Abstract
Objective
Titin gene (TTN) mutations have been described in 8 families with hereditary myopathy with early respiratory failure (HMERF). Some of the original patients had features resembling myofibrillar myopathy (MFM), arguing that TTN mutations could be a much more common cause of inherited muscle disease, especially in presence of early respiratory involvement.
Methods
We studied 127 undiagnosed patients with clinical presentation compatible with MFM. Sanger sequencing for the two previously described TTN mutations in HMERF (p.C30071R in the 119th fibronectin-3 (FN3) domain, and p.R32450W in the kinase domain) was performed in all patients. Patients with mutations had detailed review of their clinical records, muscle MRI findings and muscle pathology.
Results
We identified 5 new families with the p.C30071R mutation who were clinically similar to previously reported cases, and muscle pathology demonstrated diagnostic features of MFM. Two further families had novel variants in the 119th FN3 domain (p.P30091L and p.N30145K). No patients were identified with mutations at position p.32450.
Conclusions
Mutations in TTN are a cause of MFM, and titinopathy is more common than previously thought. The finding of the p.C30071R mutation in 3.9% of our study population is likely due to a British founder effect. The occurrence of novel FN3 domain variants, although still of uncertain pathogenicity, suggests that other mutations in this domain may cause MFM, and that the disease is likely globally distributed. We suggest that HMERF due to mutations in the TTN gene be nosologically classified as MFM-titinopathy.
Keywords: titin, myofibrillar myopathy, hereditary myopathy with early respiratory failure, neuromuscular respiratory failure
Introduction
Myofibrillar myopathies (MFM) are a clinically and genetically heterogeneous group of hereditary muscle diseases, with presence of myofibrillar degradation products on myopathology. MFMs are known to be caused by mutations in DES, CRYAB, MYOT, ZASP, FLNC, DNAJB6 and BAG3 genes, although about 50% of cases do not have identifiable genetic defects.(1)
Hereditary myopathy with early respiratory failure (HMERF) is a rare and possibly under-recognized condition showing clinical and pathological overlap with MFM, being characterized by proximal and/or distal muscle weakness, early respiratory muscle weakness, and myofibrillar abnormalities.(2,3) Recently, two novel titin (TTN) gene mutations, affecting residues in the A-band of the protein at p.C30071R(2,3) and p.R32450W(4) (reference sequence: Genebank AJ277892) have been identified in 8 apparently unrelated HMERF families. We therefore screened a cohort of patients with undiagnosed MFM or overlapping conditions to verify if these A-band TTN mutations could also be responsible for these more common phenotypes.
Methods
Clinical assessment
Patients were recruited from the Newcastle Muscle Centre at the Institute of Genetic Medicine, Newcastle-upon-Tyne, as part of the specialized diagnostic service (National Specialist Commissioning Team) for limb girdle muscular dystrophies (LGMD). Institutional clinical research ethics board approval was obtained and all participants provided written informed consent for research. Patients were selected based on the following features: phenotype compatible with MFM; CK values up to a maximum of 3000 IU/L; adult age at onset; sporadic or suggestive/putative autosomal dominant inheritance; MRI(5,6) and/or muscle biopsy findings compatible with MFM. 127 probands were selected, 72 of which had a clinical diagnosis of MFM (based on histopathology in 45 cases and on compatible phenotype in 27 patients after exclusion of other aetiologies), 52 LGMD/proximal myopathy and 3 distal myopathy. Possible alternative diagnoses (such as FSHD), were excluded as appropriate. All included patients had a muscle biopsy at time or during the study and 38 had a muscle MRI investigation. Clinical details for patients with TTN mutation were collected retrospectively and affected/unaffected family members of these subjects were assessed where possible.
Molecular analysis
Sanger sequencing was performed for the 119th fibronectin 3 (FN3) domain of TTN (which contains the p.30071 position) and a region of the kinase domain directly flanking the p.32450 position. Polymerase chain reaction was performed with Immolase (Bioline), and sequencing with BigDye, (Applied Biosystems), according to the manufacturer’s protocol with an ABI 3130XL sequencer. For patients carrying the p.C30071R mutation, we sequenced single nucleotide variants flanking the disease mutation which defined the shared haplotype of 2.9 Mbp as described in three prior HMERF families.(2) Data from 170 individuals of UK origin within the 1000 genomes dataset(7) were used to investigate the background on which the p.C30071R mutation occurred. The program PHASE(8) provided probabilistic estimates of haplotypes for our families using the 340 phased haplotypes from the 1000 genomes data as known haplotypes to inform our reconstructions. Missing data are imputed by PHASE.
Magnetic Resonance Imaging (MRI)
Muscle MRI was performed in 4 patients with TTN variants. Axial and coronal planes of the pelvis and lower limbs were obtained using conventional T1-weighted spin echo and STIR (short tau inversion recovery) sequences.
Histological examination
Muscle biopsies of 9 TTN positive patients within the current cohort were reviewed retrospectively. Muscle histopathology was assessed by hematoxylin-eosin (H&E) and Gomori trichrome (G-Tri) staining. All muscle biopsies were processed for immunohistochemistry (IHC) and multiplex Western Blot (WB). Immunostaining of unfixed frozen tissue for both procedures was performed using antibodies relating to diagnosis of LGMD and MFM as previously described.(9) Muscle tissue had not been processed using methods to permit ultrastructural evaluation.
Results
Molecular results
Sequencing identified the heterozygous g.274375T>C mutation (p.C30071R) in 5/127 unrelated subjects (F.1A, F.2A, F3, F.4B and F.5) with a total detection rate of 3.9% in the entire cohort. Four of these patients had a clinical diagnosis of MFM, giving a detection rate within this group of 5.5%. Two additional subjects had novel heterozygous variants in the FN3 domain (g.274436C>T, p.P30091L in F.6 and g.274599C>G, p.N30145K in F.7). Sequencing for the TTN kinase domain mutation (4) revealed wild-type sequence in all subjects.
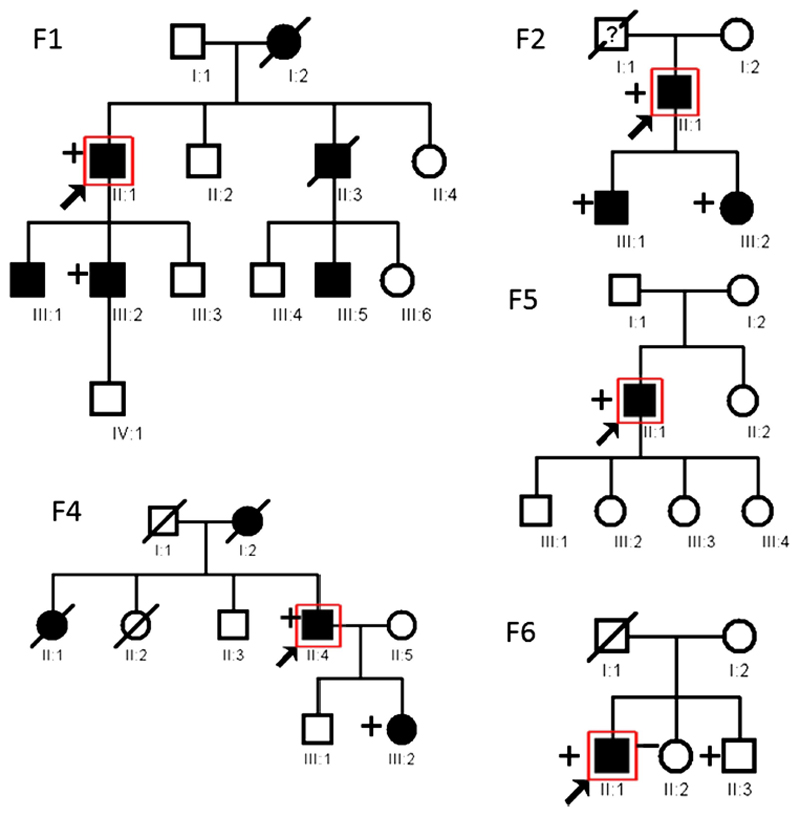
The TTN gene changes co-segregated with the disease in families F.1, F.2, and F.4 (Figure 1). Analysis of family F.6 revealed that the 61 year old brother of the proband is a healthy carrier of the g.274436C>T/p.P30091L variant. No segregation analysis was possible for families F.3, F.5 and F.7.
Figure 1. Pedigree structure of the reported families.
Pedigree structure for the reported families, where available. The presence of a “+” symbol at the left of a pedigree symbol indicates that DNA tested positive for the p.C30071R mutation in TTN for F1-F5, and p.P30091L mutation for F6. The “-” symbol indicates that the patient was tested and no mutation in TTN was found. Pedigree structure for Families F.2 and F.7 was not available at the time of the study.
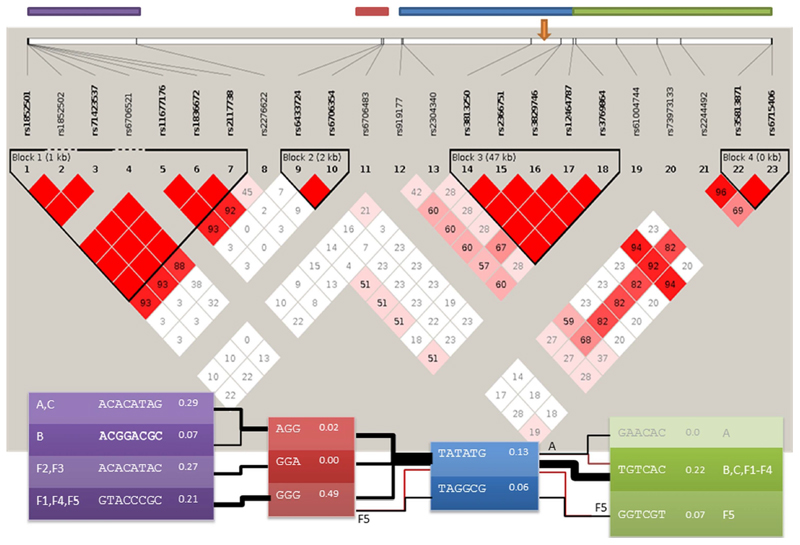
Analysis of polymorphic markers on chromosome 2 in probands from families F.1-5 and the previously reported 3 families with the p.C30071R mutation(2) showed a common haplotype that included rs6706354, rs6706483, rs919177, and the p.C30071R mutation, indicating a core region of 171 kbp.
Haplotype analysis of these 8 families was compared with 170 UK control subjects from the 1000 genomes database of phased genotypes,(10) and 23 shared SNPs were present (Figure 2). The core region of 171 kbp is the most common inferred disease haplotype for seven of our families, and this disease haplotype is consistent with genotypes observed in family F.5. Across the wider region, two pairs of families share likely disease haplotypes of over 790kb. While the core region has a population frequency of 13%, it is highly improbable (<0.002) that all three extended families and F.5 had independent mutations. Families F.2 and F.3 share a rare haplotype block that is not seen in the population of 170 genotypes, further evidence that haplotypes are identical by descent.
Figure 2. Haplotype analysis of 8 families with the p.C30071R mutation.
Haplotype structure around the p.C30071R mutation. The plot on a grey background gives the LD structure and estimates LD between SNPs for UK population data from the 1000 genomes project. The orange arrow on this plot indicates the position of the p.C30071R mutation (g.179410829A>G on chromosome 2, using GRCh37 as the reference sequence). The top four coloured bars give the regions underlying the four haplotype blocks for the disease haplotypes with matching colours. For the haplotype block, the left and rightmost give the family labels, other columns give the estimated haplotype sequence and frequency of haplotype block in the UK 1000 genomes samples. The line joining haplotype blocks indicate the most likely estimated haplotypes for each family (estimated using PHASE). Line thicknesses are proportional to the number of families underlying the line. Where the most likely haplotype for a family is not the overall consensus, yet the consensus is supported by the data, a red line joins to the consensus.
Clinical results
Clinical information of the 5 patients with the p.C30071R mutation and their affected relatives are summarized in Table 1, and pedigree structure reported in Figure 1. Onset was typically in the 4th or 5th decades, and symptoms included tripping and falling, myalgia and walking difficulties. CK values ranged from normal levels to 1195 IU/L (average 730 IU/L). The natural history appears to be characterised by slow progression, with involvement of the proximal, distal, and axial musculature. Ambulation was retained in all but subject F.2A who lost ambulation after a disease course of about 25 years Ten of 11 patients had abnormal pulmonary function tests indicative of respiratory muscle weakness, and requirement for nocturnal noninvasive ventilation occurred approximately 20-30 years after disease onset in 4 patients.
Table 1. clinical and molecular features of patients with TTN gene variants.
| Pt N | Mutation (amino acidic change) | age last seen * | onset | Pattern of muscle involvement at last assessment | Extra skeletal involvement | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age * | symptom | CK IU/L | Ambulant | UL prox | UL distal | LL prox | LL distal | Neck flex | ankle DF | muscle bulk | Scapular winging | Contractures | spine | Cardiac exam and ECG | Respiratory function (FVC) | |||||
| F.1A | n.a. | 65 | late 30s | proximal weakness | 5X | yes | ++ | - | +++ | + | +++ | n.a. | n.a. | no | TAs | n.a. | normal | NIV | ||
| F.1B | C30071R | 44 | 33 | foot drop tripping | 700 | yes | + | - | +++ | + | +++ | ++++ | Calf hypertrophy | yes (AS) | TAs | mild rigidity | normal | 89% sitting 81% lying | ||
| F.1C | C30071R | 44 | 30s | tripping | 563 | yes | + | - | +++ | + | + | ++ | Quads hypertrophy | yes (AS) | no | mild rigidity CS | normal | 65% sitting 51% lying | ||
| F.2A | C30071R | 63 | 30s | Falling finger weakness | n.a. | no (late 50s) | ++ | +++ | ++ | +++ | + | ++++ | Quads hypertrophy (mild); distal atrophy | yes | no | no | mild LV impairment, palpitations | NIV | ||
| F.2B | C30071R | 38 | 36 | proximal weakness | 1096 | yes | + | - | + | ++ | + | - | Normal | no | TAs | mild rigidity | normal | 77% sitting 63% lying | ||
| F.2C | C30071R | 36 | 36 | mild myalgia | 189 | yes | +/- | +/ | +/- | +/ | +++ | no | Normal | no | no | mild rigidity | normal | 76% sitting 73% lying | ||
| F.3 | C30071R | 44 | 30s | foot drop | 1195 | yes | - | - | ++ | +++ | - | ++++ | Normal | no | no | thoracic kyphosis | normal | 68% sitting 51% lying | ||
| F.4A | n.a. | 58 * | n.a. | n.a. | n.a. | n.a. | - | - | +++ | - | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | NIV | ||
| F.4B | C30071R | 57 | 20s | foot drop | 551 | yes | + | +/-∞ | ++ | +++ | + | ++++ | Calf hypertrophy | mild (AS) | right elbow | no | SVT | NIV | ||
| F.4C | C30071R | 34 | 30s | myalgia | 550 | yes | + | n.a. | + | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | normal | 105% sitting 96% lying | ||
| F.5 | C30071R | 39 | late 20s | Myalgia tripping | 1019 | yes | + | +/- | +/- | +++ | - | ++++ | Normal | no | no | normal | normal | 69% sitting 61% lying | ||
| F.6 | P30091L | 57 | 30s | difficulty lifting LL | 204 | yes | ++ | - | +++ | +/- | + | - | Calf hypertrophy | AS | no | normal | normal | 52% sitting | ||
| F.7 | N30145K | 39 | 30s | n.a. | 290 | yes | + | + | + | + | n.a. | +++ | Deltoid atrophy. | mild | no | no | normal | normal | ||
Legend: Pt: patient; N: number; *: indicated in years; CK: creatine Kinase; prox: UL: upper limbs; proximal; LL: lower limbs; DF: dorsiflexion; FVC: forced vital capacity; n.a.: data not available; +, ++, +++ etc: severity of muscle weakness; AS: asymmetric; TAs: achille’s tendons; NIV: non invasive ventilation; CS: cervical spine; LV: left ventricular; ∞ Finger extension 3-; SVT: supraventricular tachycardia
Previously undocumented examination findings in this condition included scapular winging, often asymmetrical, elbow and/or achilles tendon (TA) contractures and a variable degree of spinal rigidity. Cardiac function was affected in 2 patients, one showing mild left ventricular impairment (F.2A, possibly related to respiratory dysfunction) and the other supraventricular tachycardia (F.4B). Two subjects (F.2C and F.4C) only reported mild mylagias and showed mild/very mild muscle weakness, reduction of FVC (F.2C) and raised CK (F.4C) Subjects F.6 and F.7 carrying novel TTN variants presented with clinical features compatible with MFM (Table 1). Both showed adult age at onset, mildly raised CK values and proximal upper and lower limb involvement with severe ankle dorsiflexion weakness in patient F.7. Patient F.6 showed mild neck weakness and calf hypertrophy. Scapular winging was present in both patients, and patient F.6 had respiratory involvement. Patient F.7, originally from Brazil, has several affected family members but none were available for clinical assessment.
Muscle MRI findings
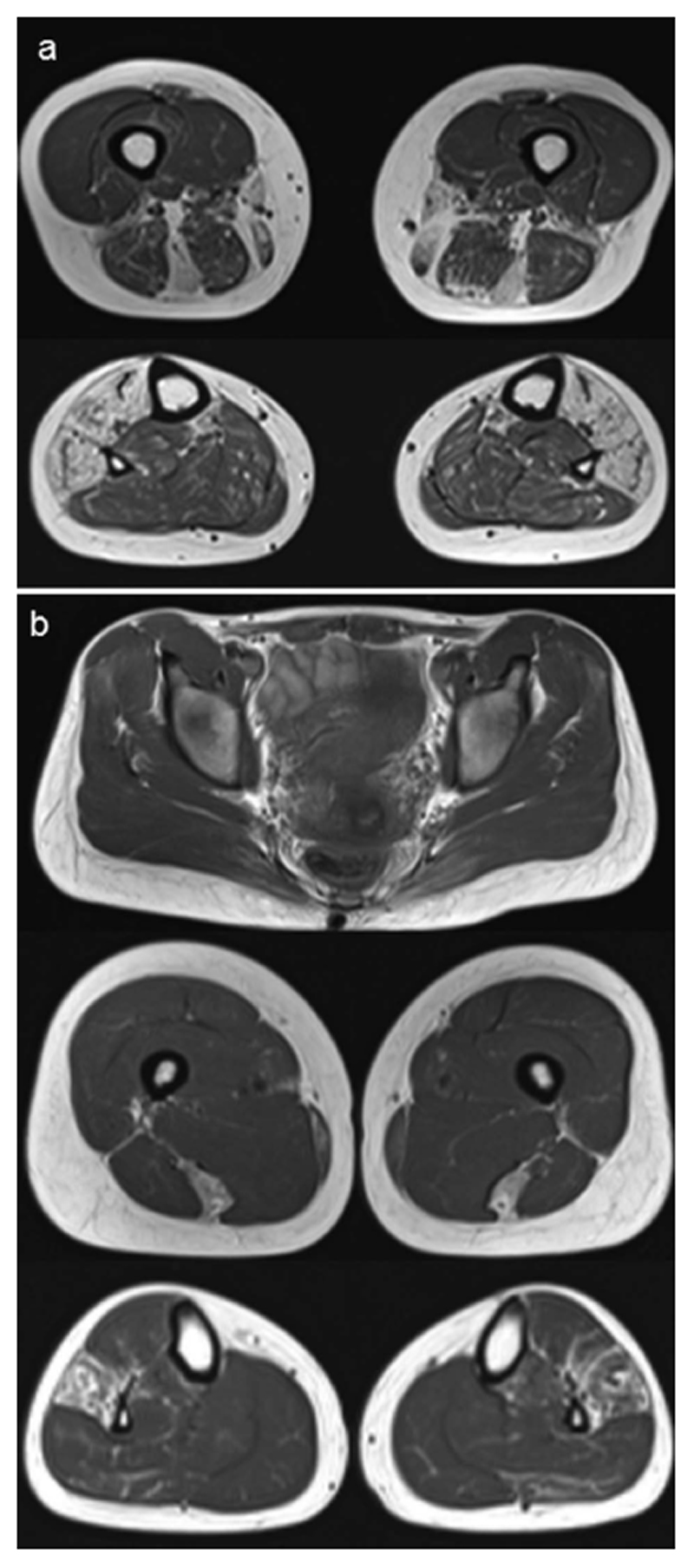
Muscle MRI was performed in 3 subjects with the p.C30071R mutation (F.2C, F.4B and F.5) and in subject F.7 carrying the novel p.N30145K variant. Findings were consistent with previous reports, with early involvement of the semitendinosus and peroneus longus muscles.(2,11) Patient F.7 showed major involvement of the semitendinosus muscle bilaterally, asymmetric involvement of the semimembranosus and variable involvement of the gracilis and sartorius muscles, while the lower leg showed fatty infiltration of the tibialis anterior and peroneus longus muscles (Figure 3).
Figure 3. Muscle MRI findings.
(a) Imaging of the patient from F.7, having the novel p.N30145K variant. The upper leg (above) demonstrates signal abnormality predominantly in the semitendinosus muscle, and in the lower leg (below) the tibialis anterior and peroneus longus are affected by fatty infiltration. (b) Imaging of patient F.2C. This presymptomatic individual with the p.C30071R mutation has the characteristic damage pattern of fatty infiltration in the peroneus longus and semitendinosus muscles.
Histological findings
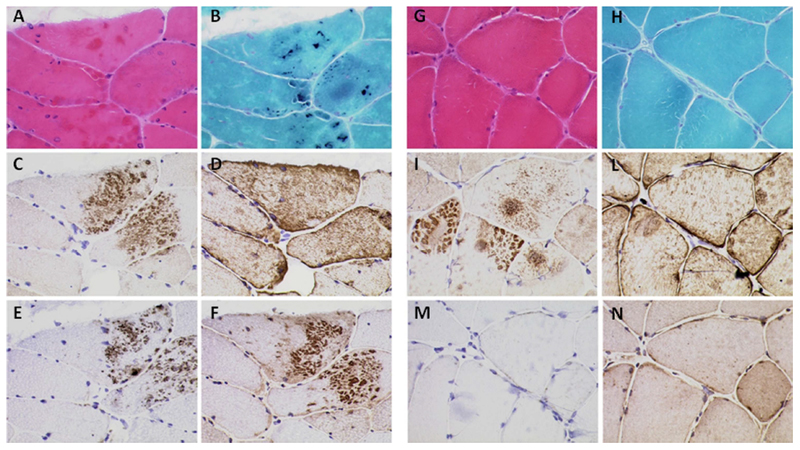
Nine patients had a muscle biopsy, with one having a repeat biopsy (Table 2). Biopsies of patients with the p.C30071R TTN mutation displayed myopathic or dystrophic changes, with variation in fiber size, central nuclei, fiber splitting and increase in perimysial and endomysial connective tissue (Supplemental Figure). Rimmed vacuoles were observed in 3 biopsies, while eosinophilic inclusions and dark blue inclusions on G-Tri staining were present in 3 biopsies (Figure 4, Table 2, and Supplemental Figure). Variable degrees of cytoplasmic accumulation of desmin, myotilin, P62 and ubiquitin was present in 6/7 biopsies. In three biopsies (F.1B, F.4A and F.5), cytoplasmic aggregates showed a “cheetah skin appearance”, particularly with myotilin staining (Figure 4). In patient F.4A we observed accumulation of further proteins but in particular of gamma-sarcoglycan and dystrophin (Supplemental Figure). Cytoplasmic inclusions observed in H&E and G-Tri did not fully overlap with regions of protein accumulation described above (Figure 4).
Table 2. muscle histopathology and immunohistochemistry findings of patients with TTN gene variants.
| Pt N | Muscle biopsy | ||||||
|---|---|---|---|---|---|---|---|
| site | age at biopsy (yrs) | Histology | Myotilin | Desmin | P62 | Ubiquitin | |
| F.1A | n.a. | 50 | mildly myopathic | rare fibres with diffuse cytoplasmic accumulation | rare fibres with diffuse cytoplasmic accumulation | normal | normal |
| F.1B | triceps | 37 | mildly myopathic, eosinophilic inclusions on H&E in a group of fibres, also labelled with G-Tri | small dense inclusion with “cheetah skin” appearance, mostly not overlapping eosinophilic inclusions | diffuse cytoplasmic accumulation | small dense inclusion with “cheetah skin” appearance | normal |
| F.3 | n.a. | 44 | severe end stage pathology with rimmed vacuoles, fibre splitting and rare eosinophilic inclusions. Some inclusions also positive on G-Tri | rare fibres with diffuse cytoplasmic accumulation | rare fibres with diffuse cytoplasmic accumulation | rare fibres with diffuse cytoplasmic accumulation | one fibre showing dense cytoplasmic accumulation |
| F.4A | Quad-riceps | 55 | dystrophic changes with rimmed vacuoles | fibres showing dense accumulation, some with ”cheetah skin” appearance | few fibres showing dense cytoplasmic accumulation | few fibres showing dense cytoplasmic accumulation | very mild diffuse cytoplasmic accumulation in occasional fibres |
| F.4B | n.a. | 52 | very subtle myopathic changes | normal | normal | normal | normal |
| F.4C | Quad-riceps | 14 | poor biopsy condition, probably very mildly myopathic | rare fibres with subsarcolemmal and cytoplasmic accumulation | rare fibres with subsarcolemmal and cytoplasmic accumulation | rare fibres with subtle cytoplasmic accumulation | not done |
| F.5 | Quad-riceps | 38 | very mild myopathic changes, some fibres showing dark, bluish areas on G-Tri, eosinophilic on H&E | small dense inclusion with “cheetah skin” appearance, some of which show enhanced H&E and G-Tri staining | normal | normal | normal |
| F.6 | Deltioid (quad-riceps) | 53 | dystrophic features, rimmed vacuoles, fibre splitting, few fibres showing dark blue areas on G-Tri | rare fibres with cytoplasmic accumulation | very rare fibres | very rare fibres with cytoplasmic accumulation | normal |
| F.7 | n.a. | 34 | poor biopsy condition, probably very mildly myopathic | normal | normal | normal | normal |
| F.7 | n.a. | 38 | myopathic changes, rimmed vacuoles, eosinophilic accumulation, cytoplasmic bodies positive on G-Tri | fibres with cytoplasmic accumulation | cytoplasmic accumulation | cytoplasmic accumulation | normal |
Legend: Pt: patient; N: number; n.a.: data not available
Figure 4. Histopathologic features.
Figure legend: Histological and Immunohistochemical findings for patients F.1B (A-F) and F.5 (G-N). A, G: H&E staining. B, H: Gomori Trichrome staining. Immunolabelling for myotilin (C, I), Desmin (D, L), P62 (E, M) and VCP (H, N). Note the presence of basophilic inclusion on H&E (A) and dark blue inclusions on G-Tri staining (B) and cheetah-like aggregates with labelling for Mytolin (C, I), P62 (E) and VCP (F).
Muscle biopsy analysis of patients F.6 and F.7 showed findings compatible with a diagnosis of MFM, with rimmed vacuoles, dark blue areas and cytoplasmic bodies with G-Tri and cytoplasmic accumulation of myofibrillar proteins, in particular myotilin and P62 (Table 2).
Discussion
We report 5 new British families with the previously reported p.C30071R mutation in the TTN gene, and two unrelated patients with novel heterozygous changes affecting nearby residues within the same FN3 domain of the titin A-band. Analysis of clinical, MRI and histological features of these 7 subjects and their affected relatives supported a diagnosis of MFM in all of them. Our results therefore indicate that TTN mutations are an additional cause of MFM, and that HMERF caused by the p.C30071R mutation in TTN should be classified as MFM-titinopathy.
The condition described in the 11 families with the p.C30071R TTN mutation ((2,3) and present series) is compatible with a MFM phenotype, including autosomal dominant inheritance, onset in adulthood, and weakness in proximal, distal, axial, and/or respiratory muscles. Pelvic girdle weakness, foot drop and neck weakness are the main symptoms at onset, but ultimately the weakness usually involves the proximal compartment of both upper and lower limbs. The weakness pattern is nearly always symmetric. New observations include variable degrees of TA contractures, spinal rigidity and muscle hypertrophy.
Respiratory involvement might be the presenting symptom(2) and often leads to requirement for NIV support. MFM-titinopathy does not appear to be associated with major cardiac involvement, but in view of titin’s role in myocardial function(12) we recommend cardiac surveillance for all patients with TTN mutations.
Haplotype analysis indicated a shared region of 171 kbp between all 8 families with the p.C30071R mutation ((2) and current report), and that the probability of this shared haplotype occurring in 8 unrelated families by chance is less than 10-6. Larger haplotype blocks are shared respectively between families A, B and C(2); families F.2 and F.3; and families F.1, F.4 and F.5 indicating that these families are more recently connected (Figure 2). However, sharing of haplotype blocks between these families did not strictly correspond to their place of origin in the UK (for example, families A, B, C and F2 are from the Northumberland region). Estimation of the age of this mutation is unreliable on account of the haplotype differences in the patient F.5. This pattern of sharing around the disease haplotype with erosion of the shared haplotypes is expected with a single origin for the disease haplotype with subsequent rearrangement by recombination over time. The very small size of the shared haplotype suggests an ancient mutation, or that (less likely) the mutation occurred more than once in time on similar haplotype backgrounds of approximately 171 kbp. In either situation, this analysis suggests that this mutation may be more common than was previously believed and is potentially present in other geographic locations.
In this study we also identified two novel TTN variants, both at highly conserved residues within the same FN3 domain (Supplemental Table 1), which were absent in 400 ethnically-matched control chromosomes and publically available databases. The p.P30091L variant found in subject F.6 was also identified in an unaffected brother, suggesting that p.P30091L is a neutral variant. However, it remains possible that this is a pathogenic variant with variable penetrance and/or expressivity, also in view of the very late onset age (up to age 71 years) reported in one patient with the p.C30071R mutation (Patient A-III:3(2)). In the absence of segregation analysis, we cannot further comment on the p.N30145K variant identified in subject F.7, although MRI findings were supportive of a diagnosis of MFM-titinopathy (Figure 3).
Notably, our experience with exome sequencing indicates that TTN is a highly polymorphic gene. Exome data from 239 subjects at our centre discovered 71 novel TTN coding changes of uncertain significance. The distribution of the variants across the various TTN domains is represented in Supplemental Table 2. Analysis using chi-squared goodness of fit against expected values in each domain was not significant. A Kolmogorov Smirnov test failed to reject the hypothesis that the distribution of the mutations across the TTN transcript was uniform, suggesting these variants were randomly distributed in the gene. None of these variants appeared in the 119th FN3 domain (amino acid positions 30068-30160), although these data still indicate that novel coding variants in TTN should be interpreted with caution. Muscle biopsy analyses of the p.C30071R patients revealed characteristic protein aggregates with a cheetah-skin appearance (particularly for myotilin) and absence of correlation between inclusions (Figure 4). Although this pattern is highly characteristic we cannot at present comment on the specificity of these findings to MFM-titinopathy. Overall, most of the p.C30071R positive patients meet pathological criteria for MFM, with myofibrillar aggregates (as demonstrated by the protein accumulations on immunohistochemistry) and dark cytoplasmic deposits, although as in the present series a degree of histopathological variability is expected given the focal nature of abnormalities in MFMs. (1,13,14) Ultrastructural examination was not possible in our patients, although previous work on patients with the p.C30071R mutation has demonstrated Z-disc streaming, cytoplasmic bodies, and evidence of myofibrillar aggregation,(3) further evidence that this condition represents a subtype of MFM.
An important consideration is whether other families previously designated as HMERF also fall under the category of MFM-titinopathy. The 3 families reported by Ohlsson et al with the p.C30071R mutation have clinical and pathologic features compatible with MFM.(3) Available clinical and pathological information of further 13 HMERF cases (15–19) are suggestive for MFM, and these subjects may also carry the same TTN mutation. Of note, two of these families were later found to have the p.R32450W mutation in the kinase domain.(4) Two reports described cases with clinical similarity to MFM, although muscle pathology was nonspecific (20,21) and 5 further patients previously designated as HMERF are probably affected by a different disease, on account of their childhood onset (22–24) and significant cardiac involvement.(23,25) Whether the allelic conditions tibial muscular dystrophy and LGMD2J caused by M-line TTN mutations, may also represent subtypes of MFM might need further investigations, although so far sarcomeric or Z-disc abnormalities have not been demonstrated in these conditions.(26)
Our results indicate that the p.C30071R TTN mutation causes MFM and sequencing of this FN3 domain of TTN should be performed as part of the diagnostic workup for MFM, particularly in the presence of early respiratory involvement and supportive muscle MRI. The p.C30071R mutation is more common than anticipated, identified in 5.5% of undiagnosed MFM patients from the UK. This value appears comparable to reported mutation frequencies for other MFM genes (4-15%)(27) and similar to what is observed at our diagnostic service in Newcastle (9% for MYOT, 3%, for DES and ZASP, 2% for FLNC, and <1% for CRYAB, DNAJB6 and BAG3 genes). These data suggest that TTN mutations are a relatively common cause of MFM in our population, possibly due to a founder effect. Although the status of the newly identified variants is still uncertain, they suggest that a repertoire of TTN mutations may cause MFM-titinopathy, indicating that this disease may be globally distributed.
Supplementary Material
Acknowledgements
The authors would like to thank the patients who took part in this study. GP is the recipient of a Bisby fellowship from the Canadian Institutes of Health Research. PFC is an Honorary Consultant Neurologist at Newcastle upon Tyne Foundation Hospitals NHS Trust, is a Wellcome Trust Senior Fellow in Clinical Science (084980/Z/08/Z), and a UK NIHR Senior Investigator. PFC receives additional support from the Wellcome Trust Centre for Mitochondrial Research (096919Z/11/Z), the Medical Research Council (UK) Centre for Translational Muscle Disease research, the Association Française contre les Myopathies, and EU FP7 TIRCON, and the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre based at Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Diagnostic facilities at the Newcastle Muscle Centre are supported by the National Specialised Commissioning Team (NSCT) for rare neuromuscular disorders. The Institute of Genetic Medicine in Newcastle is part of the MRC Centre for Neuromuscular Diseases and the TREAT-NMD Alliance (www.treat-nmd.eu).
Footnotes
Disclosure statement
All authors (GP, RB, IW, SAH, HG, JH, HRE, AVR, ARadunovic, JW, SV, ARaman, JW, SV, MB, MEF, AM, CE, EH, RH, VS, KB, HL, PFC, AS) report no competing interests.
References
- (1).Selcen D. Myofibrillar myopathies. Neuromuscul Disord. 2011 Mar;21(3):161–171. doi: 10.1016/j.nmd.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Pfeffer G, Elliott HR, Griffin H, et al. Titin mutation segregates with hereditary myopathy with early respiratory failure. Brain. 2012;135(Pt 6):1695–1713. doi: 10.1093/brain/aws102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ohlsson M, Hedberg C, Bradvik B, et al. Hereditary myopathy with early respiratory failure associated with a mutation in A-band titin. Brain. 2012;135(Pt 6):1682–1694. doi: 10.1093/brain/aws103. [DOI] [PubMed] [Google Scholar]
- (4).Lange S, Xiang F, Yakovenko A, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308(5728):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- (5).Fischer D, Kley RA, Strach K, et al. Distinct muscle imaging patterns in myofibrillar myopathies. Neurology. 2008;71(10):758–765. doi: 10.1212/01.wnl.0000324927.28817.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wattjes MP, Kley RA, Fischer D. Neuromuscular imaging in inherited muscle diseases. Eur Radiol. 2010;20(10):2447–2460. doi: 10.1007/s00330-010-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pogue R, Anderson LV, Pyle A, et al. Strategy for mutation analysis in the autosomal recessive limb-girdle muscular dystrophies. Neuromuscul Disord. 2001;11(1):80–87. doi: 10.1016/s0960-8966(00)00154-1. [DOI] [PubMed] [Google Scholar]
- (10).1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Birchall D, von der Hagen M, Bates D, Bushby KM, Chinnery PF. Subclinical semitendinosus and obturator externus involvement defines an autosomal dominant myopathy with early respiratory failure. Neuromuscul Disord. 2005;15(9–10):595–600. doi: 10.1016/j.nmd.2005.05.002. [DOI] [PubMed] [Google Scholar]
- (12).Herman DS, Lam L, Taylor MR, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Walter MC, Reilich P, Huebner A, et al. Scapuloperoneal syndrome type Kaeser and a wide phenotypic spectrum of adult-onset, dominant myopathies are associated with the desmin mutation R350P. Brain. 2007;130(Pt 6):1485–1496. doi: 10.1093/brain/awm039. [DOI] [PubMed] [Google Scholar]
- (14).Schroder R, Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol. 2009;19(3):483–492. doi: 10.1111/j.1750-3639.2009.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kinoshita M, Satoyoshi E, Suzuki Y. Atypical myopathy with myofibrillar aggregates. Arch Neurol. 1975;32(6):417–420. doi: 10.1001/archneur.1975.00490480083010. [DOI] [PubMed] [Google Scholar]
- (16).Chapon F, Viader F, Fardeau M, et al. Familial myopathy with “cytoplasmic body” (or “spheroid”) type inclusions, disclosed by respiratory insufficiency] Rev Neurol (Paris) 1989;145(6–7):460–465. [PubMed] [Google Scholar]
- (17).Edstrom L, Thornell LE, Albo J, Landin S, Samuelsson M. Myopathy with respiratory failure and typical myofibrillar lesions. J Neurol Sci. 1990;96(2–3):211–228. doi: 10.1016/0022-510x(90)90134-9. [DOI] [PubMed] [Google Scholar]
- (18).Abe K, Kobayashi K, Chida K, Kimura N, Kogure K. Dominantly inherited cytoplasmic body myopathy in a Japanese kindred. Tohoku J Exp Med. 1993;170(4):261–272. doi: 10.1620/tjem.170.261. [DOI] [PubMed] [Google Scholar]
- (19).Tasca G, Mirabella M, Broccolini A, et al. An Italian case of hereditary myopathy with early respiratory failure (HMERF) not associated with the titin kinase domain R279W mutation. Neuromuscul Disord. 2010;20(11):730–734. doi: 10.1016/j.nmd.2010.07.269. [DOI] [PubMed] [Google Scholar]
- (20).Winter JH, Neilly JB, Henderson AF, et al. Life-threatening respiratory failure due to a previously undescribed myopathy. Q J Med. 1986;61(236):1171–1178. [PubMed] [Google Scholar]
- (21).Evangelista T, Ferro J, Pereira P, de Carvalho M. A case of asymptomatic cytoplasmic body myopathy revealed by sinvastatin. Neuromuscul Disord. 2009;19(1):66–68. doi: 10.1016/j.nmd.2008.10.008. [DOI] [PubMed] [Google Scholar]
- (22).Jerusalem F, Ludin H, Bischoff A, Hartmann G. Cytoplasmic body neuromyopathy presenting as respiratory failure and weight loss. J Neurol Sci. 1979;41(1):1–9. doi: 10.1016/0022-510x(79)90134-5. [DOI] [PubMed] [Google Scholar]
- (23).Patel H, Berry K, MacLeod P, Dunn HG. Cytoplasmic body myopathy. Report on a family and review of the literature. J Neurol Sci. 1983;60(2):281–292. doi: 10.1016/0022-510x(83)90069-2. [DOI] [PubMed] [Google Scholar]
- (24).Bertini E, Ricci E, Boldrini R, et al. Involvement of respiratory muscles in cytoplasmic body myopathy--a pathology study. Brain Dev. 1990;12(6):798–806. doi: 10.1016/s0387-7604(12)80010-6. [DOI] [PubMed] [Google Scholar]
- (25).Baeta AM, Figarella-Branger D, Bille-Turc F, Lepidi H, Pellissier JF. Familial desmin myopathies and cytoplasmic body myopathies. Acta Neuropathol. 1996;92(5):499–510. doi: 10.1007/s004010050552. [DOI] [PubMed] [Google Scholar]
- (26).Udd B. Genetics and pathogenesis of distal muscular dystrophies. Adv Exp Med Biol. 2009;652:23–38. doi: 10.1007/978-90-481-2813-6_3. [DOI] [PubMed] [Google Scholar]
- (27).Selcen D, Engel AG. Myofibrillar myopathies. In: Pagon RA, Bird TD, Dolan CR, et al., editors. Genereviews. Seattle (WA): University of Washington, Seattle; 1993. [accessed November 2, 2012]. 2005 Jan 28 [Updated 2012 Oct 29], [ http://www.ncbi.nlm.nih.gov/books/NBK1499/] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.