Abstract
Background
Chronic heavy alcohol drinking (CHD) leads to significant organ damage, increased susceptibility to infections, and delayed wound healing. These adverse outcomes are believed to be mediated by alterations in the function of myeloid cells; however, the mechanisms underlying these changes are poorly understood.
Methods
We determined the impact of CHD on the phenotype of splenic macrophages using flow cytometry. Changes in functional responses to LPS were measured using luminex and RNA-Seq. Finally, alterations in chromatin accessibility were uncovered using ATAC-Seq.
Findings
A history of CHD led to increased frequency of splenic macrophages that exhibited a heightened activation state at resting. Additionally, splenic macrophages from CHD animals generated a larger inflammatory response to LPS, both at protein and gene expression levels. Finally, CHD resulted in increased levels of H3K4me3, a histone mark of active promoters, as well as chromatin accessibility at promoters and intergenic regions that regulate inflammatory responses.
Interpretation
These findings suggest that a history of CHD alters the immune fitness of tissue-resident macrophages via epigenetic mechanisms.
Fund
National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institutes of Health (NIH) - R24AA019431, U01 AA13641, U01 AA13510, R21AA021947, and R21AA025839.
Keywords: Chronic heavy drinking, Ethanol, Splenic macrophages, Rhesus macaques, Chromatin accessibility, LPS
Research in context.
Evidence
Chronic heavy drinking (CHD) is associated with increased susceptibility to infection and poor wound healing, suggesting a compromised immune system. Several lines of evidence indicate that CHD disproportionately affects the myeloid cells of the innate immune system. Specifically, data from mouse models and monocytic cell lines suggest that chronic ethanol exposure triggers heightened inflammatory responses and poor functional outcomes. However, the impact of CHD on tissue-resident macrophages is still unknown. Furthermore, mechanisms underlying changes in myeloid cell function are not well understood.
Added value of this study
Here, we used a rhesus macaque model of voluntary ethanol self-administration to study the impact of a history of CHD on phenotype and function of splenic macrophages. This macaque model mimics human patterns of drinking while obviating confounding variables associated with mouse models and human studies. We show that CHD is associated with transcriptional reprogramming of macrophages, resulting in a heightened inflammatory response to LPS stimulation. This exaggerated response can be linked to significant changes in the epigenome; specifically, to alterations in the chromatin accessibility of promoters and intergenic regions that regulate genes critical for triggering an inflammatory response.
Implications of all the available evidence
This study provides novel insights into the epigenetic basis of CHD-mediated alterations in myeloid cell function. These findings are crucial in linking a history of voluntary CHD in a nonhuman primate model with clinical outcomes seen in patients suffering from Alcohol Use Disorder (AUD). Consequently, these studies will facilitate future studies into using anti-inflammatory therapeutics targeted at the epigenetic level.
Alt-text: Unlabelled Box
1. Introduction
Both acute and chronic heavy alcohol consumption significantly hamper the body's defenses against pathogens by reducing mucosal barrier integrity and interfering with several aspects of the immune system. Consequently, a history of chronic heavy drinking (CHD) results in increased susceptibility to respiratory diseases such as pneumonia [1,2] and tuberculosis [3,4]; as well as viral infections, notably HIV [5] and HCV [6,7]. Additionally, CHD is associated with heightened risk for cardiovascular disease [8,9] and cancer [10,11]. Furthermore, CHD has been associated with poor gastric, dermal, and bone wound healing outcomes [12], increasing the risks for morbidity and mortality following surgery or trauma [13]. Several lines of evidence suggest that this defect is primarily mediated by excessive inflammation associated with CHD, impairing the function of innate immune cells and thus compromising the process of tissue repair [14].
Data from several studies indicate that CHD disproportionately affects the innate branch of the immune system. Ethanol consumption or exposure can alter the function of innate immune cells, notably monocytes, within hours to days in a time and dose-dependent manner. Acute exposure to ethanol in vitro attenuates the ability of human monocytes, macrophage cells lines, and monocyte-derived macrophages to respond to LPS, a ligand for TLR4, as evidenced by reduced production of canonical inflammatory cytokines such as TNFα and IL-6 [15]. This phenomenon is mediated through increased expression of negative regulators of TLR signalling [[16], [17], [18], [19]] as well as prevention of NFκB translocation to the nucleus [20]. This phenomenon is not limited to signalling through TLR4 as acute exposure to ethanol inhibits type-I interferon signalling and promotes IL-10 production in human monocytes following stimulation with TLR8 ligands [21,22]. Finally, both in vivo and ex vivo ethanol exposure in rodent models reduces production of inflammatory cytokines IL-6 and IL-12 while increasing production of anti-inflammatory IL-10 in response to TLR2/TLR6, TLR4, TLR5, TLR7 (R-848), and TLR9 ligands [[23], [24], [25]]. In contrast to acute exposure, prolonged ethanol treatment enhances TNFα production by human monocytes and macrophage cells lines following LPS or PMA stimulation [22,26]. This switch to a pro-inflammatory response occurs via increased phosphorylation of the NFκB p65 subunit, increased NFκB translocation to the nucleus, and greater induction of responsive genes [16]. Similar observations are reported in murine models where in vivo chronic ethanol consumption results in a hyper-inflammatory response by circulating monocytes [27]. A single binge episode in alcohol-experienced (but not CHD) human volunteers (men and women) increased TNFα production after 20 min and IL-10 production at 2 and 5 h [28]. The few studies that have examined the impact of CHD on tissue-resident myeloid cells found a hyper-responsive phenotype of Kupffer cells in the liver [29], alveolar macrophages in the lung [30], and microglia in the brain [31].
Although in vitro studies with human monocytes and in vivo rodent models provide invaluable insights into the impact of acute and chronic ethanol consumption, they also present significant limitations. The human studies primarily use monocytes obtained from healthy donors (and not CHD subjects) and transformed human macrophage cell lines that are treated in vitro with ethanol. Clinical studies, on the other hand, can be hard to interpret in the presence of complex confounding factors such as smoking, use of recreational or illicit drugs, nutritional deficiencies, and unreliable drinking data [32]. The limitations of the rodent models include reliance on forced gavage feedings or access to only ethanol liquid diets for a relatively short period (weeks), raising the possibility of stress-induced inflammation. Additionally, rodent models fail to recapitulate the complexity or chronicity of voluntary and erratic patterns of drinking exhibited by humans. Consequently, our understanding of the mechanisms by which CHD alters function and phenotype of tissue-resident macrophages remains incomplete.
To answer these questions, we leveraged a nonhuman primate model of voluntary ethanol self-administration [33,34]. This model allows us to interrogate the physiological impact of CHD in vivo in an outbred translational primate model of voluntary ethanol self-administration while avoiding confounders that complicate clinical studies such as housing, diet, smoking, and use of illicit drugs. Recent studies using this model have reported that CHD induces significant transcriptional differences in PBMC [35,36]. Importantly, subsequent bioinformatics analyses strongly suggest that chronic ethanol consumption exerts its biggest impact on circulating myeloid cells [36]. These analyses also revealed significant changes in the expression of chromatin remodelling enzymes and histone molecules with CHD. However, it is still unclear if CHD results in functional and epigenetic reprogramming of myeloid cells.
In this study, we investigated the impact of a history of voluntary CHD on phenotypic and functional responses on long-lived tissue resident macrophages. Our studies indicate that CHD is associated with increased macrophage numbers in the spleen. The transcriptional profiles of these cells indicate alterations in their differentiation program. Functional assays suggest that CHD leads to a hyper-inflammatory response to LPS both at the protein and transcriptional levels. Moreover, we demonstrate that higher activation is mediated by greater chromatin accessibility at loci critical for the inflammatory response. These studies are the first to investigate the impact of chronic, voluntary ethanol consumption on the function and epigenetic landscape of tissue-resident macrophages in the in vivo setting of CHD using the highly translational nonhuman primate model.
2. Materials and methods
2.1. Experimental model and subject details
We used a schedule-induced polydipsia protocol followed by an “open access” to both 4% (w/v) ethanol and water 22 h/day to establish voluntary ethanol consumption in rhesus macaques as previously described [35,36]. A total of 7 controls (5 males and 2 females) and 10 animals with a history of CHD (5 males and 5 females) were included in this study (Fig. 1a). These animals came from three separate cohorts (6a, 7a, and 10) that followed two variations of the voluntary consumption protocol (detailed at MATRR.com). Briefly, spleen samples were collected at necropsy either at the end of the approximately 12 months voluntary consumption or a continuation of “open access drinking” with intermittent 1-month periods of forced abstinence over an additional year (details in [37]). The age of the subjects ranged from 5.5–8.4 years of age at the time of sample collection. For comparison across cohorts, the average daily ethanol intake (g/kg) was calculated during the first twelve months of ethanol open-access and ranged from 2.9 to 5.1 g/kg as detailed in Supplementary Fig. S1. For the purposes of these studies, CHD is defined as an average 12-month intake >2.8 g/kg and at least 330 days of ethanol intake (Table 1). Due to sample limitations, only subsets of these animals were included in individual assays (Table 1).
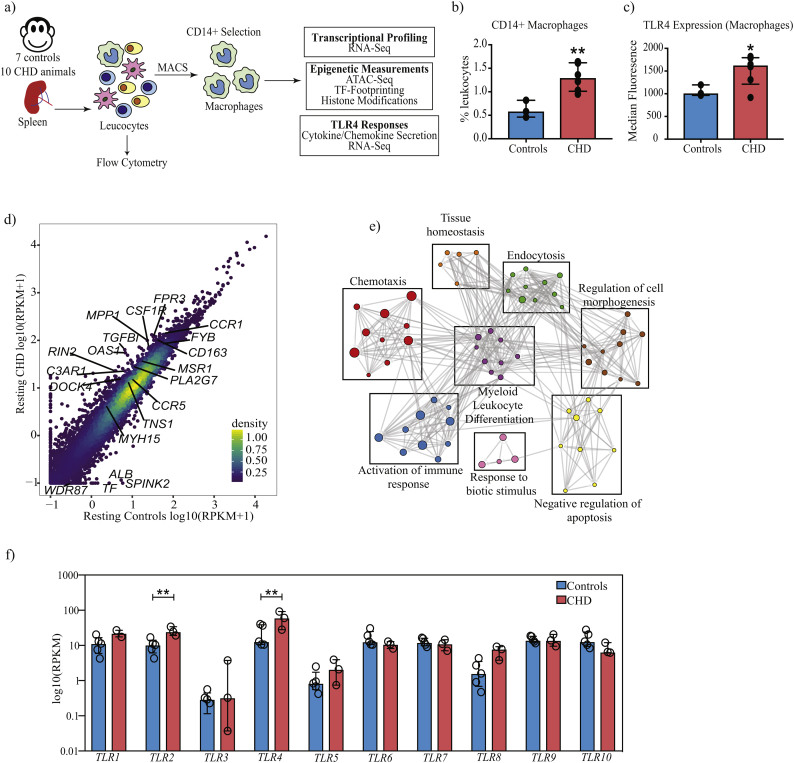
Fig. 1.
CHD is associated with transcriptional reprogramming of splenic macrophages.
(a) Schematic of the experimental design. Splenocytes from 7 control and 10 CHD animals in total were used in this study. Except for flow cytometry, all other experiments were performed on purified CD14+ splenic macrophages. (b) Bar graph depicting the percentage of CD14+ macrophages in controls (n = 3) and CHD (n = 6) animals derived from the singlet gate of splenocytes. (** - p < 0·01, unpaired t-test with Welch's correction) (c) Bar graph showing differences in TLR4 surface expression within the CD14+ splenic macrophage populations of controls (n = 3) and CHD (n = 6) animals measured using flow cytometry. Median Fluorescence Intensity (MFI) is depicted on the Y-axis. (* - p < 0·05, unpaired t-test with Welch's correction) (d) Scatterplot comparing average normalised transcript counts (RPKM) from resting splenic macrophages in response to CHD. Select significantly differentially expressed genes (DEG) outside the confidence interval of the linear fit are highlighted. A total of 5 controls and 3 CHD animals were used in the RNA-Seq experiment. (e) Functional enrichment of DEG detected in resting splenic macrophages in response to CHD was carried out using Metascape. Each circle denotes a Gene Ontology (GO) term. The size of the circle indicates the number of DEG enriching to the GO term in question. Closely related (GO) terms are clustered into groups labelled with the most statistically significant GO term within that group. Grey lines indicate the relationship between different GO terms based on shared DEG. (f) Bar graphs showing normalised transcript counts (RPKM) of Toll-like receptors genes (TLR) (** - FDR <0·01, exact test in edgeR followed by FDR correction using Benjamini-Hochberg method).
Table 1.
Description of samples used in this study.
| Sample ID | MATRR ID | Sex | Flow cytometry | Stimulation assay | RNA-Seq | ATAC-Seq |
|---|---|---|---|---|---|---|
| Control-1 | 10076 | Female | X | X | X | |
| Control-2 | 10071 | Female | X | X | ||
| Control-3 | 10222 | Male | X | X | ||
| Control-4 | 10221 | Male | X | X | X | |
| Control-5 | 10220 | Male | X | X | X | |
| Control-6 | 10093 | Male | X | |||
| Control-7 | 10096 | Male | X | |||
| CHD-1 | 10080 | Female | X | |||
| CHD-2 | 10081 | Female | X | X | X | |
| CHD-3 | 10069 | Female | X | |||
| CHD-4 | 10070 | Female | X | |||
| CHD-5 | 10078 | Female | X | |||
| CHD-6 | 10214 | Male | X | X | X | X |
| CHD-7 | 10215 | Male | X | X | X | |
| CHD-8 | 10088 | Male | X | |||
| CHD-9 | 10098 | Male | X | |||
| CHD-10 | 10097 | Male | X |
2.2. Ethics statement
This study was performed in strict accordance with the recommendations made in the Guide for Care and Use of Laboratory Animals of the National Institutes of Health, the Office of Animal Welfare and the United States Department of Agriculture. The ONPRC Institutional Animal Care and Use Committee approved all animal work.
2.3. Tissue collection and processing
Spleens from rhesus macaques were collected at necropsy (euthanasia method: 8 mg/kg ketamine followed by Pentobarbital injection) in RPMI supplemented with 10% fetal bovine serum (FBS), streptomycin/penicillin, and l-glutamine. Splenic leukocytes were isolated by mechanical disruption and filtration through a 70 μm cell strainer followed by red blood cell lysis. Cells were cryopreserved in 10% DMSO in Fetalplex Animal Serum Complex (Gemini Bio-Products, Sacramento CA).
2.4. Flow cytometry
1 to 2 × 106 freshly thawed spleen cells were surface stained using a cocktail of fluorescent-labelled antibodies – PerCP/Cy5.5-CD3 (Clone SP34-2, BD Biosciences), BV510-CD20 (Clone 2H7, Biolegend), APC/Cy7-HLA-DR (Clone L243, Biolegend), AF700-CD14 (Clone M5E2, Biolegend), APC-TLR4 (Clone HTA125, Biolegend), PB-CD16 (Clone 3G8, Biolegend), PE-CX3CR1 (Clone 2A9-1, Biolegend), PerCP-CCR2 (Clone 48,607, R&D Systems), BV605-CD40 (Clone 5C3, Biolegend), and PE-Dazzle 594-CD80 (Clone 2D10, Biolegend). All samples were acquired with the Attune NxT Flow Cytometer (ThermoFisher Scientific, Waltham MA) and analysed using FlowJo software (Ashland OR). Median Fluorescence Intensities (MFI) for all markers within the CD14+ macrophage gate were extracted in FlowJo and tested for significant differences using an unpaired t-test with Welch's correction on Prism 5 (GraphPad, San Diego CA).
2.5. Purification of splenic macrophages
Splenic CD14+ macrophages were purified from freshly thawed splenic leukocytes using CD14 antibodies conjugated to magnetic microbeads per the manufacturer's recommendations (Miltenyi Biotec, San Diego CA). Positive selection of macrophages was chosen to ensure high purity of the population, which was assessed using flow cytometry and was on average ≥ 90%.
2.6. TLR4 stimulation assay
2 × 105 CD14+ purified splenic macrophages were cultured in RPMI supplemented with 10% FBS with or without 100 ng/mL LPS (TLR4 ligand, E.coli 055:B5; Invivogen, San Diego CA) for 16 h in 96-well tissue culture plates at 37C in a 5% CO2 environment. Plates were spun down: supernatants were used to measure production of immune mediators using Luminex technology, and cell pellets were resuspended in Qiazol (Qiagen) for RNA extraction. Both cells and supernatants were stored in −80C until they could be processed as a batch.
2.7. RNA extraction and library preparation
Total RNA was isolated from CD14+ purified splenic macrophages using mRNeasy kit (Qiagen, Valencia CA) and quality assessed using Agilent 2100 Bioanalyzer. Following ribosomal RNA depletion using Ribo-Gone rRNA removal kit (Clontech, Mountain View CA), libraries were generated using SMARTer Stranded RNA-Seq kit. Briefly, rRNA depleted RNA was fragmented, converted to double-stranded cDNA and ligated to adapters. The roughly 300 bp-long fragments were then amplified by PCR and selected by size exclusion. Libraries were multiplexed using unique barcodes to facilitate sequencing of several biological samples. Following quality control for size, quality, and concentrations, libraries of 100 bp read lengths were sequenced on single-end mode.
2.8. Luminex assay
Circulating immune mediators were measured using nonhuman primate Cytokine/Chemokine/Growth Factor (eBioscience, San Diego CA) 29-plex panel measuring levels of cytokines (IFNγ, IL-1β, IL-2, IL4, IL-5, IL-6, IL-12, IL-15, IL-17, TNFα, IL-1RA, IL-10, and MIF), chemokines (MCP-1, MIP-1α, MIP-1β, RANTES, Eotaxin, MDC, IL-8, MIG, and I-TAC), and growth factors (EGF, FGF, G-CSF, GM-CSF, HGF, and VEGF-A). Standard curves were generated using 5-parameter logistic regression and differences in analyte levels between resting and post stimulation samples were tested using a paired t-test. Differences in levels of spontaneously produced analytes were tested using unpaired t-test with Welch's correction. Principal Component Analysis of overall mediator profiles was generated using ggbiplot function in R.
2.9. ATAC-Seq library preparation
105 purified CD14+ splenic macrophages were lysed in lysis buffer (10 mM Tris-HCl (pH 7·4), 10 mM NaCl, 3 mM MgCl2, and NP-40 for 10 min on ice to prepare the nuclei. Immediately after lysis, nuclei were spun at 500 g for 5 min to remove the supernatant. Nuclei were then incubated with Tn5 transposase and tagmentation buffer at 37C for 30 min. Stop buffer was then added directly into the reaction to end the tagmentation. PCR was performed to amplify the library for 15 cycles using the following PCR conditions: 72C for 3 min; 98C for 30s and thermocycling at 98C for 15 s, 60C for 30s and 72C for 3 min; following by 72C 5 min. Libraries were then purified with AMPure (Beckman Coulter, Brea CA) beads and quantified on the Bioanalyzer (Agilent Technologies, Santa Clara CA).
2.10. Histone ELISA
Nuclear extracts from 2 × 105 purified CD14+ splenic macrophages were isolated per manufacturer's instructions (Abcam, Cambridge UK) and quantified using a micro BCA assay protein assay kit (ThermoFisher Scientific, Waltham MA). Histone modifications were measured using a Histone H3 modification Multiplex Assay (Abcam, Cambridge UK). Input was normalised based on total protein concentration, and 20 ng of nuclear extract was added to each well. Given limited sample availability, only a subset of histone methylation marks was assayed (H3K4me1, H3K4me2, H3K4me3, H3K9me3). Optical density was measured at 450 nm.
2.11. RNA-Seq analysis
RNA-Seq reads were quality checked using FASTQC, adapter and quality trimmed using TrimGalore, retaining reads at least 35 bp long. Reads were aligned to Macaca mulatta genome based on annotations available on ENSEMBL (Rhemac3) using Tophat2 (version 2.1.1) (Supplementary Table 1). Aligned reads were counted gene-wise using GenomicRanges, counting reads in a strand-specific manner. Read counts were normalised using RPKM method for generation of PCA and heatmaps. Raw counts were used for testing differentially expressed genes (DEG) using edgeR using an exact test. DEG was defined as genes with at least two-fold up or down-regulation and an FDR controlled at 5%. Only genes with known macaque gene annotations were included in all downstream analyses. Functional enrichment of DEG was performed using Metascape (www.metascape.org) and InnateDB (https://www.innatedb.com). Networks of functional enrichment terms generated using MetaScape were visualised in Cytoscape (https://cytoscape.org).
2.12. ATAC-Seq – read preprocessing and alignment
Paired ended reads from sequencing were quality checked using FASTQC and trimmed to retain reads with a quality threshold of at least 20 and minimum read lengths of 50. Trimmed paired reads were aligned to the Macaca mulatta genome (Mmul_8.0.1) using Bowtie2 (−X 2000 –k 1 –very-sensitive –no-discordant –no-mixed). Reads aligning to mitochondrial genome and allosomes were removed using samtools. PCR duplicate artefacts were then removed using Picard. Finally, unmapped reads, supplementary alignment, potential PCR/optical duplicates, poor quality alignments and improperly mapped reads were filtered using samtools (samtools view –q 20 –F 3844). All samples had at least 25 million non-duplicate non-mitochondrial reads, with >95% alignment rates and > 0·9 non-redundant fraction (Supplementary Table 2). BAM files were repositioned using ATACseqQC package in R to reflect the accurate read start site due to Tn5 insertion. The positive and negative strands were offset by +4 bp and -5 bp respectively as previously described [38].
2.13. ATAC-Seq – peak calling
Accessible chromatin peaks were called for each sample from only paired reads using MACS2 (--BAMPE –g 2·4e9 –nomodel –q 0.01 –B –SPMR) callpeak function. Q-values were generated by Benjamini-Hochberg correction of p-values generated from a dynamic Poisson distribution in MACS2. Sample QC and statistics were generated using ATACseqQC package. For all downstream purposes, we tightened the signal 200 bp around the predicted summit (Supplementary Fig. S4a), without dramatically changing the annotation profiles (Supplementary Fig. S4b). PCA and sample clustering was performed on these regions following normalisation using DiffBind. Only samples with a FRiP score of at least 0·1 were retained in the analysis.
2.14. Profiling open chromatin regions in splenic macrophages
To identify open chromatin profile of splenic macrophages under homeostatic conditions, consensus profiles of macrophages from control animals were built using DiffBind using summits. Peaks were restricted to 200 bp on either end of the summits providing a list of 10,417 400 bp-peaks. Gene candidate list was further narrowed to those with at least two peaks overlapping promoters or exons and functional enrichment was carried out using Metascape (http://metascape.org) [39].
2.15. Identifying differential accessibility regions (DAR)
Differentially accessible peaks/regions between CHD and control animals were identified around both narrow peaks and summits (+/− 200 bp) using DESeq2 module in DiffBind. Statistical significance of differential peaks was tested using Wald-test followed by an FDR correction. For all downstream analyses, we define Differentially Accessible Regions (DAR) as 400 bp regions (200 bp on either end of the summits) that have at least fold increase or decrease of 1·5 on a log2 scale, and a False Discovery Rate (FDR) of ≥1% were included in the analysis.
2.16. Promoter and intergenic peak annotations
Genomic annotation of open chromatin regions in splenic macrophages and DAR with CHD was assigned using ChIPSeeker. Promoters were defined as follows: −1000 bp to +100 bp around the transcriptional start site (TSS) and then compared against a background of a million 400 bp macaque genomic regions generated using bedtools. Genes with no annotations were excluded from downstream analyses.
Due to the lack of available macaque annotation databases, distal intergenic regions (within 100 KB window) from macaque assembly were converted to the human genome (hg19) coordinates using UCSC liftOver tool. This strategy is a viable alternative when studying nonhuman primates since recent studies suggest that quantitative and not qualitative differences in enhancer activity are the prevalent source of regulatory landscape divergence among closely related species such as primates [40]. 1633 identified open chromatin regions with at least 50% overlap after lifting over from hg18 to hg19 genome builds using liftOver were subsequently compared to published human macrophage active enhancers (H3K27Ac) (GSE43036) and inactive/poised enhancers (H3K4me1) (GSE66595) peaks [41] using bedtools. Cis-Regulatory roles of these putative enhancer regions were identified using GREAT (http://great.stanford.edu/public/html/) using the default definition of gene regulatory domain (5 kb upstream, 1 kb downstream, and up to 1000 kb distal relative to TSS). Curated regulatory domains were excluded from the analysis. Bedgraphs were generated from BAM files using bedtools, converted to TDF using IGVtools and visualised in IGV (Integrated Genome Viewer).
2.17. Transcription factor footprinting analyses
To identify particular transcription factor binding sites (TFBS) that are over-represented in the differentially accessible promoter regions, genes were compared to CisRed, a database of genome-wide regulatory module and element predictions from JASPAR, TRANSFAC, and ORegAnno. Potential over-representative transcription factor binding sites within intergenic regions were identified using HOMER (http://homer.ucsd.edu/homer/). Statistically significant motifs in these regions were identified using findMotifsGenome.pl application.
For footprinting analyses, we used the highest-ranking hits from transcription factor (TF) analyses in both promoter and intergenic DAR. Position Weight Matrices (PWM) for each TF was taken from version 1.02 of CISBP database (http://cisbp.ccbr.utoronto.ca/) and converted to MEME format using the matrix2meme function in the MEME suite. Binding sites were then calculated using CENTIPEDE (http://centipede.uchicago.edu/), using the rhesus genome and narrowPeaks from pooled (Controls and CHD) BAM files called using MACS2. The posterior probability of TF binding sites for each peak was used to filter low confidence sites.
2.18. Nucleosome occupancy analyses
Due to limitations in coverage required for nucleosome analysis, reads were pooled within groups, and broad peaks were established using MACS2 (-- broad). Nucleosome Free Region (NFR) and nucleosome models were built using NucleoATAC using default settings [42].
2.19. Data integration – accessibility-gene expression association
Promoter regions for every annotated macaque gene were defined in ChIP-Seeker as −1000 bp to +100 bp relative to the transcription start site. Pileups were generated for these regions using featureCounts using pooled bam files for each group and normalised to total numbers of mapped reads. ATAC-Seq promoter pileups were tested for association with median RPKM of the gene using Spearman correlation analysis. For other comparisons, scatterplots of promoter ATAC-Seq pileups and RPKM were generated using ggplot, and Pearson correlations were calculated using cor.test in R and slopes compared to a y = x line using slope.test in smatr package in R.
3. Statistical methods
Two group comparisons were carried out using an unpaired t-test with Welch's correction. Differences between four groups were tested using one-way ANOVA (α = 0·05) with Sidak's multiple comparisons corrections. Statistical significance of functional enrichment was established using hypergeometric tests. Linear association between ATAC-Seq readouts was established using Pearson's correlation. Association between ATAC-Seq and RNA-Seq was tested using Spearman's correlation test. Slopes were compared using F-tests and significance of Venn overlaps established using a hypergeometric test.
3.1. Data deposition and materials sharing
The RNA-Seq and ATAC-Seq data files are available on NCBI's GEO under project accession PRJNA525851.
4. Results
4.1. Chronic heavy drinking (CHD) alters the frequency, phenotype, and transcriptional profile of spleen resident macrophages
We compared frequency and phenotype of resting macrophages within splenic leukocyte populations obtained from rhesus macaques classified as controls and CHD animals (Fig. 1a, Supplementary Fig. S1, and Table 1). Flow analyses showed an increased frequency of CD14+ macrophages in the spleen of CHD animals (Supplementary Fig. S2a and Fig. 1b). Analysis of surface expression of CD163 and CD16, which delineate tissue-resident macrophages from circulating monocytes [43] indicate that CD14+ cells are predominantly (97% average) splenic macrophages (Supplementary Fig. S2b). Additionally, splenic macrophages from CHD animals expressed higher surface levels of TLR4 (Fig. 1c and Supplementary Fig. S2c), in the absence of any differences in surface expression of CD16, HLA-DR, CCR2, or CX3CR1 (Supplementary Fig. S2c-e). Similarly, basal levels of activation markers CD40 and CD80 were comparable between the two groups (Supplementary Fig. S2c and S2f).
RNA-Seq analyses revealed a significant impact of CHD on the transcriptional profiles of tissue-resident macrophages with 63 up-regulated and 11 down-regulated genes (Fig. 1d and Supplementary Fig. S3a). These differentially expressed genes (DEG) play a role in immune activation as evidenced by their enrichment to gene ontology (GO) terms such as “chemotaxis” and “myeloid leukocyte differentiation” (Fig. 1e). In line with our flow cytometry data, we observed higher levels of transcripts associated with TLR4 as well as other pattern recognition receptors (e.g. TLR2, and FPR3); chemokine and complement receptors (CCR1, CCR5, and C3AR1); and regulators of myeloid differentiation (CSF1R, MSR1, CD163) in CHD group (Fig. 1d and Fig. 1f) suggesting that splenic macrophages from CHD animals are potentially poised to respond differently to stimulation.
4.2. Chronic HD enhances splenic macrophage response to LPS
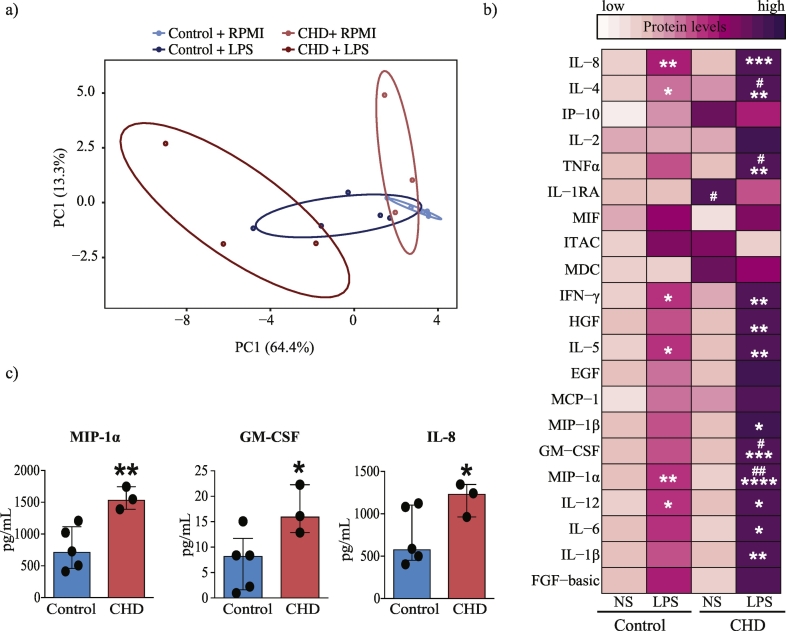
To assess the impact of CHD on splenic macrophage responses purified splenic CD14+ macrophages from control and CHD rhesus macaques were stimulated with LPS for 16 h, and secreted immune mediators were measured using Luminex (Fig. 1a and Supplementary Table 3). Principal Component Analysis (PCA) of the immune mediators (cytokines chemokines and growth factors) produced following LPS stimulation suggests distinct response profiles in CHD animals compared to controls (Fig. 2a). More specifically, in contrast to the pre- and post-LPS samples from the control group, which occupied overlapping space in the PCA, the pre- and post-LPS samples from the CHD group form distinct clusters in the PCA, suggesting a substantially larger response by the CHD animals. This heightened response is evident when comparing the production of specific analytes (Fig. 2b). Statistically significant induction of TNFα, MIP-1β, GM-CSF, IL-6, and IL-1β in response to LPS (Fig. 2b) was observed only in the CHD group. Levels of IL-4, TNFα, GM-CSF, and MIP-1α were higher in the CHD group compared to controls following LPS stimulation (Fig. 2b). After correcting for background production, levels of MIP-1α, IL-8, and GM-CSF were also higher in CHD animals relative to controls (Fig. 2c).
Fig. 2.
CHD is associated with a hyper-inflammatory response to LPS in splenic macrophages.
(a) Principal Component Analysis (PCA) of cytokines, chemokines, and growth factors produced by splenic macrophages from control (n = 5) and CHD (n = 3) animals in response to LPS and measured using Luminex. (b) Heatmap of individual secreted cytokines, chemokines, and growth factors produced spontaneously (NS) and in response to LPS stimulation (pg/mL). Significant increase in response to LPS stimulation in both groups is denoted by “*”; significant differences in mediators' levels between groups are denoted by “#” (p-values: */# - p < 0·05; **/## - p < 0·01; *** - p < 0·001; *** - p < 0·0001. Statistical analysis was carried out using ordinary one-way ANOVA (α = 0·05) with Sidak's multiple comparison tests. (c) Bar graphs of analytes with significantly higher levels of induction following LPS stimulation after correction for spontaneous production (NS) in the CHD group. (* - p < 0·05; ** - p < 0·01, unpaired t-test with Welch's correction)
4.3. CHD associated transcriptional responses to LPS mirror protein responses
Using RNA-Seq, we measured and compared transcript levels 16 h post LPS stimulation in both groups (Fig. 1a). As described for immune mediator production, we detected a larger number of differentially expressed genes (DEG) in CHD group relative to control animal following LPS stimulation (Fig. 3a, Supplementary Fig. S3a, and S3b) albeit with significant overlap (368 genes, p < 0·001) (Fig. 3a). Overall, DEG detected in both groups enriched to biological processes associated with canonical inflammatory responses to LPS, such as “cytokine production”, “myeloid cell activation” and “leukocyte migration” (Fig. 3b). However, the number of DEG enriching to these processes was higher in the CHD group (Fig. 3b).
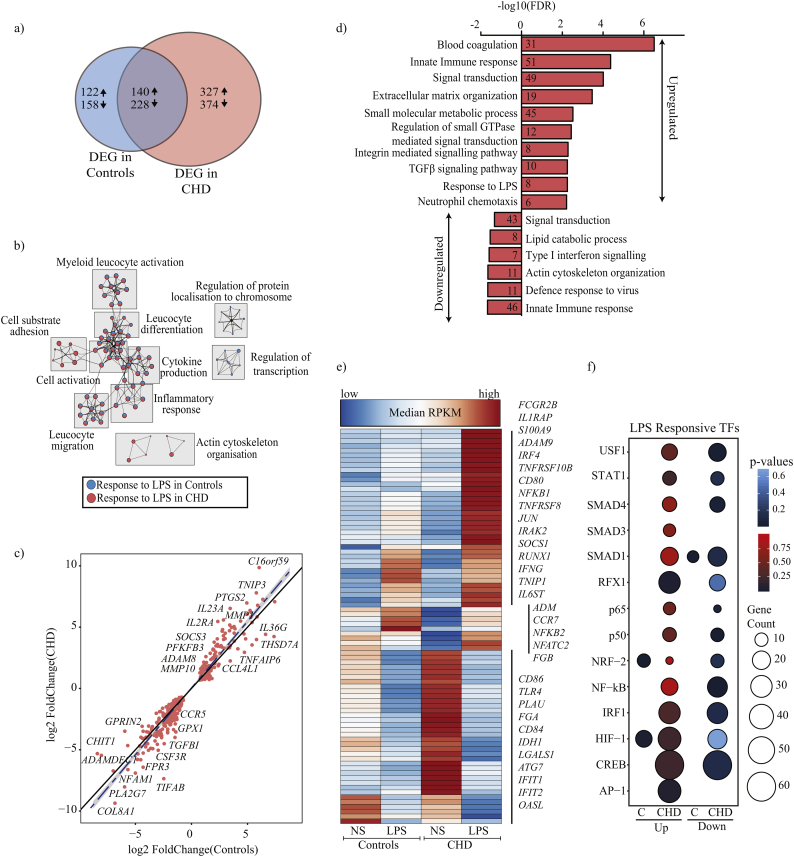
Fig. 3.
CHD leads to heightened transcriptional responses to LPS in splenic macrophages.
(a) Venn of DEG that were up- and down-regulated in splenic macrophages from control (n = 5) and CHD animals (n = 3) in response to LPS stimulation. Numbers and arrows within each circle represent the DEG numbers and direction of change relative to no stim samples respectively. (b) A network of functional enrichment of DEG detected in both groups following LPS stimulation identified by Metascape. Each box delineates a group of highly related GO terms; each GO term is shown as a pie chart that depict the relative transcriptional contributions from control and CHD groups (blue-controls; red-CHD). The size of the pie chart is indicative of the number of DEG enriching to that GO term, and the lines indicate relationship between GO terms based on shared DEG. (c) Scatter plot comparing fold changes of a subset of DEG detected in both groups in response to LPS. Innate immune genes outside the confidence interval (grey highlight) of the linear fit (dashed blue line) are annotated. (d) Functional enrichment of up and down-regulated DEG detected exclusively in CHD samples following LPS stimulation. (e) Clustered heatmap of up- or down-regulated DEG detected only in CHD samples post LPS exposure that enriched to GO term “innate immune response”. Colours on the heatmap represent scaled median RPKMs ranging from low (blue) to high (red) expression. (f) Bubble plots of up- (red) and down-regulated (blue) genes regulated by LPS responsive transcription factors (TF) as predicted by CisRed. Size and colour of the bubble represent the number of genes mapped and statistical significance (hypergeometric test) respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The magnitude of fold change of DEG detected in both groups showed a strong correlation with each other with few exceptions (e.g. IL23A, ADAM8, FPR3, CCR5, CSF3R, SOCS3, and IL2RA) (Fig. 3c). Several genes that play a critical role in the innate immune response to LPS were exclusively dysregulated with CHD (Fig. 3d). This list includes genes involved in sustaining a pro-inflammatory innate immune response to LPS (FCGR2B, CD80, NFKB1, JUN, IFNG, IL6ST), as well as genes associated with host defence (LAMP1, IFIT1, IFIT2, ATG7, OASL, and TLR4) (Fig. 3d and Fig. 3e). In contrast, genes exclusively dysregulated in controls play a role in chemotaxis, lipid metabolism, apoptosis, and cytokine secretion (CCR2, C3, CR1, CCR6, VEGFB) (Supplementary Fig. S3c and S3d).
Transcription factor (TF)-mediated regulation of the gene expression changes was detected following LPS stimulation using InnateDB and cisRed databases with a focus on macrophage-specific TF [44]. Analysis of canonical LPS responsive TFs (Fig. 3f) suggests genes regulated by RFX-1, AP-1, STAT1, NFκB, and IRF1 were only differentially expressed in the CHD group following LPS stimulation (Fig. 3f). Given that the transcriptional response to LPS was assessed at 16 h post-stimulation, we also assessed TFs that regulate the resolution of inflammation and differentiation program following LPS exposure (Supplementary Fig. S3e). Interestingly, several genes regulated by resolving factors PPARγ, NRF-1, and ATF3 (Supplementary Fig. S3e) regulated genes were up-regulated exclusively in the CHD group. Furthermore, a number of NF-κB and PPARγ regulated genes were down-regulated in both groups, in line with the resolution of the inflammatory phase at 16 h post stimulation.
4.4. CHD alters the chromatin landscape of splenic macrophages
The epigenetic basis for functional and transcriptional changes in splenic macrophages induced by CHD was assessed using an unbiased approach to profile the chromatin landscape of splenic macrophages from controls and CHD animals using the assay for transposase-accessible chromatin sequencing (ATAC-Seq) (Fig. 4a). First, we assessed the accessible chromatin regions in splenic macrophages in control animals under homeostatic conditions. Because larger genomic loci with higher number of mapped reads can bias differential accessibility measurements, all readouts were tightened to 200 bp around each peak (Supplementary Fig. S4a). Genomic annotations of open chromatin profile in macrophages from control animals indicate a predominance of promoter (−1000 bp to +100 bp) (4464 genes) and distal intergenic loci (Supplementary Fig. S4b) relative to a background of randomly regenerated 400 bp regions (Supplementary Fig. S4c). A core set of 90 genes with at least two accessible peaks were identified, which play roles in: “myeloid leukocyte differentiation”; “apoptosis”; “phagocytosis”, and “iron transport” (Fig. S4d).
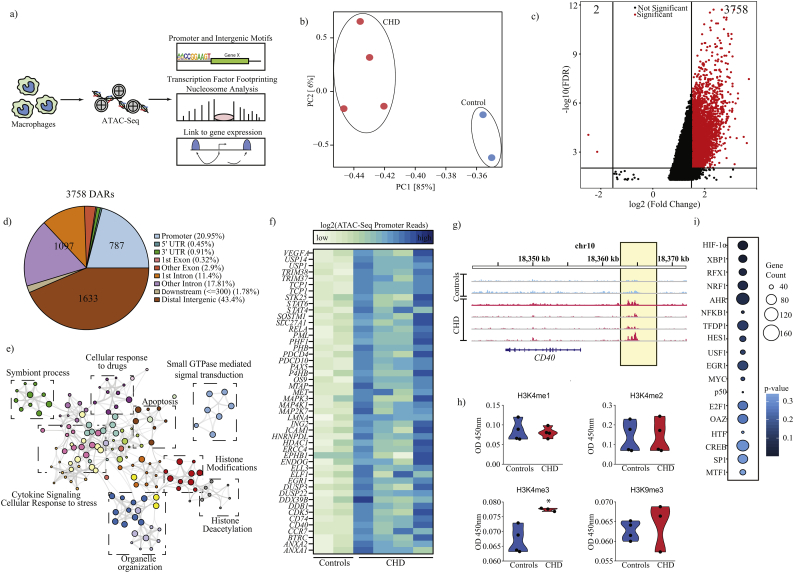
Fig. 4.
Chronic heavy ethanol consumption alters chromatin accessibility.
(a) Experimental design for ATAC-Seq and downstream in silico analyses. (b) PCA of chromatin accessibility profiles (peaks) of splenic macrophages from control (n = 2) and CHD animals (n = 4). (c) Volcano plot showing cutoffs used to identify differentially accessible regions (DAR) in macrophages from CHD compared to control animals. X and Y-axes represent fold change and FDR (−log10) respectively. DAR were defined as those with |FC| ≥ 1·5 and FDR ≤ 0·01 (Wald test). (d) Genomic contexts of DAR showing preferential mapping to intergenic and promoter regions. (e) A network of biological processes to which genes regulated by promoters that are significantly more accessible with CHD enrich generated using Metascape. Each coloured circle represents a GO term and the size of the circle indicated the number of DEG enriching to it. Closely related GO terms are clustered within a group labelled with the name of the most significant GO term in that cluster. Grey lines indicate associations between the different GO terms based on shared DEG. (f) Heatmap of ATAC-Seq read pileups overlapping promoters of genes that enriched to GO terms “cellular response to stress”, “cytokine signalling”,” histone modifications”, and “histone deacetylation”. (g) Representative pileups demonstrating increased accessibility (highlighted in yellow) upstream of CD40 promoter with CHD. (h) Box plots representing differences in histone activation and repressive marks in controls (n = 4) and CHD animals (n = 3) (* - p-value <0·05, unpaired t-test with Welch's correction). (i) Bubble plots summarising transcriptional factor binding site (TFBS) enrichment of genes regulated by the promoters that are more accessible in splenic macrophages from CHD animals predicted by cisRed. Size and colour of the bubble represent the number of genes mapped and statistical significance (hypergeometric test) respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The PCA of accessible chromatin regions indicated that samples from the CHD and control groups form distinct clusters separated by loadings from PC1 (Fig. 4b). Differential accessibility analysis of these summits using DiffBind (internally running DESeq2) confirmed enhanced chromatin accessibility (higher number of open chromatin regions) in macrophages from CHD animals relative to controls, defined as those with a fold change in accessibility >1.5 and an FDR <0.01 (Fig. 4c and Supplementary Fig. S4e). This filtering approach identified 3760 genomic elements (3758 open regions vs. 2 closed regions) with altered chromatin accessibility with CHD (Fig. 4c and Supplementary Fig. S4e).
Genomic annotations of these differentially accessible regions (DAR) suggest an over-representation of promoter regions (Fig. 4d) relative to background (Fig. S4c). Moreover, 43% of the DAR (1633) mapped to distal intergenic regions (Fig. 4d), indicating the potential impact on cis-regulatory networks as well. The 787 DAR that mapped to promoter regions (defined as −1000 bp to +100 bp around TSS) mapped to 644 unique annotated genes, which play roles in “cytokine signalling pathways”, “cellular response to stress”; and “apoptosis” (Fig. 4e). In particular, we observed increased promoter accessibility of markers of myeloid cell activation (e.g. CD40) (Fig. 4f and Fig. 4g). Interestingly, a number of genes regulated by promoter regions differentially accessible with CHD are involved in histone deacetylation and chromatin remodelling (Fig. 4e). Given the large changes in promoter accessibility with CHD, we measured the expression of a subset of active and repressive histone marks in nuclear extracts using a multiplexed ELISA. Our analysis showed no significant differences in mono or di-methyl marks on histone H3K4 (associated with enhancers); however, we detected a significant increase (p < .05) in global tri-methyl levels (H3K4me3), an active promoter mark while levels of H3K9me3 a repressive mark remained unchanged with CHD (Fig. 4h).
Finally, we investigated if enhanced chromatin accessibility is associated with concomitant changes in nucleosome positioning. Using a conservative approach to model Nucleosome Free Regions (NFR) and nucleosomes [42], our analysis indicates a drop in relative frequencies of nucleosomes in CHD relative to controls (Supplementary Fig. S5a and Fig. S5b).
4.5. CHD enhances the accessibility of stress-responsive transcription factors in splenic macrophages
Next, we carried out a TF binding site (TFBS) analysis using InnateDB and cisRed to gain a better understanding of how chromatin accessibility changes impact TF binding within the promoter regions. This approach predicted increased availability of binding sites for cellular stress-responsive TF such as hypoxia-inducible HIF-1α, oxidative stress-responsive NRF-1 and AHR; and ER stress-responsive XBP1; as well as master regulators of inflammation NFKB1 and subunit p50 (Fig. 4i). Approximately 43% of the increased accessible regions mapped to intergenic elements of the macaque genome. To identify functional attributes to these changes, we transformed the genomic coordinates from macaque assembly to human assembly (hg19, see methods) and identified potential cis-regulation using GREAT (Fig. 5a and Fig. 5b). We further restricted our analysis of intergenic changes to characterised enhancers from human monocyte-derived macrophages [41] (see methods). Assessment of changes overlapping H3K4me1 and H3K27Ac rich regions demonstrate cis-regulatory roles in host defence (e.g. “Immune System Process” and “regulation of cytokine production”) (Fig. 5c and Fig. 5d).
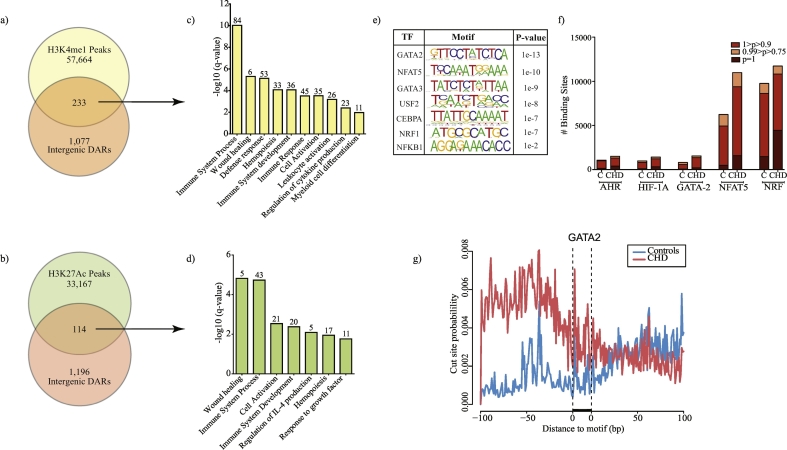
Fig. 5.
CHD associated changes in the regulatory landscape of macrophage chromatin.
(a, b) Venn of distal intergenic differences observed with CHD compared with (a) poised and (b) active enhancer regions in monocyte-derived macrophages [41]. Comparisons were made following liftOver of macaque genomic loci to human hg19 reference. (c, d) Functional enrichment of genes associated with intergenic (c) poised and (d) active enhancer regions more significantly accessible in CHD animals using GREAT. (e) Summary of transcription factors (TF) with over-represented motifs overlapping differentially accessible intergenic regions identified using HOMER (hypergeometric test). (f) Bar graph showing the number of binding sites for a subset of TFs predicted to bind to promoter and intergenic regions using InnateDB (Fig. 4I) and HOMER (Fig. 5e) respectively determined using TF footprinting tool CENTIPEDE. (g) CENTIPEDE derived binding probability graph around GATA2 motif in macrophages from controls (blue) and CHD (red) animals. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Further unbiased TF motif-based analysis using HOMER's findMotifsGenome tool in intergenic regions identified binding sites for TF essential for macrophage phagocytosis (GATA-2) [45], activation (CEBPA) [46], pro-inflammatory cytokine production (GATA-2, NRF-1, NFAT5) [47], and polarization (GATA-3 and NFAT5) [47,48] (Fig. 5e). The putative TF binding sites for GATA2, NFAT5, and NRF (intergenic candidates) as well as HIF-1α and AHR (promoter candidates) were confirmed by binding probability measurements using CENTIPEDE (Fig. 5f and Fig. 5g).
4.6. Integrating CHD-associated chromatin changes with differential gene expression
Direct comparisons of gene expression with promoter accessibility using CHD samples suggested a modest but significant positive association (r2 = 0·32; p < 2·2e-16, Spearman correlation) (Fig. 6a). A comparison of the list of genes regulated by the differentially accessible promoters (644 genes) and the list of genes differentially expressed with CHD at resting (using FDR <0·1–117 genes) showed a poor overlap (Supplementary Fig. S6). Therefore, we next investigated the association between the degree of chromatin accessibility and gene expression. To that end, we compared normalised ATAC-Seq readouts at the promoters of the 117 genes differentially expressed with CHD at resting in controls and CHD samples (using FDR <0.1). This analysis revealed a significant linear association (r2 = 0·953; p < 2·2e-16, Pearson correlation), and more importantly, a significant shift (p < 0·001, relative to the slope of 1) strongly indicating enhanced chromatin accessibility for several genes (Fig. 6b), notably C3AR1, TLR4, LAMP1, and CCL2.
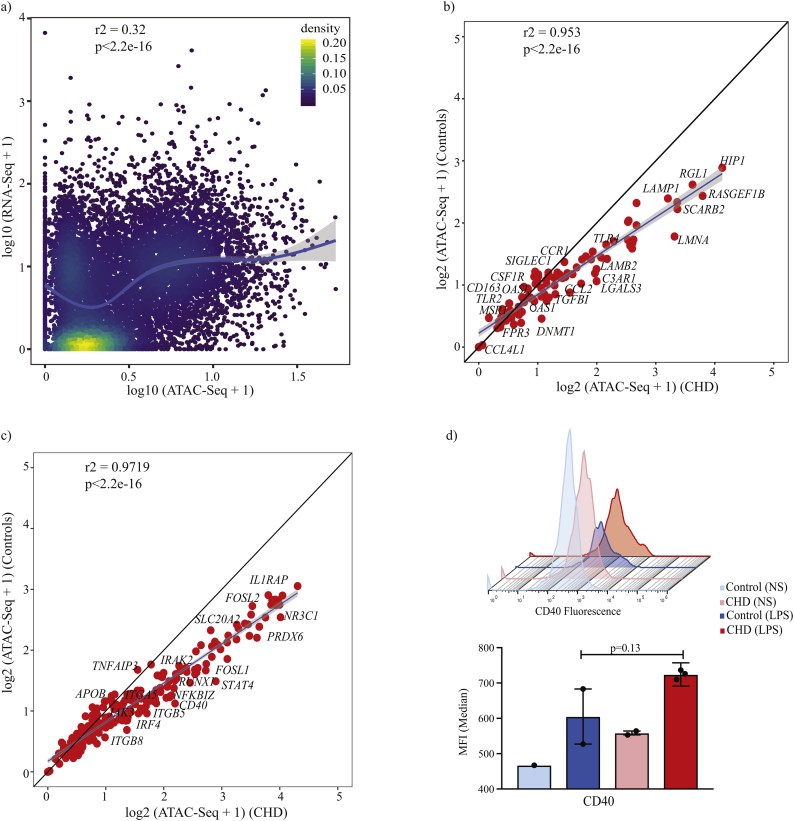
Fig. 6.
Integration of RNA-Seq and ATAC-Seq links CHD associated epigenetic changes with post LPS responses.
(a) Scatter plots comparing expression (median RPKM) and normalised promoter ATAC-Seq pileups of all the genes in the CHD group. Association was tested using Spearman correlation test and curve fit using polynomial loess regression. Each dot is a gene and colours of the dots represent the relative overlap of the two distributions. (b) Scatter plot comparing ATAC-Seq readouts at promoters of genes differentially expressed with CHD at resting from control and CHD animals. Linear regression and association were tested using Pearson's correlation test. (c) Scatter plot comparing ATAC-Seq readouts from control and CHD animals at promoters of genes differentially expressed exclusively with CHD in response to LPS. Linear regression and association were tested using Pearson's correlation test. (d) Histogram and bar graphs comparing surface CD40 induction with 16 h LPS exposure in macrophages from controls (post stim, n = 2) and CHD (post stim, n = 3) animals (p-values from unpaired t-test with Welch's correction).
Next, we next compared normalised ATAC-Seq readouts at the promoters of the 327 genes (Fig. 3a) that were exclusively upregulated in splenic macrophages following LPS stimulation in controls and CHD samples (Fig. 6c). A similar linear trend (r2 = 0.9719, p < 0·0001, Pearson correlation) and a shift in slopes of linear distributions were also observed (p < 0·001, relative to the slope of 1) notably for TF STAT4, IRF4, and RUNX1; signalling molecules IRAK2, FOSL1/FOSL2, and TNFAIP3 (Fig. 6c). Among the genes with significantly increased promoter accessibility with CHD is CD40 (Fig. 4g) where we observed a significantly higher induction of its mRNA expression (Fold change 0·85 for controls, 1·4 for CHD, FDR < 0·05, exact test following by Benjamini-Hochberg correction) and a higher surface expression with CHD following LPS stimulation (Fig. 6d) but not at resting. This observation suggesta a potentially significant contribution of chromatin changes observed at resting to post stimulatory gene expression patterns.
5. Discussion
While it is well accepted that long term ethanol exposure significantly impacts the function of peripheral and tissue-resident myeloid cells [49], our understanding of the mechanisms underlying these changes remains limited. The current study using a rhesus macaque model of chronic voluntary ethanol self-administration in vivo provides insights into the mechanism of myeloid cell dysfunction associated with chronic heavy alcohol drinking (CHD) (males and females included). First, the data show that a history of CHD is associated with increased frequency of macrophages in the spleen. This phenotype has previously only been reported in the liver, under conditions of alcoholic liver disease (ALD) [50]. Since this model does not produce overt signs of tissue damage in liver but rather a metabolic syndrome of oxidative stress [[51], [52], [53]], our finding suggests that chronic drinking can alter the frequency of tissue-resident myeloid cells even in the absence of overt organ damage. Secondly, the characterisation of purified splenic macrophages from CHD monkeys, included responses to LPS both at protein and gene expression levels, and changes in chromatin accessibility allowing for a deeper understanding of inflammatory reprogramming. Our data indicate that a history of CHD is associated with higher expression of TLR4 (protein and gene expression level) as well as other pattern recognition receptors (FPR3, TLR2), chemokine receptors (CCR1, CCR5), differentiation, and activation markers (CD163). These findings are in line with earlier studies that reported alterations in expression of pro-inflammatory cytokines [30], surface expression of pathogen sensors, adhesion molecules, activation markers [54], and impaired phagocytosis [55] on human alveolar macrophages with alcohol use disorder. Both TLR2 and TLR4 are implicated in attenuation or augmentation of inflammation by acute alcohol exposure in monocytes [56], peritoneal macrophages [57,58], and the CNS [59]. The present analyses provide evidence that this phenotype holds for chronic exposure as well.
The enhanced activation of macrophages following CHD can be hypothesised to occur via two different mechanisms. First, the oxidative stress induced by ethanol or its metabolites (acetyl-CoA) and acetaldehyde adducts can exert a significant impact on cellular transcriptional profiles [60,61]. Specifically, these molecules can serve as cofactors and substrates within signalling pathways that result in epigenetic changes via histone modifications [62], potentially altering chromatin accessibility and regulating gene expression. In support of this hypothesis, we have reported changes in post-transcriptional networks, particularly of microRNAs that modulate immune function [63] as well as transcriptional changes that have the potential to alter chromatin organisation in PBMC from CHD macaques [36]. Similarly, alterations in DNA methylation and gene expression in the brain were also reported in this model [64,65]. Finally, clinical studies have also reported alterations in histone modifications and DNA methylation with CHD in neurons that contribute to memory deficits and addiction [[66], [67], [68], [69], [70]].
Secondly, CHD results in an impaired gut barrier and potential translocation of endotoxins into circulation [71,72]. This dysbiosis contributes to the development of ALD [73,74], acute lung injury [75], and addictive behaviour [72]. Indeed, we recently reported a dose-dependent increase in levels of circulating IgM-bound endotoxin in male rhesus macaques with a history of CHD [76]. Similarly, recent studies reported a positive correlation between the amounts of ethanol consumed, serum endotoxin levels, and markers of monocyte activation in humans [49]. Therefore, higher circulating levels of bacterial pathogen-associated molecular patterns (PAMPs) could result in the activation of tissue-resident macrophages. It is, however, still unclear if the impact of the PAMPs is limited to mature myeloid cells or can reprogram precursor cells in the bone marrow.
Interestingly, the higher basal activation state of splenic macrophages did not result in LPS tolerance. Instead, stimulation with LPS resulted in heightened inflammatory responses in CHD animals. Immune mediator secretion profile indicated a significantly larger response with higher levels of canonical LPS-induced factors such as MIP-1α, TNFα, GM-CSF, and IL-8. This hyperinflammatory response was paralleled with an exacerbated inflammatory response to LPS at the transcriptional level. Alterations in responses to LPS can be mediated by cellular signalling perturbations or nuclear/epigenetic defects. Given that metabolites of ethanol (e.g. acetyl-CoA and acetaldehyde) regulate several aspects of chromatin biology [77,78], we next determined if CHD is associated with reprogramming of the splenic macrophage epigenome.
ATAC-Seq analysis of resting macrophage revealed global increases in chromatin accessibility of regulatory elements, notably promoters and intergenic enhancer regions that regulate the expression of genes critical for immune activation, cellular stress responses and gene expression. Interestingly, we detected increased accessibility of regulatory elements harbouring transcription factors that respond to oxidative stress and hypoxia as well as those that regulate inflammatory responses. Additional analyses revealed a weak but significant association between chromatin accessibility and gene expression levels. Findings from the integration approach strongly suggest that CHD-induced differences in gene expression at resting and post-LPS stimulation can be explained by higher promoter accessibility for a subset of the genes. The genes that do not show a change in promoter accessibility may be regulated by additional mechanisms such as changes in TF translocation to the nucleus, recruitment of Pol II, and cis-regulatory factors, and post-transcriptional modification. Within the genes that showed increased promoter accessibility with CHD is CD40, which showed higher protein and gene expression following LPS stimulation.
To our knowledge, this is the first comprehensive analysis of the genomic and functional impact of voluntary CHD (for at least 12 months) on tissue-resident macrophages in nonhuman primates, albeit in a small sample. Given the significant similarity between human and nonhuman primate physiologic and immunologic systems, these findings have a high translational value. Although experiments in this study included both males and females, we are underpowered to draw any conclusions regarding the impact of sex on ethanol-associated immune dysregulation, which remains a high public health priority. Future experiments should determine whether implicated changes in the chromatin can alter the effector functions of macrophage (e.g. pathogen killing, phagocytosis, wound healing), thereby linking experimental observations with clinical outcomes of patients with AUD. Further studies will also address chromatin accessibility and the role of histone modifications in mediating this pro-inflammatory phenotype seen with CHD, including alterations in TF access. Ultimately, in addition to studying mature tissue resident cells, future endeavours will have to focus on understanding the impact of CHD on precursor myeloid cells in the bone marrow, which populate cells in the periphery and large portions of cells in the tissue.
The following are the supplementary data related to this article.
Alignment Statistics for RNA-Seq data.
Alignment Statistics for ATAC-Seq data.
Protein measurements from Luminex.
Supplementary material
Acknowledgments
Acknowledgements
The authors wish to thank the members of the Grant lab and Oregon National Primate Research Center for the animal work and sample collection. We thank Ms. Nicole Walters for critical reading of the manuscript and Ms. Maham Rais (University of California Riverside) for assistance with cell separations and ATAC-Seq library preparation.
Funding sources
This work was funded by National Institutes for health (NIH) and National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants - NIH 8P51 ODO11092-533, NIH/NIAAA R24AA019431, U01 AA13641, U01 AA13510 and NIH/NIAAA R21AA021947 and R21AA025839. The funding agencies had no role in the study design, data collection, data analysis, interpretation, or writing of the manuscript.
Conflicts of interests
The authors declare no competing interests.
Author contributions
I.M and K.G designed the study. S·S, S.B.·N, and C·S performed the experiments. S·S, S.B.·N, and B.J.L analysed the data. S·S, I.M, and K.G wrote the manuscript. All authors have read and approved the final version of the manuscript.
References
- 1.Schmidt W., De Lint J. Causes of death of alcoholics. Q J Stud Alcohol. 1972;33(1):171–185. [PubMed] [Google Scholar]
- 2.Saitz R., Ghali W.A., Moskowitz M.A. The impact of alcohol-related diagnoses on pneumonia outcomes. Arch Intern Med. 1997;157(13):1446–1452. [PubMed] [Google Scholar]
- 3.Sabot G., Vendrame G. Incidence of pulmonary tuberculosis in alcoholics. Study based on investigations made at the Ospedale Psichiatrico Provinciale di Udine in the decade 1958-1967. Minerva Med. 1969;60(101):5190–5194. [PubMed] [Google Scholar]
- 4.Hudolin V. Tuberculosis and alcoholism. Ann N Y Acad Sci. 1975;252:353–364. doi: 10.1111/j.1749-6632.1975.tb19179.x. [DOI] [PubMed] [Google Scholar]
- 5.Baum M.K., Rafie C., Lai S., Sales S., Page J.B., Campa A. Alcohol use accelerates HIV disease progression. AIDS Res Hum Retroviruses. 2010;26(5):511–518. doi: 10.1089/aid.2009.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya R., Shuhart M.C. Hepatitis C and alcohol: interactions, outcomes, and implications. J Clin Gastroenterol. 2003;36(3):242–252. doi: 10.1097/00004836-200303000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Szabo G., Saha B., Bukong T.N. Alcohol and HCV: implications for liver cancer. Adv Exp Med Biol. 2015;815:197–216. doi: 10.1007/978-3-319-09614-8_12. [DOI] [PubMed] [Google Scholar]
- 8.Mukamal K.J., Rimm E.B. Alcohol's effects on the risk for coronary heart disease. Alcohol Res Health. 2001;25(4):255–261. [PMC free article] [PubMed] [Google Scholar]
- 9.Djousse L., Mukamal K.J. Alcohol consumption and risk of hypertension: does the type of beverage or drinking pattern matter? Rev Esp Cardiol. 2009;62(6):603–605. doi: 10.1016/s1885-5857(09)72223-6. [DOI] [PubMed] [Google Scholar]
- 10.Fedirko V., Tramacere I., Bagnardi V., Rota M., Scotti L., Islami F. Alcohol drinking and colorectal cancer risk: an overall and dose-response meta-analysis of published studies. Ann Oncol. 2011;22(9):1958–1972. doi: 10.1093/annonc/mdq653. [DOI] [PubMed] [Google Scholar]
- 11.Grewal P., Viswanathen V.A. Liver cancer and alcohol. Clin Liver Dis. 2012;16(4):839–850. doi: 10.1016/j.cld.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Jung M.K., Callaci J.J., Lauing K.L., Otis J.S., Radek K.A., Jones M.K. Alcohol exposure and mechanisms of tissue injury and repair. Alcohol Clin Exp Res. 2011;35(3):392–399. doi: 10.1111/j.1530-0277.2010.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radek K.A., Ranzer M.J., DiPietro L.A. Brewing complications: the effect of acute ethanol exposure on wound healing. J Leukoc Biol. 2009;86(5):1125–1134. doi: 10.1189/jlb.0209103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosa D.F., Sarandy M.M., Novaes R.D., Freitas M.B., do Carmo Gouveia Peluzio M., Goncalves R.V. High-fat diet and alcohol intake promotes inflammation and impairs skin wound healing in Wistar rats. Mediators Inflamm. 2018;2018:4658583. doi: 10.1155/2018/4658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szabo G., Saha B. Alcohol's effect on host Defense. Alcohol Res. 2015;37(2):159–170. [PMC free article] [PubMed] [Google Scholar]
- 16.Mandrekar P., Bala S., Catalano D., Kodys K., Szabo G. The opposite effects of acute and chronic alcohol on lipopolysaccharide-induced inflammation are linked to IRAK-M in human monocytes. J Immunol. 2009;183(2):1320–1327. doi: 10.4049/jimmunol.0803206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandrekar P., Catalano D., Jeliazkova V., Kodys K. Alcohol exposure regulates heat shock transcription factor binding and heat shock proteins 70 and 90 in monocytes and macrophages: implication for TNF-alpha regulation. J Leukoc Biol. 2008;84(5):1335–1345. doi: 10.1189/jlb.0407256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muralidharan S., Ambade A., Fulham M.A., Deshpande J., Catalano D., Mandrekar P. Moderate alcohol induces stress proteins HSF1 and hsp70 and inhibits proinflammatory cytokines resulting in endotoxin tolerance. J Immunol. 2014;193(4):1975–1987. doi: 10.4049/jimmunol.1303468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norkina O., Dolganiuc A., Catalano D., Kodys K., Mandrekar P., Syed A. Acute alcohol intake induces SOCS1 and SOCS3 and inhibits cytokine-induced STAT1 and STAT3 signaling in human monocytes. Alcohol Clin Exp Res. 2008;32(9):1565–1573. doi: 10.1111/j.1530-0277.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandrekar P., Jeliazkova V., Catalano D., Szabo G. Acute alcohol exposure exerts anti-inflammatory effects by inhibiting IkappaB kinase activity and p65 phosphorylation in human monocytes. J Immunol. 2007;178(12):7686–7693. doi: 10.4049/jimmunol.178.12.7686. [DOI] [PubMed] [Google Scholar]
- 21.Norkina O., Dolganiuc A., Shapiro T., Kodys K., Mandrekar P., Szabo G. Acute alcohol activates STAT3, AP-1, and Sp-1 transcription factors via the family of Src kinases to promote IL-10 production in human monocytes. J Leukoc Biol. 2007;82(3):752–762. doi: 10.1189/jlb.0207099. [DOI] [PubMed] [Google Scholar]
- 22.Pang M., Bala S., Kodys K., Catalano D., Szabo G. Inhibition of TLR8- and TLR4-induced type I IFN induction by alcohol is different from its effects on inflammatory cytokine production in monocytes. BMC Immunol. 2011;12:55. doi: 10.1186/1471-2172-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruett S.B., Zheng Q., Fan R., Matthews K., Schwab C. Ethanol suppresses cytokine responses induced through toll-like receptors as well as innate resistance to Escherichia coli in a mouse model for binge drinking. Alcohol. 2004;33(2):147–155. doi: 10.1016/j.alcohol.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Pruett S.B., Fan R., Zheng Q., Schwab C. Differences in IL-10 and IL-12 production patterns and differences in the effects of acute ethanol treatment on macrophages in vivo and in vitro. Alcohol. 2005;37(1):1–8. doi: 10.1016/j.alcohol.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Dai Q., Pruett S.B. Ethanol suppresses LPS-induced toll-like receptor 4 clustering, reorganization of the actin cytoskeleton, and associated TNF-alpha production. Alcohol Clin Exp Res. 2006;30(8):1436–1444. doi: 10.1111/j.1530-0277.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z., Bagby G.J., Stoltz D., Oliver P., Schwarzenberger P.O., Kolls J.K. Prolonged ethanol treatment enhances lipopolysaccharide/phorbol myristate acetate-induced tumor necrosis factor-alpha production in human monocytic cells. Alcohol Clin Exp Res. 2001;25(3):444–449. [PubMed] [Google Scholar]
- 27.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8(3):627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afshar M., Richards S., Mann D., Cross A., Smith G.B., Netzer G. Acute immunomodulatory effects of binge alcohol ingestion. Alcohol. 2015;49(1):57–64. doi: 10.1016/j.alcohol.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maraslioglu M., Oppermann E., Blattner C., Weber R., Henrich D., Jobin C. Chronic ethanol feeding modulates inflammatory mediators, activation of nuclear factor-kappaB, and responsiveness to endotoxin in murine Kupffer cells and circulating leukocytes. Mediators Inflamm. 2014;2014 doi: 10.1155/2014/808695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Halloran E.B., Curtis B.J., Afshar M., Chen M.M., Kovacs E.J., Burnham E.L. Alveolar macrophage inflammatory mediator expression is elevated in the setting of alcohol use disorders. Alcohol. 2016;50:43–50. doi: 10.1016/j.alcohol.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman L.G., Jr., Crews F.T. Innate immune signaling and alcohol use disorders. Handb Exp Pharmacol. 2018;248:369–396. doi: 10.1007/164_2018_92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stampfer M.J., Colditz G.A., Willett W.C., Manson J.E., Arky R.A., Hennekens C.H. A prospective study of moderate alcohol drinking and risk of diabetes in women. Am J Epidemiol. 1988;128(3):549–558. doi: 10.1093/oxfordjournals.aje.a115002. [DOI] [PubMed] [Google Scholar]
- 33.Baker E.J., Farro J., Gonzales S., Helms C., Grant K.A. Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcohol Clin Exp Res. 2014;38(11):2835–2843. doi: 10.1111/acer.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jimenez V.A., Grant K.A. Studies using macaque monkeys to address excessive alcohol drinking and stress interactions. Neuropharmacology. 2017;122:127–135. doi: 10.1016/j.neuropharm.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barr T., Girke T., Sureshchandra S., Nguyen C., Grant K., Messaoudi I. Alcohol consumption modulates host Defense in rhesus macaques by altering gene expression in circulating leukocytes. J Immunol. 2016;196(1):182–195. doi: 10.4049/jimmunol.1501527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sureshchandra S., Rais M., Stull C., Grant K., Messaoudi I. Transcriptome profiling reveals disruption of innate immunity in chronic heavy ethanol consuming female rhesus macaques. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen D.C., Gonzales S.W., Grant K.A. Effect of repeated abstinence on chronic ethanol self-administration in the rhesus monkey. Psychopharmacology (Berl) 2018;235(1):109–120. doi: 10.1007/s00213-017-4748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prescott S.L., Srinivasan R., Marchetto M.C., Grishina I., Narvaiza I., Selleri L. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163(1):68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt S.V., Krebs W., Ulas T., Xue J., Bassler K., Gunther P. The transcriptional regulator network of human inflammatory macrophages is defined by open chromatin. Cell Res. 2016;26(2):151–170. doi: 10.1038/cr.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schep A.N., Buenrostro J.D., Denny S.K., Schwartz K., Sherlock G., Greenleaf W.J. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25(11):1757–1770. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagelkerke S.Q., Bruggeman C.W., den Haan J.M.M., Mul E.P.J., van den Berg T.K., van Bruggen R. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of fc-gamma receptors. Blood Adv. 2018;2(8):941–953. doi: 10.1182/bloodadvances.2017015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pope S.D., Medzhitov R. Emerging principles of gene expression programs and their regulation. Mol Cell. 2018;71(3):389–397. doi: 10.1016/j.molcel.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Bresnick E.H., Katsumura K.R., Lee H.Y., Johnson K.D., Perkins A.S. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40(13):5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee B., Qiao L., Lu M., Yoo H.S., Cheung W., Mak R. C/EBPalpha regulates macrophage activation and systemic metabolism. Am J Physiol Endocrinol Metab. 2014;306(10):E1144–E1154. doi: 10.1152/ajpendo.00002.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tellechea M., Buxade M., Tejedor S., Aramburu J., Lopez-Rodriguez C. NFAT5-regulated macrophage polarization supports the proinflammatory function of macrophages and T lymphocytes. J Immunol. 2018;200(1):305–315. doi: 10.4049/jimmunol.1601942. [DOI] [PubMed] [Google Scholar]
- 48.Li H., Jiang T., Li M.Q., Zheng X.L., Zhao G.J. Transcriptional regulation of macrophages polarization by MicroRNAs. Front Immunol. 2018;9:1175. doi: 10.3389/fimmu.2018.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liangpunsakul S., Toh E., Ross R.A., Heathers L.E., Chandler K., Oshodi A. Quantity of alcohol drinking positively correlates with serum levels of endotoxin and markers of monocyte activation. Sci Rep. 2017;7(1):4462. doi: 10.1038/s41598-017-04669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karakucuk I., Dilly S.A., Maxwell J.D. Portal tract macrophages are increased in alcoholic liver disease. Histopathology. 1989;14(3):245–253. doi: 10.1111/j.1365-2559.1989.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 51.Ivester P., Shively C.A., Register T.C., Grant K.A., Reboussin D.M., Cunningham C.C. The effects of moderate ethanol consumption on the liver of the monkey, Macaca fascicularis. Alcohol Clin Exp Res. 2003;27(11):1831–1837. doi: 10.1097/01.ALC.0000095633.26284.FA. [DOI] [PubMed] [Google Scholar]
- 52.Ivester P., Roberts L.J., 2nd, Young T., Stafforini D., Vivian J., Lees C. Ethanol self-administration and alterations in the livers of the cynomolgus monkey, Macaca fascicularis. Alcohol Clin Exp Res. 2007;31(1):144–155. doi: 10.1111/j.1530-0277.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 53.Lebold K.M., Grant K.A., Freeman W.M., Wiren K.M., Miller G.W., Kiley C. Individual differences in hyperlipidemia and vitamin E status in response to chronic alcohol self-administration in cynomolgus monkeys. Alcohol Clin Exp Res. 2011;35(3):474–483. doi: 10.1111/j.1530-0277.2010.01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guth A.M., Janssen W.J., Bosio C.M., Crouch E.C., Henson P.M., Dow S.W. Lung environment determines unique phenotype of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2009;296(6):L936–L946. doi: 10.1152/ajplung.90625.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta A.J., Yeligar S.M., Elon L., Brown L.A., Guidot D.M. Alcoholism causes alveolar macrophage zinc deficiency and immune dysfunction. Am J Respir Crit Care Med. 2013;188(6):716–723. doi: 10.1164/rccm.201301-0061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oak S., Mandrekar P., Catalano D., Kodys K., Szabo G. TLR2- and TLR4-mediated signals determine attenuation or augmentation of inflammation by acute alcohol in monocytes. J Immunol. 2006;176(12):7628–7635. doi: 10.4049/jimmunol.176.12.7628. [DOI] [PubMed] [Google Scholar]
- 57.Dai Q., Pruett S.B. Different effects of acute and chronic ethanol on LPS-induced cytokine production and TLR4 receptor behavior in mouse peritoneal macrophages. J Immunotoxicol. 2006;3(4):217–225. doi: 10.1080/15476910601080156. [DOI] [PubMed] [Google Scholar]
- 58.Pascual M., Fernandez-Lizarbe S., Guerri C. Role of TLR4 in ethanol effects on innate and adaptive immune responses in peritoneal macrophages. Immunol Cell Biol. 2011;89(6):716–727. doi: 10.1038/icb.2010.163. [DOI] [PubMed] [Google Scholar]
- 59.Pandey S.C. TLR4-MyD88 signalling: a molecular target for alcohol actions. Br J Pharmacol. 2012;165(5):1316–1318. doi: 10.1111/j.1476-5381.2011.01695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cederbaum A.I. Alcohol metabolism. Clin Liver Dis. 2012;16(4):667–685. doi: 10.1016/j.cld.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heymann H.M., Gardner A.M., Gross E.R. Aldehyde-induced DNA and protein adducts as biomarker tools for alcohol use disorder. Trends Mol Med. 2018;24(2):144–155. doi: 10.1016/j.molmed.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G. Acetyl coenzyme a: a central metabolite and second messenger. Cell Metab. 2015;21(6):805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Asquith M., Pasala S., Engelmann F., Haberthur K., Meyer C., Park B. Chronic ethanol consumption modulates growth factor release, mucosal cytokine production, and microRNA expression in nonhuman primates. Alcohol Clin Exp Res. 2014;38(4):980–993. doi: 10.1111/acer.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cervera-Juanes R., Wilhelm L.J., Park B., Grant K.A., Ferguson B. Alcohol-dose-dependent DNA methylation and expression in the nucleus accumbens identifies coordinated regulation of synaptic genes. Transl Psychiatry. 2017;7(1):e994. doi: 10.1038/tp.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cervera-Juanes R., Wilhelm L.J., Park B., Grant K.A., Ferguson B. Genome-wide analysis of the nucleus accumbens identifies DNA methylation signals differentiating low/binge from heavy alcohol drinking. Alcohol. 2017;60:103–113. doi: 10.1016/j.alcohol.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marutha Ravindran C.R., Ticku M.K. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res Mol Brain Res. 2004;121(1–2):19–27. doi: 10.1016/j.molbrainres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 67.Pandey S.C., Ugale R., Zhang H., Tang L., Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiang M., Denny A., Lieu M., Carreon S., Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics. 2011;6(9):1095–1104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponomarev I., Wang S., Zhang L., Harris R.A., Mayfield R.D. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manzardo A.M., Henkhaus R.S., Butler M.G. Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene. 2012;498(1):5–12. doi: 10.1016/j.gene.2012.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mutlu E.A., Gillevet P.M., Rangwala H., Sikaroodi M., Naqvi A., Engen P.A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leclercq S., Matamoros S., Cani P.D., Neyrinck A.M., Jamar F., Starkel P. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 2014;111(42):E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaffert C.S., Duryee M.J., Hunter C.D., Hamilton B.C., 3rd, DeVeney A.L., Huerter M.M. Alcohol metabolites and lipopolysaccharide: roles in the development and/or progression of alcoholic liver disease. World J Gastroenterol. 2009;15(10):1209–1218. doi: 10.3748/wjg.15.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szabo G., Bala S. Alcoholic liver disease and the gut-liver axis. World J Gastroenterol. 2010;16(11):1321–1329. doi: 10.3748/wjg.v16.i11.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Massey V.L., Poole L.G., Siow D.L., Torres E., Warner N.L., Schmidt R.H. Chronic alcohol exposure enhances lipopolysaccharide-induced lung injury in mice: potential role of systemic tumor necrosis factor-alpha. Alcohol Clin Exp Res. 2015;39(10):1978–1988. doi: 10.1111/acer.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barr T., Sureshchandra S., Ruegger P., Zhang J., Ma W., Borneman J. Concurrent gut transcriptome and microbiota profiling following chronic ethanol consumption in nonhuman primates. Gut Microbes. 2018:1–19. doi: 10.1080/19490976.2018.1441663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zakhari S. Alcohol metabolism and epigenetics changes. Alcohol Res. 2013;35(1):6–16. [PMC free article] [PubMed] [Google Scholar]
- 78.Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153(1):56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment Statistics for RNA-Seq data.
Alignment Statistics for ATAC-Seq data.
Protein measurements from Luminex.
Supplementary material