Abstract
Introduction
There is observational evidence that low circulating levels of vitamin B12 are associated with an increased risk of Alzheimer's disease
Methods
We used a two-sample summary-statistics–based Mendelian randomization design to assess the relationship of genetic factors contributing to vitamin B12 with late-onset Alzheimer's disease risk.
Results
Our results do not support a causal role of decreased vitamin B12 levels on Alzheimer's disease risk.
Discussion
This work encourages research on other modifiable biomarkers for Alzheimer's disease risk.
Keywords: Alzheimer's disease, Vitamin B12, Mendelian randomization, Biomarker, Dementia
1. Introduction
Vitamin B12 (denoted here as B12) is a water-soluble compound involved in many biological pathways, such as the formation of DNA nucleotides and in DNA methylation. In the clinic, low levels of circulating B12 are defined as <148 pmol/L [1]. Deficiency can result in anemia, neurological problems, and possibly cardiovascular disease. B12 deficiency occurs worldwide across all age groups in both males and females [1]. The prevalence in the United States, for instance, ranges from 3% to 26% [2].
There is conflicting evidence in the literature as to whether B12 deficiency is associated with an increase in Alzheimer's disease risk (Table 1). Two observational studies found evidence of individuals with Alzheimer's or cognitive impairment having lower levels of B12 [3], [4]. On the other hand, a large meta-analysis of B12, conducted in an Icelandic population, did not find a correlation with Alzheimer's disease using their phenotype database [5]. In addition, a previous Mendelian randomization study assessing B12-associated variants identified in the aforementioned Icelandic B12 meta-analysis also did not find a relationship between the two traits [6].
Table 1.
Mixed evidence on low vitamin B12 levels and Alzheimer's disease risk in the literature
| Study | Sample | Method | Association evidence for low B12 and Alzheimer's |
|---|---|---|---|
| Clarke et al. 1998 [3] | 240 AD cases + 108 controls | Observational | Y |
| Moore et al. 2014 [4] | 1354 cognitive impairment | Observational | Y |
| Grarup et al. 2013 [5] | 45K Icelandic + Danish | B12 GWAS | N |
| Larsson et al. 2017 [6] | 17K AD cases + 37K controls | Mendelian Randomization | N |
Abbreviation: AD, Alzheimer's disease.
Several genome-wide studies have been published assessing the associations between genetic variants and B12 levels (Table 2). By making use of publicly available B12 and recently published Alzheimer's disease genetic association summary-level data, we aimed to provide further evidence either for or against the genetic role of B12 levels in Alzheimer's risk, which was addressed through summary-statistics–based Mendelian randomization.
Table 2.
Previously published genetic studies assessing vitamin B12 levels
| Study | Samples | N | Genomewide significant loci |
|---|---|---|---|
| Studies included in the current analysis | |||
| Tanaka et al. 2009 [7] | Discovery: SardiNIA + InCHIANTI + BLSA Replication: Progetto Nutrizione study |
N∼3000 + ∼700 in replication | FUT2 |
| Hazra et al. 2009 [8] | European Meta-analysis: women in NHS-CGEMS + men & women in SHARe | N∼4700 | FUT2, CUBN, TCN1, MUT |
| Grarup et al. 2013 [5] | Discovery: Icelandic (1K WGS; impute rest) + Danish exome |
N∼37K WGS + N∼8K Danish exome | FUT2, CD320, TCN1, TCN2, ABCD4, MMAA, MMACHC, MUT, MTHFR |
| Keene et al. 2014 [9] | Stroke patients in the Vitamin Intervention for Stroke Prevention trial | N ∼2100 | CUBN, TCN1 |
| Studies not included due to non-European ancestry samples | |||
| Lin et al. 2012 [10] | Discovery: Chinese men Replication: independent Chinese population |
N∼2000 + ∼1500 in replication | MS4A3, CLYBL, FUT6, 5q32, FUT2 |
| Nongmaithem et al. 2017 [11] | Discovery: Pune Maternal Nutrition Study Replication: Other Indian cohorts |
N∼1000 + ∼3400 in replication | FUT6, FUT2, TCN1, TCN2, CUBN, MMAA |
Abbreviation: SHARe, Framingham SNP Health Association Resource.
2. Methods
In a Mendelian randomization experiment, we test whether “instruments” (in our case B12-associated genetic variants), which directly affect the “exposure” (B12 levels), are associated with the “outcome” (Alzheimer's disease risk) only through the exposure. There are three key assumptions. First, the B12 variants are indeed associated with B12 levels. Second, the variants are not associated to confounding factors, and finally, the variants are related to Alzheimer's disease risk only through their association with B12 levels. We are interested in the effect of B12 levels on Alzheimer's disease risk, assuming that the genetic variants do not have pleiotropic effects on the outcome, which will bias the resulting estimates. To obtain the estimate of association between B12 and Alzheimer's disease risk, we performed a two-sample Mendelian randomization experiment by combining the Wald estimate (the ratio of genetic association with Alzheimer's disease risk with B12) for each uncorrelated variant, using inverse weighting with a fixed effects model.
2.1. Source of Alzheimer's disease genetic association summary statistics (“outcome”)
We used the largest (71,880 cases, 383,378 controls) and most recent Alzheimer's meta-analysis, based on European-ancestry samples, which includes proxy cases from the UK Biobank in addition to the traditional case control design [12].
2.2. Source of vitamin B12 genetic association summary statistics (“Exposure”) and selection of vitamin B12 variants (“Instruments”)
We downloaded genetic association summary-level results for “vitamin B12 measurement” (EFO ID 0004620) from the NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) downloaded on December 20, 2018, keeping results published in 2009 or more recent [13]. Results reported from studies carried out in non-European cohorts were removed because of possible population stratification confounding between these estimates and the European-ancestry Alzheimer's disease study. This filter removed two B12 studies: Lin et al. [10] (GWAS carried out in Chinese men) and Nongmaithem et al. [11] (GWAS carried out in Indian cohorts). We then referred to the original articles to determine the standard error of the β (SE) and the effect alleles if not provided in the catalog (or if provided, to verify that they were correctly reported). If only the effect allele, and not the other allele, was reported in the original publication, we looked up the variant in the NHLBI Trans-Omics for Precision Medicine (TOPMed) high-depth whole-genome sequencing effort (Freeze5, https://www.nhlbiwgs.org) to determine the other allele and verify the accuracy of the reported effect frequency. This freeze of the TOPMed data set reports 463 million variants across 62,784 individuals and was accessed through the BRAVO browser (https://bravo.sph.umich.edu). Variants from the study by Grarup et al. [5] were missing from the catalog, and so the significantly associated variants identified in this study (from their Table 1) were manually added. When the effect size (β), SE, or both were not provided in the study, we used the approximation in Zhu et al. [14] (in the following) to compute those statistics.
where Z = Z statistic, MAF = minor allele frequency, and N = sample size.
Only variants with a reported significance P value of <5E-8 were retained. In cases where the reported effect allele from the B12 study did not match the effect allele in the Alzheimer's study, the allele was flipped in the B12 study and the opposite sign was assigned to the β. None of the variants were palindromic with an effect allele frequency close to 0.5, and thus we were able to confidently verify that the effect allele was correctly assigned.
2.3. Mendelian randomization primary analysis
We used nine independent B12 variants as instrumental variables (Table 3). Five were reported in a single (the largest) study (rs1131603: TCN2, rs2270655: MMAA, rs2336573: CD320, rs3742801: ABCD4, rs41281112: CLYBL), and four were reported in two or more independent studies. For the four variants reported as significant in multiple B12 studies (rs602662: FUT2, rs1801222: CUBN, rs34324219: TCN1, rs1141321: MUT), we conducted a standard-error–based fixed effects meta-analysis in METAL [15]. Note that as rs9473555, another reported variant in MUT, is in perfect linkage disequilibrium in European populations with rs1141321 (assessed using LDlink [16]), we used the former as a proxy for the latter. The resulting effect size (β), standard error of the β (SE), and the average effect allele frequency for these four variants were used in the Mendelian randomization alongside the summary-level data for the remaining five variants that only appeared in one study with genomewide significance.
Table 3.
Summary of selected instrumental variables
| SNP | β outcome | SE outcome | Effect allele outcome | Other allele outcome | P value outcome | Chr:pos outcome | Other allele exposure | Effect allele exposure | Eaf exposure | β exposure | SE exposure | Wald | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1131603 | 0.0053 | 0.0048 | C | T | 0.2739 | 22:31018975 | T | C | 0.055 | 0.159 | 0.01 | 0.03 | TCN2 |
| rs1141321 | 0.0022 | 0.0022 | T | C | 0.3108 | 6:49412433 | C | T | 0.365 | −0.127 | 0.006 | −0.02 | MUT |
| rs1801222 | 0.0015 | 0.0022 | A | G | 0.5012 | 10:17156151 | G | A | 0.363 | −0.132 | 0.006 | −0.01 | CUBN |
| rs2270655 | 0.0019 | 0.0047 | C | G | 0.6812 | 4:146576418 | C | G | 0.941 | −0.103 | 0.01 | −0.02 | MMAA |
| rs2336573 | −0.0013 | 0.0050 | T | C | 0.7940 | 19:8367709 | C | T | 0.031 | 0.288 | 0.02 | −0.005 | CD320 |
| rs34324219 | −0.0014 | 0.0035 | A | C | 0.6786 | 11:59623378 | C | A | 0.118 | −0.185 | 0.01 | 0.008 | TCN1 |
| rs3742801 | 0.0005 | 0.0021 | T | C | 0.7992 | 14:74759006 | C | T | 0.294 | 0.053 | 0.007 | 0.01 | ABCD4 |
| rs41281112 | 0.0001 | 0.0074 | T | C | 0.9838 | 13:100518634 | C | T | 0.948 | −0.160 | 0.007 | −0.001 | CLYBL |
| rs602662 | −0.0055 | 0.0022 | G | A | 0.0120 | 19:49206985 | A | G | 0.416 | −0.051 | 0.006 | 0.11 | FUT2 |
Abbreviations: Eaf, effect allele frequency; SE, standard error of β; Wald, Wald test statistic Human genome build 38 chromosomal coordinates are reported.
We verified that each selected SNP was also reported in the Alzheimer's study, was not associated with Alzheimer's (either in the Alzheimer's disease meta-analysis or through a PubMed search of the variant rsID) to circumvent possible pleiotropic effects. We conducted an inverse weighting Mendelian randomization [17] as our primary analysis using the MendelianRandomization and metafor packages in R version 3.5.0 (http://www.r-project.org/) [18].
2.4. Sensitivity analyses
Beyond the inverse weighting Mendelian randomization as our primary analysis, as sensitivity analyses, we used weighted median, MR-Egger [19], and MR-PRESSO [20] to assess bias from invalid instruments when using more than one genetic variant and the consistency of estimates.
3. Results
3.1. Mendelian randomization primary analysis
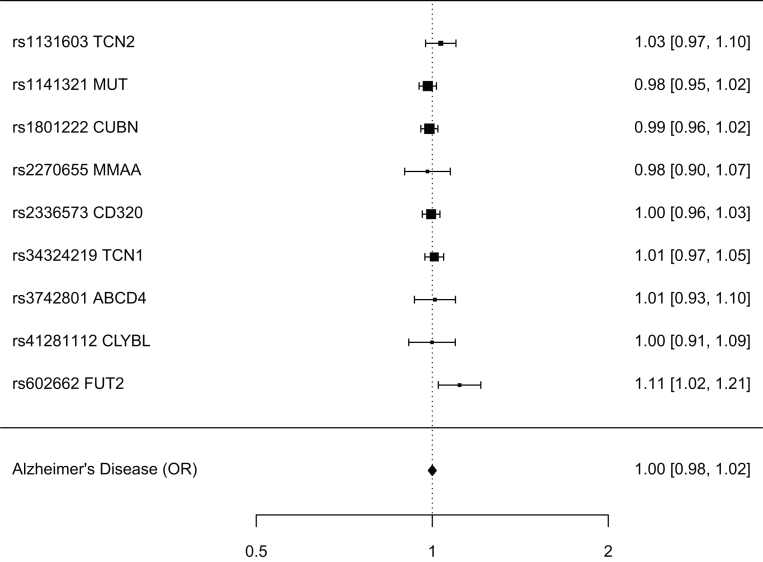
Based on the 9 selected SNPs, B12 was unrelated to Alzheimer's disease risk (95% confidence interval [CI] −0.016, 0.016, P value = .99). A forest plot of inverse weighting Mendelian randomization with fixed effects (Fig. 1) showed that none of the effect sizes of the variants were outliers (outlying effect sizes could suggest pleiotropic effects of the variant between the exposure and the outcome).
Fig. 1.
Associations of vitamin B12 variants with Alzheimer's disease risk.
3.2. Sensitivity analyses
To account for the potential bias from invalid instruments, we conducted numerous types of Mendelian randomization experiments to assess consistency. The results among the experiments were consistent, suggesting soundness in the statistical techniques for creating the estimates. The 95% CI for the weighted median method was −0.026, 0.013 (P value = .52), −0.057, 0.012 (P value = .20) for MR-Egger, and −0.008, 0.009 (P value = .36) for MR-PRESSO.
4. Discussion
Discordant with observational studies, but consistent with a previously reported Mendelian randomization experiment [6], we did not find evidence of B12 levels being associated with late-onset Alzheimer's disease risk with a more comprehensively examined set of genetic variants for vitamin B12 and a much larger Alzheimer's disease data set.
Statistical power for a Mendelian randomization analysis can be estimated by computing the power for a simple regression divided by the squared correlation of the genetic instrumental variables with the exposure (vitamin B12). However, the variance explained by the genetic associations for B12 was not available because we amalgamated results from several studies together, which prevented us from obtaining a precise calculation. With power in mind, we only selected variants strongly associated with B12 (either reported in numerous independent studies or highly significant in a single study), and are in functionally relevant genes.
We took care to undertake sensitivity analyses and to avoid confounding, but in Mendelian randomization experiments one cannot completely discount the existence of extraneous factors, which could influence the estimates. The validity of results relies on the assumptions for the instrumental variables being met, and on the underlying integrity of the underlying genetic association results such as that case-control studies are free from selection bias.
Although we cannot completely rule out the role of low vitamin B12 levels on Alzheimer's disease risk, the findings presented here do not support the observational evidence of a relationship between the two.
Research in context.
-
1.
Systematic review: There is conflicting evidence in the literature as to whether lower levels of vitamin B12 increases one's risk for developing Alzheimer's disease. We used a Mendelian randomization framework making use of the most recent Alzheimer's disease (2019) association summary statistics, and of a meta-analysis of vitamin B12 associated variants.
-
2.
Interpretation: We did not find evidence of a causal relationship between low vitamin B12 levels and Alzheimer's disease risk.
-
3.
Future directions: Research into relationships between Alzheimer's disease risk and modifiable biomarkers may be important in Alzheimer's prevention or intervention.
Acknowledgments
The authors thank Danielle Posthuma's group for making available the Alzheimer's disease summary statistics, which can be accessed at https://ctg.cncr.nl/software/summary_statistics.
Authors' contributions: S.A.G.T. planned and carried out all analyses and wrote the manuscript.
Footnotes
Conflict of interest: None.
References
- 1.Green R., Allen L.H., Bjørke-Monsen A.-L., Brito A., Guéant J.-L., Miller J.W. Vitamin B12 deficiency. Nat Rev Dis Primer. 2017;3:17040. doi: 10.1038/nrdp.2017.40. [DOI] [PubMed] [Google Scholar]
- 2.Bailey R.L., Carmel R., Green R., Pfeiffer C.M., Cogswell M.E., Osterloh J.D. Monitoring of vitamin B-12 nutritional status in the United States by using plasma methylmalonic acid and serum vitamin B-121234. Am J Clin Nutr. 2011;94:552–561. doi: 10.3945/ajcn.111.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke R., Smith A.D., Jobst K.A., Refsum H., Sutton L., Ueland P.M. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- 4.Moore E.M., Ames D., Mander A.G., Carne R.P., Brodaty H., Woodward M.C. Among vitamin B12 deficient older people, high folate levels are associated with worse cognitive function: combined data from three cohorts. J Alzheimers Dis. 2014;39:661–668. doi: 10.3233/JAD-131265. [DOI] [PubMed] [Google Scholar]
- 5.Grarup N., Sulem P., Sandholt C.H., Thorleifsson G., Ahluwalia T.S., Steinthorsdottir V. Genetic architecture of vitamin B12 and folate levels uncovered applying deeply sequenced large datasets. Plos Genet. 2013;9:e1003530. doi: 10.1371/journal.pgen.1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsson S.C., Traylor M., Malik R., Dichgans M., Burgess S., Markus H.S. Modifiable pathways in Alzheimer's disease: Mendelian randomisation analysis. BMJ. 2017;359:j5375. doi: 10.1136/bmj.j5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M.G., Usala G. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet. 2009;84:477–482. doi: 10.1016/j.ajhg.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazra A., Kraft P., Lazarus R., Chen C., Chanock S.J., Jacques P. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet. 2009;18:4677–4687. doi: 10.1093/hmg/ddp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keene K.L., Chen W.-M., Chen F., Williams S.R., Elkhatib S.D., Hsu F.-C. Genetic Associations with Plasma B12, B6, and Folate Levels in an Ischemic Stroke Population from the Vitamin Intervention for Stroke Prevention (VISP) Trial. Front Public Health. 2014;2:112. doi: 10.3389/fpubh.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X., Lu D., Gao Y., Tao S., Yang X., Feng J. Genome-wide association study identifies novel loci associated with serum level of vitamin B12 in Chinese men. Hum Mol Genet. 2012;21:2610–2617. doi: 10.1093/hmg/dds062. [DOI] [PubMed] [Google Scholar]
- 11.Nongmaithem S.S., Joglekar C.V., Krishnaveni G.V., Sahariah S.A., Ahmad M., Ramachandran S. GWAS identifies population-specific new regulatory variants in FUT6 associated with plasma B12 concentrations in Indians. Hum Mol Genet. 2017;26:2551–2564. doi: 10.1093/hmg/ddx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. 2019;1 doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Causal associations between risk factors and common diseases inferred from GWAS summary data | Nature Communications. https://www.nature.com/articles/s41467-017-02317-2 [DOI] [PMC free article] [PubMed]
- 15.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4626747/ [DOI] [PMC free article] [PubMed]
- 17.Burgess S., Scott R.A., Timpson N.J., Davey Smith G., Thompson S.G. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeMeur N., Lawrence M., Bar M., Tewari M., Gentleman R. R and bioconductor packages in bioinformatics: towards systems biology. In: Lee J.K., editor. Stat. Bioinforma. John Wiley & Sons, Inc.; 2010. pp. 309–338. [Google Scholar]
- 19.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verbanck M., Chen C.-Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]