Abstract
Background
Accurate laboratory diagnosis of HIV is essential to reduce the risk of HIV-positive individuals transmitting HIV-1 infection. The goal of this study was to identify and assess a panel of host derived plasma miRNAs that could to serve as a prognostic and predictive biomarker to detect early/acute HIV-1 infection.
Methods
A total of 372 microRNAs were analyzed in nine plasma samples from HIV-1 infected individuals in the early phase of infection and three healthy controls using the miRNA PCR-array. Seventeen microRNAs were selected and validated in 80 plasma samples from HIV-1 infected individuals in the early phase of infection (20 each of eclipse stage, RNA+ stage, Ag + stage, and Ag + Ab+ stage of HIV-1 patients) and 25 healthy controls. Using the validation study results a plasma miRNA panel was developed and evaluated to detect early/acute HIV-1 infection in 49 blinded samples.
Finding
We identified an miRNA panel (PeHIV-1) containing four differentially expressed miRNAs (miR-16-5p, miR-20b-5p, miR-195-5p, and miR-223-3p) that could distinguish early HIV-1 infection from healthy controls with high AUC (1·000[1·00–1·00]), sensitivity (100%), and specificity (100%).We also found that miR-223-3p demonstrates 100% sensitivity and specificity (AUC 1·00[1·00–1·00]) and could distinguish eclipse stage of HIV-1 infection from healthy controls. To detect eclipse stage of HIV-1 infection we also developed a four-miRNA based (miR-16-5p, miR-206, let-7 g-3p, and miR-181c-3p) panel (PE) with AUC 0·999 (0·995–1·000), 100% sensitivity and 95·8% specificity.
Interpretation
The miRNA panel, PeHIV-1 is a potential biomarker for detecting early/acute stage of HIV-1infection and could help initiate early antiretroviral treatment, thus preventing the spread of HIV-1 infection.
Keywords: Early HIV-1, Acute HIV-1, Plasma miRNA and circulatory biomarker
Research in context.
Evidence before this study
We systematically searched PubMed, without date restriction or limitation to English language publications, for research articles with the following terms: “circulating” AND “miRNA” AND “HIV-1” AND “diagnosis” AND “detection”. This search identified only one study where miRNA expression profiles were determined in plasma from HIV elite controllers and chronically infected individuals. This study proposed that plasma miRNA profiling might be used as a diagnostic or a prognostic marker in studying HIV-1 pathogenesis and disease progression. The report did not verify the diagnostic performance of circulating miRNAs in the nine cases of chronic HIV-1 infection. Our search did not identify publications that have evaluated the diagnostic performance of circulating miRNAs to detect early-stage or acute stage or eclipse stage of HIV-1 infection.
Added value of study
We did a large-scale study to assess the performance of plasma miRNA for detection of early/acute HIV-1 infection. The results revealed that a four-miRNA based panel (PeHIV-1) had significantly higher sensitivity (100%) than any single miRNA to distinguish early HIV-1 infection from healthy controls. PeHIV-1 was also able to identify viral RNA-negative early stage HIV-1 infection. Importantly, the blinded panel validation study with blinded specimens showed that PeHIV-1 could correctly identify samples from early stage of HIV-1 infection in patients with undetectable viral markers (RNA and HIV-1 p24). Results with two unrelated viruses HBV and HCV indicate that the modulation of these miRNAs is specific to HIV-1 infection. However, with plasma samples from HIV-2 infected individuals considerable cross-reactivity was observed. These findings indicate that PeHIV-1 is a promising panel for the detection of early HIV-1 infection, especially for eclipse stage of infection. To our knowledge, this is the first attempt to assess the value of circulating miRNA for the detection of early stage (including eclipse stage) of HIV-1 infection.
Implication of all the available evidence
Our findings highlight the potential of circulating miRNA in the detection of both early and eclipse stage of HIV-1 infection, which may help to reduce window period, and facilitate early detection and initiation of therapy. Initiation of ART treatment during this stage would greatly reduce HIV-1 transmission.
Alt-text: Unlabelled Box
1. Introduction
Although low levels of viral markers (HIV-1 RNA and HIV-1 p24) might be present immediately after HIV-1 infection, these markers are consistently undetectable during this stage of infection. This stage is also termed as the eclipse stage or window period [1]. Generally, HIV-1 viral markers become detectable after 10 days post infection, when HIV-1 RNA becomes detectable by NAT assays [[2], [3], [4], [5], [6]]. Subsequently, after 15–22 days post infection HIV-1 p24 antigen can be detected. Acute HIV infection can be defined as the time from HIV acquisition until seroconversion. Acute HIV infection is a highly infectious phase where viral load often reaches peak levels, sometimes >1 × 107copies/mL [7]. Furthermore, the high viral load and the absence of antibody contribute disproportionately to HIV transmission [[8], [9], [10], [11]]. Next, immunoglobulin (IgG and IgM) antibodies are expressed which can be detected by 3rd and 4th generation immunoassays [[12], [13], [14], [15]]. There is a high possibility that false negative results could be detected during the window period of infection [16]. Accurate laboratory diagnosis of HIV is essential to reduce the risk of HIV-positive individuals transmitting HIV-1 infection. Thus, there is a need to add host derived prognostic and predictive biomarkers to the current diagnostic strategy which could be achieved with the panel of validated host microRNA (miRNA) biomarkers described in this report.
Individuals with acute HIV infection are often unaware of their infection and can be involved in high-risk behaviors facilitating HIV transmission. Therefore [17], early diagnosis is a key strategy for preventing HIV transmission. The Fiebig staging system provides a useful benchmark for prognostic definition and establishment of treatment strategies [18]. It was found that antiretroviral treatment (ART) initiated during the acute phase of HIV infection enhanced recovery of CD4+ T cells, preservation of immunity and reduction of HIV reservoir size [[19], [20], [21]] reduce epidemic spread [22], and potentially achieve long-term control of plasma viremia without the use of long-term antiviral treatment [23]. In addition to these benefits, ART initiated during acute and very early HIV infection rapidly decreases viral load and infectivity [24] leading to a reduction in transmission. In addition, increasing use of pre-exposure prophylaxis (PrEP) has resulted in undetectable levels of HIV in the blood of individuals who may present at blood and plasma donor centers without prior knowledge of their infection. Such persons may be classified as HIV negative based on undetectable viremia by NAT or antigen test results. Therefore, the identification of host miRNA biomarkers that are specific for HIV-1 infection would be of use in the early diagnosis of HIV-1 infection.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNAs that regulate gene expression post-transcriptionally. MiRNAs contribute to the pathophysiology of important human diseases [25,26]. MiRNAs can be detected outside the cell. Circulating miRNAs mirror physiological and pathophysiological conditions and have high stability in stored patient samples, allowing them to serve as biomarkers for various diseases [27]. Moreover, one of the most important advantages of using circulating miRNAs as biomarkers, apart from being easily measured in blood samples, is their remarkable stability in plasma and serum, where they are most likely protected from RNase degradation due to their binding to Argonaut proteins [28,29] and residing in microvesicles, exosomes, and microparticles [30]. Emerging evidence shows that circulating miRNAs could be biomarkers for infectious diseases, like Dengue, Ebola and HIV-1 [[31], [32], [33]]. Although a few investigations [33,34] have aimed to identify circulating miRNAs that distinguish individuals with HIV-1 infection from those who are free from HIV-1, most studies have had limitations, including too few miRNAs examined, a small study population, and no independent validation. So far, very few studies have screened and verified the diagnostic performance of circulating miRNAs in HIV-1 infection [[33], [34], [35]]. All previous studies reported the differential expression of host miRNAs in PBMCs from HIV-1 infected patients, but none assessed whether circulating miRNAs could detect acute/early-stage of the infection. In this study, our goal was to analyze miRNA expression profiles from acute/early stage of HIV-1 infected individuals to develop a miRNA-based predictor for this stage of infection.
2. Materials and methods
The methods for plasma sample preparation, RNA isolation, PCR arrays and quantitative real-time PCR are described in the supporting supplementary information.
2.1. Patient characteristics and study design
Nine seroconversion panels were obtained from SeraCare Life Sciences, Gaithersburg, MD USA (47 HIV-1 positive plasma samples) and two seroconversion panels from ZeptoMetrix Corp., Buffalo, NY USA (18 HIV-1 positive plasma samples). Two additional AccuSet HIV-1 performance panels were purchased from SeraCare Life Sciences, Gaithersburg, MD USA (41 HIV-1 positive plasma samples). All other HIV-1 plasma samples representing early infection were obtained from the American Red Cross Society Gaithersburg, MD USA (10 HIV-1 positive plasma samples), and Discovery Life Sciences Los Osos, CA USA (10 HIV-1 positive plasma samples). Samples from HIV-1 infected individuals in the early stage of infection were classified based on the presence of viral RNA, p24 antigen, antibody, and specific bands on Western blot and categorized as eclipse, RNA+, Ag+, Ag + Ab+/seroconverted, and acute HIV-1 infection/pre-seroconversion. All the eclipse stage samples had viral loads below the detection limit of the assay, antigen non reactive and antibody non reactive, RNA+ samples had a detectable viral load, antigen non reactive and antibody non reactive. Ag + samples had a detectable viral load, antigen reactive and antibody non reactive. Ag + Ab+ samples had a detectable viral load, antigen reactive, antibody reactive and Western blot indeterminant or positive without p31 bands. Acute HIV-1 infected samples were defined to include all samples until seroconversion (includes all eclipse, RNA+ and Ag + specimens, supplementary Table S1). Healthy donor plasma samples obtained from apparently healthy volunteers with no history of any infection or illness were provided by the National Institutes of Health (NIH) Blood bank, Bethesda, MD USA (40 plasma sample). All volunteers were confirmed to be healthy by physical examination and HIV-1, HBV, and HCV negative.
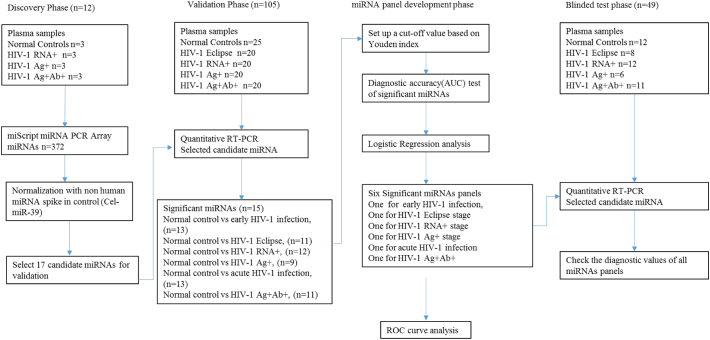
The study consisted of four phases (Fig. 1): discovery phase, validation phase, miRNA panel development phase and blinded test phase. In the discovery phase, samples were screened using miRNA PCR Arrays and potential miRNAs were identified and selected for further investigation. Candidate microRNAs were further evaluated in the validation phase. The validated miRNAs were grouped together to create a diagnostic panel and these miRNAs were evaluated as potential biomarkers. In addition, the predictive performance was investigated in the validation phase. In the blinded test phase, the clinical diagnostic performance of the miRNA diagnostic panels was tested with samples whose clinical diagnostic information was blinded to the investigators. After unblinding test results, clinical diagnostic information, sensitivity and specificity of the logistic models in discriminating early HIV-1, eclipse, RNA+, Ag+, acute HIV-1, and seroconverted, from healthy control subjects were calculated.
Fig. 1.
Study design. The microRNA (miRNA) profiles of 117 plasma samples from 89 HIV-1 infected individuals and 28 healthy controls were used to generate outcomes in 2 different phases. The candidate miRNAs identified in 12 plasma samples using miRNA PCR arrays were validated in 105 plasma samples using quantitative RT-PCR. The logistic regression and ROC curve analyses were performed in the validation panel development stage. The panels were further validated in the blinded test phase with 49 blinded plasma samples.
2.2. Identification of differentially expressed of host miRNAs
In the discovery phase, differential expression of host miRNAs was identified using miRNA PCR array (MIHS-3106Z, Qiagen, USA) with total RNA isolated from plasma samples from HIV-1 infected patients in the early phase of infection. A total of nine plasma samples from HIV-1 infected patients (three RNA+, three Ag+, and three from Ag + Ab+/seroconverted individuals) and three healthy normal controls were used. This PCR array allows for the analysis of 372 human miRNAs. The cycle threshold (Ct) values were analyzed using the Qiagen web based software. Because no standard reference miRNA has been established for normalization of circulating miRNAs in plasma, we had to first determine the normalization reference [36]. Although the miScript PCR Array provides SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A, and RNU6-6P as internal controls, which are often used for normalization for cellular miRNAs, these internal controls were not useful for normalizing the levels of circulating miRNAs, since the levels of these miRNAs were very low or exhibited a high degree of sample to sample variation. We therefore normalized the Ct values using cel-miR-39 as an external spike-in control. The normalized PCR array data sets were then analyzed using significant analysis to identify candidate miRNAs. To determine whether these candidate miRNAs could serve as effective biomarkers, we performed several pairwise comparisons (control vs RNA+, control vs Ag+, control vs Ag + Ab+/seroconverted, control vs acute HIV-1 infection and control vs early HIV-1). Seventeen miRNAs that were differentially expressed in these comparisons were chosen as potential candidates for further investigation.
In the biomarker validation phase, we assessed the relative levels of the 17 candidate miRNAs in 80 plasma samples from early stage of HIV-1 infected individuals (including 20 eclipse stage, 20 RNA+ stage, 20 Ag + stage, and 20 Ag + Ab+ stage), and 25 normal controls. In this phase, we included 20 eclipse stage plasma samples to identify potential biomarkers that could be used as diagnostic biomarkers to narrow the window period just after infection and between the first appearance of detectable levels of viral RNA. Plasma miRNAs from the validation stage were analyzed by quantitative real-time PCR with a non-human spike-in miRNA, cel-miR-39, as a reference. Relative quantitation was used and the levels of miRNA in plasma samples were normalized against the reference miRNA (cel-miR-39) and the results were presented as median-normalized Ct values.
2.3. miRNA panel construction and testing
To construct the miRNA panel that could differentiate between early HIV-1 infection and uninfected controls and categorize the different stages of HIV-1 infection compared to uninfected controls, the median-normalized Ct-value of the significant miRNAs was applied in the logistic regression analysis. The ideal miRNA panel, denoted as PeHIV-1, to differentiate early HIV-1 infected individuals from healthy controls was established using a logistical regression model. The predicted probability of categorizing samples as early HIV-1 infection by PeHIV-1 was calculated as: logit (p = early HIV-1) = 0·033 + 0·7*miR-223-3p + 0·7*miR-16-5p – 0·267*miR-195-5p – 0·167*miR-20b-5p. In this equation, the miRNA symbol was replaced with the discretized value one when the median normalized Ct value of the miRNA was higher than the corresponding cutoff; otherwise, it was replaced with the discretized value of zero when the median normalized Ct values of the miRNA was below the cutoff. If the result of logit [p = early HIV-1] was greater than or equal to 0·733, the detected sample was predicted to be in the early stage of HIV-1 infection.
The clinical diagnostic performance of the miRNA panel established in the validation phase was further tested in the blinded test phase. In this phase, the investigators were blinded to the clinical diagnostic test results of the forty nine plasma samples. After unblinding the clinical diagnostic information, sensitivity and specificity of the logistic models in discriminating early HIV-1 infection, acute HIV-1 infection, eclipse stage, RNA+ stage, Ag + stage, Ag + Ab+ stage from healthy subjects were calculated. The samples used in the validation phase and blinded test phase were independent of the discovery phase and were also independent of each other.
2.4. Statistical analysis
A basic power analysis calculation was performed to assess the control of type-II errors during miRNA screening and validation in plasma samples. Power calculations were conducted in the discovery stage with nine HIV-1 positive samples and three controls for different microRNAs using a 1·5fold change as the cut-off. In the validation stage, results from eighty HIV-1 positive plasma and twenty-five normal control individuals indicated that a 99% observed power for each miRNA was achieved.
Significance analysis of the data obtained from the PCR arrays was used to identify the differentially expressed miRNAs between HIV-1 infected individuals and healthy uninfected individuals. Student's t-test was used to compare differences in plasma miRNA expression levels between the HIV-1 infected group and the healthy uninfected group. Receiver-operator characteristic (ROC) curves and the area under the ROC curve (AUC) were used to assess the performance of the selected miRNAs to serve as diagnostic tools/biomarkers for detecting early-stage HIV-1 infection. The optimal cutoff point for the plasma miRNAs expression level was determined by Youden index [37]. Logistic regression was used to develop a linear combination of miRNAs. The AUC was used as an accuracy index for evaluating the diagnostic performance of the selected microRNA panels. All statistical analyses were performed using normalized data. Statistical analyses were performed using SPSS software, version 17·0 (SPSS Inc., Chicago, IL) and GraphPad Prism 5·0 (GraphPad Software, Inc., California). All statistical tests were two-sided, and a p-value <0·05 was considered statistically significant.
3. Results
In this study we have tested a total of 117 plasma samples from HIV-1 infected patients in early phase infection and normal controls to identify circulating miRNA that could serve as biomarkers for detecting early HIV-1 infection (Fig. 1). To identify potential plasma miRNA biomarkers specific for early HIV-1 infection, plasma RNA from three normal controls, and nine HIV-1 positive plasma samples in the early phase of infection (three each of RNA+ stage, Ag + stage, and Ag + Ab+ stage) were pre-amplified, and the expression levels of 372 miRNAs were profiled using the miScript miRNA PCR array (MIHS-3106Z, Qiagen, USA). We performed several pairwise comparisons in the individual 12 samples tested and identified the differentially expressed miRNAs levels between the groups (Supplementary Fig. S1 and Supplementary Table S2). Details about age, sex, race and other confounders that may influence the study results were not available for the plasma samples from HIV-1 infected individuals supplied by vendors and thus were not included in the analysis. Although the impact of confounders like age, sex, and race on miRNA expression is not known, many studies have shown that gender differences play a minimal role in modulating miRNA expression [[38], [39], [40]].
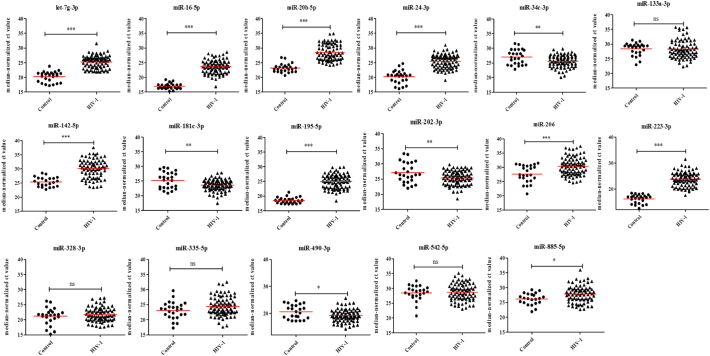
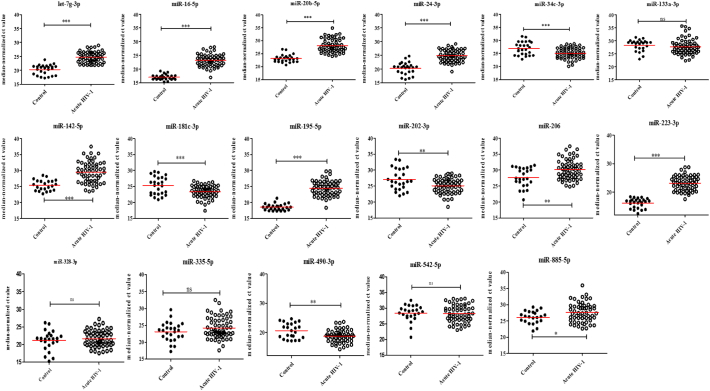
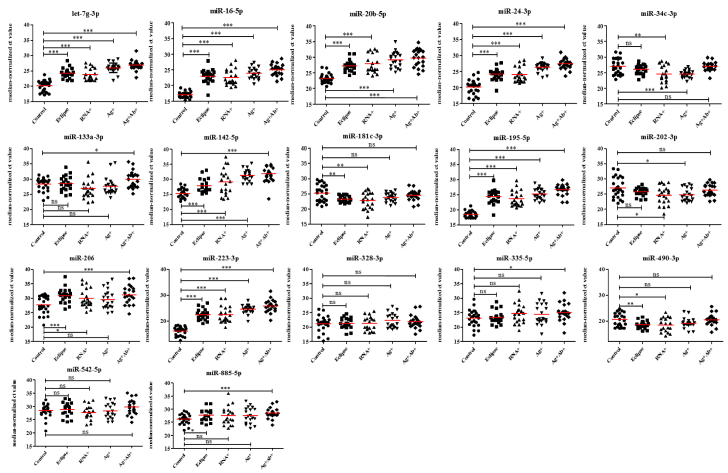
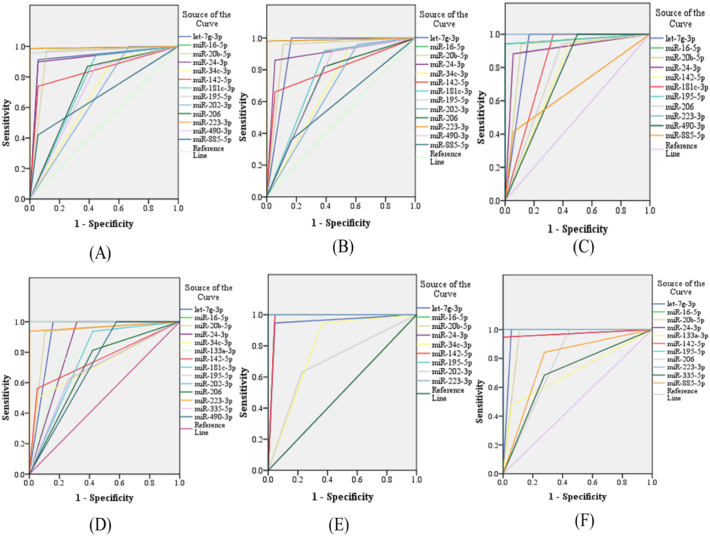
Since our goal was to identify potential biomarkers to detect early HIV-1 infection in clinical applications, we focused on the 17 miRNAs (Supplementary Table S2) that were differentially expressed in HIV-1 infected patients for further validation by qPCR. These miRNAs were selected based on their fold change difference between the analyzed groups and their low Ct values, indicating that these miRNAs might be abundant and easily detected in plasma. Next, the selected miRNAs were validated in a larger sample size (80 plasma samples from HIV-1 infected individuals in the early phase of infection and 25 normal controls) using quantitative RT-PCR and cel-miR-39 as an internal control normalizer. In this validation stage, we included eclipse stage samples. Eclipse stage is defined as the length of time between exposure and reliable detection of infection. Comparison of the miRNA expression profiles in plasma samples from HIV-1 infected individuals in the early phase of infection with the uninfected controls indicated that the expression of nine miRNAs were significantly decreased and four miRNAs were significantly increased in the plasma from HIV-1 infected individuals (Fig. 2 and Supplementary Table S3). In acute HIV-1 infection, nine miRNAs were significantly down regulated, and four miRNAs were significantly up regulated when compared to uninfected controls (Fig. 3 and Supplementary Table S3). Likewise, 11 miRNAs (9 down and 2 up) in the eclipse stage, 12 miRNAs (8 down and 4 up) in the RNA+ stage, nine miRNAs (7 down and 2 up) in the Ag + stage, and 11 miRNAs (11 down) in the Ag + Ab+ stage were differentially expressed compared to uninfected controls (Fig. 4 and Supplementary Table S3). The sensitivity and specificity of these miRNAs were evaluated and the optimal cutoff point was set up based on the Youden index [37] for diagnosing early HIV-1 infection, acute HIV-1 infection, eclipse stage, RNA+ stage, Ag + stage, and Ag + Ab+ stage using ROC analysis (Supplementary Table S4). Results from the validation phase for let-7 g-3p, miR-16-5p, miR-20b-5p, miR-24-3p, miR-34c-3p, miR-142-5p, miR-181c-3p, miR-195-5p, miR-202-3p, miR-206, miR-223-3p, miR-409-3p, and miR-885-5p indicated that these miRNAs were significantly modulated and were able to discriminate individuals with early HIV-1 infection from normal uninfected controls. These miRNAs had an AUC value of 0·929 (95% CI 0·854–1·000), 0·993 (95% CI 0·977–1·000) 0·930 (95% CI 0·841–1·000), 0·921 (95% CI 0·847–0·996), 0·679 (95% CI 0·523–0·834), 0·842 (95% CI 0·751–0·933), 0·749 (95% CI 0·600–0·898), 0·978 (95% CI 0·948–1·000), 0·667 (95% CI 0·505–0·828), 0·740 (95% CI 0·596–0·884), 0·929 (95% CI 0·854–1·000), 0·713 (95% CI 0·564–0·861), and 0·682 (95% CI 0·561–803) respectively (Fig. 5 and Supplementary Table S4). The results from the validation phase for let-7 g-3p, miR-16-5p, miR-20b-5p, miR-24-3p, miR-34c-3p, miR-142-5p, miR-181c-3p, miR-195-5p, miR-202-3p, miR-206, miR-223-3p, and miR-409-3p indicated that these miRNAs could significantly distinguish acute HIV-1 infection from uninfected controls and the AUC value for these miRNAs was 0·917 (95% CI 0·813–1·000), 0·990 (95% CI 0·966–1·000), 0·924 (95% CI 0·834–1·000), 0·902 (95% CI 0·818–0·987), 0·692 (95% CI 0·533–0·851), 0·803 (95% CI 0·695–0·910), 0·766 (95% CI 0·619–0·910), 0·990 (95% CI 0·966–1·000), 0·674 (95% CI 0·513–0·836), 0·716 (95% CI 0·567–0·864), 0·990 (95% CI 0·966–1·000), and 0·738 (95% CI 0·586–0·889) with a sensitivity of 100%, 98·33%, 96·49%, 88·33%, 93·22%, 65·22%, 90%, 98·33%, 95%, 80%, 98·33%, 89·66%, and specificity of 84%, 100%, 88%, 88%, 44%, 95·45%, 52%, 96%, 36%, 54·17%,100%, and 47·83% respectively (Fig. 5 and Supplementary Table S4). Results indicated that several miRNAs were able to discriminate between eclipse stage, RNA+ stage, Ag + stage, and Ag + Ab+ stage of infection from the uninfected controls (Supplementary Table S4). MiR-223-3p had an AUC value of 1·000 (95% CI 1·000–1·000) with 100% sensitivity, and specificity, and could discriminate between eclipse stage, Ag + stage and Ag + Ab+ stage of infection from the uninfected controls (Fig. 5, Supplementary Table S4). MiR-16-5p and miR-195-5p had an AUC value of 1·000 (95% CI 1·000–1·000) with sensitivity and specificity of 100% and could discriminate between RNA+ stage, Ag + stage, and Ag + Ab+ stages of HIV-1 infection from normal uninfected controls (Fig. 5, Supplementary Table S4).
Fig. 2.
Expression level of plasma miRNA candidates as early HIV-1 infection markers in the validation stage. The level of plasma miRNAs in 80 plasma samples from individuals in the early stage of HIV-1infection and 25 healthy control plasma samples were examined using real-time RT-PCR and normalized with cel-miR-39 as the control. Comparison of the miRNA expression profiles in plasma samples from HIV-1 infected individuals in the early phase of infection with the uninfected controls indicated that the expression of nine miRNAs were significantly decreased and four miRNAs were significantly increased in the plasma from HIV-1 infected individuals. Each sample was run in triplicate. (*P < 0·05; **P < 0·01, ***P < 0·0001 and ns = non-significant as calculated using Student's t-test).
Fig. 3.
Expression level of plasma miRNA candidates as biomarkers for acute HIV-1 infection in the validation stage. The level of plasma miRNAs in 60 HIV-1 positive plasma samples in the acute stage infection and 25 healthy controls plasma were examined using real-time RT-PCR and normalized with cel-miR-39 as the control. Comparison of the miRNA expression profiles in plasma samples from HIV-1 infected patients in the acute phase of infection with the uninfected controls indicated that, nine miRNAs were significantly down regulated, and four miRNAs were significantly up regulated when compared to uninfected controls. Each sample was run in triplicate. (*P < 0·05; **P < 0·01, ***P < 0·0001 and ns = non-significant, as calculated using Student's t-test).
Fig. 4.
Expression levels of plasma miRNA candidates in different stages of HIV-1 infection from the validation stage. The level of plasma miRNAs from eclipse (n = 20), RNA+ (n = 20), Ag + (n = 20) and Ag + Ab+ (n = 20) and 25 healthy control plasma samples were examined using real-time RT-PCR and normalized with cel-miR-39 as the control. 11 miRNAs (9 down and 2 up) in the eclipse stage, 12 miRNAs (8 down and 4 up) in the RNA+ stage, 9 nine miRNAs (7 down and 2 up) in the Ag + stage and 11 miRNAs (11 down) in the Ag + Ab+ stage were deregulated compared to uninfected controls. Each sample was run in triplicate. (*P < 0·05; **P < 0·01, ***P < 0·0001 and ns = non-significant, as calculated using Student's t-test).
Fig. 5.
Receiver operating characteristic (ROC) curve analysis of differentially expressed miRNAs in the validation stage. ROC curves for individual miRNAs in (A) early HIV-1 vs healthy controls, (B) acute HIV-1 vs. healthy controls, (C) eclipse vs. healthy controls, (D) RNA+ vs. healthy controls, (E) Ag + vs. healthy controls, and (F) Ag + Ab+ vs. healthy controls.
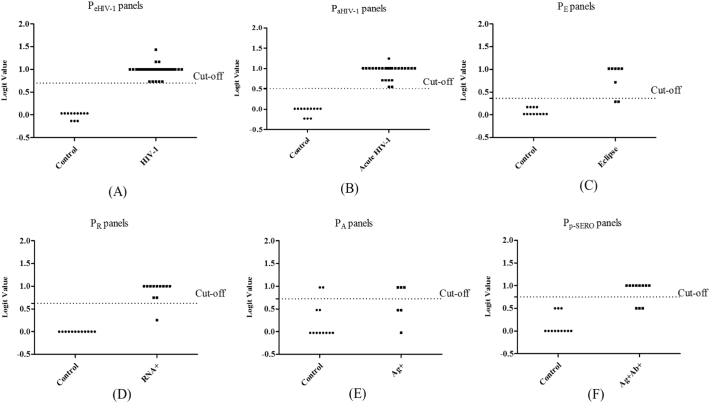
A logistic regression analysis was employed to determine the best combination of miRNAs to diagnose early HIV-1 infection. The results of the analysis indicated that a linear combination of expression levels of miR-16-5p, miR-20b-5p, miR-195-5p, and miR-223-3p produced the best model to diagnose early infection. The predicted probability of being detected as early HIV-1 infection by PeHIV-1 was calculated by: logit (p = early HIV-1) = 0·033 + 0·7*miR-223-3p + 0·7*miR-16-5p – 0·267*miR-195-5p – 0·167*miR-20b-5p. The four miRNA signature panel showed significant diagnosis of early HIV-1 infection and this panel increased the AUC value to 1·00 (95% CI, 1·00–1·00, P < 0·00001; Supplementary Fig. S2) with 100% sensitivity and specificity, compared with the use of each miRNA alone (Table 1 and Supplementary Table S4). Likewise, those miRNAs that were significantly different between eclipse stage vs normal control, RNA+ stage vs normal control, Ag + stage vs normal control, acute HIV-1 vs normal control, and post seroconversion/Ag + Ab+ stage vs normal control (Fig. 5 and Supplementary Table S4) were included in the logistic regression analysis (except those miRNAs that have an AUC value = 1·00). The selected miRNAs were grouped into panels designated as PE, PR, PA, Pa HIV-1, and PP-SERO for eclipse, RNA+, Ag+, acute HIV-1, and post seroconverted HIV-1 diagnosis (Table 1). The cut-off values of the diagnostic performances of these models were determined based on the maximum of Youden index (PE ≥ 0·363 for eclipse stage vs uninfected control, P ≥ 0·625 for RNA+ stage vs uninfected control, P ≥ 0·725 for Ag + stage vs uninfected control, P ≥ 0·509 for acute HIV-1 vs uninfected control, P ≥ 0·750 for Ag + Ab+ vs uninfected control). 100%, 95%, 100%, 100%, 95% sensitivity, and 95·8%, 100%, 100%, 100% and 100% specificity were demonstrated between eclipse stage, RNA+ stage, Ag + stage, acute HIV-1, and Ag + Ab+ stage respectively compared with uninfected controls (Table 1). In the eclipse stage miR-223-3p had AUC value = 1·00 and in RNA+ stage, miR-16-5p and miR-195-5p had AUC value = 1·00. Thus, we considered miR-223-3p with the PE, miR-16-5p and miR-195-5p with PR as a classifier for eclipse stage and RNA+ stage respectively. MiR-16-5p, miR-195-5p and miR-223-3p had AUC value = 1·00 for diagnosis of Ag + stage and seroconverted HIV-1 group. Therefore, the PA, and PP-SERO panel can be used with these miRNAs for the diagnosis of Ag + stage and post seroconverted HIV-1 individuals.
Table 1.
MiRNA panels for diagnosis of HIV-1 in validation datasets.
| miRNA panel | Targeted disease/stage vs control | Model | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|---|
| PeHIV-1 | Early HIV-1 | Logit (P = eHIV-1) = 0·033 + 0·7*miR-223-3p + 0·7*miR-16-5p – 0·267*miR-195-5p – 0·167*miR-20b-5p | >0·733 | 100% | 100% |
| Pa HIV-1 | Acute HIV-1 | Logit (P = aHIV-1) = 0·014 + 0·535*miR-223-3p + 0·535*miR-195-5p – 0·240*miR-20b-5p + 0·162* let-7 g-3p | >0·509 | 100% | 100% |
| PE | Eclipse | Logit (P = eclipse) = − 0·133 + 0·579*miR-16-5p + 0·270*let-7 g-3p + 0·151*miR-206 + 0·151*miR-181c-3p | >0·363 | 100% | 95·8% |
| PR | RNA+ | Logit (P = RNA+) = − 2·558E-16 + 0·750*miR-223-3p + 0·250*let-7 g-3p | >0·625 | 95% | 100% |
| PA | Ag+ | Logit (P = Ag+) = − 0·024 + 0·499*miR-142-5p + 0·499*miR-20b-5p | >0·725 | 100% | 100% |
| PP-SERO | Ag + Ab+/post seroconverted | Logit(P = Ag + Ab+) = − 4·103E-16 + 0·5*let-7 g-3p + 0·5*miR-142-5p | >0·750 | 95% | 100% |
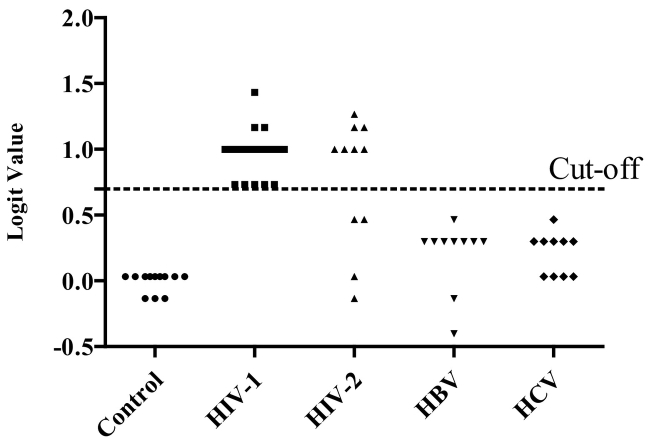
To validate the accuracy of the previously built diagnostic model we tested in a blinded study another 37 plasma samples from HIV-1 infected patients in the early phase of infection and 12 healthy controls. In this study, the patients' clinical data was not known to us. The sensitivity and specificity were calculated based on the models established in the validation stage. The results showed that, a signature composed of the four miRNAs (miR-16-5p, miR-20b-5p, miR-195-5p, and miR-223-3p) correctly discriminated early HIV-1 infection from uninfected controls with 100% sensitivity and specificity (Fig. 6A). The results also showed that all the eclipse stage, RNA+ stage, Ag + stage, and post seroconversion/Ag + Ab+ samples were detected from normal control with an accuracy of 90.00%, 95.83%, 72.22%, and 86.96% respectively (Fig. 6C-6F). In addition, our data suggest that the plasma miRNA panel Pa HIV-1 has excellent sensitivity and specificity as a biomarker for detecting acute HIV-1 infection (Fig. 6B). To determine the specificity of the four miRNA signature panel, plasma samples from HIV-1 infected individuals (n = 37), HIV-2 infected individuals (n = 11), HBV infected individuals (n = 10) and HCV infected individuals (n = 10) were tested. Results indicated the four miRNA signature panel was specific to HIV-1 infection and the plasma samples from HBV or HCV infected individuals had signatures similar to uninfected control samples (Fig. 7). The two unrelated viruses HBV and HCV had no impact on the detection of this panel of miRNAs denoting 100% specificity (Fig. 7). However, with plasma samples from HIV-2 infected individuals considerable cross-reactivity was observed. Seven out of eleven plasma samples from HIV-2 infected individuals were scored as reactive, however with a lowered sensitivity when compared to samples from HIV-1 infected individuals (sensitivity of 63.64%). This is not surprising since HIV-2 is a closely related retrovirus, similar to HIV-1 and shares many common traits like modes of transmission, viral replication and pathogenesis.
Fig. 6.
Logistic regression analysis- logit values from the blinded samples. The level of plasma miRNAs from 37 HIV-1 positive plasma from the early stage of infection (eclipse stage n = 8, RNA+ stage n = 12, Ag + stage n = 6 and Ag + Ab+ stage n = 11) and 12 healthy controls plasma were examined as blinded samples using real-time RT-PCR and normalized with cel-miR-39 as the control. Logit values were derived from regression analysis (A) PeHIV-1 for detection of early HIV-1 infection. The results showed that early HIV-1 were detected from normal control with an 100% accuracy (B) Pa HIV-1 for detection of acute stage of HIV-1 infection. The results showed that acute HIV-1 were detected from normal control with an 100% accuracy (C) PE for eclipse stage. The results showed that eclipse stage was detected from normal control with an 90.00% accuracy (D) PR, for RNA+ stage. The results showed that RNA+ stage was detected from normal control with an 95.83% accuracy (E) PA for Ag + stage. The results showed that Ag + were detected from normal control with a 72.22% accuracy and (F) PP-SERO for Ag + Ab+ or post-seroconverted stage of HIV-1 infection. The results showed that Ag + Ab+ stage was detected from normal control with an 86.96% accuracy.
Fig. 7.
Specificity of the PeHIV-1 miRNA panel. Expression of the PeHIV-1 miRNA signature [Logit (P = eHIV-1 = 0·033 + 0·7*miR-223-3p + 0·7*miR-16-5p – 0·267*miR-195-5p – 0·167*miR-20b-5p] in HIV-1(n = 37), HIV-2 (n = 11), HBV (n = 10) and HCV (n = 10) positive plasma samples and healthy donors (n = 12).
4. Discussion
According to the World Health Organization (WHO) 36·7 million people are now living with HIV throughout the world. After three decades of HIV discovery, while there is currently no cure, we are now capable of controlling the severity of disease progression and improving the lifespan of HIV infected individuals. But still 1·8 million people were newly infected and nearly 150,000 people were newly infected in USA in 2016. Early diagnosis of HIV-1 infection reduces morbidity and mortality and minimizes potential transmission of HIV-1. During the time-course of HIV-1 infection, plasma HIV-1 RNA is first detected about 7–10 days after initial exposure. The window period between initial exposure and the detection of HIV-1 RNA remains of some concern in ensuring the safety of blood used for transfusion and particularly in the prevention of HIV-1 transmission. Here we report the development of a sensitive, host based miRNA assay to detect early/acute HIV-1 infection in the absence of any detectable viral markers.
In this study, after the primary screen by miRNA PCR array, 17 miRNAs were selected for subsequent validation. Through the validation process, 15 potential miRNAs were identified that were differentially expressed in early HIV-1 infection, eclipse stage, RNA+ stage, Ag + stage, acute and Ag + Ab+ stages of HIV-1 infection compared with uninfected normal controls. MiRNAs that had an AUC value <1·00, and < 100% sensitivity and specificity were included in the multivariate analysis. A combined panel of miRNAs significantly increased the accuracy of detection and improved the specificity and sensitivity of diagnosis [41]. Thus, the miRNA-based test that we have designed employs a panel of miRNAs to detect eclipse stage, RNA+ stage, Ag + stage, Ag + Ab+ stage, and differentiates between early HIV-1 infection from uninfected control samples.
Our studies have been devoted to identifying host based miRNAs as biomarkers for early HIV-1 infection to improve diagnosis and individualized treatment; however, most of the reported work on HIV-1 diagnosis is focused on viral markers [15,16,42]. The potential viral biomarkers reported for detecting acute HIV-1 infection have several drawbacks because, approximately 3% to 5% of people with HIV will have a negative RNA test result during the window period of their HIV infection [16]. Furthermore, during the initial stages of HIV infection there is relatively low abundance of HIV-1 p24 antigen that may render its detection difficult. However, circulating miRNAs originating from cell-derived microvesicles, exosomes, apoptotic vesicles bodies and other unknown pathways [43,44] that can be detected by qPCR could serve as potential biomarkers of HIV infection. A few studies have identified circulating miRNAs that could serve as biomarkers for HIV-1infection [[33], [34], [35]]. All the previous reports have done case-control studies with patients who have been clinically diagnosed with HIV-1 but none so far have assessed the capacity of circulating miRNA to detect the eclipse stage of HIV-1 infection as well as the early stage of HIV-1 infection. Our findings highlight the potential of using miRNA panels for detecting early HIV-1 infection when viral markers may or may not be present. Diagnosing acute HIV infection has been considered a rare event; data from the US National HIV Surveillance System on a subset of new HIV diagnoses showed that only 3·1% of HIV infections were diagnosed in the acute phase from 2008 through 2012 [45]. In this study, we have developed a panel of miRNAs (PaHIV-1) that can detect acute HIV-1 infections with 100% sensitivity and specificity.
We have developed a model equation using the miRNAs in panel PeHIV-1 (PeHIV-1 = 0·033 + 0·7*miR-223-3p + 0·7*miR-16-5p – 0·267*miR-195-5p – 0·167*miR-20b-5p) and determined the threshold value (≥0·733) for detecting early HIV-1 infection. In blinded test phase, we have successfully predicted the occurrence of early HIV-1 infection by using the miRNA panels. We correctly identified 37 out of the 37 early HIV-1 infected samples (100% sensitivity), and 12 out of 12 uninfected/normal control samples (100% specificity) by using the miRNA panel PeHIV-1. Thus, in the future, after additional large-scale validation studies we may be able to develop a new miRNA panel and set a threshold value to detect early HIV-1 infection based on host miRNAs when viral markers may not be detected.
In this study, the levels of plasma miR-16-5p, miR-20b-5p, miR-195-5p, and miR-223-3p were significantly decreased in patients in eclipse stage, RNA+ stage, Ag + stage and post seroconversion stage of HIV-1 infection compared with healthy normal controls. It was previously reported that miR-16-5p and miR-223-3p were down regulated in plasma from HIV-1 infected individuals [[33], [34], [35]]. We have identified several unique circulating miRNAs in individuals with HIV-1 infection in our study as previously reported [[33], [34], [35]]. The panel (PeHIV-1) composed of four miRNAs had significantly higher sensitivity than any single miRNAs to distinguish individuals with early HIV-1 infection from the controls. In the validation and blinded stage of the study, PeHIV-1 had 100% sensitivity and specificity to detect early stage of HIV-1 infection. In addition, PeHIV-1 could detect RNA-negative HIV-1 infected plasma. Most importantly, PeHIV-1 could identify the eclipse stage of infection before HIV-1 RNA is detected in plasma. These findings highlight the potential use of PeHIV-1 as a non-invasive assessment for early HIV-1 at the eclipse stage of infection.
With the widespread use of pre-exposure prophylaxis (PrEP) and post exposure prophylaxis (PEP) as treatment options, host biomarker based detection assays may have an advantage over conventional HIV-1 marker based detection assays to detect early HIV-1 infection. Antiretroviral medication used in PrEP and PEP can suppress HIV RNA and delay seroconversion thereby making it difficult to detect HIV antibody or HIV RNA in persons who become HIV infected [46].
In summary, our data indicates that the panels of circulating miRNAs described in this report are able to detect early stages of HIV-1 infection and efficiently discriminate each stage of HIV-1 infection (eclipse, RNA+, Ag+, acute HIV-1, and post seroconverted stages) from normal controls. All samples in the discovery set were analyzed for only 372 miRNAs using PCR Arrays. However, >2600 human mature miRNAs are currently listed in the miRBase (http://www.mirbase.org/, accessed November 6, 2018) and there may be additional miRNAs that could serve as useful markers of HIV-1 infection. In the future if other key miRNAs are identified, they can be tested and integrated into our miRNA panels and be used to construct a clinically available model for stratification of early HIV-1 infection or disease progression.
Authors' contributions
All authors read and approved the final manuscript. S.B. and K.D. performed the experiments; S.B., K.D., and I.K.H. contributed to designing research, analyzing data, and discussing findings; S.B., K.D. and I.K.H. wrote the manuscript; M.H., and S.L. sample preparation; I.K.H. supervised the work.
Additional information
The findings and conclusions in this paper are an informal communication and represent my own best judgment. These comments do not bind or obligate FDA.
Conflicts of interest
All the authors have nothing to disclose.
Acknowledgements
This work is supported by CBER Intramural Research Fund for Biomarker of HIV Disease. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.04.023.
Contributor Information
Indira Hewlett, Email: Indira.Hewlett@fda.hhs.gov.
Krishnakumar Devadas, Email: Krishnakumar.Devadas@fda.hhs.gov.
Appendix A. Supplementary data
Supplementary material
References
- 1.Fiebig E.W., Heldebrant C.M., Smith R.I., Conrad A.J., Delwart E.L., Busch M.P. Intermittent low-level viremia in very early primary HIV-1 infection. J Acquir Immune Defic Syndr. 2005;39(2):133–137. [PubMed] [Google Scholar]
- 2.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H.Y., Giorgi E.E., Keele B.F. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261(2):341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindback S., Karlsson A.C., Mittler J. Viral dynamics in primary HIV-1 infection. Karolinska Institutet Primary HIV Infection Study Group. AIDS. 2000;14(15):2283–2291. doi: 10.1097/00002030-200010200-00009. [DOI] [PubMed] [Google Scholar]
- 5.Lindback S., Thorstensson R., Karlsson A.C. Diagnosis of primary HIV-1 infection and duration of follow-up after HIV exposure. Karolinska Institute primary HIV infection study group. AIDS. 2000;14(15):2333–2339. doi: 10.1097/00002030-200010200-00014. [DOI] [PubMed] [Google Scholar]
- 6.Vermeulen M., Coleman C., Mitchel J. Comparison of human immunodeficiency virus assays in window phase and elite controller samples: viral load distribution and implications for transmission risk. Transfusion. 2013;53(10):2384–2398. doi: 10.1111/trf.12117. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinn T.C. Acute primary HIV infection. JAMA. 1997;278(1):58–62. [PubMed] [Google Scholar]
- 8.Pilcher C.D., Tien H.C., Eron J.J., Jr. Brief but efficient: acute HIV infection and the sexual transmission of HIV. J Infect Dis. 2004;189(10):1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- 9.Brenner B.G., Roger M., Routy J.P. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195(7):951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 10.Wawer M.J., Gray R.H., Sewankambo N.K. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191(9):1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 11.Lewis F., Hughes G.J., Rambaut A., Pozniak A., Leigh Brown A.J. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5(3) doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masciotra S., McDougal J.S., Feldman J., Sprinkle P., Wesolowski L., Owen S.M. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(Suppl. 1):S17–S22. doi: 10.1016/j.jcv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Fiebig E.W., Wright D.J., Rawal B.D. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 14.Owen S.M., Yang C., Spira T. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol. 2008;46(5):1588–1595. doi: 10.1128/JCM.02196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomaras G.D., Yates N.L., Liu P. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westheimer E., Fu J., Radix A. An HIV-1 RNA test following a reactive fourth-generation antigen/antibody combination assay confirms a high proportion of HIV infections. J Clin Virol. 2014;61(4):623–624. doi: 10.1016/j.jcv.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Powers K.A., Ghani A.C., Miller W.C. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet. 2011;378(9787):256–268. doi: 10.1016/S0140-6736(11)60842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DoHaHS. D . Office of AIDS Research Advisory Council; 2016. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. [Google Scholar]
- 19.Le T., Wright E.J., Smith D.M. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N Engl J Med. 2013;368(3):218–230. doi: 10.1056/NEJMoa1110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananworanich J., Schuetz A., Vandergeeten C. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien M., Markowitz M. Should we treat acute HIV infection? Curr HIV/AIDS Rep. 2012;9(2):101–110. doi: 10.1007/s11904-012-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen M.S., Smith M.K., Muessig K.E., Hallett T.B., Powers K.A., Kashuba A.D. Antiretroviral treatment of HIV-1 prevents transmission of HIV-1: where do we go from here? Lancet. 2013;382(9903):1515–1524. doi: 10.1016/S0140-6736(13)61998-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill A.L., Rosenbloom D.I., Fu F., Nowak M.A., Siliciano R.F. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A. 2014;111(37):13475–13480. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoenigl M., Chaillon A., Little S.J. CD4/CD8 cell ratio in acute HIV infection and the impact of early antiretroviral therapy. Clin Infect Dis. 2016;63(3):425–426. doi: 10.1093/cid/ciw293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiat D., Olson E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123(1):11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devadas K., Biswas S., Haleyurgirisetty M. Identification of host micro RNAs that differentiate HIV-1 and HIV-2 infection using genome expression profiling techniques. Viruses. 2016;8(5) doi: 10.3390/v8050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grasedieck S., Sorrentino A., Langer C. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121(25):4977–4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 28.Arroyo J.D., Chevillet J.R., Kroh E.M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell P.S., Parkin R.K., Kroh E.M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 31.Duy J., Koehler J.W., Honko A.N. Circulating microRNA profiles of Ebola virus infection. Sci Rep. 2016;6 doi: 10.1038/srep24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang X., Jiang X., Gu D. Dysregulated serum MiRNA profile and promising biomarkers in dengue-infected patients. Int J Med Sci. 2016;13(3):195–205. doi: 10.7150/ijms.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuhua Qi H.H., Guo Hongxiong, Xu Peng, Shi Zhiyang, Huan Xiping, Zhu Zheng. MicroRNA profiling in plasma of HIV-1 infected patients: potential markers of infection and immune status. J Public Health Emerg. 2017;1:13. [Google Scholar]
- 34.Reynoso R., Laufer N., Hackl M. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Sci Rep. 2014;4:5915. doi: 10.1038/srep05915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thapa D.R., Hussain S.K., Tran W.C. Serum microRNAs in HIV-infected individuals as pre-diagnosis biomarkers for AIDS-NHL. J Acquir Immune Defic Syndr. 2014;66(2):229–237. doi: 10.1097/QAI.0000000000000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heneghan H.M., Miller N., Kerin M.J. Systemic microRNAs: novel biomarkers for colorectal and other cancers? Gut. 2010;59(7):1002–1004. doi: 10.1136/gut.2009.200121. [author reply 4] [DOI] [PubMed] [Google Scholar]
- 37.Akobeng A.K. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 2007;96(5):644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 38.Qi Y., Cui L., Ge Y. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. 2012;12:384. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z., Qi Y., Ge A. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza a (H7N9) virus infection in humans. Viruses. 2014;6(4):1525–1539. doi: 10.3390/v6041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Ba Y., Ma L. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 41.Huang W., Hu J., Yang D.W. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med. 2012;186(11):1160–1167. doi: 10.1164/rccm.201203-0534OC. [DOI] [PubMed] [Google Scholar]
- 42.Owen M.P.P., Wesolowski L. 16th conference on retroviruses and opportunistic infections. Montreal, Canada. 2009. Evaluation of the Abbott ARCHITECT Ag/Ab combo assay, an antigen/antibody combination test: Implications for US HIV testing programs. [Google Scholar]
- 43.Reid G., Kirschner M.B., van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80(2):193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Zen K., Zhang C.Y. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32(2):326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 45.Linley G.K., Oster A., Hernandez A. Conference on retroviruses and opportunistic infections (CROI). Seattle,WA. 2015. Detection of acute HIV infection, US national HIV surveillance system, 2008–2012. [Google Scholar]
- 46.Grant R.M., Lama J.R., Anderson P.L. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363(27):2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material