Abstract
Background
Diffuse intrinsic pontine glioma (DIPG) is a rare and fatal pediatric brain cancer without cure. Seeking therapeutic strategies is still a major challenge in DIPG research. Previous study has shown that dysregulation of G1/S cell cycle checkpoint was common in DIPG and this dysregulation is even more enriched in the H3.3K27 M mutant subgroup. Here we assess potential anti-tumor efficacy of palbociclib, a specific and cytostatic inhibitor of CDK4/6, on high grade H3.3-K27 M-mutant DIPGs in vitro and in vivo.
Methods
We established patient-derived cell lines from treatment-naïve specimens. All the lines have H3.3K27 M mutation. We used a range of biological in vitro assays to assess the effect of palbociclib on growth of DIPGs. Palbociclib activity was also assayed in vivo against three independent DIPG orthotropic xenografts model.
Findings
Dysregulation of G1/S cell cycle checkpoint is enriched in these DIPGs. Then, we showed that depletion of CDK4 or CDK6 inhibits DIPG cells growth and blocks G1/S transition. Furthermore, palbociclib effectively repressed DIPG growth in vitro. Transcriptome analysis showed that palbociclib not only blocks G1/S transition, it also blocks other oncogenic targets such as MYC. Finally, palbociclib activity was assayed in vivo against DIPG orthotropic xenografts to demonstrate the high efficiency of blocking tumor growth.
Interpretation
Our findings thus revealed that palbociclib could be the therapeutic strategy for treatment-naïve DIPG with H3.3K27 M mutation.
Fund
Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support, Beijing Municipal Natural Science Foundation, Ministry of Science and Technology of China, and National Natural Science Foundation of China.
Keywords: DIPG, Cell cycle, Palbociclib, H3.3-K27 M mutation
Research in context.
Evidence before this study
The integrated analysis of over 1000 cases of pediatric high-grade glioma and DIPG has shown that dysregulation of G1/S cell cycle checkpoint was common in DIPG and this dysregulation is even more enriched in the H3.3K27 M mutant subgroup. Study showed that H3.3K27 M mediated epigenetic silencing of P16INK4A, the inhibitor of CDK4/6, and the therapy targeting on EZH2 was shown to be effective due to the induction of P16. Therefore, G1/S cell cycle checkpoint could be the potential therapeutic target for DIPG. Palbociclib (PD0332991, Pfizer), a specific, cytostatic inhibitor of cyclin-dependent kinase CDK4/6 had been approved by the United States (US) Food and Drug Administration (FDA) for treating breast cancer. Previous study showed that palbociclib prolongs survival in a PDGF-B driven, Ink4a-ARF, p53 deficient genetically engineered mouse model of DIPG. Combination use of CDK4/6 and mTOR inhibitors induce synergistic growth arrest of DIPG cells in vitro.
Added value of this study
In this report, we established patient-derived primary DIPG cell lines from treatment-naïve patients' stereotactic biopsy or open biopsy samples, which provide better materials to be investigated to reveal the cause of the disease and explore the potential therapeutic strategies for DIPG. We used these cell lines to demonstrate the anti-tumor efficacy of palbociclib both in vitro and in vivo. To our knowledge, this is the first time to show palbociclib has anti-tumor efficacy in vivo with DIPG orthotropic xenograft model. The transcriptome analysis showed that palbociclib not only blocks G1/S transition, it also blocks other oncogenic targets such as MYC. Furthermore, we showed that combination of CDK4/6 and EGFR inhibitiors in a EGFR highly expressed DIPG cell line synergistically arrested cancer cell growth both in vitro and in vivo.
Implications of all the available evidence
In conclusion, we have confirmed G1/S cell cycle checkpoint could be the potential therapeutic target for DIPG with H3.3-K27 M-mutation and have demonstrated that palbociclib could provide a therapeutic benefit to high grade treatment-naïve H3.3-K27 M-mutant DIPGs.
Alt-text: Unlabelled Box
1. Introduction
Diffuse intrinsic pontine glioma (DIPG) is a rare and deadly pediatric brain cancer with short survival of 9–12 months [1]. Various radiation therapies, while relieving the neurological symptoms, do little to improve overall survival [2]. Serveral small molecule chemicals targeting the oncogenic H3K27 M mutation have been reported to inhibit the growth of DIPG in vitro and in vivo [[3], [4], [5], [6]]. Plus, CDK7 inhibition, combination inhibition of PI3K/AKT and MEK/ERK pathways, dual targeting of NOTCH and MYCN, and blocking BMP pathway, all showed antitumor efficacy for DIPG [[7], [8], [9], [10]]. Furthermore, immunotherapy is also a promising option for treatment [11]. However, currently there are no clinical reports of effective treatment to improve survival. Therefore, finding new therapeutic strategies is still a major challenge in DIPG research.
One of the molecular signatures of DIPG is recurrent histone mutation H3K27M, which is believed to be one of the drivers of the tumorigenesis [12]. DIPG with the H3.3K27 M mutation are associated with the poorest outcome [13]. The integrated analysis of over 1000 cases of pediatric high-grade glioma and DIPG has shown that dysregulation of G1/S cell cycle checkpoint was common in DIPG and this dysregulation is even more enriched in the H3.3K27 M mutant subgroup [14]. Another study showed that H3.3K27 M mediated epigenetic silencing of P16INK4A, an inhibitor of CDK4/6, plays important roles in the pathogenesis of DIPG. Meanwhile, the therapy targeting on EZH2 was shown to be effective due to the induction of P16INK4A [5,15]. Therefore, G1/S cell cycle checkpoint could be a potential therapeutic target for DIPG.
Palbociclib (PD0332991) is a specific and cytostatic inhibitor of CDK4/6 at low nanomolar concentration, which binds the ATP-binding pocket of CDK4/6 blocking the phosphorylation of RB and subsequently promotes cell cycle arrest at G1 phase [16]. It has been approved by the US Food and Drug Administration (FDA) to treat patients with hormone receptor (HR)-positive, human epidermal growth factor receptor 2 (HER2) negative advanced or metastatic breast cancer combined with other drugs [16,17]. Previous study in GBM (glioblastoma multiform) orthotopic xenograft mouse model demonstrated that palbociclib could penetrate blood brain barrier (BBB) and has antitumor activity [[18], [19], [20]]. Another study also showed that palbociclib prolongs survival in a PDGF-B driven, Ink4a-ARF, p53 deficient genetically engineered mouse model of DIPG [21]. Combination use of CDK4/6 and mTOR inhibitors induce synergistic growth arrest of DIPG cells in vitro [22].
In this report, we established eight patient-derived DIPG cell lines with H3.3K27 M mutation from treatment-naïve specimens and used these cell lines to test the anti-tumor efficacy of palbociclib both in vitro and in vivo. First, we demonstrated that there is high expression of CDK4/6 in a cohort of pre-treatment DIPG tumors and panel of eight patient derived DIPG cell lines compared to normal fetal pons progenitor cells (PPCs). Plus, most of the DIPG cell lines have lower expression of P16INK4A compared to PPCs. Then, we showed that depletion of CDK4 or CDK6 inhibits DIPG cells growth and blocks G1/S transition. Furthermore, palbociclib effectively repressed all eight cell lines self-renewal, proliferation and cell cycle progression from G1 to S phase in vitro with much lower concentration compared to previous report. The transcriptome analysis showed that palbociclib not only blocks G1/S transition, it also blocks other oncogenic targets such as MYC. Finally, its activity was assayed in vivo with three DIPG orthotropic xenograft models. Our findings revealed that palbociclib effectively suppresses the growth of RB-proficient DIPG cells in vitro and in vivo.
2. Materials and methods
2.1. Ethics statement
For all human tissue studies, informed consent was obtained and Institutional Review Board approval was granted by Tsinghua University, Beijing, China and Beijing Tiantan Hospital, Capital Medical University, Beijing, China (KY2014–021-02 and KY2018–042-02). Both protocols were approved by the human research ethics committee of Tsinghua University and Beijing Tiantan Hospital. The written informed consent was obtained from the subjects or their guardians.
2.2. Primary DIPG cell lines establishment and propagation
The patient derived DIPG cell lines were established as previously described [23]. The cell lines TT10630, TT10714, TT10728 and TT11201 were renamed as TT150630, TT150714, TT150728 and TT151201 to mark the year when they were established. All these primary DIPG cells we used are within 20 passages. DIPG17 (SU-DIPG17), which was reported previously [11], was a kind gift from Dr. Yujie Tang in Shanghai Jiao Tong University. DIPG cells were cultured in matrigel (356,243, BD, 1%, 4-12 h at 37 °C) coated plates with serum-free medium containing DMEM (C11995500BT, Invitrogen), B27 (1:50), N2 (1100), insulin (20 μg/ml), bFGF (20 ng/ml), EGF (20 ng/ml), PDGF-AB (20 ng/ml) (PeproTech).
2.3. Establishment of primary pontine neural progenitor cell lines (PPCs)
The primary PPC cell lines were established as previously reported [24]. In brief, aborted human embryos (9–12 weeks) of either sex were obtained from Beijing Tiantan Hospital. The embryos were stored on ice until sent to the lab within 24 h after abortion. The hindbrain of the fetus was soaked in Hibernation buffer and the meninges was removed. The tissue was dissociated mechanically into pieces and incubated in DMEM supplemented with Papain (Worthington, 20 units/ml) and DNase I (Sigma, 5 Kunitz units/ml) at 37 °C for 30 min. Pipet up and down every 10 min until a single-cell suspension was formed. The supernatant was removed after centrifuging at 300g for 5 min. Washed the pellet with DMEM twice and resuspended the pellet in DMEM supplemented with N2 (Gibco, 1:100), B27 (Gibco, 1:50), EGF (PeproTech, 20 ng/ml), bFGF (PeproTech, 20 ng/ml), and penicillin streptomycin (Gibco, 1:100). The cells were then plated into dishes coated with matrigel (BD). Medium was changed 24 h after plating, and the cells were passaged using 0.05% trypsin every 3–5 days. Half of the medium was changed every 2–3 days to maintain culture condition. All the cells we used are within 5 passages.
2.4. Sphere formation assay
Sphere formation assay was performed by seeding 1000 DIPG cells per well in 96-well plates (Excellbio) treated with vehicle or 100 nM palbociclib with triplicates for each condition. Cells were cultured for 10 days. The images for each well were taken by Opera Phenix (Perkin Elmer) with 5× objective lens in the brightfield. The images were analyzed by the software of HARMONY.
2.5. DIPG xenograft mouse models and administration of palbociclib
All animal experiments were done in accordance with the guidelines provided by the Tsinghua University Animal Care and Use Committee. Orthotopic xenograft mouse model were established using 4-week female NOD-Prkdcscid1l2rgtm1/Bcgen mice (Biocytogen). Luciferase engineered DIPG cells were resuspended in 5 μl PBS at the density of 20,000 cells/μl and implanted into the brainstem of immunodeficient mouse as previously described [23]. Two weeks after the implantation, the tumor burden was measured by bioluminescence imaging once a week. In brief, mice were anesthetized with isoflurane before bulbus oculi injection and imaging began 1 min after administration of D-luciferin (Goldbio). Anesthesia was maintained during imaging by nose cone delivery of isoflurane as necessary. Once the luminescence signal was detected, the tumor bearing mice were assigned to palbociclib or vehicle treated groups randomly, then treated with palbociclib at 150 mg/kg/day or vehicle intragastrically for 21 days continuously. For the combination of palbociclib and Erlotinib treatment, palbociclib were administered at 150 mg/kg/day for 14 days, followed by 75 mg/kg palbociclib with 37.5 mg/kg erlotinib for 7 days. If the body weight loss of the mouse reached 5% per day, the administration of the drugs would be suspended for one day. For the survival study, tumor-bearing mice were monitored daily until animals become moribund, as evidenced by a 20% loss of body weight or other indicators, for example, inability to ambulate, agonal breathing, and loss of consciousness, and then euthanized. Bioluminescence imaging was performed once a week. Bioluminescence imaging was taken by IVIS Spectrum (PerkinElmer). Bioluminescence signal was measured using the ROI tool in Living Image 4.4 software (PerkinElmer).
2.6. Plasmids and reagents
The shRNA-expressing Lentivirus system was described previously [25]. To generate pLKO plasmids containing short hairpins against CDK4 and CDK6, the pLKO vector was ligated with the annealed oligos. The sequences of shRNA oligo pairs are shown in Supplementary Table 2. Luciferase-GFP were cloned into pLEX based lentivirus vector, which was used to establish DIPG cell lines with luciferase-GFP [26].
The antibodies used in this study were anti-H3K27 M (ABE219, Millipore), anti-H3K27me3 (61,017, active motif), anti-H3K27Ac (ab191, Abcam), anti-H3 (BE3015, Easybio), anti-CDK4 (AC251, Beyotime), anti-CDK6 (AC256, Beyotime), anti-P-RB-S780 (ab173289, Abcam), anti-Ki67-PE conjugate antibody (12–5698-80, eBioscience), anti-CD133 (ab19898, abcam), anti-Nestin (Rat-401,DSHB), anti-GFAP (Z033401–2, DAKO), anti-Olig2 (AB9610, Milipore) and anti-RB (ab181616, Abcam). Secondary antibodies for western blot (Goat anti mouse/rabbit) were from Jackson. Secondary antibodies for flow cytometry (Goat anti mouse/rabbit) were from Huaxingbio. Enhanced chemiluminescence system (Tanon 5200) was used for detection. Palbociclib (PD0332991) were purchased from MedChemExpress.
2.7. Western blot analyses and RT-qPCR analyses
The acid extraction of histone was performed as previously described [27]. The cell lysates were prepared using RIPA lysis buffer plus phosphatase inhibitor cocktail (Roche) and proteinase inhibitor cocktail (Sigma). Western immunoblotting was done as previously described [28]. The dilution conditions of primary antibodies used in this study are phosphorylated-RB S780 (1:10000), RB (1:1000), GAPDH (1:2000). H3K27me3 (1:1000), H3K27ac (1:1000), H3K27 M (1:1000), H3 (110000).
Total RNA was extracted by using Total RNA Purification Kit (GeneMark, Taiwan), followed by reverse transcription using a Thermoscript RT-PCR kit (Invitrogen). qRT-PCR was performed on a ViiA 7 Real-Time PCR system (Life Technologies). Primer sequences were designed using primer3 software. Sequences of the synthesized primers used for qRT-PCR assays are shown below. qRT-PCR conditions were 40 cycles at 95 °C for 30s, 60 °C for 30s, 72 °C for 30s, and 72 °C for 10 min. RT-qPCR primers used in this research are listed below:
CDK4: Forward: 5′ATCTTTGACCTGATTGGGCT3′;
Reverse: 5′CATCTCAGGTACCACCGAC3′.
CDK6: Forward: 5′ACCTACTTCTGAAGTGTTTGAC3′;
Reverse: 5′TCCTGGAAGTATGGGTGAG3′.
CCNA: Forward: 5′GCCATCAGTTATTGCTGGA3′;
Reverse: 5′CGTATTAATGATTCAGGCCAG3′.
CDKN2A:Forward: 5′ GAAGGTCCCTCAGACATCC3′;
Reverse: 5′ GTAGGACCTTCGGTGACTG 3′.
2.8. Cell viability assay
Cell viability was measured using CellTiter-Glo (G7572, Promega) according to the manufacturer's protocol. DIPG cells were treated with palbociclib at the concentration of 6.25 nM, 25 nM, 100 nM, 400 nM, 1600 nM, 6400 nM, and 25600 nM or 0.01% DMSO. Cell viabilities were assessed before treatment (T0) and 72 h after treatment (T72). Relative viability was calculated as T72-T0, and the value for DMSO-treated cells was set to be 100%. For each concentration, triplicate wells were used. IC50 values were calculated using Prism 6 (Graphpad). Combination Index(CI) were calculated by the software of CompuSyn (ComboSyn, Inc). The CI < 1, =1, and > 1 indicates synergism, additive effect and antagonism, respectively [29].
2.9. Cell cycle analysis and flow cytometry
Cell cycle analysis: 106 DIPG cells treated with vehicle or palbociclib (100 nM) were treated with 0.05% trypsin EDTA, then washed with ice-cold PBS twice and incubated in 70% ethanol at 4 °C overnight. The cell pellets were collected and washed twice with cold PBS. The cell pellets were then resuspended and incubated in staining buffer containing RNase A (100 μg/ml; Transgen biotech) and propidium iodide (50 μg/ml; Leagene) at 37 °C for 30 min. After centrifugation, discard the staining buffer and resuspend the pellet in cold PBS, then the cell cycle was analyzed by flow cytometry(BD FACS Calibur) and the data were analyzed using Modfit software (Verity Software House).
Flow cytometry: Lineage marker, which include CD133, Nestin, GFAP, Olig2, and Ki67, staining followed by flow cytometry was performed using monoclonal Ki67-PE conjugate antibody, CD133, Nestin, GFAP, Olig2 primary antibody and coresponding secondary antibody. In brief, 1 × 106 DIPG cells were washed in 1 ml cold PBS twice and fixed in 70% ethanol at 4 °C overnight. Cells were then washed with cold PBS and permeabilized by washing in PBS containing 0.3% Triton X-100 for 3 times. The cells were blocked by incubating in PBS containing 5% BSA for 30 min at 4 °C. After incubation, cells were exposed to 100ul diluted primary antibody (followed the instructions of the antibody manufactures) for 30 min at 4 °C. After washing in PBS twice, the percentage of Ki67 positive cells were analyzed by flow cytometry. For other lineage markers staining, the cells were incubated in the corresponding secondary antibodies, then analyzed by flow cytometry (BD FACS Calibur). The data were analyzed by FlowJo V10 (FlowJo, LLC).
2.10. Apoptosis analysis
Cells were harvested and stained using the AnnexinV/PI Apoptosis Detection Kit as described by the manufacturer (BD). FACS analysis was performed after staining to count the proportion of cells undergoing apoptosis. The original data were analyzed using FlowJo V10 (FlowJo, LLC).
2.11. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC)
Mouse xenograft pons tissues were fixed in 4% paraformaldehyde (PFA), followed by dehydration in ethanol gradients, permeabilization with xylene and paraffin embedding. The paraffin embedded pons tissues were sliced into many sections, on a microtome and float in a 40 °C water bath with distill water. The sections were transferred onto glass slides suitable for H&E staining and IHC. Allow the slides to dry overnight.
H&E staining: Sections with 5 μm thickness were deparaffinized with xylene for twice, 3 min each. Transfer slides to 100% alcohol and then transfer once through 95%, 70%, and 50% alcohols respectively for 3 min each. The slides were stained with H&E.
IHC: The deparaffinized sections were washed 3 times with 100% alcohol and then transfer once through 95%, 70%, and 50% alcohols respectively for 3 min each. The slides were boiled with Antigen Retrieval Solution (ORIGENE) in microwave oven for 15 min. Tissue sections were blocked in 5% BSA in PBS for 1 h. Then apply 100 μl appropriately diluted primary antibody to the sections on the slides for 12 h in a humidified chamber. Rinse the slides with PBS twice. Then the sections were incubated with biotinylated goat anti-rabbit IgG (HRP/DAB Detection IHC Kit, ORIGENE) for 1 h. Wash slides with PBS twice. Add 20 μl DAB plus chromogen to 1 ml of DAB plus substrate, mix by swirling and apply to tissues. Incubate for 5 min. Rinse 4 times with PBS. Add enough drops of hematoxylin to cover the section. Incubate for 1 min. Rinse 7–8 times in tap water followed by dehydration in ethanol gradients. Add mounting medium to cover the section.
H&E staining and IHC sections were analyzed by Axio Scan. Z1 (Zeiss).
2.12. RNA-seq library preparation and sequencing
Total RNA was extracted by using Total RNA Purification Kit (GeneMark, Taiwan). RNA was prepared for sequencing using the Illumina TruSeq Stranded Total RNA Library Prep Kit. The libraries were sequenced on the Illumina HiSeq X platform (Novogen, China).
2.13. RNA-seq analysis
For RNA-seq data analysis, we first evaluated RNA-seq reads quality using FastQC (version 0.10.1). Total RNA-seq reads were trimmed from the 3′ end until the final base had a quality score > 30, using Trimmomatic. Then, RNA-seq was mapped to the human reference genome (hg38) with HISAT2 and summarized as gene level fragments per kilobase per million reads sequenced (FPKM) using cufflinks. Differential testing and log2 fold change calculation was performed using cuffdiff with two biological replicates. Gene ontology (GO) analysis and gene set enrichment analysis (GSEA) were performed as previously described [30,31].
2.14. Statistical analysis
All the experiments were performed at least three times. Graphpad prism 6 was used for statistical analysis. t-test were used to determine the significance of differences between groups. P < .05 was considered to be statistically significant.
3. Results
3.1. Patient-derived DIPG cell lines with H3.3K27 M mutation have high CDK4/6 expression
The limitation of tumor tissues and experimental models used to be the major barrier for DIPG research. Recent advances in establishment of patient derived cell lines and orthotopic xenograft mouse models provided opportunity to explore the origin of the disease and biological features of DIPG [3,32,33]. Initially the DIPG patient derived primary cell lines were established from autopsy samples, for which the patients may have been treated with radiotherapy or chemotherapy. These cell lines are not the ideal materials for testing drug efficacy because radiation and chemotherapy could induce extra mutations.
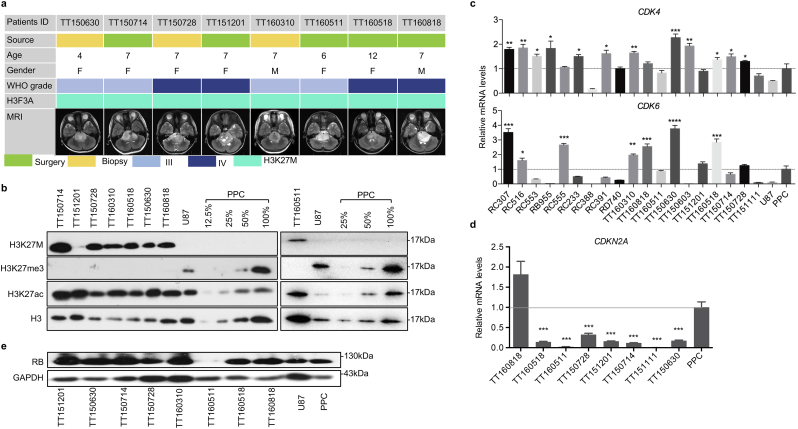
We established patient-derived primary DIPG cell lines from treatment-naïve patients' stereotactic biopsy or open biopsy samples, which provide better materials to be investigated to reveal the cause of the disease and explore the potential therapeutic strategies for DIPG. The clinical and molecular characteristics of the patients are summarized in Fig. 1a, Supplementary Fig. S1, Table. S1 and S2. Patients' brain magnetic resonance imaging (MRI) reveal infiltrative tumors in pons. All the cancer cell lines and tumors have H3.3K27 M mutation. Most of cell lines have high percentage of CD133, Nestin, and Olig2 positive cells [23] (Supplementary Table S2), suggesting that these DIPG cell lines are similar to neural progenitor cells (NPCs) or oligodendrocyte precursor cells (OPCs). Given the unavailability of para-carcinoma tissues from DIPG patients, we also established fetal pons primary neural progenitor cell lines (PPCs) from the hindbrain of aborted 10-week fetuses as normal control for DIPG cells, which has high percentage expression of neural progenitor markers (Supplementary Table S2). Western blot analyses indicate that our DIPG cell lines show decreased levels of H3K27me3 and modestly increased levels of H3K27ac compared to U87 (Uppsala 87 Malignant Glioma) and PPCs, which share the similar global changes in key histone modifications with previously reported H3K27 M DIPG cell lines [12] (Fig. 1b). The majority of DIPG tumor tissues and cell lines express higher levels of CDK4/6 than do PPCs (Fig. 1c), plus most of the DIPG cell lines have much lower expression of CDK4/6 inhibitor P16INK4A (Fig. 1d), suggesting high levels of CDK4/6 kinase activity in patient-derived DIPG cell lines with the H3.3K27 M mutation. Retinoblastoma tumor suppressor protein (RB) in DIPG cell lines remain intact (Fig. 1e). Phosphorylation of RB by CDK4/6 leads to E2F activating transcription of a plethora of genes involved in cell cycle progression from G1 to S phase.
Fig. 1.
Patient-derived DIPG cell lines with H3.3K27 M mutation have high CDK4/6 expression.
(a) Clinical information of the treatment-naïve patients, from which the DIPG cell lines were established. The pathology of the tumor tissues reveals high grade glioma. T2 weighted axial image of magnetic resonance imaging show the infiltrative tumors in pons.
(b) Western blot analysis of acid-extracted histones from DIPG cells and pontine neural progenitor cells (PPCs) with indicated Histone H3 antibodies (n = 3). The percentage 12.5%, 25%,50% and 100% respresent the titrated loading volume of the cell lysis to setup a scale reference.
(c) qRT-qPCR analysis of CDK4/6 mRNA levels in DIPG tumor tissues, primary DIPG cell lines, U87, and PPCs. The CDK4/6 mRNA levels in various DIPG cell lines and tumors are significantly higher than those in PPCs. Unpaired t-tests were performed. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) (n = 3).
(d) qRT-qPCR analysis of CDKN2A(coding for P16 protein)mRNA levels in primary DIPG cell lines and PPCs. The CDKN2A mRNA levels in various DIPG cell lines are significantly lower than those in PPC. Unpaired t-tests were performed. (***p < 0r.001) (n = 3).
(e) Western blot analysis of RB expression in primary DIPG cell lines (n = 3).
3.2. CDK4/6 is a potential therapeutic target for DIPG in vitro
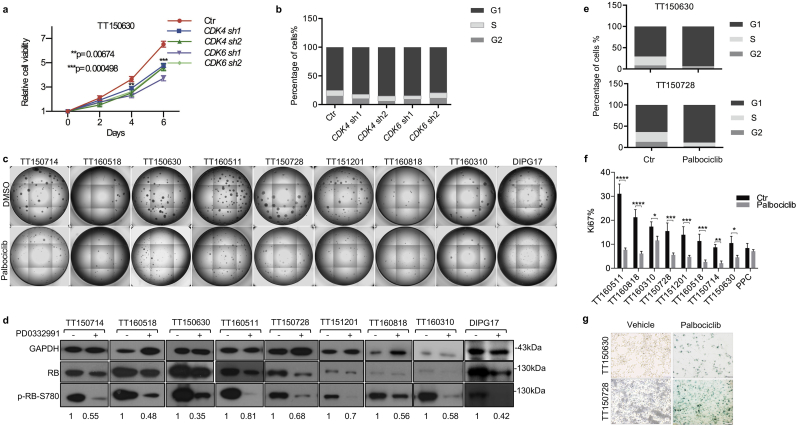
Targeting CDK4/6 activity is one of the options to block G1/S cell cycle transition, which has been shown to induce cell cycle arrest in cancer therapy [34]. Depletion of CDK4 or CDK6 in our patient-derived cell line TT150630 blocks the progression of cell cycle from G1 to S phase and thereby inhibits the growth of DIPG cells in vitro (Fig. 2a, b and Supplementary Fig. S2a). This suggests that CDK4/6 is a potential therapeutic target in DIPG. Palbociclib is a specific and cytostatic inhibitor of CDK4/6. We further investigated anti-tumor efficacy of palbociclib in our treatment naïve patient-derived DIPG cell lines. Sphere formation assay was used to evaluate the self-renewal ability of DIPG cells. Palbociclib uniformly inhibits sphere formation in all nine patient-derived DIPG cell lines (Fig. 2c and Supplementary Fig. S2e). The IC50 values, calculated for all DIPG cell lines, H3.3 wild type GBM cell line U87 and normal control PPC cells (Supplementary Fig. 2b and Supplementary Table. S4). And 100 nM of palbociclib was sufficient to repress expression of the E2F target gene CCNA in all DIPG cells (Supplementary Fig. S2c and S2d) and to block phosphorylation of RB at Ser780 in all nine DIPG cell lines (Fig. 2d). Palbociclib treatment also leads to G1 arrest (Fig. 2e and Supplementary Fig. S3a). Meanwhile, proliferation-associated marker Ki67 decreases in all DIPG cell lines treated with palbociclib (Fig. 2f). In addition, senescence-associated (SA)-β-galactosidase activity is increased in palbociclib-treated TT150630 and TT150728 cells (Fig. 2g). We did not detect significant apoptosis in cells treated with palbociclib (Supplementary Fig. S3b). In conclusion, palbociclib inhibits the growth of DIPG cells in vitro by blocking G1/S transition and inducing senescence but not apoptosis.
Fig. 2.
CDK4/6 is a potential therapeutic target for DIPG in vitro.
(a) Knockdown of CDK4 or CDK6 significantly blocks the growth of DIPG cells in vitro. 6-day growth curve of CDK4 or CDK6 depleted TT150630 is compared to control TT150630. The plots represent CellTiter-Glo quantification measured every other day (day0 to day6) and normalized to day 0 (one representative of two independent experiments). Each point is an average of cell-culture triplicates (n = 3). Data are shown as mean ± standard deviation (SD) (**p = 0.00674, ***p = 0.000498); two-tailed t-tests were performed. Results reveal a significant difference of cell viability between treated and control cells on day 4 or day 6.
(b) Depletion of CDK4 or CDK6 induces cell cycle G1 arrest. Cell cycle analyses were performed on TT150630 cells, CDK4 or CDK6 depleted v.s. control (n = 3).
(c) Sphere formation assays of indicated patient-derived DIPG cell lines with control (0.01% DMSO) or 100 nM palbociclib for 10 days (n = 6). Sphere formation assays were performed in 96-well plates.
(d) Palbociclib blocks RB phosphorylation at Ser780. Western blots of RB phosphorylation at Ser780 were performed in indicated DIPG cell lines treated with vehicle (0.1% DMSO) or 100 nM palbociclib (n = 3). Densitometry analysis was performed using ImageJ software by normalizing the densitometry of ph-RB by total RB levels. The normalized p-RB densitometry were labeled under the bands.
(e) Palbociclib treatment induces cell cycle G1 arrest in DIPG cell lines TT150630 and TT150728. DIPG cell lines TT150630 and TT150728 were treated with 100 nM palbociclib for 48 h. Cell cycle analysis to show G1 arrest in DIPGs with palbociclib treatment (n = 3).
(f) Quantification of Ki-67 FACS staining in patient-derived DIPG lines treated with 100 nM palbociclib or vehicle (0.1% DMSO) for 48 h. The experiments were performed in triplicate. Data are shown as mean ± SD (*p < 0.05, **p < 0.01, ***p < 0.001) (n = 3).
(g) Pabociclib treatment induces senescence in DIPG cell lines. DIPG cell lines TT150630 and TT150728 were treated with 100 nM palbociclib for 10 days. SA-β-Gal staining reveals SA-β-Gal activity. (Scale bar = 50 μm) (n = 3).
3.3. Genome wide impact of palbociclib to DIPG
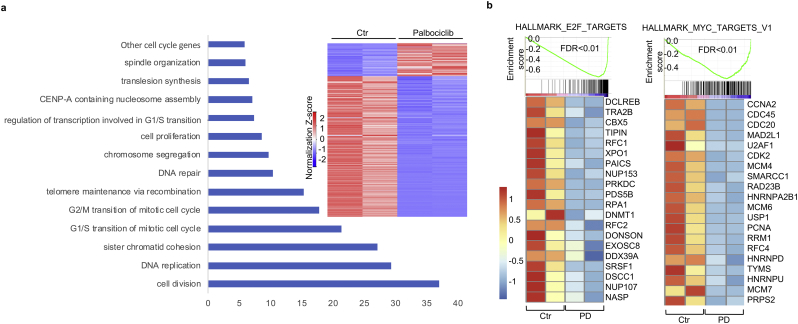
Next, we explored the biological impact of palbociclib treatment in DIPG cells at the whole-transcriptome level by high-throughput sequencing (RNA-seq). 306 of the genes are down-regulated whereas 65 genes are upregulated (two folds higher or 50% lower than control, FDR < 0.05, fragment per kilo base per million reads (FPKM) > 1 in control) (Fig. 3a), which suggests that palbociclib mainly represses gene expression. Gene ontology (GO) analysis indicates that most of the affected genes are cell cycle-related genes (Fig. 3a). Gene set enrichment analysis (GSEA) shows that the E2F target genes are significantly down-regulated in DIPG cells with FDR < 0.01 (Fig. 3b). Some of these down-regulated genes are involved in mitotic spindle and G2/M transition, both of which are cell cycle-related (Supplementary Fig. S4). Interestingly, MYC target genes were found in the down-regulated gene list with FDR < 0.01 (Fig. 3b). MYC is an oncogene that contributes to pathogenesis of most human cancers. Therefore, this finding suggests that palbociclib could inhibit the growth of DIPG via downregulation of MYC target genes in addition to affecting E2F pathway.
Fig. 3.
Genome wide impact of palbociclib on DIPG.
(a) Heatmaps of palbociclib affected genes (Right panel) and Gene Ontology analysis for genes in the heatmap (Left panel). Heatmaps show that 306 of the genes are down-regulated, whereas 65 genes are upregulated (log2 fold change >1 or < −1, FDR < 0.05, FPKM >1 in control). Gene Ontology (GO) analysis demonstrates the pathways enriched in the differentially expressed genes (n = 2).
(b) Gene Set Enrichment Analysis (GSEA) of palbociclib-repressed genes (FDR < 0.01, p < 0.001). Heatmap shows the RNA-seq enrichment (FPKM) for genes repressed by palbociclib treatment. Color bar represents signal density distribution (n = 2).
3.4. Palbociclib inhibits the growth of DIPG in orthotopic xenograft mouse models
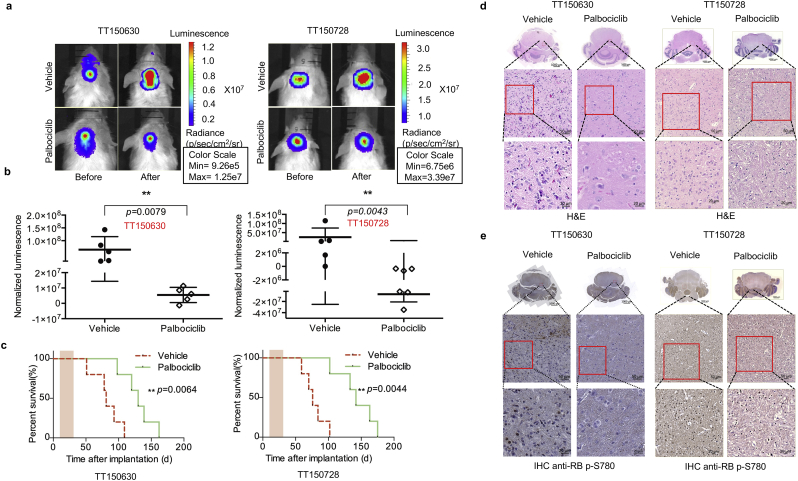
To further investigate the anti-tumor efficacy of palbociclib in vivo, we established luciferase-modified TT150630 and TT150728 xenografts by injecting tumor cells into the pons of a cohort of B-NDG (B: Biocytytogen; N: NOD background; D: DNAK (Prkdc) null; G: IL2rg knockout) mice. Tumor formation was detected two weeks after the implantation. The mice bearing the similar volume of tumor were randomly divided into vehicle and palbociclib-treated groups. 150 mg/kg palbociclib or vehicle was administered orally every day to the tumor-bearing mice. In vivo bioluminescence imaging was performed every week to monitor tumor growth (Fig. 4a). The control mice exhibited faster xenograft growth than the mice treated with palbociclib (n = 5 control mice, 5 palbociclib-treated mice for TT150630; p = 0.0079 and n = 5 control mice, 6 palbociclib-treated mice for TT150728; p = 0.0043, Fig. 4b). The overall survival of the palbociclib-treated group was significantly longer than the control group (p = 0.0064 for TT150630 and p = 0.0044 for TT150728, Fig. 4c). Hematoxylin and eosin (H&E) staining of tumor sections of palbociclib-treated mice shows fewer pleomorphic tumor cells compared to the control mice (Fig. 4d). The immunohistochemistry (IHC) of phosphorylated RB (Ser-780) demonstrates that RB phosphorylation was efficiently blocked in palbociclib-treated tumor sections (Fig. 4e). The shrinkage of DIPG tumors, down-regulation of phosphorylated-RB at Ser780, and longer survival in palbociclib-treated xenografted mice suggest a direct and strong anti-tumor effect of palbociclib treatment on DIPG in vivo. We tested one more patient-derived DIPG brainstem orthotopic xenograft mouse models (TT151201, Supplementary Fig. S5, a-d) using the same protocol and observed the similar anti-tumor effect of palbociclib treatment on DIPG in vivo.
Fig. 4.
Potent anti-tumor efficacy of palbociclib in H3.3 K27 M-mutant DIPG cells in vivo.
(a&b) Effects of palbociclib treatment on DIPG in vivo. Engineered TT150630 and TT150728 DIPG cells with luciferase were xenografted to the B-NDG mice pons and allowed to engraft for 1 month prior to treatment. Mice with established orthotopic TT150630 DIPG tumor were randomly assigned to vehicle (n = 5) or palbociclib (n = 5) treated groups. TT150728 DIPG tumor were randomly assigned to vehicle (n = 5) or palbociclib (n = 6) treated groups. Palbociclib was orally administered for 21 days consecutively at the dose of 150 mg/kg every day. Bioluminescence imaging was taken once a week, and the luminescence imaging showed the growth of the tumor at the end of the 21-days palbociclib treatment. The heatmap superimposed over the mouse heads represents the degree of photon emission by DIPG cells expressing firefly luciferase. Left panel is TT150630 DIPG cells and right one is TT150728 DIPG cells.
(c) Palbociclib treatment prolongs the survival in a DIPG orthotopic xenograft model. Kaplan-Meier survival curve are plotted; n = 5 per group of TT150630 DIPG cells; vehicle (n = 5) or palbociclib (n = 6) treated groups of TT150728 DIPG cells; shaded area are indicated for pacbociclib treatment time length; log-rank test is performed (p = 0.0064). Left panel is TT150630 DIPG cells and right one is TT150728 DIPG cells.
(d) Representative H&E staining in the tumor sections from pons of palbociclib or vehicle treated mice at the end of 21-day treatment. Left panel is TT150630 DIPG cells and right one is TT150728 DIPG cells.
(e) Representative RB (phosphor-RB Ser780) staining for vehicle or palbociclib treated tumors, from mice euthanized at the end of 21-day treatment. Left panel is TT150630 DIPG cells and right one is TT150728 DIPG cells.
We further used TT150728 to test whether palbociclib could be combined with other drugs for more efficient treatment. There was relative high expression of EGFR in TT150728, hence we examined whether combination of palbociclib with EGFR inhibitor, erlotinib, arrest tumor growth in vitro and in vivo. Palbociclib and erlotinib treatment synergistically inhibited self renewal and proliferation of TT150728 and TT151201 in a dosage-dependent manner (Supplementary Fig. S6, a-c). Anti-tumor effect of combination of palbociclib with erlotinib was also observed in orthotopic xenograft mouse model of TT150728 (Supplementary Fig. S6d). This further confirmed the feasibility of palbociclib treatment on DIPG.
4. Discussion
4.1. Establishment of treatment-naïve DIPG cell lines
DIPG are currently the number one cause of brain tumor-related death in children [35,36]. No effective treatment available for DIPG makes it urgent to explore effective therapeutic targets. In our study, all the cell lines were established from biopsy of treatment-naïve patients. We also established neural progenitor cells from human fetal hindbrain (PPCs) as normal control cells. We found that our eight cell lines are more sensitive to palbociclib treatment in vitro compared to previous report [22]. Using these cell lines for study will lead to better understanding of the mechanisms involved in tumorigenesis and eventually to achieve the goal of individualized drug therapy. Our treatment-naïve patient derived DIPG cell lines showed higher sensitivity to palbociclib treatment in vitro than previously reported (IC50 = 100–500 nM vs 8000–16,000 nM) [22]. This might suggest to treat H3.3 mutant DIPG patients with palbociclib prior to other treatments (radiotherapy or chemotherapy).
4.2. Palbociclib as a potential therapeutic strategy for DIPG
Palbociclib was originally developed by David Fry and Peter Toogood and is a specific inhibitor to CDK4/6 kinases [16,37]. Since then the antitumor activity of palbociclib has been tested in multiple cancers both in cell based or human cancer xenograft model [16,18,[38], [39], [40], [41], [42], [43], [44]]. These intensive studies of palbociclib eventually led to approval from the US Food and Drug Administration (FDA) in February 2015 and in February 2016 for using palbociclib to treat postmenopausal patients with estrogen receptor-positive, HER2 - negative metastatic breast cancer [16,17]. So far there has been >50 clinical trials using palbociclib as treatment involving a broad spectrum of cancers [45]. A clinical trial (NCT02255461), which recruited patients with recurrent pediatric brain stem glioma, has already completed on March 20, 2019. The pharmacokinetics data were available for a variety of species in a pharmacology review for FDA approval application of palbociclib (please see the link at the end of this paragraph). In this review, when the palbociclib was administrated orally at the dose of 200 mg/kg in a rat model, the Cmax was 2240 ng/ml.
(https://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/207103Orig1s000PharmR.pdf)
Our study confirmed the efficacy of palbociclib in three patient derived xenograft DIPG mouse models for the first time. The decreased p-RB at Ser780 in IHC section of the tumor tissue confirmed the penetration of palbociclib to the blood brain barrier (BBB) of brainstem, which is commonly deemed to be more intact than the hemispheric BBB [46]. The shrinkage of the tumor further demonstrated that palbociclib could efficiently repress DIPG xenograft tumor. All these evidences suggest that CDK4/6 specific inhibitor, palbociclib, could be promising option for DIPG treatment. Palbociclib has been reported to be more effective when it was combined with other treatments, such as mTOR inhibitor [47], PI3K inhibitor [48] or EGFR inhibitor [49]. In this study, we showed that combination of CDK4/6 and EGFR inhibitors in a EGFR highly expressed DIPG cell line TT150728 arrested tumor cells growth synergistically both in vitro and in vivo.
4.3. Targets of palbociclib in DIPG
Genome-wide analyses have identified recurrent amplifications of cell-cycle regulatory genes in DIPG, which include CDK4/6 and CCND1/2/3 [14,50]. We showed that high E2F activity was prominent in patient derived DIPG cells or tissues compared to PPCs, which is less common in adult GBM [51]. This further confirmed the notion that the pathogenesis of pediatric high grade glioma was different from adult hemispheric GBM [52]. The administration of CDK4/6 inhibitor palbociclib inhibited DIPG derived cells self-renewal, proliferation and cell cycle progression from G1 to S phase in vitro. Palbociclib treatment also induced senescence, which was demonstrated by increased senescence-associated (SA)-β-galactosidase activity, and has been reported in several types of cancer cells treated with palbociclib [18,[53], [54], [55]]. In our RNA-seq data, the palbociclib treatment dramatically decreased the mRNA levels of FOXM1, which was reported to be a substrate of CDK4/6 and played the role of anti-senescence in the pathogenesis of cancers [53]. The upstream regulators of FOXM1 related to CDK4/6 inhibition were needed to be identified. There was no obvious evidence of apoptosis followed by palbociclib treatment, which was also not observed in adult GBM [51].
Interestingly, transcriptome profile analysis shows palbociclib treatment significantly down-regulated genes expression. Palbociclib treatment not only down-regulated E2F activity, it also repressed “MYC target genes” dramatically. Inhibition of CDK4/6 by palbociclib was reported to be a highly effective strategy for the treatment of SHH and MYC-amplified group 3 medulloblastoma [56]. Previous study grouped DIPG into three subgroups: MYCN, silent and H3K27M. The amplification or gain of MYC is exclusive to the H3K27 M subgroup [57]. In addition to the cell cycle G1 arrest, we also observed that G2/M phase-associated genes were also affected by palbociclib treatment. These observation was not reported in other palbociclib related researches. We also found the global epigenome change after palbociclib treatment (data not shown), for example, the H3K27me3 level was increased and H3K27ac was subtlely decreased in several DIPG lines. The global change of modification of histone H3 may contribute to the transcriptome shift beyond CDK4/6-RB-E2F signaling. However, this need further be validated. Study has reported the immune response triggered by CDK4/6 inhibitor, which was attributed to the increased activity of antigen presentation [58]. However, in our immunodeficient mouse model, this effect of palbociclib was abolished by the defect immune system. As a result, to some extent, the efficacy of palbociclib may be underestimated from the data of this mouse model.
Acknowledgments
Acknowledgements
We would like to thank all of the patients and families for donating tissue for this research. We also would like to thank Hanqiu Zheng for luciferase-GFP plasmid; Yujie Tang for DIPG17 cells; Qingran Bai, Xiaoqing Liu, and Qin Shen for establishment of primary cell lines TT150630, TT150714, TT150728, TT151201, PPC and helpful discussions; and Yu Rao for valuable discussion.
Funding sources
Liwei Zhang is supported by Beijing municipal administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201608) to L.Z. Beijing Municipal Natural Science Foundation (7161004) to L.Z. and National Natural Science Foundation of China (81872048) to L.Z. Qiaoran Xi is supported by National Natural Science Foundation of China (31471229, 31771622, and 91540108) to Q.X., Center for Life Sciences (to Q.X.), and Ministry of Science and Technology of China (2018YFA0107702)to Q.X.
Declaration of interests
Dr. Zhang reports In addition, Dr. Zhang has a patent PIDC3175771A issued to Tsinghua University and Beijing Tiantan hospital.
Dr. Xi reports In addition, Dr. Xi has a patent PIDC3175771A issued to Tsinghua University and Beijing Tiantan hospital.
Dr. Yu Sun reports In addition, Dr. Sun has a patent PIDC3175771A issued to Tsinghua University and Beijing Tianan hospital.
Dr. Ye Sun reports In addition, Dr. Sun has a patent PIDC3175771A issued to Tsinghua University and Beijing Tiantan hospital.
Author contributions
YuS., YeS., and Z.L. performed most of the experiments. YuS., C.X., Y.G., C.P., X.C., and L.Z. provided all the biopsy samples and clinical data of DIPG patients. C.X and Y.G. established primary DIPG cell lines. YuS. established PPC cell lines. YeS. performed RNA-seq experiment. Y.K. performed RNA-seq analysis. Q.X. and L.Z. designed the work. Q.X. L.Z. YuS. and YeS. contributed to the experimental design. All authors contributed ideas to the project. Q.X. and L.Z. supervised the project and Q.X. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Accession numbers
The raw RNA-seq data for this manuscript are available at GEO under the accession number GSE115976.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.04.043.
Contributor Information
Liwei Zhang, Email: Zhangliweittyy@163.com.
Qiaoran Xi, Email: xiqiaoran@tsinghua.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Jones C., Karajannis M.A., Jones D.T.W., Kieran M.W., Monje M., Baker S.J. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153–161. doi: 10.1093/neuonc/now101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanan M.I., Eisenstat D.D. DIPG in children - what can we learn from the past? Front Oncol. 2015;5:237. doi: 10.3389/fonc.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasso C.S., Tang Y., Truffaux N., Berlow N.E., Liu L., Debily M.A. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(6):555–559. doi: 10.1038/nm.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashizume R., Andor N., Ihara Y., Lerner R., Gan H., Chen X. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammad F., Weissmann S., Leblanc B., Pandey D.P., Hojfeldt J.W., Comet I. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23(4):483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 6.Piunti A., Hashizume R., Morgan M.A., Bartom E.T., Horbinski C.M., Marshall S.A. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23(4):483–492. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor K.R., Mackay A., Truffaux N., Butterfield Y.S., Morozova O., Philippe C. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–461. doi: 10.1038/ng.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y.L., Maachani U.B., Schweitzer M., Singh R., Wang M., Chang R. Dual inhibition of PI3K/AKT and MEK/ERK pathways induces synergistic antitumor effects in diffuse intrinsic Pontine Glioma cells. Transl Oncol. 2017;10(2):221–228. doi: 10.1016/j.tranon.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor I.C., Hutt-Cabezas M., Brandt W.D., Kambhampati M., Nazarian J., Chang H.T. Disrupting NOTCH slows diffuse intrinsic Pontine Glioma growth, enhances radiation sensitivity, and shows combinatorial efficacy with Bromodomain inhibition. J Neuropathol Exp Neurol. 2015;74(8):778–790. doi: 10.1097/NEN.0000000000000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagaraja S., Vitanza N.A., Woo P.J., Taylor K.R., Liu F., Zhang L. Transcriptional dependencies in diffuse intrinsic Pontine Glioma. Cancer Cell. 2017;31(5) doi: 10.1016/j.ccell.2017.03.011. (635-52.e6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mount C.W., Majzner R.G., Sundaresh S., Arnold E.P., Kadapakkam M., Haile S. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat Med. 2018;24(5):572–579. doi: 10.1038/s41591-018-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender S., Tang Y., Lindroth A.M., Hovestadt V., Jones D.T., Kool M. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Khuong-Quang D.A., Buczkowicz P., Rakopoulos P., Liu X.Y., Fontebasso A.M., Bouffet E. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackay A., Burford A., Carvalho D., Izquierdo E., Fazal-Salom J., Taylor K.R. Integrated molecular meta-analysis of 1,000 Pediatric high-grade and diffuse intrinsic Pontine Glioma. Cancer Cell. 2017;32(4):520–537 e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordero F.J., Huang Z., Grenier C., He X., Hu G., McLendon R.E. Histone H3.3K27M represses p16 to accelerate Gliomagenesis in a murine model of DIPG. Mol Cancer Res. 2017;15(9):1243–1254. doi: 10.1158/1541-7786.MCR-16-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry D.W., Harvey P.J., Keller P.R., Elliott W.L., Meade M., Trachet E. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3(11):1427–1438. [PubMed] [Google Scholar]
- 17.Beaver J.A., Amiri-Kordestani L., Charlab R., Chen W., Palmby T., Tilley A. FDA approval: Palbociclib for the treatment of postmenopausal patients with Estrogen receptor-positive, HER2-negative metastatic breast Cancer. Clin Cancer Res. 2015;21(21):4760–4766. doi: 10.1158/1078-0432.CCR-15-1185. [DOI] [PubMed] [Google Scholar]
- 18.Michaud K., Solomon D.A., Oermann E., Kim J.S., Zhong W.Z., Prados M.D. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cen L., Carlson B.L., Schroeder M.A., Ostrem J.L., Kitange G.J., Mladek A.C. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 2012;14(7):870–881. doi: 10.1093/neuonc/nos114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiedemeyer W.R., Dunn I.F., Quayle S.N., Zhang J., Chheda M.G., Dunn G.P. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107(25):11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barton K.L., Misuraca K., Cordero F., Dobrikova E., Min H.D., Gromeier M. PD-0332991, a CDK4/6 inhibitor, significantly prolongs survival in a genetically engineered mouse model of brainstem glioma. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asby D.J., Killick-Cole C.L., Boulter L.J., Singleton W.G., Asby C.A., Wyatt M.J. Combined use of CDK4/6 and mTOR inhibitors induce synergistic growth arrest of diffuse intrinsic pontine glioma cells via mutual downregulation of mTORC1 activity. Cancer Manag Res. 2018;10:3483–3500. doi: 10.2147/CMAR.S167095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C., Liu X., Geng Y., Bai Q., Pan C., Sun Y. Patient-derived DIPG cells preserve stem-like characteristics and generate orthotopic tumors. Oncotarget. 2017;8(44):76644–76655. doi: 10.18632/oncotarget.19656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y., Xu C., Pan C., Chen X., Geng Y., Wu Y. Diffuse intrinsic Pontine Gliomas exhibit cell biological and molecular signatures of Fetal hindbrain-derived neural progenitor cells. Neurosci Bull. 2019;35(2):216–224. doi: 10.1007/s12264-018-00329-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Root D.E., Hacohen N., Hahn W.C., Lander E.S., Sabatini D.M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 26.Zheng H., Bae Y., Kasimir-Bauer S., Tang R., Chen J., Ren G. Therapeutic antibody targeting tumor- and Osteoblastic niche-derived Jagged1 sensitizes bone metastasis to chemotherapy. Cancer Cell. 2017;32(6) doi: 10.1016/j.ccell.2017.11.002. [731-47 e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat Protoc. 2007;2(6):1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 28.Sapkota G., Alarcon C., Spagnoli F.M., Brivanlou A.H., Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25(3):441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N., Fu J.N., Chou T.C. Synergistic combination of microtubule targeting anticancer fludelone with cytoprotective panaxytriol derived from panax ginseng against MX-1 cells in vitro: experimental design and data analysis using the combination index method. Am J Cancer Res. 2016;6(1):97–104. [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monje M., Mitra S.S., Freret M.E., Raveh T.B., Kim J., Masek M. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108(11):4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashizume R., Smirnov I., Liu S., Phillips J.J., Hyer J., McKnight T.R. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. J Neurooncol. 2012;110(3):305–313. doi: 10.1007/s11060-012-0973-6. [DOI] [PubMed] [Google Scholar]
- 34.O'Leary B., Finn R.S., Turner N.C. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 35.Freeman C.R., Perilongo G. Chemotherapy for brain stem gliomas. Childs Nerv Syst. 1999;15(10):545–553. doi: 10.1007/s003810050542. [DOI] [PubMed] [Google Scholar]
- 36.Maria B.L., Rehder K., Eskin T.A., Hamed L.M., Fennell E.B., Quisling R.G. Brainstem glioma: I. pathology, clinical features, and therapy. J Child Neurol. 1993;8(2):112–128. doi: 10.1177/088307389300800203. [DOI] [PubMed] [Google Scholar]
- 37.Garber K. The cancer drug that almost wasn't. Science. 2014;345(6199):865–867. doi: 10.1126/science.345.6199.865. [DOI] [PubMed] [Google Scholar]
- 38.Dean J.L., Thangavel C., McClendon A.K., Reed C.A., Knudsen E.S. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29(28):4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 39.Saab R., Bills J.L., Miceli A.P., Anderson C.M., Khoury J.D., Fry D.W. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol Cancer Ther. 2006;5(5):1299–1308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 40.Baughn L.B., Di Liberto M., Wu K., Toogood P.L., Louie T., Gottschalk R. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66(15):7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 41.Wang L., Wang J., Blaser B.W., Duchemin A.M., Kusewitt D.F., Liu T. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110(6):2075–2083. doi: 10.1182/blood-2007-02-071266. [DOI] [PubMed] [Google Scholar]
- 42.Nemoto A., Saida S., Kato I., Kikuchi J., Furukawa Y., Maeda Y. Specific Antileukemic activity of PD0332991, a CDK4/6 inhibitor, against Philadelphia chromosome-positive lymphoid Leukemia. Mol Cancer Ther. 2016;15(1):94–105. doi: 10.1158/1535-7163.MCT-14-1065. [DOI] [PubMed] [Google Scholar]
- 43.Eilers G., Czaplinski J.T., Mayeda M., Bahri N., Tao D., Zhu M. CDKN2A/p16 loss implicates CDK4 as a therapeutic target in Imatinib-resistant Dermatofibrosarcoma Protuberans. Mol Cancer Ther. 2015;14(6):1346–1353. doi: 10.1158/1535-7163.MCT-14-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn R.S., Dering J., Conklin D., Kalous O., Cohen D.J., Desai A.J. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veringa S.J., Biesmans D., van Vuurden D.G., Jansen M.H., Wedekind L.E., Horsman I. In vitro drug response and efflux transporters associated with drug resistance in pediatric high grade glioma and diffuse intrinsic pontine glioma. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olmez I., Brenneman B., Xiao A., Serbulea V., Benamar M., Zhang Y. Combined CDK4/6 and mTOR inhibition is synergistic against Glioblastoma via multiple mechanisms. Clin Cancer Res. 2017;23(22):6958–6968. doi: 10.1158/1078-0432.CCR-17-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrera-Abreu M.T., Palafox M., Asghar U., Rivas M.A., Cutts R.J., Garcia-Murillas I. Early adaptation and acquired resistance to CDK4/6 inhibition in Estrogen receptor-positive breast Cancer. Cancer Res. 2016;76(8):2301–2313. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Tang Y., Yuan X., Yuan D., Liu J., Li B. Inhibition of Rb and mTOR signaling associates with synergistic anticancer effect of palbociclib and erlotinib in glioblastoma cells. Invest New Drugs. 2018;36(6):961–969. doi: 10.1007/s10637-018-0575-z. [DOI] [PubMed] [Google Scholar]
- 50.Paugh B.S., Broniscer A., Qu C., Miller C.P., Zhang J., Tatevossian R.G. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaud K., Solomon D.A., Oermann E., Kim J.S., Zhong W.Z., Prados M.D. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grobner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 53.Anders L., Ke N., Hydbring P., Choi Y.J., Widlund H.R., Chick J.M. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20(5):620–634. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thangavel C., Dean J.L., Ertel A., Knudsen K.E., Aldaz C.M., Witkiewicz A.K. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer. 2011;18(3):333–345. doi: 10.1530/ERC-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovatcheva M., Liu D.D., Dickson M.A., Klein M.E., O'Connor R., Wilder F.O. MDM2 turnover and expression of ATRX determine the choice between quiescence and senescence in response to CDK4 inhibition. Oncotarget. 2015;6(10):8226–8243. doi: 10.18632/oncotarget.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook Sangar M.L., Genovesi L.A., Nakamoto M.W., Davis M.J., Knobluagh S.E., Ji P. Inhibition of CDK4/6 by Palbociclib significantly extends survival in Medulloblastoma patient-derived Xenograft mouse models. Clin Cancer Res. 2017;23(19):5802–5813. doi: 10.1158/1078-0432.CCR-16-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buczkowicz P., Hoeman C., Rakopoulos P., Pajovic S., Letourneau L., Dzamba M. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. doi: 10.1038/ng.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goel S., DeCristo M.J., Watt A.C., BrinJones H., Sceneay J., Li B.B. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material