Abstract
Maintaining gradients of solvated protons and alkali metal ions such as Na+ and K+ across membranes is critical for cellular function. Over the last few decades, both the interactions of protons and alkali metal ions with phospholipid membranes have been studied extensively and the reported interactions of these ions with phospholipid headgroups are very similar, yet few studies have investigated the potential interdependence between proton and alkali metal ion binding at the water–lipid interface. In this short review, we discuss the similarities between the proton–membrane and alkali ion–membrane interactions. Such interactions include cation attraction to the phosphate and carbonyl oxygens of the phospholipid headgroups that form strong lipid–ion and lipid–ion–water complexes. We also propose potential mechanisms that may modulate the affinities of these cationic species to the water–phospholipid interfacial oxygen moieties. This review aims to highlight the potential interdependence between protons and alkali metal ions at the membrane surface and encourage a more nuanced understanding of the complex nature of these biologically relevant processes.
Keywords: Protons, Hydronium ions, Alkali ions, Ion lipid interactions, Membranes, Lipid bilayers
Introduction
Both protons and alkali metal ions are critical in a wide range of cellular processes such as energy production and metabolism, the import of nutrients, osmotic regulation, nerve conduction and cell signalling. Maintaining a proton gradient across membranes in an energy-efficient manner requires the protons to be confined to a restricted space close to the bilayer preventing them from diffusing to extra- or intracellular space. The molecular mechanism of (solvated) protons moving along membrane surfaces has been investigated for many decades (Agmon et al. 2016; Gutman and Nachliel 1990; Heberle 2000; Mulkidjanian and Cherepanov 2006; Mulkidjanian et al. 2006). Similarly, the interactions of alkali ions with membranes have been studied extensively (Leontidis 2017). While there is still an ongoing controversy whether alkali ions interact with membranes at physiological concentrations (Catte et al. 2016), there is growing evidence that they can alter the morphology and fluidity of membranes (Binder and Zschornig 2002; Böckmann et al. 2003; Garcia-Manyes et al. 2005; Vorobyov et al. 2014) and thus indirectly affect cellular processes.
Both protons and alkali ions are small cations that have been shown to bind to the water–lipid interface of phospholipid membranes and, through these water–ion–lipid interactions, have the capacity to change the structure and physico-chemical properties of membranes (Böckmann et al. 2003; Cordomí et al. 2009; Cranfield et al. 2016; Deplazes et al. 2017; Garcia-Manyes et al. 2005; Garcia-Manyes et al. 2006; Garcia et al. 2019; Koynova and Caffrey 1998; Petelska and Figaszewski 2002; Piantanida et al. 2017; Reif et al. 2017; Vácha et al. 2009). As we will outline in this review, in many cases, the reported findings on how protons and alkali ions interact with phospholipid membranes demonstrate remarkable similarities. Yet, to the best of our knowledge, there is very little research connecting these findings and even less on understanding the potential interdependence between the binding of protons and alkali ions to membranes. Note that the focus of this review is to highlight the interplay of protons and alkali ions at the water–lipid interface. A detailed discussion of the many, and often conflicting, studies investigating the mechanism of proton diffusion along membranes, as well as the challenges in measuring this process, is beyond the scope of this short review. For this, the reader is referred to reviews dedicated to these topics (Agmon et al. 2016; Leontidis 2017; Medvedev and Stuchebrukhov 2011; Mulkidjanian and Cherepanov 2006; Mulkidjanian et al. 2006).
The remainder of this review is structured as follows. In sections “Interactions of (solvated) protons with membranes” and “Interactions of alkali ions with membranes”, we separately discuss the interaction of protons and alkali ions to phospholipid membranes with a particular focus on the molecular mechanism and the ‘site’ of interaction. In the section “Competing for the same space? The relationship between proton-membrane and ion-membrane interactions and its implications”, we propose a competitive relationship between protons and alkali ions at the water–lipid interface and describe several potential mechanisms for how this competition may manifest itself. In addition, we discuss the implications of this competitive binding for experiments aimed at understanding proton migration or alkali ion–membrane interactions.
Interactions of (solvated) protons with membranes
Migration of protons along membranes has been observed in purple membranes and membranes reconstituted with bacteriorhodopsin (Alexiev et al. 1995; Brändén et al. 2006; Heberle and Dencher 1992; Heberle et al. 1994; Nachliel et al. 1996; Scherrer et al. 1994) as well as phospholipid monolayers, bilayers and phospholipid vesicles (Amdursky et al. 2019; Antonenko and Pohl 2008; Gabriel et al. 1994; Gabriel and Teissie 1996; Ojemyr et al. 2010; Sandén et al. 2010; Serowy et al. 2003). It has also been suggested that compared to the movement along the surface, the transfer of protons into the bulk is delayed (Alexiev et al. 1995; Cherepanov et al. 2004; Nachliel and Gutman 1996; Scherrer et al. 1994; Zhang et al. 2012). However, what constitutes the main energy barrier for surface-to-bulk release of protons remains unclear (Agmon et al. 2016). While the detailed mechanism of this proton migration is yet to be determined, studies have suggested that the process is facilitated by one or more of the following: (i) ionisable groups on the membrane surface, which can be either phospholipid headgroups or residues on proteins, collectively referred to as immobile buffers (Ädelroth and Brzezinski 2004; Alexiev et al. 1995; Brändén et al. 2006; Heberle and Dencher 1992; Heberle et al. 1994; Jones and Jackson 1989; Junge and McLaughlin 1987; Nachliel and Gutman 1996; Scherrer et al. 1994; Tocanne and Teissié 1990); (ii) interfacial water and other molecules acting as mobile buffers (Cherepanov et al. 2004; Springer et al. 2011; Tocanne and Teissié 1990). As discussed in Agmon et al. (2016) and demonstrated by Springer et al. (2011), proton transfer does not necessarily require the presence of immobile buffers, yet proton diffusion is affected by the phospholipid composition of the membrane (Amdursky et al. 2019).
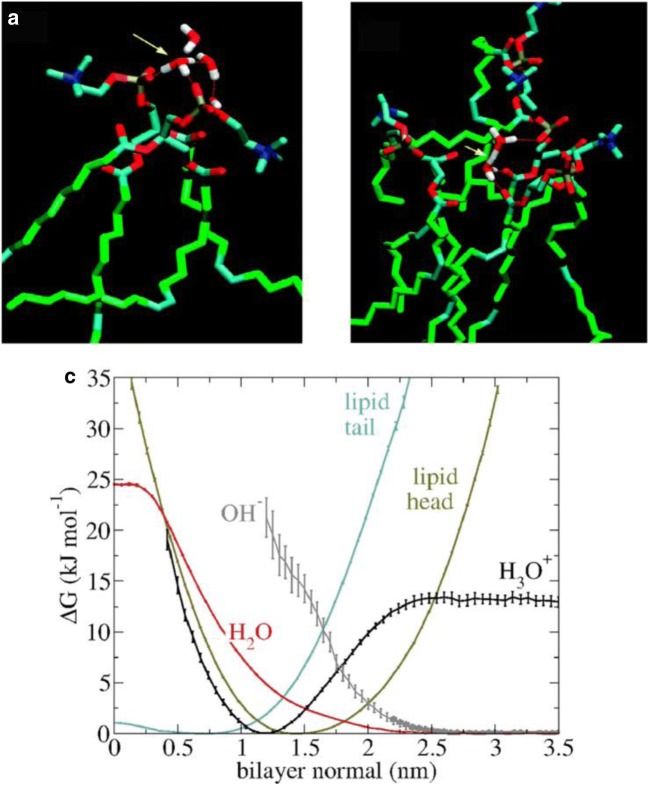
Independent of the detailed mechanisms, proton migration involves the spatial restriction of the proton to the membrane surface. The solvated proton can exist in a number of forms including the hydronium cation (H3O+) where the proton is located on a single water molecule, the Zundel cation (H5O2+) where the excess proton is shared by two water molecules and the Eigen cation (H9O4+) where the H3O+ forms hydrogen bonds with three surrounding water molecules. The affinity of (solvated) protons for the surface of phospholipid membranes has been demonstrated both with “wet-lab” experiments (Brändén et al. 2006; Cranfield et al. 2016; Sandén et al. 2010; Springer et al. 2011; Weichselbaum et al. 2017) and simulations (Smondyrev and Voth 2002; Wolf et al. 2014; Yamashita and Voth 2010). It is also worth noting that simulations of hydrated protons at the water–lipid interface are one of the most complex and challenging tasks in computational biophysics and the predictions of proton diffusivity often differ by orders of magnitudes from experimental values. Nevertheless, data from simulations have provided molecular level insight into the structure of lipid–ion complexes formed. Combined results from these simulation studies suggest that the hydronium and Zundel cation can interact with the headgroups of phospholipids and sit in the cavities formed by neighbouring phospholipid molecules (Deplazes et al. 2017; Smondyrev and Voth 2002; Wolf et al. 2014; Yamashita and Voth 2010). For example, Yamashita and Voth (2010) investigated the configurations of solvated protons at the water–phospholipid interface of both zwitterionic and anionic phospholipid bilayers. In both types of membranes, hydronium ions are coordinated by phosphate oxygens as well as water molecules sitting in the lipid headgroups (Fig. 1A). In comparison, distorted Zundel-like cations are located deeper in the interfacial region (i.e. closer to the hydrophobic core of the membrane) where they bind to phosphate and carbonyl oxygens in the phospholipid headgroup (Fig. 1B). Lipid–ion complexes of similar configurations were demonstrated by other simulations of solvated protons with zwitterionic phospholipid bilayers (Deplazes et al. 2017; Smondyrev and Voth 2002; Wolf et al. 2014). In addition, Wolf et al. (2014) showed that the hydronium ion can also reside in lipid binding cavities in a less tightly bound configuration where the cation is surrounded by clusters of water molecules that are bridging the cation to the phosphate and carbonyl oxygens. The authors also used free energy calculations to demonstrate that the hydronium ion is attracted to the phospholipid headgroup with energy minima closer to the hydrophobic core compared to the energy minima of water molecules (Fig. 1C). The affinity of hydronium ions has also been shown in other simulations where hydronium ions that were randomly placed in the bulk solution accumulated at the water–lipid interface where they formed strong interactions with phospholipid headgroups (Deplazes et al. 2017). Interestingly, these simulations also showed that the interactions of hydronium ions with the phospholipid headgroups resulted in a reduced area per phospholipid and increased membrane thickness, consistent with experimental data (Cranfield et al. 2016). In addition, a number of studies have demonstrated that low pH alters the physico-chemical properties of membranes including phase transition temperature (Koynova and Caffrey 1998), mechanical and electrical properties (Zhou and Raphael 2007), interfacial tension (Petelska and Figaszewski 2002) and morphology (Cranfield et al. 2016).
Fig. 1.
Interactions of protons with phospholipid membranes from simulations studies. (A) Structure of a hydronium ion coordinated by phosphate oxygen and water. (B) Structure of a Zundel cation coordinated by phosphate and carbonyl oxygen; (A, B) adapted with permission from Yamashita and Voth (2010). Copyright 2010 American Chemical Society. (C) Free energy profiles of the hydronium ion (H3O+), hydroxyl ion (OH−), water and the phospholipid head and tail along the bilayer normal associated with the upper leaflet of the membrane. 0 nm is the centre of the membrane (i.e. the centre of the hydrophobic core) and 3.5 nm is the water layer of the outer lipid headgroups. The free energy, ΔG, was obtained from ΔG(z) = − RT ln p(z), where p(z) is the normalised number density of a given system component at position z along the bilayer normal, calculated from MD simulations of a proton near the surface of a hydrated DMPC lipid bilayer. Reprinted from Wolf et al. (2014) with permission from Elsevier
In summary, both experimental measurements and simulations show that solvated protons show strong affinity for water–lipid interfaces where they interact with the phosphate and carbonyl oxygen in the phospholipid headgroups and form stable lipid–ion and lipid–ion–water complexes.
Interactions of alkali ions with membranes
The extent of the interaction between zwitterionic phospholipid membranes and monovalent ions is still disputed. Generally, it is considered that interactions between monovalent cations and zwitterionic phospholipid bilayers are relatively weak. This was supported by several experimental studies reporting little effect on physico-chemical properties of phospholipid membranes at sub-molar ion concentrations (Akutsu and Seelig 1981; Binder and Zschornig 2002; Brown and Seelig 1977; Clarke and Lupfert 1999; Cunningham et al. 1986, 1988; Gottlieb and Eanes 1972; Petrache et al. 2006). However, there are divergent findings using different experimental techniques demonstrating significant changes in the properties of the bilayer such as hydration at the interface, changes in bilayer mechanical strength and the phase transition induced by sub-molar concentrations of monovalent ions (Garcia-Manyes et al. 2005; Garcia-Manyes et al. 2006; Garcia et al. 2019; Piantanida et al. 2017). In addition, simulations have demonstrated changes to the area per lipid, order parameters, diffusion coefficients and orientations of the headgroup dipole (Böckmann et al. 2003; Cordomí et al. 2008; Gurtovenko and Vattulainen 2008; Reif et al. 2017; Vácha et al. 2009).
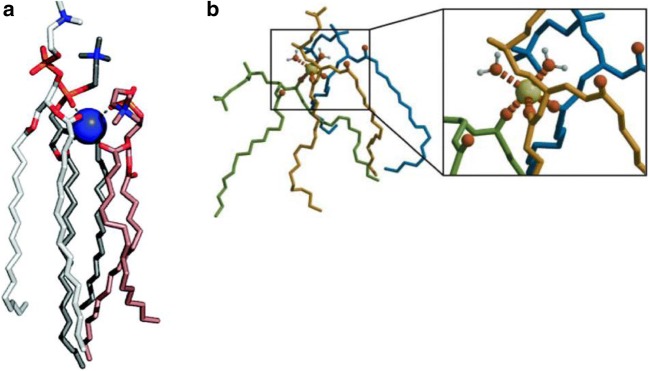
Similar to the study of proton–membrane interactions, molecular details on where and how alkali ions bind to phospholipids have predominantly been obtained from MD simulations. A large number of studies demonstrated the binding of Na+ and/or K+ ions to the water–lipid interface of zwitterionic and anionic phospholipid bilayers at physiologically relevant salt concentrations (Böckmann et al. 2003; Cordomí et al. 2008, 2009; Gambu and Roux 1997; Gurtovenko and Vattulainen 2008; Javanainen et al. 2017; Jurkiewicz et al. 2012; Klasczyk and Knecht 2011; Lee et al. 2008; Mao et al. 2013; Mukhopadhyay et al. 2004; Pabst et al. 2007; Pandit et al. 2003; Reif et al. 2017; Vácha et al. 2010, 2009; Vorobyov et al. 2014). Combined results from these studies suggest that alkali ions form stable interactions with the phosphate and carbonyl groups in the phospholipid headgroups (Böckmann et al. 2003; Cordomí et al. 2008, 2009; Gambu and Roux 1997; Javanainen et al. 2017; Jurkiewicz et al. 2012; Lee et al. 2008; Mukhopadhyay et al. 2004; Pandit et al. 2003; Vácha et al. 2010). In much the same way as protons, alkali ions are found in the binding cavity formed by neighbouring phospholipids and can interact with the carbonyl or phosphate oxygens either directly or via bridging water molecules (Fig. 2). In studying these interactions using MD simulations, the exact binding mode (i.e. the number of phospholipids or water molecules coordinated to the ion) and the preference for the phosphate or carbonyl oxygen depend on the force field and other simulation parameters used. However, there is a consensus that the alkali ions accumulate deep into the interfacial region, close to phosphate and carbonyl groups, while the corresponding negative counter ions (usually Cl−) reside in the regions adjacent to the bulk water.
Fig. 2.
Interactions of Na+ ions with phospholipid membranes from simulation studies. (A) Structure of a Na+−lipid complex where the ion is coordinated by phosphate and carbonyl oxygens from three lipids. Reprinted with permission from Cordomí et al. (2009). Copyright 2009 American Chemical Society. (B) Typical structure of a Na+ ion coordinated by the carbonyl oxygen of two lipids and water molecules. Reprinted from Böckmann et al. (2003) with permission from Elsevier
Simulations have also been used to predict the relative binding affinities of Na+ and K+ for phospholipid bilayers (Gambu and Roux 1997; Klasczyk and Knecht 2011; Mao et al. 2013; Melcr et al. 2018; Vácha et al. 2009). The accurate calculation of binding affinities is challenging and, in the case of alkali ions, the predicted affinities are often overestimated (Catte et al. 2016). This is further complicated by conflicting results from experimental measures (Melcr et al. 2018). In some studies, Na+ appears to show stronger binding and forms more stable lipid–ion complexes than K+ (Cordomí et al. 2008; Gurtovenko and Vattulainen 2008; Mao et al. 2013; Vácha et al. 2009, 2010). However, some studies also reported no significant difference in the affinities of the two ions (Vorobyov et al. 2014), while other simulations predicted that water molecules are preferred to Na+ (and Cl−) at the water–lipid interface (Melcr et al. 2018). The lower binding affinity of K+ has been attributed to the larger size, smaller ionic surface charge and less ordered hydration shell (Gurtovenko and Vattulainen 2008). Irrespective of its predicted lower affinity, K+ reportedly associates with the negatively charged carbonyl and phosphate oxygen moieties, much like Na+ (Gambu and Roux 1997). It should be noted that an accurate prediction of binding affinities for biomolecular systems is very challenging. The choice of force fields for the phospholipids and the water model used, as well as other simulations parameters, significantly affect the predicted affinities (Cordomí et al. 2009; Gurtovenko and Vattulainen 2008; Melcr et al. 2018).
Another similarity between the alkali ions and protons is their capacity to alter the properties of the membrane. Studies in the presence of alkali ions at physiologically relevant concentrations have reported significant changes to the physical properties of membranes (Binder and Zschornig 2002; Böckmann et al. 2003; Garcia-Manyes et al. 2005; Vorobyov et al. 2014). Generally, ion binding to phospholipid membranes increases lipid ordering (Garcia-Manyes et al. 2005), area per phospholipid (Cornell and Separovic 1983) and area compressibility, with a concurrent reduction in elasticity (Reif et al. 2017). Much like the reported difference in binding affinities for Na+ and K+ to the lipid interface, relative to Na+, K+ induces a weaker effect on membrane structure (Petrache et al. 2006). The authors reported no structural alterations in the phosphatidylcholine bilayer with K+, which they determined was due to minimal headgroup binding. However, effects of membrane fluidity in the presence of K+ and Na+ were reported to be indistinguishable (Kagawa et al. 2013; Zimmermann et al. 2012). As in MD simulations, the alkali ions seem to modulate the physical properties of membranes, but determining precise binding affinities and ion-specific modulation of the physical properties of the membrane remains an experimental challenge.
In summary, although both experiment and simulations have demonstrated that Na+ and K+ interact with phospholipid bilayers at sub-molar concentrations, and induce subsequent alterations to the physico-chemical properties of the bilayer, there remains significant debate regarding these assertions. Nevertheless, across a large number of studies spanning more than two decades, there appears at least a consensus that Na+ and K+ are attracted to the water–lipid interface, where they coordinate with the phosphate and the carbonyl oxygen moieties to form ion–lipid or ion–lipid–water complexes.
Competing for the same space? The relationship between proton–membrane and ion–membrane interactions and its implications
The studies outlined above were collected using a wide range of experimental techniques, membrane structures (e.g. multilamellar and unilamellar vesicles, supported bilayers) and simulations. They demonstrate that protons and alkali ions accumulate at the interfacial region of phospholipid membranes. In addition, results from simulations suggest that both protons and alkali ions form ion–lipid or ion–lipid–water complexes where the ion can be coordinated by the phosphate or carbonyl oxygens from neighbouring phospholipid headgroups as well as water molecules. Based on this evidence, it is conceivable that protons and alkali ions would participate in competitive binding.
To the best of our knowledge, the paper Tocanne and Teissié (1990) published almost 30 years ago is one of the few papers that provides an in-depth discussion of both proton migration and alkali ion–membrane interactions. The authors also point to the complex interplay between these processes noting that the ionisation state of phospholipids at the water–lipid interface depends, among other parameters, on the alkali–ion lipid binding constants. The paper also reports the pKa values for a range of phospholipids from various studies showing that the pKa depends on the type and concentration of salt used as a buffer.
Interestingly, many studies reporting the affinity or the effect of alkali ions on zwitterionic phospholipid bilayers fail to buffer their solutions to ensure a neutral pH. Similarly, studies investigating proton migration do not consider whether the measurements depend on the type or concentration of the salt buffer. Buffering solutions may be unsuited to the experimental techniques reported in these articles, but one is unable to ignore the fact that these reports have been used as evidence for limited monovalent cation binding to zwitterionic phospholipid bilayers (Catte et al. 2016). Considering the likelihood of proton accumulation at the surface of the phospholipid bilayer, it is impossible not to question the relevance of this experimental confounder, especially when determining alkali ion interactions at physiologically relevant concentrations.
The idea that a simple competitive relationship may exist at the water–lipid interface between monovalent cations and solvated protons may constrain other potential aspects that are associated with proton accumulation at the surface. One of these is the capacity to alter the water structure at the interface (Nguyen et al. 2018; Vácha et al. 2007). The deliberate arrangement of the water structure by specific ions has been previously described in terms of ‘structure makers’ and ‘structure breakers’ (Collins 1995). While it is believed to be a short-range effect, local changes in the structure of interfacial water due to the accumulation of solvated protons at the membrane surface may impose a barrier to free diffusion of ions that do not form complementary water structures. This potential higher energy cost associated with ion movement from bulk water into the lipid binding cavity will exist when the water structure around the ion differs from the interfacial water structure at the membrane surface. In this case, modulation of ion interactions with the phospholipid bilayer surface could also be associated with potential changes in the interfacial water structure (Boström et al. 2005).
Direct competition and localised structural changes in water are unable to exist without concomitant changes in the structural properties of the phospholipid headgroups. The fact that bulk pH has the capacity to modulate the packing density of a phospholipid bilayer (Cranfield et al. 2016; Deplazes et al. 2017, 2018) may introduce a structural component to the affinity of alkali ions to the membrane. The coordination of alkali ions at the lipid–water interface involves the carbonyl and phosphate oxygens of the lipid headgroup indicating that structural changes to this coordination pocket such as rigidification or changes to the radial size of the binding cavity itself can occur. It is reasonable to propose that the capacity of the solvated proton to increase phospholipid packing would produce a far more constrained binding cavity, both in terms of flexibility and size, within the headgroup region. This in itself may alter the comparative affinity of the monovalent ions at the phospholipid surface.
A further consideration is any effects due to presence of counter ions at or near the lipid–water interface. One could speculate that the counter ion would have a significant effect on the affinity of both the alkali metal ion and quite possibly protons with the phospholipid bilayer. It may also distort the relative affinities of each of these with respect to the phospholipid interface. Though beyond the scope of this article, there would be value in designing future experiments to determine just how important the presence of counter ions is to the interplay of protons and alkali metal ions at the interface.
Conclusion
A new perspective has emerged in which understanding of proton and ion interactions at membrane surface are interwoven. The potential interdependence between protons and alkali ions at the membrane surface, which has so far mostly been overlooked, means that studies disregarding bulk pH may be obscuring the interactions of physiologically relevant ions at the water–phospholipid interface. This may well lead to a significant underestimation of the affinity of these ions to the membrane interface. In turn, the capacity of protons to migrate along the phospholipid bilayer surface may be modulated by other ionic species at the surface, if they are competing for the same ionisable groups. The interdependence of pH and ion–membrane interactions is an important topic of investigation that may reconcile some of the disparate reported results and provide a significant step forward towards a richer understanding of the complex nature of the water–lipid interface.
Acknowledgements
The authors wish to acknowledge Adj Prof Bruce Cornell (UTS), Associate Prof Ron Clarke (University of Sydney), Dr Stephen Holt (Australian Nuclear Science and Technology Organisation) and Dr Paul Duckworth (eDAQ Pty Ltd) for valuable discussions on these topics.
Compliance with ethical standards
Funding
ED and AG are supported by the UTS Chancellor’s Postdoctoral Research Fellowship scheme.
Conflicts of interest
Evelyne Deplazes declares that she has no conflict of interest. Jacqueline White declares that she has no conflict of interest. Christopher Murphy declares that he has no conflict of interest. Charles G Cranfield declares that he has no conflict of interest. Alvaro Garcia declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Evelyne Deplazes, Email: evelyne.deplazes@uts.edu.au.
Alvaro Garcia, Email: alvaro.garcia@uts.edu.au.
References
- Ädelroth P, Brzezinski P. Surface-mediated proton-transfer reactions in membrane-bound proteins. BBA-Bioenergetics. 2004;1655:102–115. doi: 10.1016/j.bbabio.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Agmon N, et al. Protons and hydroxide ions in aqueous systems. Chem Rev. 2016;116:7642–7672. doi: 10.1021/acs.chemrev.5b00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu H, Seelig J. Interaction of metal ions with phosphatidylcholine bilayer membranes. Biochemistry. 1981;20:7366–7373. doi: 10.1021/bi00529a007. [DOI] [PubMed] [Google Scholar]
- Alexiev U, Mollaaghababa R, Scherrer P, Khorana HG, Heyn MP. Rapid long-range proton diffusion along the surface of the purple membrane and delayed proton transfer into the bulk. Proc Natl Acad Sci U S A. 1995;92:372–376. doi: 10.1073/pnas.92.2.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdursky N, Lin Y, Aho N, Groenhof G. Exploring fast proton transfer events associated with lateral proton diffusion on the surface of membranes. Proc Natl Acad Sci U S A. 2019;116:2443. doi: 10.1073/pnas.1812351116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonenko YN, Pohl P. Microinjection in combination with microfluorimetry to study proton diffusion along phospholipid membranes. Eur Biophys J. 2008;37:865–870. doi: 10.1007/s00249-008-0295-y. [DOI] [PubMed] [Google Scholar]
- Binder H, Zschornig O. The effect of metal cations on the phase behavior and hydration characteristics of phospholipid membranes. Chem Phys Lipids. 2002;115:39–61. doi: 10.1016/S0009-3084(02)00005-1. [DOI] [PubMed] [Google Scholar]
- Böckmann RA, Hac A, Heimburg T, Grubmüller H. Effect of sodium chloride on a lipid bilayer. Biophys J. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström M, Kunz W, Ninham BW. Hofmeister effects in surface tension of aqueous electrolyte solution. Langmuir. 2005;21:2619–2623. doi: 10.1021/la047437v. [DOI] [PubMed] [Google Scholar]
- Brändén M, Sandén T, Brzezinski P, Widengren J. Localized proton microcircuits at the biological membrane-water interface. Proc Natl Acad Sci U S A. 2006;103:19766–19770. doi: 10.1073/pnas.0605909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MF, Seelig J. Ion-induced changes in head group conformation of lecithin bilayers. Nature. 1977;269:721–723. doi: 10.1038/269721a0. [DOI] [Google Scholar]
- Catte A, et al. Molecular electrometer and binding of cations to phospholipid bilayers. Phys Chem Chem Phys. 2016;18:32560–32569. doi: 10.1039/C6CP04883H. [DOI] [PubMed] [Google Scholar]
- Cherepanov DA, Junge W, Mulkidjanian AY. Proton transfer dynamics at the membrane/water interface: dependence on the fixed and mobile pH buffers, on the size and form of membrane particles, and on the interfacial potential barrier. Biophys J. 2004;86:665–680. doi: 10.1016/s0006-3495(04)74146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RJ, Lupfert C. Influence of anions and cations on the dipole potential of phosphatidylcholine vesicles: a basis for the Hofmeister effect. Biophys J. 1999;76:2614–2624. doi: 10.1016/s0006-3495(99)77414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KD. Sticky ions in biological systems. Proc Natl Acad Sci U S A. 1995;92:5553. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordomí A, Edholm O, Perez JJ. Effect of ions on a dipalmitoyl phosphatidylcholine bilayer. A molecular dynamics simulation study. J Phys Chem B. 2008;112:1397–1408. doi: 10.1021/jp073897w. [DOI] [PubMed] [Google Scholar]
- Cordomí A, Edholm O, Perez JJ. Effect of force field parameters on sodium and potassium ion binding to dipalmitoyl phosphatidylcholine bilayers. J Chem Theory Comput. 2009;5:2125–2134. doi: 10.1021/ct9000763. [DOI] [PubMed] [Google Scholar]
- Cornell BA, Separovic F. Membrane thickness and acyl chain length. Biochim Biophys Acta. 1983;733:189–193. doi: 10.1016/0005-2736(83)90106-2. [DOI] [PubMed] [Google Scholar]
- Cranfield CG, et al. Evidence of the key role of H3O+ in phospholipid membrane morphology. Langmuir. 2016;32:10725–10734. doi: 10.1021/acs.langmuir.6b01988. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Shimotake JE, Tamura-Lis W, Mastran T, Kwok WM, Kauffman JW, Lis LJ. The influence of ion species on phosphatidylcholine bilayer structure and packing. Chem Phys Lipids. 1986;39:135–143. doi: 10.1016/0009-3084(86)90107-6. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Gelerinter E, Lis LJ. Monovalent ion-phosphatidylcholine interactions: an electron paramagnetic resonance study. Chem Phys Lipids. 1988;46:205–211. doi: 10.1016/0009-3084(88)90023-0. [DOI] [PubMed] [Google Scholar]
- Deplazes E, Poger D, Cornell B, Cranfield CG. The effect of hydronium ions on the structure of phospholipid membranes. Phys Chem Chem Phys. 2017;20:357–366. doi: 10.1039/c7cp06776c. [DOI] [PubMed] [Google Scholar]
- Deplazes E, Poger D, Cornell B, Cranfield CG. The effect of H(3)O(+) on the membrane morphology and hydrogen bonding of a phospholipid bilayer. Biophys Rev. 2018;10:1371–1376. doi: 10.1007/s12551-018-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel B, Teissie J. Proton long-range migration along protein monolayers and its consequences on membrane coupling. Proc Natl Acad Sci U S A. 1996;93:14521–14525. doi: 10.1073/pnas.93.25.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel B, Prats M, Teissié J. Proton lateral conduction along a lipid monolayer spread on a physiological subphase. BBA-Bioenergetics. 1994;1186:172–176. doi: 10.1016/0005-2728(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Gambu I, Roux B. Interaction of K+ with a phospholipid bilayer: a molecular dynamics study. J Phys Chem B. 1997;101:6066–6072. doi: 10.1021/jp9640134. [DOI] [Google Scholar]
- Garcia A, Zou H, Hossain KR, Xu QH, Buda A, Clarke RJ. Polar interactions play an important role in the energetics of the main phase transition of phosphatidylcholine membranes. ACS Omega. 2019;4:518–527. doi: 10.1021/acsomega.8b03102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manyes S, Oncins G, Sanz F. Effect of ion-binding and chemical phospholipid structure on the nanomechanics of lipid bilayers studied by force spectroscopy. Biophys J. 2005;89:1812–1826. doi: 10.1529/biophysj.105.064030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manyes S, Oncins G, Sanz F. Effect of pH and ionic strength on phospholipid nanomechanics and on deposition process onto hydrophilic surfaces measured by AFM. Electrochim Acta. 2006;51:5029–5036. doi: 10.1016/j.electacta.2006.03.062. [DOI] [Google Scholar]
- Gottlieb MH, Eanes ED. Influence of electrolytes on the thicknesses of the phospholipid bilayers of lamellar lecithin mesophases. Biophys J. 1972;12:1533–1548. doi: 10.1016/S0006-3495(72)86180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtovenko AA, Vattulainen I. Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. J Phys Chem B. 2008;112:1953–1962. doi: 10.1021/jp0750708. [DOI] [PubMed] [Google Scholar]
- Gutman M, Nachliel E. The dynamic aspects of proton transfer processes. BBA-Bioenergetics. 1990;1015:391–414. doi: 10.1016/0005-2728(90)90073-D. [DOI] [Google Scholar]
- Heberle J. Proton transfer reactions across bacteriorhodopsin and along the membrane. BBA-Bioenergetics. 2000;1458:135–147. doi: 10.1016/S0005-2728(00)00064-5. [DOI] [PubMed] [Google Scholar]
- Heberle J, Dencher NA. Surface-bound optical probes monitor protein translocation and surface potential changes during the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 1992;89:5996. doi: 10.1073/pnas.89.13.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370:379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- Javanainen M, Melcrová A, Magarkar A, Jurkiewicz P, Hof M, Jungwirth P, Martinez-Seara H. Two cations, two mechanisms: interactions of sodium and calcium with zwitterionic lipid membranes. Chem Commun. 2017;53:5380–5383. doi: 10.1039/C7CC02208E. [DOI] [PubMed] [Google Scholar]
- Jones MR, Jackson JB. Proton release by the quinol oxidase site of the cytochrome b/c1 complex following single turnover flash excitation of intact cells of Rhodobacter capsulatus. Biochim Biophys Acta Biomembr. 1989;975:34–43. doi: 10.1016/S0005-2728(89)80198-7. [DOI] [Google Scholar]
- Junge W, McLaughlin S. The role of fixed and mobile buffers in the kinetics of proton movement. Biochim Biophys Acta. 1987;890:1–5. doi: 10.1016/0005-2728(87)90061-2. [DOI] [PubMed] [Google Scholar]
- Jurkiewicz P, Cwiklik L, Vojtiskova A, Jungwirth P, Hof M. Structure, dynamics, and hydration of POPC/POPS bilayers suspended in NaCl, KCl, and CsCl solutions. Biochim Biophys Acta. 2012;1818:609–616. doi: 10.1016/j.bbamem.2011.11.033. [DOI] [PubMed] [Google Scholar]
- Kagawa R, Hirano Y, Taiji M, Yasuoka K, Yasui M. Dynamic interactions of cations, water and lipids and influence on membrane fluidity. J Membr Sci. 2013;435:130–136. doi: 10.1016/j.memsci.2013.02.006. [DOI] [Google Scholar]
- Klasczyk B, Knecht V. Validating affinities for ion–lipid association from simulation against experiment. J Phys Chem A. 2011;115:10587–10595. doi: 10.1021/jp202928u. [DOI] [PubMed] [Google Scholar]
- Koynova R, Caffrey M. Phases and phase transitions of the phosphatidylcholines. BBA Biomembranes. 1998;1376:91–145. doi: 10.1016/S0304-4157(98)00006-9. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Song Y, Baker NA. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophys J. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontidis E. Investigations of the Hofmeister series and other specific ion effects using lipid model systems. Adv Colloid Interf Sci. 2017;243:8–22. doi: 10.1016/j.cis.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Mao Y, Du Y, Cang X, Wang J, Chen Z, Yang H, Jiang H. Binding competition to the POPG lipid bilayer of Ca2+, Mg2+, Na+, and K+ in different ion mixtures and biological implication. J Phys Chem B. 2013;117:850–858. doi: 10.1021/jp310163z. [DOI] [PubMed] [Google Scholar]
- Medvedev ES, Stuchebrukhov AA. Proton diffusion along biological membranes. J Phys Condens Matter. 2011;23:234103. doi: 10.1088/0953-8984/23/23/234103. [DOI] [PubMed] [Google Scholar]
- Melcr J, Martinez-Seara H, Nencini R, Kolafa J, Jungwirth P, Ollila OHS. Accurate binding of sodium and calcium to a POPC bilayer by effective inclusion of electronic polarization. J Phys Chem B. 2018;122:4546–4557. doi: 10.1021/acs.jpcb.7b12510. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Monticelli L, Tieleman DP. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys J. 2004;86:1601–1609. doi: 10.1016/s0006-3495(04)74227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Cherepanov DA. Probing biological interfaces by tracing proton passage across them. Photochem Photobiol Sci. 2006;5:577–587. doi: 10.1039/b516443e. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Heberle J, Cherepanov DA. Protons @ interfaces: implications for biological energy conversion. Biochim Biophys Acta. 2006;1757:913–930. doi: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Nachliel E, Gutman M. Quantitative evaluation of the dynamics of proton transfer from photoactivated bacteriorhodopsin to the bulk. FEBS Lett. 1996;393:221–225. doi: 10.1016/0014-5793(96)00870-8. [DOI] [PubMed] [Google Scholar]
- Nachliel E, Gutman M, Kiryati S, Dencher NA. Protonation dynamics of the extracellular and cytoplasmic surface of bacteriorhodopsin in the purple membrane. Proc Natl Acad Sci U S A. 1996;93:10747. doi: 10.1073/pnas.93.20.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Zhang C, Weichselbaum E, Knyazev DG, Pohl P, Carloni P. Interfacial water molecules at biological membranes: structural features and role for lateral proton diffusion. PLoS One. 2018;13:e0193454. doi: 10.1371/journal.pone.0193454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojemyr LN, Lee HJ, Gennis RB, Brzezinski P. Functional interactions between membrane-bound transporters and membranes. Proc Natl Acad Sci U S A. 2010;107:15763–15767. doi: 10.1073/pnas.1006109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst G, Hodzic A, Strancar J, Danner S, Rappolt M, Laggner P. Rigidification of neutral lipid bilayers in the presence of salts. Biophys J. 2007;93:2688–2696. doi: 10.1529/biophysj.107.112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SA, Bostick D, Berkowitz ML. Molecular dynamics simulation of a dipalmitoylphosphatidylcholine bilayer with NaCl. Biophys J. 2003;84:3743–3750. doi: 10.1016/S0006-3495(03)75102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petelska AD, Figaszewski ZA. Effect of pH on the interfacial tension of bilayer lipid membrane formed from phosphatidylcholine or phosphatidylserine. BBA Biomembranes. 2002;1561:135–146. doi: 10.1016/S0005-2736(01)00463-1. [DOI] [PubMed] [Google Scholar]
- Petrache HI, Tristram-Nagle S, Harries D, Kucerka N, Nagle JF, Parsegian VA. Swelling of phospholipids by monovalent salt. J Lipid Res. 2006;47:302–309. doi: 10.1194/jlr.M500401-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantanida L, Bolt HL, Rozatian N, Cobb SL, Voitchovsky K. Ions modulate stress-induced nanotexture in supported fluid lipid bilayers. Biophys J. 2017;113:426–439. doi: 10.1016/j.bpj.2017.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif Maria, Kallies Christopher, Knecht Volker. Effect of Sodium and Chloride Binding on a Lecithin Bilayer. A Molecular Dynamics Study. Membranes. 2017;7(1):5. doi: 10.3390/membranes7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandén T, Salomonsson L, Brzezinski P, Widengren J. Surface-coupled proton exchange of a membrane-bound proton acceptor. Proc Natl Acad Sci U S A. 2010;107:4129–4134. doi: 10.1073/pnas.0908671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer P, Alexiev U, Marti T, Khorana HG, Heyn MP. Covalently bound pH-indicator dyes at selected extracellular or cytoplasmic sites in bacteriorhodopsin. 1. Proton migration along the surface of bacteriorhodopsin micelles and its delayed transfer from surface to bulk. Biochemistry. 1994;33:13684–13692. doi: 10.1021/bi00250a019. [DOI] [PubMed] [Google Scholar]
- Serowy S, Saparov SM, Antonenko YN, Kozlovsky W, Hagen V, Pohl P. Structural proton diffusion along lipid bilayers. Biophys J. 2003;84:1031–1037. doi: 10.1016/S0006-3495(03)74919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smondyrev AM, Voth GA. Molecular dynamics simulation of proton transport through the influenza A virus M2 channel. Biophys J. 2002;83:1987–1996. doi: 10.1016/s0006-3495(02)73960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A, Hagen V, Cherepanov DA, Antonenko YN, Pohl P. Protons migrate along interfacial water without significant contributions from jumps between ionizable groups on the membrane surface. Proc Natl Acad Sci U S A. 2011;108:14461–14466. doi: 10.1073/pnas.1107476108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocanne J-F, Teissié J. Ionization of phospholipids and phospholipid-supported interfacial lateral diffusion of protons in membrane model systems. BBA Biomembranes. 1990;1031:111–142. doi: 10.1016/0304-4157(90)90005-W. [DOI] [PubMed] [Google Scholar]
- Vácha R, Buch V, Milet A, Devlin JP, Jungwirth P. Autoionization at the surface of neat water: is the top layer pH neutral, basic, or acidic? Phys Chem Chem Phys. 2007;9:4736–4747. doi: 10.1039/B704491G. [DOI] [PubMed] [Google Scholar]
- Vácha R, et al. Effects of alkali cations and halide anions on the DOPC lipid membrane. J Phys Chem A. 2009;113:7235–7243. doi: 10.1021/jp809974e. [DOI] [PubMed] [Google Scholar]
- Vácha R, et al. Mechanism of interaction of monovalent ions with phosphatidylcholine lipid membranes. J Phys Chem B. 2010;114:9504–9509. doi: 10.1021/jp102389k. [DOI] [PubMed] [Google Scholar]
- Vorobyov I, Olson TE, Kim JH, Koeppe RE, 2nd, Andersen OS, Allen TW. Ion-induced defect permeation of lipid membranes. Biophys J. 2014;106:586–597. doi: 10.1016/j.bpj.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum E, et al. Origin of proton affinity to membrane/water interfaces. Sci Rep. 2017;7:4553. doi: 10.1038/s41598-017-04675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MG, Grubmuller H, Groenhof G. Anomalous surface diffusion of protons on lipid membranes. Biophys J. 2014;107:76–87. doi: 10.1016/j.bpj.2014.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Voth GA. Properties of hydrated excess protons near phospholipid bilayers. J Phys Chem B. 2010;114:592–603. doi: 10.1021/jp908768c. [DOI] [PubMed] [Google Scholar]
- Zhang C, Knyazev DG, Vereshaga YA, Ippoliti E, Nguyen TH, Carloni P, Pohl P. Water at hydrophobic interfaces delays proton surface-to-bulk transfer and provides a pathway for lateral proton diffusion. Proc Natl Acad Sci U S A. 2012;109:9744–9749. doi: 10.1073/pnas.1121227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Raphael RM. Solution pH alters mechanical and electrical properties of phosphatidylcholine membranes: relation between interfacial electrostatics, intramembrane potential, and bending elasticity. Biophys J. 2007;92:2451–2462. doi: 10.1529/biophysj.106.096362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Küttner D, Renner L, Kaufmann M, Werner C. Fluidity modulation of phospholipid bilayers by electrolyte ions: insights from fluorescence microscopy and microslit electrokinetic experiments. J Phys Chem A. 2012;116:6519–6525. doi: 10.1021/jp212364q. [DOI] [PubMed] [Google Scholar]