Abstract
Generalized-ensemble algorithms are powerful techniques for investigating biomolecules such as protein, DNA, lipid membrane, and glycan. The generalized-ensemble algorithms were originally developed in the canonical ensemble. On the other hand, not only temperature but also pressure is controlled in experiments. Additionally, pressure is used as perturbation to study relationship between function and structure of biomolecules. For this reason, it is important to perform efficient conformation sampling based on the isothermal–isobaric ensemble. In this article, we review a series of the generalized-ensemble algorithms in the isothermal–isobaric ensemble: multibaric–multithermal, pressure- and temperature-simulated tempering, replica-exchange, and replica-permutation methods. These methods achieve more efficient simulation than the conventional isothermal–isobaric simulation. Furthermore, the isothermal–isobaric generalized-ensemble simulation samples conformations of biomolecules from wider range of temperature and pressure. Thus, we can estimate physical quantities more accurately at any temperature and pressure values. The applications to the biomolecular system are also presented.
Keywords: Generalized-ensemble algorithm, Molecular simulation, High pressure, Protein folding
Introduction
Molecular dynamics (MD) and Monte Carlo (MC) simulations play a crucial role to obtaining properties of biomolecules from the atomic level. In many cases, however, MD and MC simulations sample conformations only in a few local-minimum free-energy states because of its time scale and degrees of freedom. This hampers accurate estimation of physical quantities such as free-energy differences. The specially designed hardware has extended upper limit of the time scale up to milliseconds (Shaw et al. 2010; Lindorff-Larsen et al. 2011). On the other hand, highly optimized software such as GENESIS (Jung et al. 2015; Kobayashi et al. 2017) and MODYLAS (Andoh et al. 2013) have extended upper limit of the system size. Yet, it remains difficult to sample sufficient conformations in the phase space.
Generalized-ensemble algorithms (Mitsutake et al. 2001) were developed to overcome the sampling difficulties. In a multicanonical ensemble (Berg and Neuhaus 1991, 1992b; Hansmann et al. 1996; Nakajima et al. 1997), the target system performs a free one-dimensional random walk in the potential-energy space; thus, the target system does not get trapped in local-minimum free-energy states. In a simulated tempering method (Lyubartsev et al. 1992; Marinari and Parisi 1992), temperature is treated as a dynamic variable so that a random walk in the temperature space can be realized. These methods introduce non-Boltzmann weight factors, which are not known a priori, and have to be determined before simulations. A replica-exchange (Hukushima and Nemoto 1996; Sugita and Okamoto 1999) and a replica-permutation (Itoh and Okumura 2012) methods are other generalized-ensemble algorithms, which are not necessary to determine the weight factor in advance. We prepare non-interacting copies (or replicas) of the target system at different temperatures and perform simulation in parallel. During the simulation, temperature exchange/permutation is conducted among the replicas. Each replica performs a random walk in the temperature space, which helps the target system to escape from a local-minimum free-energy state.
The generalized-ensemble algorithms were originally proposed in the canonical ensemble. In the canonical ensemble, the volume of the simulation box is constant and pressure is not controlled. In contrast, pressure as well as temperature is controlled in experiments; thus, physical quantities estimated from experiments follow isothermal–isobaric ensemble. Based on this point, the isothermal–isobaric ensemble simulation is important to compare the simulation results with experiments.
In this review article, we focus on a series of the generalized-ensemble algorithms in the isothermal–isobaric ensemble, namely multibaric–multithermal, pressure- and temperature-simulated tempering, replica-exhange, and replica-permutation methods. These methods achieve the efficient conformational sampling in the wider range of temperature and pressure. We also present applications of the generalized-ensemble algorithms in the isothermal–isobaric ensemble to the biomolecule systems: chignolin, AK16 peptide, C-peptide, and ubiquitin.
This review is organized as follows. First, we introduce the basics of the isothermal–isobaric ensemble and then we review the generalized-ensemble algorithms in the isothermal–isobaric ensemble. In the following section, the applications of the isothermal–isobaric generalized-ensemble algorithms to biomolecular systems are presented. The final section is devoted to conclusions.
Basics of isothermal–isobaric ensemble
In this section, we briefly review the fundamentals of thermodynamics and statistical mechanics for the isothermal–isobaric ensemble as described in many textbooks for example, McQuarrie and Simon (1997).
Isothermal–isobaric ensemble
In the isothermal–isobaric ensemble, the number of particles N, pressure P, and temperature T are treated as parameters. Let us consider N-particle system in a box with volume V. The system is specified by the coordinates q ≡{q1,⋯ ,qN}, momenta p ≡{p1,⋯ ,pN}, and volume of the box V. The potential energy of the system is given as a function of the coordinates and volume: U(q,V ). The probability distribution PNPT(U,V ;T,P) is given by the product of density of states n(U,V ) and distribution function WNPT(U,V ; T,P), as follows:
| 1 |
The distribution function in the isothermal–isobaric ensemble is given by
| 2 |
where β = 1/(kBT) is the inverse temperature (kB is the Boltzmann constant) and is the enthalpy. Scaled coordinates are used in the isothermal–isobaric molecular simulation (McDonald 1972). When an isotropic box is used, the coordinates are scaled as s = V− 1/3q. In this case, the distribution function is transformed as
| 3 |
Thermodynamic quantities
The difference in the Gibbs free-energy ΔG between states A and B is calculated as follows:
| 4 |
where R is the gas constant, fA and fB are fractions of the states A and B, respectively. In the case of evaluating protein stability, for example, fA and fB are probabilities of the folded and unfolded states, respectively. The differences in the partial molar enthalpy ΔH, the heat capacity ΔCP, and the partial molar volume ΔV are calculated as follows:
| 5 |
| 6 |
| 7 |
The differences in the internal energy ΔU and the entropy ΔS are calculated as follows:
| 8 |
| 9 |
The temperature and pressure dependence of ΔG can be fitted by the following equation proposed by Hawley (1971):
| 10 |
Here, T0 and P0 are the reference temperature and pressure. ΔG0, ΔS0, and ΔV0 are the differences in the Gibbs free-energy, entropy, and partial molar volume between states A and B, respectively, at T0 and P0. Δα and Δβ are the differences in the thermal expansivity factor and compressibility factor between states A and B, respectively; these factors are related to, but are not identical to the thermal expansion coefficient and compressibility in thermodynamics, respectively. By substituting (10) into (5) and (7), the temperature and pressure dependence of ΔH and ΔV are obtained:
| 11 |
| 12 |
Generally, from Le Chatelier’s principle, the volume of a substance becomes smaller by applying pressure. In other words, the probability of the smaller volume state increases as pressure increases.
Generalized-ensemble algorithms in isothermal–isobaric ensemble
In this section, we first introduce the general view of Markov chain Monte Carlo algorithms (Berg 2004). Then, we review formulations on a series of generalized-ensemble algorithms in the isothermal–isobaric ensemble: multibaric–multithermal, pressure- and temperature-simulated tempering, replica-exchange, and replica-permutation methods.
Markov chain Monte Carlo algorithm
Let us consider the Markov chain Monte Carlo (MCMC) algorithm. We assume here that the number of candidates for a state transition is n. Each state i has a weight wi and transits to state j with a transition probability P(i → j). To converge the unique distribution, Markov chain should satisfy ergodicity (that is, the transitions are regular and non-periodic). In addition, the following equations are imposed to obtain the required equilibrium distribution (Manousiouthakis and Deem 1999) such as isothermal–isobaric ensemble:
| 13 |
| 14 |
Equation 13 is the normalization condition and Eq. 14 is the balance condition. These equations are summarized by using a stochastic flow from state i to state j defined by v(i → j) = wiP(i → j), as follows:
| 15 |
The Metropolis algorithm (Metropolis et al. 1953) (its generalization is called Metropolis–Hastings algorithm (Hastings 1970)) and heat bath (Gibbs sampler) algorithm (Geman and Geman 1984) are the two most practical implementations of the MCMC algorithms. These algorithms only satisfy the detailed balance condition, which is the sufficient condition for Eq. 15:
| 16 |
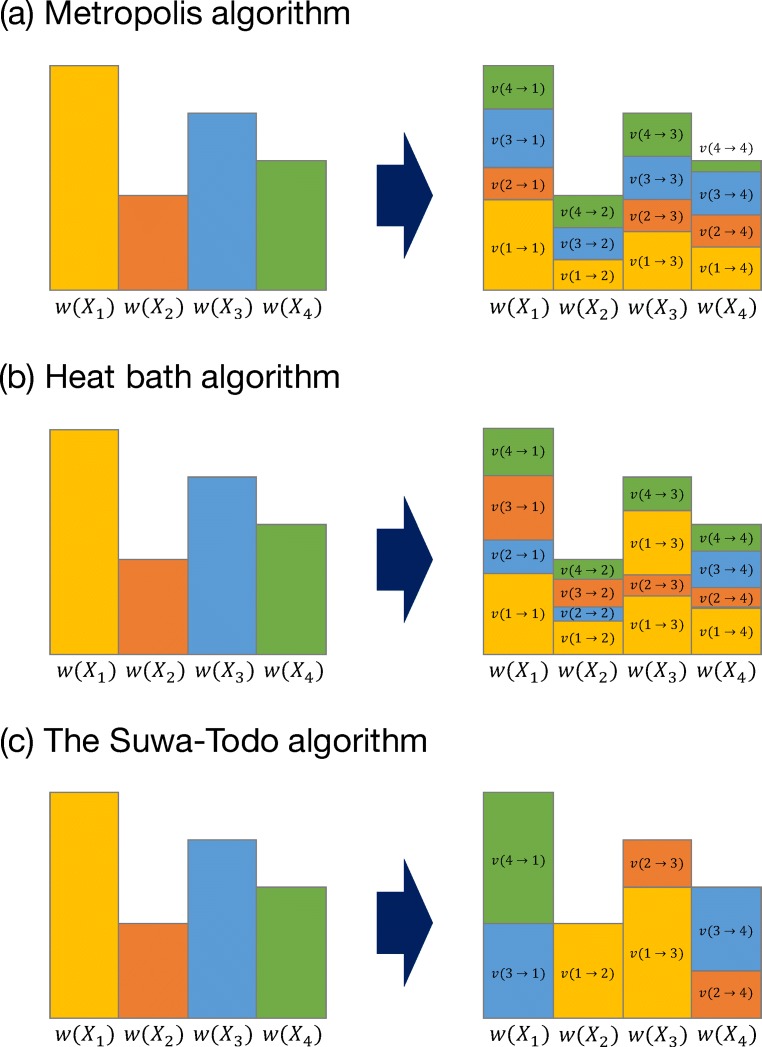
In the Metropolis algorithm, the amount of stochastic flow from state i to state j is given as follows (see Fig. 1a):
| 17 |
where the coefficient 1/(n − 1) arises from the random selection of state j from n − 1 candidates except state i. In the heat bath algorithm, the amount of stochastic flow from state i to state j is given as follows (see Fig. 1b):
| 18 |
Fig. 1.
Schematic figures of the a Metropolis, b heat bath, and c Suwa–Todo algorithms
Recently, Suwa and Todo proposed a new type of MCMC algorithm (Suwa and Todo 2010), which satisfies (15) without imposing the detailed balance condition (16). This method succeeds in minimizing the rejection ratio, and in many cases, the rejection ratio becomes zero. The amount of stochastic flow is given as follows (see Fig. 1c):
| 19 |
where
and wk is a maximum weight among n states. Note that the Suwa–Todo algorithm is identical to the Metropolis algorithm when n = 2.
Multibaric–multithermal method
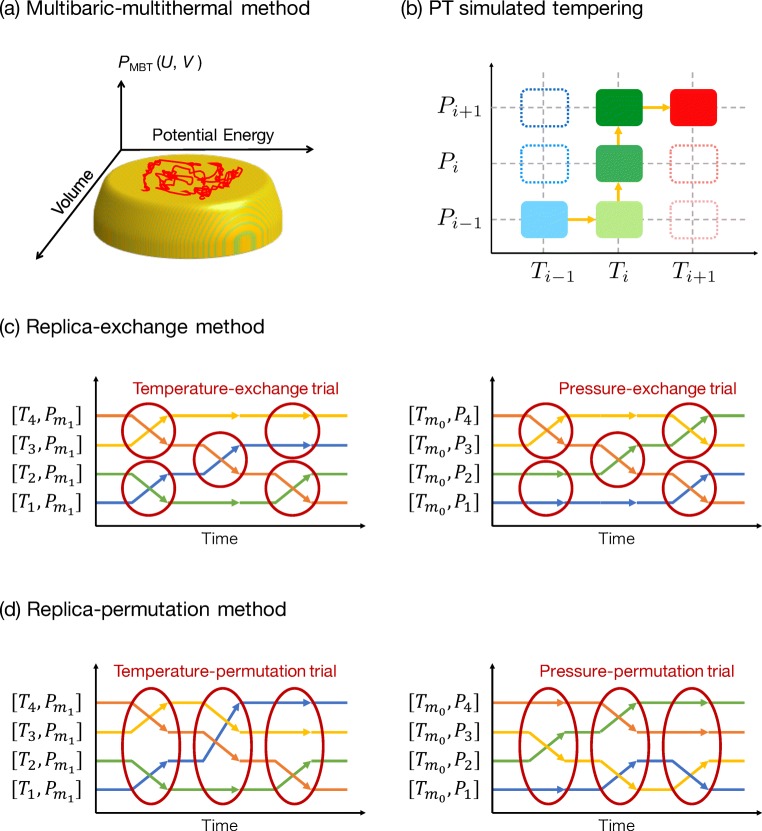
A multibaric–multithermal (MBT) method (Okumura and Okamoto 2004a, b, c, 2006) was proposed as an extension of the multicanonical method. In the MBT method, the uniform distribution in the potential energy and volume spaces is realized so that the system can perform a random walk in the potential energy and volume spaces, as shown in Fig. 2a. Hence, we can obtain the isothermal–isobaric ensemble in a wide range of temperature and pressure values from one MBT simulation.
Fig. 2.
Schematic figures of generalized-ensemble algorithms in isothermal–isobaric ensemble: a multibaric–multithermal, b pressure- and temperature-simulated tempering, c replica-exchange, and d replica-permutation methods
In the MBT method, a non-Boltzmann weight factor
| 20 |
is used instead of the usual distribution function in the isothermal–isobaric ensemble given in Eq. 2. Here, β0 = 1/(kBT0) is a reference temperature and is the multibaric–multithermal enthalpy. is determined so as to realize the uniform probability distribution of the potential energy and volume:
To perform the MBT simulation, the non-Boltzmann weight factor (or multibaric–multithermal enthalpy) must be determined in advance. An iterative procedure of trial simulations (Berg and Celik 1992a; Lee 1993; Okamoto and Hansmann 1995; Mitsutake et al. 2001) or Wang–Landau method (Wang and Landau 2001a, 2001b) is used to obtain the weight factor (also see Refs. Okumura and Okamoto 2008, 2012). By using the obtained weight factor, we perform one long multibaric–multithermal simulation. For MC simulation, the transition probability is calculated using multibaric–multithermal enthalpy instead of the enthalpy (Okumura and Okamoto 2004b, 2004c). For MD simulation, the enthalpy term in the equations of motion is substituted by multibaric–multithermal enthalpy (Okumura and Okamoto 2004a, 2006).
Pressure- and temperature-simulated tempering method
Let us introduce a simulated tempering (ST) method associated with pressure and temperature (PTST) (Mori and Okamoto 2010). The PTST realizes a uniform distribution in the temperature and pressure spaces by treating those as dynamic variables. Temperature and pressure as well as the coordinates and momenta are changed during the PTST simulation.
The weight factor for the PTST is given by
| 21 |
Here, g(T,P) is introduced to realize the uniform distribution in the temperature and pressure spaces. g(T,P) is determined so as to satisfy the following equation:
| 22 |
and thus, g(T,P) is written as
| 23 |
We find that g(T,P) corresponds to a dimensionless Gibbs free-energy omitting a constant. In practice, the temperature and pressure are discretized. Let us consider using M0 temperatures and M1 pressures. Here, we introduce indices m0 and m1 that indicate one of the M0 temperatures and one of M1 pressures, respectively. The weight factor and the dimensionless Gibbs free-energy at and are given as follows:
| 24 |
| 25 |
The weight factor (i.e., the dimensionless Gibbs free-energy) should be determined before performing the PTST simulation. The weight factor is determined by an iterative procedure of short trial simulations (Irbäck and Potthast 1995; Hansmann and Okamoto 1997; Mitsutake et al. 2001) or short replica-exchange simulation combining with reweighting techniques (Kumar et al. 1992; Shirts and Chodera 2008). On-the-fly weight determination scheme during the ST simulation was also proposed (Nguyen et al. 2013).
The PTST simulation is performed as follows (schematic figure is shown in Fig. 2b): (i) isothermal–isobaric MD or MC simulation at temperature and is performed for certain steps, (ii) calculate the transition probability from to another set of temperature and pressure value in M0 temperatures and M1 pressures, and (iii) update the temperature and pressure following the obtained transition probability. Usually, the transition probability is calculated by the Metropolis algorithm. Instead of the Metropolis algorithm, the Gibbs sampler (heat bath algorithm) or Suwa–Todo algorithm is also applicable to enhance acceptance ratio (Chodera and Shirts 2011; Mori and Okumura 2015).
Isothermal–isobaric replica-exchange method
In an isothermal–isobaric replica-exchange method (TP-REM) (Okabe et al. 2001; Mori and Okamoto 2010), simulations at different thermodynamic states, characterized by a set of temperature and pressure , are performed in parallel. M0 different temperatures and M1 different pressures are assigned to M(= M0 × M1) non-interacting replicas (copies of the system). A state in the TP-REM is expressed by a combination of the thermodynamic state indices and the replica indices: , where i(m) is a permutation function from the thermodynamic state index to replica index and m(i) is the inverse.
The weight factor for the state Xα is given by the product of the weight factor of each replica i (or each thermodynamic state m):
| 26 |
In the TP-REM, exchange trials for a pair of parameters
| 27 |
are performed during the simulation (Fig. 2c). Typically, neighboring parameters are selected as a pair of replica exchange. The temperature exchange and pressure exchange trials are performed alternately or randomly. The Metropolis algorithm is employed to calculate the amount of stochastic flow v(Xα → Xγ) and transition probability P(Xα → Xγ). Note that the weight factors wi and wj in Eq. 17 are replaced by wR(Xα) and wR(Xα), respectively.
Isothermal–isobaric replica-permutation method
An isothermal–isobaric replica-permutation method (TP-RPM) (Yamauchi and Okumura 2017, 2019) is an improved alternative to the TP-REM. In the TP-RPM, the parameter permutation among more than two replicas is performed during the simulation (Fig. 2d):
| 28 |
where m and n are permutation functions. All possible permutations between replicas and parameters are considered; thus, γ takes values from 1 to M!. However, it becomes impractical to consider all possible permutations as the number of replicas increases, especially exceeding ten replicas. To avoid this problem, the temperature permutations and the pressure permutations are performed alternately. If the number of permutation candidates is still large, the sets of temperatures or pressures are divided into several subsets. Usually, subsets are set so that the number of replicas in each subset is the same and the temperature or pressure labels in each subset are sequential and cyclic. Parameter permutation is performed among the replicas in each subset.
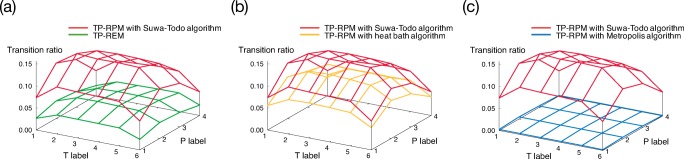
The weight factor for state Xα is identical to the REM as in Eq. 26. The Suwa–Todo algorithm is employed to calculate the amount of stochastic flow v(Xα → Xγ) and transition probability P(Xα → Xγ). Of course, instead of the Suwa–Todo algorithm, the Metropolis or heat bath algorithm is also able to be employed. However, sampling efficiency, such as transition ratio of the parameters, is lower than the Suwa–Todo algorithm (Yamauchi and Okumura 2017). The comparison of the transition ratio of temperature and pressure values between TP-RPM with the Suwa–Todo, Metropolis, heat bath algorithms, and TP-REM is presented in Figure 3. Here, the transition ratio at a parameter label (i.e., a set of temperature and pressure labels) is defined as a probability with which the replica at that parameter label is transferred to other parameter labels. These results clearly show that the TP-RPM with the Suwa–Todo algorithm achieves the highest transition ratio, indicating that it is the most efficient among the four methods.
Fig. 3.
Comparison of the transition ratio between TP-RPM with the Suwa–Todo algorithm and a TP-REM, b TP-RPM with the heat bath algorithm, and c TP-RPM with the Metropolis algorithm
Estimation of physical quantities: reweighting techniques
After performing the generalized-ensemble simulation, we can obtain statistical averages of physical quantities by using reweighting techniques.
For the MBT method, a single-histogram reweighting technique (Ferrenberg and Swendsen 1988, 1989a, Okumura and Okamoto 2004c) is employed to obtain the density of states and probability distribution at any temperature and pressure values.
For the PTST, TP-REM, and TP-RPM, the arithmetic mean of the trajectory data from the one equilibrium state gives the isothermal–isobaric average at the set temperature and pressure values for the simulation. On the other hand, a weighted histogram analysis method (WHAM) (Kumar et al. 1992; Chodera et al. 2007), which is also called multiple-histogram reweighting techniques (Ferrenberg and Swendsen 1989b), and a multistate Bennett acceptance ratio (MBAR) method (Shirts and Chodera 2008) are widely applied to calculate the isothermal–isobaric averages of the physical quantities. These methods enable us to evaluate the physical quantities more precisely because of considering all trajectory data from multiple equilibrium states. The WHAM gives the density of states, whereas the MBAR cannot directly calculate. However, the histogram construction of the WHAM makes bias due to the binning, and is a time-consuming process. The MBAR does not require making the histogram; thus, the MBAR provides the weight for each trajectory and the direct estimation of statistical errors.
A generic transition-based reweighting analysis method (TRAM) (Wu et al. 2016) has been developed, which is a general formalization of the WHAM and MBAR. The TRAM takes advantage of the Markov states model. The key assumption of the TRAM is that the data are sampled from the local equilibrium distributions, while the WHAM and MBAR assume those from the global equilibrium distribution of each ensemble. The TRAM provides not only statistical average of the physical quantities, but also the transition probability from one state to the other states. Thus, we can obtain information about kinetics of the target system.
Application of isothermal–isobaric generalized methods to biomolecules
Pressure dependence for β-hairpin formation peptide: chignolin
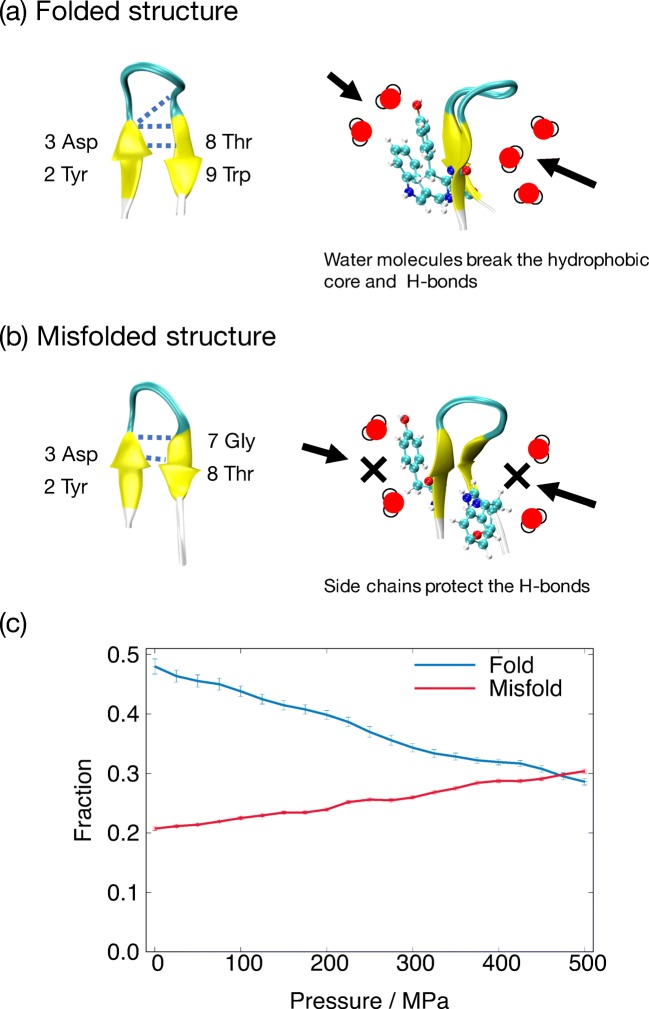
Chignolin is an artificially designed peptide comprised of ten amino acid residues, and its amino acid sequence is GYDPETGTWG (Honda et al. 2004). Chignolin folds into a β-hairpin structure. Molecular dynamics simulation studies revealed that chignolin is able to form two types of β-hairpin structure: one is a folded structure and the other is a misfolded structure (see Fig. 4a and b) (van der Spoel and Seibert 2006; Satoh et al. 2006; Suenaga et al. 2007; Harada and Kitao 2011; Kührová et al. 2012).
Fig. 4.
Typical conformations of the a folded and b misfolded chignolin. c The pressure dependence of fraction for folded, misfolded, and unfolded chignolin
To investigate the folding–unfolding mechanism of chignolin, the multibaric–multithermal molecular dynamics simulation was performed (Okumura 2012). Folding and unfolding processes were discussed by calculating free-energy landscapes. The unfolding process begins from only the C terminus or both C and N termini of the β-hairpin structures. The intermediate structures, for example, 310-helix and α-helix, can be formed under the unfolding pathway. The folding is the reverse process. Chignolin is denatured by not only temperature but also pressure. The pressure denaturation occurs because water molecules approach the hydrophobic residues with increasing pressure, and the water molecules break the hydrophobic contacts among side chains.
The isothermal–isobaric replica-permutation MD simulation was also performed (Yamauchi and Okumura 2017), revealing that the fraction of the misfolded structure increases while that of folded structure decreases with increasing pressure (see Fig. 4c). The partial molar volume difference was estimated as ΔV = Vmisfold − Vfold = − 4.6 ± 0.2 cm3/mol at 300 K, indicating that the misfolded structure has smaller partial volume. The molecular mechanism of the different stability between folded and misfolded structures under high pressure arises from different orientation of the Tyr2 and Trp9 side chains (Fig. 4a and b). For the folded structure, as pressure increases, water molecules approach not only hydrophobic core, but also intra-hydrogen bonds from the opposite side of the hydrophobic core. Therefore, the water molecules break the intra-hydrogen bonds as well as the hydrophobic core. On the other hand, in the misfolded structure, the hydrogen bonds that are important to form the misfolded structure are covered with the Try2 and Trp9 side chains. The side chains protect the hydrogen bonds from the approaching water molecules. Therefore, the misfolded structure becomes more stable with compressing its structure under high-pressure condition.
Pressure dependence for helix formation peptide: AK16
AK16 peptide is comprised of 16 amino acid residues, and its amino acid sequence is YGAAKAAAAKAAAAKA. AK16 peptide forms α-helix conformation at room temperature and atmospheric pressure (Chakrabartty and Baldwin 1995). FT-IR spectroscopic studies investigated pressure dependence of the alanine-rich peptide including AK16 peptide, revealing that the fraction of α-helix conformation increases with increasing pressure (Takekiyo et al. 2005; Imamura and Kato 2009).
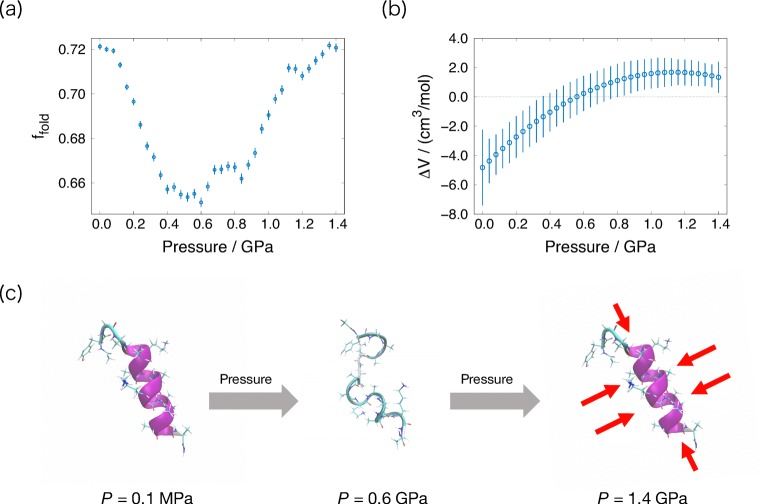
To understand the molecular mechanism for the pressure-induced conformation changes of AK16 peptide, the ST simulations associated with pressure were performed (Mori and Okumura 2013, 2014). The pressure dependence of the fraction for helical conformation and the partial molar volume difference between folded and unfolded structures, ΔV = Vunfold − Vfold, were calculated, as presented in Fig. 5a, b. In the high-pressure condition, especially more than 0.6 GPa, the fraction for the helical conformation increases and the ΔV takes a positive value. These results are consistent with experimental results (Takekiyo et al. 2005). The typical conformations of the AK16 peptide at various pressures are illustrated in Fig. 5c. As pressure increases, the diameter of the helix structure and the distance between hydrogen bonds that form the helix structure become short. These results indicate that the helical structure at high pressure is more compressed from the longitudinal and transversal directions than that at atmospheric pressure. This is also the reason why the ΔV takes positive value at high pressure.
Fig. 5.
Pressure dependence of the a fraction for the folded AK16 peptide and b partial molar volume difference. c Typical conformations of the AK16 peptide at P = 0.1 MPa, 0.6 GPa, and 1.4 GPa
Pressure dependence for helix formation peptide: C peptide
C-peptide analogs are polypeptides that exist at the N terminus of ribonuclease A. Here, we only discuss the C-peptide analog comprised of thirteen amino acid residues (AETAAAKFLRAHA). Hereafter, we refer to this C-peptide analog as simply C-peptide. The C-peptide forms α-helix conformation with stabilization of a salt bridge (ionic interaction) between Glu2 and Arg10 (Scholtz and Baldwin 1992).
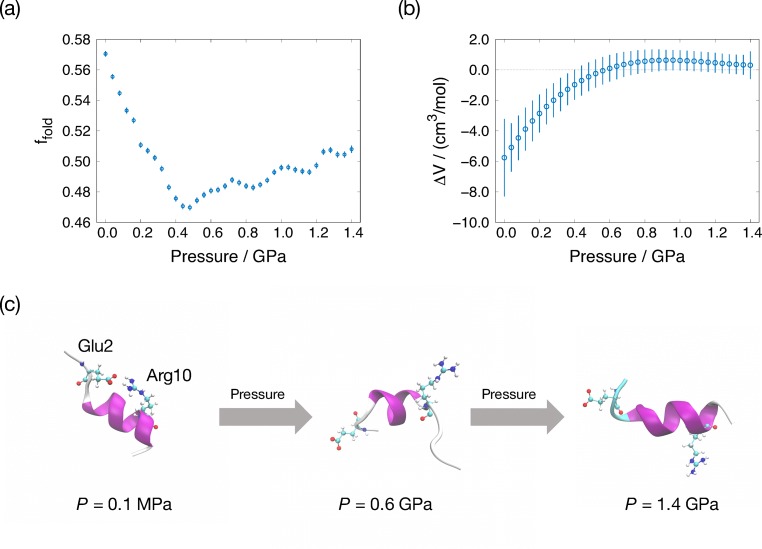
To reveal the pressure-induced conformation changes of the C-peptide, the ST simulations associated with pressure were performed (Mori and Okumura 2014). Figure 6a presents pressure dependence of the fraction for the helical conformation. Below 0.6 GPa, the fraction of the helical conformation of the C-peptide decreases as pressure increases. In contrast, above 0.6 GPa, the fraction of the helical conformation increases. Figure 6b presents partial molar volume difference between folded and unfolded structures: ΔV = Vunfold − Vfold. The partial molar volume difference takes a positive value at high-pressure conditions. The typical conformations of the C-peptide at various pressures are illustrated in Fig. 6c. It is noteworthy that the helical conformation of the C-peptide at high pressure does not form the salt bridge. This is a different point from the helical conformation at atmospheric pressure. The salt bridge contributes to the stabilization of the helical conformation at room temperature and atmospheric pressure. As the pressure increases up to 0.6 GPa, the salt bridge is broken by the hydration of water molecules. This is the reason why the fraction for helical conformation takes minimum value around 0.6 GPa. As further pressure is applied, the volume of the helical conformation becomes smaller than the helical conformation at the atmospheric pressure and the unfolded structure; thus, the helical conformation is stable under high-pressure conditions. Indeed, the diameter of the helix structure and distance between hydrogen bonds that form the helix structure gradually become short with increasing pressure. These results reflect compression of the helical conformation under the high-pressure condition.
Fig. 6.
Pressure dependence of the a fraction for the folded C-peptide and b partial molar volume difference. c Typical conformations of the C-peptide at P = 0.1 MPa, 0.6 GPa, and 1.4 GPa
Ubiquitin
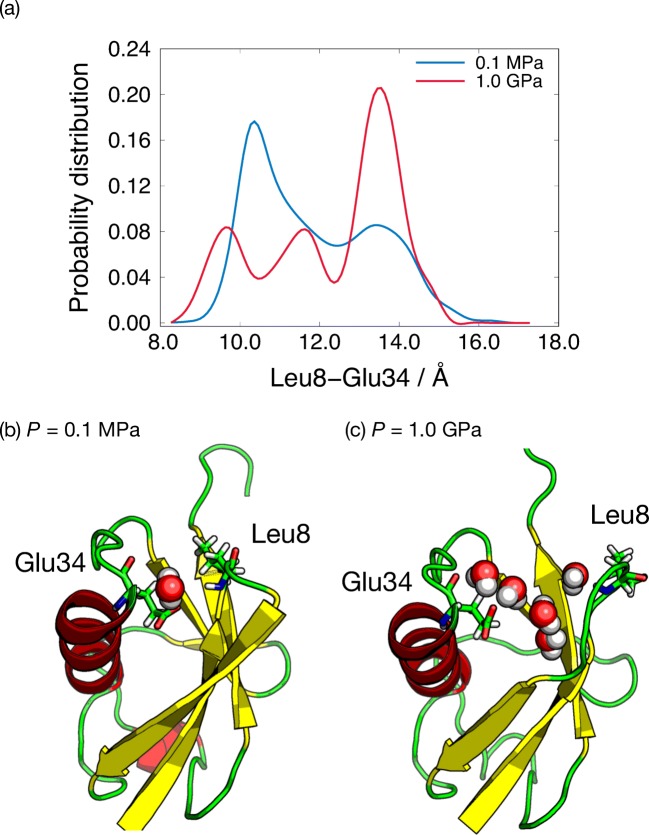
Ubiquitin (UB), consisting of 76 amino acid residues, is involved in many cellular phenomena such as proteasomal degradation of damaged proteins. Its structure, functions, and protein folding have been studied thoroughly both in experimental and theoretical aspects. NMR experiments detected conformational change of the UB induced by pressure perturbation (Kitahara et al. 2001, 2003, 2005, Nisius and Grzesiek 2012). The pressure-simulated tempering (PST) simulations also demonstrated the pressure-induced conformational changes of the UB (Mori and Okamoto 2017). In particular, larger fluctuations were found between 7–9 and 30–40 amino acid residues during the simulations. As presented in Fig. 7a, the peak position of the distribution for the distance between Leu8 and Glu34 is shifted with increasing pressure from 0.1 MPa to 1.0 GPa, which is consistent with the constant pressure MD simulation (Imai and Sugita 2010). The conformations at 0.1 GPa and 1.0 GPa are illustrated in Fig. 7b, c. The water molecules penetrate into the inside of the UB in the high-pressure condition, which induces denaturation. The trajectory data from the PST simulations were also employed to 15N NMR chemical shift estimation (La Penna et al. 2016). This technique is useful to integrate experiment and simulation results.
Fig. 7.
a The probability distribution of the distance between Leu8 and Glu34. Typical conformation of ubiquitin at bP = 0.1 MPa and cP = 1.0 GPa
Conclusions
In this review, we outlined the generalized-ensemble algorithms in the isothermal–isobaric ensemble and its applications to the biomolecule system. The generalized-ensemble simulation in the isothermal–isobaric ensemble enables us to sample conformations of the target system at various temperature and pressure values. Furthermore, a random walk in the parameter spaces enhances the sampling efficiency in the conformational space; thus, physical quantities are estimated more precisely.
We also reviewed the application of the generalized-ensemble algorithms in the isothermal–isobaric ensemble to the following biomolecular systems: chignolin, AK16 peptide, C-peptide, and ubiquitin. The physical quantities, such as ΔV and 15N chemical shift, were estimated from the simulations. We can directly compare these quantities from simulations with those from experiments. Furthermore, the generalized-ensemble simulation in the isothermal–isobaric ensemble enables us not only to elucidate the mechanism behind the conformational change, but also to predict the stable conformation under high-pressure conditions. We believe that integrating experimental and theoretical results will lead to deep understanding of life system.
Although we only focused on the conformational change induced by pressure pertubation in this review, the generalized-ensemble methods in the isothermal–isobaric ensemble are also powerful tools to investigate the molecular mechanism of the conformational change induced by cold temperature (Privalov 1989). To access higher pressure and lower temperature region, more efficient generalized-ensemble algorithms are necessary. The post analysis methods such as reweighting become key techniques for not only estimating physical quantities more accurately but also extracting more information from the generalized-ensemble simulations. Furthermore, it should be emphasized that using accurate force field, especially water model, is crucial for quantitative estimation of physical quantities at various temperature and pressure values (Yang et al. 2014; Best et al. 2014).
Acknowledgments
This work was supported by JSPS KAKENHI (no. JP16H00790) and the Okazaki Orion Project of National Institutes of Natural Sciences.
Compliance with Ethical Standards
Conflict of interest
Masataka Yamauchi declares that he has no conflict of interest. Yoshiharu Mori declares that he has no conflict of interest. Hisashi Okumura declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue dedicated to the “2018 Joint Conference of the Asian Biophysics Association and Australian Society for Biophysics” edited by Kuniaki Nagayama, Raymond Norton, Kyeong Kyu Kim, Hiroyuki Noji, Till Böcking, and Andrew Battle.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andoh Y, Yoshii N, Fujimoto K, Mizutani K, Kojima H, Yamada A, Okazaki S, Kawaguchi K, Nagao H, Iwahashi K, Mizutani F, Minami K, Ichikawa S, Komatsu H, Ishizuki S, Takeda Y, Fukushima M. Modylas: a highly parallelized general-purpose molecular dynamics simulation program for large-scale systems with long-range forces calculated by fast multipole method (fmm) and highly scalable fine-grained new parallel processing algorithms. J Chem Theory Comput. 2013;9(7):3201–3209. doi: 10.1021/ct400203a. [DOI] [PubMed] [Google Scholar]
- Berg BA, Neuhaus T. Multicanonical algorithms for first order phase transitions. Phys Lett B. 1991;267(2):249–253. doi: 10.1103/PhysRevLett.68.9. [DOI] [PubMed] [Google Scholar]
- Berg Bernd A., Celik Tarik. New approach to spin-glass simulations. Physical Review Letters. 1992;69(15):2292–2295. doi: 10.1103/PhysRevLett.69.2292. [DOI] [PubMed] [Google Scholar]
- Berg Bernd A., Neuhaus Thomas. Multicanonical ensemble: A new approach to simulate first-order phase transitions. Physical Review Letters. 1992;68(1):9–12. doi: 10.1103/PhysRevLett.68.9. [DOI] [PubMed] [Google Scholar]
- Berg BA. Markov chain Monte Carlo simulations and their statistical analysis. Singapore: World Scientific; 2004. [Google Scholar]
- Best RB, Miller C, Mittal J. Role of solvation in pressure-induced helix stabilization. J Chem Phys. 2014;141(22):12B621_1. doi: 10.1063/1.4901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabartty A, Baldwin RL. Stability of α-helices. Adv Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- Chodera JD, Swope WC, Pitera JW, Seok C, Dill KA. Use of the weighted histogram analysis method for the analysis of simulated and parallel tempering simulations. J Chem Theory Comput. 2007;3(1):26–41. doi: 10.1021/ct0502864. [DOI] [PubMed] [Google Scholar]
- Chodera JD, Shirts MR. Replica exchange and expanded ensemble simulations as gibbs sampling: Simple improvements for enhanced mixing. J Chem Phys. 2011;135(19):194110. doi: 10.1063/1.3660669. [DOI] [PubMed] [Google Scholar]
- Ferrenberg AM, Swendsen RH. New monte carlo technique for studying phase transitions. Phys Rev Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- Ferrenberg Alan M., Swendsen Robert H. New Monte Carlo Technique for Studying Phase Transitions. Physical Review Letters. 1989;63(15):1658–1658. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- Ferrenberg Alan M., Swendsen Robert H. Optimized Monte Carlo data analysis. Physical Review Letters. 1989;63(12):1195–1198. doi: 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- Geman S, Geman D. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE T Pattern Anal. 1984;6(6):721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- Hansmann UH, Okamoto Y, Eisenmenger F. Molecular dynamics, langevin and hydrid monte carlo simulations in a multicanonical ensemble. Chem Phys Lett. 1996;259(3-4):321–330. [Google Scholar]
- Hansmann UHE, Okamoto Y. Numerical comparisons of three recently proposed algorithms in the protein folding problem. J Comput Chem. 1997;18(7):920–933. [Google Scholar]
- Harada R, Kitao A. Exploring the folding free energy landscape of a β-hairpin miniprotein, chignolin, using multiscale free energy landscape calculation method. J Phys Chem B. 2011;115(27):8806–8812. doi: 10.1021/jp2008623. [DOI] [PubMed] [Google Scholar]
- Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57(1):97–109. [Google Scholar]
- Hawley SA. Reversible pressure-temperature denaturation of chymotrypsinogen. Biochemistry. 1971;10(13):2436–2442. doi: 10.1021/bi00789a002. [DOI] [PubMed] [Google Scholar]
- Honda S, Yamasaki K, Sawada Y, Morii H. 10 residue folded peptide designed by segment statistics. Structure. 2004;12(8):1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Hukushima K, Nemoto K. Exchange monte carlo method and application to spin glass simulations. J Phys Soc Jpn. 1996;65(6):1604–1608. [Google Scholar]
- Imai T, Sugita Y. Dynamic correlation between pressure-induced protein structural transition and water penetration. J Phys Chem B. 2010;114(6):2281–2286. doi: 10.1021/jp909701j. [DOI] [PubMed] [Google Scholar]
- Imamura H, Kato M. Effect of pressure on helix-coil transition of an alanine-based peptide: an ftir study. Proteins. 2009;75(4):911–918. doi: 10.1002/prot.22302. [DOI] [PubMed] [Google Scholar]
- Irbäck A, Potthast F. Studies of an off-lattice model for protein folding: Sequence dependence and improved sampling at finite temperature. J Chem Phys. 1995;103(23):10298–10305. [Google Scholar]
- Itoh SG, Okumura H. Replica-permutation method with the suwa–todo algorithm beyond the replica-exchange method. J Chem Theory Comput. 2012;9(1):570–581. doi: 10.1021/ct3007919. [DOI] [PubMed] [Google Scholar]
- Jung J, Mori T, Kobayashi C, Matsunaga Y, Yoda T, Feig M, Sugita Y. Genesis: a hybrid-parallel and multi-scale molecular dynamics simulator with enhanced sampling algorithms for biomolecular and cellular simulations. Wiley Interdiscip Rev: Comput Mol Sci. 2015;5(4):310–323. doi: 10.1002/wcms.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara R, Yamada H, Akasaka K. Two folded conformers of ubiquitin revealed by high-pressure nmr. Biochemistry. 2001;40(45):13556–13563. doi: 10.1021/bi010922u. [DOI] [PubMed] [Google Scholar]
- Kitahara R, Akasaka K. Close identity of a pressure-stabilized intermediate with a kinetic intermediate in protein folding. Proc Natl Acad Sci USA. 2003;100(6):3167–3172. doi: 10.1073/pnas.0630309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara R, Yokoyama S, Akasaka K. Nmr snapshots of a fluctuating protein structure: ubiquitin at 30 bar–3 kbar. J Mol Bio. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- Kobayashi C, Jung J, Matsunaga Y, Mori T, Ando T, Tamura K, Kamiya M, Sugita Y. Genesis 1.1: a hybrid-parallel molecular dynamics simulator with enhanced sampling algorithms on multiple computational platforms. J Comput Chem. 2017;38(25):2193–2206. doi: 10.1002/jcc.24874. [DOI] [PubMed] [Google Scholar]
- Kührová P, De Simone A, Otyepka M, Best RB. Force-field dependence of chignolin folding and misfolding: comparison with experiment and redesign. Biophys J. 2012;102(8):1897–1906. doi: 10.1016/j.bpj.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. The weighted histogram analysis method for free-energy calculations on biomolecules. i. the method. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
- La Penna G, Mori Y, Kitahara R, Akasaka K, Okamoto Y. Modeling 15n nmr chemical shift changes in protein backbone with pressure. J Chem Phys. 2016;145(8):085104. doi: 10.1063/1.4961507. [DOI] [PubMed] [Google Scholar]
- Lee J. New monte carlo algorithm: Entropic sampling. Phys Rev Lett. 1993;71:211–214. doi: 10.1103/PhysRevLett.71.211. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- Lyubartsev AP, Martsinovski AA, Shevkunov SV, Vorontsov-Velyaminov PN. New approach to monte carlo calculation of the free energy: Method of expanded ensembles. J Chem Phys. 1992;96(3):1776–1783. [Google Scholar]
- Manousiouthakis VI, Deem MW. Strict detailed balance is unnecessary in monte carlo simulation. J Chem Phys. 1999;110(6):2753–2756. [Google Scholar]
- Marinari E, Parisi G. Simulated tempering: a new monte carlo scheme. Europhys Lett. 1992;19(6):451. [Google Scholar]
- McDonald I. Npt-ensemble monte carlo calculations for binary liquid mixtures. Mol Phys. 1972;23(1):41–58. [Google Scholar]
- McQuarrie DA, Simon JD (1997) Physical chemistry:a molecular approach. University Science Books
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21(6):1087–1092. [Google Scholar]
- Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60(2):96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Mori Y, Okamoto Y. Generalized-ensemble algorithms for the isobaric–isothermal ensemble. J Phys Soc Jpn. 2010;79(7):074003. [Google Scholar]
- Mori Y, Okumura H. Pressure-induced helical structure of a peptide studied by simulated tempering molecular dynamics simulations. J Phys Chem Lett. 2013;4(12):2079–2083. doi: 10.1021/jz400769w. [DOI] [PubMed] [Google Scholar]
- Mori Y, Okumura H. Molecular dynamics of the structural changes of helical peptides induced by pressure. Proteins. 2014;82(11):2970–2981. doi: 10.1002/prot.24654. [DOI] [PubMed] [Google Scholar]
- Mori Y, Okumura H. Simulated tempering based on global balance or detailed balance conditions: S uwa–t odo, heat bath, and m etropolis algorithms. J Comput Chem. 2015;36(31):2344–2349. doi: 10.1002/jcc.24213. [DOI] [PubMed] [Google Scholar]
- Mori Y, Okamoto Y. Conformational changes of ubiquitin under high pressure conditions: a pressure simulated tempering molecular dynamics study. J Comput Chem. 2017;38(15):1167–1173. doi: 10.1002/jcc.24767. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Nakamura H, Kidera A. Multicanonical ensemble generated by molecular dynamics simulation for enhanced conformational sampling of peptides. J Phys Chem B. 1997;101(5):817–824. [Google Scholar]
- Nguyen PH, Okamoto Y, Derreumaux P. Communication: Simulated tempering with fast on-the-fly weight determination. J Chem Phys. 2013;138(6):061102. doi: 10.1063/1.4792046. [DOI] [PubMed] [Google Scholar]
- Nisius L, Grzesiek S. Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network. Nat Chem. 2012;4(9):711. doi: 10.1038/nchem.1396. [DOI] [PubMed] [Google Scholar]
- Okabe T, Kawata M, Okamoto Y, Mikami M. Replica-exchange monte carlo method for the isobaric–isothermal ensemble. Chem Phys Lett. 2001;335(5-6):435–439. [Google Scholar]
- Okamoto Y, Hansmann UH. Thermodynamics of helix-coil transitions studied by multicanonical algorithms. J Phys Chem. 1995;99(28):11276–11287. [Google Scholar]
- Okumura Hisashi, Okamoto Yuko. Molecular dynamics simulations in the multibaric–multithermal ensemble. Chemical Physics Letters. 2004;391(4-6):248–253. [Google Scholar]
- Okumura H, Okamoto Y (2004b) Monte carlo simulations in generalized isobaric-isothermal ensembles. Phys Rev E 70(2):026702 [DOI] [PubMed]
- Okumura Hisashi, Okamoto Yuko. Monte Carlo simulations in multibaric–multithermal ensemble. Chemical Physics Letters. 2004;383(3-4):391–396. [Google Scholar]
- Okumura H, Okamoto Y. Multibaric–multithermal ensemble molecular dynamics simulations. J Comput Chem. 2006;27(3):379–395. doi: 10.1002/jcc.20351. [DOI] [PubMed] [Google Scholar]
- Okumura H, Okamoto Y. Temperature and pressure dependence of alanine dipeptide studied by multibaric-multithermal molecular dynamics simulations. J Phys Chem B. 2008;112(38):12038–12049. doi: 10.1021/jp712109q. [DOI] [PubMed] [Google Scholar]
- Okumura H. Temperature and pressure denaturation of chignolin: Folding and unfolding simulation by multibaric-multithermal molecular dynamics method. Proteins. 2012;80(10):2397–2416. doi: 10.1002/prot.24125. [DOI] [PubMed] [Google Scholar]
- Privalov P. Thermodynamic problems of protein structure. Annu Rev Biophys Biophys Chem. 1989;18(1):47–69. doi: 10.1146/annurev.bb.18.060189.000403. [DOI] [PubMed] [Google Scholar]
- Satoh D, Shimizu K, Nakamura S, Terada T. Folding free-energy landscape of a 10-residue mini-protein, chignolin. FEBS Lett. 2006;580(14):3422–3426. doi: 10.1016/j.febslet.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Scholtz JM, Baldwin RL. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct. 1992;21(1):95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330(6002):341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- Shirts MR, Chodera JD. Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys. 2008;129(12):124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga A, Narumi T, Futatsugi N, Yanai R, Ohno Y, Okimoto N, Taiji M. Folding dynamics of 10-residue β-hairpin peptide chignolin. Chem Asian J. 2007;2(5):591–598. doi: 10.1002/asia.200600385. [DOI] [PubMed] [Google Scholar]
- Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314(1-2):141–151. [Google Scholar]
- Suwa H, Todo S. Markov chain monte carlo method without detailed balance. Phys Rev Lett. 2010;105(12):120603. doi: 10.1103/PhysRevLett.105.120603. [DOI] [PubMed] [Google Scholar]
- Takekiyo T, Shimizu A, Kato M, Taniguchi Y. Pressure-tuning ft-ir spectroscopic study on the helix–coil transition of ala-rich oligopeptide in aqueous solution. Biochim Biophys Acta. Proteins Proteomics. 2005;1750(1):1–4. doi: 10.1016/j.bbapap.2005.02.014. [DOI] [PubMed] [Google Scholar]
- van der Spoel D, Seibert MM. Protein folding kinetics and thermodynamics from atomistic simulations. Phys Rev Lett. 2006;96(23):238102. doi: 10.1103/PhysRevLett.96.238102. [DOI] [PubMed] [Google Scholar]
- Wang F, Landau DP (2001a) Determining the density of states for classical statistical models: a random walk algorithm to produce a flat histogram. Phys Rev E 64:056101 [DOI] [PubMed]
- Wang Fugao, Landau D. P. Efficient, Multiple-Range Random Walk Algorithm to Calculate the Density of States. Physical Review Letters. 2001;86(10):2050–2053. doi: 10.1103/PhysRevLett.86.2050. [DOI] [PubMed] [Google Scholar]
- Wu H, Paul F, Wehmeyer C, Noé F. Multiensemble markov models of molecular thermodynamics and kinetics. Proc Natl Acad Sci USA. 2016;113(23):E3221–E3230. doi: 10.1073/pnas.1525092113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M, Okumura H. Development of isothermal-isobaric replica-permutation method for molecular dynamics and monte carlo simulations and its application to reveal temperature and pressure dependence of folded, misfolded, and unfolded states of chignolin. J Chem Phys. 2017;147(18):184107. doi: 10.1063/1.4996431. [DOI] [PubMed] [Google Scholar]
- Yamauchi M, Okumura H (2019) Replica sub-permutation method for molecular dynamics and Monte Carlo simulations. J Comput Chem submitted [DOI] [PubMed]
- Yang C, Jang S, Pak Y. A fully atomistic computer simulation study of cold denaturation of a β-hairpin. Nat Commun. 2014;5:5773. doi: 10.1038/ncomms6773. [DOI] [PubMed] [Google Scholar]