Abstract
Glucosamine (GlcN) and its acetylated derivative N-acetylglucosamine (GlcNAc) are widely used in the pharmaceutical industries. Here, we attempted to achieve efficient production of GlcNAc via genomic engineering of Corynebacterium glutamicum. Specifically, we ligated the GNA1 gene, which converts GlcN-6-phosphate to GlcNAc-6-phosphate by transferring the acetyl group in Acetyl-CoA to the amino group of GlcN-6-phosphate, into the plasmid pJYW4 and then transformed this recombinant vector into the C. glutamicum ATCC 13032, ATCC 13869, ATCC 14067, and S9114 strains, and we assessed the GlcNAc titers at 0.5 g/L, 1.2 g/L, 0.8 g/L, and 3.1 g/L from each strain, respectively. This suggested that there were likely to be significant differences among the key genes in the glutamate and GlcNAc synthesis pathways of these C. glutamicum strains. Therefore, we performed whole genome sequencing of the S9114 strain, which has not been previously published, and found that there are many differences among the genes in the glutamate and GlcNAc synthesis pathways among the four strains tested. Next, nagA (encoding GlcNAc-6-phosphate deacetylase) and gamA (encoding GlcN-6-phosphate deaminase) were deleted in C. glutamicum S9114 to block the catabolism of intracellular GlcNAc, leading to a 54.8% increase in GlcNAc production (from 3.1 to 4.8 g/L) when grown in a shaker flask. In addition, lactate synthesis was blocked by knockout of ldh (encoding lactate dehydrogenase); thus, further increasing the GlcNAc titer to 5.4 g/L. Finally, we added a key gene of the GlcN synthetic pathway, glmS, from different sources into the expression vector pJYW-4-ceN, and the resulting recombinant strain CGGN2-GNA1-CgglmS produced the GlcNAc titer of 6.9 g/L. This is the first report concerning the metabolic engineering of C. glutamicum, and the results of this study provide a good starting point for further metabolic engineering to achieve industrial-scale production of GlcNAc.
Keywords: C. glutamicum, Complete genome, N-acetylglucosamine, Metabolic engineering
1. Introduction
N-acetylglucosamine (GlcNAc) is widely present in nature as a derivative of glucose. This monosaccharide is mainly used as an additive in medicine, food, health care, and other related fields, and has been shown to effectively treat arthritis and participate in liver and kidney detoxification and liver protection [1,2]. Moreover, GlcNAc has been widely used as a food ingredient in many developed countries. At present, the production methods of GlcNAc mainly include chitin hydrolysis, biotransformation, and microbial synthesis. Compared with the former two, the production method via microbial synthesis has many advantages, including a short production time, high yield, high efficiency, and limited environmental impacts.
To date, the strains used for glucosamine (GlcN) and GlcNAc production include the fungi Aspergillus sp. BCRC 31742 [2,3] as well as recombinant Escherichia coli [4,5] and Bacillus subtilis [[6], [7], [8]] strains. By pathway design and modular optimization, the titer of GlcNAc can reach as high as 17 g/L and 13.2 g/L by recombinant E. coli and B. subtilis in shake flasks [[6], [7], [8]]. For fermentation of filamentous fungal, GlcN production from the fungal cell wall requires acid hydrolysis and a long culture period; the low productivity of this approach weakens its economic competitiveness when compared to conventional extraction approaches. In addition to filamentous fungal fermentation, GlcN and GlcNAc can also be produced using engineered E. coli or B. subtilis. E. coli is not generally recognized as safe (GRAS) for production of food additives, which limits the applications of the GlcN and GlcNAc produced from this bacteria in regard to the nutraceutical and pharmaceutical industries [9]. Moreover, B. subtilis typically produces spores in the late stage of fermentation, and this affects the fermentation production process [10]. Therefore, in this study, we selected an alternative host bacterium, Corynebacterium glutamicum, and engineered these strains to generate production levels of GlcNAc.
C. glutamicum is currently the most widely used corynebacterium in industrial fermentation for the production of amino acids, including the strains ATCC 13032, ATCC 13869, ATCC 14067, and other subspecies [11,12]. In recent years, the entire genomes of C. glutamicum ATCC 13032, C. glutamicum ATCC 13869, and C. glutamicum ATCC 14067 have been sequenced, creating favorable conditions for the metabolic engineering of C. glutamicum in regard to high-yield amino acid production [[13], [14], [15], [16]]. However, none of the three strains have the capability to produce glutamate, which is the prerequisite for GlcNAc production [[17], [18], [19], [20], [21], [22], [23]]. Therefore, we selected the strain S9114, with its high-yield glutamate capability, for comparison with the three previously mentioned strains. This strain has been registered and preserved in the China Center for Type Culture Collection, and the collection number is CCTCCAB2019183. However, the complete genome sequence of S9114 has not been previously released.
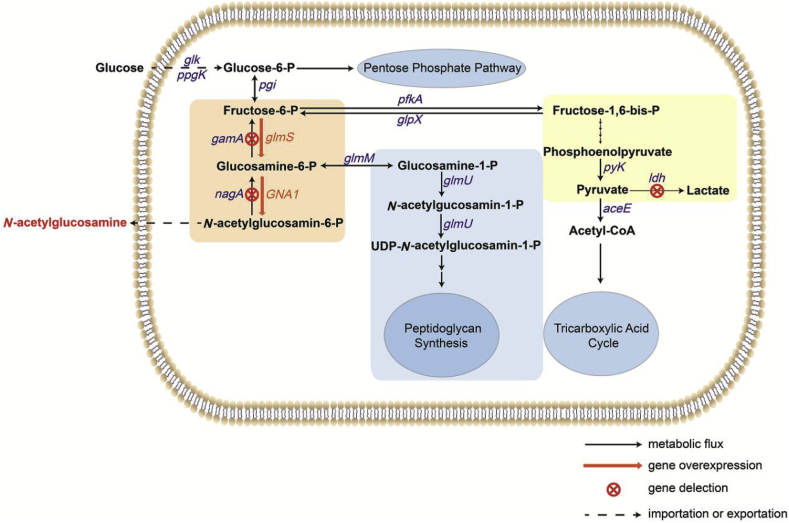
Although C. glutamicum can absorb glucose as a carbon source, the pathway for glucose synthesis of GlcNAc in C. glutamicum is incomplete. Therefore, we first exogenously expressed the GNA1 gene (encoding glucosamine-6-phosphate acetyltransferase, which converts GlcN-6-phosphate to GlcNAc-6-phosphate by transferring the acetyl group in Acetyl-CoA to the amino group of GlcN-6-phosphate, and not present in C. glutamicum genome) from Caenorhabditis elegans, making C. glutamicum capable of producing GlcNAc. The GNA1 gene was ligated into the plasmid pJYW4 [20], and the recombinant vector was verified. We transformed this recombinant vector into the C. glutamicum ATCC 13032, ATCC 13869, ATCC 14067, and S9114 strains, and initially selected the host strain expressing the highest levels of GlcNAc. Second, in order to improve the expression levels of glmS (encoding GlcN-6-phosphate synthase, which is the first rate-limiting enzyme in GlcNAc synthesis pathway and converts fructose-6-phosphate to GlcN-6-phosphate with glutamine acting as an amino donor), we added glmS genes from different sources downstream of the GNA1 gene and selected for the glmS gene with the highest expression. Third, in order to better perform genome editing of the S9114 strain, we performed genome-wide sequencing. The nagA (encoding GlcNAc-6-phosphate deacetylase) and gamA (encoding GlcN-6-phosphate deaminase) genes were knocked out, as based on their genomic sequences, to block the degradation of GlcNAc by the C. glutamicum host strain. In addition, to eliminate the formation of major acidic by-products, we completely blocked the lactate synthesis by knocking out ldh (encoding lactate dehydrogenase). After a series of transformations, we successfully introduced a heterologous synthetic pathway for GlcNAc and optimized the endogenous GlcNAc related metabolic pathways of C. glutamicum (Fig. 1). Importantly, this study is the first to reveal the whole genome sequence of C. glutamicum S9114, and this will prove useful for the metabolic engineering of C. glutamicum S9114 for the production of other products in the future.
Fig. 2.
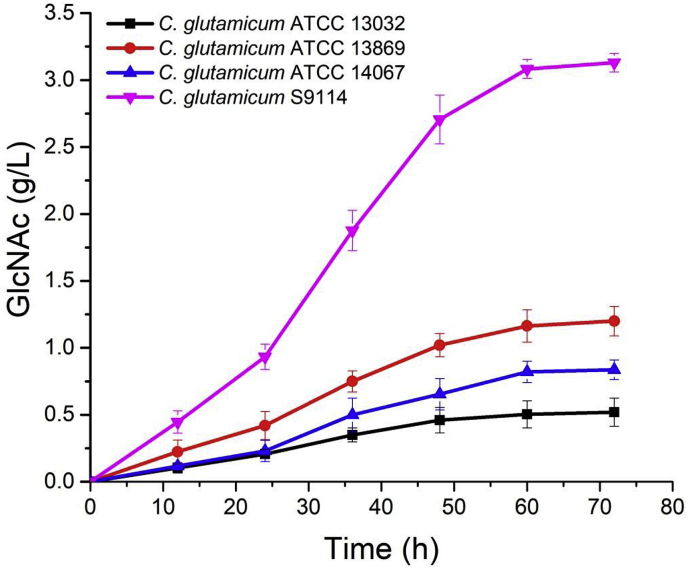
The GlcNAc titers of C. glutamicum ATCC 13032-GNA1 (black line), C. glutamicum ATCC 13869-GNA1 (red line), C. glutamicum ATCC 14067-GNA1 (blue line), and C. glutamicum S9114-GNA1 (pink line). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 1.
The metabolic pathway of N-acetylglucosamine (GlcNAc) synthesis in C. glutamicum.
2. Materials and methods
2.1. Bacterial strains, plasmids, and materials
The primers, strains, and plasmids used in this study are shown in Table 1, Table 2. The C. glutamicum ATCC 13032, ATCC 13869, ATCC 14067, and S9114 strains were used as initial host bacteria. C. glutamicum ATCC 13032, ATCC 13869, and ATCC 14067 are laboratory-preserved strains, and S9114 strain was obtained from Dr. Liming Liu (State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, P. R. China). Plasmid pJYW-4 was a gift from Dr. Xiaoyuan Wang (State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, P. R. China). The recombinant plasmid pJYW-4-ceN, which was used as the expression vector, was constructed as detailed in our previous work [19].
Table 1.
Primers used in this study.
| Primer | Sequence (5’→3′) |
|---|---|
| na-galoxPU F | ATCTCGAGGGTCAAAGGTGATTTCACCGGCGAATT |
| na-galoxPU R | ACCTCTAGAGGAAACGGCCACATCGCTTTCAATGAGC |
| na-galoxPD F | ACGGATCCAGGAAACGCGCCACCGTTTCC |
| na-galoxPD R | CCGGAATTCTCCTTGGTGCCTGCAAGAACGCCA |
| kan-loxp-F | ACCTCTAGAGCGCAATTAACCCTCACTAAAG |
| kan-loxp-R | ATGGATCCAATACGACTCACTATAGGGCG |
| ldhloxPU F | ATCTCGAGGACCTTAATTGCATCGACTGCTTGT |
| ldhloxPU R | ACCTCTAGACCCCAACCCATTACGTTGGTG |
| ldhloxPD F | ACGGATCCCGGCGAATTAACCCAGCAC |
| ldhloxPD R | CCGGAATTCCTTGCTTGGGAGTTTTCAACCATTC |
| loxpy F | AACCGCTGGCAGGTCCGTCGATA |
| loxpy R | CCAGCAGTTACGGGAACGCGG |
| C.gl-glmS F | GCGCTTTTAAGCTGCAACTTAATTATGGTCCTCCC |
| C.gl-glmS R | CTTTGCTAGTTTATTCGACGGTGACAGACTTTGC |
| B.su-glmS F | CAGCGCTTTTAACCAAAAAACATGTAGGAGGGGACG |
| B.su-glmS R | TCCTTTGCTAGTTTACTCCACAGTAACACTCTTCGCAAG |
| E.co-glmS F | GCGCTTTTAAATCCCGCGAAATTAATACGACTCAC |
| E.co-glmS R | TCCTTTGCTAGTTTACTCAACCGTAACCGATTTTGCC |
| P4–C.gl-glmS F | CGTCGAATAAACTAGCAAAGGAGAAGAAAAGCCG |
| P4–C.gl-glmS R | TAAGTTGCAGCTTAAAAGCGCTGGGTCATAAAATTACAGTCA |
| P4–B.su-glmS F | CTGTGGAGTAAACTAGCAAAGGAGAAGAAAAGCCGGA |
| P4–B.su-glmS R | CATGTTTTTTGGTTAAAAGCGCTGGGTCATAAAATTACAGTCA |
| P4-E.co-glmS F | CGGTTGAGTAAACTAGCAAAGGAGAAGAAAAGCCGGA |
| P4-E.co-glmS R | TTCGCGGGATTTAAAAGCGCTGGGTCATAAAATTACAGT |
Table 2.
Strains and plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| Strains | ||
| C. glutamicum ATCC 13032 | Wild type C. glutamicum | Laboratory stock |
| C. glutamicum ATCC 13869 | Wild type C. glutamicum | Laboratory stock |
| C. glutamicum ATCC 14067 | Wild type C. glutamicum | Laboratory stock |
| C. glutamicum S9114 | Wild type C. glutamicum | Laboratory stock |
| C. glutamicum ATCC 13032-GNA1 | C. glutamicum ATCC 13032 derivate, overexpression of C. elegans GNA1 gene | This work |
| C. glutamicum ATCC 13869-GNA1 | C. glutamicum ATCC 13869 derivate, overexpression of C. elegans GNA1 gene | This work |
| C. glutamicum ATCC 14067-GNA1 | C. glutamicum ATCC 14067 derivate, overexpression of C. elegans GNA1 gene | This work |
| C. glutamicum s9114-GNA1 | C. glutamicum S9114 derivate, overexpression of C. elegans GNA1 gene | This work |
| CGGN1 | C. glutamicum S9114 derivate: ΔnagA-ΔgamA | This work |
| CGGN1-GNA1 | CGGN1 derivate, overexpression of C. elegans GNA1 gene | This work |
| CGGN2 | CGGN1 derivate: ΔnagA-ΔgamA-Δldh | This work |
| CGGN2-GNA1 | CGGN2 derivate, overexpression of C. elegans GNA1 gene | This work |
| CGGN2-GNA1-CgglmS | CGGN2-GNA1 derivate, overexpression of C. glutamicum glmS gene | This work |
| CGGN2-GNA1-BsglmS | CGGN2-GNA1 derivate, overexpression of B. subtilis glmS gene | This work |
| CGGN2-GNA1-EcglmS | CGGN2-GNA1 derivate, overexpression of E. coli glmS gene | This work |
| Plasmids | ||
| pJYW-4-ceN | pJYW-4 derivate, Ptac-GNA1,Kanr | [19] |
| pTYW-4-ceN-C.glglmS | pJYW-4-ceN derivate with C.gglmS cloned | This work |
| pTYW-4-ceN-B.suglmS | pJYW-4-ceN derivate with B, sglmS cloned | This work |
| pTYW-4-ceN-E.coglmS | pJYW-4-ceN derivate with E.cglmS cloned | This work |
| pBluescript IISK(+) | Cloning vector | [18] |
| pDTW-109 | Vector carry Cre | [18] |
| pDTW-202 | pBluescript IISK(+) carry the segment loxp-kan-loxp, Ampr | [18] |
| pDTW-NG | pBluescript IISK(+) carry the nagA/gamA genes knockout cassette | This work |
| pDTW-LDH | pBluescript IISK(+) carry the ldh gene knockout cassette | This work |
Ampr, ampicillin resistance; Kanr, kanamycin resistance.
All microorganisms were grown at 30 °C in LBB broth (Luria–Bertani broth with 18.5 g/L brain heart infusion) or on LBB agar plates. LB consists of 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl). The seed medium contained the following components in units of g/L: glucose, 25.0; corn syrup, 20.0; KH2PO4, 1.0; (NH4) 2SO4, 0.5; and urea, 1.25; and was adjusted to a final pH of 7.0. The fermentation medium contained the following components in units of g/L: glucose, 100.0; corn syrup, 10.0; KH2PO4, 1.0; (NH4) 2SO4, 20.0; MgSO4, 0.5; CaCO3, 20.0; and FeSO4, 0.18 adjusted to a final pH of 7.0. A total of 25 mg/L kanamycin was added to all media for detection of transformants or for recombinant culture. A single colony grown on the LBB plate was inoculated into the seed culture medium. After 16–18 h of culture, the bacterial culture medium of the seed culture was inoculated into the fermentation medium at an initial OD562 of 1.6 and grown for 72 h. Samples were taken every 12 h.
The E. coli strain JM109 was used as the host for plasmid construction. The PrimeSTAR HS DNA polymerase, restriction endonucleases, PCR reagents, Genomic Extraction Kit, Blunting Kination Ligation (BKL) Kit, and DNA purification kit were purchased from Takara (Dalian, China). The ClonExpress II One Step Cloning Kit was purchased from Vazyme (Nanjing, China). All other chemicals and reagents were of analytical grade. Sequence determinations were completed by Sangon Biological Engineering Technology & Services Co., Ltd (Shanghai, China).
2.2. DNA manipulation and construction of plasmids
The isolation and manipulation of recombinant DNA were performed using standard protocols. C. glutamicum transformation was performed via electrotransformation. The modified DNA fragments and plasmids were sequenced by Sangon Biotech Co., Ltd. (China). The C. glutamicum ATCC 13032, ATCC 13869, ATCC 14067, and S9114 strains were prepared into competence according to the preparation method of C. glutamicum competence. The recombinant plasmid pJYW-4-ceN, containing the Caenorhabditis elegans gene GNA1, was transformed into competent C. glutamicum cells, and transformants were selected to perform the fermentation. After 72 h of fermentation, we assessed the accumulation of GlcNAc in the different strains over time during the fermentation process.
2.3. De novo genome assembly of C. glutamicum S9114
The genome of C. glutamicum S9114 was sequenced by integrating Illumina paired-end sequencing and PacBio single-molecule real-time sequencing technologies. Terminal degrees and long readings were obtained on the Illumina HiSeq 2000 platform and the PacBio sequencing platform, respectively. The Illumina readings were processed using NGS QC Toolkit [24] version 2.3 software for quality inspection and low-quality data filtering. The PacBio reads were corrected with high-quality Illumina reads using PacBioToCA [25] and then assembled using Celera Assembler [26] version 8.0. All coding genes were annotated using Prokaryotic Genome Annotation Pipeline (PGAP) version 2.9 software at the NCBI (http://www.ncbi.nlm.nih.gov/genome/annotation prok/) and Rapid Annotation using the Subsystem Technology (RAST; http://rast.nmpdr.org/) server [27]. Additional functional annotations were performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) database [28]. The metabolic pathways were established using the KEGG automatic annotation server (KAAS; http://www.genome.jp/tools/kaas/).
2.4. Gene knockouts
The method for constructing knockout mutations in C. glutamicum is described in Hu et al. [29]. The pBluscriptII SK (+) plasmid was preserved in the laboratory as a starting plasmid. This plasmid contained only E. coli replicons and no C. glutamicum replicon. Therefore, the plasmid could not be autonomously replicated in C. glutamicum, facilitating the removal of the plasmid in subsequent procedures.
First, we designed knockout cassettes for the nagA and gamA genes to block the pathway by which the host strain C. glutamicum degrades GlcNAc. To increase the knockout efficiency, the length of the front and back regions was designed to be approximately 1000 bp. Using the C. glutamicum S9114 genomic sequence as a template, primers na-galoxPU F, na-galoxPU R, na-galoxPD F, and na-galoxPD R were used in PCR to amplify the homologous arm fragments na-gaU and na-gaD of the nagA-gamA gene.
The DNA fragment was recovered using a gel recovery kit. The plasmid pDTW202 carrying the Kana resistance gene fragment loxp-kan-loxp cassette was used as a template, and the kan-loxp-F and kan-loxp-R primers were used to amplify the loxp-kan-loxp cassette. The E. coli-derived pBluscriptII SK(+) plasmid was extracted using a plasmid extraction kit, and the DNA concentration was determined. The fragment na-gaU was digested with XhoI and XbaI, the fragment loxp-kan-loxp cassette was digested with XbaI and BamHl, the fragment na-gaD was digested with BamHI and EcoRI, and the plasmid pBluscriptII SK(+) was digested with XhoI and EcoRI. All the digested fragments were recovered using a gel recovery kit, and the DNA concentrations were determined. The enzymatically purified fragments na-gaU, na-gaD, and loxp-kan-loxp cassette were ligated to the pBluscriptII SK(+) plasmid using T4 DNA ligase at a molar ratio of 1:3 (plasmid: fragment). The ligation was then transformed into competent E. coli JM109 cells and plated onto Kana-resistant LB plates for screening. The resulting correct transformants carried the plasmid pDTW-NG, which harbored the nagA/gamA knockout cassette.
The correctly sequenced pDTW-NG plasmid was transformed into the C. glutamicum S9114 strain and then coated onto Kana-resistant LBB plates. The upstream and downstream homologous arms of nagA/gamA underwent homologous recombination with the upstream and downstream homologous arms of the C. glutamicum S9114 genome nagA/gamA genes, with the loxp-kan-loxp cassette replacing the nagA/ gamA gene fragment within the genome, thereby achieving the goal of deletion of these genes in the genome. The obtained recombinant genome carried the loxp-kan-loxp cassette fragment, and the lox sequence at both ends of the Kana resistance gene could be specifically recognized and cleaved by the Cre enzyme. The plasmid pDTW-109 carrying the Cre gene was further electroporated into the mutant strain. In addition to carrying the Cre gene, this plasmid also carried a temperature-sensitive replicon (normal copy at 25 °C, cannot copy at 37 °C or higher) and a chloramphenicol resistance gene. LBB plates containing kanamycin and chloramphenicol were separately streaked at 25 °C, and transformants that grew only on the chloramphenicol plates were selected and further cultured in a 37 °C incubator to remove the pDTW-109 plasmid. Finally, primers loxpy F and loxpy R were used to verify the strains containing both homologous arms bound to the genome. The resulting correct transformant was named C. glutamicum S9114-ΔnagA-ΔgamA (CGGN1).
The ldh gene was knocked out using the same method as for the nagA and gamA genes. The primers for amplifying the left and right arms of the ldh gene knockout frame were: ldhloxPU F, ldhloxPU R, ldhloxPD F, and ldhloxPD R. The plasmid with the ldh gene knockout cassette was named pDTW-LDH. The transformant with the further knock out of the ldh gene was named C. glutamicum S9114-ΔnagA-ΔgamA-Δldh (CGGN2).
2.5. Enhancing gene expression
In our previous work, the expression vector pTYW-4 was used to clone the gene encoding glucosamine deacetylase, GNA1, of C. elegans, which was ligated into plasmid pTYW-4-ceN, which was constructed as an expression vector for the GNA1 gene. Additionally, in order to increase the expression level of another key gene, glmS, in the GlcNAc synthesis pathway, we added the glmS gene derived from C. glutamicum S9114, B. subtilis 168, or E. coli K-12 MG1655 to the 3′ untranslated region of the GNA1 gene, allowing it to be co-expressed with GNA1 in the same expression frame.
The glmS genes were amplified from C. glutamicum S9114, B. subtilis 168, and E. coli K-12 MG1655 using the primers C.gl-glmS F and C.gl-glmS R, B.su-glmS F and B.su-glmS R, and E.co-glmS F and E.co-glmS R, respectively. The linearized pTYW-4-ceN vectors were amplified with the primers P4–C.gl-glmS F and P4–C.gl-glmS R, P4–B.su-glmS F and P4–B.su-glmS R, and P4-E.co-glmS F and P4-E.co-glmS R. The glmS genes and the linearized pTYW-4-ceN vector were then ligated together via Gibson assembly, yielding the recombinant plasmids pTYW-4-ceN-C.glglmS, pTYW-4-ceN-B.sulglmS, and pTYW-4-ceN-E.coglmS. All of the recombinant plasmids were then transformed into the C. glutamicum S9114-ΔnagA-gamA strain via electroporation.
2.6. Analytic methods
The concentrations of GlcNAc and lactate in the fermentation broth were measured via high-performance liquid chromatography on an instrument equipped with an HPX-87H column (Bio-Rad Hercules, CA, USA) and a refractive index detector. High-performance liquid chromatography was carried out with 5 mM H2SO4 as the mobile phase with a flow rate of 0.6 mL/min at 35 °C. To assess for intracellular GlmS and GNA1 activities, cells were harvested during fermentation via centrifugation at 10,000×g and 4 °C for 10 min and then disrupted with ultrasonic oscillation (Ultrasonic processor, Sonics Vibra-Cell, Ultrasonic processor VCX 750 W; Sonics & Materials, Newtown, CT, USA; amplitude 30%, time 30 min). After centrifugation at 10,000×g and 4 °C for 10 min, the supernatant was used to determine the GlmS activity levels [8]. One unit of GlmS activity was defined as the amount of enzyme that produced 1 pmoL/min of GlcN-6-phosphate under the conditions assayed. Protein concentrations were detected using the protein concentration kit SK3041 from Sangon Biotech Co., Ltd. (China). The glucose concentration in the supernatant was measured using a glucose-glutamate analyzer (SBA-40C; Biology Institute of Shandong Academy of Sciences, Jinan, China).
2.7. Statistical analysis
All the experiments were performed independently at least three times and the results were expressed in means ± standard deviations (SD).
3. Results
3.1. Production of GlcNAc by the different C. glutamicum strains
After culture of the different C. glutamicum strains, it was found that the GlcNAc production of the C. glutamicum ATCC 13032, ATCC 13869, ATCC 14067, and s9114 strains were 0.5 g/L, 1.2 g/L, 0.8 g/L, and 3.1 g/L, respectively, after 72 h (Fig. 3B). Since the GlcNAc production of C. glutamicum S9114 was quite different from that of the other three strains, we would like to have compared and analyzed the genomic differences between the S9114 strain and the ATCC 13032, ATCC 13869, ATCC 14067 strains. However, the complete genome sequence of strain S9114 has not been previously published. As such, we sequenced the complete genome of strain S9114 by way of de novo genome assembly.
Fig. 3.
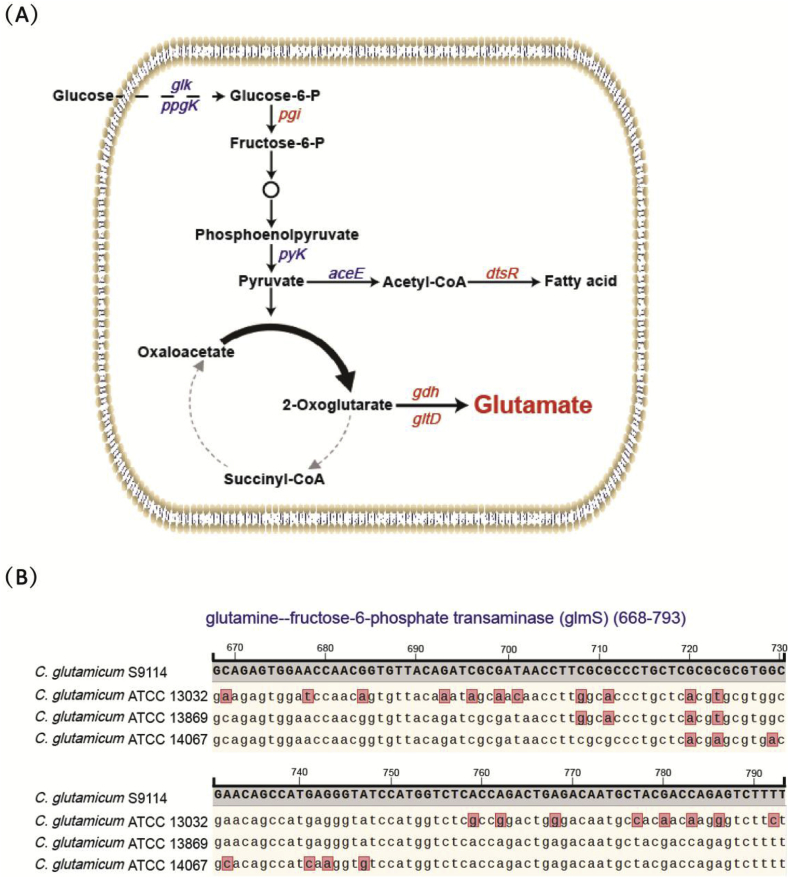
Key genes in the glutamate synthesis pathway in C. glutamicum and a homologous alignment of glmS genes. (A) Overview of the glutamate production mechanism in C. glutamicum. (B) A partial nucleotide sequence alignment map of the glmS genes from different sources. Bases within the red box represent non-conserved bases.
3.2. Whole genome sequencing of C. glutamicum S9114
The whole genome sequences of C. glutamicum ATCC 13032, ATCC 13869, and ATCC 14067 are all known, but the continuous whole genome sequence of C. glutamicum strain S9114 has not been published, making subsequent genome editing of the C. glutamicum strain S9114 difficult. Therefore, we utilized the new generation of bacterial genome de novo sequencing for generating the genome sequence of the S9114 strain. Bacterial genome de novo sequencing involves the de novo assembly of the bacterial genome after sequencing. The procedure first involves randomly cleaving the stained DNA of the bacteria into fragments of a certain relative molecular mass range and then to construct two sets of libraries for large-scale sequencing. The gaps in the sequence are filled after preliminary assembly. Gap filling is a key step, and it usually takes several combinations of assembly methods to complete, which can be time-consuming and require significant effort. The ability to complete the gap filling can often be the key to a complete genome-wide sequence. The final assembly level is based on the needs of the study and the characteristics of the bacteria itself. The highest indicator is a contig, which is the complete sequence of the genome. On the basis of the assembly, subsequent genomic component analysis, functional annotation, and other analyses are performed to determine whether an ORF is a real protein coding sequence as well as to check for functional sites, analyze consensus or characteristic sequences, assess for single gene or inter-gene interactions, and to define expression regulation and other functions. Bacterial de novo sequencing has replaced other traditional methods and is an important tool for studying the genetic mechanisms of bacterial evolution and identifying key functional genes.
The complete genome of C. glutamicum S9114 consists of a circular chromosome of 3,353,693 bp with a GC content of 54.86%. In total, 3097 identified protein-coding sequences (CDS) were encoded by 86.52% of the whole genomic DNA. A total of 59 tRNAs and 18 rRNA gene copies were also detected (Table 3). The genome data have been deposited in the NCBI Sequence Read Archive (SRA) (accession number: SRP198722).
Table 3.
Basic features of the C. glutamicum S9114 genome.
| Features | Chromosome |
|---|---|
| Length [bp] | 3,353,693 |
| Total number of genes | 3162 |
| G + C content [%] | 54.86% |
| CDS | 3097 |
| 5s_rRNA (De novo) | 6 |
| 16s_rRNA (De novo) | 6 |
| 23s_rRNA (De novo) | 6 |
| sRNA | 7 |
| tRNA | 59 |
We made a brief comparative analysis of genome sequence of ATCC 13032, ATCC 13869, ATCC 14067, and C. glutamicum S9114. The preliminary results showed that some notable sequences of some genes related to the glutamate, lysine, valine, serine and arginine synthesis pathway differences in these C. glutamicum strains, which might lead to difficulties in amino acid production rates. Genome analysis revealed that the genome of C. glutamicum S9114 had a complete set of genes for glutamate biosynthesis. Many base alterations were found within the key genes potentially relevant to GlcNAc synthesis, including key genes in the GlcNAc biosynthesis pathway (pgi, glmS), cell wall biosynthesis and glutamate transport (murE, ftsI, and dtsR), NADPH generation (zwf, gnd, malE,ppnk, etc.), biosynthesis pathways (pyc, pdh, pepC, odhA, and gltB), and key genes in the glutamate biosynthesis pathway (gdh, gltD, etc.) (Fig. 3A). According to the alignment of the genome, it was determined that the nucleotide sequences proximal the catalytic center of glmS of the three C. glutamicum strains, ATCC 13032, ATCC 13869, and ATCC 14067, differed from those of C. glutamicum S9114 (Fig. 3B). In particular, glmS of C. glutamicum ATCC 13032 and C. glutamicum S9114 have distinctly different sequences, and at least some of the base changes detected in this analysis may contribute to the high glutamate production capacity and high GlcNAc production capacity of S9114.
3.3. Effects of nagA/gamA deletion on the accumulation of GlcNAc in C. glutamicum
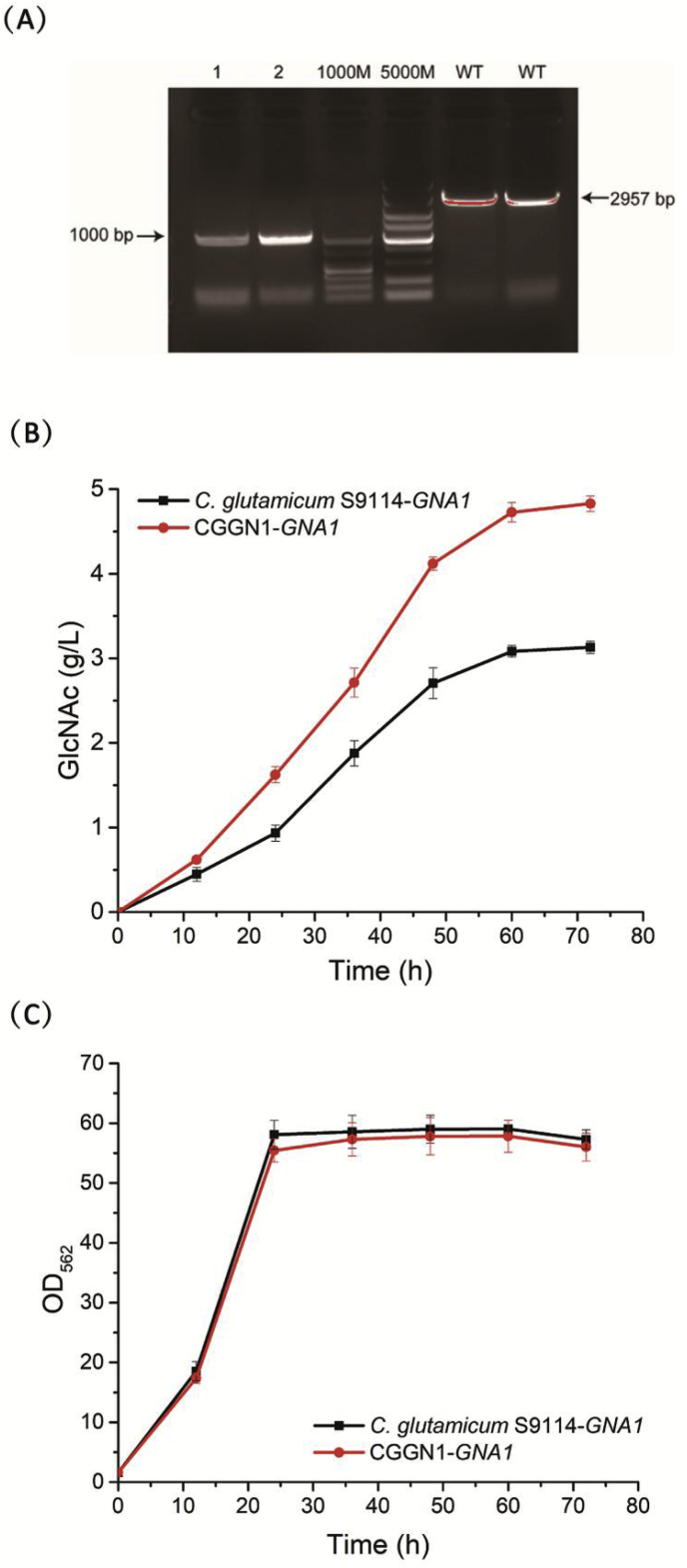
In C. glutamicum S9114, intracellular GlcNAc could still be degraded by N-deacetylation and deaminated by GlcNAc deacetylase (NagA) and GlcN deaminases (GamA) (Fig. 1) [6]. These intracellular catabolic reactions would hamper GlcNAc accumulation and should be eliminated to block GlcNAc catabolism. Therefore, we deleted the nagA and gamA genes in S9114 strain by homologous recombination, yielding the recombinant strain CGGN1, in which the GlcNAc and GlcN catabolic pathways were completely deleted. The deletion mutant strain CGGN1 was verified by colony PCR using the corresponding primers loxpy F and loxpy R (Fig. 4A), and the vector pJYW-4-ceN was transformed into CGGN1 to strengthen the GlcNAc biosynthetic pathway during the fermentation process. Fig. 4B shows the GlcNAc production of the metabolically engineered strains C. glutamicum S9114-GNA1 and CGGN1-GNA1, and it is apparent that the deletion of the nagA and gamA genes significantly enhanced GlcNAc production, indicating that deletion of amino sugar catabolism was highly effective for promoting the accumulation of GlcNAc. Deletion of the nagA and gamA genes thoroughly blocked the N-deacetylation and deamination processes and improved the GlcNAc production of CGGN1-GNA1 to 4.8 g/L, which was 54.8% higher than that produced by C. glutamicum S9114. In addition, it was determined that there was only a slight decrease in the OD562 with the knockout of the nagA and gamA genes, indicating that cell growth was not significantly impacted when the degradation pathway for GlcNAc was blocked in C. glutamicum (Fig. 4C).
Fig. 4.
Effects of the knockout of nagA and gamA genes on cell growth and GlcNAc production. (A) Confirmation of the knockout of nagA and gamA. 1000 M: DL1000 DNA marker; 5000 M: DL5000 DNA marker; all samples used loxpy F/loxpy R as validation primers; 1, 2: CGGN1ΔnagAΔgamA:kan as the template; WT: original S9114 strain as the template. (B) GlcNAc titers of C. glutamicum S9114-GNA1 (black line) and CGGN1-GNA1 (red line). (C) Cell growth of C. glutamicum S9114-GNA1 (black line) and CGGN1-GNA1 (red line).
3.4. Effects of blocking lactate formation on GlcNAc production
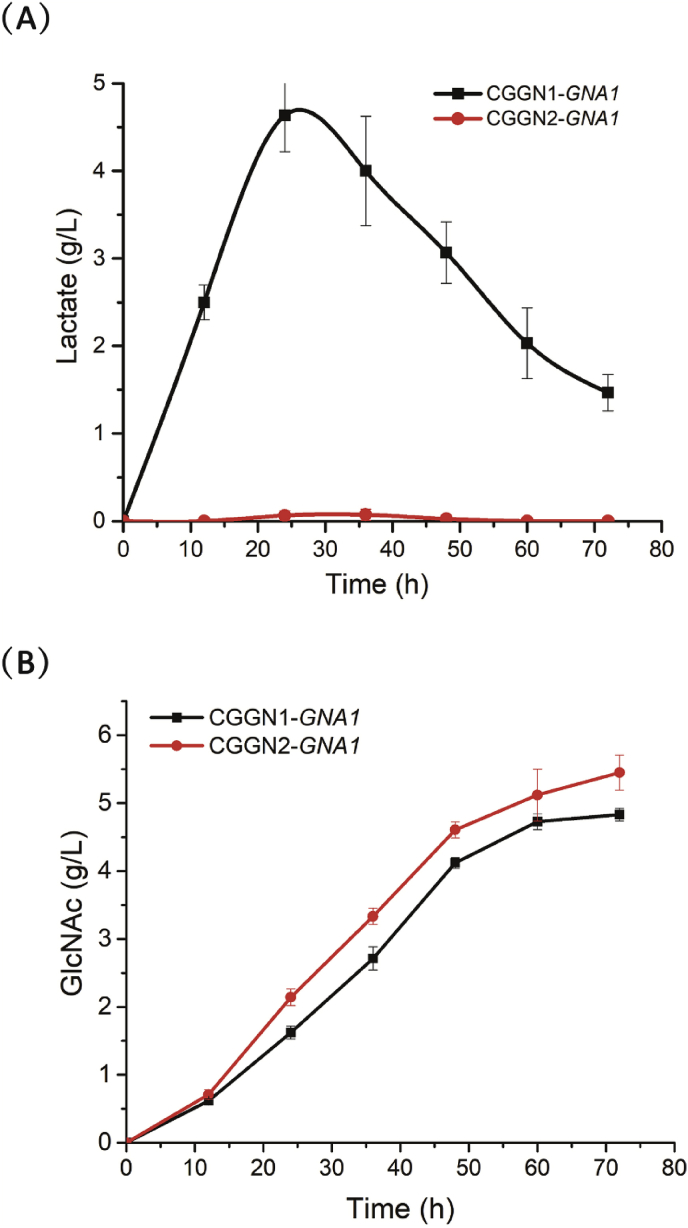
The accumulation of lactic acid, a by-product of the GlcNAc synthesis pathway, is toxic to cellular growth and competes with the synthesis of GlcNAc for carbon resources. To enhance GlcNAc synthesis, we used gene knockout to block the formation of lactate in C. glutamicum completely. First, the ldh gene encoding l-lactate dehydrogenase, which catalyzes the formation of lactate from pyruvate, was knocked out. As expected, no lactate was detected during fermentation of CGGN2-GNA1 (Fig. 5A). After blocking the lactate synthesis pathway, the OD562 was significantly enhanced, and the titer of GlcNAc was also increased to 5.4 g/L (Fig. 5B).
Fig. 5.
Effects of the knockout of ldh gene on GlcNAc production. (A) Concentration of the acidic by-product lactate during CGGN1-GNA1 (black line) and CGGN2-GNA1 (red line) culture. (B) GlcNAc titers of CGGN1-GNA1 (black line) and CGGN2-GNA1 (red line). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Influence of GNA1 and glmS overexpression on GlcN and GlcNAc production
Our laboratory has employed metabolic engineering to construct a recombinant C. glutamicum that can produce GlcNAc. Our previous study inserted the GNA1 gene into the pJYW-4 plasmid. In this study, the glmS gene, a key enzyme of the GlcN synthesis pathway, was added to this expression vector. Recombinant C. glutamicum CGGN2 strain was constructed by over-expressing GlmS from different sources, and the accumulation of the target product GlcNAc was examined. The three different source genes that express GlmS that were assessed in this experiment were CgglmS (from C. glutamicum, NCBI CP004048.1), BsglmS (from B. subtiiis, NCBI NC_ 000964.3), and EcglmS (from E. coli. K12, NCBI CP032667.1). All of the glmS genes were amplified from the genome of their corresponding strains. Finally, the glmS genes were each individually inserted into the 3′ untranslated region of the GNA1 gene.
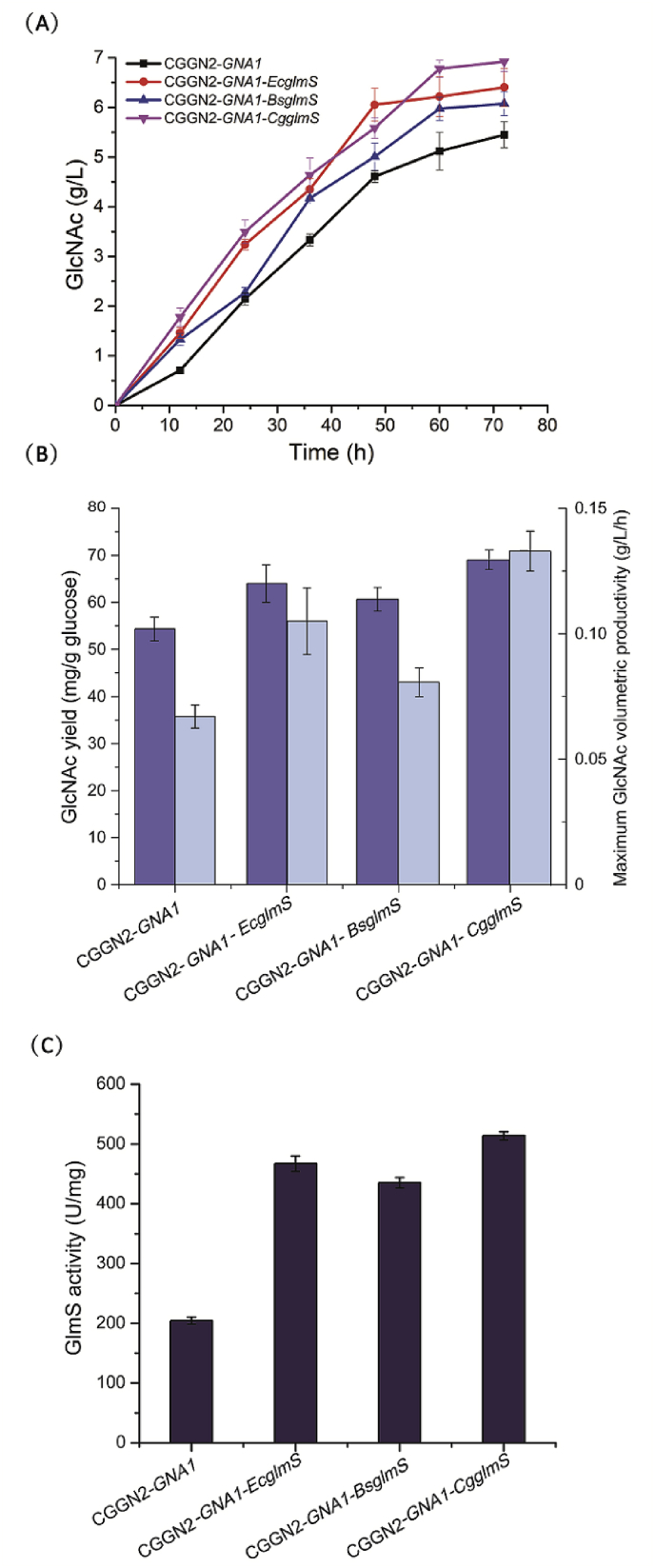
Three recombinant strains CGGN2-GNA1-CgglmS, CGGN2-GNA1-BsglmS, and CGGN2-GNA1-EcglmS were cultured in shaker flasks, and the yields of GlcNAc were sampled and tested. After fermentation, the GlcNAc titer was substantially enhanced after blockage of the lactate synthesis pathway (Fig. 6A). During the entire fermentation process, the GlcNAc titer of the three recombinant strains increased, and the accumulation of extracellular GlcNAc reached a maximum in the three recombinant strains at 72 h. It can be seen from Fig. 6 that the titer of GlcNAc in the CGGN2-GNA1-CgglmS strain increased rapidly in the early stage, and reached the maximum value of 6.9 g/L at 72 h, and the GlcNAc yields of GlcNAc on glucose is 69 mg/g. The titer of GlcNAc and the GlcNAc yields of GlcNAc on glucose in the recombinant strain CGGN2-GNA1-CgglmS were 7.8%, 13.4%, and 27.8% higher than that of CGGN2-GNA1-EcglmS (6.4 g/L, 64 mg/g), CGGN2-GNA1-BsglmS (6.1 g/L, 61 mg/g), and CGGN2-GNA1 (5.4 g/L, 54 mg/g) strains, respectively. The maximum GlcNAc volumetric productivity of strain CGGN2-GNA1, CGGN2-GNA1-EcglmS, CGGN2-GNA1-BsglmS and CGGN2-GNA1-CgglmS was 0.07 g/L/h, 0.11 g/L/h, 0.08 g/L/h, 0.13 g/L/h, respectively (Fig. 6B). Together, these observations indicated that the recombinant CGGN2-GNA1-CgglmS strain had the best cumulative production effect on the target product GlcNAc.
Fig. 6.
Shake flask fermentation of recombinant strains containing glmS from different sources. (A) GlcNAc titers of CGGN2-GNA1 (black line), CGGN2-GNA1-CgglmS (red line), CGGN2-GNA1-BsglmS (blue line), and CGGN2-GNA1-EcglmS (pink line). (B) Yields of GlcNAc on glucose (dark blue bar) and maximum GlcNAc volumetric productivity (wathet blue bar) in the recombinant strains. (C) The comparison of GlmS activity. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The intracellular fractions collected after cell lysis of the three recombinant strains were subjected to enzyme activity analyses. The results of the GlmS enzyme activity assays are shown in Fig. 6C. The GlmS activity of the recombinant strain CGGN2-GNA1-CgglmS was 513.8 U/mg, which was higher than the recombinant strains CGGN2-GNA1-EcglmS (467.1 U/mg), CGGN2-GNA1-BsglmS (435.4 U/mg), or CGGN2-GNA1 (204.6 U/mg), demonstrating an increase of 10%, 18%, and 104.6%, respectively. Together, these results demonstrated that the recombinant enzyme CgglmS from the CGGN2-GNA1-CgglmS strain demonstrated the best catalytic efficiency.
4. Discussion
C. glutamicum is a Gram-positive bacterium and has been widely used for the industrial production of amino acids, including monosodium glutamate, and lysine, and it can also utilize carbohydrate derivatives such as N-acetyl-glucosamine and sialic acid as carbon sources [13,30]. In recent years, with the gradual improvement of the genomic information and genome editing tools related to C. glutamicum, this bacterium has developed into a model microorganism capable of producing a variety of bio-based chemicals, materials, and fuels, including polygalacturonic acid, 2-ketoisovalerate, and succinate [31,32]. By way of metabolic engineering, C. glutamicum is also capable of producing amino acid derivatives, including amino sugars. Additionally, the production of non-amino–acid compounds by C. glutamicum has also been explored by a number of labs in recent years.
Table 4 shows the progress of GlcNAc production in different strains in recent years. E. coli produces endotoxin, which is not suitable for food and medicine, and lacks post-translational modification mechanism. Compared with E. coli, C. glutamicum has more advantages in food safety index and its metabolic pathway is simpler. In addition, the clarification of metabolic regulation mechanisms and the improvement of related vector systems in recent years have enabled us to carry out genome transformation of C. glutamicum by molecular biology methods. Although B. subtilis is a food safety strain, its physiological characteristics also have some deficiencies in industrial production. First of all, B. subtilis can form spores in the stable period, which increases the difficulty of controlling engineering bacteria in the production process and is easy to cause environmental pollution by engineering bacteria. Compared with B. subtilis, C. glutamicum has the advantages of rapid growth, no spore production, no secretion of extracellular protease, stable transcriptome, no odor generation during fermentation, etc. It is considered as a safe expression host and suitable for large-scale production and fermentation [33]. Compared to B. subtilis, C. glutamicum also has a superior ability to synthesize glutamate, and glutamate is an essential precursor of GlcNAc [17,34]. Therefore, this report details the construction of a C. glutamicum strain that can produce GlcNAc, and this was achieved via metabolic modification and optimization of genes expression.
Table 4.
Comparison of GlcNAc production by different strains.
| Strains | Culture method | Titer (g/L) | Source |
|---|---|---|---|
| E. coli 7107-18 | fed batch fermentation in 1-L fermentor | 110 | [9] |
| Aspergillus sp. BCRC 31742 | batch fermentation in 3-L fermentor | 14.37 | [3] |
| Bacillus subtilis 168 | fermentation in 500 mL shake flask culture | 5.19 | [6] |
| B. subtilis strain BSGN6-P xylA -glmS | fermentation in 500 mL shake flask culture | 13.2 | [7] |
| B. subtilis strain BNX122 | fed batch fermentation in 3-L fermentor | 103.1 | [40] |
| B. subtilis strain BSGN13 | fed batch fermentation in 3-L fermentor | 82.5 | [41] |
| C. glutamicum CGGN2-GNA1-CgglmS | fermentation in 500 mL shake flask culture | 6.9 | This work |
The most commonly used C. glutamicum strains mainly include the three subspecies: ATCC 13032, ATCC 13869, and ATCC 14067. In recent years, the C. glutamicum ATCC 13032, C. glutamicum ATCC 13869, and C. glutamicum ATCC 14067 genomes have been sequenced and published. However, due to differences in their genomes, they each have the ability to produce different amino acids: ATCC 13032 has the ability to produce lysine [35], ATCC 13869 has the ability to produce valine and serine [15], and ATCC 14067 is capable of high-yield arginine production [16]. Since the synthesis goal of this study was GlcNAc, which requires glutamate to provide amino groups, we utilized a high-yield glutamate strain, S9114, as one possible expression host. After fermentation, it was apparent that the S9114 strain has the strongest ability to produce GlcNAc among the strains tested. By comparing the nucleotide sequences of the glmS genes, a key gene in GlcNAc synthesis, among the four strains mentioned above, we found that there were many different nucleotides encoded at its key catalytic sites, resulting in obvious differences in the amino acid composition of GlmS among the different strains, which was likely one of the reasons for the differences in GlcNAc production among these strains. Based on genomic sequence comparison, we also found that some synthesis pathways involved in amino acid production in these four strains, such as glutamate, lysine, and other amino acids, also had sequence differences in important key genes, which would likely lead to different amino acid production capacities. These results provide indications regarding the importance of choosing the most appropriate starting strain in selection breeding experiments for the production of amino acids or related substances. In the future, we intend to produce other derivatives of amino acids or amino acids in C. glutamicum, and it will be important to rationally choose the starting strain according to the level of expression of key genes among the selected strains.
In previous studies, C. glutamicum has been used to produce amino acids and to produce other products, such as vitamins and succinic acid [12]. Because C. glutamicum has a good molecular basis for efficient transformation and a wide range of metabolic networks, this study attempted to produce saccharides using C. glutamicum. Based on alterations to the metabolic network, including knocking out the nagA and gamA genes, we were able to block the degradation of GlcNAc by the host strain, allowing intracellular GlcNAc to accumulate at increased levels.
In addition to the strength of the GlcNAc synthesis module, the formation of lactate and the competitive effects of glycolysis and peptidoglycan synthesis also affected the GlcNAc titer and yield. Therefore, only engineering the GlcNAc pathway was insufficient to enhance GlcNAc production substantially. For lactate synthesis pathways, which promote nonessential metabolism that competes for carbon resources and energy, we took a gene knockout approach to block the formation of acidic by-products completely. Reducing non-essential metabolism is a demonstrably effective approach for boosting cellular properties in optimized prokaryotic cell factories [36,37]. Therefore, further deletion of dispensable regions of C. glutamicum is a promising strategy for enhancing GlcNAc production. In follow-up experiments, we intend to continue to search for the GlcNAc-competitive pathways and knock out the non-essential genes of the growth of the bar in an effort to reduce production of by-products and, thus, optimize the production of GlcNAc.
Finally, overexpression of key genes is an important aspect of metabolic engineering. In B. subtilis, the metabolic pathway for GlcN production is strictly regulated, and the expression of GlmS, a key enzyme of the GlcN synthesis pathway, is controlled by a riboswitch [38,39]. When intracellular GlcN-6-P accumulates, GlcN-6-P binds to the GlmS riboswitch, and the transcription product of the GlmS-encoding gene is degraded, reducing the expression of GlmS. However, it is not clear whether a GlmS riboswitch exists in C. glutamicum, so we introduced additional glmS genes from different sources downstream of the GNA1 gene in the pJYW-4-ceN expression vector. Ultimately, it was found that the GlcNAc yield following the introduction of glmS from C. glutamicum was the highest. Additionally, comparing the structures of glmS from different sources allowed us to further our understanding regarding the key aspects of its catalysis. By comparing the nucleotide sequences of the glmS genes, we found that there were many different nucleotides encoded at its key catalytic sites, resulting in obvious differences in the amino acid composition of glmS among the different strains, which was likely one of the reasons for the differences in GlcNAc production among these strains. Readers can compare the similarities and differences of related genes according to their needs.In subsequent research, we intend to determine if the GlmS riboswitch is functional in C. glutamicum.
In conclusion, this study provided a food-safe bacteria strain for microbial production of non-shellfish GlcNAc, and this may widen the application of GlcNAc in the nutraceutical and pharmaceutical industries. Moreover, additional genome comparisons may provide additional avenues for altering C. glutamicum S9114 for further optimization of GlcNAc production.
Competing financial interests
The authors declare that they have no competing financial interests.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (31622001, 31671845, 31600068), the Natural Science Foundation of Jiangsu Province (BK20160176), and the 111 Project (111-2-06).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Kubomura D., Ueno T., Yamada M., Nagaoka I. Evaluation of the chondroprotective action of N-acetylglucosamine in a rat experimental osteoarthritis model. Exp Ther Med. 2017;14:3137–3144. doi: 10.3892/etm.2017.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sitanggang A.B., Wu H.S., Wang S.S., Ho Y.C. Effect of pellet size and stimulating factor on the glucosamine production using Aspergillus sp. BCRC 31742. Bioresour Technol. 2010;101:3595–3601. doi: 10.1016/j.biortech.2009.12.084. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., Liu L., Li J., Du G., Chen J. Enhanced glucosamine production by Aspergillus sp. BCRC 31742 based on the time-variant kinetics analysis of dissolved oxygen level. Bioresour Technol. 2012;111:507–511. doi: 10.1016/j.biortech.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Liu L., Li J., Du G., Chen J. Improved glucosamine and N-acetylglucosamine production by an engineered Escherichia coli via step-wise regulation of dissolved oxygen level. Bioresour Technol. 2012;110:534–538. doi: 10.1016/j.biortech.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Liu L., Li J., Liu J., Du G., Chen J. Optimization of glucose feeding approaches for enhanced glucosamine and N-acetylglucosamine production by an engineered Escherichia coli. J Ind Microbiol Biotechnol. 2012;39:359–365. doi: 10.1007/s10295-011-1046-0. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Liu L., Shin H.D., Chen R.R., Li J., Du G., Chen J. Pathway engineering of Bacillus subtilis for microbial production of N-acetylglucosamine. Metab Eng. 2013;19:107–115. doi: 10.1016/j.ymben.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Gu Y., Deng J., Liu Y., Li J., Shin H.D., Du G., Chen J., Liu L. Rewiring the glucose transportation and central metabolic pathways for overproduction of N-acetylglucosamine in Bacillus subtilis. Biotechnol J. 2017;12 doi: 10.1002/biot.201700020. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Zhu Y., Li J., Shin H.D., Chen R.R., Du G., Liu L., Chen J. Modular pathway engineering of Bacillus subtilis for improved N-acetylglucosamine production. Metab Eng. 2014;23:42–52. doi: 10.1016/j.ymben.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Deng M.D., Severson D.K., Grund A.D., Wassink S.L., Burlingame R.P., Berry A., Running J.A., Kunesh C.A., Song L., Jerrell T.A., Rosson R.A. Metabolic engineering of Escherichia coli for industrial production of glucosamine and N-acetylglucosamine. Metab Eng. 2005;7:201–214. doi: 10.1016/j.ymben.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Ma W., Liu Y., Shin H.D., Li J., Chen J., Du G., Liu L. Metabolic engineering of carbon overflow metabolism of Bacillus subtilis for improved N-acetyl-glucosamine production. Bioresour Technol. 2018;250:642–649. doi: 10.1016/j.biortech.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Drager A., Kronfeld M., Ziller M.J., Supper J., Planatscher H., Magnus J.B., Oldiges M., Kohlbacher O., Zell A. Modeling metabolic networks in C. glutamicum: a comparison of rate laws in combination with various parameter optimization strategies. BMC Syst Biol. 2009;3:5. doi: 10.1186/1752-0509-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto S., Gunji W., Suzuki H., Toda H., Suda M., Jojima T., Inui M., Yukawa H. Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol. 2012;78:4447–4457. doi: 10.1128/AEM.07998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B.J., Gaigalat L., Goesmann A., Hartmann M., Huthmacher K., Krämer R., Linke B., McHardy A.C., Meyer F., Möckel B., Pfefferle W., Pühler A., Rey D.A., Rückert C., Rupp O., Sahm H., Wendisch V.F., Wiegräbe I., Tauch A. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen C., Li Y., Hu J., Dong X., Wang X. Metabolic engineering of Corynebacterium glutamicum ATCC13869 for L-valine production. Metab Eng. 2015;29:66–75. doi: 10.1016/j.ymben.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi Y., Date M., Itaya H., Matsui K., Wu L.F. Functional analysis of the twin-arginine translocation pathway in Corynebacterium glutamicum ATCC 13869. Appl Environ Microbiol. 2006;72:7183–7192. doi: 10.1128/AEM.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Y., Liao J., Wu Z., Han S., Lin Y., Zheng S. Genome sequence of Corynebacterium glutamicum ATCC 14067, which provides insight into amino acid biosynthesis in coryneform bacteria. J Bacteriol. 2012;194:742–743. doi: 10.1128/JB.06514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei J., Xu N., Ye C., Liu L., Wu J. Reconstruction and analysis of a genome-scale metabolic network of Corynebacterium glutamicum S9114. Gene. 2016;575:615–622. doi: 10.1016/j.gene.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Dorfmueller H.C., Fang W., Rao F.V., Blair D.E., Attrill H., van Aalten D.M. Structural and biochemical characterization of a trapped coenzyme A adduct of Caenorhabditis elegans glucosamine-6-phosphate N-acetyltransferase 1. Acta Crystallogr D Biol Crystallogr. 2012;68:1019–1029. doi: 10.1107/S0907444912019592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng C., Lv X., Li J., Liu Y., Du G., Amaro R.L., Liu L. Synthetic repetitive extragenic palindromic (REP) sequence as an efficient mRNA stabilizer for protein production and metabolic engineering in prokaryotic cells. Biotechnol Bioeng. 2019;116:5–18. doi: 10.1002/bit.26841. [DOI] [PubMed] [Google Scholar]
- 20.Hu J., Li Y., Zhang H., Tan Y., Wang X. Construction of a novel expression system for use in Corynebacterium glutamicum. Plasmid. 2014;75:18–26. doi: 10.1016/j.plasmid.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira F.M., Mendoza-Hernandez G., Castaneda-Bueno M., Aparicio R., Fischer H., Calcagno M.L., Oliva G. Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli. J Mol Biol. 2006;359:308–321. doi: 10.1016/j.jmb.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Zeng L., Burne R.A. NagR differentially regulates the expression of the glmS and nagAB genes required for amino sugar metabolism by Streptococcus mutans. J Bacteriol. 2015;197:3533–3544. doi: 10.1128/JB.00606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elustondo P.A., White A.E., Hughes M.E., Brebner K., Pavlov E., Kane D.A. Physical and functional association of lactate dehydrogenase (LDH) with skeletal muscle mitochondria. J Biol Chem. 2013;288:25309–25317. doi: 10.1074/jbc.M113.476648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai M., Thompson R.C., Maher C., Contreras-Galindo R., Kaplan M.H., Markovitz D.M., Omenn G., Meng F. NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genomics. 2010;11(Suppl 4):S7. doi: 10.1186/1471-2164-11-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koren S., Schatz M.C., Walenz B.P., Martin J., Howard J.T., Ganapathy G., Wang Z., Rasko D.A., McCombie W.R., Jarvis E.D., Adam M.P. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denisov G., Walenz B., Halpern A.L., Miller J., Axelrod N., Levy S., Sutton G. Consensus generation and variant detection by Celera Assembler. Bioinformatics. 2008;24:1035–1040. doi: 10.1093/bioinformatics/btn074. [DOI] [PubMed] [Google Scholar]
- 27.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J., Tan Y., Li Y., Hu X., Xu D., Wang X. Construction and application of an efficient multiple-gene-deletion system in Corynebacterium glutamicum. Plasmid. 2013;70:303–313. doi: 10.1016/j.plasmid.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Costa-Riu N., Maier E., Burkovski A., Krämer R., Lottspeich F., Benz R. Identification of an anion-specific channel in the cell wall of the Gram-positive bacterium Corynebacterium glutamicum. Mol Microbiol. 2003;50:1295–1308. doi: 10.1046/j.1365-2958.2003.03754.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee J. Development and characterization of expression vectors for Corynebacterium glutamicum. J Microbiol Biotechnol. 2014;24:70–79. doi: 10.4014/jmb.1310.10032. [DOI] [PubMed] [Google Scholar]
- 32.Krause F.S., Blombach B., Eikmanns B.J. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yim S.S., An S.J., Choi J.W., Ryu A.J., Jeong K.J. High-level secretory production of recombinant single-chain variable fragment (scFv) in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;98:273–284. doi: 10.1007/s00253-013-5315-x. [DOI] [PubMed] [Google Scholar]
- 34.Yang J., Yang S. Comparative analysis of Corynebacterium glutamicum genomes: a new perspective for the industrial production of amino acids. BMC Genomics. 2017;18:940. doi: 10.1186/s12864-016-3255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanase M., Aikoh T., Sawada K., Ogura K., Hagiwara T., Imai K., Wada M., Yokota A. Pyruvate kinase deletion as an effective phenotype to enhance lysine production in Corynebacterium glutamicum ATCC13032: redirecting the carbon flow to a precursor metabolite. J Biosci Bioeng. 2016;122:160–167. doi: 10.1016/j.jbiosc.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 36.Manabe K., Kageyama Y., Morimoto T., Ozawa T., Sawada K., Endo K., Tohata M., Ara K., Ozaki K., Ogasawara N. Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874. Appl Environ Microbiol. 2011;77:8370–8381. doi: 10.1128/AEM.06136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morimoto T., Kadoya R., Endo K., Tohata M., Sawada K., Liu S., Ozawa T., Kodama T., Kakeshita H., Kageyama Y., Manabe K., Kanaya S., Ara K., Ozaki K., Ogasawara N. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res. 2008;15:73–81. doi: 10.1093/dnares/dsn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu T., Liu Y., Li J., Koffas M., Du G., Alper H.S., Liu L. Engineering a glucosamine-6-phosphate responsive glmS ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of N-acetylglucosamine. ACS Synth Biol. 2018;7:2423–2435. doi: 10.1021/acssynbio.8b00196. [DOI] [PubMed] [Google Scholar]
- 39.Brooks K.M., Hampel K.J. Rapid steps in the glmS ribozyme catalytic pathway: cation and ligand requirements. Biochemistry. 2011;50:2424–2433. doi: 10.1021/bi101842u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y., Chen T., Liu Y., Lv X., Li J., Du G., Ledesma-Amaro R., Liu L. CRISPRi allows optimal temporal control of N-acetylglucosamine bioproduction by a dynamic coordination of glucose and xylose metabolism in Bacillus subtilis. Metab Eng. 2018;49:232–241. doi: 10.1016/j.ymben.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Ma W., Liu Y., Lv X., Li J., Du G., Liu L. Combinatorial pathway enzyme engineering and host engineering overcomes pyruvate overflow and enhances overproduction of N-acetylglucosamine in Bacillus subtilis. Microb Cell Factories. 2019;18:1. doi: 10.1186/s12934-018-1049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]