Abstract
Integrons are genetic elements consisting of a functional platform for recombination and expression of gene cassettes (GCs). GCs usually carry promoter-less open reading frames (ORFs), encoding proteins with various functions including antibiotic resistance. The transcription of GCs relies mainly on a cassette promoter (PC), located upstream of an array of GCs. Some integron GCs, called ORF-less GCs, contain no identifiable ORF with a small number shown to be involved in antisense mRNA mediated gene regulation. In this study, the promoter activity of ORF-less GCs, previously recovered from the oral metagenome, was verified by cloning them upstream of a gusA reporter, proving they can function as a promoter, presumably allowing bacteria to adapt to multiple stresses within the complex physico-chemical environment of the human oral cavity. A bi-directional promoter detection system was also developed allowing direct identification of clones with promoter-containing GCs on agar plates. Novel promoter-containing GCs were identified from the human oral metagenomic DNA using this construct, called pBiDiPD. This is the first demonstration and detection of promoter activity of ORF-less GCs from Treponema bacteria and the development of an agar plate-based detection system will enable similar studies in other environments.
Subject terms: Bacterial genetics, Bacterial genetics
Introduction
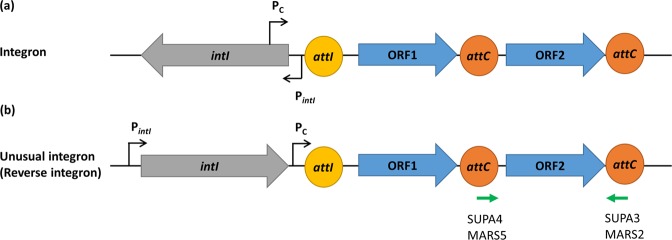
Integrons are bacterial genetic elements able to integrate and express genes present on gene cassettes (GCs)1–3. They consist of two main components; a functional platform and a variable array of GCs. The functional platform, located on the 5′ end of an integron, consists of an integrase gene (intI), and its promoter (PintI), an attI recombination site and a constitutive cassette promoter (PC) for the expression of GCs4. IntI is a site-specific tyrosine integrase that catalyses the insertion and excision of GCs via recombination mainly at attI and the attC, the latter located on circularised GCs. The integrase gene; intI, is normally transcribed in the opposite direction to GCs within an integron (Fig. 1a). However, some integrons have integrase genes transcribed in the same directions as their GCs. These are called unusual integrons or reverse integrons (Fig. 1b), and have been identified in Treponema denticola, Acinetobacter baumannii, Chlorobium phaeobacteroides and Blastopirellula marina5,6.
Figure 1.
A generalised structure of (a) usual integrons and (b) unusual, or reverse integrons. The green arrows indicate the primer binding sites on the unusual integron structure of T. denticola. The grey and blue open arrowed boxes represent integrase gene (intI) and the open reading frames (ORFs), respectively, pointing in the direction of transcription. The promoters, PintI and PC, were represented by black arrows. The recombination sites, attI and attC, were represented by yellow and orange circles, respectively.
The second part of an integron is an array of GCs. Each usually contains a single promoterless open reading frame (ORF) and an attC recombination site7. The proteins encoded by GCs are diverse in which most GCs are either have no known homologues in the database or predicted to encode hypothetical proteins, while the remaining showed homology to proteins associated with antibiotic resistance, virulence, metabolism and etc.2,8. When a GC is excised from integron, it forms a non-replicative mobile genetic element, which can be a substrate for integrase mediated recombination between attI (on the integrons) and attC (on the circular GC). This directionality of recombination is favoured over attC:attC recombination, resulting in the usual insertion of a newly integrated GC immediately next to the PC promoter in the first position of the GC array, ensuring maximal expression9–11.
The expression of integron integrases is controlled via the SOS response; there is a LexA-binding site located in the PintI12. In the absence of stress, the transcriptional repressor LexA binds to PintL and prevents the transcription of intI. The SOS response is activated upon the accumulation of single-strand DNA (ssDNA), generated during DNA damage, DNA repair, transformation, conjugation and certain antibiotic exposure e.g. trimethoprim and fluoroquinolones13–15. RecA recognises ssDNA and polymerises into RecA nucleofilaments, which induce autocleavage of LexA, releasing PintI from repression and allowing intI transcription12,16. By controlling the expression of IntI, bacteria can reshuffle their GCs at the precise moments of need (stress), generating genetic diversity and rapid adaptation to selective pressures, thereby avoiding accumulation of random recombination events that could be deleterious to the host cell17–19.
As most of the GCs do not contain a promoter, their expression usually relies on the PC promoter. The level of expression of GCs varies depending on the distance from PC, as the strength of expression decreases when GCs are located further from PC20. This ensures that a recently acquired GC will be immediately expressed. There are also some GCs that contain their own promoters, ensuring constitutive expression of their genes regardless of the PC promoter and their position within the integron array; examples include cmlA1 (chloramphenicol resistance), qnrVC1 (quinolone resistance), ere(A) (erythromycin resistance) and many of the GCs encoding toxin-antitoxin (TA) systems21–24.
Integron GCs have been identified from environments such as soils, marine sediments, seawater and more recently from human oral metagenomes25–29. In our previous study on the detection of integron GCs in the human oral metagenome, we found 13 ORF-less GCs out of 63 identified GCs (20%)29. ORF-less GCs have been shown to encode regulatory RNAs, for example the trans-acting small RNA (sRNA)-Xcc1, encoded by the ORF-less GC of a Xanthomonas campestris pv. campestris integron, which is involved in regulation of virulence30. Whilst promoter activity of ORF-less GCs has been discussed, this has not been experimentally demonstrated8.
In this study, we performed in silico analysis to identify promoter sequences in the GCs identified in our previous study on the oral metagenome. Promoter activity was experimentally determined by cloning the selected GCs upstream of the gusA reporter gene and measuring β-glucuronidase enzyme activity. Furthermore, we devised a GC-based promoter detection strategy utilising PCR and subsequent cloning between divergently orientated reporter genes. With this system, the successful cloning of amplicons from promoter-containing GCs can be visualised directly on agar plates, allowing the direct isolation of GC PCR amplicons with promoter activity from metagenomic DNA.
Results
Determination of promoter activity of the ORF-less GCs using the β-glucuronidase assay
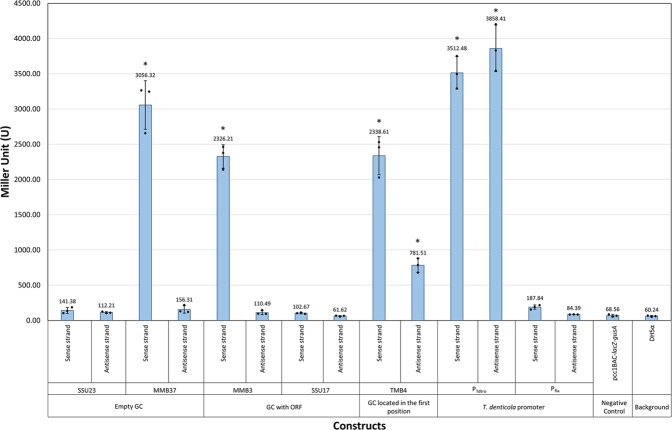
Among 63 GCs previously identified from human oral metagenomic DNA, 13 were predicted to be ORF-less GCs29. Five GCs were chosen for experimental expression analysis. GC TMB4 (amplified with primers targeting intI and attC) was selected as it is ORF-less and located in the first position of the integron array29. ORF-less GCs MMU23 and MMB37 were selected as they had the highest overall score predicted by BPROM promoter prediction software (Supplementary Table S1). Finally, GCs SSU17 and MMB3 were selected as controls, to represent GCs with an ORF. In this study, we have defined the sense strand as the same strand containing the PC promoter (Fig. 1).
As BPROM predicted putative promoter sequences on both strands, promoter activity of the selected GCs was determined by directionally cloning upstream of a promoterless β-glucuronidase (gusA) gene on pCC1BAC-lacZα-gusA (Fig. 2) in both directions. As the selected GCs were likely derived from Treponema spp., two experimentally verified T. denticola promoters, PTdTro and PFla, were also included as controls showing that T. denticola promoters can be recognised in our E. coli host31,32. PFla and PTdtro were selected as they rely on different sigma factors. PTdtro is recognised by sigma factor 70 (σ70) that is responsible for the transcription of most genes during growth in both E. coli and Treponema spp.31,33, while PFla is recognised by sigma factor 28 (σ28), involved in the expression of flagella-related genes in motile bacteria32,34. This will determine the limitations of our assay in recognising promoters associated with different types of sigma factors.
Figure 2.
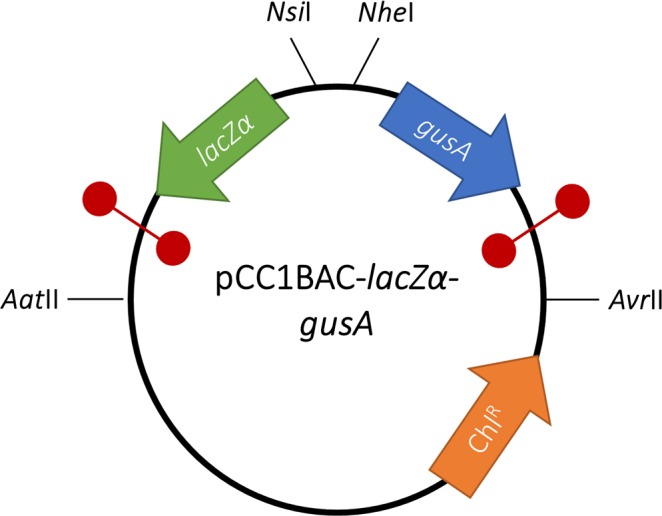
The structure of pCC1BAC-lacZα-gusA plasmid. The green, blue and orange open arrowed boxes represent lacZα, gusA and chloramphenicol resistance gene, respectively, pointing in the direction of transcription. The black lines indicate the position of restriction sites on the plasmid. The red circles indicate bidirectional transcriptional terminators.
The results showed that MMB37 and MMB3 had promoter activity on one strand, while MMU23 and SSU17 had no promoter activity, compared to the negative control, while the TMB4 GC showed promoter activities on both strands (Fig. 3). The PTdtro from T. denticola showed strong promoter activity on both sense and antisense strands, while PFla showed no promoter activity, suggesting that σ70 promoters, but not σ28 promoters, from T. denticola can be recognised by E. coli.
Figure 3.
The promoter activity of ORF-less GCs estimated by β-glucuronidase enzyme assays. Error bars indicate the standard errors of the means from three replicates. The scatter plots indicate the result from each replicate and the average Miller units for each construct were shown above the bars. The asterisks (*) indicate the constructs were statistically significantly different from the negative control group (pCC1BAC-lacZα-gusA) with the p-value < 0.05 by using ordinary one-way ANOVA followed by Dunnett’s multiple comparison tests.
Determination of synergy between PC and the ORF-less promoter GC in first position using the β-glucuronidase assay
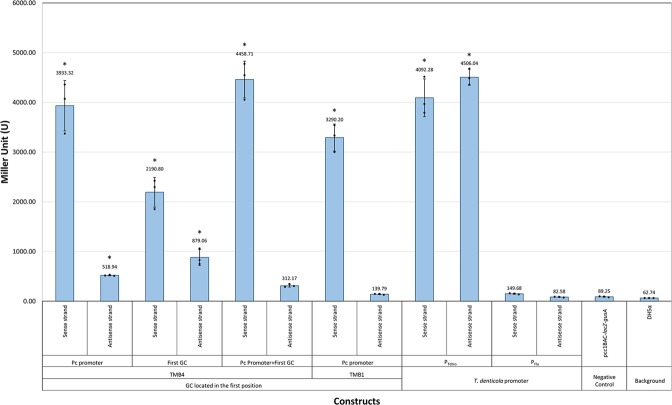
Previously, coupling PC with another promoter has been shown to significantly increase the expression of GCs, such as the presence of a second promoter (P2) downstreaming from PC and the presence of two PC in class 2 integrons (PC2A and PC2B), could result in a significantly higher expression of GCs35–37. As the promoter activities of the TMB4 GC, located in the first position of GC array, were confirmed in the previous section, another two constructs (TMB4 PC promoter and TMB4 PC-GC constructs) were therefore constructed to determine the synergy effect between PC and the ORF-less promoter GC. As the TMB4 PC promoter was not identical to the PC of T. denticola integron38, the PC of another integron GC; TMB129, which was identical to it39, was also included.
The results show that the TMB4 PC and TMB4 PC-GC showed promoter activities on both directions. However, coupling promoter GC in the first position (TMB4 PC-GC) did not significantly increase the expression of reporter genes, compared to the presence of only TMB4 PC (p-value > 0.99 by using ordinary one-way ANOVA followed by Bonferroni’s post-hoc) (Fig. 4). As the PC promoter sequences on TMB1 and TMB4 samples were different at several nucleotides, it was shown that TMB4-PC had higher promoter activities than the TMB1-PC in both directions (Fig. 4).
Figure 4.
The synergy effect between PC and the TMB4 ORF-less GC in the first position estimated by β-glucuronidase enzyme assays. Error bars indicate the standard errors of the means from three replicates. The scatter plots indicate the result from each replicate and the average Miller units for each construct were shown above the bars. The asterisks (*) indicate the constructs were statistically significantly different from the negative control group (pCC1BAC-lacZα-gusA) with the p-value < 0.05 by using ordinary one-way ANOVA followed by Dunnett’s multiple comparison tests.
Detection of promoter-containing GCs from oral metagenome
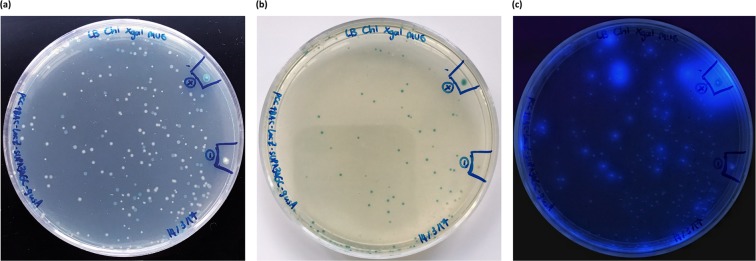
The pCC1BAC-lacZα-gusA plasmid, developed for the above enzymatic assay, had the potential to be used in an agar plate-based detection strategy to detect amplified integron GCs with promoter activity on either strand of DNA. This construct is called the Bi-Directional Promoter Detection plasmid (pBiDiPD). To verify the utility of pBiDiPD, and also to detect novel GCs containing promoter sequences in the human oral metagenome, integron GCs were amplified with SUPA4-NsiI/SUPA3-NheI and MARS5-NsiI/MARS2-NheI primers29, and cloned into pBiDiPD. The clones with GCs containing a promoter on the sense strand showed blue fluorescence when visualised under UV light, reflecting the activity of β-glucuronidase enzymes catalysing MUG to yield the blue-fluorescent 4-methylumbelliferyl. Clones with promoter activity on the antisense strand resulted in blue colonies as a result of β-galactosidase enzymes catalysing X-Gal into a blue insoluble pigment 5,5′-dibromo-4,4′-dichloro-indigo (Fig. 5).
Figure 5.
The detection of the integron GCs by using pBiDiPD. (a) pBiDiPD Transformants on LB agar supplemented with chloramphenicol, X-gal/IPTG and 4-methylumbelliferyl β-D-glucuronide (MUG), (b) Blue-white screening to detect for the clones with promoter activity on the antisense strand, (c) Exposing the colonies under the UV light to detect clones with promoter activity on the sense strand. The positive (+) and negative (−) colonies were the E. coli containing pCC1BAC-lacZα-TMB4-PC-gusA (with experimentally proven promoter activities on either strand of DNA and pCC1BAC-lacZα-gusA (no promoter activity), respectively.
After screening clones from both amplicon libraries (amplified with SUPA3-SUPA4 and MARS2-MARS5 primers), described in materials and methods, 23 different GCs with promoter activities were identified (Table 1 and Supplementary Table S2). Fourteen of these were similar to the GCs identified in the previous study with >86% nucleotide identity29. Among the recovered promoter-containing GCs, 9 out of 23 were novel including sample SSU-Pro-20, SSU-Pro-27, SSU-Pro-32, SSU-Pro-46, SSU-Pro-65, MMU-Pro-5, MMU-Pro-24, and MMU-Pro-53.
Table 1.
Summary of human oral integron GCs containing promoter sequences detected by pBiDiPD.
| Characteristics | Number of unique gene cassettes | ||||
|---|---|---|---|---|---|
| Primers SUPA3-SUPA4 | Primers MARS5-MARS2 | Total | |||
| Sequence analysis of GC amplicons | Number of screened GC amplicons | 62 | 58 | 120 | |
| Discarded amplicons | Chimeric amplicons | 23 | 11 | 34 | |
| Incompete GCs | 13 | 2 | 15 | ||
| Number of GCs followed all criteria | 26 | 45 | 71 | ||
| Number of unique GCs with promoter activities | 12 | 11 | 23 | ||
| Homologues | GCs found in previous study29 | 7 | 7 | 14 | |
| Novel GCs | 5 | 4 | 9 | ||
| Orientations and Identities of GCs | ORF-less GCs | 1 | 6 | 7 | |
| Toxin-Antitoxin GCs | 10 | 3 | 13 | ||
| Others | — | 3 | 3 | ||
| Directions of promoters | Sense strand (gusA) | 11 | 3 | 14 | |
| Antisense strand (lacZ) | — | 3 | 3 | ||
| Both directions (gusA and lacZ) | 1 | 5 | 6 | ||
The GCs can be categorised into two groups, one predicted to encode toxin-antitoxin systems in 12 out of 23 GCs, including plasmid stabilization protein (toxin)-prevent-host-death protein (antitoxin), BrnT (toxin)-BrnA (antitoxin), VapC (toxin)-AbrB/MazE/SpoVT family protein (antitoxin), RelE/ParE family (toxin)-XRE transcriptional regulator (antitoxin). The second group contained ORF-less GCs, which could be found in 7 samples, all reported in the previous study, except sample MMU-Pro-53. Most of the samples (14 out of 23 GCs) showed the promoter activity only on the sense strand. Samples with promoter activity only on the antisense strand were MMU-Pro-6, MMU-Pro-63, and MMU-Pro-65, while 6 out of 23 GCs exhibited promoter activity on both strands.
Discussion
Integrons are important disseminators of antimicrobial resistance genes in which more than 130 distinct GCs carrying antimicrobial resistance genes, covering most classes of antibiotics, have been identified40,41. Therefore, it is important to understand the diversity of GCs and how their expression is controlled. Even though ORF-less GCs have been found in the previous studies25,28,29,42,43, their functions have not been fully understood. Our findings here confirmed the promoter activities of ORF-less GCs from Treponema spp., which could be important for the expression of other GCs in integrons.
In this study, we determined promoter activity from GCs isolated by PCR from metagenomic DNA by measuring promoter activity from multiple GC containing constructs. As the ORF-less GCs were recovered from the oral metagenome, there is little information regarding the original host. Therefore, we chose to test the promoter activities by using an E. coli surrogate. Nucleotide sequence analysis suggested that these GCs were likely to be derived from Treponema spp., therefore, the ability of E. coli to utilise T. denticola promoter sequences was determined by including the experimentally verified T. denticola promoter, PTdTro31 which showed high activity on both sense and antisense strands, providing confidence that E. coli could be used. However, as no promoter activity was detected from PFla, it suggested that our enzymatic assay cannot detect promoters associated with σ28 from Treponema spp., which could be due to an inability for the E. coli host to recognise the Treponema σ28 promoter sequence. Therefore, constructs with no promoter activity in our enzymatic assay could also carry Treponema promoters associated with other sigma factors that cannot be recognised by the E. coli host like the σ28 PFla promoter.
Promoter activities of the ORF-less GCs were confirmed and quantified by using a β-glucuronidase enzymatic assay. This is the first time that the promoter activity of ORF-less GCs has been demonstrated in vitro, as shown by the activity on the sense strand of the MMB37 and both strands of the TMB4. A study on the Vibrio integron, containing a 116-cassette array, showed that most of the GCs are transcribed44. Therefore, ORF-less GCs could be responsible for transcription of the other GCs not transcribed by PC.
For the TMB4 GC (ORF-less GC in the first position), it was initially hypothesised that the promoter could help increase the expression of the downstream GCs. The constructs of TMB4 PC and TMB4 PC + GC were therefore included in the assay to determine whether having a promoter GC at the first position could help promote the expression of downstream GCs. However, the presence of promoter sequences in TMB4 GC did not significantly increase the expression of gusA. The lack of additive promoter activity can be explained by more competition for enzymes involved in transcription such as RNA polymerases (RNAP) or sigma factors to be available for transcription from each promoter, resulting in a lower transcriptional level45. Another, not mutually exclusive possibility is transcriptional interference (TI) between the four promoters on the TMB4 Pc + GC construct. We have experimentally shown promoter activity of TMB4 PC and TMB4 GC constructs on both strands, indicating convergent TI is a possibility.
In integrons, PC is in intI, which is convergent to the integron integrase promoter PIntI downstream (Fig. 1), resulting in TI. The TI between PC and PIntI has been shown to control the expression of integrase and the subsequent recombination of GCs. The weaker strength of PC could result in higher expression of integrase, which increases recombination of GCs46,47.
Promoter activity from the antisense strand in the first position of an integron, could potentially increase the expression of intI in integrons depending on the patterns of TI. Convergent expression between PC and a reverse GC promoter in an integron could relieve the repression of the PintI due to TI between PintI and PC. This could increase recombination at attI due to more integrase being produced catalysing the recruitment of a new GC at the first position (Supplementary Fig. 1). With reverse integrons, a relationship with increased expression of the integrase with a promoter cassette in the first position is more difficult to envisage. However, based on our enzymatic assay, it was shown that, in the presence of TMB4 GC, there was a decrease in antisense promoter activity in TMB4 (compared to TMB4 PC), therefore, it could result in lower TI with PintI, resulting in a higher expression of intI and subsequent catalysis of recruitment of a new GC to the first position (Supplementary Fig. 2).
Previously, only a few of ORF-less GCs have been reported in the first position of integrons. For example, only 1 out of 42 GCs and 1 out of 5 GCs in the first GC position were identified as ORF-less GCs in marine28 and oral metagenomes29, respectively. This could be because when ORF-less GCs with antisense promoters are located in the first position, integrase expression and the recombination events are increased, leading to an insertion of new GCs into the first position. When the new GC is inserted in the first position, it will push the promoter GC further down the array, which will lead to previous, lower levels of integrase expression, preserving the new GC in the first position.
As integrons contain highly diverse GCs that can be shuffled to different position within their GC arrays, ORF-less GCs, other than TMB4 GC, could be shuffled to the first position next to PC. Therefore, there is a possibility for some ORF-less GCs act in synergy with their cognate PC when they are shuffled into the first position and some, such as the one we have tested in TMB4 do not.
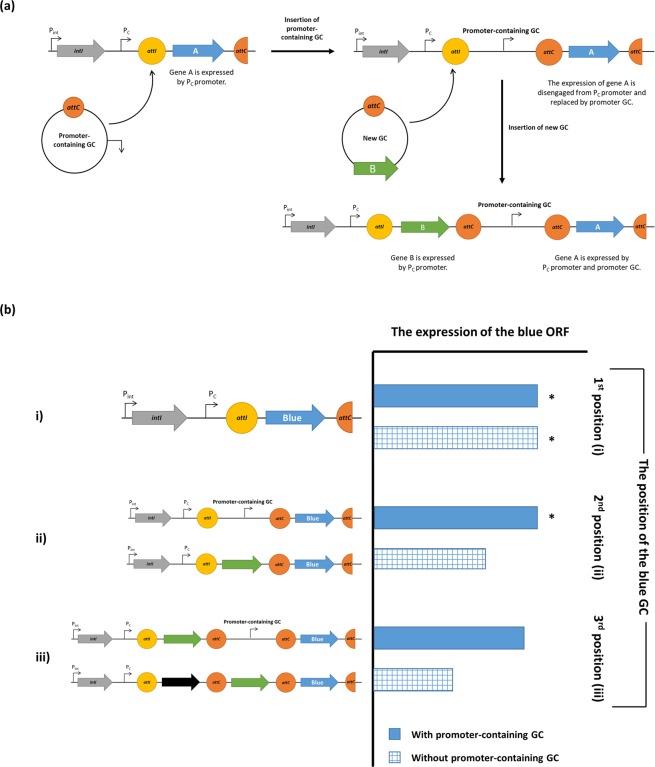
The expression level of cassette genes located further down in the array normally decreases due to the formation of a stem-loop structure on mRNA at attC sites, which impede the progression of the ribosome48. It was previously shown that the level of streptomycin resistance was reduced four-fold, when the aadA2-containing GC was located in the second position49. However, our data shows that the insertion of an ORF-less, promoter-containing GC in the first position did not decrease the gusA expression significantly (considered as a proxy for the expression of gene(s) in the second GC), i.e. comparing the data for TMB4 PC and TMB4 PC + GC. Therefore, we hypothesised that promoter-containing GCs could act as a genetic clutch, where the expression of the original first GC is partially disengaged from the PC promoter and replaced by the one on the ORF-less promoter containing GC (Fig. 6a). This can prevent a significant change in expression of the first GC while a new, first GC is sampled from the pool of GCs in order to adapt to an additional stress concurrent with the selective pressure requiring expression of the first GC. This system would work as a genetic clutch with the insertion of any GC containing a promoter in the same direction as PC, so it could be the insertion of either ORF-less GCs such as TMB4 GC, or other promoter-containing GCs such as the multiple TA-containing GCs we have identified; providing another selective advantage to retaining them and explaining their varied position within the GC array.
Figure 6.
The proposed genetic clutch. (a) When a promoter-containing GC inserts into the first position, it can act as a genetic clutch by disengaging the original first GC (blue arrow) from PC promoter and replaced with the one on promoter GC. When a new GC (green arrow) inserts, it can be expressed by PC promoter, while the blue GC is expressed by promoter-containing GC and PC promoter. (b) The expression level of gene cassettes with and without a genetic clutch. The estimated levels of expression of the blue ORF in (i) the first, (ii) the second and (iii) the third position were shown in the bar chart. The solid bars represent the situation when promoter-containing GC was inserted upstream of the blue GC, while the gridded bars represent the situation when no promoter-containing GC was inserted. The asterisks indicate the experimentally verified expression level, suggested by the results in Fig. 4 (TMB4 PC and TMB4 PC + GC). The expression of the blue ORF was hypothesised to be decreased when more GCs are inserted without the presence of a promoter-containing GC as a genetic clutch (gridded bars), based on the data from previous study49.
A genetic clutch within an integron can be of benefit to bacteria when they are exposed to multiple environmental stresses such as two different antibiotics simultaneously. The first resistance gene (green ORF in Fig. 6biii) can be expressed by the PC promoter, while the second resistance gene (blue GC), located in the third position, is expressed by PC and the promoter GC. Therefore, allowing bacteria to survive in the presence of both drugs.
As the other ORF-less GC MMU23 showed no promoter activity it may have other functions or carry a promoter that can be recognised in its native host but not in E. coli, or require other sigma factors. For the ORF-containing GC MMB3 sample, the promoter activity was found on the sense strand. This GC was predicted to carry toxin-antitoxin (TA) ORFs, including the PIN toxin and ribbon-helix-helix antitoxin domains, which were shown to contain their own promoter. Sample SSU17 and MMU23, which showed no promoter activity, can be considered as a control; illustrating that not all of GCs amplified from the oral metagenome exhibited promoter activity within our assay.
The pCC1BAC-lacZα-gusA plasmid, developed for the enzymatic assay, also had potential to be used for the detection of promoter activity in either direction from GCs. To verify the application of pCC1BAC-lacZα-gusA plasmids as promoter detection system, integron GCs were amplified from the human saliva metagenome by using SUPA3-SUPA4 and MARS2-MARS5 primers, which were the only two primer pairs that were verified for the amplification of integron GCs from the oral metagenome29. After cloning the amplified GCs between both reporter genes, two groups of GCs were identified with promoter activities: ORF-less GCs and TA-containing GCs. By detecting 7 clones containing ORF-less GCs with promoter activity it further supported that one of the functions of ORF-less GCs in integrons is to provide promoter activities.
TA-containing GCs are abundant in sedentary chromosomal integrons (SCIs), which were suggested to have a role in preventing random deletion of GCs and stabilising the large arrays SCIs23,50–52. TA systems normally encode a stable toxin and a labile antitoxin53, therefore TA cassettes have to carry their own promoters to ensure their expression. These were found in SCIs of Treponema spp., such as the HicA-HicB TA-containing GC in the fourth position within the GC array (Accession number NC_002967) in the SCI from T. denticola38. As most of the GCs amplified with our primers were homologous with Treponema spp., these TA-containing GCs should be present in our oral metagenome and were detected by our pBiDiPD based on their promoter activities.
Two of the GCs, SSU-Pro-9 and MMU-Pro-18, were similar to the MMB3 and MMB37 GCs, respectively, which were shown by the β-glucuronidase enzyme assay to have promoter activity on the sense strand. The phenotypes of SSU-Pro-9 and MMU-Pro-18 colonies also showed only a blue fluorescence phenotype, reflecting the promoter activity on the sense strand, which corresponded with the enzymatic assay results of MMB3 and MMB37.
To summarise, the promoter activities of the Treponema ORF-less integron GCs were experimentally demonstrated by using a robust β-glucuronidase enzyme assay, confirming that one of the functions of ORF-less GCs is to provide promoters for the expression of ORF containing GCs, in addition to expression from PC. This could be extended to ORF-less GCs from other bacteria, which should be determined further. The dual reporter plasmid; pBiDiPD, was developed for the direct visualisation of clones containing gene cassettes with promoter activity on agar plates. This can be applied as a detection system for promoter activity for any other DNA fragments.
Materials and Methods
In silico analysis of the human oral cavity gene cassettes and the construction of pCC1BAC-lacZα-GC-gusA constructs
All of the ORF-less GCs and some of the GCs containing ORFs, identified in the previous study29, were predicted for putative promoter sequences by using the web-based software BPROM in the Softberry package39.
Construction of pUC19-GC-gusA and pCC1BAC-lacZα-GC-gusA constructs
To determine the promoter activity of the selected GCs, the constructs were initially cloned into the EcoRI and KpnI restriction sites on pUC19-Ptet(M)-gusA plasmid54. The selected GCs were amplified from the pGEM-T easy vector containing the GC amplicon from a previous study29, as shown in Supplementary Fig. 3, by using primer listed in Supplementary Table S3.
Due to a significant difference in the plasmid copy number in some constructs of the pUC19-GC-gusA, new constructs were prepared based on a low copy number CopyControl™ pCC1BAC™ vector (Epicenter, UK) as it will be maintained in E. coli cell as one plasmid per cell and enable us to control the plasmid copy number to be similar between each construct. The construct was designed to contain two reporter genes, β-galactosidase lacZα and β-glucuronidase gusA genes (Fig. 2 and Supplementary Fig. 4). As lacZα on pCC1BAC contained T7 promoter sequence, it was first deleted by using Q5® Site-Directed Mutagenesis Kit (New England Biolabs, UK). The backbone of pCC1BAC was amplified with pCC1BAC-delLacZ-F1 and pCC1BAC-delLacZ-R1, and the amplified products were treated with a Kinase-Ligase-DpnI (KLD) enzyme mix, following the instructions from the manufacturer. The KLD-treated product was then transformed into E. coli α-Select Silver Efficiency competent cells (Bioline, UK) following the instructions from the manufacturer. The pCC1BAC-delLacZ plasmid was then extracted from E. coli by using QIAprep Spin Miniprep Kit (Qiagen, UK), following the manufacturer’s instructions.
The lacZα reporter gene was amplified from the pUC19 vector (New England Biolabs, UK) with LacZ-F1 and LacZ-R1 primers. For gusA reporter gene, it was amplified from pUC19-Ptet(M)-gusA with gusA-F1 and gusA-R1 primers. A bidirectional terminator, modified from lux operon, was added to LacZ-F1 and gusA-R1 primers, resulting in two bi-directional terminators flanking the lacZα-gusA reporter genes55. This was done to prevent transcriptional read-through from the promoter in the plasmid backbone and to also prevent promoters from the inserts interfering with the expression of genes on the plasmid backbone. The lacZα and gusA amplicons were digested with NsiI restriction enzymes (New England Biolabs, UK) and ligated together by using T4 DNA ligase (New England Biolabs, UK). The lacZα-gusA ligated product was directionally cloned into the pCC1BAC-delLacZ plasmid by digesting them with AatII and AvrII restriction enzymes and ligated together, resulting in pCC1BAC-lacZα-gusA plasmid.
The selected GCs were amplified from each pUC19-GC-gusA constructs by using primer listed in Supplementary Table S3. The amplicons were double digested with NsiI and NheI and directionally cloned into a pre-digested pCC1BAC-lacZα-gusA plasmid, then transformed into E. coli α-Select Silver Efficiency competent cells.
Determination of β-glucuronidase enzymatic activity
The β-glucuronidase enzymatic assay was performed to measure the promoter activity based on the expression of gusA, following the protocol described previously with some modifications56. The overnight cultures of E. coli containing the reporter constructs were prepared in LB broth supplemented with 12.5 µg/mL chloramphenicol. The OD600 of each overnight culture was measured. An aliquot of 1 mL of the overnight culture was centrifuged at 3000 × g for 10 min and discarded the supernatant. The cell pellets were incubated at −70 °C for 1 hr and resuspended in 800 µl of pH 7 Z buffer (50 mM 2-mercaptoethanol, 40 mM NaH2PO4·H2O, 60 mM Na2HPO4·7H2O, 10 mM KCl, and 1 mM MgSO4·7H2O) and 8 µl of toluene. The mixture was transferred to a 2 ml cryotube containing glass beads (150–212 μm in diameter) (Sigma, UK) and vortexed twice for 5 min each with an incubation on ice for 1 min in between. The glass beads were then removed by centrifugation at 3000 × g for 3 min. One-hundred microliters of cell lysate were mixed with 700 µl of Z-buffer, then incubated at 37 °C for 5 min. One-hundred sixty microliters of 6 mM ρ-nitrophenyl-β-D-glucuronide (PNPG) was then added to the reaction and incubated at 37 °C for 5 min. The reactions were stopped by adding 400 µl of 1 M Na2CO3 and centrifuged at 3000 × g for 10 min to remove cell debris and glass beads. The absorbance of the supernatant was measured with a spectrophotometer at the wavelength of 405 nm. Three biological replicates of the β-glucuronidase enzymatic assay were performed. The β-glucuronidase Miller units were calculated from57
Statistical analysis
The average and standard deviation of β-glucuronidase concentration were calculated from three biological replicates, which were used for the columns and error bars in Fig. 3, respectively. The statistical comparisons between the negative control (pCC1BAC-lacZ-gusA) to the other constructs were performed by using ordinary one-way ANOVA with either Dunnett’s post-hoc test (to compare each construct with a negative control) or Bonferroni’s post-hoc test (to compare constructs between themselves). The groups with statistically significantly difference from the control had the p-value of less than 0.05.
Recovery of promoter-containing GCs from the human oral metagenome
The integron GCs were amplified from the human oral metagenome by using as described previously with SUPA4-NsiI-SUPA4-NheI and MARS5-NsiI-MARS2-NheI primers29. The human oral metagenomic DNA was previously extracted from the saliva samples collected from 11 volunteers in the Department of Microbial Diseases, UCL Eastman Dental Institute29. The ethical approval for the collection and uses of saliva samples was obtained from University College London (UCL) Ethics Committee (Project number 5017/001). The written informed consents were obtained from all volunteers prior to the collection of saliva samples. All procedures performed in this study were in accordance with the ethical guidelines and regulations from the UCL Ethics Committee.
The amplified products were purified and digested with NsiI and NheI and ligated with the pre-digested pCC1BAC-lacZα-gusA plasmid. The ligated products were transformed into E. coli α-Select Silver Efficiency competent cells by heat shock. Cells were spread on LB agar supplemented with 12.5 µg/mL chloramphenicol, 80 µg/mL X-gal, 50 µM IPTG, and 70 µg/mL 4-methylumbelliferyl-β-D-glucuronide (MUG). After incubation at 37 °C for 18 hr, the colonies with β-galactosidase activity from lacZ was detected by blue-white screening on the agar plate, and the β-glucuronidase activity from gusA was visualisation under UV light. Colonies exhibiting either activity were selected and subcultured on fresh agar plates. The inserts were amplified by colony PCR using lacZ-F2 and gusA-F2 primers and sequenced by sequencing service from Genewiz (Genewiz, UK).
Sequence analysis and nomenclature of promoter-containing GC amplicons
DNA sequences were visualised and analysed by using BioEdit version 7.2.0 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The contigs from sequencing reactions were combined by using CAP contig function in the software58. The sequences were then matched to the nucleotide and protein database by using BlastN and BlastX (version 2.8.0) from the National Centre for Biotechnology Information (NCBI), respectively59. ORF finder (https://www.ncbi.nlm.nih.gov/orffinder) and BlastP (version 2.8.0) were used for the identification of ORFs in each GC60. The criteria for the sequence analysis of integron GC were the same as described in the previous study29. Artefactual PCRs were discounted by detecting the consensus R′ (1 R) core sites [GTTN1N2N3N4] and the complementary R″ (1 L) core sites [N′4N′3N′2N′1AAC] of attC located downstream from the attC forward primers and upstream from the attC reverse primers, respectively, where N and N′ are complmentary nucleotides and the number indicated their positions (Supplementary Table S4)29,61,62. While the conserved complimentary GTT/AAC triplet in each core site was essential, we accepted 50% non-complementarity within N1–N4. Two additional criteria for the verification of GCs detected with pCC1BAC-lacZα-gusA were included. Any clones containing incomplete GCs, caused by digestion at internal NsiI and NheI restriction sites on the GCs, were excluded from the dataset. Also chimeric inserts, which were the ligation products between digested amplicons, were also excluded.
The promoter-containing GCs were named as described in the previous study29. The first and second letters represented the forward primer and reverse primer used in the amplification. The third letter represents the source of the human oral metagenomic DNA which is U for the United Kingdom. This was followed by term “Pro”, indicating the presence of promoter activity, and the number of the clone. For instance, SSU-Pro-1 stands for the first clone amplified from the UK oral metagenome by using SUPA3 and SUPA4 primers. The sequences of these GCs were deposited in the DNA database with the accession number from MH536747 to MH536769.
Supplementary information
Acknowledgements
We would like to thank Drs Md. Ajijur Rahman (University of Rajshahi) and Azmiza Jasni (Universiti Putra Malysia) for plasmids and advice, and Prof Pål Johnsen and the MicroPop group (UiT The Arctic University of Norway) for helpful discussions.
Author Contributions
S.T. performed the experiments and wrote the initial draft of the manuscript, S.T. and A.P.R. conceived the study. All authors designed the experiments, analysed the data and contributed to the writing of the manuscript. All authors reviewed and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44640-2.
References
- 1.Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 1998;1:109–119. doi: 10.1016/S1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 2.Cambray G, Guerout AM, Mazel D. Integrons. Annual review of genetics. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 3.Michael CA, et al. Mobile gene cassettes: a fundamental resource for bacterial evolution. The American naturalist. 2004;164:1–12. doi: 10.1086/421733. [DOI] [PubMed] [Google Scholar]
- 4.Escudero, J. A., Loot*, C., Nivina, A. & Mazel, D. The Integron: Adaptation On Demand. Microbiology spectrum3, 10.1128/microbiolspec.MDNA3-0019-2014 (2015). [DOI] [PubMed]
- 5.Boucher Yan, Labbate Maurizio, Koenig Jeremy E., Stokes H.W. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends in Microbiology. 2007;15(7):301–309. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y-W, Doak TG, Ye Y. The gain and loss of chromosomal integron systems in the Treponema species. BMC evolutionary biology. 2013;13:1–9. doi: 10.1186/1471-2148-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazel Didier. Integrons: agents of bacterial evolution. Nature Reviews Microbiology. 2006;4(8):608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 8.Gillings MR. Integrons: past, present, and future. Microbiology and molecular biology reviews: MMBR. 2014;78:257–277. doi: 10.1128/mmbr.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collis CM, Hall RM. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Molecular microbiology. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 10.Partridge SR, et al. Definition of the attI1 site of class 1 integrons. Microbiology (Reading, England) 2000;146(Pt 11):2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 11.Collis CM, Recchia GD, Kim M-J, Stokes HW, Hall RM. Efficiency of Recombination Reactions Catalyzed by Class 1 Integron Integrase IntI1. Journal of bacteriology. 2001;183:2535–2542. doi: 10.1128/JB.183.8.2535-2542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerin E, et al. The SOS response controls integron recombination. Science (New York, N.Y.) 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 13.Baharoglu Z, Bikard D, Mazel D, Conjugative DNA. transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLOS Genetics. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baharoglu Z, Krin E, Mazel D. Connecting environment and genome plasticity in the characterization of transformation-induced SOS regulation and carbon catabolite control of the Vibrio cholerae integron integrase. Journal of bacteriology. 2012;194:1659–1667. doi: 10.1128/jb.05982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baharoglu Z, Mazel D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrobial agents and chemotherapy. 2011;55:2438–2441. doi: 10.1128/aac.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-D. [DOI] [PubMed] [Google Scholar]
- 17.Harms K, Starikova I, Johnsen PJ. Costly Class-1 integrons and the domestication of the the functional integrase. Mobile genetic elements. 2013;3:e24774. doi: 10.4161/mge.24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starikova I, et al. A trade-off between the fitness cost of functional integrases and long-term stability of integrons. PLoS Pathog. 2012;8:e1003043. doi: 10.1371/journal.ppat.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelstädter Jan, Harms Klaus, Johnsen Pål J. The evolutionary dynamics of integrons in changing environments. The ISME Journal. 2016;10(6):1296–1307. doi: 10.1038/ismej.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyne S, Guigon G, Courvalin P, Perichon B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrobial agents and chemotherapy. 2010;54:333–340. doi: 10.1128/aac.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stokes HW, Hall RM. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991;26:10–19. doi: 10.1016/0147-619X(91)90032-R. [DOI] [PubMed] [Google Scholar]
- 22.da Fonseca EL, Vicente AC. Functional characterization of a Cassette-specific promoter in the class 1 integron-associated qnrVC1 gene. Antimicrobial agents and chemotherapy. 2012;56:3392–3394. doi: 10.1128/aac.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Molecular microbiology. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 24.Biskri L, Mazel D. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrobial agents and chemotherapy. 2003;47:3326–3331. doi: 10.1128/AAC.47.10.3326-3331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokes HW, et al. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Applied and environmental microbiology. 2001;67:5240–5246. doi: 10.1128/aem.67.11.5240-5246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsaied H, et al. Novel and diverse integron integrase genes and integron-like gene cassettes are prevalent in deep-sea hydrothermal vents. Environmental microbiology. 2007;9:2298–2312. doi: 10.1111/j.1462-2920.2007.01344.x. [DOI] [PubMed] [Google Scholar]
- 27.Koenig JE, et al. Integron Gene Cassettes and Degradation of Compounds Associated with Industrial Waste: The Case of the Sydney Tar Ponds. PloS one. 2009;4:e5276. doi: 10.1371/journal.pone.0005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsaied H, et al. Marine integrons containing novel integrase genes, attachment sites, attI, and associated gene cassettes in polluted sediments from Suez and Tokyo Bays. The ISME journal. 2011;5:1162–1177. doi: 10.1038/ismej.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tansirichaiya S, Rahman MA, Antepowicz A, Mullany P, Roberts AP. Detection of novel integrons in the metagenome of human saliva. PloS one. 2016;11:e0157605. doi: 10.1371/journal.pone.0157605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X-L, et al. sRNA-Xcc1, an integron-encoded transposon- and plasmid-transferred trans-acting sRNA, is under the positive control of the key virulence regulators HrpG and HrpX of Xanthomonas campestris pathovar campestris. RNA Biology. 2011;8:947–953. doi: 10.4161/rna.8.6.16690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brett PJ, Burtnick MN, Fenno JC, Gherardini FC. Treponema denticola TroR is a manganese- and iron-dependent transcriptional repressor. Molecular microbiology. 2008;70:396–409. doi: 10.1111/j.1365-2958.2008.06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limberger, R. J., Slivienski, L. L., Izard, J. & Samsonoff, W. A. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. Journal of bacteriology181, 3743-3750 (1999). [DOI] [PMC free article] [PubMed]
- 33.Paget MSB, Helmann JD. The σ70 family of sigma factors. Genome Biology. 2003;4:203–203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koo B-M, Rhodius VA, Campbell EA, Gross CA. Mutational analysis of Escherichia coli σ28 and its target promoters reveal recognition of a composite–10 region, comprised of an “extended–10 motif” and a core-10 element. Molecular microbiology. 2009;72:830–843. doi: 10.1111/j.1365-2958.2009.06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papagiannitsis CC, Tzouvelekis LS, Miriagou V. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrobial agents and chemotherapy. 2009;53:277–280. doi: 10.1128/aac.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integron. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 37.Jové T, Da RS, Tabesse A, Gassama-Sow A, Ploy M-C. Gene Expression in Class 2 Integrons Is SOS-Independent and Involves Two Pc Promoters. Frontiers in microbiology. 2017;8:1499–1499. doi: 10.3389/fmicb.2017.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coleman N, Tetu S, Wilson N, Holmes A. An unusual integron in Treponema denticola. Microbiology (Reading, England) 2004;150:3524–3526. doi: 10.1099/mic.0.27569-0. [DOI] [PubMed] [Google Scholar]
- 39.Solovyev, V. & Salamov, A. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies (ed. Li, R. W.) 61–78 (Nova Science Publishers, 2011).
- 40.Deng Y, et al. Resistance integrons: class 1, 2 and 3 integrons. Annals of clinical microbiology and antimicrobials. 2015;14:45–45. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clinical microbiology reviews31, 10.1128/cmr.00088-17 (2018). [DOI] [PMC free article] [PubMed]
- 42.Boucher Y, et al. Recovery and evolutionary analysis of complete integron gene cassette arrays from Vibrio. BMC evolutionary biology. 2006;6:3. doi: 10.1186/1471-2148-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Shi L, Yang W, Li L, Yamasaki S. New array of aacA4-catB3-dfrA1 gene cassettes and a noncoding cassette from a class-1-integron-positive clinical strain of Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy. 2006;50:2278–2279. doi: 10.1128/aac.01378-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michael CA, Labbate M. Gene cassette transcription in a large integron-associated array. BMC genetics. 2010;11:82. doi: 10.1186/1471-2156-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh SS, et al. Widespread suppression of intragenic transcription initiation by H-NS. Genes & development. 2014;28:214–219. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jové T, Da RS, Denis F, Mazel D, Ploy M-C. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLOS Genetics. 2010;6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerin E, Jove T, Tabesse A, Mazel D, Ploy MC. High-level gene cassette transcription prevents integrase expression in class 1 integrons. Journal of bacteriology. 2011;193:5675–5682. doi: 10.1128/jb.05246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacquier H, Zaoui C, Sanson-le Pors MJ, Mazel D, Bercot B. Translation regulation of integrons gene cassette expression by the attC sites. Molecular microbiology. 2009;72:1475–1486. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 49.Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrobial agents and chemotherapy. 1995;39:155–162. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerout AM, et al. Characterization of the phd-doc and ccd toxin-antitoxin cassettes from Vibrio superintegrons. Journal of bacteriology. 2013;195:2270–2283. doi: 10.1128/jb.01389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome research. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loot, C. et al. Differences in Integron Cassette Excision Dynamics Shape a Trade-Off between Evolvability and Genetic Capacitance. mBio8, 10.1128/mBio.02296-16 (2017). [DOI] [PMC free article] [PubMed]
- 53.Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLOS genetics. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seier-Petersen MA, et al. Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. The Journal of antimicrobial chemotherapy. 2014;69:343–348. doi: 10.1093/jac/dkt370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swartzman A, Kapoor S, Graham AF, Meighen EA. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. Journal of bacteriology. 1990;172:6797–6802. doi: 10.1128/jb.172.12.6797-6802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Molecular microbiology. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 57.Miller, J. H. Experiments in molecular genetics. (Cold Spring Harbor, 1972).
- 58.Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/S0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Federhen S., Feolo M., Fingerman I. M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Lu Z., Madden T. L., Madej T., Maglott D. R., Marchler-Bauer A., Miller V., Mizrachi I., Ostell J., Panchenko A., Phan L., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Slotta D., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Wang Y., Wilbur W. J., Yaschenko E., Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2010;39(Database):D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cury J, Jové T, Touchon M, Néron B, Rocha EP. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic acids research. 2016;44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stokes HW, O’Gorman DB, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Molecular microbiology. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.