with concentrations of a mixture of ceramides C16, C22, and C24:1, mimicking those detected in the plasma of MS patients with normal (Low) or high BMI (High) in the absence or presence of the DNA methylation inhibitor, 5-aza-2′-deoxycytidine (5-aza). DNA was harvested and processed for Sequenom Mass Array EpiTyper, in order to quantify differential methylation of CpGs (Y axis) at the indicated genomic locations (X axis). The gene name is shown on top of each graph. Overall methylation differences for the entire region of interest for each gene were assessed using two-way ANOVA (*p < .05, ***p < .0001) (n = 3/group, n = 2 replicates/group). (d) Transcript levels of NRXN1, FZD7, and TP63 were assessed using real time-PCR and normalized to the levels of thehousekeeping gene, RPLP0. Cer C16 groups were compared to Untreated groups using Student's t-test, while the High and High +5-aza groups were compared to the Low group, using Dunnett's multiple comparisons test (*p < .05, **p < .01, ***p < .001) (n = 3 independent experiments, n = 3 biological replicates/experiment, n = 3 technical replicates/biological replicate). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
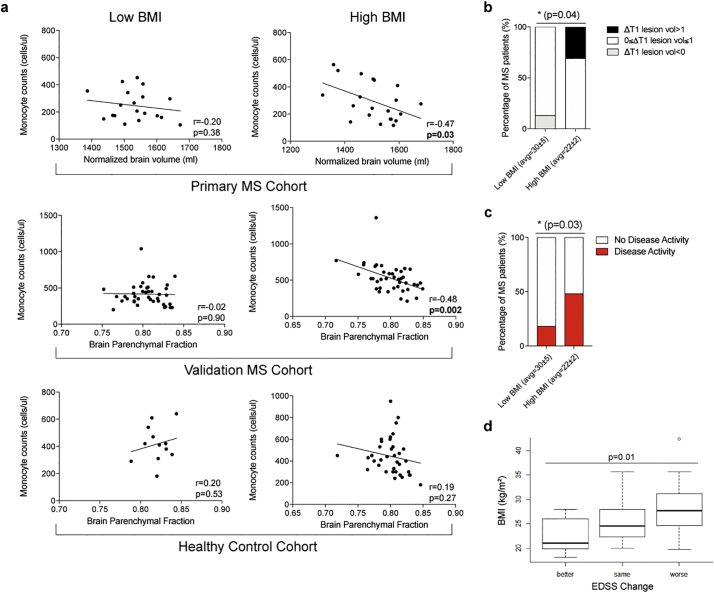
3.6. High monocytic counts, decreased brain volume and worsened disability in MS patients with high BMI
The potential link between monocytes and neurological disability in MS patients with high BMI was initially suggested by a negative correlation between MRI brain volume measurements and monocyte cell counts in our discovery cohort at baseline, a finding that was confirmed in the validation cohort of therapy naïve MS patients and was absent in the healthy cohort (Fig. 7A). These results led to the hypothesis that MS patients with high BMI would be characterized by greater brain atrophy, a correlate and predictor of clinical disability [[77], [78], [79]]. While differences in brain volume between the two-year follow-up compared to baseline could not be calculated due to incomparable sequences between the two time points, we were able to measure changes in T1 lesion volume (ΔT1 = T1 lesion volume at follow-up – T1 lesion volume at baseline), an indicator of irreversible CNS damage and axonal loss and a comparable correlate of brain atrophy and clinical disability [77,[80], [81], [82], [83]]. While the majority of patients showed small positive changes in T1 lesion volume (0 ≤ ΔT1 ≤ 1), suggestive of stable status or mild worsening of T1 lesion load, a greater proportion of MS patients with high BMI (n = 4/13) had more severe worsening (ΔT1 > 1) than those with normal BMI. Conversely, a greater proportion of MS patients with normal BMI (n = 2/15) showed better outcome at the two-year follow-up (ΔT1 < 0) (Fig. 7B). Similar trends were identified for disease activity (defined as either new MRI activity or clinical relapse) and clinical disability (as measured by EDSS), irrespective of therapy (Fig. S6). A significantly greater portion of high BMI MS patients (n = 12/25) compared to those with normal BMI (n = 4/22) showed signs of increased disease activity within the two-year period (Fig. 7C). Worsened disability in the high BMI group, was also suggested by ordinal logistic regression of the differential clinical score (after adjusting for age, sex, and race), which was calculated by comparing the EDSS score at the two-year follow-up to that at baseline (ΔEDSS = EDSS at follow-up – EDSS at baseline) and defined as better (<Δ-0.5), same (−Δ0.5 to Δ0.5) or worse (>Δ0.5) (Fig. 7D). Overall these data indicate the association between high BMI and worse disease course, both with respect to ongoing inflammatory disease activity and overall level of clinical disability.
Fig. 7.
Monocytes uniquely correlate with reduced brain volume in overweight/obese MS patients, who exhibit worsened disease course at the two-year follow up. (a) Brain volumetric analysis was performed on T1-weighted images from MRI scans and correlated with CD14+ monocyte counts from fresh whole blood, which had been obtained by flow cytometry, in a primary MS cohort (n = 41), validation MS cohort (n = 80) and a healthy cohort (n = 48) using linear regression, p < .05 considered significant. (b) Differences in the proportion of patients who showed changes in T1 lesion load (ΔT1) <1 ml, between 0 and 1 ml, and >1 ml during a 2 year period, between MS patients with normal or high BMI were assessed by chi-squared test (*p < .05). (c) Differences in the proportion of patients who had disease activity, defined by clinical relapse or new lesions, during a 2 year period, between MS patients with normal or high BMI were assessed by chi-squared test (*p < .05). (d) Change in EDSS in a 2 year period was categorized as better (<−0.5), same (−0.5 to 0.5), or worse (>0.5) and correlated with BMI using ordinal regression, p < .05 considered significant.
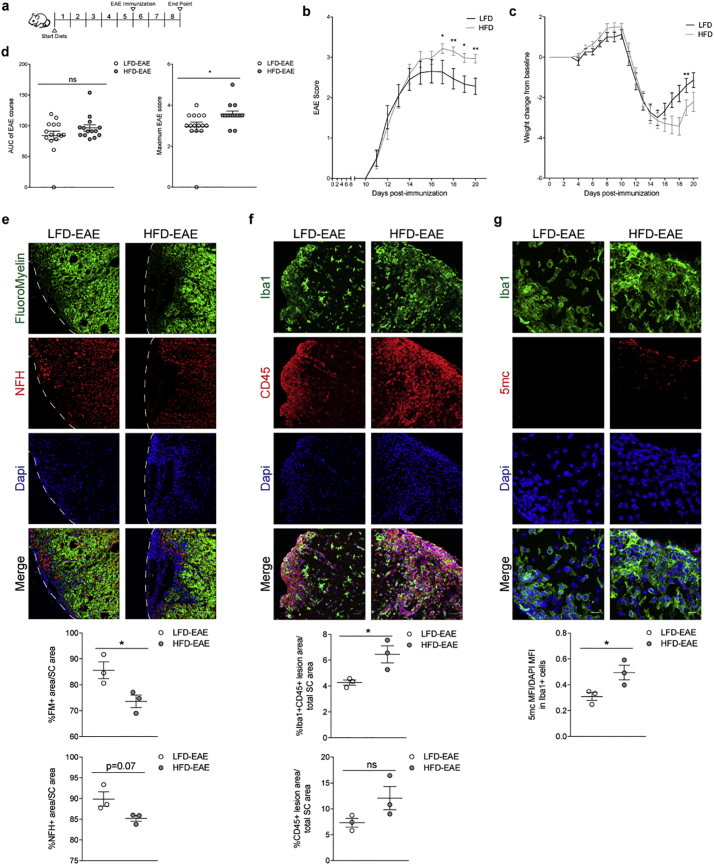
3.7. Elevated monocytic counts and worse disease course in experimental MS in mice on high fat diet
To further validate the relationship between body mass, number of monocytes and clinical disability, we immunized mice with a myelin oligodendrocyte glycoprotein peptide (MOG35–55) to induce Experimental Autoimmune Encephalitis (EAE), a model of MS. Body weight pre-induction was increased by feeding mice a high fat diet, while controls were kept on a low-fat diet. After 5 weeks on high fat diet, mice gained weight compared to controls, while retaining normal blood levels of glucose (Fig. S7A). Immunization was performed after the 5 weeks of diet, which was also kept constant for the entire duration of the experiment (Fig. 8A). Mice were scored on a clinical scale of 0 to 5 to provide a quantitative measurement of the ascending paralysis over time. Mice with higher body mass at the time of induction (HFD-EAE) showed greater clinical disability compared to controls (LFD-EAE), and this was supported by a significant weight change from the weight at immunization to the peak of disease (Fig. 8B-C). While the overall disease duration did not differ between the two groups, the maximum clinical score was higher in mice with higher body weight, thereby suggesting greater disease severity (Fig. 8D). Comparative histological analysis at the peak of disease revealed reduced Fluoromyelin+ (FM+) area and a trend for reduced Neurofilament H+ (NFH+) area in the spinal cord of mice with higher body mass, indicative of increased demyelination and axonal loss, respectively (Fig. 8E). The greater clinical disability and CNS lesion load detected in high fat diet mice were associated with increased monocytic infiltration, as measured by Iba1+CD45+ area within the demyelinated lesions, further supporting the suggested correlation between higher monocytic numbers and myelin and axonal loss (Fig. 8F). Notably, non-immunized mice fed a high fat diet for 5 weeks were characterized by increased levels of ceramide species in spinal cord (data not shown) and a trend for increased monocytic infiltration, further supporting a link between elevated ceramides and monocyte counts (Fig. S7B—C). DNA methylation levels, as assessed by 5mc intensity, were also higher in Iba1+ cells in high fat diet EAE mice compared to controls, supporting the association observed in patients (Fig. 8G). Thus, these results further support the link between greater disease severity and higher body weight in animal models of MS.
Fig. 8.
High-fat diet fed mice show increased monocytic infiltration and greater neurological deficits in an experimental model of MS. (a) Female C57/bl6 mice were fed either a low-fat (LFD) or high-fat diet (HFD) for 5 weeks, and then immunized with 200 ng MOG35–55 using a protocol to induce EAE. (b) Ascending paralysis was graded on a scale of 0 to 5 with high scores indicating increased clinical disability (black = LFD-EAE mice, gray = HFD-EAE mice). Differences in EAE scores were evaluated between LFD-EAE and HFD-EAE mice at each time point using Mann-Whitney U test, respectively (*p < .05, **p < .01) (n = 15/group). (c) Weight change from baseline (i.e. day of immunization) was calculated for each time point and differences between LFD-EAE and HFD-EAE mice were evaluated by Sidak's multiple comparisons test (**p < .01). (d) Area under the curve of the EAE disease course and maximum EAE score were also used to evaluate differences in disease course and statistical analysis was performed using Mann-Whitney U test (*p < .05). (e-g) Mice were sacrificed at the peak of disease and spinal cord tissue sectioned for histological analysis. Spinal cord sections were stained with Fluoromyelin (FM)to stain myelin and lesion areas and Neurofilament H (NFH) to stain axons. Images were acquired using the Zeiss LSM 800 confocal microscope and the 20× objective (scale bar = 50um). Myelin and axonal content were quantified as the area of FM or NFH positivity, respectively, in relation to the whole spinal cord area. Spinal cord sections were also stained with Iba1 and CD45 to mark peripheral monocytes/activated microglia (Iba1+ CD45+) and leukocytes (CD45+) and counterstained with DAPI, to identify nuclei. Confocal images were acquired using the Zeiss LSM 800 and the 20× objective (scale bar = 25um). Iba1 + CD45+ area and CD45+ area was quantified within the lesion to enhance representation of peripheral monocytes and peripheral leukocytes, respectively, and was calculated as a percentage in relation to the total spinal cord area. Spinal cord sections were stained with 5mc and Iba1 to assess DNA methylation levels in monocytes. Images were acquired using the Zeiss LSM 800 at 63× (scale bar = 10um). 5mc intensity was normalized to DAPI intensity in Iba1+ cells within the lesion. Statistical differences between LFD-EAE (white) and HFD-EAE (gray) groups were assessed by Student's t-test (*p < .05) (n = 3 mice/group, n = 2 sections/mouse, n = 3 images/section).
4. Discussion
This study identifies a mechanistic link between high BMI, ceramide-induced DNA methylation in monocytes and disease course in Multiple Sclerosis patients and mouse models. Our data suggest that the higher monocytic counts detected in high BMI MS patients compared to those with normal BMI might be consequent to ceramide-dependent hypermethylation of DNA on anti-proliferative genes, resulting in decreased expression levels, favoring proliferation. MS patients with high BMI also showed greater disease activity and worsened clinical disability at the two-year clinical follow up visit. Previous studies reported an association between obesity and increased pro-inflammatory monocyte subsets [[84], [85], [86]]. While we did not detect this increase in healthy controls, we cannot exclude the possibility that our inability to identify differential monocytic counts in the control cohort could be due to lack of sufficient power and possibly to the inclusion of several overweight subjects. It is also worth mentioning that the fasting/feeding state, to which monocytes are highly responsive [87], was not taken into consideration in our study. The differential abundance of ceramide species detected in MS patients with high BMI was not detected in healthy individuals grouped by BMI within a similar range. This finding is in agreement with previous results reporting elevated plasma ceramide levels in obese subjects with metabolic disorders and diabetes [[88], [89], [90], [91]]. Overall, this cumulative evidence supports the concept of elevated ceramide species in MS might be specific to the disease state, a concept which deserves further investigation.
Ceramides belong to the family of sphingolipids, whose role has been reported in a wide variety of pathological states, ranging from colorectal and breast cancer [[92], [93], [94], [95], [96]] to dermatological conditions [97] and osteoarthritis [98,99]. Compelling data on ceramides have been published in patients with cardiovascular diseases [100] and the ratio of specific ceramide species has been evaluated as an indicator of cardiovascular disease activity and a predictive index of clinical outcome [101,102], thereby suggesting ceramides as better predictors of death by cardiovascular diseases than other lipid biomarkers [38,39,103]. A series of lipidomic studies in patients with neurological disorders identified differential abundance of ceramides in plasma and cerebrospinal fluid of patients with Parkinson's disease [[104], [105], [106], [107]] or Alzheimer's disease [108,109]. The levels of these sphingolipids were associated with the detection of early cognitive impairment [110,111] and increased risk of disease development [112]. Elevated ceramide species, C16 and C24, were also reported in the cerebrospinal fluid [36] and plasma of patients with MS [37,72]. Our study is in agreement with these previous findings, while providing evidence for additional mechanisms of action for ceramides in MS. Besides the previously reported neurotoxic role of ceramides [36], we propose a nuclear function for ceramides as inducers of DNA hypermethylation on anti-proliferative genes in monocytes, resulting in elevated monocytic cell counts. We also define the importance of BMI as an important determinant of worsened clinical disability in MS, a concept which deserves further investigation and validation by other groups.
Ceramides are polar lipids formed by a sphingoid backbone conjugated to acyl-chains of different lengths and can be synthesized de novo from available dietary fats or through recycling of membrane lipids from damaged cells [113,114]. Importantly, their de novo synthesis has been related to the type of fats in the diet as they can be generated from saturated, but not unsaturated, dietary fats [115]. In MS patients, the immune-mediated destruction of the myelin membrane, which is highly enriched in sphingomyelin [116] likely results in increased recycling of this molecule into ceramide, which is consistent with previously reported alterations of sphingolipids in post-mortem studies on MS brains [34,35] and further supported by the negative correlations between the gray matter-enriched C18 and the white matter-enriched C24:1 [117] and brain volume. While the importance of dietary saturated fats as a source of ceramides requires further investigation, the detection of differentially abundant ceramide species in the plasma of MS patients highlights the contribution of immune-mediated demyelination to the salvage pathway and provides an explanation for the higher abundance of ceramides in high BMI patients as resulting from the combination of de novo synthesis from dietary fats and recycling of lipids from damaged myelin.
While the emphasis on the role of sphingolipids in Multiple Sclerosis has principally focused on the role of sphingosine and its receptors [33,118,119], this study addresses the mechanistic role of plasma ceramides. Ceramides represent a class of bioactive lipids that has gained recent interest due to their highly cell-specific effect on survival and proliferation [120,121]. Our data identify DNA methylation in monocytes as responsive to exogenous ceramides in a dose-dependent manner, leading to DNA hypermethylation and transcriptional repression of negative regulators of cell proliferation and overall increased proliferation. These findings are consistent with a model by which ceramide levels (either synthesized de novo from dietary lipids or recycled from damaged myelin) signal to the nucleus of monocytes and modify their epigenome and transcriptional program. The concept of lipid mediators as signals for the induction of genome-wide epigenetic changes is consistent with recent studies in cultured monocytes treated with free fatty acids in vitro [[66], [67], [68], [69], [70]]. However, the detection of nuclear and cytoplasmic localization of ceramide, after uptake in monocytes, suggests that, besides methylation, ceramide could modulate the activity in other manners [122]. Several mechanisms of action for these sphingolipids have been proposed, including the modulation of intracellular signaling pathway, due to reorganization of lipid rafts on the plasma membrane [123], or their effect on bioenergetics, due to incorporation into the mitochondrial membrane with consequently impact its integrity and function [124,125]. Future studies are needed to address the role of ceramides with acyl chains of different length on subcellular localization and function.
Finally, the correlation between increased monocytic counts and reduced brain volume in MS patients with high BMI highlighted the importance of monocytes in neurodegeneration, a concept which we validated in mice by manipulating their weight with a high fat diet and by using an experimental model of immune-mediated demyelination. Overall, the functional relationship between increased monocyte counts and neurodegeneration is an emerging concept in neuroscience [[126], [127], [128]]. In MS, this concept is supported by the detection of infiltrating monocytes/macrophages at sites of contact with damaged neurons as detected in post-mortem human MS brains and related murine models [[129], [130], [131], [132]]. It is also consistent with the protective effect of their depletion on axonal damage in preclinical models of MS [133]. This study builds on these findings and identifies ceramide-induced changes in DNA methylation as the mechanistic link which underlies increased monocytic cell counts in MS patients with high BMI.
A potential limitation of the current study is the relatively small size of the patient cohort, which was dictated by stringent inclusion/exclusion criteria and by additional difficulties in performing longitudinal studies. While we consider our results in the discovery cohort very exciting and mechanistic, we acknowledge that the potential consideration of ceramide levels as biomarkers for disease progression in MS would require validation of our findings by other groups using larger cohorts with a longitudinal and/or cross-sectional design. It will also be important to evaluate the effectiveness of dietary intervention (with an emphasis on the reduction of specific classes of saturated fats), as potential modulator of plasma ceramide levels and possibly of disease course in MS patients.
Acknowledgments
Acknowledgments
We are grateful to Yadira Bencosme and all the clinical neurologists and staff at the Corinne Goldsmith Center for Multiple Sclerosis for all the help with Patients' recruitment; to Tamjeed Sikder, Jessica Zheng, Benjamin Inbar and Payal Naik for the isolation of DNA from CD14+ cells and plasma separation. We thank Dr. Jimmy Huynh and Dr. Andrew Sharp for initial discussions on DNA methylation.
Funding sources
This work was supported by funds from the Friedman Brain Institute (PC), the National Institute for Neurological Disorders and Stroke, to PC (R37-NS042925) and to KC (F31-NS098575), and the Multiple Sclerosis Society (IK). Funders had no role in study design, data collection, data analysis, interpretation or writing of this manuscript.
Declaration of interests
The authors declare no competing interests.
The authors have declared that no conflict of interest exists.
Author contributions
KC, PC, IK designed the study. IK was responsible for patients' identification, recruitment and clinical evaluation. PK and BB provided the data on the validation RRMS cohort and healthy control cohort for immunophenotyping and MRI measurements. EC and MAK provided the lipidomic measurements. JM and DT synthesized the NBD-conjugated Ceramide C16. AN and CTW performed analysis of the Illumina 450 K data set and AN analyzed the follow-up clinical data. MA performed the EAE experiments. MI and MP were responsible for MRI data interpretation and measurements in the discovery cohort. KC collected all the data, conducted all the experiments on the THP1 cells, experiments on animal tissue, and conducted the statistical analysis.
KC and PC organized the data, prepared the figures and wrote the manuscript. All co-authors critically reviewed the manuscript for content.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.087.
Appendix A. Supplementary data
References
- 1.Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of childhood and adult obesity in the United States, 2011-2012. JAMA. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aune D., Sen A., Prasad M., Norat T., Janszky I., Tonstad S. BMI and all cause mortality: systematic review and non-linear dose-response meta-analysis of 230 cohort studies with 3.74 million deaths among 30.3 million participants. BMJ. 2016;353 doi: 10.1136/bmj.i2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masters R.K., Reither E.N., Powers D.A., Yang Y.C., Burger A., Link B.G. The impact of obesity on US mortality levels: the importance of age and cohort factors in population estimates. Am J Public Health. 2013;103:1895–1901. doi: 10.2105/AJPH.2013.301379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peitz G.W., Troyer J., Jones A.E., Shapiro N.I., Nelson R.D., Hernandez J. Association of body mass index with increased cost of care and length of stay for emergency department patients with chest pain and dyspnea. Circ Cardiovasc Qual Outcomes. 2014;7:292–298. doi: 10.1161/CIRCOUTCOMES.113.000702. [DOI] [PubMed] [Google Scholar]
- 5.Teuner C.M., Menn P., Heier M., Holle R., John J., Wolfenstetter S.B. Impact of BMI and BMI change on future drug expenditures in adults: results from the MONICA/KORA cohort study. BMC Health Serv Res. 2013;13 doi: 10.1186/1472-6963-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dee A., Kearns K., O'Neill C., Sharp L., Staines A., O'Dwyer V. The direct and indirect costs of both overweight and obesity: a systematic review. BMC Res Notes. 2014;7 doi: 10.1186/1756-0500-7-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beydoun M.A., Beydoun H., Wang Y. Obesity and central obesity as risk factors for incident dementia and its sub-types: a systematic review and meta-analysis. Obes Rev Off J Int Assoc Study Obes. 2008;9:204–218. doi: 10.1111/j.1467-789X.2008.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogrodnik M., Zhu Y., Langhi L.G.P., Tchkonia T., Krüger P., Fielder E. Obesity-induced cellular senescence drives anxiety and impairs neurogenesis. Cell Metab. 2019 doi: 10.1016/j.cmet.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitmer R.A., Gustafson D.R., Barrett-Connor E., Haan M.N., Gunderson E.P., Yaffe K. Central obesity and increased risk of dementia more than three decades later. Neurology. 2008;71:1057. doi: 10.1212/01.wnl.0000306313.89165.ef. [DOI] [PubMed] [Google Scholar]
- 10.Abbott R.D., Ross G.W., White L.R., Nelson J.S., Masaki K.H., Tanner C.M. Midlife adiposity and the future risk of Parkinson's disease. Neurology. 2002;59:1051. doi: 10.1212/wnl.59.7.1051. [DOI] [PubMed] [Google Scholar]
- 11.Hu G., Jousilahti P., Nissinen A., Antikainen R., Kivipelto M., Tuomilehto J. Body mass index and the risk of Parkinson disease. Neurology. 2006;67:1955. doi: 10.1212/01.wnl.0000247052.18422.e5. [DOI] [PubMed] [Google Scholar]
- 12.Hedström A.K., Olsson T., Alfredsson L. High body mass index before age 20 is associated with increased risk for multiple sclerosis in both men and women. Mult Scler J. 2012;18:1334–1336. doi: 10.1177/1352458512436596. [DOI] [PubMed] [Google Scholar]
- 13.Munger K.L., Chitnis T., Ascherio A. Body size and risk of MS in two cohorts of US women. Neurology. 2009;73:1543–1550. doi: 10.1212/WNL.0b013e3181c0d6e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wesnes K., Riise T., Casetta I., Drulovic J., Granieri E., Holmøy T. Body size and the risk of multiple sclerosis in Norway and Italy: the EnvIMS study. Mult Scler J. 2015;21:388–395. doi: 10.1177/1352458514546785. [DOI] [PubMed] [Google Scholar]
- 15.Frohman E.M., Racke M.K., Raine C.S. Multiple sclerosis—the plaque and its pathogenesis. N Engl J Med. 2006;354:942–955. doi: 10.1056/NEJMra052130. [DOI] [PubMed] [Google Scholar]
- 16.Hedström A.K., Olsson T., Alfredsson L. Body mass index during adolescence, rather than childhood, is critical in determining MS risk. Mult Scler J. 2016;22:878–883. doi: 10.1177/1352458515603798. [DOI] [PubMed] [Google Scholar]
- 17.Langer-Gould A., Brara S.M., Beaber B.E., Koebnick C. Childhood obesity and risk of pediatric multiple sclerosis and clinically isolated syndrome. Neurology. 2013;80:548–552. doi: 10.1212/WNL.0b013e31828154f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munger K.L., Bentzen J., Laursen B., Stenager E., Koch-Henriksen N., Sørensen T.I. Childhood body mass index and multiple sclerosis risk: a long-term cohort study. Mult Scler J. 2013;19:1323–1329. doi: 10.1177/1352458513483889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitnis T., Graves J., Weinstock-Guttman B., Belman A., Olsen C., Misra M. Distinct effects of obesity and puberty on risk and age at onset of pediatric MS. Ann Clin Transl Neurol. 2016;3:897–907. doi: 10.1002/acn3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianfrancesco M.A., Acuna B., Shen L., Briggs F.B., Quach H., Bellesis K.H. Obesity during childhood and adolescence increases susceptibility to multiple sclerosis after accounting for established genetic and environmental risk factors. Obes Res Clin Pract. 2014;8:e435–e447. doi: 10.1016/j.orcp.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bove R., Musallam A., Xia Z., Baruch N., Messina S., Healy B.C. Longitudinal BMI trajectories in multiple sclerosis: sex differences in association with disease severity. Mult Scler Relat Disord. 2016;8:136–140. doi: 10.1016/j.msard.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Kappus N., Weinstock-Guttman B., Hagemeier J., Kennedy C., Melia R., Carl E. Cardiovascular risk factors are associated with increased lesion burden and brain atrophy in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2015;87:181–187. doi: 10.1136/jnnp-2014-310051. [jnnp–2014]. [DOI] [PubMed] [Google Scholar]
- 23.Mowry E.M., Azevedo C.J., McCulloch C.E., Okuda D.T., Lincoln R.R., Waubant E. Body mass index, but not vitamin D status, is associated with brain volume change in MS. Neurology. 2018;91:e2256–e2264. doi: 10.1212/WNL.0000000000006644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira S.R., Simão A.N.C., Kallaur A.P., de Almeida E.R.D., Morimoto H.K., Lopes J. Disability in patients with multiple sclerosis: influence of insulin resistance, adiposity, and oxidative stress. Nutrition. 2014;30:268–273. doi: 10.1016/j.nut.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Tettey P., Simpson S., Taylor B.V., van der Mei I.A. Vascular comorbidities in the onset and progression of multiple sclerosis. J Neurol Sci. 2014;347:23–33. doi: 10.1016/j.jns.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Manouchehrinia A., Hedström A.K., Alfredsson L., Olsson T., Hillert J., Ramanujam R. Association of pre-Disease Body Mass Index with Multiple Sclerosis Prognosis. Front Neurol. 2018;9 doi: 10.3389/fneur.2018.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paz-Ballesteros WC, Monterrubio-Flores EA, de Jesús Flores-Rivera J, Corona-Vázquez T, Hernández-Girón C. Cigarette smoking, alcohol consumption and overweight in multiple sclerosis: disability progression. Arch Med Res 2017;48:113–20. doi: 10.1016/j.arcmed.2017.03.002. [DOI] [PubMed]
- 28.Tonks K.T., Coster A.C., Christopher M.J., Chaudhuri R., Xu A., Gagnon-Bartsch J. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity. 2016;24:908–916. doi: 10.1002/oby.21448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodpaster B.H., Wolf D. Skeletal muscle lipid accumulation in obesity, insulin resistance, and type 2 diabetes. Pediatr Diabetes. 2004;5:219–226. doi: 10.1111/j.1399-543X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 30.Hla T., Dannenberg A.J. Sphingolipid signaling in metabolic disorders. Cell Metab. 2012;16:420–434. doi: 10.1016/j.cmet.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowart L.A. Sphingolipids: players in the pathology of metabolic disease. Trends Endocrinol Metab. 2009;20:34–42. doi: 10.1016/j.tem.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Wymann M.P., Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 33.Halmer R., Walter S. Fa\s sbender K. Sphingolipids: important players in multiple sclerosis. Cell Physiol Biochem. 2014;34:111–118. doi: 10.1159/000362988. [DOI] [PubMed] [Google Scholar]
- 34.Cumings J.N., Goodwin H. Sphingolipids and phospholipids of myelin in multiple sclerosis. The Lancet. 1968;292:664–665. doi: 10.1016/s0140-6736(68)92510-5. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler D., Bandaru V.V.R., Calabresi P.A., Nath A., Haughey N.J. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain. 2008;131:3092–3102. doi: 10.1093/brain/awn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidaurre O.G., Haines J.D., Sand I.K., Adula K.P., Huynh J.L., McGraw C.A. Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain. 2014;137:2271–2286. doi: 10.1093/brain/awu139. awu139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurz J., Brunkhorst R., Foerch C., Blum L., Henke M., Gabriel L. The relevance of ceramides and their synthesizing enzymes for multiple sclerosis. Clin Sci. 2018 doi: 10.1042/CS20180506. CS20180506. [DOI] [PubMed] [Google Scholar]
- 38.Kurz J., Parnham M.J., Geisslinger G., Schiffmann S. Ceramides as novel disease biomarkers. Trends Mol Med. 2019;25:20–32. doi: 10.1016/j.molmed.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Summers S.A. Could ceramides become the new cholesterol? Cell Metab. 2018;27:276–280. doi: 10.1016/j.cmet.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Hla T., Kolesnick R. C16: 0-ceramide signals insulin resistance. Cell Metab. 2014;20:703–705. doi: 10.1016/j.cmet.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 43.Han S., Lin Y.C., Wu T., Salgado A.D., Mexhitaj I., Wuest S.C. Comprehensive immunophenotyping of cerebrospinal fluid cells in patients with neuroimmunological diseases. J Immunol Baltim Md. 2014;1950(192):2551–2563. doi: 10.4049/jimmunol.1302884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielekova B., Catalfamo M., Reichert-Scrivner S., Packer A., Cerna M., Waldmann T.A. Regulatory CD56bright natural killer cells mediate immunomodulatory effects of IL-2Rα-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci. 2006;103:5941–5946. doi: 10.1073/pnas.0601335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao F., McDaniel J., Chen E.Y., Rockwell H., Lynes M.D., Tseng Y.-H. Monoacylglycerol analysis using MS/MS(ALL) quadruple time of flight mass spectrometry. Metabolites. 2016;6 doi: 10.3390/metabo6030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X., Gross R.W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 47.Simons B., Kauhanen D., Sylvanne T., Tarasov K., Duchoslav E., Ekroos K. Shotgun Lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2012;2:195–213. doi: 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W., Laird P.W., Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45:e22. doi: 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordlund J., Bäcklin C.L., Wahlberg P., Busche S., Berglund E.C., Eloranta M.L. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Y., Morris T.J., Webster A.P., Yang Z., Beck S., Feber A. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33:3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huynh J.L., Garg P., Thin T.H., Yoo S., Dutta R., Trapp B.D. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17:121–130. doi: 10.1038/nn.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson C.T., Roussos P., Garg P., Ho D.J., Azam N., Katsel P.L. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer's disease. Genome Med. 2016;8 doi: 10.1186/s13073-015-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., V Lord R. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8 doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 57.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith S.M., Zhang Y., Jenkinson M., Chen J., Matthews P.M., Federico A. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- 61.Shiee N., Bazin P.-L., Ozturk A., Reich D.S., Calabresi P.A., Pham D.L. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage. 2010;49:1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES cell self-renewal, and ICM specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haines J.D., Vidaurre O.G., Zhang F., Riffo-Campos Á.L., Castillo J., Casanova B. Multiple sclerosis patient-derived CSF induces transcriptional changes in proliferating oligodendrocyte progenitors. Mult Scler Houndmills Basingstoke Engl. 2015;21:1655–1669. doi: 10.1177/1352458515573094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller S.D., Karpus W.J., Davidson T.S. Experimental autoimmune encephalomyelitis in the mouse. Curr Protoc Immunol Ed John E Coligan Al. 2007 doi: 10.1002/0471142735.im1501s88. CHAPTER:Unit-15.1. [DOI] [PubMed] [Google Scholar]
- 65.Pino P.A., Cardona A.E. Isolation of brain and spinal cord mononuclear cells using Percoll gradients. J Vis Exp JoVE. 2011 doi: 10.3791/2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrès R., Osler M.E., Yan J., Rune A., Fritz T., Caidahl K. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 67.Dekkers K.F., van Iterson M., Slieker R.C., Moed M.H., Bonder M.J., van Galen M. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flores-Sierra J., Arredondo-Guerrero M., Cervantes-Paz B., Rodríguez-Ríos D., Alvarado-Caudillo Y., Nielsen F.C. The trans fatty acid elaidate affects the global DNA methylation profile of cultured cells and in vivo. Lipids Health Dis. 2016;15 doi: 10.1186/s12944-016-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de la Rocha C., Pérez-Mojica J.E., León S.Z.-D., Cervantes-Paz B., Tristán-Flores F.E., Rodríguez-Ríos D. Associations between whole peripheral blood fatty acids and DNA methylation in humans. 2016;6 doi: 10.1038/srep25867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva-Martinez G.A., Rodriguez-Rios D., Alvarado-Caudillo Y., Vaquero A., Esteller M., Carmona F.J. Arachidonic and oleic acid exert distinct effects on the DNA methylome. Epigenetics. 2016;11:321–334. doi: 10.1080/15592294.2016.1161873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammad S.M., Pierce J.S., Soodavar F., Smith K.J., Gadban M.M.A., Rembiesa B. Blood sphingolipidomics in healthy humans: impact of sample collection methodology. J Lipid Res. 2010;51:3074–3087. doi: 10.1194/jlr.D008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ottenlinger F.M., Mayer C.A., Ferreirós N., Schreiber Y., Schwiebs A., Schmidt K.G. Interferon-Beta increases plasma ceramides of specific chain length in multiple sclerosis patients, unlike Fingolimod or Natalizumab. Front Pharmacol. 2016;7 doi: 10.3389/fphar.2016.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 74.Kinzfogl J., Hangoc G., Broxmeyer H.E. Neurexophilin 1 suppresses the proliferation of hematopoietic progenitor cells. Blood. 2011;118:565–575. doi: 10.1182/blood-2010-12-325381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manni I., Artuso S., Careccia S., Rizzo M.G., Baserga R., Piaggio G. The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. FASEB J. 2009;23:3957–3966. doi: 10.1096/fj.09-131847. [DOI] [PubMed] [Google Scholar]
- 76.Chakrabarti R., Wei Y., Hwang J., Hang X., Andres Blanco M., Choudhury A. ΔNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol. 2014;16:1004–1015. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bermel R.A., Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol. 2006;5:158–170. doi: 10.1016/S1474-4422(06)70349-0. [DOI] [PubMed] [Google Scholar]
- 78.Fisher E., Rudick R.A., Cutter G., Baier M., Miller D., Weinstock-Guttman B. Relationship between brain atrophy and disability: an 8-year follow-up study of multiple sclerosis patients. Mult Scler J. 2000;6:373–377. doi: 10.1177/135245850000600602. [DOI] [PubMed] [Google Scholar]
- 79.Popescu V., Agosta F., Hulst H.E., Sluimer I.C., Knol D.L., Sormani M.P. Brain atrophy and lesion load predict long term disability in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2013;84:1082–1091. doi: 10.1136/jnnp-2012-304094. [DOI] [PubMed] [Google Scholar]
- 80.Giorgio A., Stromillo M.L., Bartolozzi M.L., Rossi F., Battaglini M., De Leucio A. Relevance of hypointense brain MRI lesions for long-term worsening of clinical disability in relapsing multiple sclerosis. Mult Scler J. 2014;20:214–219. doi: 10.1177/1352458513494490. [DOI] [PubMed] [Google Scholar]
- 81.Radue E.-W., Barkhof F., Kappos L., Sprenger T., Häring D.A., de Vera A. Correlation between brain volume loss and clinical and MRI outcomes in multiple sclerosis. Neurology. 2015;84:784–793. doi: 10.1212/WNL.0000000000001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sailer M., Losseff N.A., Wang L., Gawne-Cain M.L., Thompson A.J., Miller D.H. T1 lesion load and cerebral atrophy as a marker for clinical progression in patients with multiple sclerosis. A prospective 18 months follow-up study. Eur J Neurol. 2001;8:37–42. doi: 10.1046/j.1468-1331.2001.00147.x. [DOI] [PubMed] [Google Scholar]
- 83.Simon J.H., Jacobs L., Simonian N. T1-Hypointense lesions (T1 black holes) in mild-to-moderate disability relapsing multiple sclerosis. In: Hommes O.R., Comi G., editors. Early Indic Early Treat Neuroprotection Mult Scler. Springer Milan; Milano: 2004. pp. 135–139. [Google Scholar]
- 84.Christine Poitou, Elise Dalmas, Mariana Renovato, Vanessa Benhamo, Froogh Hajduch, Meriem Abdennour. CD14dimCD16+ and CD14+CD16+ monocytes in obesity and during weight loss. Arterioscler Thromb Vasc Biol. 2011;31:2322–2330. doi: 10.1161/ATVBAHA.111.230979. [DOI] [PubMed] [Google Scholar]
- 85.Devevre E.F., Renovato-Martins M., Clement K., Sautes-Fridman C., Cremer I., Poitou C. Profiling of the three circulating monocyte subpopulations in human obesity. J Immunol Baltim Md. 2015;1950(194):3917–3923. doi: 10.4049/jimmunol.1402655. [DOI] [PubMed] [Google Scholar]
- 86.Wouters K., Gaens K., Bijnen M., Verboven K., Jocken J., Wetzels S. Circulating classical monocytes are associated with CD11c+ macrophages in human visceral adipose tissue. Sci Rep. 2017;7 doi: 10.1038/srep42665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hyson D.A., Paglieroni T.G., Wun T., Rutledge J.C. Postprandial Lipemia is associated with platelet and monocyte activation and increased monocyte cytokine expression in Normolipemic men. Clin Appl Thromb. 2002;8:147–155. doi: 10.1177/107602960200800211. [DOI] [PubMed] [Google Scholar]
- 88.Haus J.M., Kashyap S.R., Kasumov T., Zhang R., Kelly K.R., Defronzo R.A. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 2009;58:337–343. doi: 10.2337/db08-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hilvo M., Salonurmi T., Havulinna A.S., Kauhanen D., Pedersen E.R., Tell G.S. Ceramide stearic to palmitic acid ratio predicts incident diabetes. Diabetologia. 2018;61:1424–1434. doi: 10.1007/s00125-018-4590-6. [DOI] [PubMed] [Google Scholar]
- 90.Lopez X., Goldfine A.B., Holland W.L., Gordillo R., Scherer P.E. Plasma ceramides are elevated in female children and adolescents with type 2 diabetes. J Pediatr Endocrinol Metab JPEM. 2013;26:995–998. doi: 10.1515/jpem-2012-0407. [DOI] [PubMed] [Google Scholar]
- 91.Wigger L., Cruciani-Guglielmacci C., Nicolas A., Denom J., Fernandez N., Fumeron F. Plasma Dihydroceramides are diabetes susceptibility biomarker candidates in mice and humans. Cell Rep. 2017;18:2269–2279. doi: 10.1016/j.celrep.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 92.Braicu E.I., Darb-Esfahani S., Schmitt W.D., Koistinen K.M., Heiskanen L., Pöhö P. High-grade ovarian serous carcinoma patients exhibit profound alterations in lipid metabolism. Oncotarget. 2017;8:102912–102922. doi: 10.18632/oncotarget.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen L., Chen H., Li Y., Li L., Qiu Y., Ren J. Endocannabinoid and ceramide levels are altered in patients with colorectal cancer. Oncol Rep. 2015;34:447–454. doi: 10.3892/or.2015.3973. [DOI] [PubMed] [Google Scholar]
- 94.Knapp P., Bodnar L., Błachnio-Zabielska A., Świderska M., Chabowski A. Plasma and ovarian tissue sphingolipids profiling in patients with advanced ovarian cancer. Gynecol Oncol. 2017;147:139–144. doi: 10.1016/j.ygyno.2017.07.143. [DOI] [PubMed] [Google Scholar]
- 95.Kozar N., Kruusmaa K., Bitenc M., Argamasilla R., Adsuar A., Goswami N. Metabolomic profiling suggests long chain ceramides and sphingomyelins as a possible diagnostic biomarker of epithelial ovarian cancer. Clin Chim Acta. 2018;481:108–114. doi: 10.1016/j.cca.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 96.Separovic D., Shields A.F., Philip P.A., Bielawski J., Bielawska A., Pierce J.S. Altered levels of serum ceramide, sphingosine and sphingomyelin are associated with colorectal Cancer: a retrospective pilot study. Anticancer Res. 2017;37:1213–1218. doi: 10.21873/anticanres.11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Radner F.P.W., Marrakchi S., Kirchmeier P., Kim G.-J., Ribierre F., Kamoun B. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003536. e1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kosinska M.K., Liebisch G., Lochnit G., Wilhelm J., Klein H., Kaesser U. Sphingolipids in human synovial fluid - a Lipidomic study. Plos One. 2014;9 doi: 10.1371/journal.pone.0091769. e91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kosinska M.K., Mastbergen S.C., Liebisch G., Wilhelm J., Dettmeyer R.B., Ishaque B. Comparative lipidomic analysis of synovial fluid in human and canine osteoarthritis. Osteoarthr Cartil. 2016;24:1470–1478. doi: 10.1016/j.joca.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 100.Jiang X., Paultre F., Pearson T.A., Reed R.G., Francis C.K., Lin M. Plasma sphingomyelin level as a risk factor for coronary artery disease. Arterioscler Thromb Vasc Biol. 2000;20:2614–2618. doi: 10.1161/01.atv.20.12.2614. [DOI] [PubMed] [Google Scholar]
- 101.Laaksonen R., Ekroos K., Sysi-Aho M., Hilvo M., Vihervaara T., Kauhanen D. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J. 2016;37:1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meeusen Jeffrey W., Donato Leslie J., Bryant Sandra C., Baudhuin Linnea M., Berger Peter B., Jaffe Allan S. Plasma Ceramides. Arterioscler Thromb Vasc Biol. 2018;38:1933–1939. doi: 10.1161/ATVBAHA.118.311199. [DOI] [PubMed] [Google Scholar]
- 103.Yu J., Pan W., Shi R., Yang T., Li Y., Yu G. Ceramide is upregulated and associated with mortality in patients with chronic heart failure. Can J Cardiol. 2015;31:357–363. doi: 10.1016/j.cjca.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 104.Abbott S.K., Li H., Muñoz S.S., Knoch B., Batterham M., Murphy K.E. Altered ceramide acyl chain length and ceramide synthase gene expression in Parkinson's disease. Mov Disord. 2014;29:518–526. doi: 10.1002/mds.25729. [DOI] [PubMed] [Google Scholar]
- 105.Guedes L.C., Chan R.B., Gomes M.A., Conceição V.A., Machado R.B., Soares T. Serum lipid alterations in GBA-associated Parkinson's disease. Parkinsonism Relat Disord. 2017;44:58–65. doi: 10.1016/j.parkreldis.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 106.Mielke M.M., Maetzler W., Haughey N.J., Bandaru V.V.R., Savica R., Deuschle C. Plasma ceramide and glucosylceramide metabolism is altered in sporadic Parkinson's disease and associated with cognitive impairment: a pilot study. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xing Y., Tang Y., Zhao L., Wang Q., Qin W., Ji X. Associations between plasma ceramides and cognitive and neuropsychiatric manifestations in Parkinson's disease dementia. J Neurol Sci. 2016;370:82–87. doi: 10.1016/j.jns.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 108.Filippov V., Song M.A., Zhang K., Vinters H.V., Tung S., Kirsch W.M. Increased ceramide in brains with Alzheimer's and other neurodegenerative diseases. J Alzheimers Dis JAD. 2012;29:537–547. doi: 10.3233/JAD-2011-111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim M., Nevado-Holgado A., Whiley L., Snowden S.G., Soininen H., Kloszewska I. Association between plasma ceramides and phosphatidylcholines and hippocampal brain volume in late onset Alzheimer's disease. J Alzheimers Dis. 2017;60:809–817. doi: 10.3233/JAD-160645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mielke M.M., Haughey N.J., Bandaru V.V.R., Schech S., Carrick R., Carlson M.C. Plasma ceramides are altered in mild cognitive impairment and predict cognitive decline and hippocampal volume loss. Alzheimers Dement. 2010;6:378–385. doi: 10.1016/j.jalz.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mielke M.M., Haughey N.J., Bandaru V.V.R., Weinberg D.D., Darby E., Zaidi N. Plasma sphingomyelins are associated with cognitive progression in Alzheimer's disease. J Alzheimers Dis JAD. 2011;27:259–269. doi: 10.3233/JAD-2011-110405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mielke M.M., Bandaru V.V.R., Haughey N.J., Xia J., Fried L.P., Yasar S. Serum ceramides increase the risk of Alzheimer disease. Neurology. 2012;79:633. doi: 10.1212/WNL.0b013e318264e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hannun Y.A., Obeid L.M. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 114.Hannun Y.A., Obeid L.M. Many ceramides. J Biol Chem. 2011;286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chavez J.A., Summers S.A. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys. 2003;419:101–109. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 116.O'Brien J.S., Sampson E.L. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J Lipid Res. 1965;6:537–544. [PubMed] [Google Scholar]
- 117.O'Brien J.S., Sampson E.L. Fatty acid and fatty aldehyde composition of the major brain lipids in normal human gray matter, white matter, and myelin. J Lipid Res. 1965;6:545–551. [PubMed] [Google Scholar]
- 118.Jana A., Pahan K. Sphingolipids in multiple sclerosis. Neuromolecular Med. 2010;12:351–361. doi: 10.1007/s12017-010-8128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kihara Y., Matsushita T., Kita Y., Uematsu S., Akira S., Kira J. Targeted lipidomics reveals mPGES-1-PGE2 as a therapeutic target for multiple sclerosis. Proc Natl Acad Sci. 2009;106:21807–21812. doi: 10.1073/pnas.0906891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jana A., Hogan E.L., Pahan K. Ceramide and neurodegeneration: susceptibility of neurons and oligodendrocytes to cell damage and death. J Neurol Sci. 2009;278:5–15. doi: 10.1016/j.jns.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mencarelli C., Martinez–Martinez P. Ceramide function in the brain: when a slight tilt is enough. Cell Mol Life Sci. 2013;70:181–203. doi: 10.1007/s00018-012-1038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bieberich E. Ceramide signaling in cancer and stem cells. Future Lipidol. 2008;3:273–300. doi: 10.2217/17460875.3.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kolesnick R. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest. 2002;110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Samanta S., Stiban J., Maugel T.K., Colombini M. Visualization of ceramide channels by transmission Electron microscopy. Biochim Biophys Acta. 2011;1808:1196–1201. doi: 10.1016/j.bbamem.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stiban J., Caputo L., Colombini M. Ceramide synthesis in the endoplasmic reticulum can permeabilize mitochondria to proapoptotic proteins. J Lipid Res. 2008;49:625–634. doi: 10.1194/jlr.M700480-JLR200. [DOI] [PubMed] [Google Scholar]
- 126.Baufeld C., O'Loughlin E., Calcagno N., Madore C., Butovsky O. Differential contribution of microglia and monocytes in neurodegenerative diseases. J Neural Transm. 2018;125:809–826. doi: 10.1007/s00702-017-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lund H., Pieber M., Harris R.A. Lessons learned about neurodegeneration from microglia and monocyte depletion studies. Front Aging Neurosci. 2017;9 doi: 10.3389/fnagi.2017.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rawji K.S., Mishra M.K., Michaels N.J., Rivest S., Stys P.K., Yong V.W. Immunosenescence of microglia and macrophages: impact on the ageing central nervous system. Brain. 2016;139:653–661. doi: 10.1093/brain/awv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bitsch A., Schuchardt J., Bunkowski S., Kuhlmann T., Bruck W. Acute axonal injury in multiple sclerosis. Correlation with demyelination and inflammation. Brain J Neurol. 2000;123:1174–1183. doi: 10.1093/brain/123.6.1174. Pt 6. [DOI] [PubMed] [Google Scholar]
- 130.Mishra M.K., Yong V.W. Myeloid cells — targets of medication in multiple sclerosis. Nat Rev Neurol. 2016;12:539–551. doi: 10.1038/nrneurol.2016.110. [DOI] [PubMed] [Google Scholar]
- 131.Nikić I., Merkler D., Sorbara C., Brinkoetter M., Kreutzfeldt M., Bareyre F.M. A reversible form of axon damage in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat Med. 2011;17:495–499. doi: 10.1038/nm.2324. [DOI] [PubMed] [Google Scholar]
- 132.Trapp B.D., Peterson J., Ransohoff R.M., Rudick R., Mörk S., Bö L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 133.Moreno M.A., Burns T., Yao P., Miers L., Pleasure D., Soulika A.M. Therapeutic depletion of monocyte-derived cells protects from long-term axonal loss in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2016;290:36–46. doi: 10.1016/j.jneuroim.2015.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data deposited in NCBI's Gene Expression Omnibus Series accession number GSE103929.