Abstract
Dynamic susceptibility contrast enhanced magnetic resonance imaging (DSC MRI) is widely used for studying blood perfusion in brain tumors. While the time-dependent change of MRI signals related to the concentration of the tracer is used to derive the hemodynamic parameters such as regional blood volume and flow into tumors, the tissue-specific information associated with variations in profiles of signal time course is often overlooked. We report a new approach of combining model free independent component analysis (ICA) identification of specific signal profiles of DSC MRI time course data and extraction of the features from those time course profiles to interrogate time course data followed by calculating the region specific blood volume based on selected individual time courses. Based on the retrospective analysis of DSC MRI data from 38 patients with pathology confirmed low (n = 18) and high (n = 20) grade gliomas, the results reveal the spatially defined intra-tumoral hemodynamic heterogeneity of brain tumors based on features of time course profiles. The hemodynamic heterogeneity as measured by the number of independent components of time course data is associated with the tumor grade. Using 8 selected signal profile features, machine-learning trained algorithm, e.g., logistic regression, was able to differentiate pathology confirmed low intra-tumoral and high grade gliomas with an accuracy of 86.7%. Furthermore, the new method can potentially extract more tumor physiological information from DSC MRI comparing to the traditional model-based analysis and morphological analysis of tumor heterogeneity, thus may improve the characterizations of gliomas for better diagnosis and treatment decisions.
Keywords: Magnetic resonance imaging, Dynamic susceptibility contrast, Brain tumor, Cerebral blood volume, Feature extraction, Signal time course
Abbreviations: AUC, area under curve; BET, brain extraction tool; CBF, cerebral blood flow; CBV, cerebral blood volume; rCBV, regional cerebral blood volume; ΔrCBV, change of regional cerebral blood volume; DFC, defined filtering criterion; DSC, dynamic susceptibility contrast; FLAIR, flow attenuated inversion recovery; FLIRT, Linear Image Registration Tool; FWHM, full width at half maximum; IDH, isocitrate dehydrogenase; IV, intravenous; ICA, independent component analysis; IC, independent component; LAP, laplace approximation; MRI, magnetic resonance imaging; MTT, mean transit time; TTP, time to peak; MTE, mean time to enhance; PSR, percent of signal recovery; ROC, receiver operating curve; SUSAN, univalent segment assimilating nucleus; VEGF, vascular endothelial growth factor; WHO, World Health Organization
Highlights
-
•
Signal profiles of dynamic susceptibility contrast MRI data of brain tumors reflect hemodynamic properties of tumor tissue.
-
•
Features in signal profiles extracted by machine learning methods revealed the hemodynamic heterogeneity of the gliomas.
-
•
The reported approach is a new strategy to characterize the intra-tumor heterogeneity and physiological properties of gliomas.
1. Introduction
Glioblastoma is one of the most deadly cancers in adults which accounts for 52% of all parenchymal brain tumor cases and 20% of all intracranial tumors (Stupp et al., 2005). Clinical decisions regarding the treatment of brain tumors depend greatly on tumor characteristics (Fossati et al., 2015) which display substantial intra-tumoral heterogeneity in virtually all distinguishable phenotypic features (Marusyk and Polyak, 2010). Brain tumor patients typically undergo extensive magnetic resonance imaging (MRI) scans for classifying the tumor type, determining the tumor grade, and monitoring the treatment response and tumor recurrence. Unlike histopathologic analysis of tumor tissue samples collected from biopsy or surgical resection, which cannot reveal intra-tumoral heterogeneity due to limitations in obtaining tissue samples, different MRI techniques can be performed repeatedly and non-invasively to provide both morphological and physiological information of the tumor with high spatial resolution. Among the methods widely used for brain tumor imaging, dynamic susceptibility contrast enhanced (DSC) MRI uses signal intensity changes associated with the passage of intravenously injected (i.v.) paramagnetic gadolinium (Gd) chelate contrast agent (e.g., gadopentetate dimeglumine), through the micro-vascular system to characterize the hemodynamic properties of tissues. By tracking time-dependent MRI signal changes in the brain and tumor tissue after injection of the contrast agent, hemodynamic properties, including cerebral blood volume (CBV), cerebral blood flow (CBF), time to peak (TTP) and mean transit time (MTT), vascular permeability, can be derived from the signal time course data using given pharmacokinetic models (Jiang et al., 2014). These hemodynamic parameters can serve as surrogate markers for tumor angiogenesis associated with tumor progression and recurrence (Benner et al., 1997; Law et al., 2004; Stadler et al., 2017) as well as responses to anti-angiogenesis therapies (Fayed and Modrego, 2005; Fuss et al., 2001; Jarnum et al., 2010; Lehmann et al., 2010; Weber et al., 2006), such as bevacizumab (Avastin®), a clinically available vascular endothelial growth factor (VEGF) antagonist (Friedman et al., 2009; Vredenburgh et al., 2007; Zhang et al., 2012). Conventional methods for analyzing DSC MRI time course data mostly use a computational model to follow the first pass of the contrast agent bolus in different tissue compartments in reference of the arterial input of the blood to measure the hemodynamic parameters (Bjornerud et al., 2011; Boxerman et al., 2012). However, these methods mostly use empirical models with the assumption of a homogeneous distribution of the Gd-contrast agent in a single intra- and extra-cellular compartment (Winchester, 2008) while the brain tumors, especially high grade glioblastoma, are highly spatially heterogeneous (Marusyk and Polyak, 2010). Tumor heterogeneity depends heavily on tumor biology and has significant implications for diagnosis and treatment. In this report, we demonstrate that the hemodynamic properties of brain tumors are also highly heterogeneous, and the hemodynamic heterogeneity, as well-known morphological heterogeneity, potentially adds new information for better tumor characterization. Using a model-free and data-driven approach with the assistance of independent component analysis (ICA) (Stone, 2004), specific types of signal time courses of DSC MRI data were identified as independent components (ICs) which were further used to extract significant features of signal profiles. This new approach can better capture the spatial and hemodynamic heterogeneity of gliomas without depending on a priori model. Since ICA was commonly used in analyzing 4D resting-state functional MRI time course data, to extract features (Ribeiro de Paula et al., 2017; Tian et al., 2013), similarly, we focus on applying ICA to extract spatial and temporal features of 4D DSC data. Furthermore, by taking into account of tissue-specific variations of signal time courses extracted by ICA, more accurate rCBV maps can be calculated.
2. Materials and methods
2.1. Characteristics of selected patients
This retrospective study was approved by the institutional review board (IRB). Clinical MRI data from thirty patients (16 males, 14 females) who had biopsy or tumor resection after routine MRI exams were selected for retrospective analysis. The data from the clinical diagnosis, grading, and classifications of the tumor were collected and used. For the low grade glioma group, we first used histopathology based on 2007 criteria of World Health Organization (WHO) glioma grading criteria (Louis et al., 2007) to select the patients. We then followed 2016 revised WHO glioma classification criteria, including mutation status of isocitrate dehydrogenase 1 and 2 (IDH1/2) and chromosome 1p19q to further evaluate and confirm the tumor grade. Among 30 patients, based on pathology grading, 16 cases were initially diagnosed as high grade gliomas (WHO III-IV) and 14 were low grade gliomas (WHO II). The locations of tumors were seen mostly in the frontal or temporal regions of the brain. Among 30 cases, 18 were assessed with tumor proliferation MIB-1 index based on the Ki-67 assay. Table 1 summarizes the clinical and demographic information of the patients. Additional 8 cases with 4 high grade and 4 low grade gliomas with clinically diagnosis were then included for re-testing the developed classification algorithms.
Table 1.
Demographic information of enrolled patients.
| Characteristics or variables | High grade |
Low grade |
|---|---|---|
| (n = 16) | (n = 14) | |
| Age | 67 (54–70) | 55 (45–64) |
| Gender | ||
| Female | 6 | 3 |
| Male | 10 | 11 |
| Cell Types | ||
| Glioblastoma | 7 | – |
| Anaplastic astrocytoma | 4 | – |
| Anaplastic oligoastrocytoma | 5 | |
| Astrocytoma | – | 8 |
| Oligodendroglioma | – | 6 |
| Mutation Status | ||
| IDH | 4 | 5 |
| 1p19q | – | 6 |
| Wide type IDH | 7 | 2 |
| IDH mutation unknown | 5 | 1 |
| MIB Index (Ki-67) | ||
| <10% | – | 5 |
| 10%–25% | 5 | 2 |
| >25% | 6 | – |
| N/A | 5 | 7 |
2.2. MRI data acquisition
All patients underwent MRI exams on a 3 T MRI scanner (Magnetom Tim/Trio; Siemens, Germany) with a routine brain tumor protocol which includes the following sequences: pre and post-contrast enhanced T1 weighted spin-echo imaging, T2 weighted (axial) fluid-attenuated inversion recovery (FLAIR) imaging, diffusion weighted imaging and DSC MRI. FLAIR images, which were used for segmentation of tumors, were obtained by following parameters: repetition time (TR) = 6000, echo time (TE) = 93 ms, flip angle = 130°, inversion time = 2030 ms, 25 axial slices (thickness = 5 mm), field of view (FOV) = 240 × 240 mm2, and matrix of 512, giving spatial resolution of 0.469 × 0.469 × 5 mm.
DSC MRI data were acquired using a gradient echo planar sequence with TR/TE = 1990/30 ms, time points = 50 or 70, FOV: 220–240 mm, matrix: 128 × 128, 19 axial slices and slice thickness of 5 mm. A bolus of gadolinium (Gd) contrast agent (0.05–0.15 mmol/kg) was injected at a rate of 3 mL/s at 20 s after starting the acquisition of dynamic data.
2.3. Data analysis
4-dimensional (4D) DSC MRI images from each case were pre-processed with motion correction and realignment before being used to extract temporal and spatial features using ICA. Briefly, the noise reduction was applied first using Smallest Univalue Segment Assimilating Nucleus (SUSAN) (Smith and Brady, 1997). Subsequently, Linear Image Registration Tool (FLIRT) was used for image alignment to minimize the motion artifacts (Jenkinson et al., 2002; Jenkinson and Smith, 2001). Finally, skull stripping was performed on T2 weighted FLAIR images using the brain extraction tool (BET) of the FMRIB Software Library (FSL) for subsequent co-registration of anatomic images and maps of hemodynamic parameters calculated from the DSC MRI data analysis. After all preprocessing steps, the 4D DSC MRI data were analyzed following the procedure illustrated in Fig. 1.
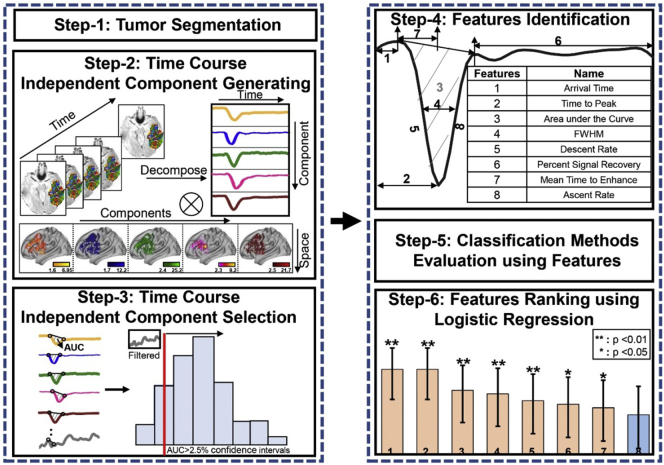
Fig. 1.
An overview of the data analysis process. Step 1: Extracting tumor regions from T2 weighted FLAIR images and transforming segmented regions to DSC image space. Step 2: Applying spatial ICA to 4-D DSC MRI time course data to obtain independent components of signal profiles and corresponding spatial maps of the components. Step 3: Using defined filtering criterion to remove unwanted components and artifacts by generating the histogram of area under the curve (AUCIC) of the signal time course and using 2.5% confidence intervals of distributions as a cut-off threshold. An example of the removed component is presented in the top-left corner of AUCIC value histogram. Step 4: Identifying signal profile features with the order according to the importance. Step 5: Evaluating classification methods. Step 6: Ranking featuresusing logistic regression.
2.3.1. Tumor segmentation
Tumor regions defined as volume of interest (VOI) were segmented in reference to FLAIR images using K-means clustering with Silhouette Criterion (Kodinariya and Makwana, 2013) and neighborhood connectivity information. We used FLAIR images for tumor segmentation because of their better delineation of the tumor given both high spatial resolution and high contrast between the tumor and surrounding tissue. Contrast enhanced T1 weighted spin echo images were also reviewed and used as the reference for evaluating the tumor location, size and shape as well as the grade, however, they were not used for segmentation as most low grade gliomas do not always have post-contrast enhancement. K-means clustering used for segmentation is a data mining approach that partitions data into mutually exclusive k clusters in which each pixel belongs to the cluster with the nearest mean. This method has been often employed for image analysis (Jose et al., 2014; Patel et al., 2013), including subdividing tumors (Selvakumar et al., 2012).
Prior to K-means clustering, the number of clusters k was chosen according to the Silhouette Criterion (Rousseeuw, 1987; Vidal et al., 2017) with the highest silhouette index value between −1 and 1. The given criterion was based on how close each voxel in one cluster are from voxels in other clusters.
Neighborhood connectivity information of the pixels (Cheng et al., 2009) is used to extract the maximum cluster, e.g., tumor region. After identifying the tumor region from each image to obtain the tumor volume, the segmented volume was transformed from the FLAIR image space into the DSC image space using both linear (FLIRT) and non-linear (FNIRT) algorithms.
2.3.2. Profile feature extraction and analysis
We used the ICA algorithm with Laplace approximation (LAP) criteria to extract temporal features of time course profiles of DSC MRI data. Briefly, the segmented tumor volume for each patient was binarized as a mask and then transformed to 4D DSC data. ICA was then performed to extract ICs of time course profiles within a defined tumor region using FSL-MELODIC package (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MELODIC). ICA is a statistical method that identifies maximally independent specific features/patterns from the source data by minimization of mutual information or maximization of non-Gaussianity (Calhoun and Adali, 2012). Given a 4D DSC MRI time course data, spatial ICA is applied to obtain independent component signals including spatial maps and time courses in pairs (Chen and Calhoun, 2018; Chen et al., 2018). The resultant maps and corresponding time courses with signal intensity were normalized to z-scores (Agarwal et al., 2017) by calculating the difference between a value in the sample and the mean, and then dividing it by the standard deviation.
LAP is a default method used in MELODIC software (Minka, 2001) given by Gaussian approximation to a continuous distribution with the highest approximation value to determine the optimal number of ICs. To select ICs for further analysis instead of using the entire set of ICs for this proof-of-principle study, we used the DFC based on the concept similarly to the selection of ICs (Wang and Li, 2015) in the resting state functional MRI data which are also time course based 4D image datasets. to remove the components, such as those time courses not affected by the contrast agent, and to retain the time course profiles resemble the typical signal time course of in-and-out first-pass contrast bolus. In this criterion, the area under curve (AUCIC) value of the signal time course was calculated by doing a definite integral under the signal time curve between the start time of first-pass contrast bolus and the end of first-pass contrast bolus. After determining the AUC value of the signal time course associated with each IC, the histogram (distribution) of each AUC of the signal time course was generated and the cut-off threshold was set to 2.5% confidence intervals of distributions. If the AUC value of the component is less than the cut-off threshold, the component would be removed as an unwanted component. For those selected ICs of signal time courses in the segmented tumor volume, contrast agent concentration changes over the time were converted from the signal intensities at different time points to further calculate the relative cerebral blood volume (rCBV) in the region (Knutsson et al., 2010). The conversion formula (Østergaard, 2005) is shown as follow:
| (A.1) |
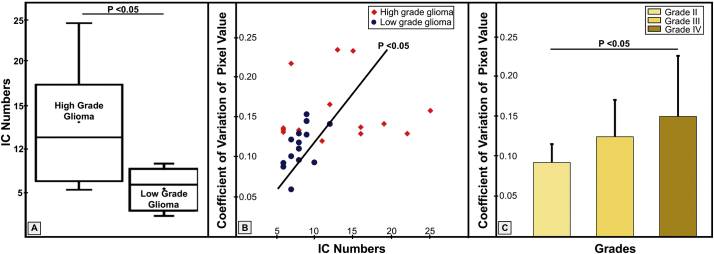
where C(t) denotes the tumor tissue contrast agent concentration, k is the constant of proportionality. In most practical applications, the proportionality constant k assumed to be equal for both tissue and blood (Knutsson et al., 2010), S(t0) represents the baseline signal before the arrival of the contrast bolus, S(t) refers to the signal intensity of each time point associated with a time course profile of a selected IC, and TE is echo time. C(t) is considered to be proportional to rCBV. The variation of rCBV or ΔrCBV which is defined as the difference between rCBV of each selected IC and the mean rCBV in the tumor was also used to show the intra-tumor heterogeneity. Finally, we calculated the variation of pixel intensity in the segmented tumor regions from T2 FLAIR images which we use as a representation of the morphological heterogeneity of the tumor using the coefficient variation (CV) value to describe the dispersion which presents the degree of morphological heterogeneity, comparing high grade gliomas to that of low grade gliomas which were shown in Fig. 3B, C. The formula is presenred as (A.2):
| (A.2) |
where σ is the standard deviation, μ is the mean.
Fig. 3.
The DSC MRI data derived intra-tumoral heterogeneity of high grade gliomas and low grade gliomas. The relationship between the coefficient of variation of pixel intensity and IC number or tumor grade was analyzed. A: the numbers of ICs in high grade glioma vs. low grade glioma patients (p < 0.05). B: the coefficient of variation of pixel intensity vs. IC number (p < 0.05). C: the coefficient of variation of pixel intensity in different tumor grades (p < 0.05).
The conventional single compartment models only use selected time course profiles in reference to the arterial input and wash-out of the contrast agent to calculate hemodynamic parameters using the scanner (Siemens) build-in toolbox and neglect other potentially useful but uncharacteristic profiles that may also contain physiologic information. The steps for the conventional way include selection of volume of interests to define the “standard” signal profiles of the time course data in the blood vessel or normal tissue, and then calculate CBV from each voxel by fitting the time course data and the area under curves (AUC) using the formula reported in the literature. Finally, Values of rCBV map were calculated as the ratio of maximum CBV from the tumor area to CBV from the contralateral normal area. In contrast, we expand the selection of the time course profile patterns for analysis based on a criteria of eight parameters, including bolus arrival time, mean transition time (MTT), time to peak (TTP), signal descent rate defined as the signal intensity difference between the arrival time and the time to peak divided by the time between these two time points, signal ascent rate defined as signal intensity difference between time to peak and the endpoint where the signal recovers to the baseline divided by the time between two time points, full width at half maximum (FWHM), percent signal recovery (PSR) and AUC. A logistic regression algorithm, fitting a logistic function to the entire data and further measuring the relationship between the classification label and features, was used to sort features according to contributions for classifying groups. The formula is given by the Eq. (A.3),
| (A.3) |
where pi corresponds to probability at observation i, βj refers to the jth regression coefficient, xij is the jth variable observation i.
2.3.3. Evaluation of the performance of classification methods
To test the performance of the reported approach in differentiating low and high grade gliomas using current data sets and identified features, we compared several commonly used machine learning based classification methods, including Gaussian native Bayes which predicts the probability of each class based on features with a prior distribution of the probability (Wang et al., 2007), decision tree which uses a tree-like graph and possible consequence to classify features (Friedl and Brodley, 1997), wrapper-based logistic regression (forward) (Kurt et al., 2008), discriminant analysis which relies on measured characteristics of known clusters to classify new samples (Liu and Wechsler, 2002) and support vector machine (SVM) which find optimal segmentation hyperplane to separate different clusters (Hearst et al., 1998). We split the initial data of 30 cases (16 high grad glioma cases and 14 low grade glioma cases) into the training data set with 24 cases and a testing dataset with 6 cases. The training set and testing set were randomly partitioned using a five-fold cross-validation scheme proposed by others (Wong, 2015) with a grid search (Hsu et al., 2003) to tune/select the hyperparameters. This process was then performed with 100 times repetition (Lee et al., 2015). We then averaged 100 results to produce a single estimation of classification performance. To minimize the effect of overfit and overestimate when using the same data set to train and test the machine-learning algorithm, we then used the trained model to test the algorithms with a independent testing dataset of 8 cases with 4 high grade and 4 low grade gliomas cases.
2.4. Statistical analysis
Statistical analyses were conducted in Matlab using the statistics toolbox (The MathWorks Inc., MA). Demographic variables were compared between groups (Chi-square or two-tailed independent samples t-test) for descriptive purposes. The numbers of selected ICs from different groups (i.e., high grade or low grade gliomas) were compared using independent sample t-test with a threshold of p < 0.05 which was considered as statistically significant. In the meantime, the correlation coefficient value was calculated between the coefficient of variation of pixel intensity and IC numbers with a threshold of p < 0.05. In addition, one-factor ANOVA analysis was applied to test the relationship between coefficient of variation of pixel intensity and the tumor grade in the cases of p < 0.05. Finally, independent sample t-test was conducted again for the group comparison of ΔrCBV in the cases of p < 0.05.
3. Results
3.1. Patients and related clinical information
We divided the patients into high and low grade glioma groups based on the clinical information collected from the patients for analyzing data, testing and validating the results. Among 14 low grade glioma patients, we found five patients carrying the IDH mutation, six having 1p19q co-deletion, two carrying wild type IDH, and one without IDH information. Among 16 patients in the high grade glioma group, we found four patients carrying the IDH mutation, seven carrying wild type IDH and five patients without IDH information. The prognostic relevance of tumor proliferation (Ki-67/MIB-1) index was also obtained for the patients and used as the reference when evaluating the tumor grade. More detailed clinical information on selected patients used in selecting and characterizing features is summarized in Table 1.
3.2. ICA-derived signal profile features and their spatial distributions
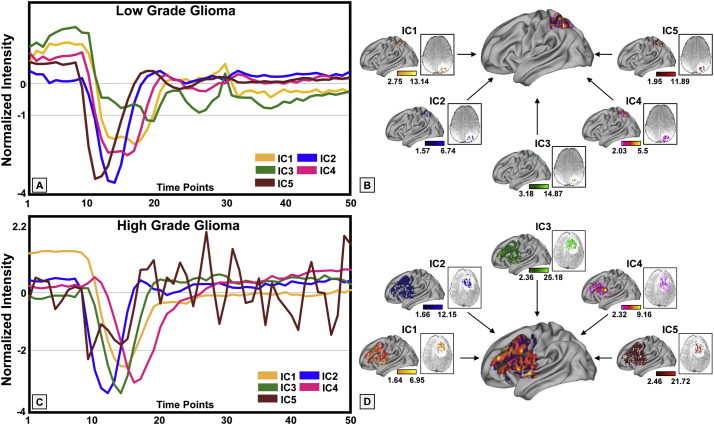
Using K-means clustering with Silhouette Criterion and neighborhood connectivity information to segment T2 weighted FLAIR images, tumor regions were successfully segmented (colored in red in Fig. 1) as shown in the example of a high grade glioma case and a low grade glioma case. Different ICs representing different profiles of signal time course from the segmented tumor volume were extracted from DSC MRI data using ICA. After calculating histogram of AUCIC values of signal profiles of each IC, a cut-off threshold of 7.71 was applied in the DFC to reduce the number of ICs to be used for analysis. The current DFC simplified for this proof-of-concept study allows for selecting ICs exhibiting various signal profile patterns of both high and low grade gliomas. The examples shown in Fig. 2 demonstrate that signal profiles of DSC MRI time course data in the low and high gliomas reveal different hemodynamic properties in different tissue types. More non-conventional appearing time course profiles were observed in high grade gliomas than those in low grade gliomas, indicating the greater hemodynamic variations of the tumor tissue in high grade gliomas. In addition, the hemodynamic heterogeneity can be spatially defined in high resolution based on the individual signal profiles of the time course with examples presented in different colors in the 3D rendered brain. Hemodynamic characteristics such as arrival time, time to peak, PSR, signal descent rate, signal ascent rate, mean time to enhance (MTE) (Essig et al., 2013), which is the average time for the entire bolus of injected contrast to pass through a region of tumor tissue, defined in Fig. 2A were highly variable among signal profiles of selected ICs. Furthermore, these different patterns can be seen in the different tumor regions (Fig. 2), explicitly demonstrating spatially heterogeneous blood perfusion patterns in both low grade and high-grade gliomas. It should be noted that because these time course profiles were selected based on the DFC, they can be further analyzed with similar computational models used in the conventional analysis of DSC MRI data.
Fig. 2.
Examples of the time courses of independent components (ICs) from low and high-grade gliomas. The ICs were selected from the segmented tumor regions of low-grade glioma (A) generally have less variations than those of high-grade glioma (C). The corresponding spatial locations of each IC were shown in the colored maps overlaid on the 3D brain (B, D).
3.3. Hemodynamic heterogeneity of low and high grade gliomas
We then further investigated intra-tumoral hemodynamic heterogeneity by examining the number of ICs selected from each tumor (Fig. 3), which may be used to quantify the hemodynamic heterogeneity of the tumor. The number of ICs found in high grade glioma patients was significantly higher than those from patients with low grade glioma (p < 0.05, Fig. 3A).
The observed different levels of hemodynamic heterogeneity in low and high grade gliomas are correlated with the levels morphological heterogeneity calculated based on the coefficient of variation of signal intensities measured from the FLAIR images of the tumors.
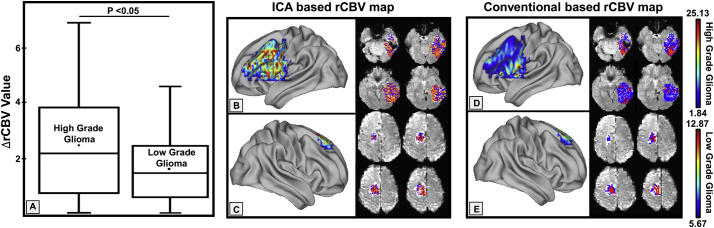
We further estimated ΔrCBV values from the time course of each selected IC. As shown in Fig. 4A, ΔrCBV values, which characterizes the degree of neovascularization (Di Stefano et al., 2014), found in patients with high-grade glioma were significantly higher than those from patients with low-grade glioma (p < 0.05). This result is in agreement with the typical findings reported in the literature (Hakyemez et al., 2005). In addition, we evaluated other parameters including mean transition (MTT) and time to peak (TTP) and found there are significant differences in MTT values (p < 0.05) and TTP (p < 0.01) between high grade gliomas and low grade gliomas.
Fig. 4.
rCBV maps of high grade gliomas and low grade gliomas calculated based on ICA derived signal profiles of the time course data. A: Comparison of ΔrCBV values in high grade glioma patients and low grade glioma (p < 0.05) with high grade tumors exhibiting higher rCBV. B, C: rCBV maps of a high and a low grade tumor based on the ICA derived data. D, E: rCBV maps from the same patients obtained by using a conventional method for calculating rCBV.
Worth noting, conventional computational methods for rCBV maps limit the computation of the regional blood volume to the fairly defined time course patterns, however, may not reveal heterogeneity of blood supplies to the tumor in details. When using individual time course patterns obtained from ICA with a model free approach, we were able to calculate the rCBV maps with inclusion of more variations of time courses as shown in Fig. 4B, C which were different from those obtained from the conventional method (Fig. 4D, E). Therefore, the reported approach may expand the quantification of hemodynamic parameters to more complex and heterogeneous patterns.
3.4. Classification methods performance
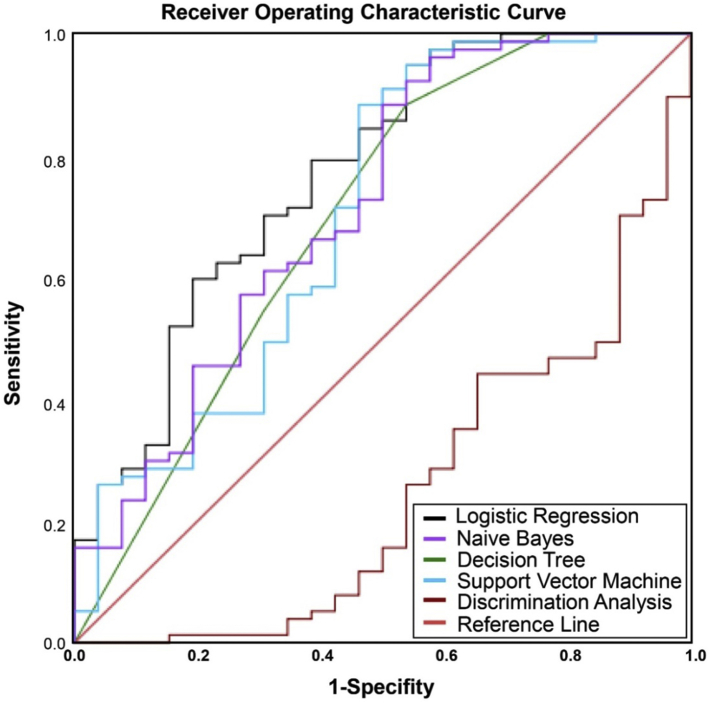
Using AUC ROC to evaluate the classification performance of different algorithms based on the 8 features, logistic regression algorithm was able to differentiate high grade gliomas from low grade gliomas among all machine learning classifiers with the highest AUCROC value (0.77). The algorithm was followed by Naïve Bayes, Decision Tree, Support Vector Machine (SVM), and Discriminant Analysis which have an AUCROC of 0.716, 0.703, 0.697, and 0.297, respectively, as shown in Fig. 5. When we used the trained model to re-test 8 cases, it was found that the logistic regressions algorithm still could reach the highest performance with AUCROC of 0.72.
Fig. 5.
The evaluation of machine learning based classification. The AUCROC values of different machine learning algorithms are: 0.77 (logistic regression), 0.716 (naive Bayes), 0.703 (decision tree), 0.697 (support vector machine), and 0.249 (discrimination analysis).
3.5. Contribution of features in differentiating tumors grades
Using the wrapper-based logistic regression (forward), we evaluated the performance of 8 signal profile features in their contributions to the accuracy of differentiating low and high grade gliomas based on increasing order of p-value. Simultaneously, we calculated the variance inflation factor (VIF) to assess correlation between these predictors as shown in Table 2 below.
Table 2.
Feature specific values of variance inflation factor.
| Features | VIF value |
|---|---|
| Area | 4.996 |
| Time to Peak | 20.667 |
| Mean_Transition_Time | 14.292 |
| Decreasing_Speed | 0.677 |
| FWHM | 5.965 |
| Increase_Speed | 11.569 |
| Arrival_Time | 12.709 |
| Slope | 0.639 |
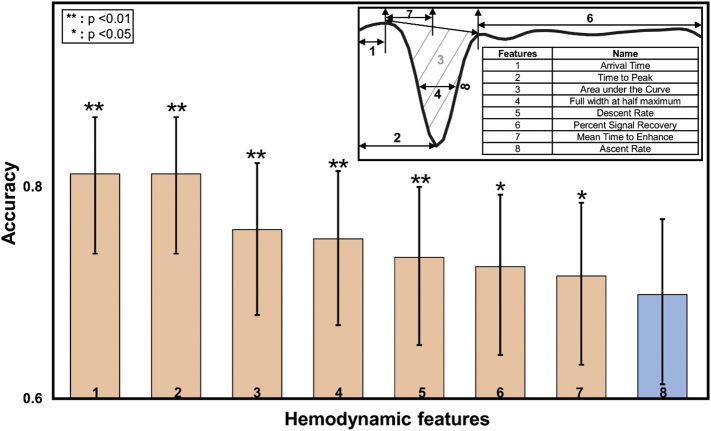
The results show that 6 features including Area, Time to peak, Mean transition time, FWHM, Increasing speed and Arrival time are highly correlated with each other (VIF > 1). Thus we used the logistic regression while treating the variables with high VIFs as control variables to minimize the multicollinearity of covariates. Based on the performances of 8 signal profile features, i.e., bolus arrival time, mean time to enhance, time to peak, signal descent rate, ascent rate, FWHM, PSR and AUC, all factors except for ascent rate were statistically significant in contributing to differentiating low and high grade gliomas (p < 0.05) as shown in Fig. 6. By calculating 95% confidence interval for a proportion, which implies the degree of accuracy, the performance of first seven features that contributed to differentiate high grade gliomas from low grade gliomas are 83.5, 83.5, 77.7, 76.7, 74.8, 73,8, and 72.8 with statistically significant p values at 0.000, 0.000, 0.002, 0.005, 0.006, 0.02 and 0.045, respectively. Furthermore, combining these seven features together for classification of tumor grades can achieve the best performance with the overall accuracy of 86.7%.
Fig. 6.
The performances of hemodynamic features in tumor classification as evaluated by accuracy. The features used for differentiating high and low grade gliomas in the present work are arrival time, time to peak, PSR, descent rate, ascent rate, mean time to enhance as demonstrated in the inserted time course profile.
4. Discussion
In the current study, we show that the model free ICA can be applied to DSC MRI of brain tumors to help discriminate high and low grade gliomas based on the hemodynamic heterogeneity associated with the tumor vasculature and blood supplies. Although it needs to be validated with larger patient cohorts and better correlation with the results from histopathological or molecular analysis, the widely available DSC MRI method combining with machine-learning selection of features in time course signal profiles has potential to improve the assessment of treatment responses and tumor progression. In this case, hemodynamic heterogeneity derived from this approach may be applied as a biomarker to predict responses to the antiangiogenic treatment commonly used in clinical management of brain tumors (LaViolette et al., 2013, 2014).
Previously, the model free ICA has been applied successfully in brain functional mapping and functional connectivity studies using fMRI time course data for analyzing specific patterns of oscillation signals related to the resting state or task-invoked low-frequency brain activities (Calhoun et al., 2001; Griffanti et al., 2014). Separation of the mixed dynamic time course signals of DSC MRI into independent source signals provides a better measurement of spatiotemporal variations in local cerebral blood perfusion (Laiwalla et al., 2017). The present study suggests that the model-free pattern extracting methods, such as ICA, can be applied on DSC MRI data to characterize the brain tumor vasculature heterogeneity. DSC MR imaging is a standard imaging method integrated into the brain tumor imaging protocol. The application of ICA to DSC MRI data may be beneficial for characterizing brain tumors and monitoring tumor response to a treatment in addition to better distinguishing tumors from the normal tissue. However, further investigation is warranted to prove these potential benefits.
We also observed statistically significant positive correlations between the coefficient of variation of pixel intensity, IC numbers, and histological grades. The observation of hemodynamic variations is consistent with the notion that high-grade gliomas have a high level of heterogeneity obtained from analyzing morphological properties of the tumor tissue based on anatomical and diffusion weighted MRI (Jiang et al., 2014), which is supported by the results from histological, analysis (Karim et al., 2016; Nakajima et al., 2012; Rebetz et al., 2008). The result from the current study indicates that hemodynamic heterogeneity, as measured by IC numbers, might be a potential indicator of prognosis in addition to morphological heterogeneity. Therefore, hemodynamic properties derived from analyzing the signal profiles of the DSC MRI time course data adds a new perspective and potential parameters for physiologically characterizing brain tumors. Furthermore, distinctive features of signal profiles of DSC MRI time course data and the number of ICs derived from the reported analysis can extract more details and information. Thus, such model free approach can be potentially used to extract a wide range of time course profiles for quantitative measurements of tumor vascular physiology and biology.
Several limitations to this study should be considered. First, we segmented tumors based only on FLAIR images. Thus, areas of non-tumor edema were likely included in some of the segmentations, while subtle signal changes from the infiltrative tumor tissue may have been excluded. As a retrospective study, there is not enough information to derive direct correlation between hemodynamic heterogeneity and histology data to validate our results. Direct correlation between imaging results with those from image-guided biopsy of multiple sites based on the observed hemodynamic heterogeneity could potentially be performed in the future to confirm the image biomarkers and measurements. In addition, we only extracted features from the time course data of DSC MRI. Other quantitative image features from additional advanced MRI sequences, such as susceptibility and diffusion weighted imaging, used in clinical brain tumor MRI exams could potentially be incorporated to increase diagnostic, prognostic, and predictive power.
5. Conclusions
The heterogeneous spatial characteristics of brain tumors on morphologic imaging and histology are well known. However, the results of the current study revealed that hemodynamic properties as measured by DSC MRI are also heterogeneous within a tumor and can be mapped spatially based on a model free analysis of DSC MRI time course data. Fewer numbers of ICs in low grade gliomas compared to high grade gliomas indicates that tumors with lower grades are less hemodynamically heterogeneous. It is demonstrated that the model free approach assisted by the feature extraction technique allows for identifying and expanding more blood perfusion information from the DSC MRI data comparing to the model based conventional DSC data analysis. Therefore, this new strategy of identifying and interrogating various forms and shapes of DSC MRI time course data may allow us to develop computational and analytical tools to investigate the tumor vascular features of the tumor tissues and regions and to better quantify imaging markers, such as rCBV and other hemodynamic parameters, for more accurate and sensitive monitoring of tumor progression and responses to the treatment.
Acknowledgments
This study is supported in parts by the grants (R01CA203388 and R01CA169937 to HM) from the National Institutes of Health.
Contributor Information
Liya Wang, Email: lwang30@emory.edu.
Hui Mao, Email: hmao@emory.edu.
References
- Agarwal S., Sair H.I., Pillai J.J. Limitations of resting-state functional MR imaging in the setting of focal brain lesions. Neuroimaging Clin. 2017;27:645–661. doi: 10.1016/j.nic.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner T., Heiland S., Erb G., Forsting M., Sartor K. Accuracy of gamma-variate fits to concentration-time curves from dynamic susceptibility-contrast enhanced MRI: influence of time resolution, maximal signal drop and signal-to-noise. Magn. Reson. Imaging. 1997;15:307–317. doi: 10.1016/s0730-725x(96)00392-x. [DOI] [PubMed] [Google Scholar]
- Bjornerud A., Sorensen A.G., Mouridsen K., Emblem K.E. T1-and T* 2-dominant extravasation correction in DSC-MRI: part I—theoretical considerations and implications for assessment of tumor hemodynamic properties. J. Cereb. Blood Flow Metab. 2011;31:2041–2053. doi: 10.1038/jcbfm.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxerman J., Prah D., Paulson E., Machan J., Bedekar D., Schmainda K. The role of preload and leakage correction in gadolinium-based cerebral blood volume estimation determined by comparison with MION as a criterion standard. Am. J. Neuroradiol. 2012;33:1081–1087. doi: 10.3174/ajnr.A2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V.D., Adali T. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Trans. Biomed. Eng. 2012;5:60–73. doi: 10.1109/RBME.2012.2211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V.D., Adali T., Pearlson G.D., Pekar J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Calhoun V. Effect of spatial smoothing on task fMRI ICA and functional connectivity. Front. Neurosci. 2018;2018:12–15. doi: 10.3389/fnins.2018.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Caprihan A., Damaraju E., Rachakonda S., Calhoun V. Functional brain connectivity in resting-state fMRI using phase and magnitude data. J. Neurosci. Methods. 2018;293:299–309. doi: 10.1016/j.jneumeth.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Cheng C., Peng G., Hwang W. Mathworks: pixel connectivity. IEEE Trans. Image Process. 2009;18:52–62. doi: 10.1109/TIP.2008.2007067. [DOI] [PubMed] [Google Scholar]
- Di Stefano A.L., Bergsland N., Berzero G., Farina L., Rognone E., Gastaldi M., Aquino D., Frati A., Tomasello F., Ceroni M. Facing contrast-enhancing gliomas: perfusion MRI in grade III and grade IV gliomas according to tumor area. Biomed. Res. Int. 2014;2014:154350–154354. doi: 10.1155/2014/154350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essig M., Nguyen T.B., Shiroishi M.S., Saake M., Provenzale J.M., Enterline D.S., Anzalone N., Dörfler A., Rovira À., Wintermark M. Perfusion MRI: the five most frequently asked clinical questions. Am. J. Roentgenol. 2013;201:495–510. doi: 10.2214/AJR.12.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayed N., Modrego P.J. The contribution of magnetic resonance spectroscopy and echoplanar perfusion-weighted MRI in the initial assessment of brain tumours. J. Neuro-Oncol. 2005;72:261–265. doi: 10.1007/s11060-004-2180-6. [DOI] [PubMed] [Google Scholar]
- Fossati N., Trinh Q.-D., Sammon J., Sood A., Larcher A., Sun M., Karakiewicz P., Guazzoni G., Montorsi F., Briganti A. Identifying optimal candidates for local treatment of the primary tumor among patients diagnosed with metastatic prostate cancer: a SEER-based study. Eur. Urol. 2015;67:3–6. doi: 10.1016/j.eururo.2014.08.056. [DOI] [PubMed] [Google Scholar]
- Friedl M.A., Brodley C.E. Decision tree classification of land cover from remotely sensed data. Remote Sens. Environ. 1997;61:399–409. [Google Scholar]
- Friedman H.S., Prados M.D., Wen P.Y., Mikkelsen T., Schiff D., Abrey L.E., Yung W.A., Paleologos N., Nicholas M.K., Jensen R. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J. Clin. Oncol. 2009;27:4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- Fuss M., Wenz F., Essig M., Muenter M., Debus J., Herman T.S., Wannenmacher M. Tumor angiogenesis of low-grade astrocytomas measured by dynamic susceptibility contrast-enhanced MRI (DSC-MRI) is predictive of local tumor control after radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2001;51:478–482. doi: 10.1016/s0360-3016(01)01691-1. [DOI] [PubMed] [Google Scholar]
- Griffanti L., Salimi-Khorshidi G., Beckmann C.F., Auerbach E.J., Douaud G., Sexton C.E., Zsoldos E., Ebmeier K.P., Filippini N., Mackay C.E., Moeller S., Xu J., Yacoub E., Baselli G., Ugurbil K., Miller K.L., Smith S.M. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakyemez B., Erdogan C., Ercan I., Ergin N., Uysal S., Atahan S. High-grade and low-grade gliomas: differentiation by using perfusion MR imaging. Clin. Radiol. 2005;60:493–502. doi: 10.1016/j.crad.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Hearst M.A., Dumais S.T., Osuna E., Platt J., Scholkopf B. Support vector machines. IEEE Intell. Syst. App. 1998;13:18–28. [Google Scholar]
- Hsu C.-W., Chang C.-C., Lin C.-J. Department of Computer Science and Information Engineering, National Taiwan University; Taipei, Taiwan: 2003. A Practical Guide to Support Vector Classification. (www.csie.ntu.edu.tw/∼cjlin/papers/guide/guide.pdf) [Google Scholar]
- Jarnum H., Steffensen E.G., Knutsson L., Frund E.T., Simonsen C.W., Lundbye-Christensen S., Shankaranarayanan A., Alsop D.C., Jensen F.T., Larsson E.M. Perfusion MRI of brain tumours: a comparative study of pseudo-continuous arterial spin labelling and dynamic susceptibility contrast imaging. Neuroradiology. 2010;52:307–317. doi: 10.1007/s00234-009-0616-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jiang J., Zhao L., Zhang Y., Zhang S., Yao Y., Qin Y., Wang C.-Y., Zhu W. Comparative analysis of arterial spin labeling and dynamic susceptibility contrast perfusion imaging for quantitative perfusion measurements of brain tumors. Int. J. Clin. Exp. Pathol. 2014;7:2790–2799. [PMC free article] [PubMed] [Google Scholar]
- Jose A., Ravi S., Sambath M. Brain tumor segmentation using k-means clustering and fuzzy c-means algorithms and its area calculation. Int. J. Innov. Res. Sci. Eng. Technol. 2014;2:3496–3501. [Google Scholar]
- Karim R., Palazzo C., Evrard B., Piel G. Nanocarriers for the treatment of glioblastoma multiforme: current state-of-the-art. J. Control. Release. 2016;227:23–37. doi: 10.1016/j.jconrel.2016.02.026. [DOI] [PubMed] [Google Scholar]
- Knutsson L., Ståhlberg F., Wirestam R. Absolute quantification of perfusion using dynamic susceptibility contrast MRI: pitfalls and possibilities. Magn. Reson. Mater. Phy. 2010;23:1–21. doi: 10.1007/s10334-009-0190-2. [DOI] [PubMed] [Google Scholar]
- Kodinariya T.M., Makwana P.R. Review on determining number of cluster in K-means clustering. Int. J. 2013;1:90–95. [Google Scholar]
- Kurt I., Ture M., Kurum A.T. Comparing performances of logistic regression, classification and regression tree, and neural networks for predicting coronary artery disease. Expert Syst. Appl. 2008;34:366–374. [Google Scholar]
- Laiwalla A.N., Kurth F., Leu K., Liou R., Pamplona J., Ooi Y.C., Salamon N., Ellingson B.M., Gonzalez N.R. Evaluation of Encephaloduroarteriosynangiosis efficacy using probabilistic independent component analysis applied to dynamic susceptibility contrast perfusion MRI. Am. J. Neuroradiol. 2017;38:507–514. doi: 10.3174/ajnr.A5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaViolette P.S., Cohen A.D., Prah M.A., Rand S.D., Connelly J., Malkin M.G., Mueller W.M., Schmainda K.M. Vascular change measured with independent component analysis of dynamic susceptibility contrast MRI predicts bevacizumab response in high-grade glioma. Neuro-Oncology. 2013;15:442–450. doi: 10.1093/neuonc/nos323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaViolette P.S., Daun M.K., Paulson E.S., Schmainda K.M. Effect of contrast leakage on the detection of abnormal brain tumor vasculature in high-grade glioma. J. Neuro-Oncol. 2014;116:543–549. doi: 10.1007/s11060-013-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., Yang S., Babb J.S., Knopp E.A., Golfinos J.G., Zagzag D., Johnson G. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. Am. J. Neuroradiol. 2004;25:746–755. [PMC free article] [PubMed] [Google Scholar]
- Lee M., Lee J.H., Oh S., Jang Y., Lee W., Lee H.J., Yoo J.j., Choi W.M., Cho Y.Y., Cho Y. CLIF-SOFA scoring system accurately predicts short-term mortality in acutely decompensated patients with alcoholic cirrhosis: a retrospective analysis. Liver Int. 2015;35:46–57. doi: 10.1111/liv.12683. [DOI] [PubMed] [Google Scholar]
- Lehmann P., Monet P., de Marco G., Saliou G., Perrin M., Stoquart-Elsankari S., Bruniau A., Vallee J.N. A comparative study of perfusion measurement in brain tumours at 3 tesla MR: arterial spin labeling versus dynamic susceptibility contrast-enhanced MRI. Eur. Neurol. 2010;64:21–26. doi: 10.1159/000311520. [DOI] [PubMed] [Google Scholar]
- Liu C., Wechsler H. Gabor feature based classification using the enhanced fisher linear discriminant model for face recognition. IEEE T Image Process. 2002;11:467–476. doi: 10.1109/TIP.2002.999679. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A., Scheithauer B.W., Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A., Polyak K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta. Rev. Cancer. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minka T.P. Automatic choice of dimensionality for PCA. Adv. Neural Inf. Proces. Syst. 2001;2001:598–604. [Google Scholar]
- Nakajima T., Anayama T., Shingyoji M., Kimura H., Yoshino I., Yasufuku K. Vascular image patterns of lymph nodes for the prediction of metastatic disease during EBUS-TBNA for mediastinal staging of lung cancer. J. Thorac. Oncol. 2012;7:1009–1014. doi: 10.1097/JTO.0b013e31824cbafa. [DOI] [PubMed] [Google Scholar]
- Østergaard L. Principles of cerebral perfusion imaging by bolus tracking. J. Magn. Reson. Imaging. 2005;22:710–717. doi: 10.1002/jmri.20460. [DOI] [PubMed] [Google Scholar]
- Patel P.M., Shah B.N., Shah V. Image segmentation using K-mean clustering for finding tumor in medical application. Int. J. Comput. Trends Technol. 2013;4:1239–1242. [Google Scholar]
- Rebetz J., Tian D., Persson A., Widegren B., Salford L.G., Englund E., Gisselsson D., Fan X. Glial progenitor-like phenotype in low-grade glioma and enhanced CD133-expression and neuronal lineage differentiation potential in high-grade glioma. PLoS ONE. 2008;3:1–12. doi: 10.1371/journal.pone.0001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Paula D., Ziegler E., Abeyasinghe P.M., Das T.K., Cavaliere C., Aiello M., Heine L., di Perri C., Demertzi A., Noirhomme Q. A method for independent component graph analysis of resting-state fMRI. Brain Behav. 2017;7:1–12. doi: 10.1002/brb3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- Selvakumar J., Lakshmi A., Arivoli T. Brain tumor segmentation and its area calculation in brain MR images using K-mean clustering and fuzzy C-mean algorithm. Int. J. Innov. Res. Sci. Eng. Technol. 2012;2:186–190. [Google Scholar]
- Smith S.M., Brady J.M. SUSAN-a new approach to low level image processing. Int. J. Comput. Vis. 1997;23:45–78. [Google Scholar]
- Stadler K.L., Pease A.P., Ballegeer E.A. Dynamic susceptibility contrast magnetic resonance imaging protocol of the normal canine brain. Front. Vet. Sci. 2017;4:41–46. doi: 10.3389/fvets.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.V. MIT press; 2004. Independent Component Analysis: A Tutorial Introduction. [Google Scholar]
- Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O., European Organisation for, R, Treatment of Cancer Brain, T, Radiotherapy, G, National Cancer Institute of Canada Clinical Trials, G Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Tian L., Kong Y., Ren J., Varoquaux G., Zang Y., Smith S.M. Spatial vs. temporal features in ICA of resting-state fMRI–A quantitative and qualitative investigation in the context of response inhibition. PLoS ONE. 2013;8:1–12. doi: 10.1371/journal.pone.0066572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M.C., Sato J.R., Balardin J.B., Takahashi D.Y., Fujita A. ANOCVA in R: a software to compare clusters between groups and its application to the study of autism Spectrum disorder. Front. Hum. Neurosci. 2017;2017:11–16. doi: 10.3389/fnins.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredenburgh J.J., Desjardins A., Herndon J.E., Marcello J., Reardon D.A., Quinn J.A., Rich J.N., Sathornsumetee S., Gururangan S., Sampson J. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J. Clin. Oncol. 2007;25:4722–4729. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li T.-Q. Dimensionality of ICA in resting-state fMRI investigated by feature optimized classification of independent components with SVM. Front. Hum. Neurosci. 2015;9:1–17. doi: 10.3389/fnhum.2015.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. J. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M.A., Zoubaa S., Schlieter M., Juttler E., Huttner H.B., Geletneky K., Ittrich C., Lichy M.P., Kroll A., Debus J., Giesel F.L., Hartmann M., Essig M. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology. 2006;66:1899–1906. doi: 10.1212/01.wnl.0000219767.49705.9c. [DOI] [PubMed] [Google Scholar]
- Winchester J.F. Springer Science & Business Media; 2008. Replacement of Renal Function by Dialysis. [Google Scholar]
- Wong T.-T. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recogn. 2015;48:2839–2846. [Google Scholar]
- Zhang G., Huang S., Wang Z. A meta-analysis of bevacizumab alone and in combination with irinotecan in the treatment of patients with recurrent glioblastoma multiforme. J. Clin. Neurosci. 2012;19:1636–1640. doi: 10.1016/j.jocn.2011.12.028. [DOI] [PubMed] [Google Scholar]