Abstract
The scope of studies investigating the architecture of genomic DNA has progressed steadily since the elucidation of the structure of B-DNA. In recent years, several non-canonical DNA structures including Z-DNA, G-quadruplexes, H-DNA, cruciform DNA, and i-motifs have been reported to form in genomic DNA and are closely related to the evolution and development of disease. The ability of these structures to form in genomic DNA indicates that they might have important cellular roles and are therefore retained during evolution. Understanding the impact of the formation of these secondary structures on cellular processes can enable identification of new targets for therapeutics. In this review, we report the state of understanding of Z-DNA structure and formation and their implication in disease. Finally, we state our perspective on the potential of Z-DNA as a therapeutic target.
Keywords: Z-DNA, Disease, Z-DNA-binding protein, Therapeutic target, Transcription
Introduction
The main backbone of the human genome is comprised of double-stranded DNA (dsDNA) in the canonical B-DNA conformation with a right-handed helix (Fig. 1a). The flexibility of B-DNA allows several non-canonical conformations within the genome. Z-DNA was among the first secondary structures to be identified in dsDNA from circular dichroism studies and subsequently from crystallographic analysis (Wang et al. 1979; Ha et al. 2009) which revealed a left-handed helical structure with Watson-Crick base pairing (Fig. 1b). Unlike B-DNA where bases are in anti-conformation, Z-DNA shows alternating syn-anti conformation. This architecture gives the Z-DNA an elongated form, where the inter-base pair distance is longer than that of B-DNA, coupled with a zig-zag arrangement of its sugar-phosphate backbone (Fig. 1b). The sheared base pair stacking seen in Z-DNA tends to pull bases more toward the outside of the helix and makes the structure more rigid than B-DNA (Fig. 1c and d). In addition, Z-DNA has no major groove.
Fig. 1.
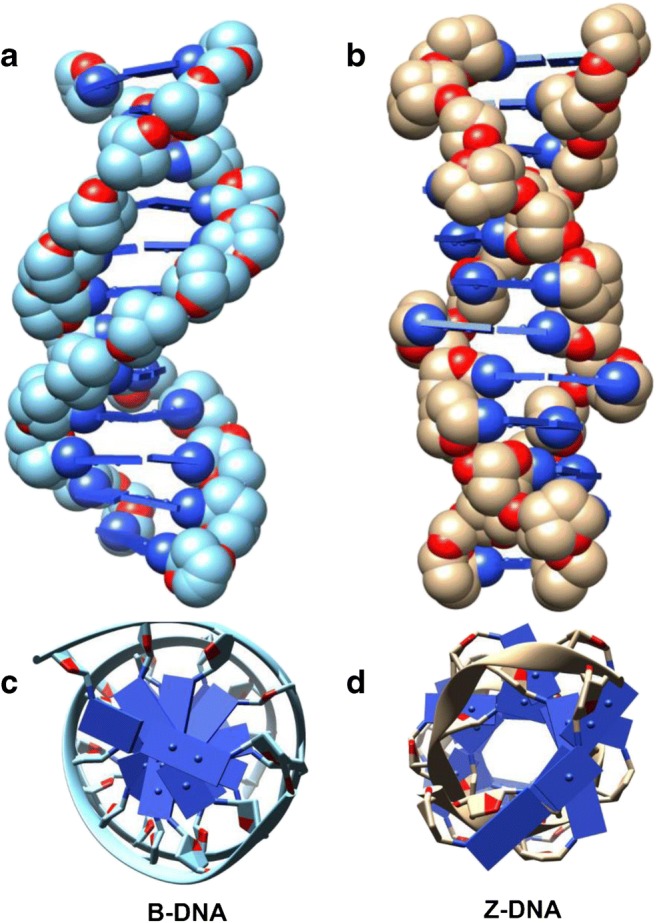
Comparison of B-DNA and Z-DNA structures. a B-DNA showing a right-handed helix. b Z-DNA showing left-handed helix and elongated structure. c, d Top view comparisons of B- and Z-DNA, respectively. B-DNA structure PDB ID: 1FQ2; Z-DNA structure PDB ID: 4LB5
An important non-canonical structure associated with Z-DNA is a BZ junction that forms where B-DNA and Z-DNA meet with an extrusion of bases at the junction between the B- and Z-DNA (Ha et al. 2005; Subramani et al. 2019). Accordingly, when Z-DNA forms in the genome, two BZ junctions flank each side of the Z-DNA-forming site (ZFS). Since Z-DNA formation is energy intensive, BZ junction formation is critical to alleviate torsional stress and stabilize the Z-DNA. Several studies have shown a clear sequence preference regarding the bases that can flip out at the junction, with A-T showing the highest propensity for extrusion (Bothe et al. 2012; Kim et al. 2018).
Conditions for Z-DNA formation
Z-DNA is a higher-order structure known to form in alternating purine-pyrimidine or pyrimidine-purine (APP) dinucleotide repeat sequences, with the propensity of Z-DNA to form in the order GC>CA>TA (Ellison et al. 1986; Ho et al. 1986; McLean et al. 1986). The formation of Z-DNA in vitro was initially tested under very high concentrations of NaCl. Additionally, several chemicals including spermine, spermidine, hexammine cobalt, and ruthenium complexes are found to induce the conversion of B-DNA to Z-DNA (Subramani et al. 2019).
Some of the Z-DNA-forming conditions that are relevant in vivo are the presence of DNA supercoiling, Z-DNA-binding proteins, and base modifications. In early in vitro studies on plasmid DNA, it was found that negative supercoiling favors Z-DNA formation. Z to B transition could be facilitated by the addition of topoisomerases that can relax negatively supercoiled DNA. This is relevant in vivo because when transcription occurs, the movement of RNA polymerase II along the DNA strand generates positive supercoiling in front of, and negative supercoiling behind, the polymerase (Wang and Vasquez 2007). Z-DNA formation could possibly influence transcription by acting as a physical barrier for polymerase progression as seen in the case of prokaryotic systems (Peck and Wang 1985). In human cells, Z-DNA was found to form in actively transcribed regions of the genome and was confirmed using ChIP-Seq (Shin et al. 2016). Three hundred ninety-one ZFS were found in the human genome during this study. Chromatin remodeling can also affect the supercoiled state of the DNA and dynamically modulate Z-DNA formation. The SWI/SNF or BAF complex of proteins can bind near the ZFS and induce changes in supercoiling, thereby favoring Z-DNA formation (Mulholland et al. 2012). Chromatin remodeling by BRG1 facilitates Z-DNA formation in the CSF1 gene promoter and regulates its expression (Liu et al. 2006). The interplay between chromatin remodeling and transcriptional induction of Z-DNA formation was observed in the case of BRG1-induced regulation of CSF1.
Z-DNA is induced and stabilized by Z-DNA-binding proteins (ZBPs) that directly interact with B-DNA. The E3L protein found in vaccinia viruses (Ha et al. 2004) and a PKR-like protein kinase (PKZ) found in fish (Kim et al. 2014) have been identified as ZBPs. Interestingly, two ZBPs found in mammals, adenosine deaminase acting on RNA 1 (ADAR1) and DNA-dependent activator of IFN-regulatory factors (DAI) (Pham et al. 2006; Wang and Vasquez 2007; Ha et al. 2008) have been identified and intensively studied. ZBPs have a conserved Z-DNA-binding domain (Zα) responsible for localization to Z-DNA. Both ADAR1 and DAI are interferon-inducible proteins that aid in nucleic acid clearance from the cytoplasm—an immune response against foreign DNA (Kim et al. 2011; Athanasiadis 2012). In vitro studies have reported that base modifications such as methylation of cytosines and guanines and deamination of adenine increase the propensity of Z-DNA stabilization (Wang et al. 1984; Xu et al. 2003). Like the effects of supercoiling, base modifications are also in vivo phenomena that act as means of epigenetic regulation. Similarly, histone acetylation also has the propensity to induce B- to Z-DNA transition (Zhang et al. 2016). Therefore, Z-DNA is a highly dynamic non-canonical DNA structure with several potent regulators.
Impact of Z-DNA formation in human disease
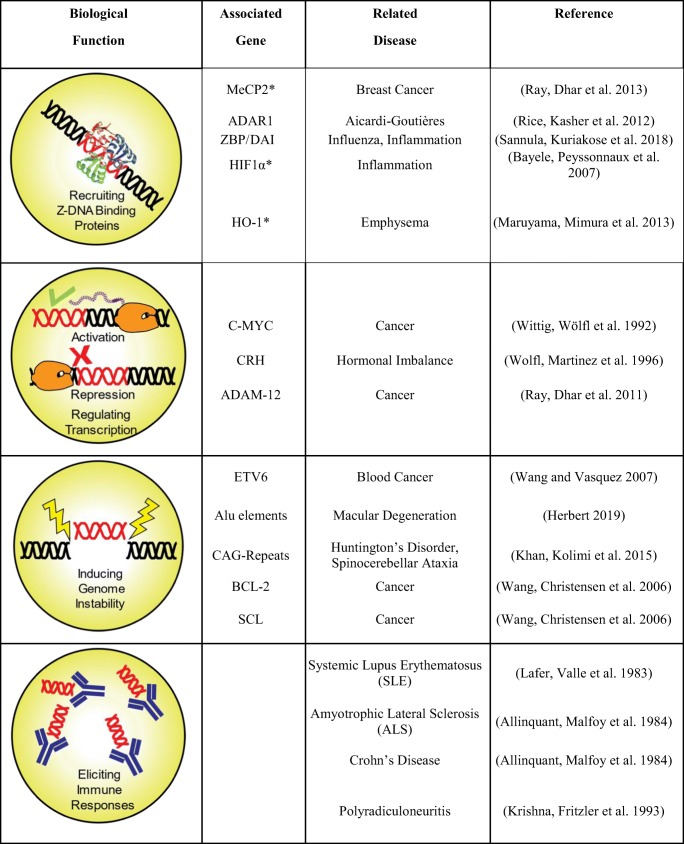
The tight regulation of Z-DNA formation within cells indicates that it plays an important role in cellular activities, such as in the recruitment of specific proteins/transcription activators or repressors, regulation of gene expression, control of genome instability, and eliciting immunogenic responses (Table 1). Specifically, in the case of humans, Z-DNA formation was associated with transcription of the C-MYC and corticotropin-releasing hormone (corticoliberin/CRH) genes, and in activation of the heme oxygenase-1 (HO-1) gene promoter (Wittig et al. 1992; Wolfl et al. 1996; Maruyama et al. 2013). The ADAM-12 promoter contains a Z-DNA-forming sequence (ZFS) that negatively regulates ADAM-12 expression in normal cells (Ray et al. 2011). Loss of this element leads to overexpression of ADAM-12, which is typically a characteristic of many human cancers. It was found that MeCP2 can bind to the ZFS, recruit NF1 transcription factor to an adjacent site, and modulate ADAM-12 repression. Lowered MeCP2 levels were found in metastatic breast cancer cell lines in addition to the loss of the ZFS, thereby underscoring the association of Z-DNA with human disease. Apart from the distinct role of Z-DNA in either activation or repression of gene expression, HIF1α-induced Z-DNA formation within a microsatellite in the SLC11A1 promoter was shown to regulate its heritable variation and allele-specific expression (Bayele et al. 2007).
Table 1.
Roles of Z-DNA formation in the genome and its association with human diseases. While Z-DNA formation (red) is directly associated with diseases in the case of genome instability and immune responses, the association of Z-DNA directly in protein binding and transcription regulation is yet to be well established
*Protein known to bind to Z-DNA forming site, but the ability to bind to Z conformation is unknown
Z-DNA is known to induce structural instability by acting as sites for large-scale deletions in mammalian cells, especially in cancer-related genes such as BCL2, C-MYC, and SCL (Wang et al. 2006). Z-DNA sequences were also found to be enriched with several immunoglobin-related genes such as ETV6 and associated with chromosomal translocations in these regions, especially in the case of blood-related cancers. ZFS overlap with recombination hotspots (Blaho and Wells 1989; Wahls et al. 1990) are also associated with retrotransposon activities and are found to be enriched in Alu elements (Herbert 2019). Z-DNA is also known to be immunogenic and has been implicated in systemic lupus erythematosus (SLE), Crohn’s disease, polyradiculoneuritis, and amyotrophic lateral sclerosis (ALS), whose patients spontaneously produce anti-Z-DNA antibodies (Lafer et al. 1983; Allinquant et al. 1984). Interestingly, ZFS with (CCTG)n (CCAGG)n repeats also have a protective effect on DNA by reducing the potential slipped-strand DNA formation in the myotonic dystrophy type 2 (DM2) gene (Edwards et al. 2009).
In addition, BZ junctions are speculated to be sites for CAG trinucleotide repeat-induced instability (Khan et al. 2015). Similarly, CGG repeats associated with a fragile X chromosome and GAC repeats associated with skeletal dysplasia can also form Z-DNA (Vorlíčková et al. 2001; Renciuk et al. 2010). There is also strong evidence of Z-DNA formation in the hippocampus of Alzheimer’s patients, implicating Z-DNA in neurodegenerative disorders (Suram et al. 2002).
Future perspectives
Z-DNA is a dynamic non-canonical DNA found within the human genome, whose formation is influenced by several molecular players. The multifarious roles that Z-DNA play in human disease present an immense opportunity to target Z-DNA by chemical modulators from either natural or synthetic sources. Therapeutic strategies using small molecules targeted against Z-DNA would theoretically have lower non-target effects due to their conformational specificity. Particularly of intrigue are the BZ junctions whose formation and role have yet to be well studied. It is possible that the extruded bases can be targeted by DNA repair enzymes, transcription factors, or small molecules. A significant opportunity lies in the discovery of novel Z-DNA inducing and repressing small molecules because the current Z-DNA modulators are unsuitable for therapeutic use or have failed in clinical trials (Fuertes et al. 2006). Hence, there is immediate need and urgency to conceptualize and design screening strategies to look for novel modulators of Z-DNA—BZ inducers, Z binders and Z disruptors, ZB inducers, and BZ inhibitors. The future, therefore, holds many exciting opportunities to further our understanding of Z-DNA formation and its targeting for therapeutic purposes.
Acknowledgements
This work was supported by the Samsung Science & Technology Foundation (SSTF-BA1301-01).
Compliance with ethical standards
Conflict of interest
Subramaniyam Ravichandran declares that he has no conflict of interest. Vinod Kumar Subramani declares that he has no conflict of interest. Kyeong Kyu Kim declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allinquant B, Malfoy B, Schuller E and Leng M (1984) Presence of Z-DNA specific antibodies in Crohn disease polyradiculoneuritis and amyotrophic lateral sclerosis.pdf. Clin Exp Immunol 58: 29–36 [PMC free article] [PubMed]
- Athanasiadis A (2012) Zalpha-domains: at the intersection between RNA editing and innate immunity. Seminars in cell & developmental biology 23: 275–280 [DOI] [PubMed]
- Bayele HK, Peyssonnaux C, Giatromanolaki A, Arrais-Silva WW, Mohamed HS, Collins H, Giorgio S, Koukourakis M, Johnson RS, Blackwell JM, Nizet V, Srai SKS. HIF-1 regulates heritable variation and allele expression phenotypes of the macrophage immune response gene SLC11A1 from a Z-DNA forming microsatellite. Blood. 2007;110:3039–3048. doi: 10.1182/blood-2006-12-063289. [DOI] [PubMed] [Google Scholar]
- Blaho JA, Wells RD. Left-handed Z-DNA and genetic recombination. Prog Nucleic Acid Res Mol Biol. 1989;37:107–126. doi: 10.1016/S0079-6603(08)60696-0. [DOI] [PubMed] [Google Scholar]
- Bothe JR, Lowenhaupt K, Al-Hashimi HM. Incorporation of CC steps into Z-DNA: interplay between B-Z junction and Z-DNA helical formation. Biochemistry. 2012;51(34):6871–6879. doi: 10.1021/bi300785b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SF, Sirito M, Krahe R, Sinden RR. A Z-DNA sequence reduces slipped-strand structure formation in the myotonic dystrophy type 2 (CCTG) x (CAGG) repeat. Proc Natl Acad Sci U S A. 2009;106:3270–3275. doi: 10.1073/pnas.0807699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison MJ, Feigon J, Kelleher RJ, 3rd, Wang AH, Habener JF, Rich A. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986;25(12):3648–3655. doi: 10.1021/bi00360a026. [DOI] [PubMed] [Google Scholar]
- Fuertes MA, Cepeda V, Alonso C, Pérez JM. Molecular mechanisms for the B−Z transition in the example of poly[d(G−C)·d(G−C)] polymers. A critical review. Chem Rev. 2006;106:2045–2064. doi: 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- Ha SC, Choi J, Hwang HY, Rich A, Kim YG, Kim KK. The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ alpha(ADAR1) Nucleic Acids Res. 2009;37(2):629–637. doi: 10.1093/nar/gkn976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SC, Kim D, Hwang HY, Rich A, Kim YG, Kim KK. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc Natl Acad Sci U S A. 2008;105(52):20671–20676. doi: 10.1073/pnas.0810463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SC, Lokanath NK, Van Quyen D, Wu CA, Lowenhaupt K, Rich A, Kim YG, Kim KK. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc Natl Acad Sci U S A. 2004;101(40):14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437(7062):1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- Herbert A. Z-DNA and Z-RNA in human disease. Commun Biol. 2019;2:7. doi: 10.1038/s42003-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986;5(10):2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Kolimi N, Rathinavelan T. Twisting right to left: a…a mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation. PLoS Comput Biol. 2015;11(4):e1004162. doi: 10.1371/journal.pcbi.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hur J, Han JH, Ha SC, Shin D, Lee S, Park S, Sugiyama H, Kim KK. Sequence preference and structural heterogeneity of BZ junctions. Nucleic Acids Res. 2018;46(19):10504–10513. doi: 10.1093/nar/gky784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hur J, Park K, Bae S, Shin D, Ha SC, Hwang HY, Hohng S, Lee JH, Lee S, Kim YG, Kim KK. Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ) Nucleic Acids Res. 2014;42(9):5937–5948. doi: 10.1093/nar/gku189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Khayrutdinov BI, Lee CK, Cheong HK, Kang SW, Park H, Lee S, Kim YG, Jee J, Rich A, Kim KK, Jeon YH. Solution structure of the Zbeta domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs. Proc Natl Acad Sci U S A. 2011;108(17):6921–6926. doi: 10.1073/pnas.1014898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer EM, Valle RP, Moller A, Nordheim A, Schur PH, Rich A, Stollar BD. Z-DNA-specific antibodies in human systemic lupus erythematosus. J Clin Invest. 1983;71:314–321. doi: 10.1172/JCI110771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 2006;26(7):2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama A, Mimura J, Harada N, Itoh K. Nrf2 activation is associated with Z-DNA formation in the human HO-1 promoter. Nucleic Acids Res. 2013;41:5223–5234. doi: 10.1093/nar/gkt243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MJ, Blaho JA, Kilpatrick MW, Wells RD. Consecutive a X T pairs can adopt a left-handed DNA structure. Proc Natl Acad Sci. 1986;83(16):5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland N, Xu Y, Sugiyama H, Zhao K. SWI/SNF-mediated chromatin remodeling induces Z-DNA formation on a nucleosome. Cell Biosci. 2012;2:3. doi: 10.1186/2045-3701-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck LJ, Wang JC. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pham HT, Park MY, Kim KK, Kim YG, Ahn JH. Intracellular localization of human ZBP1: differential regulation by the Z-DNA binding domain, Zalpha, in splice variants. Biochem Biophys Res Commun. 2006;348(1):145–152. doi: 10.1016/j.bbrc.2006.07.061. [DOI] [PubMed] [Google Scholar]
- Ray BK, Dhar S, Shakya A, Ray A. Z-DNA-forming silencer in the first exon regulates human ADAM-12 gene expression. Proc Natl Acad Sci. 2011;108:103–108. doi: 10.1073/pnas.1008831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renciuk D, Kypr J, Vorlícková M, Renčiuk D, Vorlíčková M. CGG repeats associated with fragile X chromosome form left-handed Z-DNA structure. Biopolymers. 2010;95:174–181. doi: 10.1002/bip.21555. [DOI] [PubMed] [Google Scholar]
- Shin SI, Ham S, Park J, Seo SH, Lim CH, Jeon H, Huh J, Roh TY (2016) Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome. DNA Res [DOI] [PMC free article] [PubMed]
- Subramani VK, Ravichandran S, Bansal V, Kim KK. Chemical-induced formation of BZ-junction with base extrusion. Biochem Biophys Res Commun. 2019;508(4):1215–1220. doi: 10.1016/j.bbrc.2018.12.045. [DOI] [PubMed] [Google Scholar]
- Suram A, Rao KSJ, Latha KS, Viswamitra MA. First evidence to show the topological change of DNA from B-dNA to Z-DNA conformation in the hippocampus of Alzheimer's brain. NeuroMolecular Med. 2002;2:289–297. doi: 10.1385/NMM:2:3:289. [DOI] [PubMed] [Google Scholar]
- Vorlíčková M, Kejnovská I, Tumová M, Kypr J. Conformational properties of DNA fragments containing GAC trinucleotide repeats associated with skeletal displasias. Eur Biophys J. 2001;30:179–185. doi: 10.1007/s002490000121. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990;10:785–793. doi: 10.1128/MCB.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AHJ, Hakoshima T, van der Marel G, van Boom JH, Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG) Cell. 1984;37:321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang AHJ, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci U S A. 2006;103(8):2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- Wittig B, Wölfl S, Dorbic T, Vahrson W. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfl S, Martinez C, Rich A, Majzoub JA. Transcription of the human corticotropin-releasing hormone gene in NPLC cells is correlated with Z-DNA formation. Proc Natl Acad Sci. 1996;93:3664–3668. doi: 10.1073/pnas.93.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ikeda R, Sugiyama H. 8-Methylguanosine: a powerful Z-DNA stabilizer. J Am Chem Soc. 2003;125:13519–13524. doi: 10.1021/ja036233i. [DOI] [PubMed] [Google Scholar]
- Zhang F, Huang Q, Yan J, Chen Z. Histone acetylation induced transformation of B-DNA to Z-DNA in cells probed through FT-IR spectroscopy. Anal Chem. 2016;88:4179–4182. doi: 10.1021/acs.analchem.6b00400. [DOI] [PubMed] [Google Scholar]