Abstract
Peroxisomes play vital roles in plant growth, development, and environmental stress response. During plant development and in response to environmental stresses, the number and morphology of peroxisomes are dynamically regulated to maintain peroxisome homeostasis in cells. To execute their various functions in the cell, peroxisomes associate and communicate with other organelles. Under stress conditions, reactive oxygen species (ROS) produced in peroxisomes and other organelles activate signal transduction pathways, in a process known as retrograde signaling, to synergistically regulate defense systems. In this review, we focus on the recent advances in the plant peroxisome field to provide an overview of peroxisome biogenesis, degradation, crosstalk with other organelles, and their role in response to environmental stresses.
Keywords: Arabidopsis, pexophagy, peroxisome biogenesis, retrograde signaling, stress
Introduction
Peroxisomes, which are found in all eukaryotic cells, are highly metabolic organelles surrounded by a single membrane (De Duve and Baudhuin, 1966; Hu et al., 2012). In plant cells, peroxisomes play vital roles in metabolism since they house many processes including fatty acid β-oxidation, glyoxylate cycles in seedlings, photorespiration in leaves, urate sulfite and polyamine metabolism (Hu et al., 2012), and biosynthesis of phytohormones such as indole-3-acetic acid, jasmonic acid, and salicylic acid (Reumann et al., 2004; Koo et al., 2006; Strader and Bartel, 2011; Kao et al., 2018). In addition, antioxidant enzymes and enzymes which generate reactive oxygen species (ROS) or reactive nitrogen species (RNS) are essential components in peroxisomes for maintaining redox homeostasis (Corpas et al., 2017). Metabolite exchange between peroxisomes and other organelles is important for peroxisomal function and for morphological changes which occur under specific physiological and environmental conditions (Linka and Theodoulou, 2013).
Peroxisomes are usually oxidized and damaged by metabolic byproducts or environmental stresses. Excess and/or damaged peroxisomes are removed by autophagy to ensure the normal operation of the cell. According to the environmental conditions, the number and quality of peroxisomes in cells can change rapidly and is precisely regulated to maintain peroxisomes homeostasis (Farmer et al., 2013; Yoshimoto et al., 2014; Young and Bartel, 2016). Herein, we review recent advancements in our understanding of peroxisome biogenesis including the mechanisms of import of peroxisomal membrane proteins (PMPs) and peroxisomal matrix proteins, the connection of peroxisomes with other organelles, and the dynamics of how redox status in plants regulates peroxisome homeostasis and peroxisomal-derived retrograde signaling (Figure 1).
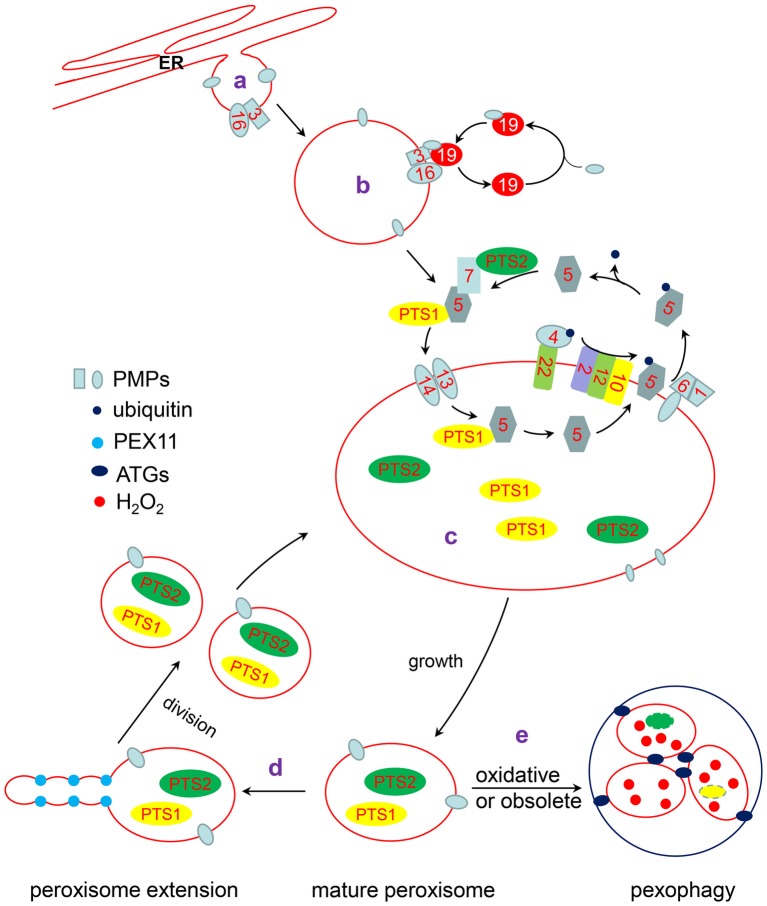
Figure 1.
The life cycle of peroxisome. a: Some PMPs are targeted to the ER and localized in special ER subdomains, where new preperoxisomes subsequently bud off. b: The remainder of PMPs are directly inserted into the peroxisomal membrane through a PEX3/PEX19-dependent pathway or another unknown pathway. c: PTS1 and PTS2 proteins interact with their receptor, PEX5 or PEX7, respectively. The receptor/cargo complex is then transported into the peroxisome lumen through the docking complex, composed of PEX13 and PEX14 in Arabidopsis. After unloading its cargo protein, PEX5 is mono-ubiquitinated by the PEX4/PEX22 (E2 ubiquitin conjugases) and PEX2/PEX12/PEX10 (E3 ligases) complexes. Subsequently, Pex5 is retrotranslocated to the cytosol by the PEX1/PEX6 complex for recycling. d: Mature peroxisomes elongate and divide into two new peroxisomes. e: Oxidative or obsoleted peroxisomes can be degraded by pexophagy under the action of ATGs.
Peroxisome Biogenesis
Origin of Peroxisomes
It has been reported that peroxin proteins encoded by PEX genes are required for peroxisome biogenesis and conserved in yeast, mammals, and plants (Ma et al., 2011; Hu et al., 2012). However, how peroxisomes form has been debated for over three decades. Early models of peroxisome biogenesis suggested that peroxisomes are autonomous organelles that divide by growth and division of pre-existing peroxisomes (Lazarow and Fujiki, 1985; Agrawal and Subramani, 2016). According to this model, a pre-existing peroxisome grows and divides asymmetrically into two new peroxisomes when a certain size threshold is reached (Akşit and van der Klei, 2018). This view was supported by the discovery of peroxisome protein import machinery and the ability of peroxisomes to divide (Lazarow and Fujiki, 1985; Kim and Hettema, 2015). Studies in plants also support the growth and division model (Hu et al., 2012). In Arabidopsis, proteins participating in peroxisomal protein targeting and peroxisome division have been identified and characterized (Woodward and Bartel, 2005; Zhang and Hu, 2008; Aung and Hu, 2011; Woodward et al., 2014).
However, the growth and division model does not account for the source of membrane lipids required for the growth of dividing peroxisomes. In Saccharomyces cerevisiae, it was shown that lipids are directly transferred from the ER to peroxisomes by a vesicle-independent pathway (Raychaudhuri and Prinz, 2008). In addition, in S. cerevisiae pex3 and pex19 cells, which are devoid of detectable peroxisomes, new peroxisomes could form when a wild-type copy of PEX3 or PEX19 was reintroduced into the corresponding mutants, signifying de novo peroxisome formation (Hohfeld et al., 1991). Research on de novo peroxisome formation in plants is scarce due to the complicacy of plant systems. However, we have gained insights from experiments in yeast. It was shown that S. cerevisiae Pex3 is delivered to the peroxisomes through the ER (Hoepfner et al., 2005). Pex3, along with Pex19, is involved in the direct insertion of some PMPs into the ER and the pre-peroxisomal vesicle membrane (Hu et al., 2012; van der Zand et al., 2012). Subsequently, theses pre-peroxisomal vesicles fuse to form a new peroxisome where these PMPs are naturally transferred into the peroxisomal membrane (van der Zand et al., 2010, 2012).
It now seems that neither de novo synthesis from the ER nor the growth and division model can fully explain all aspects of peroxisome biogenesis (Smith and Aitchison, 2013; Hettema et al., 2014). Although it is still heavily debated (Knoops et al., 2014; Wroblewska et al., 2017), it has been proposed that the generation of peroxisomes includes de novo biogenesis, during which pre-peroxisomal vesicles fuse to form a new peroxisome or fuse with preexisting peroxisomes. This is followed by growth to form mature peroxisomes, which can divide into new peroxisomes (Hu et al., 2012).
The Transport of Peroxisomal Membrane Proteins
Although the mechanism is not fully understood, there are two possible pathways by which PMPs are imported into peroxisomes (Theodoulou et al., 2013; Erdmann, 2016; Yuan et al., 2016): (1) PMPs are synthesized in the cytosol and imported directly into existing peroxisomes or (2) PMPs are integrated into the ER and subsequently targeted to peroxisomes.
PMPs directly imported into peroxisomes from cytosol need to be recognized by Pex19, a cytosolic PMP receptor that interacts with and stabilizes PMPs (Hettema et al., 2000; Rottensteiner et al., 2004). Cargo-bound Pex19 interacts with peroxisomal membrane-associated Pex3 for the subsequent docking of PMPs. Both Pex3 and Pex19 are necessary for PMP import and peroxisome biogenesis. There are two isoforms of both PEX3 and PEX19 in Arabidopsis, which are homologs of yeast Pex3 and Pex19, respectively (Hunt and Trelease, 2004). Arabidopsis deficient in PEX19 have enlarged peroxisomes (Nito et al., 2007). In mammals, PEX16 may act as a membrane receptor and, together with PEX19, mediates the direct insertion of PEX3 into peroxisome membranes (Matsuzaki and Fujiki, 2008). The transport of PEX3 to peroxisomes is reduced in mammalian cells with PEX16 knocked down (Matsuzaki and Fujiki, 2008; Aranovich et al., 2014; Hua and Kim, 2016). The null mutation of PEX16 results in non-detectable peroxisome remnants in mammalian cells (Honsho et al., 1998). In Arabidopsis, Shrunken SEed 1 (SSE1) mutants deficient in PEX16 were found to show a shrunken seeds phenotype as a result of the disordered formation of oil bodies during seed development (Lin et al., 1999, 2004). Arabidopsis pex16i mutants have enlarged peroxisomes, which are reduced in number and show functional defects (Nito et al., 2007). AtPEX16 is located on the membrane of both peroxisomes and the ER and may act as an integral membrane-bound receptor for PEX3 and other PMPs (Karnik and Trelease, 2005; Kim and Mullen, 2013). There are other PMPs, such as PMP22, which are directly inserted into the peroxisome membrane after being synthesized in the cytosol in Arabidopsis (Murphy et al., 2003). Arabidopsis PMP22 is a homolog of mouse PxMP2, which is a channel protein for small metabolites (Rokka et al., 2009). In addition, Arabidopsis PEX2 and PEX10 have been shown to insert into peroxisome membranes directly from the cytosol (Sparkes et al., 2005).
There is increasing evidence that some PMPs are transported to peroxisomes through the ER membrane. An important subgroup of these PMPS is known as the tail anchored (TA) proteins, which are anchored to the peroxisome membrane at the C-terminus and have a short luminal domain with the remaining N-terminus in the cytosol (Cross et al., 2016). In S. cerevisiae, Pex15, a TA protein containing a single transmembrane domain near its C-terminus, is targeted to the ER membrane by the guided entry of tail-anchored (Get) pathway (Okreglak and Walter, 2014). The C-terminal of yeast Pex15 contains a PEX19-binding site, which positions Pex15 on the ER membrane through the Get pathway, indicating that Pex15 is inserted into the peroxisome membrane via the ER (Halbach et al., 2006; Schuldiner et al., 2008). In plants, the initial evidence of ER-to-peroxisome trafficking emerged from in vitro experiments using cottonseed ascorbate peroxidase (APX), a TA protein targeted to the peroxisome membrane (Mullen et al., 1999). It was shown that the post-translated cottonseed APX inserts into ER microsomal membranes, but not other organelle membranes, while over-expressed cottonseed APX was located in both peroxisomes and the ER membrane (Mullen et al., 1999). Arabidopsis APX3 has also been detected in ER subdomains, indicating that APX3 is imported to the peroxisome via the ER-to-peroxisome pathway (Lisenbee et al., 2003).
Matrix Protein Entry Into the Peroxisome Lumen
Peroxisomal matrix proteins are synthesized in the cytosol and imported into the matrix of peroxisomes. The peroxisomal matrix proteins are distinguished by their peroxisomal targeting signals (PTSs), which are recognition sequences at the C-terminus (PTS1) and N-terminus (PTS2) of the respective cargo proteins. Typical PTS1 sequences consist of a highly conserved tripeptide [S/A/C]-[K/H/R]-L (Elgersma et al., 1996; Reumann et al., 2004, 2016). The consensus for a PTS2 sequence is R/K-L/V/I-X5-H/Q-L/A (Petriv et al., 2004).
In Arabidopsis, two peroxins, PEX5 and PEX7, are responsible for the recognition of PTS1 and PTS2 proteins, respectively. The disordered N-terminal region of PEX5 interacts with PEX7 and components of the docking subcomplex, such as PEX14, while the conserved C-terminal region contains a series of tetratricopeptide repeats (TPR) that bind to the PTS1 of cargo proteins (Zolman et al., 2000; Nito et al., 2002). The WD domain of PEX7 is responsible for its recognition of PTS2 proteins (Woodward and Bartel, 2005). Conserved across eukaryotes, the PTS1 import pathway is the predominant mechanism for cargo proteins to enter plant peroxisomes. However, unlike in yeast, the import of PTS1 and PTS2 proteins are interconnected in plants. In Arabidopsis, the binding of PEX7 to PTS2 proteins requires PEX5 as a co-receptor (Woodward and Bartel, 2005), which is necessary for the proper interaction of PEX7 with PTS2-cargo proteins. Mutation of PEX5 (S318L) causes PEX5 to inefficiently bind with PEX7 and disrupts the import of PTS2 proteins, indicating that the PEX5-PEX7 interaction is necessary for PTS2 protein import (Woodward and Bartel, 2005). Additionally, the import of PTS2 proteins is restored by expressing the N-terminus of PEX5 in the PEX5 (S318L) mutant, indicating that the N-terminal domain of PEX5 is necessary for PTS2 import (Khan and Zolman, 2010). Interestingly, a mutation in PEX7 (T124I) results in the reduction of the protein levels of both PEX7 and PEX5 as well as reducing import of PTS1 and PTS2 cargoes, further suggesting that the PTS1 and PTS2 targeting pathways are interconnected in plants (Ramon and Bartel, 2010).
The delivery of peroxisomal matrix proteins requires the action of a large peroxisomal membrane complex known as the importomer, which is composed of the docking subcomplex (Pex13, Pex14, and Pex17) and the RING (Really Interesting New Gene) subcomplex (Pex2, Pex10, and Pex12) bridged by Pex8 in S. cerevisiae and Pex3 in P. pastoris (Agne et al., 2003; Rayapuram and Subramani, 2006). However, it is not well established how the importomer is organized in plants. The cargo-receptor complex associates with the peroxisomal membrane through the docking subcomplex of the peroxisomal importomer. In Arabidopsis, PEX5 can interact with PEX14 but not with PEX13, and the WxxxF/Y (Trp-X-X-X-Phe/Tyr) pentapeptide repeats in PEX5 are crucial for this interaction (Nito et al., 2002). PEX13, but not PEX14, is responsible for the interaction with PEX7 in Arabidopsis (Mano et al., 2006). PEX13 interacts with PEX14 and mutation of either protein in Arabidopsis results in reduced PTS1 and PTS2 import (Mano et al., 2006; Monroe-Augustus et al., 2011; Woodward et al., 2014). The Arabidopsis pex13 null allele is lethal, but pex14 is not (Boisson-Dernier et al., 2008; Monroe-Augustus et al., 2011). In a weak pex13 mutant, pex13-4, the amount of PEX5 associated with the peroxisomal membrane is decreased, but overexpression of PEX5 can rescue the PTS2-processing defects of pex13-4 (Woodward et al., 2014), indicating, to some extent, that the role of PEX13 can be bypassed. The Arabidopsis pex14 mutant shows severe growth defects, but it is still able to complete the life cycle and produce fertile offspring, indicating that PEX14 facilitates, but is not essential for peroxisomal matrix protein import in plants (Monroe-Augustus et al., 2011; Burkhart et al., 2013).
Based on in vitro reconstitution and biochemical assays, receptor/cargo complexes are imported into the peroxisome lumen via the peroxisomal translocon mainly composed of the receptors and Pex14 in yeasts and mammals (Ma et al., 2009; Meinecke et al., 2010; Romano et al., 2019). Upon arrival in the peroxisome lumen, PTS1 proteins are released from their receptor, Pex5, with the aid of Pex8 in Hansenula polymorph (Wang et al., 2003). While in Pichia pastoris, the receptor/cargo complex dissociates as a result of a conformational change in Pex5, regulated by redox conditions within the peroxisome lumen and its interaction with Pex8 (Ma et al., 2013). However, neither the constituents of the peroxisomal translocon nor the mechanisms of how cargo is released from receptors have been characterized in plants.
Dynamic Regulation of the Peroxisomal Importomer
After cargo proteins are released within the peroxisome lumen, the receptors are recycled to the cytosol for another round of import. This process is governed by the monoubiquitination of PEX5 on a cysteine residue, which requires an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ligase (Hu et al., 2012; Kao et al., 2018). In yeasts, Pex4, as an E2 ubiquitin-conjugating enzyme, transfers ubiquitin to Pex5 at a conserved cysteine residue (monoubiquitination for recycling) or lysine residue(s) (polyubiquitination for degradation) under the catalysis of Pex2/10/12, three E3 ligases that comprise the RING subcomplex (Platta et al., 2007, 2009).
In Arabidopsis, the null mutants of pex2, pex10, and pex12 display embryo lethality at the heart stage (Fan et al., 2005; Prestele et al., 2010). Interestingly, the weak mutants of pex2-1 and pex10-2 exhibited defects only in the targeting of PTS1 but not PTS2 proteins (Burkhart et al., 2014). Although it has been demonstrated that Arabidopsis PEX2, PEX10, and PEX12 contain E3 ubiquitin ligase activity in vitro, whether PEX5 is the direct substrate of these E3 ubiquitin ligases has not been directly shown (Kaur et al., 2013). PEX10 has been implicated in PEX5 retrotranslocation because overexpression of PEX5 aggravates the PTS2-processing defects and physiological phenotype of pex10-2 (Burkhart et al., 2014).
In Arabidopsis, the function of PEX4 is enhanced by PEX22, an integral PMP, whose cytosolic domain interacts with PEX4 (El Magraoui et al., 2014). The Arabidopsis PEX4-PEX22 complex rescues the growth defect of Pex4/Pex22-deficient yeast cells, indicating that the plant ubiquitin machinery for receptor recycling or degradation is functionally similar to that in the yeast system (Zolman et al., 2005). At normal temperature (22°C), the pex4 mutant shows elevated levels of membrane-associated PEX5 without ubiquitination, suggesting that ubiquitination is required for PEX5 recycling. But at 28°C, the overall PEX5 protein levels are reduced in the pex4 mutant, probably resulting from another unknown ubiquitination enzyme promoting the degradation of PEX5 (Kao and Bartel, 2015).
After being monoubiquitinated, PEX5 is exported to the cytosol for another round of import, which requires a membrane-anchored AAA (ATPases associated with various cellular activities) ATPase complex, consisting of PEX1 and PEX6. PEX1 and PEX6 form a heterohexamer with 3 U of each. This complex is anchored to the peroxisomal membrane through a TA protein, Pex26 in yeasts and aberrant peroxisome morphology 9 (APEM9) in Arabidopsis (Goto et al., 2011; Gardner et al., 2015). The PEX1-PEX6 complex directly interacts with the ubiquitin moiety of ubiquitinated PEX5 and subsequently releases ubiquitinated PEX5 to the cytosol (Cross et al., 2016; Pedrosa et al., 2018).
In Arabidopsis, the RNA interference (RNAi) lines of pex1i, pex6i, and apem9i result in defective PTS1 and PTS2 protein import (Nito et al., 2007; Goto et al., 2011). pex1-2, a weak pex1 mutant, displays severe defects in peroxisome function, while pex1-3, a more severe mutant, is embryo lethal, indicating that Arabidopsis PEX1 is essential for embryogenesis (Rinaldi et al., 2017). Both Arabidopsis pex6 and pex26 mutants display β-oxidation deficiency, impaired matrix protein import, and low levels of PEX5 (Gonzalez et al., 2017). Interestingly, the peroxisomal defects of pex1, pex4, pex6, and pex26 are aggravated by overexpression of PEX5 in Arabidopsis (Kao and Bartel, 2015; Gonzalez et al., 2017; Rinaldi et al., 2017), indicating that PEX5 functions normally only when it can be recycled efficiently. It has been reported recently that a pex1 missense mutation, pex1-1, can alleviate the physiological defects of pex6-1 without restoring PEX5 levels, implying additional functions of the PEX1-PEX6 complex beyond PEX5 recycling (Gonzalez et al., 2018). The weak mutant pex13-1 ameliorates the growth defects of pex4-1 or pex6-1 surprisingly, although matrix protein import in pex4-1 pex13-1 and pex6-1 pex13-1 remains defective (Ratzel et al., 2011), indicating that normal peroxisomal function is dependent on the balance between import and export of peroxisomal receptors.
In Arabidopsis, SP1 (suppressor of plastid protein import locus 1), an E3 ubiquitin ligase located in the peroxisome membrane, is involved in regulating the translocation of PEX5 by participating in the degradation of PEX13 and promoting destabilization of PEX14 and the RING peroxins PEX2, PEX10, and PEX12 (Pan et al., 2016). However, SPL1 (SP1-like 1), a homolog of SP1, stabilizes PEX13 by inhibiting the function of SP1. In contrast to sp1, which suppresses the phenotypes of pex14 and pex13 mutants, spl1 enhances the phenotypes of these two mutants (Pan et al., 2018).
In summary, from docking on the peroxisome membrane to being exported to the cytosol, the translocation of Arabidopsis PEX5 is dynamically controlled by elaborate mechanisms requiring cooperation among the components of the docking subcomplex, the RING subcomplex, and the receptor recycling machinery. The import of PMPs and matrix proteins are crucial to peroxisome biogenesis. The mechanisms of PMP and matrix protein import are highly conserved but also display specificity among different organisms. The diversity and complexity of the transport system indicates that peroxisomes are highly dynamic and may be differentially regulated during plant development and other various environmental conditions.
Crosstalk Between Peroxisomes and Other Organelles
Plant peroxisomes are not isolated organelles in cells. They usually communicate and collaborate with other organelles such as lipid bodies, chloroplasts, and mitochondria to execute their multi-faceted functions in biological processes, such as lipid mobilization, photorespiration, and redox metabolism (Shai et al., 2016).
In plants, lipid bodies accumulate in seeds and cotyledons that generally store lipids as triacylglycerols (TAGs). During seed germination, fatty acids are transported from lipid bodies to glyoxysomes and subsequently metabolized through fatty acid β-oxidation and the glyoxylate cycle to supply energy for seedling establishment (Graham, 2008; Hu et al., 2012). The transport of fatty acids may be mediated by the direct contact between glyoxysomes and oil body. In Arabidopsis, the ped1 mutant, which has a defect in a thiolase gene involved in fatty acid β-oxidation, grows slowly in the dark with small cotyledons and displays abnormal glyoxysomes with tubular structures which are derived from the invagination of glyoxysomes membrane (Hayashi et al., 2001). Fatty acid-containing vesicles in the tubular compartments are formed at the contact sites of lipid bodies and glyoxysomes, suggesting that there is a direct mechanism of lipid transport from the lipid bodies to glyoxysomes (Hayashi et al., 2001).
The enzymes involved in TAG degradation can also be transferred between the peroxisome and oil body. During seed germination, TAGs are hydrolyzed by lipases to free fatty acids and glycerol, which results in the degradation of lipid bodies. During oil mobilization, Arabidopsis SDP1, a major TAG lipase, which is first localized to the peroxisome membrane, migrates to the surface of lipid bodies for oil mobilization via extensive tubulations of the peroxisome known as peroxules (Thazar-Poulot et al., 2015). The movement of SDP1 from the peroxisome to lipid bodies requires a retromer complex, which is a protein complex mediating the recycling of receptors and the retrograde transport of cargo proteins from endosomes to the trans-Golgi network (Bonifacino and Hurley, 2008). In core retromer mutants, oil body biogenesis and oil breakdown are defective, suggesting that the contact between glyoxysomes and oil body is important for oil mobilization (Thazar-Poulot et al., 2015).
In addition to oil bodies, peroxisomes are closely related to other organelles. For example, EM images showed that plant peroxisomes are localized close to chloroplasts and mitochondria (Kaur et al., 2009). The close contact between peroxisome and these two organelles plays an important role in the metabolism of plant cells. In prolonged dark conditions, free fatty acids are released from the chloroplast and subsequently metabolized via peroxisomal β-oxidation. During this process, PXA1 mediates the import of fatty acids into peroxisomes and KAT2 (PED1) is involved in β-oxidation (Germain et al., 2001; Zolman et al., 2001; Kunz et al., 2009; Hu et al., 2012).
Plant photorespiration requires the coordination of four compartments: the cytosol, chloroplasts, mitochondria, and peroxisomes. Conjunction of peroxisomes with chloroplasts and mitochondria may play an important role in photorespiration by allowing for metabolite flow between these organelles (Hu et al., 2012). Recent evidence obtained from light and transmission electron microscopy show that peroxisomes are appressed to the chloroplast envelope in Arabidopsis. The RING finger domain in PEX10 is crucial for the contact between these two organelles. Expressing endogenous levels of PEX10 with a dysfunctional RING finger domain results in more numerous, multilobed, clustered peroxisomes, which are not associated with the chloroplast envelope as they are in wild type (Schumann et al., 2007). Moreover, peroxisomes can alter their shape from spherical to elliptical to increase the interaction area with the chloroplast upon exposure of the plant to light (Oikawa et al., 2015).
Peroxisomes, chloroplast, and mitochondria not only cooperate in metabolic processes but also in division (Zhang and Hu, 2010; Aung and Hu, 2011). Peroxisome proliferation first requires the elongation of a preexisting peroxisome, followed by constriction and fission (Schrader, 2006). In Arabidopsis, the PEX11 family, consisting of five partially functionally redundant isoforms Pex11a to Pex11e (Orth et al., 2007; Desai and Hu, 2008), is involved in peroxisome elongation (Lingard and Trelease, 2006; Koch et al., 2010). Subsequently, peroxisome fission is dependent on the coordination of integral membrane-anchored proteins FIS1 (FISSION1) and dynamin-related protein (DRP). The two FIS1 homologs, FIS1A and FIS1B, are targeted to both peroxisomes and mitochondria to facilitate the division of both organelles (Scott et al., 2006; Zhang and Hu, 2008). FIS1 interacts with DRP3 to associate with the both peroxisome and mitochondria membranes. DRP3A and DRP3B, which are the constituents of DRP3, are partially redundant in mediating the division of peroxisomes and mitochondria (Mano et al., 2004; Zhang and Hu, 2008; Fujimoto et al., 2009). DRP5B is involved in the division of peroxisomes, chloroplasts, and mitochondria, which is independent of DRP3 (Aung and Hu, 2012). Another factor, a coiled-coil protein peroxisomal and mitochondrial division factor 1 (PMD1), whose function is independent of FIS1 and DRP3, is also tethered to the membranes of peroxisomes and mitochondria to regulate the division of the two organelles (Aung and Hu, 2011).
Peroxisomes Function as Sensors of Environmental Redox Changes
Peroxisomes are important for modulating redox balance between ROS production and elimination. In plants, stress conditions promote a ROS-dependent signaling pathway which triggers rapid and adaptive responses (Sandalio and Romero-Puertas, 2015). The excess oxidative stress induced in these conditions can affect the biogenesis and degradation of peroxisomes (Shibata et al. 2013; Wang et al., 2015).
Peroxisome Status Is Affected by Environmental Stresses
Because of their immobility, plants are constantly suffering from stresses caused by environmental changes. Abiotic and biotic stresses, such as drought, high and low temperatures, salinity, nutrient deficiency, and pathogens are the main factors leading to crop yield loss (Zhang et al., 2010, 2018; Ren et al., 2013; Shen et al., 2014). In general, environmental stresses induce cellular oxidative stress and as a consequence induce the ROS scavenging system (Qi et al., 2010; Li et al., 2012; Liu et al., 2017). Peroxisomes are crucial in this ROS scavenging system for controlling H2O2 levels (Mhamdi et al., 2012).
Peroxisomes are one of the main sites of H2O2 generation in plant cells. Plants peroxisomes also contain several enzymes which eliminate excess H2O2, including catalases, ascorbate peroxidases (APX), and various types of peroxiredoxins (Schrader and Fahimi, 2006; Mhamdi et al., 2010). Among these enzymes, catalases are the main eliminator of peroxisomal H2O2. In Arabidopsis, there are three catalase genes (CAT1, CAT2, and CAT3) which are involved in many plant physiological processes including abiotic stresses response (Su et al., 2018).
In regard to abiotic stresses, Arabidopsis salt overly sensitive 2 (SOS2), which is required for salt tolerance, can interact with CAT2 and CAT3 at an unknown subcellular location in response to salt stress and H2O2 metabolism (Verslues et al., 2007). A zinc finger protein lesion simulating disease 1 (LSD1) can interact with all three catalases in the cytosol to maintain their activities in regulating hypersensitive cell death (Li et al., 2013). Moreover, the chaperone no catalase activity 1 (NCA1), which interacts with all three catalases in the cytosol, is required for catalase activity during multiple stress responses, such as salt, cold, and alkaline stress, as well as for autophagy-dependent cell death (Hackenberg et al., 2013; Li et al., 2015). In addition, a peroxisome-localized small heat shock protein Hsp17.6CII interacts with CAT2 in the peroxisome and enhances CAT2 activity to protect from abiotic stresses (Li et al., 2017).
For biotic stresses, it is reported that Arabidopsis PEN2, a glycosyl hydrolase localized to both peroxisomes and mitochondria, acts as a component of an inducible preinvasion resistance system to restrict pathogen entry via the pathogen-induced enzymatic activation of indole glucosinolates (Lipka et al., 2005; Fuchs et al., 2016). Glucosinolates are secondary metabolites and are often stored in the vacuole as inactive glycoside precursors. These precursors can be activated by PEN2 when plants undergo insect feeding or necrotrophic pathogen attack (Bednarek et al., 2009). In addition, viruses can produce viral suppressors of RNA silencing (VSR) to protect themselves against RNAi, an important plant defense mechanism mediated by small interfering RNA. The peanut clump virus-encoded P15, which is one such VSR, transports the antiviral siRNA from the cytosol to the peroxisomal matrix to potentiate viral infection (Incarbone et al., 2017).
Different stresses often affect the expression of peroxisome biogenesis genes. In Arabidopsis, PEX1, PEX5, PEX10, and PEX14 are significantly induced by exogenous H2O2. In addition, pathogen attack and wounding, which trigger an oxidative burst, rapidly induce PEX1 expression, indicating that peroxisome biogenesis is directly responsive to stresses (Lopez-Huertas et al., 2000).
Various environmental conditions, which are often accompanied by ROS accumulation, can also trigger peroxisome proliferation to respond to the changes. During seedling photomorphogenesis, light induces peroxisome proliferation by upregulating PEX11b through the HYH (HY5 HOMOLOG)-mediated light signaling cascade. In this process, the activity of PEX11b depends on the far-red light receptor phytochrome A and the bZIP transcription factor HYH (Desai and Hu, 2008). In addition to directly targeting the PEX11b promoter to induce its expression, HYH can also indirectly repress the activity of FHA3 (forkhead-associated domain protein 3), which negatively regulates PEX11b expression and peroxisome division (Desai and Hu, 2008; Desai et al., 2017). In Pisum sativum L. plants, peroxisome proliferation can be induced by clofibrate, which increases activated oxygen species and lipid peroxidation of peroxisomal membranes (Palma et al., 1991). In addition, peroxisome proliferation can also be induced under clofibrate treatment in Arabidopsis (Castillo et al., 2008). Cadmium-imposed oxidative stress can also induce an increase in the number of peroxisomes and induces formation of peroxisome peroxules regulated by PEX11a (Rodríguez-Serrano et al., 2009, 2016). Peroxules can also be induced under high light stress accompanied by accumulation of H2O2 (Jaipargas et al., 2016). In addition, the number of peroxisomes in Arabidopsis is increased when exposed to high salt (Fahy et al., 2017). Under salt stress, mitogen-activated protein kinase 17 (MPK17) negatively regulates peroxisome proliferation by inhibiting PMD1 (Frick and Strader, 2018). Together, in addition to the role of ROS in signaling, these evidences suggest that ROS induced by primary environmental stress are also involved in peroxisome proliferation by regulating the expression of PEX genes.
Redox State Regulates Pexophagy and Peroxisome Remodeling
The number and activity of peroxisomes are tightly controlled and maintained in a balanced state in response to different environmental and/or physiological conditions. Peroxisome homeostasis is achieved through the coordination of peroxisome biogenesis and turnover. Pexophagy, a selective autophagy pathway to remove peroxisomes, is critical for peroxisomal quality control and requires a suite of autophagy-related (ATG) genes. These ATGs are highly conserved in yeasts, animal, and plants (Li and Vierstra, 2012).
Under oxidative stress, pexophagy is enhanced to remove oxidative, damaged peroxisomes. In Arabidopsis, the peroxisomes in atg2, atg7, and atg18 mutants aggregate together with accumulation of inactive catalases in the clustered peroxisomes, indicating these peroxisomes are ROS-damaged organelles to be degraded (Shibata et al., 2013). Aggregated peroxisomes are also observed in the cat2 mutant (Shibata et al., 2013). ATG8, the autophagosome marker, colocalizes with the aggregated peroxisomes, indicating that autophagy is involved in the selective degradation of damaged peroxisomes (Shibata et al., 2013). Pexophagy rates differ depending on the plant tissue. Increased peroxisome abundance in Arabidopsis atg2, atg5, atg7, and atg9 mutants are detected in hypocotyls and leaves but not roots (Yoshimoto et al., 2014). This phenomenon may be because the above ground part of plants is the main place for H2O2 production, resulting in more oxidatively damaged peroxisomes to be degraded by pexophagy (Yoshimoto et al., 2014).
Pexophagy also plays a crucial role in peroxisome remodeling. In plant seedlings, the peroxisomes, named glyoxysomes, contain enzymes such as thiolase involved in β-oxidation and two main glyoxylate cycle enzymes, isocitrate lyase (ICL) and malate synthase (MLS), which catalyze the conversion of fatty acids into carbohydrates (Pracharoenwattana and Smith, 2008). As the seedlings grow and photosynthesis is established, the enzymes involved in the glyoxylate cycle become unnecessary and are degraded. At the same time, redundant or obsolete glyoxysomes are degraded by pexophagy while some glyoxysomes are transformed into leaf peroxisomes harboring photorespiration enzymes (Goto-Yamada et al. 2014). In Arabidopsis atg5 and atg7 mutants, the degradation of ICL and MLS are delayed during growth in hypocotyls but not in whole seedlings, indicating pexophagy may promote degradation of peroxisomes and contribute to the degradation of peroxisome matrix proteins (Kim et al., 2013).
The degradation of ICL and MLS by pexophagy was nicely elucidated by screening for suppressors of the lon2 mutant. Lack of LON2 causes inefficient β-oxidation of indole-3-butyric acid (IBA) into indole-3-acetic acid (IAA), defective in PTS2 proteins targeting, such as thiolase and peroxisomal malate dehydrogenase (PMDH), reduced matrix protein import, and clustered peroxisomes, indicating that LON2 plays crucial roles in sustaining normal peroxisomal function (Lingard and Bartel, 2009; Farmer et al., 2013; Shibata et al., 2013; Young and Bartel, 2016). In the lon2 mutant, ICL and MLS are unstable. Mutation of autophagy genes such as ATG2, ATG3, and ATG7 in lon2 mutants result in stabilization of ICL and MLS, suggesting that pexophagy participates in degradation of peroxisomal proteins during peroxisome remodeling and is induced when LON2 is nonfunctional (Farmer et al., 2013; Goto-Yamada et al., 2014).
Damaged or obsolete ICL and MLS proteins can also be degraded through peroxisome-associated protein degradation (PexAD) pathway (Lingard et al., 2009). In the peroxisome matrix protein import mutants pex5 and pex14, ICL and MLS are stabilized, suggesting ICL and MLS must be localized in peroxisomes to be degraded (Lingard et al., 2009; Burkhart et al., 2013). Moreover, the turnover of ICL and MLS is affected by peroxisomal metabolism. In pxa1 and ped1 mutants, ICL and MLS are stabilized, potentially as a result of decreased H2O2 levels due to blocked fatty acid entry into peroxisomes and reduced β-oxidation (Kunz et al., 2009; Lingard et al. 2009; Burkhart et al., 2013). Interestingly, the degradation of ICL and MLS is enhanced in the cat2 mutant, where peroxisomes are more oxidative due to the lack of a H2O2-scavenging catalase (Lingard et al., 2009). These results indicate that redox status modulates the degradation rate of ICL and MLS. Furthermore, ICL and MLS are more stabilized in pex4, pex22, and pex6 mutants, implying that the ubiquitin-proteasome pathway is required for the degradation of ICL and MLS and implicated in peroxisome remodeling (Lingard et al., 2009; Burkhart et al., 2013).
In addition to the degradation of ICL and MLS, the conversion of glyoxysomes to leaf peroxisomes also requires the uptake of photorespiratory enzymes such as hydroxypyruvate reductases (HPR) (Lingard et al., 2009). However, the level of HPR in lon2, atg7, and lon2 atg7 mutants is similar to that in wild-type Arabidopsis, suggesting that HPR synthesis is independent of ICL and MLS abundance and the import of HPR occurs after the formation of leaf peroxisomes (Farmer et al., 2013).
The receptors for pexophagy are Atg30 (Pichia pastoris)/Atg36 (S. cerevisiae) in yeasts and NBR1 in mammals (Farre et al., 2008; Motley et al., 2012). A plant homolog of NBR1 has been identified, but there is no direct evidence to show it is a pexophagy receptor (Svenning et al., 2011; Young et al., 2019).
Redox State Regulates Retrograde Signaling From Peroxisomes to the Nucleus
Intracellular signaling from organelles to the nucleus, termed retrograde signaling, can modulate the expression of nuclear genes, which play important roles in organelle assembly, metabolism, and response to environmental changes (Sewelam et al., 2014). In plants, ROS are produced in chloroplasts, mitochondria, cytosol, peroxisomes, and the plasma membrane (Das and Roychoudhury, 2014; de Souza et al., 2017). The cellular movement and diffusion of H2O2 is proposed to be facilitated by aquaporins (Bienert and Chaumont, 2014). It has been hypothesized that H2O2 signaling could result in two kinds of responses: (1) the signaling induced by H2O2 is integrated regardless of the origin of H2O2 or (2) H2O2-induced signaling is dependent on the production site of H2O2 (Sewelam et al., 2014). Given that the production of ROS occurs in multiple organelles, it is necessary to discuss the different effects of ROS signals derived from different organelles (Figure 2).
Figure 2.
H2O2-mediated retrograde signaling. Chloroplasts, mitochondria, and peroxisomes are linked together to facilitate the shuttling of intermediate metabolites between them. When plants are subjected to environmental stresses, a large amount of H2O2 is produced in these three organelles. H2O2 can be used as a signal molecule to regulate the expression of nuclear response genes through the MAPK cascade or potentially through other unknown pathways. H2O2 in chloroplasts also promotes the dissociation of the transcription factor PTM from the chloroplast membrane to regulate nuclear gene expression. M, mitochondria; C, chloroplast; P, peroxisome; N, nucleus.
The peroxisome-derived signaling pathway has emerged by analyzing the catalase mutants which cause accumulation of H2O2 in peroxisomes. In tobacco and Arabidopsis, catalase-deficient lines display enhanced photorespiratory H2O2 levels, which trigger induction of pathogenesis-related (PR) genes and cell death (Takahashi et al., 1997; Chaouch et al., 2010). Under high light conditions, catalase-deficient plants provoke significant differences in nuclear gene expression profiles (Vandenabeele et al., 2003, 2004). In addition, large-scale transcriptomic analysis of the cat2 mutant demonstrated that H2O2 produced from peroxisomes induces prevailing protein repair responses (Queval et al., 2007). A more profound transcriptional change is observed in the cat1 cat2 cat3 triple mutant with the differentially expressed genes (DEGs) being enriched in those encoding transcription factors and receptor-like protein kinases (Su et al., 2018). In addition, expression of oxidative signal inducible 1 (OXI1), a gene encoding serine/threonine kinase, and several genes of the MAPK cascade pathway, such as MPK11 and MPK13, are dramatically changed in the cat1 cat2 cat3 triple mutant (Su et al., 2018). It has been reported that the OXI1-dependent pathway involving MPK3/6 can be activated by exogenous H2O2 (Rentel et al., 2004). Thus, it seems that signaling pathways activated by different sources of H2O2 may converge on OXI1, but the downstream signaling pathways are context-dependent.
The ROS-induced chloroplast retrograde signaling pathway is mediated by PTM, a plant homeodomain-type transcription factor with transmembrane domains which localizes in the chloroplast envelope (Sun et al., 2011). Under high light, proteolytic cleavage of PTM enables the release of the PTM N-terminus from the chloroplast envelope, which is then transferred and accumulated in the nucleus to regulate the expression of photosynthesis-associated nuclear genes including ABI4 (abscisic acid insensitive 4). In turn, this causes repression of the expression of LHCB, which encodes the photosynthesis-related light-harvesting complex II chlorophyll-a/b binding protein that is involved in photosynthetic generation of reducing power by harvesting solar energy (Sun et al., 2011). In Nicotiana benthamiana, high light induces chloroplast-derived H2O2 to be transferred to the nucleus, where they affect the nuclear redox state and gene transcription (Exposito-Rodriguez et al., 2017). During plant innate immunity responses, stromules, tubular extensions from the chloroplast, are induced and surround the nucleus to connect with it. Stromule formation is correlated with increased SA and H2O2 (Caplan et al., 2015). Chloroplast-sourced H2O2 and the chloroplast-localized defense protein NRIP1 (N receptor interacting protein 1) can move to the nucleus via stromule connections, indicating that stromules participate in chloroplast-to-nuclear retrograde signaling (Caplan et al., 2008, 2015).
As in chloroplasts, ROS is also involved in mitochondrial retrograde-signaling cascades. As a key component of chloroplast retrograde signaling pathway, ABI4 also plays a central role in mediating mitochondrial retrograde signaling by inducing alternative oxidase 1a (AOX1a). AOX1a encodes a mitochondrial inner membrane protein acting as a terminal electron acceptor of the alternative pathway in the electron transport chain. It is an important sensor of oxidation levels in cells and plays significant roles in maintaining the cellular redox state by regulating the malate valve and the antioxidative systems and sustaining photosynthesis by regulating the redox state of chloroplastic electron transport mediators and non-photochemical quenching (NPQ) in Arabidopsis (Giraud et al., 2009; Vishwakarma et al., 2014). In Arabidopsis, regulator of alternative oxidase 1 (RAO1) has been identified as the nucleus-localized cyclin-dependent kinase E1 that regulates the expression of both LHCB and AOX1a to integrate chloroplast and mitochondrial retrograde signals (Ng et al., 2013; Blanco et al., 2014). These results illustrate the coordination of these two organelles in retrograde signal pathways.
It has been reported that H2O2 generated from chloroplasts and peroxisomes induce different transcriptome profiles (Sewelam et al., 2014). H2O2 produced from chloroplasts in Arabidopsis glycolate oxidase (GO) overexpressing lines induces expression of genes involved in signaling responses, such as genes encoding transcription factors, protein/receptor kinases, and defense proteins, and genes involved in biosynthesis of secondary signaling messengers (Sewelam et al., 2014). By contrast, H2O2 produced from peroxisomes in Arabidopsis catalase-deficient mutants induces a stress recovery response by inducing genes encoding heat shock proteins (HSPs) and proteins involved in ubiquitin-dependent protein degradation (Sewelam et al., 2014). However, a group of genes are induced by H2O2 produced in both the chloroplast and peroxisome, indicating that the response induced by H2O2 can depend on either the origination of the H2O2 in specific organelles or on downstream integration of the H2O2 signals independent from the site of production (Sewelam et al., 2014). There are more evidence supports this view. According to microarray-based transcriptomic analysis, 1O2 derived in the chloroplasts of the chlorophyll b-deficient mutant chlorina1 (ch1) results in 302 genes expressed differentially compared to wild-type, far fewer than the number of DEGs (2,852 genes) observed in the cat1/2/3 mutant compared to wild-type. Interestingly, 43.7% of the DEGs in ch1 are shared with those found in the catalase triple mutant, when compared to wild type (Ramel et al., 2013; Su et al., 2018). The ROS signals derived from chloroplasts, mitochondria, and peroxisomes are proposed to connect in the cytoplasm and link to the MAPK pathway to regulate the expression of nuclear genes (Noctor and Foyer, 2016; Shumbe et al., 2016). However, to what extent these pathways connect to each other and the detailed molecular mechanisms remain unclear.
Prospects
Peroxisome homeostasis is regulated by complex regulatory systems. Recent progress has been made in understanding peroxisome biogenesis and degradation and how peroxisomes communicate with other organelles; however, many questions are still unresolved. For example, does de novo peroxisome formation exists in plants? What is the physiological importance of peroxisomes being connected to other organelles? What components comprise the membrane contact sites? What are the molecular mechanisms underlying peroxisome-derived retrograde signaling? Is ROS-mediated organellar retrograde signaling pathway integrated or organelle specific? Lastly, the pexophagy receptor in plants has yet to be identified. We hope these problems will be solved in the next few decades.
Author Contributions
TS and CM conceived and wrote the manuscript. WL and PW contributed to the revision of the manuscript. All authors read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was funded by the Foundation for Taishan Scholar from the People’s Government of Shandong Province (tshw20130962), National Natural Science Foundation of China (31470352, 31670073, and 31600204), and Natural Science Foundation of Shandong Province (ZR2014CM002 and ZR2016CB05).
References
- Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., et al. (2003). Pex8p. An intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646. 10.1016/S1097-2765(03)00062-5, PMID: [DOI] [PubMed] [Google Scholar]
- Agrawal G., Subramani S. (2016). De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta 1863, 892–901. 10.1016/j.bbamcr.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akşit A., van der Klei I. J. (2018). Yeast peroxisomes: how are they formed and how do they grow? Int. J. Biochem. Cell Biol. 105, 24–34. 10.1016/j.biocel.2018.09.019, PMID: [DOI] [PubMed] [Google Scholar]
- Aranovich A., Hua R., Rutenberg A. D., Kim P. K. (2014). PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127, 3675–3686. 10.1242/jcs.146282, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2011). The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23, 4446–4461. 10.1105/tpc.111.090142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2012). Differential roles of Arabidopsis dynamin-related proteins DRP3A, DRP3B, and DRP5B in organelle division. J. Integr. Plant Biol. 54, 921–931. 10.1111/j.1744-7909.2012.01174.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. 10.1126/science.1163732, PMID: [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Chaumont F. (2014). Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1840, 1596–1604. 10.1016/j.bbagen.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Blanco N. E., Guinea-D’ıaz M., Whelan J., Strand A. (2014). Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 369:20130231. 10.1098/rstb.2013.0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Frietsch S., Kim T. H., Dizon M. B., Schroeder J. I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 18, 63–68. 10.1016/j.cub.2007.11.067, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Hurley J. H. (2008). Retromer. Curr. Opin. Cell Biol. 20, 427–436. 10.1016/j.ceb.2008.03.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart S. E., Kao Y. T., Bartel B. (2014). Peroxisomal ubiquitin-protein ligases Peroxin2 and Peroxin10 have distinct but synergistic roles in matrix protein import and Peroxin5 retrotranslocation in Arabidopsis. Plant Physiol. 166, 1329–1344. 10.1104/pp.114.247148, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart S. E., Lingard M. J., Bartel B. (2013). Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193, 125–141. 10.1534/genetics.112.146100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J. L., Kumar A. S., Park E., Padmanabhan M. S., Hoban K., Modla S., et al. (2015). Chloroplast stromules function during innate immunity. Dev. Cell 34, 45–57. 10.1016/j.devcel.2015.05.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J. L., Mamillapalli P., Burch-Smith T. M., Czymmek K., Dinesh-Kumar S. P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132, 449–462. 10.1016/j.cell.2007.12.031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M. C., Sandalio L. M., Del Río L. A., León J. (2008). Peroxisome proliferation, wound-activated responses and expression of peroxisome-associated genes are cross-regulated but uncoupled in Arabidopsis thaliana. Plant Cell Environ. 31, 492–505. 10.1111/j.1365-3040.2008.01780.x, PMID: [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., et al. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153, 1692–1705. 10.1104/pp.110.153957, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B., Palma J. M., Rodriguez-Ruiz M. (2017). Plant peroxisomes: a nitro-oxidative cocktail. Redox Biol. 11, 535–542. 10.1016/j.redox.2016.12.033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross L. L., Ebeed H. T., Baker A. (2016). Peroxisome biogenesis, protein targeting mechanisms and PEX gene functions in plants. Biochim. Biophys. Acta 863, 850–862. 10.1016/j.bbamcr.2015.09.027 [DOI] [PubMed] [Google Scholar]
- Das K., Roychoudhury A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. 10.3389/fenvs.2014.00053 [DOI] [Google Scholar]
- De Duve C., Baudhuin P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46, 323–357. 10.1152/physrev.1966.46.2.323, PMID: [DOI] [PubMed] [Google Scholar]
- de Souza A., Wang J. Z., Dehesh K. (2017). Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 85–108. 10.1146/annurev-arplant-042916-041007, PMID: [DOI] [PubMed] [Google Scholar]
- Desai M., Hu J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 homolog, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146, 1117–1127. 10.1104/pp.107.113555, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Pan R., Hu J. (2017). Arabidopsis forkhead-associated domain protein 3 negatively regulates peroxisome division. J. Integr. Plant Biol. 59, 454–458. 10.1111/jipb.12542, PMID: [DOI] [PubMed] [Google Scholar]
- El Magraoui F., Schrotter A., Brinkmeier R., Kunst L., Mastalski T., Muller T., et al. (2014). The cytosolic domain of Pex22p stimulates the Pex4p-dependent ubiquitination of the PTS1-receptor. PLoS One 9:e105894. 10.1371/journal.pone.0105894, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., Vos A., van den Berg M., van Roermund C. W., van der Sluijs P., Distel B., et al. (1996). Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J. Biol. Chem. 271, 26375–26382. 10.1074/jbc.271.42.26375, PMID: [DOI] [PubMed] [Google Scholar]
- Erdmann R. (2016). Assembly, maintenance and dynamics of peroxisomes. Biochim. Biophys. Acta 1863, 787–789. 10.1016/j.bbamcr.2016.01.020 [DOI] [PubMed] [Google Scholar]
- Exposito-Rodriguez M., Laissue P. P., Yvon-Durocher G., Smirnoff N., Mullineaux P. M. (2017). Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 8:49. 10.1038/s41467-017-00074-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy D., Sanad M. N., Duscha K., Lyons M., Liu F., Bozhkov P., et al. (2017). Impact of salt stress, cell death, and autophagy on peroxisomes: quantitative and morphological analyses using small fluorescent probe N-BODIPY. Sci. Rep. 7:39069. 10.1038/srep46643, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139, 231–239. 10.1104/pp.105.066811, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer L. M., Rinaldi M. A., Young P. G., Danan C. H., Burkhart S. E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25, 4085–4100. 10.1105/tpc.113.113407, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre J. C., Manjithaya R., Mathewson R. D., Subramani S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376. 10.1016/j.devcel.2007.12.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick E. M., Strader L. C. (2018). Kinase MPK17 and the peroxisome division factor PMD1 influence salt-induced peroxisome proliferation. Plant Physiol. 176, 340–351. 10.1104/pp.17.01019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R., Kopischke M., Klapprodt C., Hause G., Meyer A. J., Schwarzlander M., et al. (2016). Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28, 130–145. 10.1105/tpc.15.00887, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Mano S., Kondo M., Saito C., Ueda T., et al. (2009). Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58, 388–400. 10.1111/j.1365-313X.2009.03786.x, PMID: [DOI] [PubMed] [Google Scholar]
- Gardner B. M., Chowdhury S., Lander G. C., Martin A. (2015). The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J. Mol. Biol. 427, 1375–1388. 10.1016/j.jmb.2015.01.019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V., Rylott E. L., Larson T. R., Sherson S. M., Bechtold N., Carde J. P., et al. (2001). Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28, 1–12. 10.1046/j.1365-313X.2001.01095.x, PMID: [DOI] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L. H., Whelan J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296. 10.1104/pp.109.139782, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez K. L., Fleming W. A., Kao Y. T., Wright Z. J., Venkova S. V., Ventura M. J., et al. (2017). Disparate peroxisome-related defects in Arabidopsis pex6 and pex26 mutants link peroxisomal retrotranslocation and oil body utilization. Plant J. 92, 110–128. 10.1111/tpj.13641, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez K. L., Ratzel S. E., Burks K. H., Danan C. H., Wages J. M., Zolman B. K., et al. (2018). A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling. Proc. Natl. Acad. Sci. USA 115, 3163–3172. 10.1073/pnas.1721279115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S., Mano S., Nakamori C., Nishimura M. (2011). Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23, 1573–1587. 10.1105/tpc.110.080770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Yamada S., Mano S., Nakamori C., Kondo M., Yamawaki R., Kato A., et al. (2014). Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 55, 482–496. 10.1093/pcp/pcu017, PMID: [DOI] [PubMed] [Google Scholar]
- Graham I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. 10.1146/annurev.arplant.59.032607.092938, PMID: [DOI] [PubMed] [Google Scholar]
- Hackenberg T., Juul T., Auzina A., Gwizdz S., Malolepszy A., Van Der Kelen K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25, 4616–4626. 10.1105/tpc.113.117192, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., et al. (2006). Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119, 2508–2517. 10.1242/jcs.02979, PMID: [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M., Hayashi H., Hara-Nishimura I., Nishimura M. (2001). Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218, 83–94. 10.1007/BF01288364, PMID: [DOI] [PubMed] [Google Scholar]
- Hettema E. H., Erdmann R., van der Klei I., Veenhuis M. (2014). Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 29, 25–30. 10.1016/j.ceb.2014.02.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E. H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. (2000). Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19, 223–233. 10.1093/emboj/19.2.223, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95. 10.1016/j.cell.2005.04.025, PMID: [DOI] [PubMed] [Google Scholar]
- Hohfeld J., Veenhuis M., Kunau W. H. (1991). PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114, 1167–1178. 10.1083/jcb.114.6.1167, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho M., Tamura S., Shimozawa N., Suzuki Y., Kondo N., Fujiki Y. (1998). Mutation in pex16, is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am. J. Hum. Genet. 63, 1622–1630. 10.1086/302161, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R. T., Reumann S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303. 10.1105/tpc.112.096586, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua R., Kim P. K. (2016). Multiple paths to peroxisomes: mechanism of peroxisome maintenance in mammals. Biochim. Biophys. Acta 1863, 881–891. 10.1016/j.bbamcr.2015.09.026 [DOI] [PubMed] [Google Scholar]
- Hunt J. E., Trelease R. N. (2004). Sorting pathway and molecular targeting signals for the Arabidopsis peroxin 3. Biochem. Biophys. Res. Commun. 314, 586–596. 10.1016/j.bbrc.2003.12.123, PMID: [DOI] [PubMed] [Google Scholar]
- Incarbone M., Zimmermann A., Hammann P., Erhardt M., Michel F., Dunoyer P. (2017). Neutralization of mobile antiviral small RNA through peroxisomal import. Nat. Plants 3:17094. 10.1038/nplants.2017.94, PMID: [DOI] [PubMed] [Google Scholar]
- Jaipargas E. A., Mathur N., Bou Daher F., Wasteneys G. O., Mathur J. (2016). High light intensity leads to increased peroxule-mitochondria interactions in plants. Front. Cell Dev. Biol. 4:6. 10.3389/fcell.2016.00006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y. T., Bartel B. (2015). Elevated growth temperature decreases levels of the PEX5 peroxisome-targeting signal receptor and ameliorates defects of Arabidopsis mutants with an impaired PEX4 ubiquitin-conjugating enzyme. BMC Plant Biol. 15:224. 10.1186/s12870-015-0605-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y. T., Gonzalez K. L., Bartel B. (2018). Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 176, 162–177. 10.1104/pp.17.01050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik S. K., Trelease R. N. (2005). Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 138, 1967–1981. 10.1104/pp.105.061291, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Reumann S., Hu J. (2009). Peroxisome biogenesis and function. Arabidopsis Book 7:e0123. 10.1199/tab.0123, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Zhao Q., Xie Q., Hu J. (2013). Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins (F). J. Integr. Plant Biol. 55, 108–120. 10.1111/jipb.12014, PMID: [DOI] [PubMed] [Google Scholar]
- Khan B. R., Zolman B. K. (2010). PEX5 mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 154, 1602–1615. 10.1104/pp.110.162479, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. K., Hettema E. H. (2015). Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427, 1176–1190. 10.1016/j.jmb.2015.02.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H. N., Kim S. H., Shin K. D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25, 4956–4966. 10.1105/tpc.113.117960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. K., Mullen R. T. (2013). PEX16: a multifaceted regulator of peroxisome biogenesis. Front. Physiol. 4:241. 10.3389/fphys.2013.00241, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Manivannan S., Cepinska M. N., Krikken A. M., Kram A. M., Veenhuis M., et al. (2014). Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204, 659–668. 10.1083/jcb.201310148, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J., Pranjic K., Huber A., Ellinger A., Hartig A., Kragler F., et al. (2010). PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123, 3389–3400. 10.1242/jcs.064907, PMID: [DOI] [PubMed] [Google Scholar]
- Koo A. J., Chung H. S., Kobayashi Y., Howe G. A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520. 10.1074/jbc.M607854200, PMID: [DOI] [PubMed] [Google Scholar]
- Kunz H. H., Scharnewski M., Feussner K., Feussner I., Flugge U. I., Fulda M., et al. (2009). The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749. 10.1105/tpc.108.064857, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530. 10.1146/annurev.cb.01.110185.002421, PMID: [DOI] [PubMed] [Google Scholar]
- Li Y., Chen L., Mu J., Zuo J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. 10.1104/pp.113.225805, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Li J., Hao R., Guo Y. (2017). Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genomics 44, 395–404. 10.1016/j.jgg.2017.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- Li J., Liu J., Wang G., Cha J. Y., Li G., Chen S., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27, 908–925. 10.1105/tpc.114.135095, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Pang C. H., Ding F., Sui N., Feng Z. T., Wang B. S. (2012). Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis chloroplasts enhances salt tolerance of plants. S. Afr. J. Bot. 78, 235–245. 10.1016/j.sajb.2011.09.006 [DOI] [Google Scholar]
- Li F., Vierstra R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. 10.1016/j.tplants.2012.05.006, PMID: [DOI] [PubMed] [Google Scholar]
- Lin Y., Cluette-Brown J. E., Goodman H. M. (2004). The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol. 135, 814–827. 10.1104/pp.103.036772, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Sun L., Nguyen L. V., Rachubinski R. A., Goodman H. M. (1999). The pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284, 328–330. 10.1126/science.284.5412.328, PMID: [DOI] [PubMed] [Google Scholar]
- Lingard M. J., Bartel B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151, 1354–1365. 10.1104/pp.109.142505, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M. J., Monroe-Augustus M., Bartel B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 4561–4566. 10.1073/pnas.0811329106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M. J., Trelease R. N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119, 1961–1972. 10.1242/jcs.02904, PMID: [DOI] [PubMed] [Google Scholar]
- Linka N., Theodoulou F. L. (2013). Metabolite transporters of the plant peroxisomal membrane: known and unknown. Subcell. Biochem. 69, 169–194. 10.1007/978-94-007-6889-5_10 [DOI] [PubMed] [Google Scholar]
- Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183. 10.1126/science.1119409, PMID: [DOI] [PubMed] [Google Scholar]
- Lisenbee C. S., Heinze M., Trelease R. N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol. 132, 870–882. 10.1104/pp.103.019976, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. S., Wang W. Q., Li M., Wan S. B., Sui N. (2017). Antioxidants and unsaturated fatty acids are involved in salt tolerance in peanut. Acta Physiol. Plant. 39:207. 10.1007/s11738-017-2501-y [DOI] [Google Scholar]
- Lopez-Huertas E., Charlton W. L., Johnson B., Graham I. A., Baker A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19, 6770–6777. 10.1093/emboj/19.24.6770, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Agrawal G., Subramani S. (2011). Peroxisome assembly: matrix and membrane protein biogenesis. J. Cell Biol. 193, 7–16. 10.1083/jcb.201010022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Hagstrom D., Polley S. G., Subramani S. (2013). Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 288, 27220–22731. 10.1074/jbc.M113.492694, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Schumann U., Rayapuram N., Subramani S. (2009). The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol. Biol. Cell 20, 3680–3689. 10.1091/mbc.E09-01-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38, 487–498. 10.1111/j.1365-313X.2004.02063.x, PMID: [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Nito K., Kondo M., Nishimura M. (2006). The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 47, 604–618. 10.1111/j.1365-313X.2006.02809.x, PMID: [DOI] [PubMed] [Google Scholar]
- Matsuzaki T., Fujiki Y. (2008). The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275–1286. 10.1083/jcb.200806062, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M., Cizmowski C., Schliebs W., Krüger V., Beck S., Wagner R., et al. (2010). The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277. 10.1038/ncb2027, PMID: [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525, 181–194. 10.1016/j.abb.2012.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. 10.1093/jxb/erq282, PMID: [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M., Ramon N. M., Ratzel S. E., Lingard M. J., Christensen S. E., Murali C., et al. (2011). Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol. Biol. 77, 1–15. 10.1007/s11103-011-9782-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A. M., Nuttall J. M., Hettema E. H. (2012). Atg36: the Saccharomyces cerevisiae receptor for pexophagy. Autophagy 8, 1680–1681. 10.4161/auto.21485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T., Lisenbee C. S., Miernyk J. A., Trelease R. N. (1999). Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11, 2167–2185. 10.1105/tpc.11.11.2167, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. A., Phillipson B. A., Baker A., Mullen R. T. (2003). Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133, 813–828. 10.1104/pp.103.027870, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Giraud E., Duncan O., Law S. R., Wang Y., Xu L., et al. (2013). Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 288, 3449–3459. 10.1074/jbc.M112.416727, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K., Hayashi M., Nishimura M. (2002). Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 43, 355–366. 10.1093/pcp/pcf057, PMID: [DOI] [PubMed] [Google Scholar]
- Nito K., Kamigaki A., Kondo M., Hayashi M., Nishimura M. (2007). Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 48, 763–774. 10.1093/pcp/pcm053, PMID: [DOI] [PubMed] [Google Scholar]
- Noctor G., Foyer C. H. (2016). Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 171, 1581–1592. 10.1104/pp.16.00346, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Matsunaga S., Mano S., Kondo M., Yamada K., Hayashi M., et al. (2015). Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat. Plants 1, 15035. 10.1038/nplants.2015.35, PMID: [DOI] [PubMed] [Google Scholar]
- Okreglak V., Walter P. (2014). The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA 111, 8019–8024. 10.1073/pnas.1405755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth T., Reumann S., Zhang X., Fan J., Wenzel D., Quan S., et al. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19, 333–350. 10.1105/tpc.106.045831, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma J. M., Garrido M., Rodríguez-García M. I., del Río L. A. (1991). Peroxisome proliferation and oxidative stress mediated by activated oxygen species in plant peroxisomes. Arch. Biochem. Biophys. 287, 68–74. 10.1016/0003-9861(91)90389-Z, PMID: [DOI] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Chen C., Hu J. (2018). The E3 ubiquitin ligase SP1-like 1 plays a positive role in peroxisome biogenesis in Arabidopsis. Plant J. 94, 836–846. 10.1111/tpj.13900, PMID: [DOI] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Hu J. (2016). E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, E7307–E7316. 10.1073/pnas.1613530113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa A. G., Francisco T., Bicho D., Dias A. F., Barros-Barbosa A., Hagmann V., et al. (2018). Peroxisomal monoubiquitinated PEX5 interacts with the AAA ATPases PEX1 and PEX6 and is unfolded during its dislocation into the cytosol. J. Biol. Chem. 293, 11553–11563. 10.1074/jbc.RA118.003669, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriv O. I., Tang L., Titorenko V. I., Rachubinski R. A. (2004). A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 341, 119–134. 10.1016/j.jmb.2004.05.064, PMID: [DOI] [PubMed] [Google Scholar]
- Platta H. W., Magraoui F. E., Baumer B. E., Schlee D., Girzalsky W., Erdmann R. (2009). Pex2 and Pex12 function as protein–ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29, 5505–5516. 10.1128/MCB.00388-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H. W., Magraoui F. E., Schlee D., Grunau S., Girzalsky W., Erdmann R. (2007). Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204. 10.1083/jcb.200611012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Smith S. M. (2008). When is a peroxisome not a peroxisome? Trends Plant Sci. 13, 522–525. 10.1016/j.tplants.2008.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Prestele J., Hierl G., Scherling C., Hetkamp S., Schwechheimer C., Isono E., et al. (2010). Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc. Natl. Acad. Sci. USA 107, 14915–14920. 10.1073/pnas.1009174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y. C., Liu W. Q., Qiu L. Y., Zhang S. M., Ma L., Zhang H. (2010). Overexpression of glutathione S-transferase gene increases salt tolerance of Arabidopsis. Russ. J. Plant Physiol. 57, 233–240. 10.1134/S102144371002010X [DOI] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakière B., Vanacker H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. 10.1111/j.1365-313X.2007.03263.x [DOI] [PubMed] [Google Scholar]
- Ramel F., Ksas B., Akkari E., Mialoundama A. S., Monnet F., Krieger-Liszkay A., et al. (2013). Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25, 1445–1462. 10.1105/tpc.113.109827, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon N. M., Bartel B. (2010). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21, 1263–1271. 10.1091/mbc.E09-08-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel S. E., Lingard M. J., Woodward A. W., Bartel B. (2011). Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12, 121–134. 10.1111/j.1600-0854.2010.01136.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N., Subramani S. (2006). The importomer—a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta 1763, 1613–1619. 10.1016/j.bbamcr.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S., Prinz W. A. (2008). Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 105, 15785–15790. 10.1073/pnas.0808321105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. L., Qi G. N., Feng H. Q., Zhao S., Zhao S. S., Wang Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74, 258–266. 10.1111/tpj.12123, PMID: [DOI] [PubMed] [Google Scholar]
- Rentel M. C., Lecourieux D., Ouaked F., Usher S. L., Petersen L., Okamoto H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861. 10.1038/nature02353, PMID: [DOI] [PubMed] [Google Scholar]
- Reumann S., Chowdhary G., Lingner T. (2016). Characterization, prediction and evolution of plant peroxisomal targeting signals type 1 (PTS1s). Biochim. Biophys. Acta 1863, 790–803. 10.1016/j.bbamcr.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136, 2587–2608. 10.1104/pp.104.043695, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi M. A., Fleming W. A., Gonzalez K. L., Park J., Ventura M. J., Patel A. B., et al. (2017). The PEX1 ATPase stabilizes PEX6 and plays essential roles in peroxisome biology. Plant Physiol. 174, 2231–2247. 10.1104/pp.17.00548, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M., Romero-Puertas M. C., Sanz-Fernández M., Hu J., Sandalio L. M. (2016). Peroxisomes extend peroxules in a fast response to stress via a reactive oxygen species-mediated induction of the peroxin PEX11a. Plant Physiol. 171, 1665–1674. 10.1104/pp.16.00648, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M., Romero-Puertas M. C., Sparkes I., Hawes C., del Río L. A., Sandalio L. M. (2009). Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic. Biol. Med. 47, 1632–1639. 10.1016/j.freeradbiomed.2009.09.012 [DOI] [PubMed] [Google Scholar]
- Rokka A., Antonenkov V. D., Soininen R., Immonen H. L., Pirila P. L., Bergmann U., et al. (2009). Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS One 4:e5090. 10.1371/journal.pone.0005090, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano F. B., Blok N. B., Rapoport T. A. (2019). Peroxisome protein import recapitulated in Xenopus egg extracts. J. Cell Biol. pii: jcb.201901152. 10.1083/jcb.201901152, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., et al. (2004). Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell 15, 3406–3417. 10.1091/mbc.e04-03-0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio L. M., Romero-Puertas M. C. (2015). Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 116, 475–485. 10.1093/aob/mcv074, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M. (2006). Shared components of mitochondrial and peroxisomal division. Biochim. Biophys. Acta 1763, 531–541. 10.1016/j.bbamcr.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Schrader M., Fahimi H. D. (2006). Peroxisomes and oxidative stress. Biochim. Biophys. Acta 1763, 1755–1766. 10.1016/j.bbamcr.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., et al. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645. 10.1016/j.cell.2008.06.025, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Prestele J., O’Geen H., Brueggeman R., Wanner G., Gietl C. (2007). Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc. Natl. Acad. Sci. USA 104, 1069–1074. 10.1073/pnas.0610402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Tobin A. K., Logan D. C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57, 1275–1280. 10.1093/jxb/erj096, PMID: [DOI] [PubMed] [Google Scholar]
- Sewelam N., Jaspert N., Van Der Kelen K., Tognetti V. B., Schmitz J., Frerigmann H. (2014). Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 7, 1191–1210. 10.1093/mp/ssu070, PMID: [DOI] [PubMed] [Google Scholar]
- Shai N., Schuldiner M., Zalckvar E. (2016). No peroxisome is an island—peroxisome contact sites. Biochim. Biophys. Acta, Mol. Cell Res. 1863, 1061–1069. 10.1016/j.bbamcr.2015.09.016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X. Y., Wang Z. L., Song X. F., Xu J. J., Jiang C. Y., Zhao Y. X., et al. (2014). Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 86, 303–317. 10.1007/s11103-014-0230-9 [DOI] [PubMed] [Google Scholar]
- Shibata M., Oikawa K., Yoshimoto K., Kondo M., Mano S., Yamada K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. 10.1105/tpc.113.116947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumbe L., Chevalier A., Legeret B., Taconnat L., Monnet F., Havaux M. (2016). Singlet oxygen-induced cell death in Arabidopsis under high-light stress is controlled by OXI1 kinase. Plant Physiol. 170, 1757–1771. 10.1104/pp.15.01546, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Aitchison J. D. (2013). Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14, 803–817. 10.1038/nrm3700, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. A., Hawes C., Baker A. (2005). AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol. 139, 690–700. 10.1104/pp.105.065094, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B. (2011). Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4, 477–486. 10.1093/mp/ssr006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Wang P., Li H., Zhao Y., Lu Y., Dai P., et al. (2018). The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 60, 591–607. 10.1111/jipb.12649, PMID: [DOI] [PubMed] [Google Scholar]
- Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., et al. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2:477. 10.1038/ncomms1486 [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Chen Z., Du H., Liu Y., Klessig D. F. (1997). Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11, 993–1005. 10.1046/j.1365-313X.1997.11050993.x, PMID: [DOI] [PubMed] [Google Scholar]
- Thazar-Poulot N., Miquel M., Fobis-Loisy I., Gaude T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. USA 112, 4158–4163. 10.1073/pnas.1403322112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou F. L., Bernhardt K., Linka N., Baker A. (2013). Peroxisome membrane proteins: multiple trafficking routes and multiple functions? Biochem. J. 451, 345–352. 10.1042/BJ20130078, PMID: [DOI] [PubMed] [Google Scholar]
- van der Zand A., Braakman I., Tabak H. F. (2010). Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell 21, 2057–2065. 10.1091/mbc.E10-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A., Gent J., Braakman I., Tabak H. F. (2012). Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409. 10.1016/j.cell.2012.01.054, PMID: [DOI] [PubMed] [Google Scholar]
- Vandenabeele S., Van Der Kelen K., Dat J., Gadjev I., Boonefaes T., Morsa S., et al. (2003). A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 100, 16113–16118. 10.1073/pnas.2136610100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylstecke M., Rombauts S., Langebartels C., Seidlitz H. K., et al. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58. 10.1111/j.1365-313X.2004.02105.x, PMID: [DOI] [PubMed] [Google Scholar]
- Verslues P. E., Batelli G., Grillo S., Agius F., Kim Y. S., Zhu J., et al. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell. Biol. 27, 7771–7780. 10.1128/MCB.00429-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma A., Bashyam L., Senthilkumaran B., Scheibe R., Padmasree K. (2014). Physiological role of AOX1a in photosynthesis and maintenance of cellular redox homeostasis under high light in Arabidopsis thaliana. Plant Physiol. Biochem. 81, 44–53. 10.1016/j.plaphy.2014.01.019, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., Li S., Liu Y., Ma C. (2015). Redox regulated peroxisome homeostasis. Redox Biol. 4, 104–108. 10.1016/j.redox.2014.12.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Visser N. V., Veenhuis M., van der Klei I. J. (2003). Physical interactions of the peroxisomal targeting signal 1 receptor pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 278, 43340–43345. 10.1074/jbc.M307789200, PMID: [DOI] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B. (2005). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16, 573–583. 10.1091/mbc.e04-05-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W., Fleming W. A., Burkhart S. E., Ratzel S. E., Bjornson M., Bartel B. (2014). A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol. Biol. 86, 201–214. 10.1007/s11103-014-0223-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska J. P., Cruz-Zaragoza L. D., Wei Y., Schummer A., Chuartzman S. G., de Boer R., et al. (2017). Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim. Biophys. Acta 1864, 1656–1667. 10.1016/j.bbamcr.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Shibata M., Kondo M., Oikawa K., Sato M., Toyooka K., et al. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 127, 1161–1168. 10.1242/jcs.139709, PMID: [DOI] [PubMed] [Google Scholar]
- Young P. G., Bartel B. (2016). Pexophagy and peroxisomal protein turnover in plants. Biochim. Biophys. Acta 1863, 999–1005. 10.1016/j.bbamcr.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. G., Passalacqua M. J., Chappell K., Llinas R. J., Bartel B. (2019). A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 8, 1–19. 10.1080/15548627.2019.1569915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Veenhuis M., van der Klei I. J. (2016). The birth of yeast peroxisomes. Biochim. Biophys. Acta 1863, 902–910. 10.1016/j.bbamcr.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Zhang X. C., Hu J. P. (2008). FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant 1, 1036–1047. 10.1093/mp/ssn056, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22, 431–442. 10.1105/tpc.109.071324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li Y., Zhu J. K. (2018). Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. 10.1038/s41477-018-0309-4, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Wang C., Yang C. Y., Wang J. Y., Chen L., Bao X. M., et al. (2010). The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. Plant J. 62, 539–548. 10.1111/j.1365-313X.2010.04173.x [DOI] [PubMed] [Google Scholar]
- Zolman B. K., Monroe-Augustus M., Silva I. D., Bartel B. (2005). Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17, 3422–3435. 10.1105/tpc.105.035691, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Silva I. D., Bartel B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127, 1266–1278. 10.1104/pp.010550, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156, 1323–1337. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]