Abstract
Interest in protein folding intermediates lies in their significance to protein folding pathways. The molten globule (MG) state is one such intermediate lying on the kinetic (and sometimes thermodynamic) pathway between native and unfolded states. Development of our qualitative and quantitative understanding of the MG state can provide deeper insight into the folding pathways and hence potentially facilitate solution of the protein folding problem. An extensive look at literature suggests that most studies into protein MG states have been largely qualitative. Attempts to obtain quantitative insights into MG states have involved application of high-sensitivity calorimetry (differential scanning calorimetry and isothermal titration calorimetry). This review addresses the progress made in this direction by discussing the knowledge gained to date, along with the future promise of calorimetry, in providing quantitative information on the structural features of MG states. Particular attention is paid to the question of whether such states share common structural features or not. The difference in the nature of the transition from the MG state to the unfolded state, in terms of cooperativity, has also been addressed and discussed.
Keywords: Molten globule state, Protein folding, Isothermal titration calorimetry, Differential scanning calorimetry, Thermal transitions, Thermodynamic signatures
The molten globule state
The term molten globule was coined by Ohgushi and Wada in 1983 (Ohgushi and Wada 1983), with due credit to O. B. Ptitsyn and C. Crane-Robinson about discussion on this terminology. The traditional definition of the molten globule (MG) state describes it as a compact intermediate in which the tertiary structure of the protein is lost but the secondary structure is intact or even strengthened (Kuwajima 1989). An often-used alternative definition of the MG state is “a state in which specific tertiary structure is disrupted without loss of the secondary structure” (Ptitsyn 1987; Kuwajima 1989). The MG state can therefore be considered similar to the partially folded state, sometimes observed during the unfolding of a protein. In the early 1990s, interest in the MG state arose jointly due to its presumed similarity with the intermediate state in protein folding, and as a translocation competent state in the transport of a precursor protein across biological membranes (Bychkova et al. 1988; Martin et al. 1991; Van der Goot et al. 1991). It was felt that thermodynamic characterization of MG states may provide key information in unraveling the protein folding problem.
Different models have been proposed to explain protein folding in the past, some of which are the framework model, the nucleation model, the diffusion collision model, the hydrophobic collapse model, and the ensemble new view models. The framework model proposed by Ptitsyn (1973) considered the formation of secondary structural elements followed by the formation of further advanced folding intermediate and then specific packing of the side chains (Udgaonkar and Baldwin, 1988). This model suggested the existence of several folding intermediates during the folding process (Ptitsyn and Rashin 1975; Kim and Baldwin 1982; Roder et al. 1988). A few years later, Karplus and Weaver (1976) assumed that many unstable quasiparticles constitute the intact protein. The quasiparticles are also known as microdomains which are portions of nascent secondary structures and hydrophobic clusters. In order to achieve stability, the microdomains diffuse, collide, and eventually coalesce. Hydrophobic collapse was predicted as an early event in the folding process (Go 1984; Agashe et al. 1995; Rackovsky and Scheraga 1977; Dill 1985; Arai et al. 2007) after it was recognized that the interior of the native states of proteins normally contains a hydrophobic core of nonpolar amino acid residues. On similar lines, collapse around a diffuse nucleus was considered to propose the nucleation condensation model assuming that the limiting step in the folding of a protein is nucleus formation (Fersht 1997). The nucleus formation is followed by a fast propagation of the structure. The main feature of the nucleation condensation model is that it proposes the formation of simultaneous secondary and tertiary interactions (Nölting and Agard 2008).
The new views on protein folding models propose energy landscape and folding funnel models (Bryngelson et al. 1995; Onuchic et al. 1997) based on the dependence of free energy on the coordinates that determine protein conformation. According to this most adopted model, the folding of a protein from the highest energy level disordered state flows down a funnel, passing through intermediates towards global energy minima which corresponds to its native conformation. The zipper model (Dill et al. 1993; Munoz et al. 1997) suggests a zipper-like folding process whereas the funnel model emphasizes on parallel pathways of folding (Wolynes et al. 1995; Onuchic et al. 1996). Molten globule is represented as one of the intermediate states in the funnel model (Onuchic et al. 1996).
Resemblance of the kinetic intermediates formed during protein folding with the equilibrium MG state for some proteins such as apomyoglobin and α-lactalbumin has been discussed earlier (Barrick and Baldwin 1993; Jennings and Wright 1993; Balbach et al. 1995; Forge et al. 1999). The importance of the MG state and similar other nonnative states of the protein in their transition to the MG state has been recognized (Bychkova et al. 1988; Penkett et al. 1998; Kelly 1998; Chiti et al. 1999). The role of the MG state of human α-lactalbumin in apoptosis in tumor cells has also been reported (Svensson et al. 1999).
Structural studies of MG states obtained under equilibrium conditions for a variety of proteins raised the general question of whether these intermediate states are a universal feature of protein behavior (Haynie and Freire 1993). Another term, the “pre-molten globule state” was also coined in the early 1990s (Jeng and Englander 1991). Like the MG state, the pre-molten globule had intact elements of secondary structure, but unlike the MG state, it was not considered compact. Articles did appear about the solvation dynamics (Samaddar et al. 2006) and conformation and thermodynamic stability of the pre-MG state (Khan et al. 2011) along with various models for MG states (Fink et al. 1998), but the information available from these studies remained qualitative in nature. With the passage of time, the interest in the characterization of the pre-MG state declined significantly and currently there are hardly any reports which address such pre-MG states.
Recently, Takahashi et al. (2018) have hypothesized structural heterogeneity of the unfolded proteins originating from the coupling of the local clusters and long-range distance distribution. These authors observed peak broadening in the fluorescence resonance energy transfer (FRET) efficiency plot for the unfolded proteins and suggested the significance of local heterogeneous clusters in the unfolded states of proteins. Obtaining further insights into such clusters could be important in understanding the mechanism of protein folding. FRET methods have also been applied in studies of intrinsically disordered proteins in statistical terms (Haas 2012). It was suggested that the determination of intramolecular distance distributions by ensemble and single-molecule FRET experiments enables exploration of partially folded states in the refolding of protein molecules (Haas 2005).
Sasahara et al. (2000) observed a partially unfolded equilibrium state of hen egg white lysozyme based on circular dichroism spectroscopic measurements. They further observed that the transition from intermediate to unfolded state is associated with low cooperativity and small enthalpy and entropy changes. These authors did not explicitly use the term MG state, but suggested that the observed intermediate has characteristics of kinetic intermediates observed in the refolding pathway of hen lysozyme.
Conception and progress in understanding MG states
Even though the protein folding problem was addressed as early as the 1960s (Perutz et al. 1960; Anfinsen et al. 1961; Haber and Anfinsen 1962; Dill and MacCallum 2012), the intermediate states, especially the MG state, only began receiving attention from the early 1980s; however, the number of research articles published per year was less than 10 up to the year 1990. Figure 1 suggests that the scattered information about such a state in the 1980s did not produce significant interest in the scientific community. However, a sharp increase in research on the MG state from 1990 to 1996 indicated a growing understanding of the importance of the MG conformation. From the year 1990 onwards, up to 2000, the number of research articles published on MG states increased significantly (see Fig. 1).
Fig. 1.
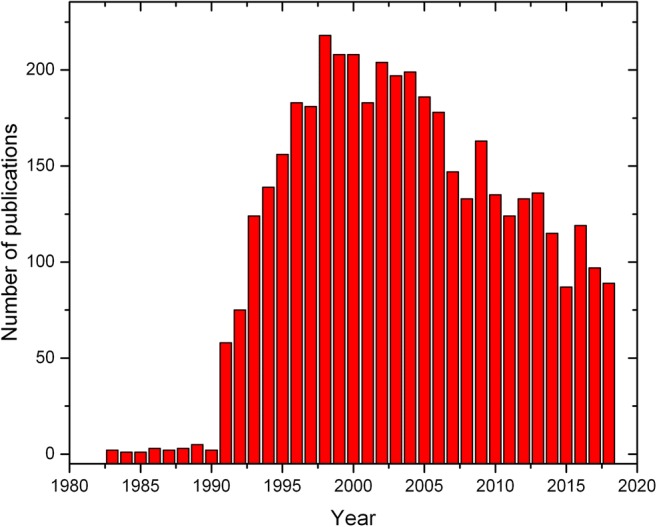
Number of publications addressing the molten globule state in proteins from the early 1980s up to 2018 representing the rise and decline in scientific output on the knowledge of intermediate states (source: Web of Science)
As judged by Fig. 1, the amount of scientific literature on the MG states started declining after the year 2000. This could mean that either a deeper understanding of the MG state is not important or it is becoming extremely difficult to obtain new information on this state over and above the existing knowledge. In view of the tough task of obtaining more insights and relevance of the MG state, rigorous efforts need to be continued to understand the role of the MG state not only in the protein folding but also in its connection to aggregation and fibrillization of the proteins (Ptitsyn 1973; Hammarströ et al. 1999; Chiti et al. 1999; Povarova et al. 2010; Skora et al. 2010).
Characterization of the MG state of the protein has mostly been done qualitatively by circular dichroism spectroscopy, a technique capable of providing direct evidence on the extent of loss of tertiary structure and retaining/strengthening of the secondary structure (Vassilenko and Uversky 2002). Measurement of the intrinsic fluorescence on proteins has also provided much supporting information on the MG state (Swaminathan et al. 1994). For dye-based fluorescence investigations, the dye, 8-anilino-naphthalene-1-sulfonic acid, has been an essential marker for identification of protein MG states. It is well established that ANS binds to the MG state of the protein with higher affinity compared to that with the native and denatured states (Gussakovsky and Haas 1995). Enhanced fluorescence emission of ANS, when excited at 295 nm, has routinely been used to characterize the MG state (Stryer 1965; Semisotnov et al. 1991). It is believed that the MG intermediate states of proteins, with different structure and function, share similarities in their characteristics in terms of the content of secondary structure, hydrophobic core, and loss of tertiary structure (Kuwajima 1989; Ptitsyn 1995).
Fluorescent probes for qualitative MG state characterization
As mentioned in the previous section, the fluorescent probe, 8-anilino-1-naphthalene sulfonic acid (ANS), has long been used to characterize the partially folded states of proteins (Semisotnov et al. 1991). Alongside ANS binding, native tryptophan fluorescence has been a key technique (whereby the fluorescence emission intensity of the tryptophan(s) in the protein increases differentially in the MG state compared to the native or unfolded state of the protein). This property has been extremely useful for characterization of MG states in protein chemistry (Redfield et al. 1994).
With the availability of high-sensitivity isothermal titration calorimetry, it has become possible to rationalize the binding of fluorescent probes to, and the nature of the native fluorescence of, the protein MG state, by comparing these fluorescence profiles against the thermodynamic signatures reflecting associated with the binding and unfolding processes. Specific binding of dyes to the MG state can be related to the environment of the exposed hydrophobic core, which is different in the MG state compared to the native and unfolded states. ANS has been used as a probe to identify molten globule states for a variety of proteins such as human serum albumin (HSA), recombinant human growth hormone, stem bromelain, and more (Hawe et al. 2008). Another dye, Nile Red, was also used to characterize the MG states—one example involved monitoring the denaturation of HSA in the presence of GdnHCl (Hawe et al. 2008). In this study, it was observed that over the guanidine hydrochloride concentration range 0.25–1.5 M, the intensity of Nile Red increased and then dropped but ANS showed maximum fluorescence intensity at 1.8 M guanidine hydrochloride which corresponds to the MG state of the protein.
In another study, ANS and pyrene were used to characterize the MG state of BSA (Hawe et al. 2008), characterized at pH 4.2 using ANS fluorescence (supplemented by CD spectroscopy, light scattering, and analytical centrifugation), but pyrene showed the same intensity at pH 4.2 and pH 7.0. In some studies, bis-ANS was also tried as a probe to study the MG state of the protein. These studies indicated that MG characterization by dyes depends on the properties of a particular dye and also on the structural conformation of the molecules (Hawe et al. 2008).
Conflicting results on the MG state of proteins
Thermodynamic characterization of the MG state of protein has produced conflicting observations. For example, based on the acidic pH-induced MG state of apo-α-lactalbumin, Kuwajima (1989) has reported that such a state does not undergo cooperative thermal transition to the unfolded state, which met with objections later on. In the years 1991 and 1992, there were conflicting reports on the thermodynamic state of the unfolded state of the protein compared to the MG state. Xie et al. (1991) demonstrated that the guanidine hydrochloride-induced MG state of apo-lactalbumin exhibited a well-defined thermal unfolding profile based on variation of heat capacity by using differential scanning calorimetry (DSC). Based on their results, it was concluded that the difference in intrinsic enthalpy between the native and MG states is 32.2 kJ mol−1 which is much lesser than that 133.1 kJ mol−1 between its unfolded and native states (Xie et al. 1991). These observations conflicted with the findings of Ptitsyn and Kuwajima (Ptitsyn 1987; Kuwajima 1989) who proposed that the MG state of a protein does not undergo a cooperative thermal transition. The heat absorption curve of holo-lactalbumin obtained by Yutani et al. (1992) showed a well-defined cooperative thermal transition centered at 61.7 °C, consistent with those reported by others. However, apo-lactalbumin under the same experimental conditions did not result in any cooperative thermal transition. According to the results of Yutani et al. (1992), the absence of cooperative thermal transition in DSC from MG to unfolded state clearly has a correlation with the complete loss of tertiary structure in apo-lactalbumin as seen in their near uv-cd spectra.
In contrast to the observations of Yutani et al. (1992), Xie et al. (1991) obtained a well-defined cooperative thermal transition in DSC centered at about 43 °C accompanied by a calorimetric enthalpy of 276 ± 13 kJ mol−1. Further, addition of guanidine hydrochloride up to a concentration of 1.5 M showed a consistent lowering of both the transition temperature and calorimetric enthalpy. The difference between calorimetric and van’t Hoff enthalpy was clearly observed by them for the MG state of α-LA indicating deviations from two-state unfolding behavior.
Emerging qualitative information on the MG state and limited quantitative information
Recognizing difficulty in obtaining thermodynamic information on the MG state of proteins, it was essential to use alternate approaches for its characterization. One such approach was isothermal titration calorimetry (Hamada et al. 1994). Cytochrome c undergoes denaturation when the pH is lowered to 1.8 due to excessive repulsion of protonated residues (Goto et al. 1990). Addition of perchlorate to the acid-unfolded state of cytochrome c pushes the protein to the MG state which was monitored by using ITC. The authors obtained exothermic interaction of perchlorate with the protein, and subsequent titration of the salt into the protein led to a cooperative behavior which agreed well with the conformational transition monitored by measuring ellipticity at 222 nm. Their observations suggested that the salt-induced conformational change from unfolded to MG state of cytochrome c can be approximated by two-state transition. The authors observed the appearance of well-defined thermal transition in the protein at pH 1.8 upon addition of various amounts of NaClO4. These observations also conflict with the observations of Yutani et al. (1992), but agree with the result of Xie et al. (1991) that the MG state of the protein does undergo cooperative thermal transition. The striking feature of their observations is that the calorimetric enthalpy of the unfolding of the MG state determined by DSC matched well with the van’t Hoff enthalpy obtained by fluorescence measurements, thereby establishing the two-state nature of the unfolding process. A nonzero, small positive value of ΔCp confirmed increased exposure of the buried or clustered hydrophobic groups of the protein in the MG state to the solvent environment. In this report, the authors could successfully ascribe quantitative numbers to the formation of the MG state.
The formation of the MG state in acid-unfolded cytochrome c induced by n-alkyl sulfates was also attempted and studied by using ITC (Chamani et al. 2003). Here also, the authors obtained exothermic enthalpies of conformational transition while recording the transition in parallel, using CD spectroscopy. During the same time period, another report described the formation of the MG state by acid-induced unfolding of cytochrome c in the presence of SDS (Moosavi-Movahedi et al. 2004). Once again, these authors reported a well-defined thermal unfolding profile in the DSC thermogram of the MG state of cytochrome c. The major point of difference in their report is that the endotherm for the unfolding of the MG state was fitted by the multistate model. The claim of these authors to assign four energetic sub-domains in the unfolding of the MG state of cytochrome c had support from an earlier report in literature (Fisher and Taniuchi 1992). Here, the authors did quantify the MG state to some extent, though differently than others.
Apomyoglobin has also been studied for an understanding of the MG state of the protein (Hamada et al. 1995). Acid-induced (pH = 2) unfolded apomyoglobin could be stabilized to the MG state by NaClO4 and Na2SO4. The approach to obtain quantitative information by employing calorimetry was the same as that used for cytochrome c. The authors found that the salt-induced conformational change in apomyoglobin from unfolded to MG state could be approximated by a two-state transition which is exothermic in nature. The quantification of thermodynamic signatures included the possibility of heat effects due to conformational changes in the protein. An essential point of consideration is how changes in conformation may produce the heat effects.
Partially unfolded states of human α-lactalbumin by molecular dynamics simulations have been explored by Paci et al. (2001). Based on their computational findings, they concluded that the unfolding of the MG state is not a cooperative process, thereby suggesting that the structural elements of the protein do not unfold simultaneously.
The protein Galectin-1, in the presence of increasing concentrations of guanidine hydrochloride, exhibited a biphasic unfolding profile—attributed to the existence of at least one stable intermediate (Iglesias et al. 2003). This intermediate was argued to belong to the molten globule type, by virtue of the fact that it retained carbohydrate binding specificity. This study, however, remained mostly qualitative with regard to the MG state; the biphasic nature of the unfolding profile suggested cooperative unfolding of the MG state to the denatured state induced by guanidine hydrochloride.
Specific quantitative information on the MG state provided by calorimetry
As discussed earlier, characterization of partially folded states, such as MG, has routinely been done by means of fluorescence spectroscopy by using ANS as a probe (Gussakovsky and Haas 1995; Stryer 1965; Semisotnov et al. 1991). It has widely been reported that enhancement in the fluorescence emission of ANS occurs when it binds specifically to the partially folded states including MG states. However, this provided only qualitative information and was mainly used like a fingerprinting method. The use of ITC in characterizing the partially folded states of proteins was looked upon with great promise (Singh and Kishore 2006). It was believed that a quantitative understanding of the interaction of ANS with the MG state of the protein might be able to elucidate the common structural features of such an intermediate state (if any existed). Thermodynamic signatures associated with the binding of dye to the MG state, such as stoichiometry, binding constant, enthalpy change, entropy change, and van’t Hoff enthalpies, can provide the nature of intermolecular interactions and mechanism of binding, and hence inform on the structural features of such states. Experiments were designed to study the binding of ANS with the native state, denatured state, and MG state of the protein α-lactalbumin (Singh and Kishore 2006). The binding of ANS with the native state was observed to be weak, and the ITC-integrated heat profile of interaction with the urea-induced denatured state did not yield any specific binding profile. However, the ITC of the binding of ANS with the A-state (also known as the MG state) of α-lactalbumin provided a valley-shaped titration profile which followed a two-site binding model with multiple ANS molecules binding at each site. These two binding sites exhibited an exothermic interaction in the range of − 21.1 kJ mol−1 to − 10.8 kJ mol−1 and affinity constants of the order of 104 and 106 M−1 at these sites, respectively (at 298.15 K). The difference of 10.3 kJ mol−1 leads to a valley-shaped ITC profile instead of a normal sigmoidal ITC-integrated heat profile. The binding of ANS at these sites was also observed to have positive entropic contributions. The results suggested that each site could accommodate 3.1 to 14.5 molecules which further indicates that such a binding does not occur at well-defined clefts of amino acid residues, but involves nonspecific binding which is predominantly exothermic in nature. It is known that ANS usually binds to hydrophobic clusters with a possibility to also interact with ionic centers due to the presence of a sulfonate group (Gussakovsky and Haas 1995, Stryer 1965, Semisotnov et al. 1991). The data discussed above suggested dominance of polar heat effects. Nonequivalence of calorimetric and van’t Hoff enthalpies associated with the binding of ANS to the MG states of proteins and absence of enthalpy/entropy compensation point out the lack of well-defined binding sites on the MG state and suggest the association to be nonspecific in nature (Singh and Kishore 2006).
Isothermal titration calorimetry was also employed to quantitatively understand the binding of ANS to the 2,2,2,-trifluorethanol (TFE)-induced MG state of concanavalin A (Banerjee and Kishore 2005). It was observed that 4 mol kg−1 TFE at pH 2.5 is able to induce the MG state in concanavalin A which was well characterized by fluorescence and circular dichroism spectroscopies. The DSC thermal profiles of concanavalin A at pH 5.2, where the protein does not exist in the MG state in the absence and presence of TFE, exhibited a well-defined cooperative transition to the unfolded state. However, the MG state of lectin at pH 2.4 lacked such cooperative thermal transition. Here also, the binding of ANS to the MG state of concanavalin A followed a two-state binding model with values of binding constants of the order of 103 and 105 M−1. Values for the enthalpy of binding ranged from − 0.20 to − 20.27 kJ mol−1 at 298.15 K. It is worth noticing here that the association of ANS with the MG state of concanavalin A is also observed to be exothermic in nature with favorable entropy change. The stoichiometry of binding at each site is more than one, varying from 3.8 to 24.8, suggesting a larger area of the binding site.
Studies have also been done with the partially folded state of α-lactalbumin induced by the mixture of hexafluoroisopropanol and guanidinium thiocyanate, the former being a helix inducer and the latter, a denaturant (Sharma and Kishore 2008). ANS binding to these partially folded states with varying levels of secondary and tertiary structures also followed a two-type binding site model, with the order of binding constants as 102 to 104 M−1, but weak endothermic enthalpy and favorable entropy change. These intermediate states exhibited thermal transition to the unfolded state, in agreement with the data provided by absorbance changes.
Binding of ANS to the TFE-induced partially folded states of myoglobin was studied by using calorimetry and spectroscopy (Talele and Kishore 2015). Here, 1.5 mol kg−1 TFE was sufficient to induce the MG state in the protein. The DSC thermal profile did not show any cooperative thermal transition, and ITC studies on the binding of ANS with the TFE-induced MG state of myoglobin yielded thermodynamic parameters of binding according to a two-site binding model. The binding constants obtained here were of the order of 105 and 107 M−1, with exothermic enthalpy of binding –(18.08 ± 0.14) kJ mol−1 and –(1.04 ± 0.05) kJ mol−1, respectively, at 298.15 K. Consistent with earlier observations, the binding of ANS with the MG state of myoglobin was also observed to be enthalpy driven. Surfactants which induced a partially folded state of α-LA also offered two sequential binding sites of ANS molecules with binding constants of the order of 103 and 104 and exothermic enthalpy of binding of –(12.1 ± 0.03) kJ mol−1 and –(35.7 ± 1.5) kJ mol−1, thereby establishing consistency of the binding model fitted to the experimental data (Misra and Kishore 2011).
Even though qualitative information on the MG states has emerged over time, the determinants of MG stability and the extent of specific packing in the MG state are still a matter of investigation. Formation of a molten globule state at acidic pH has been demonstrated for the periplasmic binding proteins [lipopolysaccharide-binding protein (LBP), leucine-isoleucine-valine-binding protein (LIVBP), maltose-binding protein (MBP), and retinol-binding protein (RBP)] (Prajapati et al. 2007). These proteins were shown to possess the ability to bind to their corresponding ligands without undergoing transition to the native state, albeit with a smaller affinity in most cases (Prajapati et al. 2007). These observations suggested that periplasmic binding proteins preserve a significant degree of long-range order even in the MG state. The DSC thermograms of all these four proteins exhibited well-defined unfolding behavior, and the thermal stability of the proteins could be raised by almost 15 °C upon binding their cognate ligand. Further, all their transitions fitted well to the two-state unfolding model. Here, the authors observed an almost complete loss of tertiary structure of the proteins, but still appreciable endotherm in the DSC profiles and significant binding to the respective ligands.
Current understanding suggests that MG states can exhibit varying degrees of compactness and solvent accessibility (Dijkstra et al. 2018). It is also argued that the nature of the MG state is highly sequence dependent and that the heat capacity versus temperature curves may exhibit heat-induced MG states. These authors further suggest that the MG state does not necessarily adopt a particular conformation and that natively disordered proteins can exhibit multiple MG-like states. With regard to the nature of DSC thermal profiles, the authors argue that MG states do show peaks in the heat capacity curves with a reduced enthalpy to that of unfolding of the native protein. For the protein staphylococcal nuclease, three different partially folded intermediates lacking rigid structure, but nevertheless containing significant tertiary structure, were observed in anion-induced refolding experiments (Uversky et al. 1998). Although mostly qualitative in nature, that study suggested that the intermediates observed represent the equilibrium counterparts of transient kinetic intermediates.
Pressure perturbation calorimetry (PPC) has also been applied to characterize the molten globule state of cytochrome c at pH 4.5. This technique evaluates the temperature dependence of the thermal expansion coefficient of the protein (Nakamura and Kidokoro 2012). The thermal unfolding curve obtained by PPC could be fitted by three-state analysis including the MG state. The partial specific volume of the MG state was found to be in between that of the N and D states. Here, the cooperative thermal transition obtained from the MG to the unfolded state is not based on heat capacity but based on volumetric parameters determined on DSC. Even though the observed thermodynamic quantity determined here is somewhat nonstandard, it demonstrated that cytochrome c in the MG state does exhibit sufficient compactness which upon heating yields a cooperative thermal transition. The change in volume of the protein as a result of conformational transitions is small and may be either positive or negative. It has been observed that the change in solution volume, per unit volume of the protein, for the unfolding of ribonuclease A, ubiquitin, lysozyme, and eglin C, all converge to a common value at high temperature (Schweiker et al. 2009). The protein cytochrome c exhibited a different value of this volumetric ratio than the above mentioned proteins. This difference was assigned to a loosely packed structure of cytochrome c compared to the other proteins. Similar differences are seen in the thermal unfolding behavior of the MG states of cytochrome c and the other mentioned proteins where the thermal transition for the former protein is cooperative and for the latter set of proteins is noncooperative (Dolgikh et al. 1985; Nakamura et al. 2007, 2011; Potekhin and Pfeil 1989; Hamada et al. 1994). The volume of the intermediate state of cytochrome c is observed to be smaller than that of the denatured state which indicates that the hydrophobic core in the protein is still retained in the MG state of the protein (Nakamura and Kidokoro 2012).
Isothermal titration calorimetry was used to determine the unfolding enthalpy of the pH 4 induced MG state of apomyoglobin (Tyagi et al. 2009). In that experiment, apomyoglobin at pH 5.5 was placed in the cell of the ITC and HCl was sequentially titrated to determine the heat of interaction. After correcting for the heat effects associated with protonation of 22 carboxylate residues, the enthalpy change associated with the unfolding of apomyoglobin to the unfolded state of the protein is zero, and on this basis, the authors argued against cooperative unfolding of the MG state. These observations are consistent with the results obtained by Griko and Privalov (1994). There have been other reports on apomyoglobin describing very shallow thermal unfolding curves for the MG state (Nishii et al. 1995). In agreement with the above ITC observations, Hamada and coworkers (Hamada et al. 1995) reported a zero enthalpy of anion-induced folding of horse heart myoglobin at 10 °C.
Calcium-binding lysozyme [canine milk lysozyme (CML)] has also been studied using DSC. CML exhibits two thermal transitions which correspond to the native to intermediate and intermediate to unfolded transitions, respectively (Koshiba et al. 2001). In contrast to the absence of a thermal transition in apo-α-LA, the intermediate state of CML showed a well-defined heat absorption peak. An interesting feature of this transition was that it occurred at a temperature higher than that of the first transition. Whether or not the transition of an MG state to the unfolded state is cooperative was previously suggested to be related to the extent of unique tertiary structure of the protein (Privalov 1979; Privalov and Gill 1988; Shakhnovich and Finkelstein 1989). Therefore, any deviations in the thermal unfolding profile of an MG state must reflect the unique packing of the amino acid residues in such a state. This in turn should be strongly dependent on the nature of the amino acid sequence (Koshiba et al. 2000).
A qualitative study on the acid-induced MG state of the prion protein was reported as it was believed that insights into the conformational aspects of the intermediate may provide crucial insights into the mechanism of oligomerization and pathogenic conversion, thereby helping in the design of new medical chaperones useful in the treatment of prion diseases (Honda et al. 2014). However, that study was largely qualitative and did not address any thermodynamic aspects. In this direction, the MG state of bovine pancreatic trypsin inhibitor was also addressed (Ferrer et al. 1995). The difference in the cooperativity of transition was correlated with the extent of α-helical or β-sheet structure in the protein.
The unfolding of N-acylamino acid amido hydrolase (aminoacylase) has been investigated in the presence of aspartate using ANS fluorescence spectra, CD, enzyme activity, and intrinsic fluorescence emission spectra. Previous studies on the denaturation of aminoacylase using urea, guanidine hydrochloride, SDS, and temperature induction methods showed a two-state transition without any indication of intermediates (Rariy and Klibanov 1997; Sato et al. 1996). Later, Bai et al. (1999) showed the existence of an intermediate state in the guanidine hydrochloride-induced unfolding of aminoacylase. Aspartate, a weak acid with pI = 2.77, was used to study the unfolding of aminoacylase (Xie et al. 2003) due to the fact that it helps to maintain the structural integrity of the protein and only weakly affects the enzyme functions (it also helps to keep the dimeric structure of the protein intact). The pH-dependent fluorescence transition of aminoacylase in the presence of aspartate indicated two distinct processes. The transition from pH 5.6 to 4.0 showed a red shift, followed by a blue shift from pH 3.7 to 3.0. This indicated a structural modification of the protein, in which the tryptophan moiety is surrounded more by hydrophobic residues. The CD results reinforced the fluorescence observations in the sense that an increase in aspartate concentration caused the secondary structure of aminoacylase to revert to the native-like state (but with an increase in ANS fluorescence intensity). This indicated the presence of an MG state during the unfolding of aminoacylase.
The anionic surfactant, sodium dodecylsulfate, has been reported to induce a MG state in the highly negatively charged ferricytochrome c at pH 12.8 (Jain et al. 2018). Of note here, even though both the protein and head groups of the surfactant molecules in the pre-micellar state carry a negative charge, the stabilization of the unfolded state to the MG state is still observed. In this study, the MG state was reported to have a native-like α-helical structure but lacked an appreciable tertiary structure. SDS-induced unfolding of the protein studied by CD and fluorescence spectroscopies provided a standard molar Gibbs energy change of 11.3 kJ mol−1 and 8.4 kcal mol−1, respectively, at 298.15 K. Though positive, these values are relatively small, and therefore suggest a lack of cooperativity in the unfolding of the MG state to an unfolded state. The authors attribute preferential stabilization of the MG state to interactions between Na+ ions and the negatively charged protein and hydrophobic interactions via the tail groups of the surfactant. It was further inferred that sub-micellar concentration of SDS leads to enhancement of thermal stability and prevents cold denaturation of the alkali-induced denatured state of cytochrome c. The information available from this study reflected more on the mechanistic details but suggested lack of cooperativity in the transition from native to MG state.
Maltose-binding protein (MBP) is a periplasmic single-chain monomeric protein of Escherichia coli having 370 amino acids and no disulfide bonds. MBP forms a MG state at pH 3. The value of ΔCp associated with the thermal unfolding of a protein can provide information on the extent of compactness of the protein. The unfolding of salt-induced MG state of yeast iso-1-cytochrome c exhibited ΔCp = 3.2 kJ K−1 mol−1; for equine-cytochrome c, the value was in the range of 1.6 kJ K−1 mol−1 to 2.4 kJ K−1 mol−1, values which are lower than the change in the heat capacity of the protein undergoing transition from native to denatured state (Kuroda et al. 1992; Hamada et al. 1994). The ΔCp value for the unfolding of the MG state of MBP was observed to be only 30% different than the native state of the protein. This study developed a quantitative understanding of the MG state of MBP in terms of change in heat capacity and its correlation to the accessible hydrophobic surface area (Sheshadri et al. 1999).
Recent qualitative studies on the MG state
A number of interesting, largely qualitative, studies on the MG state have been recently published, and some of these examples are discussed ahead. Stabilization of apo-α-lactalbumin by binding of epigallocatechin-3-gallate has been observed both with the native and MG states of the former. Binding of epigallocatechin gallate with the MG state of the protein indicated sufficient extent of structural features in the intermediate state to allow binding to the incoming ligand molecules (Radibratovic et al. 2019). Formation of the MG state in homodimeric CcD B [controller of cell death B] protein has been characterized spectroscopically (Baliga et al. 2019). That study provided insights into structural and dynamic properties of a low-pH state of CcD B. Several other reports addressing either the formation or role of the MG state have been published (Peixoto et al. 2019, Kozak et al. 2018, Kulkarni and Uversky 2018; Uversky 2018; Wirtz et al. 2018; Samanta et al. 2017, Ithychanda et al. 2017).
Pressure perturbation calorimetry was used to understand the MG state of cytochrome c, finding a high-temperature reversible oligomerization process (Zhang et al. 2017). Here, the transition from MG state to unfolded state was studied at different concentrations of the protein. The calorimetric data suggested at least a six-state unfolding process involving two MG states, the denatured state and further arrangement of the denatured state to dimeric, trimeric, and tetrameric forms, with each step being reversible. The cooperative thermal transition of the MG state of cytochrome c was also observed here. Xie et al. (1991) have reported that at neutral pH, the enthalpy of the unfolded state of α-LA differs by 100.8 kJ mol−1 at 25 °C from its MG state. Yutani et al. (1992), who observed that the enthalpy difference between the MG state and the presumed unfolded state is almost zero, pointed out this difference to the model used by Xie et al. (1991). They assigned this discrepancy to the values of enthalpy change for protein denaturation and binding of denaturant with the protein. It must be noted here that the MG state of α-LA obtained by Xie et al. was induced by GdnHCl denaturant, whereas that addressed by Yutani et al. did not involve any denaturant. A year after the report of Yutani et al. about enthalpic equivalence of the MG and unfolded state of α-LA, Xie et al. (1993) argued that the conclusion of enthalpic equivalence of these states is incorrect and assigned the absence of thermally induced transition to ionic strength dependence. It was further argued that in addition to enthalpy change, entropy change between states can also account for the observed difference.
To conclude this section, we note that calorimetry has contributed significantly in the current knowledge of the determinants of protein structure (Ladbury and Doyle 2004; Velicelebi and Sturtevant 1979). A major contribution to this understanding has come from both differential scanning calorimetry (Privalov and Dragan 2007; Mazurenko et al. 2017) and isothermal titration calorimetry (ITC) (Carra et al. 1996; Singh and Kishore 2006).
Conclusions and future perspectives
Equilibrium intermediate states, such as the MG, have resemblance to the kinetic intermediate states during the protein folding process. As discussed above and is evident from the Fig. 2, much effort has been dedicated towards elucidating a detailed understanding of MG and other partially folded states. It is also clear that majority of the efforts have yielded qualitative information; however, the quantitative information has been very limited. In such a scenario, application of high-sensitivity ITC and DSC has provided limited, but useful, quantitative information about the properties of the MG state. Still, the absence or presence of cooperative thermal transition in going from the MG state to the unfolded state remains unclear (Fig. 2). Some studies, based upon calorimetry, have indicated that equilibrium MG states of a wide variety of proteins share common structural features based upon their exhibition of similar thermodynamic signatures accompanying the binding of ANS. These common structural features have suggested two sets of nonspecific binding sites for ANS on the MG state of proteins which could be the combination of hydrophobic and ionic sites (as the former possesses both types of molecular properties). The thermodynamic parameters accompanying the binding of ANS with the MG states of different proteins suggest an exothermic nature to the binding, with significant desolvation as reflected by positive values of the change in entropy. We still do not have thermodynamic signatures accompanying the processes shown in Fig. 2 for a large number of proteins which could lead to deriving general guidelines about thermodynamic states of such intermediates. Isothermal titration calorimetry, used in combination with differential scanning calorimetry, offers great potential for providing the required insights.
Fig. 2.
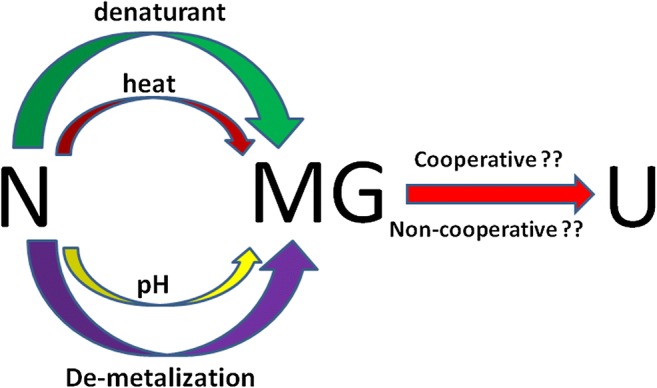
Scheme describing known methods to generate the MG state of a protein and questions to which answers are yet not clear
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eva Judy, Email: evajudy@iitb.ac.in.
Nand Kishore, Email: nandk@chem.iitb.ac.in.
References
- Agashe VR, Shastry MC, Udgaonkar JB. Initial hydrophobic collapse in the folding of barstar. Nature. 1995;377:754–757. doi: 10.1038/377754a0. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB, Haber E, Sela M, et al. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M, Kondrashkina E, Kayatekin C, et al. Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an α/β-type protein. J Mol Biol. 2007;368:219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- Bai JH, Xu D, Wang HR, et al. Evidence for the existence of an unfolding intermediate state for aminoacylase during denaturation in guanidine solutions. Biochim Biophys Acta. 1999;1430:39–45. doi: 10.1016/S0167-4838(98)00282-9. [DOI] [PubMed] [Google Scholar]
- Balbach J, Forge V, van Nuland NA, et al. Following protein folding in real time using NMR spectroscopy. Nat Struct Biol. 1995;2:865–870. doi: 10.1038/nsb1095-865. [DOI] [PubMed] [Google Scholar]
- Baliga C, Selmke B, Worobiew I, et al. CcdB at pH 4 forms a partially unfolded state with a dry core. Biophys J. 2019;116:807–817. doi: 10.1016/j.bpj.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Kishore N. 2, 2, 2-Trifluoroethanol-induced molten globule state of concanavalin A and energetics of 8-anilinonaphthalene sulfonate binding: calorimetric and spectroscopic investigation. J Phys Chem B. 2005;109:22655–22662. doi: 10.1021/jp053757r. [DOI] [PubMed] [Google Scholar]
- Barrick D, Baldwin RL. The molten globule intermediate of apomyoglobin and the process of protein folding. Protein Sci. 1993;2:869–876. doi: 10.1002/pro.5560020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryngelson JD, Onuchic JN, Socci ND, et al. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Prot Struct Funct Bioinform. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- Bychkova VE, Pain RH, Ptitsyn OB. The ‘molten globule’ state is involved in the translocation of proteins across membranes? FEBS Lett. 1988;238:231–234. doi: 10.1016/0014-5793(88)80485-X. [DOI] [PubMed] [Google Scholar]
- Carra JH, Murphy EC, Privalov PL. Thermodynamic effects of mutations on the denaturation of T4 lysozyme. Biophys J. 1996;71:1994–2001. doi: 10.1016/S0006-3495(96)79397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamani J, Moosavi-Movahedi AA, Saboury AA, et al. Calorimetric indication of the molten globule-like state of cytochrome c induced by n-alkyl sulfates at low concentrations. J Chem Thermodyn. 2003;35:199–207. doi: 10.1016/S0021-9614(02)00312-9. [DOI] [Google Scholar]
- Chiti F, Webster P, Taddei N, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra MJJ, Fokkink WJ, Heringa J, et al. The characteristics of molten globule states and folding pathways strongly depend on the sequence of a protein. Mol Phys. 2018;116:3173–3180. doi: 10.1080/00268976.2018.1496290. [DOI] [Google Scholar]
- Dill KA. Theory for the folding and stability of globular proteins. Biochem. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- Dill KA, Fiebig KM, Chan HS. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci. 1993;90:1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgikh DA, Abaturov LV, Bolotina IA, et al. Compact state of a protein molecule with pronounced small-scale mobility: bovine α-lactalbumin. Eur Biophys J. 1985;13:109–121. doi: 10.1007/BF00256531. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Barany G, Woodward C. Partially folded, molten globule and molten coil states of bovine pancreatic trypsin inhibitor. Nat Struct Biol. 1995;2:211–217. doi: 10.1038/nsb0395-211. [DOI] [PubMed] [Google Scholar]
- Fersht AR. Nucleation mechanisms in protein folding. Curr Opin Struct Biol. 1997;7:3–9. doi: 10.1016/S0959-440X(97)80002-4. [DOI] [PubMed] [Google Scholar]
- Fink AL, Oberg KA, Seshadri S. Discrete intermediates versus molten globule models for protein folding: characterization of partially folded intermediates of apomyoglobin. Fold Des. 1998;3:19–25. doi: 10.1016/S1359-0278(98)00005-4. [DOI] [PubMed] [Google Scholar]
- Fisher A, Taniuchi H. A study of core domains, and the core domain-domain interaction of cytochrome c fragment complex. Arch Biochem Biophys. 1992;296:1–16. doi: 10.1016/0003-9861(92)90538-8. [DOI] [PubMed] [Google Scholar]
- Forge V, Wijesinha RT, Balbach J, et al. Rapid collapse and slow structural reorganisation during the refolding of bovine α-lactalbumin. J Mol Biol. 1999;288:673–688. doi: 10.1006/jmbi.1999.2687. [DOI] [PubMed] [Google Scholar]
- Go N. The consistency principle in protein-structure and pathways of folding. Adv Biophys. 1984;18:149–164. doi: 10.1016/0065-227X(84)90010-8. [DOI] [PubMed] [Google Scholar]
- Goto Y, Calciano LJ, Fink AL. Acid-induced folding of proteins. Proc Natl Acad Sci. 1990;87:573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griko YV, Privalov PL. Thermodynamic puzzle of apomyoglobin unfolding. J Mol Biol. 1994;235:1318–1325. doi: 10.1006/jmbi.1994.1085. [DOI] [PubMed] [Google Scholar]
- Gussakovsky EE, Haas E. Two steps in the transition between the native and acid states of bovine α-lactalbumin detected by circular polarization of luminescence: evidence for a premolten globule state? Prot Sci. 1995;4:2319–2326. doi: 10.1002/pro.5560041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E. The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. Chem Phys Chem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- Haas E. Ensemble FRET methods in studies of intrinsically disordered proteins. In: Uversky V, Dunker A, editors. Intrinsically disordered protein analysis. Methods in molecular biology (methods and protocols) Totowa, NJ: Humana Press; 2012. [DOI] [PubMed] [Google Scholar]
- Haber E, Anfinsen CB. Side-chain interactions governing the pairing of half-cystine residues in ribonuclease. J Biol Chem. 1962;237:1839–1844. [PubMed] [Google Scholar]
- Hamada D, Kidokoro S, Fukada H, et al. Salt-induced formation of the molten globule state of cytochrome c studied by isothermal titration calorimetry. Proc Natl Acad Sci. 1994;91:10325–10329. doi: 10.1073/pnas.91.22.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada D, Fukada H, Takahashi K, et al. Salt-induced formation of the molten globule state of apomyoglobin studied by isothermal titration calorimetry. Thermochim Acta. 1995;266:385–400. doi: 10.1016/0040-6031(95)02444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarströ P, Persson M, Freskgård P-O, Mårtensson L-G, Andersson D, Jonsson B-H, Carlsson U. Structural mapping of an aggregation nucleation site in a molten globule intermediate. J Biol Chem. 1999;27:32897–32903. doi: 10.1074/jbc.274.46.32897. [DOI] [PubMed] [Google Scholar]
- Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie DT, Freire E. Structural energetics of the molten globule state. Prot Struct Funct Bioinform. 1993;16:115–140. doi: 10.1002/prot.340160202. [DOI] [PubMed] [Google Scholar]
- Honda RP, Yamaguchi KI, Kuwata K. Acid-induced molten globule state of a prion protein crucial role of strand 1-helix 1-strand 2 segment. J Biol Chem. 2014;289:30355–30363. doi: 10.1074/jbc.M114.559450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias MM, Elola MT, Martinez V, et al. Identification of an equilibrium intermediate in the unfolding process of galectin-1, which retains its carbohydrate-binding specificity. Biochim Biophys Acta Proteins Proteomics. 2003;1648:164–173. doi: 10.1016/S1570-9639(03)00119-5. [DOI] [PubMed] [Google Scholar]
- Ithychanda SS, Dou K, Robertson SP, et al. Structural and thermodynamic basis of a frontometaphyseal dysplasia mutation in filamin A. J Biol Chem. 2017;292:8390–8400. doi: 10.1074/jbc.M117.776740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Sharma D, Kumar R, et al. Structural, kinetic and thermodynamic characterizations of SDS-induced molten globule state of a highly negatively charged cytochrome c. J Biochem. 2018;165:125–137. doi: 10.1093/jb/mvy087. [DOI] [PubMed] [Google Scholar]
- Jeng MF, Englander SW. Stable submolecular folding units in a non-compact form of cytochrome c. J Mol Biol. 1991;221:1045–1061. doi: 10.1016/0022-2836(91)80191-V. [DOI] [PubMed] [Google Scholar]
- Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Sci. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/S0959-440X(98)80016-X. [DOI] [PubMed] [Google Scholar]
- Khan MKA, Rahaman H, Ahmad F. Conformation and thermodynamic stability of pre-molten and molten globule states of mammalian cytochromes-c. Metallomics. 2011;3(4):327–338. doi: 10.1039/C0MT00078G. [DOI] [PubMed] [Google Scholar]
- Kim PS, Baldwin RL. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Yao M, Kobashigawa Y, et al. Structure and thermodynamics of the extraordinarily stable molten globule state of canine milk lysozyme. Biochem. 2000;39:3248–3257. doi: 10.1021/bi991525a. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Kobashigawa Y, Demura M, et al. Energetics of three-state unfolding of a protein: canine milk lysozyme. Protein Eng. 2001;14:967–974. doi: 10.1093/protein/14.12.967. [DOI] [PubMed] [Google Scholar]
- Kozak JJ, Gray HB, Wittung-Stafshede P. Geometrical description of protein structural motifs. J Phys Chem B. 2018;122:11289–11294. doi: 10.1021/acs.jpcb.8b07130. [DOI] [PubMed] [Google Scholar]
- Kulkarni P, Uversky VN. Intrinsically disordered proteins and the Janus challenge. Biomol. 2018;8:179. doi: 10.3390/biom8040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Kidokoro SI, Wada A. Thermodynamic characterization of cytochrome c at low pH: observation of the molten globule state and of the cold denaturation process. J Mol Biol. 1992;223:1139–1153. doi: 10.1016/0022-2836(92)90265-L. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular protein structure. Prot Struct Funct Bioinformatics. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Ladbury JE, Doyle ML (eds) (2004) Biocalorimetry 2: applications of calorimetry in the biological sciences. John Wiley & Sons
- Martin J, Langer T, Boteva R, et al. Chaperonin-mediated protein folding at the surface of groEL through a ‘molten globule’-like intermediate. Nat. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Mazurenko S, Kunka A, Beerens K, et al. Exploration of protein unfolding by modelling calorimetry data from reheating. Sci Rep. 2017;7:16321. doi: 10.1038/s41598-017-16360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra PP, Kishore N. Biophysical analysis of partially folded state of a-lactalbumin in the presence of cationic and anionic surfactants. J Colloid Interface Sci. 2011;354:234–247. doi: 10.1016/j.jcis.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Moosavi-Movahedi AA, Chamani J, Gharanfoli M, et al. Differential scanning calorimetric study of the molten globule state of cytochrome c induced by sodium n-dodecyl sulfate. Thermochim Acta. 2004;409:137–144. doi: 10.1016/S0040-6031(03)00358-7. [DOI] [Google Scholar]
- Munoz V, Thompson PA, Hofrichter J, et al. Folding dynamics and mechanism of β-hairpin formation. Nat. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kidokoro SI. Volumetric properties of the molten globule state of cytochrome c in the thermal three-state transition evaluated by pressure perturbation calorimetry. J Phys Chem B. 2012;116:1927–1932. doi: 10.1021/jp209686e. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Baba T, Kidokoro SI. A molten globule-like intermediate state detected in the thermal transition of cytochrome c under low salt concentration. Biophys Chem. 2007;127:103–112. doi: 10.1016/j.bpc.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Seki Y, Katoh E, et al. Thermodynamic and structural properties of the acid molten globule state of horse cytochrome c. Biochem. 2011;50:3116–3126. doi: 10.1021/bi101806b. [DOI] [PubMed] [Google Scholar]
- Nishii I, Kataoka M, Goto Y. Thermodynamic stability of the molten globule states of apomyoglobin. J Mol Biol. 1995;250:223–238. doi: 10.1006/jmbi.1995.0373. [DOI] [PubMed] [Google Scholar]
- Nölting B, Agard DA. How general is the nucleation–condensation mechanism? Prot Struct Funct Bioinform. 2008;73:754–764. doi: 10.1002/prot.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M, Wada A. Molten-globule state: a compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;28:21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- Onuchic JN, Socci ND, Luthey-Schulten Z, et al. Protein folding funnels: the nature of the transition state ensemble. Fold Des. 1996;1:441–450. doi: 10.1016/S1359-0278(96)00060-0. [DOI] [PubMed] [Google Scholar]
- Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- Paci E, Smith LJ, Dobson CM, et al. Exploration of partially unfolded states of human α-lactalbumin by molecular dynamics simulation. J Mol Biol. 2001;306:329–347. doi: 10.1006/jmbi.2000.4337. [DOI] [PubMed] [Google Scholar]
- Peixoto PD, Trivelli X, André C, et al. Formation of β-lactoglobulin aggregates from quite, unfolded conformations upon heat activation. Langmuir. 2019;35:446–452. doi: 10.1021/acs.langmuir.8b03459. [DOI] [PubMed] [Google Scholar]
- Penkett CJ, Redfield C, Jones JA, et al. Structural and dynamical characterization of a biologically active unfolded fibronectin-binding protein from Staphylococcus a ureus. Biochem. 1998;37:17054–17067. doi: 10.1021/bi9814080. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Rossman MG, Cullis AF, et al. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-Ao. resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Potekhin S, Pfeil W. Microcalorimetric studies of conformational transitions of ferricytochrome c in acidic solution. Biophys Chem. 1989;34:55–62. doi: 10.1016/0301-4622(89)80041-9. [DOI] [PubMed] [Google Scholar]
- Povarova OI, Kuznetsova IM, Turoverov KK. Differences in the pathways of proteins unfolding induced by urea and guanidine hydrochloride: molten globule state and aggregates. PLoS One. 2010;132:e15035. doi: 10.1371/journal.pone.0015035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati RS, Indu S, Varadarajan R. Identification and thermodynamic characterization of molten globule states of periplasmic binding proteins. Biochem. 2007;46:10339–10352. doi: 10.1021/bi700577m. [DOI] [PubMed] [Google Scholar]
- Privalov P.L. Advances in Protein Chemistry Volume 33. 1979. Stability of Proteins Small Globular Proteins; pp. 167–241. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Dragan AI. Microcalorimetry of biological macromolecules. Biophys Chem. 2007;126:16–24. doi: 10.1016/j.bpc.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Privalov Peter L., Gill Stanley J. Advances in Protein Chemistry. 1988. Stability of Protein Structure and Hydrophobic Interaction; pp. 191–234. [DOI] [PubMed] [Google Scholar]
- Ptitsyn OB. Stages in the mechanism of self-organization of protein molecules. Dol Akad Nauk, SSSR. 1973;210:1213–1215. [PubMed] [Google Scholar]
- Ptitsyn OB. Protein folding: hypotheses and experiments. J Prot Chem. 1987;6:273–293. doi: 10.1007/BF00248050. [DOI] [Google Scholar]
- Ptitsyn OB. Molten globule and protein folding. Adv Prot Chem. 1995;47:83–229. doi: 10.1016/S0065-3233(08)60546-X. [DOI] [PubMed] [Google Scholar]
- Ptitsyn OB, Rashin AA. A model of myoglobin self-organization. Biophys Chem. 1975;3:1–20. doi: 10.1016/0301-4622(75)80033-0. [DOI] [PubMed] [Google Scholar]
- Rackovsky S, Scheraga HA. Hydrophobicity, hydrophilicity, and the radial and orientational distributions of residues in native proteins. Proc Nat Acad Sci. 1977;74:5248–5251. doi: 10.1073/pnas.74.12.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radibratovic M, Al-Hanish A, Minic S. Stabilization of apo α-lactalbumin by binding of epigallocatechin-3-gallate: experimental and molecular dynamics study. Food Chem. 2019;278:388–395. doi: 10.1016/j.foodchem.2018.11.038. [DOI] [PubMed] [Google Scholar]
- Rariy RV, Klibanov AM. Correct protein folding in glycerol. Proc Natl Acad Sci U S A. 1997;94:13520–13523. doi: 10.1073/pnas.94.25.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield C, Smith RA, Dobson CM. Structural characterization of a highly–ordered ‘molten globule’ at low pH. Nat Struct Mol Biol. 1994;1:23–29. doi: 10.1038/nsb0194-23. [DOI] [PubMed] [Google Scholar]
- Roder H, Elöve GA, Englander SW. Structural characterization of folding intermediates in cytochrome c by H-exchange labelling and proton NMR. Nature. 1988;335:694–699. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaddar S, Mandal AK, Mondal SK, et al. Solvation dynamics of a protein in the pre molten globule state. J Phys Chem B. 2006;110:21210–21215. doi: 10.1021/jp064136g. [DOI] [PubMed] [Google Scholar]
- Samanta HS, Zhuravlev PI, Hinczewski M, et al. Protein collapse is encoded in the folded state architecture. Soft Matt. 2017;13:3622–3638. doi: 10.1039/C7SM00074J. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Demura M, Nitta K. Partially unfolded equilibrium state of hen lysozyme studied by circular dichroism spectroscopy. Biochemistry. 2000;39:6475–6482. doi: 10.1021/bi992560k. [DOI] [PubMed] [Google Scholar]
- Sato S, Ward CL, Krouse ME, et al. Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J Biol Chem. 1996;271:635–638. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- Schweiker KL, Fitz VW, Makhatadze GI. Universal convergence of the specific volume changes of globular proteins upon unfolding. Biochem. 2009;48:10846–10851. doi: 10.1021/bi901220u. [DOI] [PubMed] [Google Scholar]
- Semisotnov GV, Rodionova NA, Razgulyaev OI, et al. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopoly Orig Res Biomol. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- Shakhnovich EI, Finkelstein AV. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first order phase transition. Biopoly Orig Res Biomol. 1989;28:1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- Sharma R, Kishore N. Isothermal titration calorimetric and spectroscopic studies on (alcohol+ salt) induced partially folded state of α-lactalbumin and its binding with 8-anilino-1-naphthalenesulfonic acid. J Chem Thermo. 2008;40:1141–1151. doi: 10.1016/j.jct.2008.02.009. [DOI] [Google Scholar]
- Sheshadri S, Lingaraju GM, Varadarajan R. Denaturant mediated unfolding of both native and molten globule states of maltose binding protein are accompanied by large ΔCp’s. Protein Sci. 1999;8:1689–1695. doi: 10.1110/ps.8.8.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Kishore N. Elucidating the binding thermodynamics of 8-anilino-1-naphthalene sulfonic acid with the A-state of α-lactalbumin: an isothermal titration calorimetric investigation. Biopoly Orig Res Biomol. 2006;83:205–212. doi: 10.1002/bip.20547. [DOI] [PubMed] [Google Scholar]
- Skora L, Becker S, Zweckstetter M. Molten globule precursor states are conformationally correlated to amyloid fibrils of human beta-2-microglobulin. J Am Chem Soc. 2010;132:9223–9225. doi: 10.1021/ja100453e. [DOI] [PubMed] [Google Scholar]
- Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin: a fluorescent probe of non-polar binding sites. J Mol Biol. 1965;13:482–495. doi: 10.1016/S0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- Svensson M, Sabharwal H, Håkansson A, et al. Molecular characterization of α–lactalbumin folding variants that induce apoptosis in tumor cells. J Biol Chem. 1999;274:6388–6396. doi: 10.1074/jbc.274.10.6388. [DOI] [PubMed] [Google Scholar]
- Swaminathan R, Periasamy N, Udgaonkar JB, et al. Molten globule-like conformation of barstar: a study by fluorescence dynamics. J Phys Chem. 1994;98:9270–9278. doi: 10.1021/j100088a030. [DOI] [Google Scholar]
- Takahashi S, Yoshida A, Oikawa H. Hypothesis: structural heterogeneity of the unfolded proteins originating from the coupling of the local clusters and the long-range distance distribution. Biophys Rev. 2018;10:363–373. doi: 10.1007/s12551-018-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele P, Kishore N. Thermodynamic analysis of partially folded states of myoglobin in presence of 2, 2, 2-trifluoroethanol. J Chem Thermo. 2015;84:50–59. doi: 10.1016/j.jct.2014.12.019. [DOI] [Google Scholar]
- Tyagi M, Bornot A, Offmann B, de Brevern AG. Analysis of loop boundaries using different local structure assignment methods. Protein Sci. 2009;18:1869–1881. doi: 10.1002/pro.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udgaonkar JB, Baldwin RL. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Bringing darkness to light: intrinsic disorder as a means to dig into the dark proteome. Proteomics. 2018;18:1800352. doi: 10.1002/pmic.201800352. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Karnoup AS, Segel DJ, et al. Anion-induced folding of staphylococcal nuclease: characterization of multiple equilibrium partially folded intermediates. J Mol Biol. 1998;278:879–894. doi: 10.1006/jmbi.1998.1741. [DOI] [PubMed] [Google Scholar]
- Van der Goot FG, Gonzalez-Manas JM, Lakey JH, et al. A molten-globule membrane-insertion intermediate of the pore-forming domain of colicin A. Nat. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- Vassilenko KS, Uversky VN. Native-like secondary structure of molten globules. Biochim Biophys Acta Prot Struct Mol Enzy. 2002;1594:168–177. doi: 10.1016/S0167-4838(01)00303-X. [DOI] [PubMed] [Google Scholar]
- Velicelebi G, Sturtevant JM. Thermodynamics of the denaturation of lysozyme in alcohol-water mixtures. Biochem. 1979;18:1180–1186. doi: 10.1021/bi00574a010. [DOI] [PubMed] [Google Scholar]
- Wirtz H, Schafer S, Hoberg C, et al. Hydrophobic collapse of ubiquitin generates rapid protein–water motions. Biochem. 2018;57:3650–3657. doi: 10.1021/acs.biochem.8b00235. [DOI] [PubMed] [Google Scholar]
- Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Sci. 1995;267:1619–1619. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- Xie D, Bhakuni V, Freire E. Calorimetric determination of the energetics of the molten globule intermediate in protein folding: apo-.alpha.-lactalbumin. Biochem. 1991;30:10673–10678. doi: 10.1021/bi00108a010. [DOI] [PubMed] [Google Scholar]
- Xie D, Bhakuni V, Freire E. Are the molten globule and the unfolded states of apo-α-lactalbumin enthalpically equivalent? J Mol Biol. 1993;232:5–8. doi: 10.1006/jmbi.1993.1364. [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo T, Wang T, et al. Aspartate-induced aminoacylase folding and forming of molten globule. Int J Biochem Cell Biol. 2003;35:1558–1572. doi: 10.1016/S1357-2725(03)00131-6. [DOI] [PubMed] [Google Scholar]
- Yutani K, Ogasahara K, Kuwajima K. Absence of the thermal transition in apo-α-lactalbumin in the molten globule state: a study by differential scanning microcalorimetry. J Mol Biol. 1992;228:347–350. doi: 10.1016/0022-2836(92)90824-4. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Yang LQ, Du BR, et al. Higher RABEX-5 mRNA predicts unfavourable survival in patients with colorectal cancer. Eur Rev Med Pharma Sci. 2017;21:2372–2376. [PubMed] [Google Scholar]


