Abstract
The hepatitis C virus (HCV) viroporin p7 oligomerizes to form ion channels, which are required for the assembly and secretion of infectious viruses. The 63-amino acid p7 monomer has two putative transmembrane domains connected by a cytosolic loop, and has both N- and C- termini exposed to the endoplasmic reticulum (ER) lumen. NMR studies have indicated differences between p7 structures of distantly related HCV genotypes. A critical question is whether these differences arise from the high sequence variation between the different isolates and if so, how the divergent structures can support similar biological functions. Here, we present a side-by-side characterization of p7 derived from genotype 1b (isolate J4) in the detergent 6-cyclohexyl-1-hexylphosphocholine (Cyclofos-6) and p7 derived from genotype 5a (isolate EUH1480) in n-dodecylphosphocholine (DPC). The 5a isolate p7 in conditions previously associated with a disputed oligomeric form exhibits secondary structure, dynamics, and solvent accessibility broadly like those of the monomeric 1b isolate p7. The largest differences occur at the start of the second transmembrane domain, which is destabilized in the 5a isolate. The results show a broad consensus among the p7 variants that have been studied under a range of different conditions and indicate that distantly related HCVs preserve key features of structure and dynamics.
Subject terms: Solution-state NMR, Molecular modelling
Introduction
Approximately 3% of the world’s population carries the hepatitis C virus (HCV), putting more than 200 million people at risk of developing severe liver diseases1–3. HCV displays high genetic heterogeneity and is classified into eight genotypes (gt 1–8) and more than a hundred subtypes4–8. The polyprotein precursor is expressed from a 9.6 kb positive-sense, single-stranded RNA genome ((+) ssRNA)) and is co- and post-translationally cleaved by cellular and viral proteases to produce at least ten viral proteins9–11. In the HCV polyprotein precursor, p7 lies between the structural and non-structural proteins. It is essential for the assembly and secretion of infectious viral particles in vitro12–16 and for virus propagation in vivo17, making it an attractive therapeutic target18.
HCV p7 is a small, hydrophobic protein comprising 63 amino acids19. The structural properties of p7 constructs derived from 1b genotypes have been widely investigated using a range of approaches18, including electron microscopy (EM)20–22, nuclear magnetic resonance spectroscopy (NMR)23–31, and molecular modeling22,24,32–36. The various NMR studies of monomeric p7 suggest a similar architecture that is in broad agreement with secondary structure prediction from its amino-acid sequence, namely two hydrophobic transmembrane (TM) regions separated by a conserved basic loop region9,19. Analytical ultracentrifugation measurements suggest that p7 self-assembly results predominantly in hexameric and heptameric viroporins, shown in electrophysiology experiments to be cation selective23. Computational modeling studies of p7 predict that the monomeric protein adopts hairpin-like structures, which then associate side-by-side with the N-terminal TM helix forming the channel pore22,24,32,34–36.
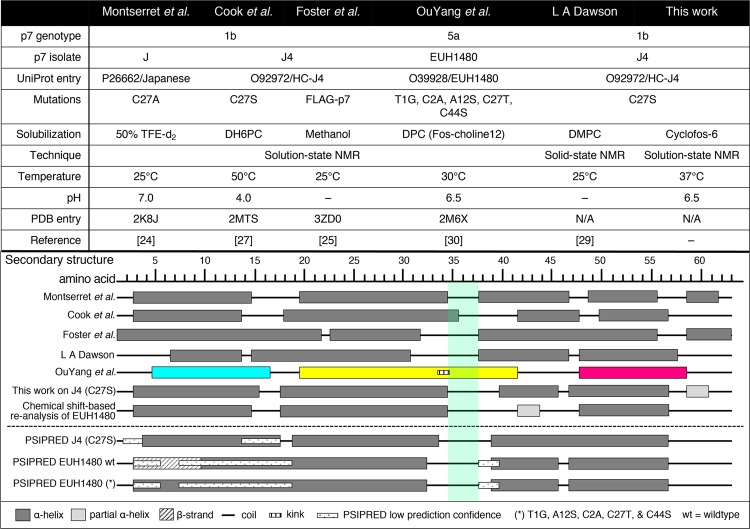
A strikingly different subunit conformation and packing arrangement was reported for a p7 construct based on a 5a genotype29. Several features of the hexameric structure determined by NMR were unexpected based on previous structural and functional studies of p7, but recent experiments indicate that the protein is not oligomeric in the detergent n-dodecylphosphocholine (DPC) used to derive the structure37. The secondary structure of the genotype 5a construct, which should be unaffected by the introduction of tertiary restraints in the structure determination, also indicated key differences compared to monomeric structures published hitherto (Fig. 1 and references therein).
Figure 1.
Comparison of p7 secondary structures. Sample conditions (top) and secondary structure determinations (bottom) reported for p7 monomers and the hexamer of OuYang et al. The sample conditions used for p7(5a/EUH1480) in this work are the same as previously reported29. For comparison, the PSIPRED secondary structure predictions based on amino acid sequences for p7(1b/J4) and both wildtype and mutated (*) p7(5a/EUH1480) are also shown82. The PSIPRED prediction for J4 with the C27S substitution is identical to wildtype J4 (not shown). For the subunit structure reported by OuYang et al., the helices are color-coded according to the original paper29. The basic loop region ~33–37, which differs notably between monomeric genotype 1b p7 and oligomeric 5a p7, is indicated by green shading23,24,26,28,29. The secondary structures were estimated directly from published data (secondary chemical shifts, NOEs, and/or dipolar waves). In the case of p7(5a/EUH1480)29, the helical regions were determined from the structure deposited in the PDB (2M6X). Plotted helical regions by residue number: Montserret et al. 2010: 3–14, 20–34, 38–46, 48–55 and 59–61; Cook et al. 2013: 3–13, 18–35, 42–47 and 50–56; Foster et al. 2014: 1–21, 23–31, 38–55 and 59–63; LA Dawson: 7–13, 15–30, 38–46 and 48–57; Ouyang et al. 2013: 5–16, 20–41 and 48–58; Oestringer et al. 2018 J4: 3–15, 18–34, 40–45, 47–56 and 59–60; Chemical shift-based re-analysis of EUH1480 using published29 chemical shifts: 3–14, 18–34, 42–43 and 48–56; PSIPRED J4: 4–17, 19–33 and 39–56; PSIPRED EUH1480: 3–32, 39–45 and 47–56; PSIPRED EUH1480 mt5: 3–9 (beta strand), 10–32, 39–45 and 47–56.
HCV has one of the highest mutation rates and genetic variability amongst RNA viruses5,38, and the unique genotype 5a p7 construct used to determine an oligomeric structure is very different from the genotype 1b constructs used to derive all other structures (Supplementary Fig. 1). Whereas the function of several genotype 1 and 2 constructs have been studied20,23,39–45, attempts at recording electrophysiological measurements of genotype 5a p7 were not successful, making it unclear how this p7 relates to the more commonly studied homologs29.
We sought to understand the extent to which sequence divergence has resulted in differences in structural preferences for p7. The structure, dynamics, and solution properties of p7 from genotype 1b (isolate J4) were studied alongside that of genotype 5a (isolate EUH1480) in phosphocholine-based detergents, using a combination of solution NMR spectroscopy and molecular dynamics (MD). The largest differences were detected in residues 40–45, which appear to adopt an unstable helical structure that may be sensitive to solution conditions. The two constructs were otherwise very similar in secondary structure and dynamics, indicating that key features of p7 structure and dynamics are conserved between distantly related HCV.
Results
Sample preparation and spectral characterization
HCV p7 from genotype 1b isolate J4 containing a C27S mutation to prevent disulfide bond formation during preparation25–27 (referred to as p7(1b/J4)) was expressed and purified from E. coli and solubilized directly into the detergent 6-cyclohexyl-1-hexylphosphocholine (Cyclofos-6) enabling solution NMR analyses of the protein at pH 6.5 and 37 °C. The spectral quality was unaffected by changes in pH between 6 and 7 or detergent concentrations between 26.8 mM and 268 mM, indicating no change in the structure or oligomeric state over these ranges.
The p7 construct based on genotype 5a isolate EUH1480 was expressed in E. coli, purified, and refolded into the detergent DPC as described in reference29. The 5a construct contained five substitutions (T1G, C2A, A12S, C27T, and C44S) to enable a direct comparison with a previously published structure29 and is referred to as p7(5a/EUH1480). The resulting 1H,15N-based spectra collected on p7(5a/EUH1480) were similar to those previously reported, confirming that the protein conformation and environment were similar to those samples used previously29 and enabling analyses of additional NMR experiments based on published chemical shift values. High quality spectra could be recorded on both p7(1b/J4) and p7(5a/EUH1480) at 37 °C using conventional, non-TROSY approaches on fully protonated protein (Fig. 2A), consistent with previously published SEC-MALS experiments indicating that the protein is monomeric under these conditions37. The spectral properties of p7(1b/J4) were similar to those of p7(5a/EUH1480) and the backbone resonances (1HN, 15N, 13Cα, 13C′) of p7(1b/J4) were assigned using conventional triple-resonance experiments.
Figure 2.
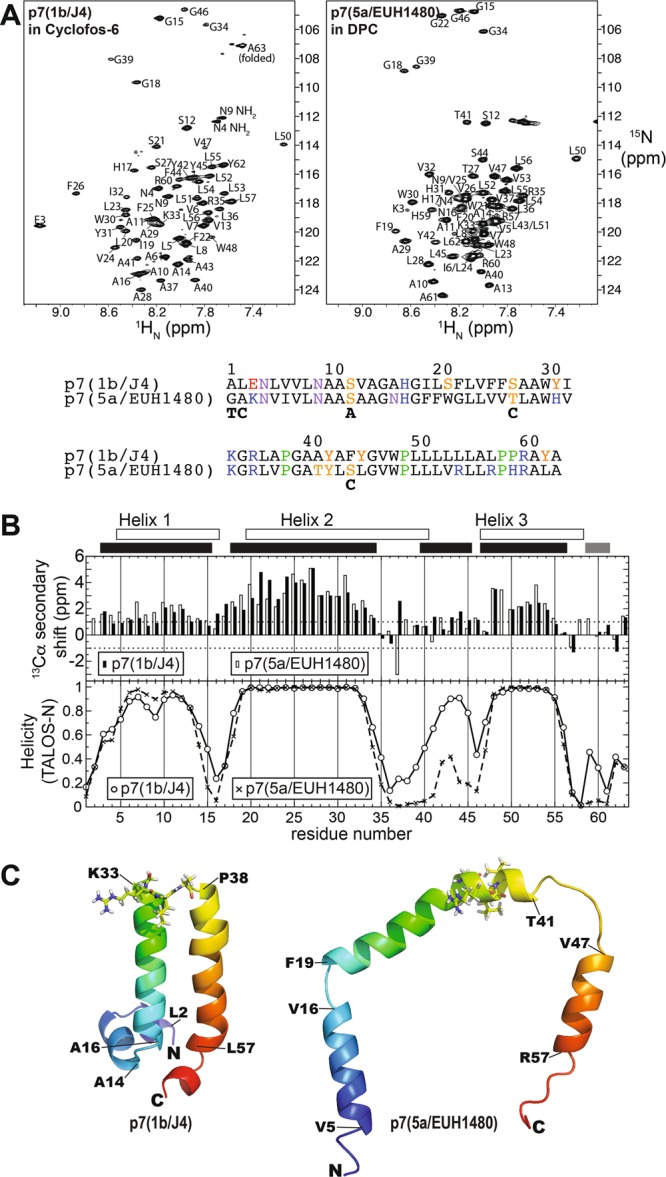
Spectral characterization of HCV p7 isolates. (A) 2D 1H-15N HSQC spectra of p7(1b/J4) in Cyclofos-6 (left), and p7(5a/EUH1480) in DPC (right), with amino acid assignments indicated. The amino acid assignments of the p7(5a/EUH1480) spectrum were transferred from29. The amino acid sequences of the constructs are shown below the spectra. Below the sequence for p7(5a/EUH1480) are indicated the native amino acids that were substituted in this construct used for experiments here and in29. (B) Top: 13Cα secondary chemical shift index for p7(1b/J4) (filled bars) and p7(5a/EUH1480) (open bars), with positive values indicative of α-helical conformation. Bottom: TALOS-N46 chemical shift-based secondary structure prediction for p7(1b/J4)(⚪) and p7(5a/EUH1480) (X). Chemical shift data for p7(5a/EUH1480) were taken from the BioMagResBank entry 1916229. Shown above the plot and indicated by unfilled and filled bars are the helical residues determined for p7(5a/EUH1480) from PDB 2M6X and those predicted from chemical shifts for p7(1b/J4), respectively. (C) Left: Membrane CS-Rosetta-based structure of p7(1b/J4). Right: A single subunit of p7(5a/EUH1480) showing the horseshoe-like conformation in the oligomeric model 2M6X29 is shown for comparison. The structures are shaded from blue (N-terminus) to red (C-terminus) and the residues 34–37 are shown as sticks. The residues at the beginning and end of each helix are indicated.
Secondary structure probabilities were determined from chemical shifts of p7(1b/J4) and p7(5a/EUH1480), with chemical shift data for p7(5a/EUH1480) taken from the BioMagResBank (entry 19162)29. The chemical shift-based predictions from TALOS-N46 were similar for the two constructs in helix 1 (residues ~3–15), helix 2 (~18–34), and the C-terminal half of helix 3 (residues ~48–56). In addition to helical breaks centered at residues 16 and 35–37, a discontinuity in helix 3 was observed in both constructs at G46, which is three residues before a proline (P49). Differences between the two constructs were apparent in the N-terminal residues of helix 3 (residues ~40–45) in p7(5a/EUH1480), which were significantly destabilized compared with p7(1b/J4) (Fig. 2B). In addition, a weak propensity for a C-terminal helix beginning at residue P59 was observed for p7(1b/J4). Surprisingly, the helical residues present in the subunits of the hexameric model of p7(5a/EUH1480) from OuYang et al. (PDB 2M6X)29 are markedly different from the secondary structure derived here from their chemical shift data (Fig. 2B).
A structural model of p7(1b/J4) was generated using chemical shift Rosetta (CS-Rosetta)47 with a membrane force field48. The structure, shown in Fig. 2C (left), is most similar to the structures determined previously by Cook et al.26, using a similar approach, and by Montserret et al.23. In the chemical shift-based Rosetta structure of p7(1b/J4), residues 16–33 (helix 2) correspond to the first TM domain, residues 34–37 form a loop, and residues 38–57 (helix 3) correspond to the second TM domain. By contrast, in the subunit structure taken from the hexameric model of p7(5a/EUH1480)29 (shown at right in Fig. 2C) residues 34–37 are helical and residues ~42–46 form a loop.
Amide backbone 15N relaxation and proton exchange
Backbone dynamics of p7(1b/J4) and p7(5a/EUH1480) were characterized by 15N R1 and R2 relaxation rates and {1H-15N} heteronuclear Overhauser effects (hetNOEs). Trimmed mean averages of the backbone amide 15N R1 and R2 values were used to calculate apparent molecular weights of the protein and detergent complex (see Experimental Methods). The derived rotational correlation times, τc, were 10.6 ns and 10.1 ns at 37 °C for p7(1b/J4) and p7(5a/EUH1480), respectively. These values correspond to apparent molecular weights of ~40 kDa, and are consistent with monomeric p7 embedded in micelles of ~30 kDa and consistent with SEC-MALS studies of p7(5a/EUH1480)37. Elevated R1 values, and depressed R2 and hetNOE values, are indicative of increased flexibility on the picosecond-nanosecond (ps-ns) timescale, as can be seen for the N- and C-termini (Fig. 3A). For both constructs, increased ps-ns timescale flexibility was apparent in helix 1, whereas the least flexible regions, as indicated by lower R1 values and higher R2 and hetNOE values, were in the middle of helices 2 and 3. Increased ps-ns flexibility is observed for both constructs in loop residues G39, A40, and A41 (T41 in p7(5a/EUH1480)). The relatively high R2 values observed for G34 of p7(1b/J4) and H31 of p7(5a/EUH1480) indicate local dynamics on the microsecond-millisecond timescale. The nonhelical residues 35–37 exhibited little or no increase in dynamics compared with the TM domain residues of helix 2 indicating they form a structured loop.
Figure 3.
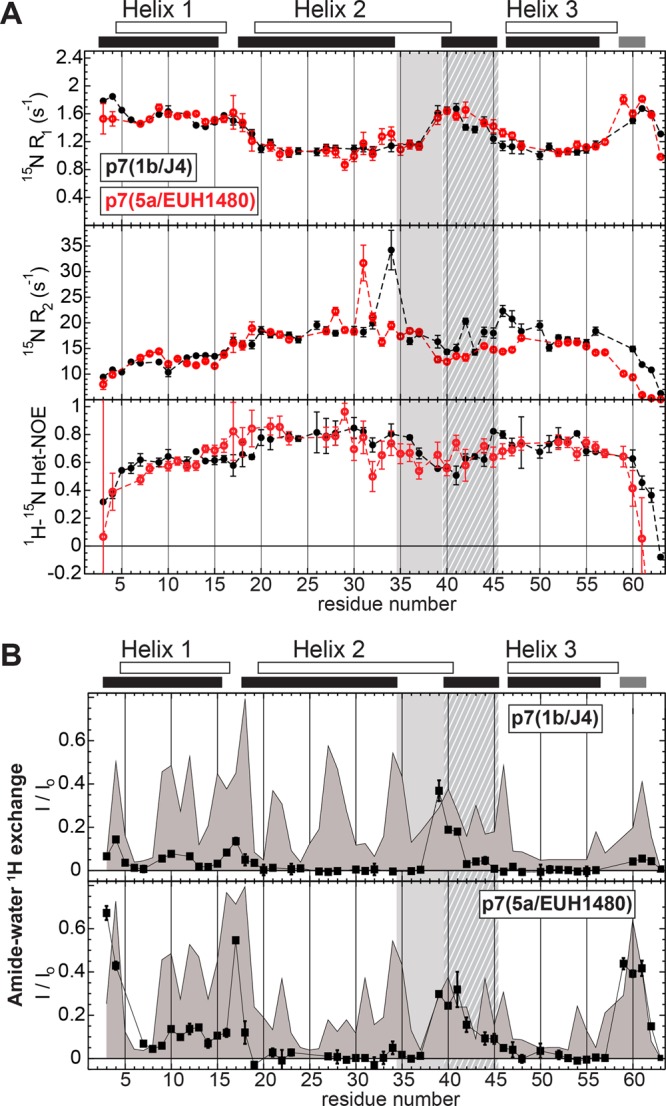
HCV p7 backbone amide dynamics and amide-water proton exchange. (A) 15N R1, 15N R2, and 1H-15N heteronuclear NOEs as a function of residue number for p7(1b/J4) (filled black circles) and p7(5a/EUH1480) (open red circles). The backbone amide heteronuclear NOE value for residue A63 of p7(5a/EUH1480) was −2.1 (data point not shown). (B) Backbone amide hydrogen exchange data for p7(1b/J4) (top) and p7(5a/EUH1480) (bottom) as a function of residue number. NMR CLEAN chemical exchange (CLEANEX) experiments83 were recorded using a 50 ms (◼) mixing time. For comparison are shown the empirically-based predictions of the magnitude of amide exchange from primary structure alone (solid line with grey shading)84. Experiments for (A,B) were recorded at 600 MHz (1H) and 37 °C. The relaxation data were collected using conventional HSQC-based pulse sequence experiments. Shown above the plots in (A,B) by unfilled and filled bars are the helical regions determined for p7(5a/EUH1480) from PDB 2M6X and those predicted from chemical shifts for p7(1b/J4), respectively. The light shading for residues 35–39 indicate helical residues in the 2M6X structural model that were found in this study to be nonhelical. The crosshatching for residues 40–45 indicate residues predicted to form an unstable helix.
While the per-residue profiles for backbone dynamics of p7(1b/J4) and p7(5a/EUH1480) were broadly similar, differences were seen in residues 39–47 (Fig. 3A). These residues are typically assigned to the start of the second TM domain (Fig. 1) but exhibited low helical propensities in both constructs (Fig. 2B), and increased 15N R1 and decreased 15N R2 in p7(5a/EUH1480) indicate that these residues in p7(5a/EUH1480) are significantly more dynamic than those in p7(1b/J4).
The rates of backbone amide proton exchange with water provide additional insights into secondary structure and dynamics since the proton exchange rates reflect hydrogen bond participation in addition to sidechain-dependent intrinsic exchange rates and solvent accessibility. Similar patterns of exchange were observed for the two constructs (Fig. 3B). In particular, H17, which is in a break between helices 1 and 2 exhibited elevated exchange rates, as well as residues 39–41. As expected, amides in the N- and C-termini also showed increased proton exchange rates for both constructs, but the magnitudes in p7(5a/EUH1480) were much higher than that of p7(1b/J4) and approached values expected for the same sequence fully unstructured in water49,50 (Fig. 3B). Several residues in helix 1 of both constructs also exhibit amide proton exchange consistent with this helix being solvent exposed or only transiently bound to the micelle surface.
MD-simulation of the p7(1b/J4) structural model in POPC bilayers
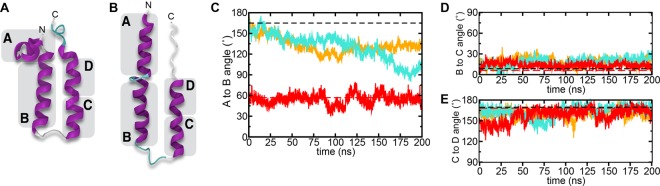
Previous atomistic MD simulations have indicated that monomeric forms of the p7(5a/EUH1480) sequence and the p7 sequence from genotype 1b isolate J (p7(1b/J)) could form stable hairpins in 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayers over a 200 ns timescale51. Identical simulations were carried out for the p7(1b/J4) Rosetta-derived structure to examine whether the hairpin conformation of p7(1b/J4) is also stable in a POPC bilayer. The results are compared with those from p7(1b/J) and p7(5a/EUH1480)51 in Fig. 4. Visual inspection of snapshots from the simulation did not reveal any large-scale alteration of the structures over the timescale of the simulation. To quantitatively analyze the conformational evolution of the monomeric protein, the structures were separated into four α-helical segments conserved throughout the simulations as observed previously51. Helix A (residues 4–14) corresponds approximately to helix 1 identified by secondary chemical shifts, helix B (20–31) corresponds to ~helix 2, and helix C (39–47) and D (48–54) correspond to ~helix 3 broken by the kink around G46 (Fig. 4). Over the 200 ns simulation the maximum variation in the angles between helices B and C, and between C and D were ~30° and ~40°, respectively, indicating that the hairpin conformations are stable over the duration of the simulations. By contrast, the variations in the angle between helices A and B were larger, and up to 80° for p7(5a/EUH1480). Variation in the angle between helices A and B arises from movements of helix A, which was not embedded in the hydrophobic portion of the membrane in the models of p7(1b/J) and p7(5a/EUH1480). Helix A in the Rosetta-derived structure of p7(1b/J4) adopts a membrane surface-bound conformation that is more stable over the duration of the simulation than the detached conformation of p7(1b/J) and p7(5a/EUH1480). The increased flexibility and solvent accessibility observed by NMR for this helix (Fig. 3) is consistent with it being outside of the micelle or loosely associated with the micelle surface.
Figure 4.
MD simulations of p7 structures. Structural evolution of p7 hairpin monomer structures in POPC bilayers measured over 200 ns MD trajectories by means of the angles formed by contiguous α-helical segments for (A) the p7(1b/J4) Rosetta-derived hairpin structure, and (B) the hairpin structure of Montserret et al. (architecture used for both p7(1b/J) and p7(5a/EUH1480) sequences). (C–E) Show the time traces of the angles between the helices for p7(1b/J4) (this work; red), p7(1b/J) from Montserret et al. (orange), and the sequence of p7(5a/EUH1480) made to adopt the hairpin conformation of Montserret et al. (cyan)51. Horizontal dashed lines correspond to the starting angles of structures based on that of Montserret et al.23 (dashed black) or of p7(1b/J4) from this study (dashed red).
Discussion
HCV p7 is a viroporin essential for infectious virus production and therefore a potential drug target52,53, with the search for a truly ‘pan-genotypic’ HCV treatment ongoing. Current standard treatment of care using the nucleotide analog sofosbuvir targeting the HCV protein NS5B is genotype-dependent in its ability to overcome innate viral resistances and generally requires combinations with other direct acting antivirals to prevent escape mutations54,55, hence several compounds targeting p7 have been investigated and could serve as anti-HCV drugs. Some of these compounds are postulated to inhibit the assembled channel whereas others are hypothesized to disassemble the oligomeric channel56 (reviewed in references18,53). Thus, detailed knowledge of the p7 monomer structure and how it assembles to form functional channels remains of great interest. However, p7 poses a challenge for structural studies because most of the protein is buried in the membrane. In addition, it forms oligomers with variable stoichiometry and may only weakly oligomerize in detergent20–23,34. Thus despite its small size only one atomic-resolution, experimental model for an assembled p7 channel has been published29. However, that structure included several unexpected features which could not be reconciled with many published structural, biochemical, and functional data and it was subsequently shown that the detergent used to solubilize the protein for those structural studies does not support p7 oligomerization37. Further questions remain about the secondary structure in that p7 model, which is not expected to be strongly affected by introducing tertiary contacts in the structure calculations. While phosphocholine detergents are strongly denaturing and can introduce artefacts57, the persistence of structural features across different solubilizing reagents, solution conditions, and sequences can provide an indication of whether those features are likely to be present in a biological membrane.
The structures of two closely related isolates from 1b (p7 from isolate J and J4; 94% identical) have been studied previously (Fig. 1). For these 1b-derived p7 constructs studied under different conditions, the largest differences were seen for an N-terminally FLAG-tagged construct in methanol that has a long N-terminal helix extending from the FLAG-tag to residue ~21 and lacks the break at position ~15 seen in other studies. By contrast, the 1b- and 5a- derived p7 sequences are two of the most distantly related among known p7 sequences (Supplementary Fig. 1). The sequences of the p7(1b/J4) and p7(5a/EUH1480) constructs studied here differ in 46% (29 of 63) of the positions, providing a good test of whether structural features of p7 are conserved.
The structure in the region around the strictly conserved dibasic motif (residues K/R33 and K/R35) is of particular interest from a functional point of view since charge neutralizing substitutions result in nonviable virus17. The dibasic motif is known to be important in ion channel activity58, and double glutamine or double alanine substitutions in the JFH-1 isolate of genotype 2a result in 100- and 1,000-fold decreases in total infectivity, respectively16. The oligomer model of the p7(5a/EUH1480) construct indicated that helix 2 extends to residue T41 with a kink of ~45° at G34, whereas the results here indicate that helix 2 ends at ~G34 in both p7(1b/J4) and p7(5a/EUH1480). Comparison with previously published work on monomeric p7 indicates a consensus that the cytosolic loop is in residues 35–37 (Fig. 1), such that R35 is within the loop and K33 is close enough to the end of the TM domain to “snorkel” into the lipid headgroup region59.
The largest structural and dynamic differences between the two distantly related constructs studied here are in residues ~40–45, which are immediately C-terminal to the cytosolic loop. The differences seem to arise from increased sequence variability and structural instability in this region. The second TM domain of p7 is typically assigned to residues ~40–56, with a discontinuity at 46 or 47. Residues 48–56 exhibit strong helicity in most studies, whereas residues 40–45 tend to be more variable. Structural instability was observed previously in this region for the p7(1b/J4) construct in different conditions26. In the work reported here, residues 40–45 exhibit moderate to high helicity in p7(1b/J4) but are mostly nonhelical in p7(5a/EUH1480). Sequence conservation in residues 40–45 is generally low (Supplementary Fig. 2), and only two of the positions in this region are conserved between 1b/J4 and 5a/EUH1480. Structural variation is also seen here between the 1b-derived p7 constructs having no or very small sequence differences but studied in different membrane mimetics (Fig. 1). The composition of residues 40–45 tends to be relatively hydrophilic for a TM domain (Supplementary Fig. 3), which can make it particularly sensitive to the membrane mimetic, and the helix here may be more stable in lipid membranes28.
A helix discontinuity at, or near, G46 was seen in both constructs and also observed in other studies23,24,26. The discontinuity is likely due in part to the proline at position 49. Prolines are often C-terminal to helical kinks and are present at higher frequency within TM helices than in helices of water-soluble domains60. Kinks can be exaggerated by the detergent environment61, but MD simulations in lipid membranes are also consistent with at least a small (~15–30°) deflection of the helix (Fig. 4E). Although G46 and P49 are not strictly conserved, one or the other is always present and substitutions in these positions are for more hydrophilic amino acids (Supplementary Fig. 2).
The published subunit structure of p7(5a/EUH1480) pointed to a helix extending from 20–41 with a kink at G3429, whereas secondary chemical shifts indicate that residues 35–41 are not helical, and 39–41 are particularly flexible (reference62 and Fig. 2B). The helical boundaries determined for the subunit structure were based on observation of characteristic NOEs, which is expected to yield similar results compared with chemical shift-based approaches. However, spectral overlap in the 1Hα region can result in information gaps with the NOE approach and chemical shift information tends to be more complete since 13C shift information (13C′, 13Cα, 13Cβ) is partially redundant. Consistent with a non-helical state, these amides exhibit proton exchange rates comparable to what would be expected for the same sequence unstructured in water (Fig. 3B).
The pattern of amide proton exchange of p7(5a/EUH1480) in DPC was similar to that of p7(1b/J4) in Cyclofos-6, and the results are consistent with the exchange rates previously reported for p7(5a/EUH1480) in similar conditions30. Some regions, including the N- and C-termini, helix 1, and H17, exhibited higher exchange rates in p7(5a/EUH1480), which may be due to differences in hydrophobicity between the constructs: The regions including residues 13–20 and 53–58 are more hydrophobic in p7(1b/J4) than p7(5a/EUH1480) (Supplementary Fig. 3), which may result in remodeling of the detergent micelle and more water exposure. Although all substitutions introduced to create p7(5a/EUH1480) decrease its hydrophobicity, the wildtype sequence is also less hydrophobic than p7(1b/J4) in the regions with large differences in amide proton exchange rates. The increased exchange rates and high flexibility in helix 1 (residues ~5–15) that have been attributed either to exposure to a hydrophilic channel pore in the oligomer30 or to membrane thinning62 may be explained more simply by the hairpin models showing that helix 2 is the TM helix, and helix 1 extends out of the membrane23,24 or possibly lies on the membrane surface26.
Finally, we provide further evidence that the hairpin conformation can be stably adopted by different p7 sequences. Most p7 modelling studies predict the monomer to adopt a closely packed helical hairpin conformation, which then assembles side-by-side into oligomers. Most studies have used p7 constructs from genotypes 1 or 2, but MD simulations have shown that p7(5a/EUH1480) can also adopt a hairpin conformation in membranes63, and that the stability is not strongly sequence dependent51. Here, we expand those results to show that the Rosetta-derived hairpin conformation of p7(1b/J4) is also stable in a lipid bilayer.
In conclusion, we find that the genetically distant genotype 1b and 5a p7 constructs behave similarly, as assessed by NMR spectroscopy in detergent and computer modelling in lipid membranes. The results for 5a p7 are largely consistent with previous reports on monomeric p7’s from other genotypes and in a range of solubilizing conditions, demonstrating that key structural and dynamic features are conserved among distantly related p7 isolates. Together, these findings provide a foundation for future studies of how p7 monomers assemble to form an oligomeric viroporin.
Experimental Section
Protein expression and purification
HCV p7 (strain J4, C27S, genotype 1b, “p7(1b/J4)”, and strain EUH1480, T1G/C2A/A12S/C27T/C44S, genotype 5a, “p7(5a/EUH1480)”) were expressed into inclusion bodies as a fusion to His9–trpΔLE using the vector pMM-LR664. The p7 genes (synthetic DNA strings) were ordered from GeneArt (Life Technologies) inserted into vector pMM-LR6 and transformed into XL10 Gold Competent Cells (NEB) for plasmid propagation and storage. The plasmid with the p7 insert was purified from high optical density (600 nm) cultures (QIAprep Spin Miniprep Kit, QIAGEN) and transformed into BL21(DE3) cells (NEB) for protein expression. Genes were confirmed by DNA sequence analysis (Source Bioscience, Oxford). A large culture in LB was grown overnight at 37 °C, followed by condensation into a smaller culture of minimal media the next morning (adapted from65). The isotopically labeled p7 peptides were purified via immobilized metal ion affinity chromatography (IMAC) and released from the fusion protein by cyanogen bromide cleavage in 70% formic acid (1 hour, 0.2 g/ml cyanogen bromide). The digest reaction was stopped with 1 N NaOH and dialyzed to water and lyophilized. The dried, cleaved HCV p7(1b/J4) was taken up in 10% SDS, sonicated for 15 minutes, mixed with the same volume of running buffer and filtered through a 0.22 μm membrane (Millex-GS, Millipore) and loaded on a Sephacryl column (HiPrep 26/60 Sephacryl S-200) and separated using an Äkta Pure FPLC system (GE Healthcare)66. An isocratic gradient eluted the protein (20 mM sodium phosphate, 1 mM EDTA, 1 mM NaN3 and 4 mM SDS, pH 8.2) and the eluate was dialyzed against water and lyophilized66,67. The lyophilized, cleaved p7(5a/EUH1480) was purified as previously described29,30 using a C18 preparative column (Proto 300 5 μm, Higgins Analytical) on a Gilson PrepLC system (321 Pump, UV/VIS-155 detector, UniPoint software). The pure eluate was freeze-dried.
The lyophilized p7(1b/J4) protein was solubilized directly by addition of Cyclofos-6 (Anatrace, Anagrade) in water at 10× (26.8 mM) or 100× (268 mM) the CMC. The efficiency of solubilization increased with detergent concentration from 26.8 to 268 mM. Insolubilized material was discarded after bench top centrifugation (5 minutes, 16,000 x g). Critically, 40 mM sodium phosphate was added after solubilization from a 200 mM stock of pH between 6.0 and 7.0 (as required), and the pH was readjusted between 6.0 and 7.0 with NaOH. Final protein monomeric concentrations were between 200 and 400 μM. All NMR samples contained 5% D2O and 0.1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). P7(5a/EUH1480) protein was prepared in a manner similar to that described previously29. The lyophilized protein was solubilized in 200 mM DPC (Anatrace, Anagrade) and 6 M guanidine hydrochloride (GuHCl), in the absence of buffer. The protein (~300 μl sample with ~300 μM polypeptide concentration) was refolded upon dialysis against two liters 25 mM MES pH 6.5 in the absence of DPC with two buffer changes at two-hour intervals. Crucially, dialysis was performed using 2 kDa MWCO slide-A-Lyzer 100 µl capacity cups (Thermo Scientific), which provide a surface area that allows full elimination of GuHCl, but retains enough DPC to maintain the protein in solution. The sample was re-equilibrated by dialysis against 30 mL of 200 mM DPC, 25 mM MES pH 6.5 for 24 hours using 10–14 kDa MWCO membranes (Pur-A-Lyzer Mini Dialysis) to allow for detergent exchange. Any insoluble material was removed by centrifugation (5 minutes, 16,000 x g). The final protein concentration was ~300 µM.
NMR spectroscopy and data analysis
All NMR spectra were recorded on NMR spectrometers with Oxford Instrument magnets with 1H frequencies between 500 to 950 MHz, equipped with home-built triple resonance probes with triple axis gradients68 or with Bruker TCI CryoProbes with single Z-axis gradients (500 and 600 MHz). Experiments were performed at 30 °C or 37 °C as indicated and pH 6.5. NMR spectra were referenced in the direct dimension against DSS at 0 ppm. NMR data were processed using NMRPipe and analyzed using NMRDraw69, Analysis70 or CARA71. The resonances of p7(J4/1b) in Cyclofos-6 could be tentatively assigned using 15N-edited NOESY-HSQCs (90 ms and 140 ms mixing times) and a 15N-edited TOCSY-HSQC (55 ms mixing time). Assignments were subsequently confirmed using a set of HSQC-based triple-resonance experiments (HNCA, HNCACB/HNCOCACB and HNCO/HNCACO). For characterization of the hydrodynamic properties of the NMR samples, the apparent molecular weights were calculated from the 15N R1 and R2 values by first estimating the rotational correlation time, τc, from the 20% trimmed means of the relaxation rates72. Trimmed means were used to exclude residues with internal motions faster or slower than the overall tumbling time. Trimmed means for R1 and R2 for p7(1b/J4) and p7(5a/EUH1480) were 1.32 s−1 and 15.92 s−1, and 1.34 s−1 and 14.80 s−1, respectively. The molecular weight was then calculated from τc using Stoke’s law73 assuming a hydration shell of 1.5 water molecules, a solution viscosity of 0.702 centipoise at 37 °C, and a protein partial specific volume of 0.73 cm3/g. To calculate the number of detergent molecules consistent with the scenario of a monomeric p7 embedded in a detergent micelle, the value of the partial specific volume was weighted according to the fraction of the complex that was protein (0.73 cm3/g) and detergent (0.94 cm3/g).
Secondary structure calculations
TALOS-N calculations of secondary structure were based on chemical shift data for the following nuclei: 1HN, 1Hα, 15N, 13C′, and 13Cα. Chemical shifts for p7(5a/EUH1480) were obtained from BioMagResBank74 entry 1916229 and corrected for deuterium isotope effects75.
Molecular dynamics
Structures for MD simulations were immersed in a fully hydrated POPC lipid bilayer of initial dimensions equal to 75 × 75 × 90 Å3. Except for the POPC aliphatic tails, which were modeled by means of a united-atom potential energy function76, use was made of an all-atom representation. The different components of the assays were described by the macromolecular CHARMM36 (chemistry at Harvard macromolecular mechanics) force field77. All the simulations reported herein were performed in the isobaric-isothermal ensemble using the NAMD simulation package78. The temperature and the pressure were maintained at 300 K and 1 atm employing, respectively, softly damped Langevin dynamics and the Langevin piston algorithm79. Periodic boundary conditions were enforced. The equations of motion were integrated using the r–RESPA multiple-time stepping scheme80 with a time step of 2 and 4 fs for short- and long-range interactions, respectively. Non-bonded van der Waals interactions were smoothly switched to zero between 10 and 12 Å. The PME algorithm81 was utilized to account for long-range electrostatic interactions. During equilibration of the membrane, soft harmonic restraints were applied to the heavy atoms of the protein, prior to their slow release after appropriate relaxation of the surroundings. Evolution of the three-dimensional structures was monitored over a simulation time of 200 ns. Each production trajectory was prefaced by equilibration of up to 50 ns. For quantitative analysis of conformational changes, the p7 helical structure was decomposed into four regions, wherein TM1 and TM2 were both split into two domains, A (residues 4 to 14), B (residues 20 to 31), C (residues 39 to 47) and D (residues 48 to 54).
Supplementary information
Acknowledgements
This work was supported by the Oxford Glycobiology Endowment. J.R.S. and J.K.C. were funded by the MRC (L018578). N.Z. is a Fellow of Merton College.
Author Contributions
B.P.O. performed protein expression, sample preparation, NMR data collection and analysis, and wrote the paper; J.H.B. performed sample preparation and wrote the paper; J.K.C. performed NMR data collection and analysis; L.A. performed Rosetta modeling; C.C. and F.D. performed MD simulations and wrote the paper; N.H. performed MD simulations; J.R.S. performed NMR data collection and analysis; J.R.S. and N.Z. conceived the study and wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jason R. Schnell, Email: Jason.Schnell@bioch.ox.ac.uk
Nicole Zitzmann, Email: Nicole.Zitzmann@bioch.ox.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44413-x.
References
- 1.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nature Rev. Gastroenterol. & Hepatol. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 2.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 3.Westbrook RH, Dusheiko G. Natural history of hepatitis C. J. Hepatol. 2014;61:S58–68. doi: 10.1016/j.jhep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Nakano T, Lau GMG, Lau GML, Sugiyama M, Mizokami M. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int. 2012;32:339–345. doi: 10.1111/j.1478-3231.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- 5.Bukh J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J. Hepatol. 2016;65:S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Borgia SM, et al. Identification of a Novel Hepatitis C Virus Genotype From Punjab, India: Expanding Classification of Hepatitis C Virus Into 8 Genotypes. J. Infect. Dis. 2018;218:1722–1729. doi: 10.1093/infdis/jiy401. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatol. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 8.Smith DB, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 10.Scheel TKH, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nature Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nature Rev. Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 12.Bentham MJ, Foster TL, McCormick C, Griffin S. Mutations in hepatitis C virus p7 reduce both the egress and infectivity of assembled particles via impaired proton channel function. J. Gen. Virol. 2013;94:2236–2248. doi: 10.1099/vir.0.054338-0. [DOI] [PubMed] [Google Scholar]
- 13.Brohm C, et al. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 2009;83:11682–11693. doi: 10.1128/JVI.00691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentzsch J, et al. Hepatitis c Virus p7 is critical for capsid assembly and envelopment. PLoS Pathogens. 2013;9:e1003355. doi: 10.1371/journal.ppat.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmann E, et al. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai A, et al. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. USA. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madan V, Bartenschlager R. Structural and functional properties of the Hepatitis C Virus p7 viroporin. Viruses. 2015;7:4461–4481. doi: 10.3390/v7082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrère-Kremer S, et al. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin SDC, et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/S0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 21.Clarke D, et al. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–68. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luik P, et al. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. USA. 2009;106:12712–12716. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montserret R, et al. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 2010;285:31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster TL, et al. Structure-guided design affirms inhibitors of hepatitis C virus p7 as a viable class of antivirals targeting virion release. Hepatol. 2014;59:408–422. doi: 10.1002/hep.26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cook GA, Opella SJ. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim. Biophys. Acta. 2011;1808:1448–1453. doi: 10.1016/j.bbamem.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook, G. A., Dawson, L. A., Tian, Y. & Opella, S. J. Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance. Biochemistry52, 5295–5303 (2013). [DOI] [PMC free article] [PubMed]
- 27.Cook GA, Opella SJ. NMR studies of p7 protein from hepatitis C virus. Eur. Biophys. J. 2010;39:1097–1104. doi: 10.1007/s00249-009-0533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson, L. A. The NMR Structure and Oligomerization State of the Membrane Associated p7 Protein from Hepatitis C Virus. 1–117 (University of California, San Diego Thesis, 2015).
- 29.OuYang B, et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dev J, Brüschweiler S, OuYang B, Chou JJ. Transverse relaxation dispersion of the p7 membrane channel from hepatitis C virus reveals conformational breathing. J. Biomol. NMR. 2015;61:369–378. doi: 10.1007/s10858-015-9912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L, et al. Structural basis of interaction between the hepatitis C virus p7 channel and its blocker hexamethylene amiloride. Protein Cell. 2016;7:300–304. doi: 10.1007/s13238-016-0256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patargias G, Zitzmann N, Dwek R, Fischer WB. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y-T, Hsu H-J, Fischer WB. Computational modeling of the p7 monomer from HCV and its interaction with small molecule drugs. Springerplus. 2013;2:324. doi: 10.1186/2193-1801-2-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler DE, Penin F, Schulten K, Chipot C. The p7 protein of hepatitis C virus forms structurally plastic, minimalist ion channels. PLoS Comput. Biol. 2012;8:e1002702. doi: 10.1371/journal.pcbi.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stgelais C, et al. Determinants of hepatitis C virus p7 ion channel function and drug sensitivity identified in vitro. J. Virol. 2009;83:7970–7981. doi: 10.1128/JVI.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y-T, Schilling R, Fink RHA, Fischer WB. Ion-dynamics in hepatitis C virus p7 helical transmembrane domains–a molecular dynamics simulation study. Biophys. Chem. 2014;192:33–40. doi: 10.1016/j.bpc.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Oestringer BP, et al. Re-evaluating the p7 viroporin structure. Nature. 2018;562:E8–E18. doi: 10.1038/s41586-018-0561-9. [DOI] [PubMed] [Google Scholar]
- 38.Geller R, et al. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat. Microbiol. 2016;1:16045. doi: 10.1038/nmicrobiol.2016.45. [DOI] [PubMed] [Google Scholar]
- 39.Pavlović D, et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmann E, et al. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C. virus. Hepatol. 2007;46:330–338. doi: 10.1002/hep.21686. [DOI] [PubMed] [Google Scholar]
- 41.Chew CF, Vijayan R, Chang J, Zitzmann N, Biggin PC. Determination of pore-lining residues in the hepatitis C virus p7 protein. Biophys. Journal. 2009;96:L10–2. doi: 10.1016/j.bpj.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitfield T, et al. The influence of different lipid environments on the structure and function of the hepatitis C virus p7 ion channel protein. Mol. Membr. Biol. 2011;28:254–264. doi: 10.3109/09687688.2011.581253. [DOI] [PubMed] [Google Scholar]
- 43.Luscombe CA, et al. A novel Hepatitis C virus p7 ion channel inhibitor, BIT225, inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-alpha-2b and nucleoside analogues. Antiviral Res. 2010;86:144–153. doi: 10.1016/j.antiviral.2010.02.312. [DOI] [PubMed] [Google Scholar]
- 44.Premkumar A, Wilson L, Ewart GD, Gage PW. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/S0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 45.Premkumar A, Wilson L, Ewart GD, Gage PW. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/S0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holzmann N, Chipot C, Penin F, Dehez F. Assessing the physiological relevance of alternate architectures of the p7 protein of hepatitis C virus in different environments. Bioorg. Med. Chem. 2016;24:4920–4927. doi: 10.1016/j.bmc.2016.07.063. [DOI] [PubMed] [Google Scholar]
- 52.Steinmann E, Pietschmann T. Hepatitis C virus p7-a viroporin crucial for virus assembly and an emerging target for antiviral therapy. Viruses. 2010;2:2078–2095. doi: 10.3390/v2092078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott C, Griffin S. Viroporins: structure, function and potential as antiviral targets. J Gen Virol. 2015;96:2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- 54.Cholongitas E, Papatheodoridis GV. Sofosbuvir: a novel oral agent for chronic hepatitis C. Ann Gastroenterol. 2014;27:331–337. [PMC free article] [PubMed] [Google Scholar]
- 55.EASL Recommendations on Treatment of Hepatitis C 2018. J. Hepatol. 69, 461–511 (2018). [DOI] [PubMed]
- 56.Foster TL, et al. Resistance mutations define specific antiviral effects for inhibitors of the hepatitis C virus p7 ion channel. Hepatol. 2011;54:79–90. doi: 10.1002/hep.24371. [DOI] [PubMed] [Google Scholar]
- 57.Chipot C, et al. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem. Rev. 2018;118:3559–3607. doi: 10.1021/acs.chemrev.7b00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffin S. D. C. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. Journal of General Virology. 2004;85(2):451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 59.Segrest JP, De Loof H, Dohlman JG, Brouillette CG, Anantharamaiah GM. Amphipathic helix motif: classes and properties. Proteins. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 60.Cordes FS, Bright JN, Sansom MSP. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 2002;323:951–960. doi: 10.1016/S0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 61.Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. Eur. Biophys. J. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W, et al. The Unusual Transmembrane Partition of the Hexameric Channel of the Hepatitis C Virus. Structure. 2018;26:627–634.e4. doi: 10.1016/j.str.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalita MM, Griffin S, Chou JJ, Fischer WB. Genotype-specific differences in structural features of hepatitis C virus (HCV) p7 membrane protein. Biochim. Biophys. Acta. 2015;1848:1383–1392. doi: 10.1016/j.bbamem.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staley JP, Kim PS. Formation of a native-like subdomain in a partially folded intermediate of bovine pancreatic trypsin inhibitor. Protein Sci. 1994;3:1822–1832. doi: 10.1002/pro.5560031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 66.Cook GA, Stefer S, Opella SJ. Expression and purification of the membrane protein p7 from hepatitis C virus. Biopolymers. 2011;96:32–40. doi: 10.1002/bip.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Claridge JK, Schnell J. Bacterial production and solution NMR studies of a viral membrane ion channel. Methods Mol. Biol. (Clifton, N.J.) 2012;831:165–79. doi: 10.1007/978-1-61779-480-3_10. [DOI] [PubMed] [Google Scholar]
- 68.Soffe N, Boyd J, Leonard M. The Construction of a High-Resolution 750 MHz Probehead. J. Magn. Res. Series A. 1995;116:117–121. doi: 10.1006/jmra.1995.1197. [DOI] [Google Scholar]
- 69.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 70.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 71.Keller, R. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. 1–1 49 (ETH Zurich Thesis, 2004).
- 72.Kay, L. E., Torchia, D. A. & Bax, A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry (1989). [DOI] [PubMed]
- 73.Cavanagh, J. et al. Protein NMR Spectroscopy. (Academic Press, 2010).
- 74.Markley JL, et al. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): new policies affecting biomolecular NMR depositions. J. Biomol. NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maltsev AS, Ying J, Bax A. Deuterium isotope shifts for backbone H, N and C nuclei in intrinsically disordered protein α-synuclein. J. Biomol. NMR. 2012;54:181–191. doi: 10.1007/s10858-012-9666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S, et al. CHARMM36 united atom chain model for lipids and surfactants. J. Phys. Chem. B. 2014;118:547–556. doi: 10.1021/jp410344g. [DOI] [PubMed] [Google Scholar]
- 77.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 78.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. doi: 10.1063/1.470648. [DOI] [Google Scholar]
- 80.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. doi: 10.1063/1.463137. [DOI] [Google Scholar]
- 81.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 82.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 83.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/A:1008276004875. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, Y.-Z. Protein and peptide structure and interactions studied by hydrogen exchanger and NMR. 1–117 (University of Pennsylvannia Thesis, 1995).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.