Abstract
Background
The finding of strong associations between certain human leukocyte antigen (HLA) genotypes and the development of severe cutaneous adverse drug reactions (SCARs), [for example, HLA-B*57:01 and abacavir (ABC), HLA-B*15:02 and carbamazepine (CBZ) and HLA-B*58:01 and allopurinol], has led to HLA screening being used to prevent SCARs. Screening has been shown to be of great benefit in a number of studies. Clinical translation from bench to bedside, however, depends upon the development of simple, rapid and cost-effective assays to detect these risk alleles. In highly populated developing countries such as Vietnam, where there is a high prevalence of HLA-B*15:02 and HLA-B*58:01 correlating with a high incidence of CBZ- and allopurinol-induced SCARs, the crucial factor in the implementation of comprehensive screening programs to detect these major risk HLA alleles is the availability of suitable assays.
Body
We have summarized the role and economic benefits of HLA screening, reviewed published HLA screening methods used currently in pharmacogenetic screening and examined the advantages and disadvantages of assays developed specifically for use in screening for risk alleles in the prevention of HLA-associated SCARs in Vietnam.
Conclusion
The optimal approach we propose may serve as a template for the development of screening programs in other emergent countries.
Keywords: Severe cutaneous adverse drug reactions, SCARs, HLA, Real-time PCR, HLA typing, HLA screening
Introduction
In 2002, Mallal and colleagues reported the close association between possession of the human leukocyte antigen (HLA)-B*57:01 allele and the occurrence of abacavir (ABC)-induced hypersensitivity in Caucasians.1, 2 Shortly thereafter, associations were described in Han Chinese for HLA-B*15:02 and carbamazepine (CBZ)-induced severe cutaneous adverse drug reactions (SCARs)3 and HLA-B*58:01 and allopurinol-induced SCARs.4 Since then it has been established that HLA-B*15:02 is the major genetic risk factor for CBZ-induced Stevens-Johnson Syndrome/toxic epidermal necrolysis (SJS/TEN) in other Asian populations such as the Thais,5 Singaporeans,6 Malaysians,7 Indians,8 Vietnamese9 and Chinese [both Han and other Chinese populations]10, 11, 12, 13], and HLA-B*58:01 has been shown to confer genetic susceptibility to allopurinol-induced SCARs across populations other than Taiwanese,4 such as the Thai,14 Japanese15 and Korean16 populations (Fig. 1).
Fig. 1.
The distribution of major HLA allele associated with severe cutaneous adverse drug reactions worldwide and a focus in South East Asia explaining the link of the high observed incidence of SCARs and the high these allele frequencies.45, 94
HLA molecules contribute to drug hypersensitivity reactions in the antigen presenting phase of T cell-mediated immune responses in the immunological synapse. The susceptible HLA proteins can present a specific medication or a metabolic product of the medication to naïve CD8 T cells by Hapten/Pro-hapten, Pharmacological Interaction (P-I) or Altered peptide repertoire models.17 Once they, as antigens, are recognized by specific T cell receptors (TCRs), the immune responses in the effector phase result in the proliferation of CD8 T cells and the release of cytokines.17, 18, 19 In the case of CBZ and allopurinol-induced SCARs, although CBZ and allopurinol and their metabolites are able to bind non-covalently to T cell receptors, consistent with the P-I model, they differ in their involvement of TCR repertoires and T cell clonotypes in generating their individual patho-mechanisms.20, 21, 22, 23, 24
Despite the positive predictive values (PPV) of the risk HLA alleles being low in some instances, 3% for HLA-B*15:02 and HLA-B*58:0125 compared with 47.9% for HLA-B*57:01,26 it can nevertheless be beneficial to screen for low PPV HLA risk alleles before prescribing the relevant drugs to prevent morbidity and mortality resulting from SCARs.27, 28 It is also cost-effective (Table 1, Table 2).27, 28 In the routine clinical setting, the availability of simple and rapid HLA screening tests for these risk alleles is crucial for widespread uptake of population screening.29
Table 1.
HLA screening tests and severe cutaneous drug adverse reactions.
| Medication-induced SCARs | Allele screening | Population | Sensitivity | Specificity | PPV | NPV | Authors |
|---|---|---|---|---|---|---|---|
| CBZ –SJS/TEN | HLA-B*15:0 2 | Chinese | 77.4 | 94.4 | 3.37 | 99.94 | Genin et al., 2014(25) |
| HLA-A*31:01 | N/A | N/A | N/A | N/A | |||
| CBZ-DRESS | HLA-B*15:02 | Chinese | N/A | N/A | N/A | N/A | |
| HLA-A*31:01 | Chinese and European | 50–75 | 95.8–96.1 | 0.59–0.89 | 99.97–99.98 | ||
| ABC-hypersensitivity | HLA-B*57:01 | Australian (Caucasians) | 100 | 96.9 | 47.9 | 100 | Mallal et al., 2008(26) |
| Allopurinol-SJS/TEN/DRESS | HLA-B*58:01 | Taiwanese | 100 | 85.2 | 2.0 | 100 | Ko et al., 2015(33) |
Table 2.
Benefits of HLA screening prior to treatment.
| Medication-induced SCARs | Allele screening | Population | Number screened | NPV | Number of cases prevented | Authors (year) |
|---|---|---|---|---|---|---|
| ABC-hypersensitivity | HLA-B*57:01 | Australian | 1956 | 100 | 39 | Mallal et al. 200826 |
| CBZ-SJS/TEN | HLA-B*15:02 | Taiwanese | 4877 | 100 | 10 | Chen et al., 2011(34) |
| CBZ-SCARs | HLA-A*31:01 | Japanese | 1187 | 100 | 16 | Mushiroda et al. 201840 |
| Allopurinol-SCARs | HLA-B*58:01 | Taiwanese | 2926 | 100 | 7 | Ko et al. 201533 |
In Vietnam, the high prevalence of HLA-B*15:02 [allele frequency (AF) 13.5%, the highest known in the world] and HLA-B*58:01 (AF 6.5%) in Kinh population, the largest population in Vietnam (86%),30 correlates with the high incidence of SCARs caused respectively by CBZ and allopurinol (Fig. 1). In 2015, we reported a strong association in Vietnamese between CBZ-induced SJS/TEN and HLA-B*15:02 but not hypersensitivity syndrome/drug reaction with eosinophilia and systemic symptoms (HSS/DRESS).9 No associations between HLA-A and CBZ-induced SCARs have been found in the Vietnamese.9, 31 HLA-B*58:01 has been shown to be a major genetic risk factor for allopurinol-induced SCARs in Vietnamese.31 In the recent investigation on phylogeographic and genome-wide of Vietnamese showed the Vietnamese population has the high genetic similarity to the Chinese population in Southern China and other populations in Laos, Cambodia and Southeast Asia.32 Therefore, we assume that SCARs caused by CBZ and allopurinol in Vietnam might then be largely preventable, especially as genetic screening programs would likely be beneficial in view of the high prevalence of HLA-B*15:02 and HLA-B*58:01, similar to that in the Taiwanese and Thai populations.33, 34 Moreover, as Vietnam is a typical developing country with a high burden of disease requiring treatment with CBZ or allopurinol (epilepsy, mental disorders, pain syndromes and hyperuricemia),35, 36, 37, 38, 39 alternative medications to CBZ or allopurinol may be too costly. Taking epilepsy as an example, the treatment gap in Vietnam (defined as the proportion of patients who are not given appropriate treatment for active epilepsy) was reported at 84.7% (compared with 56% in other countries).82 Contributing to this treatment gap is the cost of treatment,38 fear of using CBZ due to the high incidence of CBZ-induced SCARs and the lack of affordability of next generation anti-convulsant drugs. SCARs might be avoided when pharmacogenetic screening is introduced to Vietnam as CBZ and allopurinol could be used with confidence in a greater number of patients, reducing the treatment gap in diseases where CBZ or allopurinol are first line therapy and reducing the overall burden upon the healthcare system in Vietnam (see Table 1, Table 2).
In this paper, we have summarized the role and economic benefits of HLA screening, reviewed published HLA screening methods used currently in pharmacogenetic screening and examined the advantages and disadvantages of assays developed specifically for use in screening for risk alleles in the prevention of HLA-associated SCARs in Vietnam. The optimal approach we have determined may serve as a template for the development of HLA risk allele screening programs for the prevention of SCARs in other emergent countries.
The role of HLA screening
Clinical benefit
In 2007, the U.S. Food and Drug Administration (FDA)28 recommended physicians try to limit the morbidity and mortality from SCARs by screening patients with Asian backgrounds for HLA-B*15:02 prior to prescribing carbamazepine. Later, in 2012, the American College of Rheumatology27 recommended HLA-B*58:01 genotyping in individuals of Korean, Thai or Chinese ethnicity before commencing allopurinol.
Two screening studies have been undertaken in Taiwan. In 2012, Chen et al.34 genotyped 4877 patients and found 377 (7.7%) were positive for HLA-B*15:02 and noted none of the 4120 patients without HLA-B*15:02 who took CBZ developed SJS/TEN. This shows a very high negative predictive value (NPV) for HLA-B*15:02 screening (100%), and Chen and colleagues estimated that 10 cases had been prevented by screening, considering the historical prevalence of CBZ-SCARs. In 2015, Ko et al.33 screened 2910 patients who needed treatment with allopurinol, identified 571 (19.6%) individuals positive for HLA-B*58:01, and withheld allopurinol in this group whilst the rest of the patients were treated with allopurinol. No patient developed a SCAR, although 3% of those treated with allopurinol had mild and transient rashes related to allopurinol use. The authors examined the historical prevalence of allopurinol-induced SCARs, concluding pharmacogenetic screening had prevented SCARs in 7 of their patients.
In summary, Genin et al.25 estimated that one SCAR can be prevented in every 527 (NNT = 527) (Number Needed to Treat) patients screened for HLA-B*15:02 in the Han Chinese population, and 82 patients in every 1000 patients screened would be unnecessarily denied CBZ. Screening for HLA-A*31:01 to prevent DRESS is, however, much less efficient (NNT = 3334). On the other hand, the number of screened patients unnecessarily denied CBZ is estimated at only 38 per 1000 patients tested. The authors also examined the benefit of HLA screening for both HLA –A* 31:01 and HLA-B*15:02 in the Han Chinese population. They determined that if both alleles were screened for, the NNT would decrease from 527 to 455; however, the number of patients unnecessarily denied CBZ would increase from 82 to 94 in every 1000 patients tested. Genin et al. also suggested a kit screening for both HLA–B* 15:02 and HLA-A* 31:01 should be made available considering the potential clinical benefit (depending upon their prevalence in the at risk population).
In Japanese patients, screening for HLA-A*31:01 demonstrated clinical benefit in preventing SCARs caused by CBZ, avoiding approximately 16 SCARs in every 1187 cases screened.40
Economic benefits
Given the mortality and morbidity resulting from SCARs, any means by which a single case of SCAR could be prevented would be a worthwhile achievement. Its potential cost-effectiveness, however, is another powerful argument for pharmacogenetic screening. A number of studies have established the cost-effectiveness of the total cost of screening in comparison to treatment costs where no screening has been undertaken.
In Thailand, Tiamkao and colleagues41 used a computational model, based on their HLA-B*15:02 prevalence of 8.4%, to compare the total cost of CBZ treatment in 100 cases treated with CBZ with the expenditure saved if genotype screening had been performed prior to treatment. The result showed the cost savings would be approximately $3285 USD and they concluded cost–effectiveness of screening depends on risk allele prevalence. They noted their study ignored some direct costs.41 A more detailed analysis, by Dong et al.42 in Singapore examined the cost-effectiveness of HLA-B*15:02 screening and included the cost of alternate but more expensive medications in patients of different ancestries newly diagnosed with epilepsy (using a decision-tree model based on the TreeAge model), showing screening is likely to be more cost-effective in Singaporean Chinese and Singaporean Malaysian populations [which have a high prevalence of HLA-B*15:02 (more than 5%)] compared with screening in Singaporean Indians (who have a less than 2.5% prevalence of HLA-B*15:02).42 Thus, it is reasonable to assume that screening is likely to be cost-effective where the allele prevalence is greater than 2.5%.
Similarly, screening for HLA-B*58:01 appears to reduce the economic burden of allopurinol hypersensitivity. To date, there has been one study from Thailand and another from Korea investigating the cost-effectiveness of screening for HLA-B*58:01 before commencing allopurinol. A decision-analytic model was used in both studies. In the Thai study, Saokaew et al.43 proved that cost savings result from HLA-B*58:01 testing before allopurinol treatment after calculating all direct costs of gout management, alternate therapies, HLA testing, SCARs complications and the direct cost of “quality adjusted life years (QALYs) gained” of the patients. In Korea, Jin Park and co-investigators analysed the cost-benefit of HLA-B*58:01 screening in patients with gout and chronic renal insufficiency.44 The authors concluded HLA typing prior to starting allopurinol for gout is cost-effective, very likely because both the Thai and Korean populations are estimated to have a high prevalence of HLA-B*58:01 carriers (more than 10%).45 In addition, a high positive predictive value (PPV) of HLA risk allele presence in the patients with renal insufficiency (18%) in the Park study probably contributed to the cost-effectiveness of screening.
The benefit of HLA screening is dependent on three crucial elements. Firstly, the culprit medications causing SCARs have to be commonly used and are usually first-line therapy, and alternate medications are not available, less effective or more expensive. Secondly, the screening population should have a high prevalence of carriers of the risk alleles which are strongly associated with adverse drug reactions. Therefore, screening for HLA-B*15:02 and B*58:01 prior to introducing CBZ and allopurinol, respectively, should be implemented in almost all Asians. Finally, the cost of performing the screening assay should be minimized to enable widespread uptake of screening programs.
HLA and HLA typing methods for HLA screening
HLA genes are located on the short arm of chromosome 6; encoding major histocompatibility complex (MHC) molecules that have an important role in T cell-mediated immune responses. The HLA region consists of three main loci of HLA class I, II and III genes. While the HLA class II region, where HLA class II protein genes are located, is homogeneous, the HLA class I region is quite heterogeneous, containing loci for A, B and C genes, the genes that encode the structures of non-classical proteins, namely, HLA-E, F, and G and pseudo-genes (Fig. 2). The HLA class III locus comprises the cluster of genes that encodes complement proteins and some cytokines.46 The HLA region contains the highest density of genes (40 genes) and the biggest number of single nucleotide polymorphisms (SNPs) in the human genome.47 In addition, there are numerous haplotypes (clusters of genes linked together) of HLA genes. Thus, HLA genotyping methods have to take into account this characteristic of HLA genes to ensure the specificity of testing.
Fig. 2.
The Human Leukocyte Antigen Complex (HLA). Adapted from .53 Current HLA typing methods consisting of Serologic HLA typing,52 HLA typing by Sequence Specific Primers (SSP) Adapted from Marino and Fernandez,93 HLA typing by Sequence Specific oligonucleotide (SSO) and DNA sequence-based typing methods.
Standard HLA typing procedures currently in use for tissue typing and transplantation such as PCR-SSO and SBT are time consuming and expensive to carry out for screening purposes. Kwon et al.48, 49 indicated that current methods for HLA typing take an average of between 2 and 5 hours. Yeo50 determined that the average turnaround time for HLA genotyping tests is approximately 3–4 weeks. Both of these temporal factors constitute barriers to effective screening being implemented in clinical practice. The turnaround time of the test is defined as the duration of the sample cycle from the point of care to the laboratory and back to point of care. It is dependent on a number of factors, such as the time taken for sample collection and transport to the laboratory and back to the clinical point of care. Reducing the time the test takes can improve the turnaround time.
In regard to cost, Kwon et al.48, 49 estimated that the cost for reagents alone is approximately $ 3-38 USD, a cost at which it may prove prohibitive for both laboratories to perform and patients to pay, especially in countries with a low gross domestic product (GDP). Therefore, researchers have striven to develop an appropriate screening assay with the properties of both cost-effectiveness and rapidity for use in screening programs to prevent HLA-associated adverse drug reactions. In this section we shall discuss the principles in the common HLA typing methods used in drug hypersensitivity and determine which one might be the most suitable approach for use in screening to prevent HLA-associated adverse drug reactions.
Serologic assays and flow cytometry
Serologic assays are based on using a wide range of sera to detect antigens (HLA types) on the surface of lymphocytes. In the conventional serologic assay (micro-cytotoxicity), various sera with antibodies to HLA alleles that are contained in the tray wells are mixed with lymphocytes and incubated with complement. A vital dye is added to detect the presence of cell death. If a certain well has significant cell death, that means it has resulted from complement activation by the antigens on the lymphocyte surface binding to corresponding antibodies, enabling the HLA type to be identified.51 In flow cytometry, nucleated lymphocytes are mixed with fluorescence-labelled monoclonal antibody. If the lymphocyte with its surface antigen binds to the antibody, the fluorescent signal will be increased and detected by the laser beam of the flow cytometer52 (Fig. 2).
The important properties of the antibody-based methods such as serologic assays and flow cytometry are that they1 provide a rapid result and2 are able to discriminate “null” HLA alleles (eg. A*01:01:01:02N).53 The null HLA alleles are defined as being able to be detected in DNA sequences by molecular typing methods but have no expression of antigen on the cell surface.54 A disadvantage, however, is the false positive result rate due to the low resolution output.54 Therefore, molecular typing with high resolution output is needed for confirmation.55
Molecular HLA typing
Specific sequence priming (SSP)
This method utilizes the set of specific primers (sense and antisense) to amplify the sequence of the target HLA allele and allow elimination of other closely related sequences. The specificity of the primer is based on the nucleotide mismatch at the 3′ end. Accordingly, the primer is unable to be extended by Taq DNA polymerase. Therefore, to amplify an amplicon within the particular sequence HLA, the primers are designed to be complementary to the hypervariable region at the 3’ ends. In HLA genes Class I, the primers are targeted at the exon 2 and 3 where they harbor the most polymorphisms. The resolution of the specific sequence priming (SSP) method is dependent upon how the set of primers are designed and their target. Total time taken for this method, including the DNA extraction, ranges from 2 to 3 hours.56 The remarkable advantages of SSP methods are that they are less time-consuming and more cost-effective, making them suitable for use as a screening method for preventing of HLA-associated adverse drug reactions. However, SSP is unable to perform a large number of samples at the same time. In comparison with antibody-based methods, SSP as with other molecular methods, is preferable because it can genotype HLA alleles at a higher resolution, and the input sample is DNA which does not require special storage conditions. However, the major disadvantage of the traditional polymerase chain reaction (PCR)-SSP is that the method needs at least 20 ng of DNA. To adapt the method for screening, Giardina et al.57 developed successfully a fluorescent-based PCR-SSP assay through capillary electrophoresis (CE) for detecting HLA-B*57:01 and reduced the limit of detection (LOD) to 1 ng of DNA.
To enhance the specificity of PCR-SSP, Restriction Fragment Length Polymorphism (RFLP) has been employed post-PCR. The PCR products are digested using a restriction enzyme to cut specifically the amplified PCR sequence into another 2 child fragments. The fragments are detected on gel electrophoresis to identify the presence of the SNP.58
HLA typing by Sequence Specific oligonucleotide (SSO)
The HLA-SSO method has been used to type both HLA class I and II, having first beenintroduced for HLA class II genotyping.56 In the HLA-SSO method, the target HLA sequences (exons 2 and 3 in HLA class I; exon 3 in HLA class II) are amplified and then biotylinated. The PCR product, blotted firmly onto the positively charged nylon membrane, is then incubated in a hybridization solution which contains marker labelled-oligo nucleotides. Each sequence of the oligo-nucleotide is specific for a group of alleles (low to medium resolution) or for a single allele (high resolution) to allow for HLA typing (Fig. 2). The major advantage of the SSO method is being able to genotype a large number of samples (88–184 samples/one assay). However, this method is not a rapid HLA typing assay because the result can only be issued 2 days after the PCR stage,56 therefore, it is not suitable as a screening method even though it might be cost-effective. Currently, Luminex, one of the HLA-SSO methods, utilizing a series of microsphere beads, has been used routinely and commercially for HLA typing.59, 60
DNA sequence- based HLA typing method
DNA Sequence-based HLA typing, based on the Sanger approach, is a high-resolution method considered to be the “gold standard” (Fig. 2). In HLA Class I typing, the first PCR amplifies exons 1 to 4 for the HLA-A and B loci and exons 1 to 8 for the HLA-C loci as the template for sequencing. However, amplifying exons 2 and 3 is generally sufficient to discriminate the majority of HLA class I alleles.61 In the conventional Sanger method, the sequencing PCR is performed separately in four different tubes containing ddTTP, ddATP terminators, ddCTP and ddGTP terminators. The different lengths of PCR segments are amplified, terminated by terminators, and polyacrylamide gel electrophoresis (PAGE) is used to detect the target sequence and the HLA allele is able to be assigned. In the Sanger method, using capillary electrophoresis, different terminators are labelled with different fluorescent dyes, allowing detection. Unlike the conventional method, the sequencing PCR is performed in one tube and the PCR products are then cleaned to remove excess fluorescent labelled terminators. The major advantages of capillary electrophoresis are being able to genotype for a large number of samples and its ease of performance.62
Unlike SSP and SSO HLA typing methods that are based upon the database of known HLA sequences, the DNA Sequence-based HLA typing method is able to identify novel alleles.62 The major limitations of this method, however, are that it is time-consuming and expensive. Therefore, the DNA Sequence-based HLA typing method is usually reserved for use as a confirmatory assay in validation studies in HLA-associated drug adverse reactions (see Table 3).
Table 3.
Comparison of different HLA typing methods.
| HLA typing method | Principle | Sample | Output | Time | Cost | |
|---|---|---|---|---|---|---|
| Serologic assays | Conventional serological cytotoxicity method | Antibody-based method, used to be the “gold standard” method | Raw blood | Low resolution. False positive results. Need to be confirmed by molecular assays for higher resolution level. | Rapid | Cheap |
| Flow cytometry | Fluorescent-labelled monoclonal antibody-based method | Raw blood | Low resolution | Rapid | Cheap | |
| Cellular assays | Mixed lymphocyte culture | Raw blood | Low resolution, able to identify “foreign” histocompatibility antigens | Time consuming: several days for cell incubation | Expensive | |
| Molecular assays | SSP | DNA-based method | DNA | Low to high resolution | Rapid | Cheap |
| SSO | Low to High resolution | Time consuming | Cost-effective if testing a large number of samples | |||
| SBT | High resolution, able to detect a new allele. Currently the “gold standard” method | Time consuming | Expensive | |||
| PCR-RFLP | DNA | Immediate resolution | Slightly more time-consuming compared to SSP | Cheap | ||
Note: In searching for an HLA typing method with the properties of rapidity and cost-effectiveness, the serological-based methods (Flow cytometry), SPP and PCR-RFLP methods emerge as potential candidates.
Real-time PCR
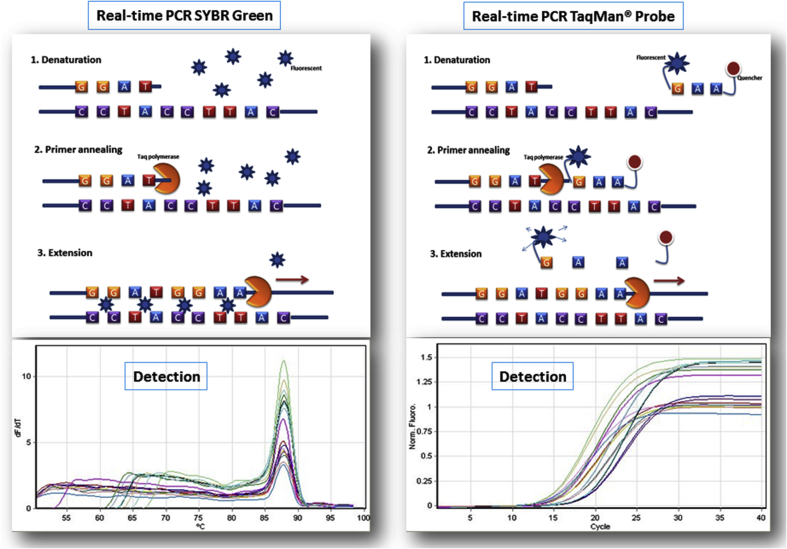
In comparing the variety of published methods (Table 4), real-time PCR (RT-PCR) (Fig. 3) appears to be the most suitable approach for screening for susceptible HLA alleles.63, 64, 65, 66, 67, 68, 69, 70, 71 RT-PCR utilizes specific dyes to detect PCR amplification instead of the post-PCR procedures of the conventional method [DNA binding dye (SYBR® green) and Fluorophore-labelled oligonucleotides (TaqMan® probe)].72 Therefore, RT-PCR using TaqMan® probes or SYBR® are the methods of choice to deliver a faster and more robust assay, able to detect the target gene at very low concentrations in DNA samples compared with conventional PCR. Furthermore, the RT-PCR assay is performed using commercial reagents that reduce the labor-intensiveness in preparing the Master Mix for PCR reactions. In addition, RT-PCR allows the performance of multiplex PCRs that means 2 or 3 target genes can be detected along with an internal control (housekeeping gene) in the TaqMan® probe.73 As a result, multiplexing is cost-effective as it reduces the amounts of reagents required, handling time and consumables used. Another advantage of multiplex PCRs is that they allow the use of internal controls (ACTB or GADH) in the same tube creating perfect conditions for ruling out any false negative results due to the presence of PCR inhibitors or low-quality input DNA. These advantages address a major limitation of the currently available commercial kits for HLA screening for prevention of SCARs which utilize RT-PCR (PG1502, PG5801 detection kits, Pharmagine Inc., Neihu Dist., Taipei City, Taiwan), the major limitation of these kits being that the internal control is run in a separate PCR tube. In addition, the recommended DNA concentration is around 20 ng/μL, so the kits are unable to detect the target HLA alleles in DNA samples extracted from collection methods such as buccal swabbing, as they yield low concentrations of DNA.65
Table 4.
Comparison of current proposed HLA typing methods for screening.
| Target allele | Method | Detection | Authors (year) | Total time of test | Reagent cost (USD) | Technical limitations |
|---|---|---|---|---|---|---|
| HLA-B*15:02 | LAMPa | HLA-B*15:02 | Cheng et al. (2009)84 | Rapid (45mins) | $3.8 | High risk of a false positive result because of the open PCR system. More handling. No internal control. |
| Nested PCR | HLA-B*15:02 | Virakul et al. (2012)85 | 240 min | $6.5 | High risk of a false positive result by cross-contamination over the second PCR. More handling. | |
| RT-PCR (TaqMan® Probe and SYBR®) | HLA-B*15:02 | Nguyen et al. (2016)69 | 238 min | $4.7 | False positive results of heterozygote of HLA-B*18 with any alleles including HLA-B*13:01; HLA-B*13:15; HLA-B*15:21 and HLA-B*15:36. | |
| HLA-A*31:01 | InvaderPlus® assay | Exon 2 of HLA-B*31:01 | Aoki et al. 201286 | 45 min | N/A, | The cost of reagents are estimated to be more expensive than the methods below. No internal control. Requires specific instrumentation. |
| Nested PCR combined with RFLP | HLA-B*31:01 | Uchiyama et al., 201458 | More time-consuming | Cheap ($2.27) | Time consuming for handling and incubation. High risk of a false positive result by cross-contamination during the second PCR. | |
| PCR-RFLP | rs1061235 linked to HLA-B*31:01 | Thorstensen et al. 201477 | Rapid | Cheap | The method has be applied in certain populations (the Norwegian population). For other populations, validation is needed. The validity of the method is dependent on the linkage between the SNP and the target HLA allele. Less specificity. More handling. | |
| LAMPa | Exon 2 of HLA-B*31:01 | Niihara et al. 2014 87 |
Rapid (40 min) | Cheap | High risk of a false positive result because of the open PCR system. More handling. No internal control. Unable to multiplex. | |
| LAMPa | Exon 2 of HLA-B*31:01 | Cheung et al. 201488 |
Rapid (40 min) | Cheap | High risk of a false positive result because of the open PCR system. More handling. No internal control. | |
| HLA-B*15:02/HLA-A*31:01 | RT-PCR (TaqMan® Probe) | Both HLA-B*15:02 and HLA-A*31:01 | Nguyen et al. 201767 | 110 | $5 | The continuous need for sequence updates to avoid any allele drop off due to any polymorphism originating at the binding sites of primers or probe. |
| HLA-B*58:01 | PCR-SSP | Exons 2 and 3 of HLA-B*58:01 | Virakul et al. 201089 | Rapid | Cheap | Using touch down PCR. High risk of false positive with HLA-B*57:01 |
| LAMP* | Exon 2 and 3 of HLA-B*58:01 | Kwok et al., 2013 49 | 60 min | Cheap | High risk of a false positive result because of the open PCR system. More handling. No internal control. | |
| PCR-RFLP | Rs9263726 of PSORS1C1 | Maekawa et al. 201276 | Rapid | Cheap | The method has been applied in the Japanese population. Need to validate in other populations. | |
| RT-PCR (TaqMan probe) | HLA-B*58:01 | Zhang et al. 201563 | Rapid | Rapid | The continuous need for sequence updates to avoid any allele drop off due to any polymorphism originating at the binding sites of primers or probe. | |
| RT-PCR (TaqMan probe) | HLA-B*58:01 | Kang et al. 201664 | Rapid | Rapid | The continuous need for sequence updates to avoid any allele drop off due to any polymorphism originating at the binding sites of primers or probe. | |
| RT-PCR (SYBR) | HLA-B*58:01 | Nguyen et al. 201665 | 120 min | $3.8 | The continuous need for sequence updates to avoid any allele drop off due to any polymorphism originating at the binding sites of primers or probe. More risk of issuing a false positive result due to the nature of method in comparison with TaqMan probe RT-PCR | |
| HLA-B*57:01/58:01 | Flow cytometry based serological test | HLA-B57/58 antigens (serological HLA-B17 group) using mAbs | Kostenko et al. 201155 | Rapid | Cheap | Low resolution needs to be confirmed by molecular methods. The initial sample is raw blood that can be a major disadvantage in comparison with other methods. |
| RT-PCR (TaqMan® probe) | HLA-B*57:01 and HLA-B*58:01 | Nguyen et al. 201671 | 110 min | Approximate $5 | False positive result of HLA-B*15:17. | |
| HLA-B*57:01 | PCR-SSP based on fluorescence detection through CEb | HLA-B*57:01 | Giardina et al. 201057 | Less time compared to PCR-SSP | Expensive 50–70 Euro/test | Although the method can increase the detection ability to 1 ng/μL of input DNA. However, it requires a sequencer (ABI 3130xL automated sequencer). More time consuming compared to real-time PCR because the PCR products have to be cleaned and run on CE (24mins). |
| The TaqMan® 5′-nuclease assay | rs2395029 of HCP5 gene linked to HLA-B*57:01 | Rodrıguez-No voa et al. 201090 | Rapid | N/A | The method can be applied in certain populations- Caucasian, Hispanic and African populations. For other populations, validation is needed. | |
| Real-time PCR | Exons 2 and 3 of HLA-B*57:01 | Dello Russo et al. 201191 | Rapid (6–8 h including DNA extraction for 30 samples) | N/A | Labour-intensive and more expensive reagents because one sample has to undergo 2 PCRs (exon 2 and exon 3). | |
| Pyro sequencing | rs2395029 (T > G) and rs3093726 (T > C) linked to HLA-B*57:01 | Sankuntaw et al. 201492 | More time-consuming than real-time PCR | N/A | In the Thai population. Validation is needed for other populations. Few laboratories use Pyrosequencing instruments. |
LAMP: loop-mediated isothermal amplification method. * In the LAMP method, the result can be visualized by naked eye.
CE: capillary electrophoresis.
Fig. 3.
Real-time PCR
HLA typing method using surrogate markers
Alternatively, typing single nucleotide polymorphisms (SNPs) which are in linkage disequilibrium (LD) with corresponding HLA alleles has been utilized as a method for HLA typing. There are two main approaches for detecting polymorphisms. One method uses a restriction enzyme (Restriction Fragment Length Polymorphism-PCR-RFLP), and the other uses TaqMan® chemistry. The major limitation of this approach is that the accuracy of the method is dependent upon the level of LD between the tagged SNPs and their corresponding HLA alleles. It is not recommended, therefore, as a diagnostic method for HLA typing. It is, however, a potential candidate for utilization in HLA screening due to its simplicity and cost-effectiveness.74 Currently, there are a number of single nucleotide polymorphisms (SNPs) which have been reported as potential surrogate markers, based on their linkage disequilibrium (LD) for detection of HLA-B*15:02,75 HLA-B*58:0176 and HLA-A*31:01,77 HLA-B*57:0178, 79 Heterogeneity has been observed, however, across different populations.80 For example, a haplotype of two SNPs, rs2844682C > T and rs3903184C > G, was proposed as a surrogate marker for HLA-B*15:02 in Han Chinese,75 although later work refuted this finding81, and rs9263726 A > G has been found to be tightly linked with HLA-B*58:01 in Japanese15 and Chinese82 but not Australians.83 This method, then, needs to be validated in each population before use in screening.
In PCR-RFLP, a set of primers is designed to amplify a sequence containing the target polymorphism (Fig. 4). The PCR products are then digested using a restriction enzyme to cut specifically the amplified PCR sequence into another 2 child fragments. The fragments are detected on gel electrophoresis to identify the polymorphism in homozygotes (1/1), heterozygotes (1/2) or wild types (2/2).
Fig. 4.
SNP genotyping using PCR-RFLP.
Developing pharmacogenetic screening methods for an emergent country: Vietnam
Considering all the advantages and disadvantages of the different methods and the context wherein the assays would be used, RT-PCR was selected as the method of choice for the development of new screening assays. This decision was based on expected test performance and cost as well as suitability of the method for use in Vietnam for screening purposes which mandates a rapid and cheap method. Therefore, we developed several RT-PCR-based assays for screening for HLA-B*57:01/58:01,71 HLA-B*58:01,70 HLA-B*15:0269 and HLA-A*31:01/HLA-B*15:02.67 These assays are robust and rapid, incorporate multiplexing, and were designed to avoid the major technical limitations of the previous assays (Table 4) and to tailor to the local conditions in Vietnam. For example, in the method developed for detection of HLA-B*58:01, SYBR® was used instead of TaqMan probe. Although the use of fluorogenic probes for detection is believed to provide greater specificity and reproducibility than SYBR® dye,72 as well as allowing a multiplex PCR to incorporate an internal control.72 Regardless of the cost-effectiveness and the sensitivity of SYBR®, it is reasonable to use it as a screening test, and it can be applied in a duplex PCR (the target gene and internal control) using melting curves.68 In addition, where there is a lack of a real-time cycler, the SYBR®-based method is able to be switched to a conventional PCR-SSP assay. This method then is more suitable for use in Vietnam where facilities for full HLA typing are not available other than in major centers and where the majority of people reside outside large cities. If the purported surrogate markers, the SNPs rs3909184, rs1061235, rs2395029 and rs9263726 are confirmed in Vietnamese as predicting the respective presence of HLA-B*15:02, HLA-A*31:01, HLA-B*57:01 and HLA-B*58:01, this also would be very advantageous for screening in Vietnam. Screening using the surrogate markers would help deliver simpler and less expensive testing at the point of care. In work prepared for publication, our results show that rs9263726 is in perfect linkage with HLA-B*58:01, whilst other SNPs are not linked with their corresponding HLA alleles. Therefore, rs9263726 can be used as a surrogate marker for detection of HLA-B*58:01 in the Vietnamese population.
In the case of CBZ-induced SCARs, a novel multiplex PCR method using TaqMan® probes for detection of HLA-B*15:02 and HLA-A*31:01 was developed and validated successfully. The method can be applied across populations, and it is more practical when dealing with a patient with an unclear ethnic background or multiple ethnicities. In the Vietnamese population, although possession of HLA-B*15:02 is the major genetic susceptibility for CBZ-induced SCARs, the high prevalence of the HLA-A*31:01 allele that is comparable to that in the Japanese and Caucasian populations (Fig. 2) indicates that screening for this allele should be beneficial. Taken together, these considerations indicate that the multiplex PCR method for detection of both HLA-B*15:02 and HLA-A*31:01 offers the most practical and cost-effective approach to pharmacogenetic screening in Vietnam.
In conclusion, the application of the multiplex PCR method in Vietnam as outlined above should prove a useful model for the development of screening methods in developing countries other than Vietnam. This model is an excellent illustration of clinical translation from the laboratory bench to the bedside of research which has established the HLA-associations in several drug-induced SCARs, which themselves originated from clinical observations at the bedside, to offer, in turn, the prospect of practical and affordable preventive screening.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Funding
No funding was applicable for the writing, submitting and publication of this paper.
Authors’ contributions
DVN reviewed the data from the literature, contributed to the concept development and wrote the manuscript. HCC, CV, SVN were major contributors to the writing and critical revision of the manuscript and the intellectual content within the paper. All authors read and approved the final manuscript.
Acknowledgments
We are grateful to the Australian Government for funding Dr Dinh Van Nguyen's Australia Awards Scholarship for his PhD (Medicine) studies at The University of Sydney. We also thank Quan Van Hong Nguyen, our resident for redrawing the figure.
References
- 1.Mallal S., Nolan D., Witt C. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359(9308):727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]; Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359(9308):727-732. [DOI] [PubMed]
- 2.Hetherington S., Hughes A.R., Mosteller M. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359(9312):1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]; Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359(9312):1121-1122. [DOI] [PubMed]
- 3.Chung W.H., Hung S.I., Hong H.S. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428(6982):486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]; Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428(6982):486. [DOI] [PubMed]
- 4.Hung S.I., Chung W.H., Liou L.B. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A. 2005;102(11):4134–4139. doi: 10.1073/pnas.0409500102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hung SI, Chung WH, Liou LB, Chu CC, Lin M, Huang HP, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):4134-4139. [DOI] [PMC free article] [PubMed]
- 5.Kulkantrakorn K., Tassaneeyakul W., Tiamkao S. HLA-B*1502 strongly predicts carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Thai patients with neuropathic pain. Pain practice. the official journal of World Institute of Pain. 2012;12(3):202–208. doi: 10.1111/j.1533-2500.2011.00479.x. [DOI] [PubMed] [Google Scholar]; Kulkantrakorn K, Tassaneeyakul W, Tiamkao S, Jantararoungtong T, Prabmechai N, Vannaprasaht S, et al. HLA-B*1502 strongly predicts carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Thai patients with neuropathic pain. Pain practice : the official journal of World Institute of Pain. 2012;12(3):202-208. [DOI] [PubMed]
- 6.Chong K.W., Chan D.W., Cheung Y.B. Association of carbamazepine-induced severe cutaneous drug reactions and HLA-B*1502 allele status, and dose and treatment duration in paediatric neurology patients in Singapore. Arch Dis Child. 2014;99(6):581–584. doi: 10.1136/archdischild-2013-304767. [DOI] [PubMed] [Google Scholar]; Chong KW, Chan DW, Cheung YB, Ching LK, Hie SL, Thomas T, et al. Association of carbamazepine-induced severe cutaneous drug reactions and HLA-B*1502 allele status, and dose and treatment duration in paediatric neurology patients in Singapore. Archives of disease in childhood. 2014;99(6):581-584. [DOI] [PubMed]
- 7.Then S.M., Rani Z.Z., Raymond A.A., Ratnaningrum S., Jamal R. Frequency of the HLA-B*1502 allele contributing to carbamazepine-induced hypersensitivity reactions in a cohort of Malaysian epilepsy patients. Asian Pac J Allergy Immunol/launched by the Allergy and Immunology Society of Thailand. 2011;29(3):290–293. [PubMed] [Google Scholar]; Then SM, Rani ZZ, Raymond AA, Ratnaningrum S, Jamal R. Frequency of the HLA-B*1502 allele contributing to carbamazepine-induced hypersensitivity reactions in a cohort of Malaysian epilepsy patients. Asian Pacific journal of allergy and immunology / launched by the Allergy and Immunology Society of Thailand. 2011;29(3):290-293. [PubMed]
- 8.Khor A.H., Lim K.S., Tan C.T., Wong S.M., Ng C.C. HLA-B*15:02 association with carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in an Indian population: a pooled-data analysis and meta-analysis. Epilepsia. 2014;55(11):e120–e124. doi: 10.1111/epi.12802. [DOI] [PubMed] [Google Scholar]; Khor AH, Lim KS, Tan CT, Wong SM, Ng CC. HLA-B*15:02 association with carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in an Indian population: a pooled-data analysis and meta-analysis. Epilepsia. 2014;55(11):e120-e124. [DOI] [PubMed]
- 9.Nguyen D.V., Chu H.C., Nguyen D.V. HLA-B*1502 and carbamazepine-induced severe cutaneous adverse drug reactions in Vietnamese. Asia Pac Allergy. 2015;5(2):68–77. doi: 10.5415/apallergy.2015.5.2.68. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nguyen DV, Chu HC, Nguyen DV, Phan MH, Craig T, Baumgart K, et al. HLA-B*1502 and carbamazepine-induced severe cutaneous adverse drug reactions in Vietnamese. Asia Pac Allergy. 2015;5(2):68-77. [DOI] [PMC free article] [PubMed]
- 10.Wu X.T., Hu F.Y., An D.M. Association between carbamazepine-induced cutaneous adverse drug reactions and the HLA-B*1502 allele among patients in central China. Epilepsy Behav : E&B. 2010;19(3):405–408. doi: 10.1016/j.yebeh.2010.08.007. [DOI] [PubMed] [Google Scholar]; Wu XT, Hu FY, An DM, Yan B, Jiang X, Kwan P, et al. Association between carbamazepine-induced cutaneous adverse drug reactions and the HLA-B*1502 allele among patients in central China. Epilepsy & behavior : E&B. 2010;19(3):405-408. [DOI] [PubMed]
- 11.Man C.B., Kwan P., Baum L. Association between HLA-B*1502 allele and antiepileptic drug-induced cutaneous reactions in Han Chinese. Epilepsia. 2007;48(5):1015–1018. doi: 10.1111/j.1528-1167.2007.01022.x. [DOI] [PubMed] [Google Scholar]; Man CB, Kwan P, Baum L, Yu E, Lau KM, Cheng AS, et al. Association between HLA-B*1502 allele and antiepileptic drug-induced cutaneous reactions in Han Chinese. Epilepsia. 2007;48(5):1015-1018. [DOI] [PubMed]
- 12.He X.J., Jian L.Y., He X.L. Association between the HLA-B*15:02 allele and carbamazepine-induced Stevens-Johnson syndrome/toxic epidermal necrolysis in Han individuals of northeastern China. Pharmacol Rep. 2013;65(5):1256–1262. doi: 10.1016/s1734-1140(13)71483-x. [DOI] [PubMed] [Google Scholar]; He XJ, Jian LY, He XL, Wu Y, Xu YY, Sun XJ, et al. Association between the HLA-B*15:02 allele and carbamazepine-induced Stevens-Johnson syndrome/toxic epidermal necrolysis in Han individuals of northeastern China. Pharmacol Rep. 2013;65(5):1256-1262. [DOI] [PubMed]
- 13.Zhang Y., Wang J., Zhao L.M. Strong association between HLA-B*1502 and carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in mainland Han Chinese patients. Eur J Clin Pharmacol. 2011;67(9):885–887. doi: 10.1007/s00228-011-1009-4. [DOI] [PubMed] [Google Scholar]; Zhang Y, Wang J, Zhao LM, Peng W, Shen GQ, Xue L, et al. Strong association between HLA-B*1502 and carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in mainland Han Chinese patients. European journal of clinical pharmacology. 2011;67(9):885-887. [DOI] [PubMed]
- 14.Tassaneeyakul W., Jantararoungtong T., Chen P. Strong association between HLA-B*5801 and allopurinol-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in a Thai population. Pharmacogenetics Genom. 2009;19(9):704–709. doi: 10.1097/FPC.0b013e328330a3b8. [DOI] [PubMed] [Google Scholar]; Tassaneeyakul W, Jantararoungtong T, Chen P, Lin PY, Tiamkao S, Khunarkornsiri U, et al. Strong association between HLA-B*5801 and allopurinol-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in a Thai population. Pharmacogenetics and genomics. 2009;19(9):704-709. [DOI] [PubMed]
- 15.Tohkin M., Kaniwa N., Saito Y. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenomics J. 2013;13(1):60–69. doi: 10.1038/tpj.2011.41. [DOI] [PubMed] [Google Scholar]; Tohkin M, Kaniwa N, Saito Y, Sugiyama E, Kurose K, Nishikawa J, et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. The pharmacogenomics journal. 2013;13(1):60-69. [DOI] [PubMed]
- 16.Kang H.R., Jee Y.K., Kim Y.S. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenetics Genom. 2011;21(5):303–307. doi: 10.1097/FPC.0b013e32834282b8. [DOI] [PubMed] [Google Scholar]; Kang HR, Jee YK, Kim YS, Lee CH, Jung JW, Kim SH, et al. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenetics and genomics. 2011;21(5):303-307. [DOI] [PubMed]
- 17.Adam J., Pichler W.J., Yerly D. Delayed drug hypersensitivity: models of T-cell stimulation. Br J Clin Pharmacol. 2011;71(5):701–707. doi: 10.1111/j.1365-2125.2010.03764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adam J, Pichler WJ, Yerly D. Delayed drug hypersensitivity: models of T-cell stimulation. British journal of clinical pharmacology. 2011;71(5):701-707. [DOI] [PMC free article] [PubMed]
- 18.Yun J., Cai F., Lee F.J., Pichler W.J. T-cell-mediated drug hypersensitivity: immune mechanisms and their clinical relevance. Asia Pac Allergy. 2016;6(2):77–89. doi: 10.5415/apallergy.2016.6.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yun J, Cai F, Lee FJ, Pichler WJ. T-cell-mediated drug hypersensitivity: immune mechanisms and their clinical relevance. Asia Pac Allergy. 2016;6(2):77-89. [DOI] [PMC free article] [PubMed]
- 19.Duong T.A., Valeyrie-Allanore L., Wolkenstein P., Chosidow O. Severe cutaneous adverse reactions to drugs. Lancet. 2017;390(10106):1996–2011. doi: 10.1016/S0140-6736(16)30378-6. [DOI] [PubMed] [Google Scholar]; Duong TA, Valeyrie-Allanore L, Wolkenstein P, Chosidow O. Severe cutaneous adverse reactions to drugs. Lancet. 2017;390(10106):1996-2011. [DOI] [PubMed]
- 20.Lichtenfels M., Farrell J., Ogese M.O. HLA restriction of carbamazepine-specific T-Cell clones from an HLA-A*31:01-positive hypersensitive patient. Chem Res Toxicol. 2014;27(2):175–177. doi: 10.1021/tx400460w. [DOI] [PubMed] [Google Scholar]; Lichtenfels M, Farrell J, Ogese MO, Bell CC, Eckle S, McCluskey J, et al. HLA restriction of carbamazepine-specific T-Cell clones from an HLA-A*31:01-positive hypersensitive patient. Chemical research in toxicology. 2014;27(2):175-177. [DOI] [PubMed]
- 21.Yun J., Mattsson J., Schnyder K. Allopurinol hypersensitivity is primarily mediated by dose-dependent oxypurinol-specific T cell response. Clin Exp Allergy : journal of the British Society for Allergy and Clinical Immunology. 2013;43(11):1246–1255. doi: 10.1111/cea.12184. [DOI] [PubMed] [Google Scholar]; Yun J, Mattsson J, Schnyder K, Fontana S, Largiader CR, Pichler WJ, et al. Allopurinol hypersensitivity is primarily mediated by dose-dependent oxypurinol-specific T cell response. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2013;43(11):1246-1255. [DOI] [PubMed]
- 22.Yun J., Marcaida M.J., Eriksson K.K. Oxypurinol directly and immediately activates the drug-specific T cells via the preferential use of HLA-B*58:01. J Immunol. 2014;192(7):2984–2993. doi: 10.4049/jimmunol.1302306. [DOI] [PubMed] [Google Scholar]; Yun J, Marcaida MJ, Eriksson KK, Jamin H, Fontana S, Pichler WJ, et al. Oxypurinol directly and immediately activates the drug-specific T cells via the preferential use of HLA-B*58:01. Journal of immunology. 2014;192(7):2984-2993. [DOI] [PubMed]
- 23.Chung W.H., Pan R.Y., Chu M.T. Oxypurinol-specific T cells possess preferential TCR clonotypes and express granulysin in allopurinol-induced severe cutaneous adverse reactions. J Investig Dermatol. 2015;135(9):2237–2248. doi: 10.1038/jid.2015.165. [DOI] [PubMed] [Google Scholar]; Chung WH, Pan RY, Chu MT, Chin SW, Huang YL, Wang WC, et al. Oxypurinol-Specific T Cells Possess Preferential TCR Clonotypes and Express Granulysin in Allopurinol-Induced Severe Cutaneous Adverse Reactions. J Invest Dermatol. 2015;135(9):2237-2248. [DOI] [PubMed]
- 24.Ko T.M., Chung W.H., Wei C.Y. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. J Allergy Clin Immunol. 2011;128(6):1266–1276. doi: 10.1016/j.jaci.2011.08.013. e11. [DOI] [PubMed] [Google Scholar]; Ko TM, Chung WH, Wei CY, Shih HY, Chen JK, Lin CH, et al. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. The Journal of allergy and clinical immunology. 2011;128(6):1266-1276 e11. [DOI] [PubMed]
- 25.Genin E., Chen D.P., Hung S.I. HLA-A*31:01 and different types of carbamazepine-induced severe cutaneous adverse reactions: an international study and meta-analysis. Pharmacogenomics J. 2014;14(3):281–288. doi: 10.1038/tpj.2013.40. [DOI] [PubMed] [Google Scholar]; Genin E, Chen DP, Hung SI, Sekula P, Schumacher M, Chang PY, et al. HLA-A*31:01 and different types of carbamazepine-induced severe cutaneous adverse reactions: an international study and meta-analysis. The pharmacogenomics journal. 2014;14(3):281-288. [DOI] [PubMed]
- 26.Mallal S., Phillips E., Carosi G. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]; Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, et al. HLA-B*5701 screening for hypersensitivity to abacavir. The New England journal of medicine. 2008;358(6):568-579. [DOI] [PubMed]
- 27.Khanna D., Fitzgerald J.D., Khanna P.P. 2012 American College of Rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia. Arthritis Care Res. 2012;64(10):1431–1446. doi: 10.1002/acr.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khanna D, Fitzgerald JD, Khanna PP, Bae S, Singh MK, Neogi T, et al. 2012 American College of Rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia. Arthritis Care Res (Hoboken). 2012;64(10):1431-1446. [DOI] [PMC free article] [PubMed]
- 28.Ferrell P.B., Jr., Carbamazepine McLeod HL. HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics. 2008;9(10):1543–1546. doi: 10.2217/14622416.9.10.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ferrell PB, Jr., McLeod HL. Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics. 2008;9(10):1543-1546. [DOI] [PMC free article] [PubMed]
- 29.Pavlos R., Mallal S., Phillips E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics. 2012;13(11):1285–1306. doi: 10.2217/pgs.12.108. [DOI] [PubMed] [Google Scholar]; Pavlos R, Mallal S, Phillips E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics. 2012;13(11):1285-1306. [DOI] [PubMed]
- 30.Hoa B.K., Hang N.T., Kashiwase K. HLA-A, -B, -C, -DRB1 and -DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue Antigens. 2008;71(2):127–134. doi: 10.1111/j.1399-0039.2007.00982.x. [DOI] [PubMed] [Google Scholar]; Hoa BK, Hang NT, Kashiwase K, Ohashi J, Lien LT, Horie T, et al. HLA-A, -B, -C, -DRB1 and -DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue antigens. 2008;71(2):127-134. [DOI] [PubMed]
- 31.Van Nguyen D., Chu H.C., Vidal C. Genetic susceptibility to carbamazepine and allopurinol–induced severe cutaneous adverse reactions in Vietnamese. J Allergy Clin Immunol. 2017;139(2) AB118. [Google Scholar]; Van Nguyen D, Chu HC, Vidal C, Nguyen NN, Quynh Do NT, Linh Tran TT, et al. Genetic Susceptibility to Carbamazepine and Allopurinol-Induced Severe Cutaneous Adverse Reactions in Vietnamese. Journal of Allergy and Clinical Immunology.139(2):AB118.
- 32.Pischedda S., Barral-Arca R., Gómez-Carballa A. Phylogeographic and genome-wide investigations of Vietnam ethnic groups reveal signatures of complex historical demographic movements. Sci Rep. 2017;7(1):12630. doi: 10.1038/s41598-017-12813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pischedda S, Barral-Arca R, Gomez-Carballa A, Pardo-Seco J, Catelli ML, Alvarez-Iglesias V, et al. Phylogeographic and genome-wide investigations of Vietnam ethnic groups reveal signatures of complex historical demographic movements. Scientific reports. 2017;7(1):12630. [DOI] [PMC free article] [PubMed]
- 33.Ko T.M., Tsai C.Y., Chen S.Y. Use of HLA-B*58:01 genotyping to prevent allopurinol induced severe cutaneous adverse reactions in Taiwan: national prospective cohort study. BMJ. 2015;351:h4848. doi: 10.1136/bmj.h4848. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ko TM, Tsai CY, Chen SY, Chen KS, Yu KH, Chu CS, et al. Use of HLA-B*58:01 genotyping to prevent allopurinol induced severe cutaneous adverse reactions in Taiwan: national prospective cohort study. Bmj. 2015;351:h4848. [DOI] [PMC free article] [PubMed]
- 34.Chen P., Lin J.J., Lu C.S. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med. 2011;364(12):1126–1133. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]; Chen P, Lin JJ, Lu CS, Ong CT, Hsieh PF, Yang CC, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. The New England journal of medicine. 2011;364(12):1126-1133. [DOI] [PubMed]
- 35.Shirasaka Y., Hinh le D., Minh P.H., Sato W. Causes of childhood epilepsy in Vietnam: cases in bach mai hospital. Pediatr Int : official journal of the Japan Pediatric Society. 2007;49(5):584–588. doi: 10.1111/j.1442-200X.2007.02432.x. [DOI] [PubMed] [Google Scholar]; Shirasaka Y, Hinh le D, Minh PH, Sato W. Causes of childhood epilepsy in Vietnam: cases in Bach Mai Hospital. Pediatrics international : official journal of the Japan Pediatric Society. 2007;49(5):584-588. [DOI] [PubMed]
- 36.Tuan N.A., Cuong le Q., Allebeck P., Chuc N.T., Persson H.E., Tomson T. The prevalence of epilepsy in a rural district of Vietnam: a population-based study from the EPIBAVI project. Epilepsia. 2008;49(9):1634–1637. doi: 10.1111/j.1528-1167.2008.01663.x. [DOI] [PubMed] [Google Scholar]; Tuan NA, Cuong le Q, Allebeck P, Chuc NT, Persson HE, Tomson T. The prevalence of epilepsy in a rural district of Vietnam: a population-based study from the EPIBAVI project. Epilepsia. 2008;49(9):1634-1637. [DOI] [PubMed]
- 37.Aydemir N., Trung D.V., Snape D., Baker G.A., Jacoby A., Team C.S. Multiple impacts of epilepsy and contributing factors: findings from an ethnographic study in Vietnam. Epilepsy Behav : E&B. 2009;16(3):512–520. doi: 10.1016/j.yebeh.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aydemir N, Trung DV, Snape D, Baker GA, Jacoby A, Team CS. Multiple impacts of epilepsy and contributing factors: findings from an ethnographic study in Vietnam. Epilepsy & behavior : E&B. 2009;16(3):512-520. [DOI] [PMC free article] [PubMed]
- 38.Tuan N.A., Tomson T., Allebeck P., Chuc N.T., Cuong le Q. The treatment gap of epilepsy in a rural district of Vietnam: a study from the EPIBAVI project. Epilepsia. 2009;50(10):2320–2323. doi: 10.1111/j.1528-1167.2009.02298.x. [DOI] [PubMed] [Google Scholar]; Tuan NA, Tomson T, Allebeck P, Chuc NT, Cuong le Q. The treatment gap of epilepsy in a rural district of Vietnam: a study from the EPIBAVI project. Epilepsia. 2009;50(10):2320-2323. [DOI] [PubMed]
- 39.Vuong D.A., Van Ginneken E., Morris J., Ha S.T., Busse R. Mental health in Vietnam: burden of disease and availability of services. Asian journal of psychiatry. 2011;4(1):65–70. doi: 10.1016/j.ajp.2011.01.005. [DOI] [PubMed] [Google Scholar]; Vuong DA, Van Ginneken E, Morris J, Ha ST, Busse R. Mental health in Vietnam: Burden of disease and availability of services. Asian journal of psychiatry. 2011;4(1):65-70. [DOI] [PubMed]
- 40.Mushiroda T., Takahashi Y., Onuma T. Association of HLA-A*31:01 screening with the incidence of carbamazepine-induced cutaneous adverse reactions in a Japanese population. JAMA neurology. 2018 Jul 1;75(7):842–849. doi: 10.1001/jamaneurol.2018.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mushiroda T, Takahashi Y, Onuma T, Yamamoto Y, Kamei T, Hoshida T, et al. Association of HLA-A*31:01 Screening With the Incidence of Carbamazepine-Induced Cutaneous Adverse Reactions in a Japanese Population. JAMA neurology. 2018. [DOI] [PMC free article] [PubMed]
- 41.Tiamkao S., Jitpimolmard J., Sawanyawisuth K., Jitpimolmard S. Cost minimization of HLA-B*1502 screening before prescribing carbamazepine in Thailand. Int J Clin Pharm. 2013;35(4):608–612. doi: 10.1007/s11096-013-9777-9. [DOI] [PubMed] [Google Scholar]; Tiamkao S, Jitpimolmard J, Sawanyawisuth K, Jitpimolmard S. Cost minimization of HLA-B*1502 screening before prescribing carbamazepine in Thailand. International journal of clinical pharmacy. 2013;35(4):608-612. [DOI] [PubMed]
- 42.Dong D., Sung C., Finkelstein E.A. Cost-effectiveness of HLA-B*1502 genotyping in adult patients with newly diagnosed epilepsy in Singapore. Neurology. 2012;79(12):1259–1267. doi: 10.1212/WNL.0b013e31826aac73. [DOI] [PubMed] [Google Scholar]; Dong D, Sung C, Finkelstein EA. Cost-effectiveness of HLA-B*1502 genotyping in adult patients with newly diagnosed epilepsy in Singapore. Neurology. 2012;79(12):1259-1267. [DOI] [PubMed]
- 43.Saokaew S., Tassaneeyakul W., Maenthaisong R., Chaiyakunapruk N. Cost-effectiveness analysis of HLA-B*5801 testing in preventing allopurinol-induced SJS/TEN in Thai population. PLoS One. 2014;9(4):e94294. doi: 10.1371/journal.pone.0094294. [DOI] [PMC free article] [PubMed] [Google Scholar]; Saokaew S, Tassaneeyakul W, Maenthaisong R, Chaiyakunapruk N. Cost-effectiveness analysis of HLA-B*5801 testing in preventing allopurinol-induced SJS/TEN in Thai population. PloS one. 2014;9(4):e94294. [DOI] [PMC free article] [PubMed]
- 44.Park D.J., Kang J.H., Lee J.W. Cost-effectiveness analysis of HLA-B5801 genotyping in the treatment of gout patients with chronic renal insufficiency in Korea. Arthritis Care Res. 2015;67(2):280–287. doi: 10.1002/acr.22409. [DOI] [PubMed] [Google Scholar]; Park DJ, Kang JH, Lee JW, Lee KE, Wen L, Kim TJ, et al. Cost-effectiveness analysis of HLA-B5801 genotyping in the treatment of gout patients with chronic renal insufficiency in Korea. Arthritis Care Res (Hoboken). 2015;67(2):280-287. [DOI] [PubMed]
- 45.Gonzalez-Galarza F.F., Takeshita L.Y., Santos E.J. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784-D788. [DOI] [PMC free article] [PubMed]
- 46.Allcock R.J.N. The major histocompatibility complex: a paradigm for study of human genome. In: Christiansen F.T., Tait B.D., editors. Immunogenetics: Methods and Applications in Clinical Practice, Methods in Molecular Biology. Spinger Science + Business Media; New York: 2012. [Google Scholar]; Allcock RJN. The Major Histocompatibility Complex: A Paradigm for Study of Human Genome. In: Christiansen FT, Tait BD, editors. Immunogenetics: Methods and Applications in Clinical Practice, Methods in Molecular Biology. New York: Spinger Science + Business Media; 2012.
- 47.Robinson J., Halliwell J.A., McWilliam H., Lopez R., Marsh S.G. IPD--the immuno polymorphism database. Nucleic Acids Res. 2013;41(Database issue):D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic acids research. 2013;41(Database issue):D1234-D1240. [DOI] [PMC free article] [PubMed]
- 48.Kwok J., Kwong K.M. Detection of HLA-B*58:01, the susceptible allele for allopurinol-induced hypersensitivity, by loop-mediated isothermal amplification. Br J Dermatol. 2013;168(3):526–532. doi: 10.1111/bjd.12097. [DOI] [PubMed] [Google Scholar]; Kwok J, Kwong KM. Detection of HLA-B*58:01, the susceptible allele for allopurinol-induced hypersensitivity, by loop-mediated isothermal amplification. The British journal of dermatology. 2013;168(3):526-532. [DOI] [PubMed]
- 49.Kwok J., Kwong K.M. Loop-mediated isothermal amplification for detection of HLA-B*58:01 allele. Tissue Antigens. 2013;81(2):83–92. doi: 10.1111/tan.12042. [DOI] [PubMed] [Google Scholar]; Kwok J, Kwong KM. Loop-mediated isothermal amplification for detection of HLA-B*58:01 allele. Tissue antigens. 2013;81(2):83-92. [DOI] [PubMed]
- 50.Yeo S.I. HLA-B*5801: utility and cost-effectiveness in the Asia-Pacific Region. International journal of rheumatic diseases. 2013;16(3):254–257. doi: 10.1111/1756-185X.12050. [DOI] [PubMed] [Google Scholar]; Yeo SI. HLA-B*5801: utility and cost-effectiveness in the Asia-Pacific Region. International journal of rheumatic diseases. 2013;16(3):254-257. [DOI] [PubMed]
- 51.Coleman W.B., Tsongalis G.J., editors. Molecular Diagnostics: For the Clinical Laboratorian. second ed. Humana Press, a part of Springer; 2006. [Google Scholar]; Coleman WB, Tsongalis GJ, editors. Molecular Diagnostics: For the Clinical Laboratorian. Second ed: Humana Press, a part of Springer; 2006.
- 52.Berger A. HLA typing. BMJ. 2001;322(7280):218. doi: 10.1136/bmj.322.7280.218. [DOI] [PMC free article] [PubMed] [Google Scholar]; Berger A. HLA typing. Bmj. 2001;322(7280):218. [DOI] [PMC free article] [PubMed]
- 53.Robinson J., Halliwell J.A., Hayhurst J.D., Flicek P., Parham P., Marsh S.G. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423-D431. [DOI] [PMC free article] [PubMed]
- 54.Tinckam K.J. Basic histocompatibility testing methods. In: Chandraker A., Sayegh M.H., Singh A.K., editors. Core Concepts in Renal Transplantation. Springer US; Boston, MA: 2012. pp. 21–42. [Google Scholar]; Tinckam KJ. Basic Histocompatibility Testing Methods. In: Chandraker A, Sayegh MH, Singh AK, editors. Core Concepts in Renal Transplantation. Boston, MA: Springer US; 2012. p. 21-42.
- 55.Kostenko L., Kjer-Nielsen L., Nicholson I. Rapid screening for the detection of HLA-B57 and HLA-B58 in prevention of drug hypersensitivity. Tissue Antigens. 2011;78(1):11–20. doi: 10.1111/j.1399-0039.2011.01649.x. [DOI] [PubMed] [Google Scholar]; Kostenko L, Kjer-Nielsen L, Nicholson I, Hudson F, Lucas A, Foley B, et al. Rapid screening for the detection of HLA-B57 and HLA-B58 in prevention of drug hypersensitivity. Tissue antigens. 2011;78(1):11-20. [DOI] [PubMed]
- 56.Dunckley H. HLA typing by SSO and SSP methods. In: Christiansen F.T., Tait B.D., editors. Immunogenetics: Methods and Applications in Clinical Practice. Humana Press; Totowa, NJ: 2012. pp. 9–25. [Google Scholar]; Dunckley H. HLA Typing by SSO and SSP Methods. In: Christiansen FT, Tait BD, editors. Immunogenetics: Methods and Applications in Clinical Practice. Totowa, NJ: Humana Press; 2012. p. 9-25. [DOI] [PubMed]
- 57.Giardina E., Stocchi L., Foti Cuzzola V. A fluorescence-based sequence-specific primer PCR for the screening of HLA-B(*)57:01. Electrophoresis. 2010;31(21):3525–3530. doi: 10.1002/elps.201000283. [DOI] [PubMed] [Google Scholar]; Giardina E, Stocchi L, Foti Cuzzola V, Zampatti S, Gambardella S, Patrizi MP, et al. A fluorescence-based sequence-specific primer PCR for the screening of HLA-B(*)57:01. Electrophoresis. 2010;31(21):3525-3530. [DOI] [PubMed]
- 58.Uchiyama K., Kubota F., Ariyoshi N., Matsumoto J., Ishii I., Kitada M. Development of a simple method for detection of the HLA-A*31:01 allele. Drug Metab Pharmacokinet. 2013;28(5):435–438. doi: 10.2133/dmpk.dmpk-12-nt-136. [DOI] [PubMed] [Google Scholar]; Uchiyama K, Kubota F, Ariyoshi N, Matsumoto J, Ishii I, Kitada M. Development of a simple method for detection of the HLA-A*31:01 allele. Drug metabolism and pharmacokinetics. 2013;28(5):435-438. [DOI] [PubMed]
- 59.Heinemann F.M. HLA genotyping and antibody characterization using the Luminex multiplex technology. Transfus Med Hemotherapy : offizielles Organ der Deutschen Gesellschaft fur Transfusionsmedizin und Immunhamatologie. 2009;36(4):273–278. doi: 10.1159/000228834. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heinemann FM. HLA Genotyping and Antibody Characterization Using the Luminex Multiplex Technology. Transfusion medicine and hemotherapy : offizielles Organ der Deutschen Gesellschaft fur Transfusionsmedizin und Immunhamatologie. 2009;36(4):273-278. [DOI] [PMC free article] [PubMed]
- 60.Trajanoski D., Fidler S.J. HLA typing using bead-based methods. In: Christiansen F.T., Tait B.D., editors. Immunogenetics: Methods and Applications in Clinical Practice. Humana Press; Totowa, NJ: 2012. pp. 47–65. [Google Scholar]; Trajanoski D, Fidler SJ. HLA Typing Using Bead-Based Methods. In: Christiansen FT, Tait BD, editors. Immunogenetics: Methods and Applications in Clinical Practice. Totowa, NJ: Humana Press; 2012. p. 47-65. [DOI] [PubMed]
- 61.Smith L.K. HLA typing by direct DNA sequencing. Methods Mol Biol. 2012;882:67–86. doi: 10.1007/978-1-61779-842-9_5. [DOI] [PubMed] [Google Scholar]; Smith LK. HLA typing by direct DNA sequencing. Methods in molecular biology (Clifton, NJ). 2012;882:67-86. [DOI] [PubMed]
- 62.Smith L.K. HLA typing by direct DNA sequencing. In: Christiansen F.T., Tait B.D., editors. Immunogenetics: Methods and Applications in Clinical Practice. Humana Press; Totowa, NJ: 2012. pp. 67–86. [Google Scholar]; Smith LK. HLA Typing by Direct DNA Sequencing. In: Christiansen FT, Tait BD, editors. Immunogenetics: Methods and Applications in Clinical Practice. Totowa, NJ: Humana Press; 2012. p. 67-86.
- 63.Zhang X., Ma H., Hu C. Detection of HLA-B*58:01 with TaqMan assay and its association with allopurinol-induced sCADR. Clin Chem Lab Med. 2015;53(3):383–390. doi: 10.1515/cclm-2014-0251. [DOI] [PubMed] [Google Scholar]; Zhang X, Ma H, Hu C, Yu B, Ma W, Wu Z, et al. Detection of HLA-B*58:01 with TaqMan assay and its association with allopurinol-induced sCADR. Clin Chem Lab Med. 2015;53(3):383-390. [DOI] [PubMed]
- 64.Kang X., Chen R., Han M. Rapid and reliable genotyping of HLA-B*58:01 in four Chinese populations using a single-tube duplex real-time PCR assay. Pharmacogenomics. 2016;17(1):47–57. doi: 10.2217/pgs.15.160. [DOI] [PubMed] [Google Scholar]; Kang X, Chen R, Han M, Liu Z, Liu J, Dai P, et al. Rapid and reliable genotyping of HLA-B*58:01 in four Chinese populations using a single-tube duplex real-time PCR assay. Pharmacogenomics. 2016;17(1):47-57. [DOI] [PubMed]
- 65.Nguyen D.V., Vida C., Chu H.C., Fulton R., Li J., Fernando S.L. Validation of a rapid, robust, inexpensive screening method for detecting the HLA-B*58:01 allele in the prevention of allopurinol-induced severe cutaneous adverse reactions. Allergy Asthma Immunol Res. 2016;8 doi: 10.4168/aair.2017.9.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nguyen DV, Vida C, Chu HC, Fulton R, Li J, Fernando SL. Validation of a Rapid, Robust, Inexpensive Screening Method for Detecting the HLA-B*58:01 Allele in the Prevention of Allopurinol-Induced Severe Cutaneous Adverse Reactions. Allergy Asthma Immunol Res. 2016;8. [DOI] [PMC free article] [PubMed]
- 66.Nguyen D.V., Vidal C., Li J., Fulton R., Fernando S.L. Validation of a rapid test for HLA-B*58:01/57:01 allele screening to detect individuals at risk for drug induced hypersensitivity. Pharmacogenomics. 2016;17(5) doi: 10.2217/pgs.15.185. [DOI] [PubMed] [Google Scholar]; Nguyen DV, Vidal C, Li J, Fulton R, Fernando SL. Validation of a rapid test for HLA-B*58:01/57:01 allele screening to detect individuals at risk for drug induced hypersensitivity. Pharmacogenomics. 2016;17(5). [DOI] [PubMed]
- 67.Nguyen D.V., Vidal C., Chi H.C. A novel multiplex polymerase chain reaction assay for detection of both HLA-A*31:01/HLA-B*15:02 alleles, which confer susceptibility to carbamazepine-induced severe cutaneous adverse reactions. HLA. 2017;90(6):335–342. doi: 10.1111/tan.13143. [DOI] [PubMed] [Google Scholar]; Nguyen DV, Vidal C, Chi HC, Do NTQ, Fulton R, Li J, et al. A novel multiplex polymerase chain reaction assay for detection of both HLA-A*31:01/HLA-B*15:02 alleles, which confer susceptibility to carbamazepine-induced severe cutaneous adverse reactions. HLA. 2017;90(6):335-342. [DOI] [PubMed]
- 68.Hammond E., Mamotte C., Nolan D., Mallal S. HLA-B*5701 typing: evaluation of an allele-specific polymerase chain reaction melting assay. Tissue Antigens. 2007;70(1):58–61. doi: 10.1111/j.1399-0039.2007.00840.x. [DOI] [PubMed] [Google Scholar]; Hammond E, Mamotte C, Nolan D, Mallal S. HLA-B*5701 typing: evaluation of an allele-specific polymerase chain reaction melting assay. Tissue antigens. 2007;70(1):58-61. [DOI] [PubMed]
- 69.Nguyen D.V., Vidal C., Chu H.C. Validation of a novel real-time PCR assay for detection of HLA-B*15:02 allele for prevention of carbamazepine - induced Stevens-Johnson syndrome/Toxic Epidermal Necrolysis in individuals of Asian ancestry. Hum Immunol. 2016;77(12):1140–1146. doi: 10.1016/j.humimm.2016.08.010. [DOI] [PubMed] [Google Scholar]; Nguyen DV, Vidal C, Chu HC, Do NT, Tran TT, Le HT, et al. Validation of a novel real-time PCR assay for detection of HLA-B*15:02 allele for prevention of carbamazepine - Induced Stevens-Johnson syndrome/Toxic Epidermal Necrolysis in individuals of Asian ancestry. Human immunology. 2016;77(12):1140-1146. [DOI] [PubMed]
- 70.Nguyen D.V., Vida C., Chu H.C., Fulton R., Li J., Fernando S.L. Validation of a Rapid, Robust, Inexpensive Screening Method for Detecting the HLA-B∗58:01 Allele in the Prevention of Allopurinol-Induced Severe Cutaneous Adverse Reactions. Allergy Asthma Immunol Res. 2017 Jan;9(1):79–84. doi: 10.4168/aair.2017.9.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nguyen DV, Vidal C, Chu HC, Fulton RB, Li J, Fernando SL. Validation of a rapid, robust and inexpensive screening method for detection of HLA-B*58:01 in prevention of allopurinol-induced severe cutaneous adverse reactions. . Allergy, Asthma and Immunology Research. 2016. [DOI] [PMC free article] [PubMed]
- 71.Nguyen D.V., Vidal C., Li J., Fulton R.B., Fernando S.L. Validation of a rapid test for HLA-B*58:01/57:01 allele screening to detect individuals at risk for drug-induced hypersensitivity. Pharmacogenomics. 2016;17(5):473–480. doi: 10.2217/pgs.15.185. [DOI] [PubMed] [Google Scholar]; Nguyen DV, Vidal C, Li J, Fulton RB, Fernando SL. Validation of a rapid test for HLA-B*58:01/57:01 allele screening to detect individuals at risk for drug-induced hypersensitivity. Pharmacogenomics. 2016;17(5):473-480. [DOI] [PubMed]
- 72.Navarro E., Serrano-Heras G., Castano M.J., Solera J. Real-time PCR detection chemistry. Clinica chimica acta; international journal of clinical chemistry. 2015;439:231–250. doi: 10.1016/j.cca.2014.10.017. [DOI] [PubMed] [Google Scholar]; Navarro E, Serrano-Heras G, Castano MJ, Solera J. Real-time PCR detection chemistry. Clinica chimica acta; international journal of clinical chemistry. 2015;439:231-250. [DOI] [PubMed]
- 73.Nguyen DV, Vidal C, Chu HC, et al. A Novel Multiplex PCR Assay for Detection of Both HLA-A*31:01/HLA-B*15:02 Alleles, Which Confer Susceptibility to Carbamazepine-Induced Severe Cutaneous Adverse Reactions. (HLA.n/a-n/a). [DOI] [PubMed]
- 74.Varney M.D., Castley A.S.L., Haimila K., Saavalainen P. Methods for diagnostic HLA typing in disease association and drug hypersensitivity. In: Christiansen F.T., Tait B.D., editors. Immunogenetics: Methods and Applications in Clinical Practice. Humana Press; Totowa, NJ: 2012. pp. 27–46. [Google Scholar]; Varney MD, Castley ASL, Haimila K, Saavalainen P. Methods for Diagnostic HLA Typing in Disease Association and Drug Hypersensitivity. In: Christiansen FT, Tait BD, editors. Immunogenetics: Methods and Applications in Clinical Practice. Totowa, NJ: Humana Press; 2012. p. 27-46. [DOI] [PubMed]
- 75.de Bakker P.I., McVean G., Sabeti P.C. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]; de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nature genetics. 2006;38(10):1166-1172. [DOI] [PMC free article] [PubMed]
- 76.Maekawa K., Nishikawa J., Kaniwa N. Development of a rapid and inexpensive assay for detecting a surrogate genetic polymorphism of HLA-B*58:01: a partially predictive but useful biomarker for allopurinol-related Stevens-Johnson syndrome/toxic epidermal necrolysis in Japanese. Drug Metab Pharmacokinet. 2012;27(4):447–450. doi: 10.2133/dmpk.dmpk-11-nt-120. [DOI] [PubMed] [Google Scholar]; Maekawa K, Nishikawa J, Kaniwa N, Sugiyama E, Koizumi T, Kurose K, et al. Development of a rapid and inexpensive assay for detecting a surrogate genetic polymorphism of HLA-B*58:01: a partially predictive but useful biomarker for allopurinol-related Stevens-Johnson syndrome/toxic epidermal necrolysis in Japanese. Drug metabolism and pharmacokinetics. 2012;27(4):447-450. [DOI] [PubMed]
- 77.Thorstensen K., Kvitland M., Shirzadi M., Helde G., Moen T., Brodtkorb E. Carbamazepine-induced cutaneous reactions: a simple assay to identify patients carrying the HLA-A*31:01 allele. Scand J Clin Lab Invest. 2014;74(7):644–647. doi: 10.3109/00365513.2014.921835. [DOI] [PubMed] [Google Scholar]; Thorstensen K, Kvitland M, Shirzadi M, Helde G, Moen T, Brodtkorb E. Carbamazepine-induced cutaneous reactions: A simple assay to identify patients carrying the HLA-A*31:01 allele. Scandinavian journal of clinical and laboratory investigation. 2014;74(7):644-647. [DOI] [PubMed]
- 78.Colombo S., Rauch A., Rotger M. The HCP5 single-nucleotide polymorphism: a simple screening tool for prediction of hypersensitivity reaction to abacavir. J Infect Dis. 2008;198(6):864–867. doi: 10.1086/591184. [DOI] [PubMed] [Google Scholar]; Colombo S, Rauch A, Rotger M, Fellay J, Martinez R, Fux C, et al. The HCP5 single-nucleotide polymorphism: a simple screening tool for prediction of hypersensitivity reaction to abacavir. The Journal of infectious diseases. 2008;198(6):864-867. [DOI] [PubMed]
- 79.Galvan C.A., Elbarcha O.C., Fernandez E.J., Beltramo D.M., Soria N.W. Rapid HCP5 single-nucleotide polymorphism genotyping: a simple allele-specific PCR method for prediction of hypersensitivity reaction to Abacavir. Clinica chimica acta. international journal of clinical chemistry. 2011;412(15-16):1382–1384. doi: 10.1016/j.cca.2011.04.010. [DOI] [PubMed] [Google Scholar]; Galvan CA, Elbarcha OC, Fernandez EJ, Beltramo DM, Soria NW. Rapid HCP5 single-nucleotide polymorphism genotyping: a simple allele-specific PCR method for prediction of hypersensitivity reaction to Abacavir. Clinica chimica acta; international journal of clinical chemistry. 2011;412(15-16):1382-1384. [DOI] [PubMed]
- 80.He Y., Hoskins J.M., Clark S. Accuracy of SNPs to predict risk of HLA alleles associated with drug-induced hypersensitivity events across racial groups. Pharmacogenomics. 2015;16(8):817–824. doi: 10.2217/pgs.15.41. [DOI] [PubMed] [Google Scholar]; He Y, Hoskins JM, Clark S, Campbell NH, Wagner K, Motsinger-Reif AA, et al. Accuracy of SNPs to predict risk of HLA alleles associated with drug-induced hypersensitivity events across racial groups. Pharmacogenomics. 2015;16(8):817-824. [DOI] [PubMed]
- 81.Zhu G.D., Brenton A.A., Malhotra A. Genotypes at rs2844682 and rs3909184 have no clinical value in identifying HLA-B*15:02 carriers. Eur J Clin Pharmacol. 2015;71(8):1021–1023. doi: 10.1007/s00228-015-1879-y. [DOI] [PubMed] [Google Scholar]; Zhu GD, Brenton AA, Malhotra A, Riley BJ, Church KE, Espin FG, et al. Genotypes at rs2844682 and rs3909184 have no clinical value in identifying HLA-B*15:02 carriers. European journal of clinical pharmacology. 2015;71(8):1021-1023. [DOI] [PubMed]
- 82.Chen Z., Zhang S., Zhang J., Zhang Y., Xue L., Miao L. rs9263726 is a specific genetic marker for allopurinol-induced severe cutaneous adverse reactions in Chinese patients. Pers Med. 2015;12(6):585–592. doi: 10.2217/pme.15.38. [DOI] [PubMed] [Google Scholar]; Chen Z, Zhang S, Zhang J, Zhang Y, Xue L, Miao L. rs9263726 is a specific genetic marker for allopurinol-induced severe cutaneous adverse reactions in Chinese patients. Personalized Medicine. 2015;12(6):585-592. [DOI] [PubMed]
- 83.Vidal C., Li J., Fulton R., Fernando S.L. A polymorphism within the psoriasis susceptibility 1 candidate 1 (PSORS1C1) gene is not linked to HLA-B*58:01 in an Australian cohort. Drug Metab Pharmacokinet. 2016;31(3):252–255. doi: 10.1016/j.dmpk.2015.08.007. [DOI] [PubMed] [Google Scholar]; Vidal C, Li J, Fulton R, Fernando SL. A polymorphism within the psoriasis susceptibility 1 candidate 1 (PSORS1C1) gene is not linked to HLA-B*58:01 in an Australian cohort. Drug metabolism and pharmacokinetics. 2016;31(3):252-255. [DOI] [PubMed]
- 84.Cheng S.H., Kwan P., Ng H.K., Ng M.H. New testing approach in HLA genotyping helps overcome barriers in effective clinical practice. Clin Chem. 2009;55(8):1568–1572. doi: 10.1373/clinchem.2009.127894. [DOI] [PubMed] [Google Scholar]; Cheng SH, Kwan P, Ng HK, Ng MH. New testing approach in HLA genotyping helps overcome barriers in effective clinical practice. Clinical chemistry. 2009;55(8):1568-1572. [DOI] [PubMed]
- 85.Virakul S., Kupatawintu P., Nakkuntod J., Kangwanshiratada O., Vilaivan T., Hirankarn N. A nested sequence-specific primer-polymerase chain reaction for the detection of HLA-B*15:02. Tissue Antigens. 2012;79(4):295–301. doi: 10.1111/j.1399-0039.2012.01836.x. [DOI] [PubMed] [Google Scholar]; Virakul S, Kupatawintu P, Nakkuntod J, Kangwanshiratada O, Vilaivan T, Hirankarn N. A nested sequence-specific primer-polymerase chain reaction for the detection of HLA-B*15:02. Tissue antigens. 2012;79(4):295-301. [DOI] [PubMed]
- 86.Aoki M., Hosono N., Takata S., Nakamura Y., Kamatani N., Kubo M. New pharmacogenetic test for detecting an HLA-A*31: 01 allele using the InvaderPlus assay. Pharmacogenetics Genom. 2012;22(6):441–446. doi: 10.1097/FPC.0b013e3283527c40. [DOI] [PubMed] [Google Scholar]; Aoki M, Hosono N, Takata S, Nakamura Y, Kamatani N, Kubo M. New pharmacogenetic test for detecting an HLA-A*31: 01 allele using the InvaderPlus assay. Pharmacogenetics and genomics. 2012;22(6):441-446. [DOI] [PubMed]
- 87.Niihara H., Kohno K., Taketani T. Simple and rapid detection of HLA-A*31:01 for prediction of carbamazepine-induced hypersensitivity using loop-mediated isothermal amplification method. J Dermatol Sci. 2014;74(1):88–92. doi: 10.1016/j.jdermsci.2013.11.013. [DOI] [PubMed] [Google Scholar]; Niihara H, Kohno K, Taketani T, Kaneko S, Ito T, Sugamori T, et al. Simple and rapid detection of HLA-A*31:01 for prediction of carbamazepine-induced hypersensitivity using loop-mediated isothermal amplification method. Journal of dermatological science. 2014;74(1):88-92. [DOI] [PubMed]
- 88.Cheung Y.K., Kwok M., Chan E., Kwan P. Rapid detection of HLA-A*31:01 allele in DNA and blood samples using loop-mediated isothermal amplification. Br J Dermatol. 2014;171(1):90–96. doi: 10.1111/bjd.12897. [DOI] [PubMed] [Google Scholar]; Cheung YK, Kwok M, Chan E, Kwan P. Rapid detection of HLA-A*31:01 allele in DNA and blood samples using loop-mediated isothermal amplification. The British journal of dermatology. 2014;171(1):90-96. [DOI] [PubMed]
- 89.Virakul S., Nakkuntod J., Kupatawintu P., Kangwanshiratada O., Paiboonkasarp S., Hirankarn N. Khonkaen University; 2010. Detection of HLA-B*5801 by In-House PCR-SSP. The 11th Graduate Research Conference; pp. 960–966. [Google Scholar]; Virakul S, Nakkuntod J, Kupatawintu P, Kangwanshiratada O, Paiboonkasarp S, Hirankarn N. Detection of HLA-B*5801 by in-house PCR-SSP. The 11th Graduate Research Conference: Khonkaen University 2010. p. 960-966.
- 90.Rodriguez-Novoa S., Cuenca L., Morello J. Use of the HCP5 single nucleotide polymorphism to predict hypersensitivity reactions to abacavir: correlation with HLA-B*5701. J Antimicrob Chemother. 2010;65(8):1567–1569. doi: 10.1093/jac/dkq204. [DOI] [PubMed] [Google Scholar]; Rodriguez-Novoa S, Cuenca L, Morello J, Cordoba M, Blanco F, Jimenez-Nacher I, et al. Use of the HCP5 single nucleotide polymorphism to predict hypersensitivity reactions to abacavir: correlation with HLA-B*5701. The Journal of antimicrobial chemotherapy. 2010;65(8):1567-1569. [DOI] [PubMed]
- 91.Dello Russo C., Lisi L., Lofaro A. Novel sensitive, specific and rapid pharmacogenomic test for the prediction of abacavir hypersensitivity reaction: HLA-B*57:01 detection by real-time PCR. Pharmacogenomics. 2011;12(4):567–576. doi: 10.2217/pgs.10.208. [DOI] [PubMed] [Google Scholar]; Dello Russo C, Lisi L, Lofaro A, Di Giambenedetto S, Federico B, Madeddu G, et al. Novel sensitive, specific and rapid pharmacogenomic test for the prediction of abacavir hypersensitivity reaction: HLA-B*57:01 detection by real-time PCR. Pharmacogenomics. 2011;12(4):567-576. [DOI] [PubMed]
- 92.Sankuntaw N., Chantarangsu S., Chantratita W., Sungkanuparph S., Kiertiburanakul S., Lulitanond V. Development of multiplex pyrosequencing for HLA-B*57:01 screening using single nucleotide polymorphism haplotype. J Clin Pharm Ther. 2014;39(5):545–550. doi: 10.1111/jcpt.12175. [DOI] [PubMed] [Google Scholar]; Sankuntaw N, Chantarangsu S, Chantratita W, Sungkanuparph S, Kiertiburanakul S, Lulitanond V. Development of multiplex pyrosequencing for HLA-B*57:01 screening using single nucleotide polymorphism haplotype. Journal of clinical pharmacy and therapeutics. 2014;39(5):545-550. [DOI] [PubMed]
- 93.Marino S.G., Jaramillo A., Fernández-Viña M.A. Handbook of Human Immunology [Internet] CRC Press-Taylor & Francis Group. Second; 2008. The human major histocompatibility complex and DNA-based typing of human leukocyte antigens for transplantation; pp. 541–564. [Google Scholar]; Marino SG, Jaramillo A, Fernandez-Viña MA. The Human Major Histocompatibility Complex and DNA-Based Typing of Human Leukocyte Antigens for Transplantation 2008. In: Handbook of Human Immunology [Internet]. CRC Press-Taylor & Francis Group. Second. [541-564].
- 94.Dos Santos EJ, McCabe A, Gonzalez-Galarza FF, Jones AR, Middleton D. Allele Frequencies Net Database: Improvements for storage of individual genotypes and analysis of existing data. Human Immunol. 2016;77:238–248. doi: 10.1016/j.humimm.2015.11.013. [DOI] [PubMed] [Google Scholar]; Dos Santos EJ, McCabe A, Gonzalez-Galarza FF, Jones AR, Middleton D.Allele Frequencies Net Database: Improvements for storage of individual genotypes and analysis of existing data. Human Immunol, 77,(2016) 238-48. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.