Abstract
Background
Investigations into the function of non-promoter DNA methylation have yielded new insights into the epigenetic regulation of gene expression. However, integrated genome-wide non-promoter DNA methylation and gene expression analyses across a wide number of tumour types and corresponding normal tissues have not been performed.
Methods
To investigate the impact of non-promoter DNA methylation on cancer pathogenesis, we performed a large-scale analysis of gene expression and DNA methylation profiles, finding enrichment in the 3’UTR DNA methylation positively correlated with gene expression. Filtering for genes in which 3’UTR DNA methylation strongly correlated with gene expression yielded a list of genes enriched for functions involving T cell activation.
Findings
The important immune checkpoint gene Havcr2 showed a substantial increase in 3’UTR DNA methylation upon T cell activation and subsequent upregulation of gene expression in mice. Furthermore, this increase in Havcr2 gene expression was abrogated by treatment with decitabine.
Interpretation
These findings indicate that the 3’UTR is a functionally relevant DNA methylation site. Additionally, we show a potential novel mechanism of HAVCR2 regulation in T cells, providing new insights for modulating immune checkpoint blockade.
Keywords: 3’UTR, Epigenetics, Immune checkpoint, Methylation
Research in context.
Evidence before this study
DNA methylation allows for differential expression of a single gene without requiring modification to the DNA sequence. This plays a critical role in both normal cellular processes, as well as in disease. Cancer cells are well known to have radically altered DNA methylation profiles. However, these changes are not isolated to the cancer cells themselves, but rather occur in various cell types within the tumour microenvironment. In particular, DNA methylation has been revealed as a major driver in why T cells become “exhausted” and no longer target cancer cells. Expression of factors that dampen T cell activity has been implicated in this phenotype, and as such, combining demethylating agents with immunotherapy has been shown to increase efficacy. HAVCR2 (TIM-3) is a critical immunoregulatory gene in which expression, especially in conjunction with PDCD1 (PD-1), induces an exhausted T cell state. Much of how DNA methylation functions in regulating HAVCR2 remains to be understood.
Added value of this study
Understanding how site-specific DNA methylation impacts expression is critical for gaining a more complete control over cellular processes, particularly in the context of cancer. T cells represent a potent tool for eliminating tumour cells, and DNA methylation is a major determinant of T cell function. This study investigates how intragenic site-specific DNA methylation across genes involved in T cell function changes based on T cell activation state.
Implications of all the available evidence
In this study, we have uncovered a strong positive correlation between 3’UTR DNA methylation of specific genes, and increased gene expression. Genes that play a role in T cell activation are enriched among those exhibiting the most robust correlations. Furthermore, 3’UTR methylation and gene expression of the immune checkpoint gene HAVCR2 increases significantly after T cell activation. Treating activated T cells with the demethylating agent decitabine, or knocking out the DNA methylating enzyme Dnmt3a in mice results in decreased 3’UTR methylation and gene expression of Havcr2. Therefore, the 3’UTR may serve as a functionally relevant site of DNA methylation. Moreover, alterations in the methylation of this region may be involved in activated and exhausted T cell phenotypes. Modulating this region may grant additional control over harnessing the immune system.
Alt-text: Unlabelled Box
1. Introduction
DNA methylation is found at nearly every region of the genome, with methylation at the gene promoter region being the most comprehensively understood. Promoter DNA methylation, typically occurring within CpG islands, results in powerful repression of transcription, primarily by recruiting repressor proteins or chromatin modifiers that enhance the binding of DNA to histones [1,2]. The interplay between epigenetic modifications on DNA and histones allows for remarkable plasticity among cells that share identical genomes by providing a means of activating or silencing genes whose expression affects the developmental state of specific cell types [3]. However, promoter DNA methylation accounts for only a small portion of the overall DNA methylation of the genome [4]. Much remains to be understood about the diverse functions of site-specific non-promoter DNA methylation on gene regulation.
Recent studies have explored the connection between non-promoter DNA methylation and the regulation of gene expression. Enhancer DNA methylation has emerged as a robust predictor of gene expression, with a fraction of genes showing a stronger link between gene expression and enhancer DNA methylation relative to promoter methylation [5,6]. Gene body DNA methylation, like promoter methylation, can repress gene expression through altered binding of regulatory proteins [7], but it is frequently associated with increased gene expression, which runs counter to its role in promoter DNA methylation [8]. Multiple mechanisms to explain this relationship have been uncovered, such as promoting genomic stability by suppressing mobile DNA elements [9,10], modulating patterns of histone methylation that stabilize transcriptional elongation [11], and regulating alternative splicing [12,13]. Importantly, a direct link between gene body DNA methylation and high expression of oncogenes has been demonstrated in colorectal cancer [14], highlighting the functional relevance of this mode of epigenetic modification in both normal and pathogenic processes.
Non-promoter DNA methylation undergoes extensive changes in disease, most strikingly in cancer [15]. The DNA methylation of enhancer regions of clinically relevant genes, particularly within super-enhancers, were drastically altered, resulting in pathogenic transcriptional output [[16], [17], [18]]. Global changes to gene body DNA methylation in Burkitt and follicular lymphoma have been observed [19]. Methylation of the ITPKA gene body is associated with increased expression in lung cancer, and the extent of gene body methylation serves as a biomarker for lung cancer progression [20]. In colon cancer, increased gene expression through gene body DNA methylation is enriched for genes activated by c-Myc, and demethylating these regions impairs the ability of tumour cells to survive and proliferate [14].
Modulating the epigenetic profiles of tumours has become a promising new route for treating cancer [21].Recent therapeutic endeavors have focused primarily on re-activation of the genes suppressed by promoter methylation; therefore, cataloging the functions of non-promoter DNA methylation may broaden our understanding of tumourigenesis and tumour progression, while offering new opportunities for clinical intervention. To address this gap in knowledge, a comprehensive analysis of how DNA methylation of regions outside the promoter region may impact cancer pathogenesis was conducted. Here, for the first time, we uncovered a relationship between 3’UTR DNA methylation and gene expression across 10 tumour types, revealing a unique association between these two phenomena that has implications for our understanding of the role of DNA methylation in normal cellular processes and disease.
2. Materials and methods
2.1. Study design
Sample size: We used all of the tumour and normal tissue samples available in the TCGA database, which includes data on 11,000 US cancer patients. There are no publication restrictions on these data according to the TCGA data policy (http://cancergenome.nih.gov/publications/publicationguidelines).
Rules for stopping data collection: N/A.
Data inclusion and exclusion criteria: Three cut-offs for the correlation coefficient were originally set: 0∙7, 0∙5, and 0∙3. The minimum was set at 0∙3. However, because this yielded a very large number of genes, greater specificity was sought. Therefore, 0∙5 was used as the final cut-off.
Outliers: No outliers were excluded from this analysis.
Selection of endpoints: N/A.
Replicates: For in-house experiments, each assay was conducted in triplicate, on the basis of previously designed methods. Each experiment was conducted at least twice, with similar results being achieved each time.
2.2. TCGA tissue selection
For this analysis, we used tumour and normal tissue samples from 10 tumour types; Illumina HiSeq RNASeqV2 and Illumina HumanMethylation 450 k data were available at the time of our initial download on March 26, 2013. The tumour types included bladder carcinoma, breast carcinoma, colon and rectal carcinoma, head and neck squamous cell carcinoma, kidney renal cell carcinoma, liver carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, prostate adenocarcinoma, thyroid carcinoma, and uterine carcinoma. The corresponding clinical data used for the survival analysis were downloaded from the TCGA data portal and were current as of January 8, 2014.
2.3. Methylation and gene expression data
According to the TCGA description file associated with the Illumina Human Methylation 450 K array data, probes with a SNP within 10 nucleotide base pairs (bp) of the interrogated CpG site or those in which 15 bp of the interrogated CpG site are overlapped with a REPEAT element are masked as NA across all samples. There are 88,058 probes that interrogate such sites (18.3% of all probes). While these beta values are not reported at level 3, the methylated and un-methylated intensity values for these probes are recorded in the level 2 data. Therefore, we used the level 2 data to reconstruct the beta values for all probes as methylated/(methylated+unmethylated); these data are also used by the TCGA.
We used the log2-transformed level 3 RNASeqV2 data to analyse gene expression. To avoid errors for RNASeq raw counts of 0, all values were offset by 1 prior to obtaining logs.
2.4. Gene methylation-expression correlation
All analyses were performed using R software, version 2∙15∙1. Using the complete set of probes targeting CpG dinucleotides, we performed a genome-wide analysis exploring the relationship between the proportion of methylation at various locations within and up to 1500 bp upstream of a gene and the corresponding log-transformed gene expression. We used the Spearman rank statistic to quantify the correlation for each pair. Because we expected these patterns to vary by the tissue source site, we calculated coefficients individually using each of the 10 tissue types for which we had data from both the 450 k methylation and RNASeq arrays.
2.5. Survival analysis
Tests for differences in survival were performed by comparing the overall survival of patients in the top and bottom quartiles of 3’UTR methylation using the “survival” package in R software for all genes; correlations of >0∙5 between gene expression and methylation were used for each tissue type.
2.6. Pathway analysis
The pathway enrichment analysis was performed using Netwalker (http://www.netwalkersuite.org) on all genes with correlations of > 0∙5 between gene expression and methylation at the 3’UTR and < 0∙5 between gene body methylation and gene expression for each tissue type. An additional pathway analysis was run using Ingenuity Pathway Analysis (http://qiagenbioinformatics.com/products/ingenuity-pathway-analysis.com), which yielded similar results.
2.7. Network identification and construction
The network nodes were obtained using Netwalker to determine how many nodes exist and how the genes interact with one another. A singular network node was identified using this method; we also isolated genes that have been shown to be associated with one another but were not identified in the Netwalker database. The node and other genes that were associated with it were exported to Cytoscape (https://www.cytoscape.org/). Connections were then manually included in Cytoscape on the basis of results in the published literature [22].
2.8. T cell isolation, activation, and de-methylation
The spleens of 4 healthy female C57Bl/6 mice were excised after cervical dislocation and then ground on a 40-μm filter while being repeatedly washed with RPMI (Sigma-Aldrich) + 10% heat-inactivated FBS (Thermo Fisher Scientific) + 1% PenStrep (Thermo Fisher Scientific) + 0∙01% β-mercaptoethanol (Sigma-Aldrich). The resulting slurry was spun down at 4 °C at 450 ×g for 5 min, the supernatant was removed, and the pellet was washed with 50 mL of PBS (HyClone). The pellet was incubated at ambient temperature for 5 min using 4 mL of ACK lysis buffer (Gibco), washed with 50 mL of PBS, and spun down at 4 °C at 450 ×g. The pellet was then resuspended in the media.
To obtain naïve T cells, cells were sent immediately for flow cytometry cell sorting. To activate the T cells, 2 μL/mL of mouse CD3e (BD Biosciences, RRID: AB_2259894) and 3.5 μL/10 mL of mouse CD28 (BioXCell, RRID: AB_1107624) activating antibodies were added to the media, along with 1:10000 ng/μL mouse recombinant IL-2 (R&D Systems). To induce demethylation, activated T cells were treated with 500 nM decitabine (Sigma-Aldrich) for 72 h or vehicle DMSO control.
2.9. Flow cytometry analysis
Mouse splenocytes were grown in the conditions outlined above and spun down at 450 ×g at 4 °C for 5 min to pellet the cells. The supernatant was removed and the cells were re-suspended in 2 mL of FACS buffer (PBS + 2% FBS). Cells were then spun at 450 ×g at 4 °C for 5 min. The supernatant was again removed, and the cells were re-suspended in 200 μL of FACS buffer +2 μL of mouse CD16/CD32 blocking antibody (BD Biosciences, RRID: AB_2740348) and incubated on ice for 10 min. After incubation, 2 μL each of mouse CD45-PE, CD4-eFluor 450, and CD8-APC-eFluor 780 (eBiosciences, RRID: AB_469717, AB_467067, AB_11180008) were added, and the cells were incubated on ice for 30 min. All cells that were CD45+ and CD4+ or CD45+ and CD8+ were sorted by flow cytometry analysis and collected for molecular analysis.
2.10. Quantitative polymerase chain reaction
Specimens were collected from the flow cytometry analysis, spun down at 450 ×g for 5 min at 4 °C, and then re-suspended in 350 μL of TRIzol (Thermo Fisher). RNA was isolated from specimens using the Direct-zol RNA isolation kit (Zymo) and quantified by NanoDrop. We used 100 ng of RNA for a cDNA template. cDNA was created using the Verso cDNA synthesis kit (Invitrogen). We combined 5 μg of cDNA with 1 μL of 100 μM forward and reverse primers and 5 μL of SYBR Green PCR Master Mix (Thermo Fisher) in each well. The resulting mixture was then run in a real-time PCR machine (Applied Biosystems) using the following program: 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min × 40 cycles. The resulting Ct values were compared, and the ΔΔCt was obtained. This was used to quantify the relative change in mRNA across samples.
2.11. Primers
Please see Supplementary Table 3 for primer sequences.
2.12. Methylation analysis
One microgram of genomic DNA was treated with sodium bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) according to the manufacturer's protocol. The samples were eluted in 40 μL of M-elution buffer, and 2 μL were used for each PCR reaction. Both bisulfite conversion and a subsequent pyrosequencing analysis were performed at the DNA Methylation Analysis Core at The University of Texas MD Anderson Cancer Center (Houston, Texas).
PCR primers for the pyrosequencing methylation analysis of Havcr2, Itk, and Vav1 were designed using Pyromark Assay Design SW 1∙0 software (Qiagen, Germany). In brief, a sequencing primer was identified within 1 to 5 base pairs near the CpG sites of interest, with an annealing temperature of 40 ± 5 °C. Forward and reverse primers were identified upstream and downstream of the sequencing primer. The optimal annealing temperatures for each of these primers were tested using gradient PCR. Controls for high methylation (SssI-treated DNA), low methylation (WGA-amplified DNA), and no-DNA template were included in each reaction. PCR reactions were performed in a total volume of 20 μL, and the entire volume was used for each pyrosequencing reaction, as previously described [23]. In brief, PCR product purification was performed using streptavidin-sepharose high-performance beads (GE Healthcare Life Sciences, Piscataway, NJ), and co-denaturation of the biotinylated PCR products and sequencing primer (3∙6 pmol/reaction) was conducted following the PSQ96 sample preparation guide. Sequencing was performed on a PyroMark Q96 ID instrument with PyroMark Gold Q96 Reagents (Qiagen, Germany), according to the manufacturer's instructions. The degree of methylation for each individual CpG site was calculated using PyroMark Q96 software. The average methylation of all sites and duplicates was reported for each sample.
2.13. Statistical analysis
Statistical analyses were performed using unpaired t-tests for comparisons between 2 groups and one-way ANOVA with Tukey's posttest for multiple comparisons for >2 groups (Graphpad Prism).
2.14. Sample size estimation
Three replicates for each experiment were prepared in order to identify and exclude any potential outliers; however, there were no outliers excluded from the in vitro experiments.
2.15. Randomization
No specific randomization procedure was employed in the in vitro experiments.
2.16. Blinding
For all conducted experiments, the experimenter that submitted the samples for methylation analysis was not blinded, but the investigator that carried out the methylation analysis was blinded to the groups, and therefore the submitter had no control or input over the results of the analysis. All other data was generated using in silico approaches.
3. Results
3.1. DNA methylation probes positively correlated with increased gene expression are enriched in 3’UTRs
To explore the relationship between poorly understood sites of DNA methylation and gene expression, we used genome-wide RNA-seq and methylation array datasets from the TCGA platform, which includes over 11,000 patient tissue samples, providing a unique opportunity to harness large-scale molecular profiling datasets across numerous tumour types [24].Initially, the proportion of probes achieving Spearman correlations of < −0∙5 or > 0∙5 between gene expression and DNA methylation were examined within each of the 6 gene regions included in the Illumina methylation probe annotation after normalizing for the total number of probes interrogating each region (Supplementary Fig. S1). The sample sizes associated with TCGA are such that these correlations were significant (p < 0∙0004, assuming n = 50 [Spearman correlation]). With sample sizes per tissue type in excess of a few hundred, absolute correlations of < −0∙5 or > 0∙5 were therefore considered highly significant in this analysis. At the probe level, there were 44,309 negative and 29,043 positive associations; at the gene level, there were 6287 negative and 3200 positive associations. The majority (> 75%) of the negatively correlated probes across all 10 tissue types were concentrated within the first exon, 5’-UTR, and upstream of the transcription start site (Fig. 1a). However, approximately a third of probes exhibiting these correlation values were positively correlated with gene expression. Based upon this observation, the specific locations of the positively correlated probes were examined to determine whether any underlying pattern exists.
Fig. 1.
3’UTR is enriched for methylation probes that are positively correlated with gene expression. Proportion heat map representing the distribution of probes in which the correlation between methylation and gene expression was < −0∙5 (a) or > 0∙5 (b) in each of the 10 tissue types. Areas with high proportions are shaded red, while regions with low proportions are shaded blue. The color bar depicts the gene structure: purple, promoter region; light green, 5’UTR; dark green, first exon; grey, body; red, 3’UTR. The negatively correlated probes were concentrated in the first exon, 5’UTR, and were < 200 bp upstream of the transcriptional start site. The positively correlated probes were found predominantly in the gene body and 3’UTR. (c) The arrows represent the directional change of the percentage of probes with negative correlations < −0∙5 compared with the percentage of probes with positive correlations >0∙5 when evaluating the entire gene. The arrows in the southeast direction indicate an increase in the percentage of positive probes and a simultaneous decrease in the percentage of negative probes. In the case of LUSC, we observed a 5% increase in the number of positively correlated probes and a 3% decrease in the number of negatively correlated probes. There was enrichment in the percentage of positively correlated probes in the 3’UTR for all 10 tissue types. BLCA = bladder carcinoma; BRCA = breast carcinoma; CORE = colon and rectal carcinoma; HNSC = head and neck squamous cell carcinoma; KIRC = kidney renal cell carcinoma; LIHC = liver carcinoma; LUAD = lung adenocarcinoma; LUSC = lung squamous cell carcinoma; PRAD = prostate adenocarcinoma; THCA = thyroid carcinoma; UCEC = uterine carcinoma. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Upon segregating the positively correlated probes on the basis of region, we noted that approximately 3% of these probes interrogated the 3’UTR. Gene body DNA methylation is a known feature of highly transcribed genes [25]; however, previous studies have grouped the 3’UTR with the rest of the gene body rather than investigating it as a distinct region. To determine whether there is a statistical rationale for separating the 3’UTR from the gene body, we calculated the proportion of probes in the 3’UTR that exhibited a positive correlation of >0∙5 between DNA methylation and gene expression (Fig. 1b). This revealed a substantial net enrichment in the proportion of positively correlated probes in the 3’UTR compared to the entire gene region across all 10 tissue types (Fig. 1c), with up to 590 genes exhibiting a correlation coefficient of >0∙5 between 3’UTR methylation and gene expression (Supplementary Table 1). These findings prompted us to evaluate how methylation of the 3’UTR relates to gene expression in particular, whether this is a passive modification and whether its association with gene expression can be attributed to another variable or whether levels of 3’UTR methylation are the key differentiators of gene expression levels.
3.2. Extent of 3’UTR methylation predicts differential gene expression between normal and tumour tissues
To determine whether other drivers of gene expression could explain the differences in gene expression observed in tumour and normal tissues, we focused on the HOXC13 gene. We chose this gene because of its strong positive correlation with 3’UTR methylation (> 0∙5 in all 10 examined tissues), its comprehensive set of probes interrogating each region of the gene, and its significantly higher expression in tumour tissues than in corresponding normal tissues (Fig. 2a).
Fig. 2.
Investigation of the drivers of HOXC13 gene expression. (a) Dotplot of log2 RNA-seq expression values across 10 TCGA tissue types; black stars indicate normal tissue samples. (b) Copy number alterations in HOXC13across 10 TCGA tissue types. (c) Heatmap representing the methylation patterns across HOXC13 in bladder carcinomas. The row color bar depicts the gene structure: purple, promoter region; light green, 5’UTR; dark green, first exon; grey, body; red, 3’UTR. The column color bar represents the top 25% gene expressers in pink and the bottom 25% expressers in black. (d) Heatmap summarizing the log10 p-value for each potential regulatory non-coding region of HOXC13 across all tissue types.BLCA = bladder carcinoma; BRCA = breast carcinoma; CORE = colon and rectal carcinoma; HNSC = head and neck squamous cell carcinoma; KIRC = kidney renal cell carcinoma; LIHC = liver carcinoma; LUAD = lung adenocarcinoma; LUSC = lung squamous cell carcinoma; PRAD = prostate adenocarcinoma; THCA = thyroid carcinoma; UCEC = uterine carcinoma. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further understand the nature of this variation, we examined common processes that are known to account for differential gene expression, primarily promoter methylation and copy number variation. First, we addressed possible allelic gain or loss using the TCGA copy number data for these tissue types (Fig. 2b). There was no significant HOXC13 gene amplification or deletion in any of the 10 tumour types. Specifically, < 5% of cases had copy number alterations in the HOXC13 gene region, making it unlikely that copy number drove the observed variation in expression.
We also considered whether the difference in HOXC13 expression between normal and tumour tissues could be due to divergent methylation. Interestingly, HOXC13 exhibited significantly higher levels of methylation at the 3’UTR (all p-values <0∙001 [unpaired t-tests) in tumour tissue than in normal tissue in bladder, breast, colorectal, head and neck, lung, and uterine tissues, suggesting that 3’UTR methylation could be a primary driver of this expression (Supplementary Fig. S2). Bladder cancer was selected as a representative example as it demonstrated the greatest variation in gene expression between tumour and normal tissue samples (Fig. 2c). <10% of samples in the lowest quartile of gene expression had evidence of methylation in the promoter region of HOXC13, strongly suggesting that differences in promoter methylation cannot account for the observed differences in expression. To systematically assess the association of methylation with expression, we fit a multivariate regression model with the potential non-coding regulatory regions; transcriptional start site, 5’UTR, and 3’UTR, as predictors of gene expression within each tissue type. Across the majority of cancer types, the 3’UTR was most strongly associated with gene expression (Fig. 2d).
3.3. Extent of 3’UTR methylation is correlated with overall survival
Considering the genome-wide changes in methylation pattern exhibited by tumours and the clinically actionable nature of these modifications because of the reversible nature of DNA methylation [15], we next examined genome-wide differences in 3’UTR methylation between tumour and normal tissues in genes with a > 0∙5 correlation coefficient between 3’UTR methylation and gene expression.
Because of the observed divergence in 3’UTR methylation between tumour and normal tissues in certain genes, we hypothesized that these genes play roles in tumourigenesis and tumour progression and thus determined whether they were associated with rate of overall survival. In 5 of the tumour types, including head and neck, lung adeno, lung squamous cell, bladder, and kidney renal cell carcinomas, survival was associated with the extent of methylation at the 3’UTR (Supplementary Fig. S3a). In kidney renal cell carcinoma, of the 38 genes with differential 3’UTR methylation, 22 were significantly associated with overall survival (p-value = 0∙05 [log-rank tests]). Methylation at the 3’UTR of the myosin 1G (MYO1G) gene was associated with gene expression (Supplementary Fig. S3b), and patients with methylation levels in the lowest 20% had significantly longer survival (p < 0∙001 [log-rank tests]) than did those with levels in the highest 20% (Supplementary Fig. S3c). The 22 genes whose 3’UTR methylation was associated with survival represent a significantly higher number of genes than the 2 genes that would have been expected to be associated with survival had the 38 genes been selected at random. In summary, the extent of 3’UTR methylation in a significant number of genes was associated with overall survival in 5 of the 10 tumour types examined.
3.4. Genes with highly correlated expression and 3’UTR methylation are enriched for T cell activation
Because the majority of genes in this analysis exhibited a positive correlation between gene body methylation and expression, those genes in which 3’UTR methylation had a > 0∙5 correlation coefficient with gene expression and a < 0.5 correlation in the gene body were selected for further analysis.
Filtering using these criteria yielded a list of 156 genes (Supplementary Table 2). These genes were then subjected to a pathway analysis, which revealed an enrichment of the genes involved in regulating various aspects of T cell activation (Fig. 3a), including T cell receptor signaling (ITK and VAV1) and T cell exhaustion (HAVCR2), as well as antigen presentation, particularly the MHC class II complex (HLA-DQA1 and HLA-DOA). These genes were then subjected to a network analysis using Netwalker, which revealed a single interconnected node of 23 genes (Fig. 3b), 7 of which were involved in T cell activation. The protein products of 3 of these genes (ITK, HAVCR2, and VAV1) interact with each other and are primarily expressed in T cells.
Fig. 3.
Genes with stronger correlation between gene expression and 3’UTR methylation relative to gene body methylation were enriched for functioning in T cell activation. (a) A pathway analysis of the 156 genes shows the correlation between gene expression and 3’UTR methylation of >0.5 and gene body methylation of <0.5. T cell activation and antigen presentation were the most overrepresented pathways. (b) A single network node with >1 interaction was identified using a Netwalker analysis. The genes in the red box represent genes that are canonically expressed in T cells and are known to be integral to T cell activation. (c) The expression of ITK, VAV1, and HAVCR2, genes that are canonically known to regulate T cell activation, was correlated with T cell counts in tumour samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As the samples analysed were tumour tissues that contain immune cells, we determined whether methylation and expression differences in T cell-related genes occurred in tumour cells or resident T cells present in the tumour samples. We plotted the expression of ITK, VAV1, and HAVCR2 against the estimated levels of T cells using previously established methods for ascertaining T cell counts in TCGA samples [24]. For each gene, a highly significant association was discovered between gene expression and the T cell count (Fig. 3c). These data support the conclusion that in these analysed genes, 3’UTR methylation was associated with gene expression in T cells. Additionally, strong correlations between neutrophil and dendritic cell infiltration, and expression of these genes, were also observed, suggesting the association between 3’UTR methylation and gene expression may be relevant in these cell types as well (Supplementary Fig. S4).
3.5. T cell activation resulted in increased 3’UTR DNA methylation and gene expression of Havcr2
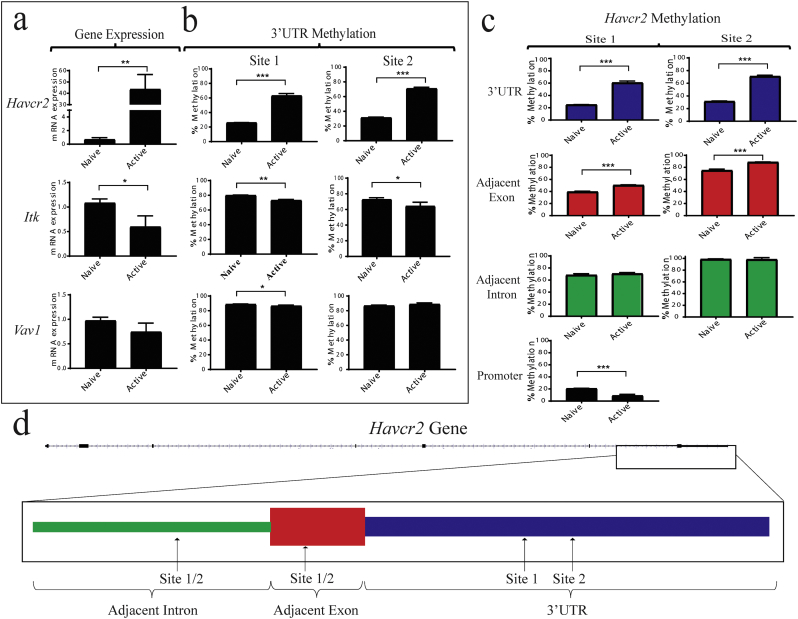
T cells are known to dynamically modulate DNA methylation when changing activation states [26], and a specific epigenetic profile is essential for proper function. Therefore, we evaluated the changes to 3’UTR methylation and the expression of Itk, Vav1, and Havcr2 in ex vivo activated T cells from c57Bl/6 mice.
Seventy-two hours after stimulation, the expression of Vav1 was unchanged, and Itk expression decreased slightly; however, Havcr2 expression increased by nearly 50-fold (Fig. 4a). Next, we examined 2 CpG sites in the 3’UTR for each gene to determine whether any changes to DNA methylation occurred after T cell activation. Similar to gene expression, Vav1 3’UTR methylation did not differ between naïve and activated T cells, whereas Itk 3’UTR methylation had decreased slightly; however, Havcr2 3’UTR methylation had increased substantially: it was around 2∙5-fold higher at both sites in activated T cells relative to naïve T cells (Fig. 4b).
Fig. 4.
Activation of T cells is accompanied by increased Havcr2 gene expression and specific 3’UTR methylation. (a) Comparing the gene expression of naïve and ex vivo-activated T cells isolated from C57Bl/6 mice. (b) Comparing the 3’UTR DNA methylation of naïve and ex vivo-activated T cells isolated from C57Bl/6 mice. (c) Difference in DNA methylation of naïve versus ex vivo-activated T cells at the promoter region, the 3’UTR, and 3’UTR-adjacent exon and intron. (d) Schematic of the Havcr2 gene, with the location of the interrogated sites in each discrete region of the gene highlighted. Statistics were performed using unpaired t-tests for comparisons between 2 groups.
To determine whether the increase in Havcr2 expression was due to changes in promoter methylation, as is the case for many genes switched on after T cell activation, we assayed the methylation of the promoter region. Additionally, a significant decrease was observed in the methylation of the promoter region after activation; very low promoter methylation was observed in the naïve state, and although the Havcr2 promoter lacks a CpG island, non-CpG island methylation has been shown to be involved in gene expression regulation, and therefore may also be functioning as a regulatory element in this context (Fig. 4c). Next, we evaluated methylation of the exon and intron immediately adjacent to the 3’UTR of Havcr2. Methylation of the intron did not change upon activation, and methylation of the adjacent exon was slightly increased; however, this increase was not nearly as substantial as that of the 3’UTR (Fig. 4c). These data demonstrate the exquisite specificity with which robust increases in methylation are targeted to the 3’UTR.
3.6. Treatment with decitabine or Dnmt3a knockout resulted in reduced Havcr2 gene expression
To determine whether increases in DNA methylation are necessary for increased Havcr2 gene expression, we activated T cells ex vivo and then treated them with the Dnmt inhibitor decitabine or DMSO vehicle as a control. After 72 h, the T cells treated with decitabine showed a significant decrease in 3’UTR methylation and a 4-fold decrease in Havcr2 gene expression relative to the DMSO-treated T cells, indicating that DNA methylation occurs upstream of upregulated Havcr2 gene expression after T cell activation (Fig. 5a, b). These data indicate that de novo DNA methylation is an important component of gene expression modulation after activation, as observed in a previous study [27].Additionally, these data suggest that any effects of increasing gene expression by decreasing promoter methylation are outweighed by decrease in DNA methylation outside the promoter region.
Fig. 5.
Blocking DNA methylation results in decreased Havcr2 expression. (a, b) The impact of treating activated T cells with the demethylating agent decitabine on Havcr2 gene expression. (c) Havcr2 expression in wild-type versus Dnmt3a-knockout naïve and chronic exhausted T cells. (d) Levels of Havcr2 DNA methylation in wild-type versus Dnmt3a-knockout T cells mapped to location on the gene. (e) Closer view of the specific loci in which Havcr2 3’UTR experienced decreased 3’UTR methylation in Dnmt3a-knockout T cells relative to wild-type T cells. Statistics were performed using unpaired t-tests for comparisons between 2 groups and one-way ANOVA with Tukey's posttest for multiple comparisons for >2 groups.
We examined the gene expression profile of exhausted T cells from Dnmt3a knockout mice using gene array and genome-wide bisulfite sequencing data obtained from Ghoneim et al [27]. Havcr2 was downregulated in exhausted CD8+ T cells lacking Dnmt3a relative to wild-type (Fig. 5c), supporting the conclusion that de novo methylation of Havcr2 3’UTR results in increased gene expression. Next, using the bisulfite array data generated from this study, we compared the gene body and 3’UTR methylation of Havcr2 in wild-type and Dnmt3a knockout CD8+ T cells. A substantial decrease in Havcr2 methylation occurred in the 3’UTR, whereas most sites in the rest of the gene body remained relatively unchanged (Fig. 5d, e). Furthermore, methylation in the promoter region was observed to be completely erased, though this change did not reach statistical significance due to already-low methylation levels. This finding independently supported the site specificity of Havcr2 methylation, as well as suggests that promoter methylation is not the most critical driver of Havcr2 expression.
4. Discussion
Ascertaining the functional and clinical effects of site-specific DNA methylation remains an important step in unraveling the many layers of epigenetic regulation. Here, we found that the 3’UTR is a functionally distinct site for epigenetic modification. DNA methylation of the gene body is known to be associated with increased gene expression, but by separately examining the 3’UTR across both normal and tumour tissue samples, we revealed an enrichment of DNA methylation sites in this region that are uniquely correlated with increased gene expression. Moreover, we identified several genes that exhibited divergent gene expression between normal and tumour tissues; they lacked significant alterations in copy number or promoter methylation that would explain the differences in expression independently of changes in 3’UTR methylation. In 5 of the 10 tumour types examined, 3’UTR methylation of a substantial number of genes was associated with patient overall survival.
Interestingly, by separating out the 3’UTR as a distinct functional region for the first time, an unexpected link between DNA methylation of this region and T cell regulation was observed. For certain genes in this category, particularly those related to T cell receptor activation (ITK and VAV1), the extent of 3’UTR DNA methylation was correlated with both the presence of T cells in a tumour and with patients' overall survival. For the progressively important immune checkpoint gene HAVCR2, DNA methylation of the 3’UTR may serve as a means by which T cell exhaustion occurs. TIM-3-expressing T cells exhibit a severely exhausted phenotype [28,29], and this protein is frequently found to be expressed in tumour-infiltrating lymphocytes [30]. In addition, the expression of TIM-3 promotes resistance to PD-1/PD-L1 blockade [31], and demethylating agents enhance sensitivity to PD-1/PD-L1 blockade when given in combination [27]. Inferring that DNA methylation inhibition and the subsequent sustained or increased expression of HAVCR2 may be an underlying reason for this observation.
These data suggest two new avenues of exploration that will broaden our understanding of this epigenetic modification. The first is ascertaining how 3’UTR methylation influences gene expression. It is well established that DNA methylation affects the binding of regulatory proteins [2]. In the case of proteins with methylation-binding domains, DNA methylation can increase binding [32,33]. On the other hand, DNA methylation can inhibit protein binding or mask sequence recognition, as is the case for many transcription factors [3]. Another potential explanation for the effect of the 3’UTR on gene expression may be differential alternative splicing and alternative polyadenylation. Gene body methylation has already been shown to affect exon inclusion [34]; however, alternative polyadenylation has not been linked with DNA methylation, but if different lengths of the 3’UTR are dependent on methylation, transcripts with shorter 3’UTRs would have greater mRNA stability and thereby higher gene expression [35]. However, gene body methylation has also been observed as a consequence of higher gene expression, rather than as a cause [36]; therefore, 3’UTR methylation may occur downstream of higher gene expression. Going forward, IP-mass spectrometry will be conducted to hone in on the mechanism of how 3’UTR methylation influences gene expression. Discovering proteins that exhibit sensitivity to binding methylated versus unmethylated 3’UTR that also impact gene expression may reveal how methylation of the 3’UTR results in increased gene expression. In addition to 3’UTR methylation, HAVCR2 promoter methylation may also affect gene expression, despite not containing a CpG island. The methylation of the Havcr2 promoter decreased significantly after ex vivo mouse T cell activation. However, demethylation using decitabine or Dnmt3a knockout resulted in decreased Havcr2 gene expression, which supports the hypothesis that promoter methylation is not the most potent driver of expression in this context.
The second question arises in regard to how 3’UTR methylation is regulated. De novo DNA methylation is deposited by the DNMT3 enzymes [37], and de novo demethylation is handled by the TET family of enzymes; therefore, these are likely to play a role in producing the differential methylation observed in tumour and normal tissues and in activated versus naïve T cells. Indeed, given the substantial decrease in Havcr2 gene expression after Dnmt3a knockout, Dnmt3a in particular seems to be involved. However, the co-factors that position these enzymes in a tightly controlled spatial and temporal context are currently unknown.
These data have multiple implications. First, these genes may play a previously unidentified role in cancer pathogenesis. Second, 3’UTR methylation of these genes may serve as a biomarker for disease presence and progression. Finally, demethylating this region may serve as a novel target for cancer therapy and how demethylating agents affect the 3’UTR should be taken into account when evaluating the mechanism and efficacy of these therapies.
Taken together, our findings indicate that the 3’UTR is a region of epigenetic importance. These data raise the possibility of a novel component of epigenetic regulation that operates during T cell development and activation. Furthermore, they shed light on a potential novel mechanism by which T cells upregulate immune checkpoint mediators. These findings lay a foundation for a broader understanding of the effect of DNA methylation on cellular processes, and most importantly, they may highlight novel components of cancer pathogenesis, opening new avenues for clinical therapy.
The following are the supplementary data related to this article.
Breakdown of 450 K methylation array probe distribution. The total number of probes in each region, and the percent of total probes that interrogate each region, are shown. While probes are enriched for the promoter region, the gene body contains the greatest number of probes overall, due to the size of this region relative to other regions. TSS1500 = region located 1500 bp from transcriptional start site; TSS200 = region located 200 bp from transcriptional start site; 5’UTR = 5′ untranslated region; Body = gene body, excluding exon; 3’UTR = 3′ untranslated region.
Investigating the drivers of HOXC13 expression. Heatmaps representing the methylation patterns across HOXC13 in all tissue types examined. The row color bar depicts the gene structure: purple, promoter region; light green, 5’UTR; dark green, first exon; grey, body; red, 3’UTR. The column color bar represents the top 25% gene expressers in pink and the bottom 25% expressers in black.
The extent of 3’UTR methylation was significantly associated with overall survival in 5 of the 10 tumour types. (a) The first row of the table represents the total number of differentially methylated genes that had significant separation in overall survival (p < .05 [log-rank tests]). The second column represents the number of genes we would have expected to be significantly associated with survival on the basis of Poisson distribution if the genes had been chosen at random. The final row summarizes the total number of differentially methylated genes that were positively correlated with a 3’UTR > 0.5. (b) Differences in overall survival on the basis of methylation status using an example gene, MYO1G, in the context of kidney renal clear cell carcinoma. The red and blue points in the dotplot (b, left panel) distinguish the samples with the highest and lowest 20% of methylation, respectively. (b, right panel) The same samples were used to evaluate the difference in overall survival between these groups. BLCA = bladder carcinoma; BRCA = breast carcinoma; CORE = colon and rectal carcinoma; HNSC = head and neck squamous cell carcinoma; KIRC = kidney renal cell carcinoma; LIHC = liver carcinoma; LUAD = lung adenocarcinoma; LUSC = lung squamous cell carcinoma; PRAD = prostate adenocarcinoma; THCA = thyroid carcinoma; UCEC = uterine carcinoma.
Correlation between HAVCR2, ITK, and VAV1 expression and infiltration of dendritic cells, T cells, B cells, neutrophils, and macrophages: These plots represent the relationship between expression of the investigated genes and amount of immune cell infiltrates across tissue types (as represented by different colored circles), as determined by Li et al. (PMID: 27549193). BLCA: green BRCA: blue CORE: brownHNSC: magenta KIRC: goldenrod LUAD: gold LUSC: turquoise PRAD: forest green THCA: sea green UCEC: tomato.
List of all genes that had a correlation coefficient of >0.5 between 3’UTR methylation and gene expression in at least 1 tissue, listed by tissue type.
Genes that matched the criteria of a correlation coefficient of <0.5 in the gene body and > 0.5 in the 3’UTR in at least 1 tissue type.
List of primers and corresponding sequences used to carry out qPCR and methylation analyses.
Conflict of interests
Section 2
At no time did the authors of this manuscript, nor the institution, receive payment or services from a third party for any aspect of the submitted work.
Section 3
Anil Sood announces the following relationships: consulting (Kiyatec, Merck); research funding (M-Trap); stock holder (BioPath). All other authors have no other financial relationships to report.
Section 4
There are no pending or issued patents broadly relevant to the work at this time.
Section 5
There are no other relationships or activities that readers could perceive to have influenced, or that give the appearance of potentially influencing, what was written in the submitted work.
Ethics approval
-
•
Ethics approval and consent to participate: All human data were obtained from the publically available TCGA database
Data accessibility
-
•
Availability of data and material: The datasets generated or analysed during the current study are available in the TCGA database repository, https://cancergenome.nih.gov/.
Author contributions
-
•
MH McGuire: Conceptualization, data curation, formal analysis, investigation, validation, visualization, methodology, and writing—original draft, review and editing.
-
•
SM Herbrich: Data curation, formal analysis, investigation, validation, visualization, and writing— original draft, review and editing.
-
•
SK Dasari: Data curation, investigation, visualization, and writing—review and editing.
-
•
SY Wu: Methodology and investigation.
-
•
Y Wang: Data curation, formal analysis, validation, and investigation.
-
•
R Rupaimoole: Validation, visualization, and methodology.
-
•
G Lopez-Berestein: Conceptualization and supervision.
-
•
KA Baggerly: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, and project administration
-
•
AK Sood: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, project administration, and writing—original draft, review, and editing.
Funding sources
-
•
Portions of the research reported in this publication were supported by the NIH (P30 CA016672, P50 CA217685, P50 CA098258, U01 CA2 13759, and R35 CA209904). Additional portions of this work were also funded by the Blanton-Davis Ovarian Cancer Research Program, American Cancer Society Research Professor Award, and the Frank McGraw Memorial Chair in Cancer Research. SW was supported by CPRIT grant RP101502 and the Foundation for Women's Cancer.
Acknowledgements
-
•
We thank Dr. Marcos Estecio and the MD Anderson DNA methylation core for performing the methylation assays. Cells were sorted by the MD Anderson Flow Cytometry and Cellular Imaging Core Facility before being used in this study (this facility is funded by NCI Cancer Center Support Grant P30 CA16672).
-
•
We would like to acknowledge Ann Sutton from MD Anderson Scientific Publications for editing this manuscript.
Contributor Information
Santosh K. Dasari, Email: skdasari@mdanderson.org.
Sherry Y. Wu, Email: sherry.wu@uq.edu.au.
Ying Wang, Email: ywang31@mdanderson.org.
Gabriel Lopez-Berestein, Email: glopez@mdanderson.org.
Keith A. Baggerly, Email: kabagger@mdanderson.org.
Anil K. Sood, Email: asood@mdanderson.org.
References
- 1.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 2.Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 3.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 4.Flintoft L. DNA methylation: looking beyond promoters. Nat Rev Genet. 2010;11(9):596. doi: 10.1038/nrg2851. [DOI] [PubMed] [Google Scholar]
- 5.Aran D., Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154(1):11–13. doi: 10.1016/j.cell.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Charlet J., Duymich C.E., Lay F.D., Mundbjerg K., Dalsgaard Sorensen K., Liang G. Bivalent regions of cytosine methylation and H3K27 acetylation suggest an active role for DNA methylation at enhancers. Mol Cell. 2016;62(3):422–431. doi: 10.1016/j.molcel.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinde B., Wu D.Y., Greenberg M.E., Gabel H.W. DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc Natl Acad Sci U S A. 2016;113(52):15114–15119. doi: 10.1073/pnas.1618737114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich M., Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5(5) doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizwana R., Hahn P.J. CpG methylation reduces genomic instability. J Cell Sci. 1999;112(Pt 24):4513–4519. doi: 10.1242/jcs.112.24.4513. [DOI] [PubMed] [Google Scholar]
- 10.Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet: TIG. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 11.Lorincz M.C., Dickerson D.R., Schmitt M., Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 12.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood A.J., Schulz R., Woodfine K., Koltowska K., Beechey C.V., Peters J. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22(9):1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baylin S.B. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl. 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 16.Bell R.E., Golan T., Sheinboim D., Malcov H., Amar D., Salamon A. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26(5):601–611. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyn H., Vidal E., Ferreira H.J., Vizoso M., Sayols S., Gomez A. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17(11) doi: 10.1186/s13059-016-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ooi W.F., Xing M., Xu C., Yao X., Ramlee M.K., Lim M.C. Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity. Nat Commun. 2016;7 doi: 10.1038/ncomms12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kretzmer H., Bernhart S.H., Wang W., Haake A., Weniger M.A., Bergmann A.K. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y.W., Ma X., Zhang Y.A., Wang M.J., Yatabe Y., Lam S. ITPKA gene body methylation regulates gene expression and serves as an early diagnostic marker in lung and other cancers. J Thorac. Oncol: Off Publ Int Assoc Stud Lung Cancer. 2016;11(9):1469–1481. doi: 10.1016/j.jtho.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahuja N., Sharma A.R., Baylin S.B. Epigenetic therapeutics: a new weapon in the war against Cancer. Annu Rev Med. 2016;67:73–89. doi: 10.1146/annurev-med-111314-035900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Weyer P.S., Muehlfeit M., Klose C., Bonventre J.V., Walz G., Kuehn E.W. A highly conserved tyrosine of Tim-3 is phosphorylated upon stimulation by its ligand galectin-9. Biochem Biophys Res Commun. 2006;351(2):571–576. doi: 10.1016/j.bbrc.2006.10.079. [DOI] [PubMed] [Google Scholar]
- 23.Estecio M.R., Yan P.S., Ibrahim A.E., Tellez C.S., Shen L., Huang T.H. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17(10):1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B., Li T., Pignon J.-C., Wang B., Wang J., Shukla S.A. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet. 2016;48:725. doi: 10.1038/ng.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 26.Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 27.Ghoneim H.E., Fan Y., Moustaki A., Abdelsamed H.A., Dash P., Dogra P. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1) doi: 10.1016/j.cell.2017.06.007. [142-57.e19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakuishi K., Jayaraman P., Behar S.M., Anderson A.C., Kuchroo V.K. Emerging Tim-3 functions in antimicrobial and tumor immunity. Trends Immunol. 2011;32(8):345–349. doi: 10.1016/j.it.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q., Munger M.E., Veenstra R.G., Weigel B.J., Hirashima M., Munn D.H. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117(17):4501–4510. doi: 10.1182/blood-2010-10-310425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Zhang S., Hu Y., Yang Z., Li J., Liu X. Targeting PD-1 and Tim-3 pathways to reverse CD8 T-cell exhaustion and enhance ex vivo T-cell responses to autologous dendritic/tumor vaccines. J Immunother (Hagerstown, Md: 1997) 2016;39(4):171–180. doi: 10.1097/CJI.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 31.Koyama S., Akbay E.A., Li Y.Y., Herter-Sprie G.S., Buczkowski K.A., Richards W.G. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7 doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanovic O., Veenstra G.J. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou X., Ma W., Solov'yov I.A., Chipot C., Schulten K. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 2012;40(6):2747–2758. doi: 10.1093/nar/gkr1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr C., Bartel D.P. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du J., Johnson L.M., Jacobsen S.E., Patel D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16(9):519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chedin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Breakdown of 450 K methylation array probe distribution. The total number of probes in each region, and the percent of total probes that interrogate each region, are shown. While probes are enriched for the promoter region, the gene body contains the greatest number of probes overall, due to the size of this region relative to other regions. TSS1500 = region located 1500 bp from transcriptional start site; TSS200 = region located 200 bp from transcriptional start site; 5’UTR = 5′ untranslated region; Body = gene body, excluding exon; 3’UTR = 3′ untranslated region.
Investigating the drivers of HOXC13 expression. Heatmaps representing the methylation patterns across HOXC13 in all tissue types examined. The row color bar depicts the gene structure: purple, promoter region; light green, 5’UTR; dark green, first exon; grey, body; red, 3’UTR. The column color bar represents the top 25% gene expressers in pink and the bottom 25% expressers in black.
The extent of 3’UTR methylation was significantly associated with overall survival in 5 of the 10 tumour types. (a) The first row of the table represents the total number of differentially methylated genes that had significant separation in overall survival (p < .05 [log-rank tests]). The second column represents the number of genes we would have expected to be significantly associated with survival on the basis of Poisson distribution if the genes had been chosen at random. The final row summarizes the total number of differentially methylated genes that were positively correlated with a 3’UTR > 0.5. (b) Differences in overall survival on the basis of methylation status using an example gene, MYO1G, in the context of kidney renal clear cell carcinoma. The red and blue points in the dotplot (b, left panel) distinguish the samples with the highest and lowest 20% of methylation, respectively. (b, right panel) The same samples were used to evaluate the difference in overall survival between these groups. BLCA = bladder carcinoma; BRCA = breast carcinoma; CORE = colon and rectal carcinoma; HNSC = head and neck squamous cell carcinoma; KIRC = kidney renal cell carcinoma; LIHC = liver carcinoma; LUAD = lung adenocarcinoma; LUSC = lung squamous cell carcinoma; PRAD = prostate adenocarcinoma; THCA = thyroid carcinoma; UCEC = uterine carcinoma.
Correlation between HAVCR2, ITK, and VAV1 expression and infiltration of dendritic cells, T cells, B cells, neutrophils, and macrophages: These plots represent the relationship between expression of the investigated genes and amount of immune cell infiltrates across tissue types (as represented by different colored circles), as determined by Li et al. (PMID: 27549193). BLCA: green BRCA: blue CORE: brownHNSC: magenta KIRC: goldenrod LUAD: gold LUSC: turquoise PRAD: forest green THCA: sea green UCEC: tomato.
List of all genes that had a correlation coefficient of >0.5 between 3’UTR methylation and gene expression in at least 1 tissue, listed by tissue type.
Genes that matched the criteria of a correlation coefficient of <0.5 in the gene body and > 0.5 in the 3’UTR in at least 1 tissue type.
List of primers and corresponding sequences used to carry out qPCR and methylation analyses.
Data Availability Statement
-
•
Availability of data and material: The datasets generated or analysed during the current study are available in the TCGA database repository, https://cancergenome.nih.gov/.