Abstract
Purpose
Internationally-adopted variant interpretation guidelines from the American College of Medical Genetics and Genomics (ACMG) are generic and require disease-specific refinement. Here we developed CardioClassifier (www.cardioclassifier.org), a semi-automated decision-support tool for inherited cardiac conditions (ICCs).
Methods
CardioClassifier integrates data retrieved from multiple sources with user-input case-specific information, through an interactive interface, to support variant interpretation. Combining disease- and gene-specific knowledge with variant observations in large cohorts of cases and controls, we refined 14 computational ACMG criteria and created three ICC-specific rules.
Results
We benchmarked CardioClassifier on 57 expertly-curated variants and show full retrieval of all computational data, concordantly activating 87.3% of rules. A generic annotation tool identified fewer than half as many clinically-actionable variants (64/219 vs 156/219, Fisher’s P=1.1x10-18), with important false positives; illustrating the critical importance of disease and gene-specific annotations.
CardioClassifier identified putatively disease-causing variants in 33.7% of 327 cardiomyopathy cases, comparable with leading ICC laboratories. Through addition of manually-curated data, variants found in over 40% of cardiomyopathy cases are fully annotated, without requiring additional user-input data.
Conclusion
CardioClassifier is an ICC-specific decision-support tool that integrates expertly curated computational annotations with case-specific data to generate fast, reproducible and interactive variant pathogenicity reports, according to best practice guidelines.
Keywords: variant interpretation, inherited cardiac conditions, clinical genomics, next-generation sequencing, bioinformatics
Introduction
Inherited cardiac conditions (ICCs) represent a major health burden with a combined prevalence of ~1%1. Genetic testing is recommended to support the management of many ICCs, with roles in diagnosis (particularly valuable for identification of at-risk relatives), prognostication, and therapeutic stratification.
The principle challenge in genetic testing across all diseases is the interpretation of identified sequence variants. This requires evaluation of data from diverse sources, including clinical observations, computational data and data derived from the literature. Although existing tools aid collection of some of these data types, scientists and clinicians must often access multiple data sources whilst interpreting a single genetic variant.
The American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) recently released guidelines that aim to standardise variant interpretation2. These guidelines outline a set of evidence criteria to assess each variant against, along with how these might be weighted and combined to reach a classification. Studies have, however, shown that even when following the ACMG/AMP guidelines, interpretation can differ between different laboratories, with discordance in excess of 10%3. One key reason for this discordance is the structure of the ACMG/AMP guidelines, which are deliberately broad and lack specific thresholds, to allow adoption across the full spectrum of genetic disorders. As a result, the challenge to individual disease domains is to incorporate expert gene and disease-specific knowledge, to optimise variant interpretation and introduce consensus. Initiatives such as the Clinical Genome Resource (ClinGen)4, are working to define such disease- and gene-specific thresholds, although these are currently limited to pilot phases for specific gene-disease pairs.
The introduction of guidelines, including the logic behind reaching each classification, opens the way for new computational solutions to facilitate their adoption and increase consistency. Indeed, publication of the guidelines has led to the emergence of interactive tools5,6, however, to date only one of these builds in automation7, and none incorporate expert disease-specific knowledge.
Here, we describe CardioClassifier, a powerful new tool that utilises the framework outlined by the ACMG/AMP guidelines, to automatically annotate variants across 17 computational criteria. Each criterion has been individually parametrised for each gene-disease pair using expert disease-specific knowledge. Automated data are integrated with interactively added case-specific information to calculate variant pathogenicity in a fully interactive web-interface that represents a comprehensive variant interpretation platform for ICCs.
Materials and Methods
The development and optimisation of CardioClassifier and is described in three sections:
Rule selection and optimisation – adapting and paramaterising ACMG/AMP criteria for ICCs
Code and implementation
Benchmarking CardioClassifier
Rule selection and optimisation
For each rule in the ACMG/AMP framework, we first evaluated whether the rule was applicable to the ICC under investigation and, where appropriate, defined more precisely the circumstances under which the rule would be activated. For seven computational criteria (PS1, PM4, PM5, PP3, BA1, BP3 and BP4), parameterisation is consistent across all gene-disease pairs. For the remaining criteria, we have incorporated expert disease, gene and variant-type specific knowledge and data to define thresholds for activation. This includes determination of robust disease-specific maximum frequency thresholds taking into account the genetic architecture of each disease 8 (BS1 and PM2; Supplementary Table 1), and using large disease cohorts to define both 'mutational hotspots'9 (PM1; Figure 2a) and variants observed more frequently in cases when compared with population controls (PS4). As part of this development process, we compared rule activation in CardioClassifier to a set of variants manually curated as part of routine clinical service at the Royal Brompton Hospital (see Supplement). Full details of how each rule is parameterised can be found in the Supplement to this manuscript.
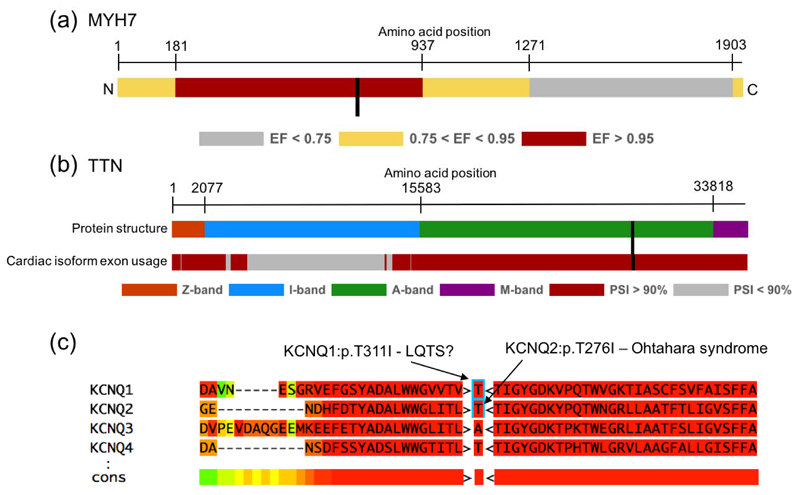
Figure 2. Examples of disease-specific optimisation of ACMG/AMP rules.
(a) Missense variants within a sub-portion of MYH7, when identified in a HCM patient, have a 97% prior probability of being Pathogenic (etiological fraction; EF=0.97). We activate PM1 for missense variants in this region. Here we use MYH7:c.2221G>T as an example (labelled with a black bar). (b) Truncating variants in TTN are only known to cause DCM when found in exons constitutively expressed in the heart (proportion spliced in (PSI) > 0.9). We activate PVS1_strong for these variants. Here we use TTN:c.86641delC as an example (labelled with a black bar). (c) Variants that have been identified as Pathogenic in paralogous genes may identify residues that are intolerant to variation. We have created two modified rules, PS1_moderate and PM5_supporting to incorporate this evidence. Here we use KCNQ1:p.T311I as an example. KCNQ2:p.T276I is associated with Ohtahara syndrome. We activate PS1_moderate for KCNQ1:p.T311I which is the equivalent missense change (i.e. same reference and alternate amino acids) in a different member of the same protein family.
As most large reference populations, such as ExAC, are not comprehensively screened for health, disease-associated alleles may be observed at low frequency. This holds true for ICCs, which can be difficult to detect even with targeted investigation, as they often manifest later in life and exhibit incomplete penetrance. We have therefore modified PM2 so as not to inappropriately discard variants seen at very low frequencies in these reference datasets.
In addition, we have created extensions to three ACMG/AMP rules, to enhance interpretation of ICC variants. Firstly, we have modified PVS1 for the titin (TTN) gene, which has a role in up to 20% of dilated cardiomyopathy (DCM) cases15. We have previously shown that only TTN truncating variants (TTNtv) in exons constitutively expressed in the heart are robustly associated with DCM15. Additionally, it is unclear that the mechanism of action for these variants is truly loss of function (LoF). Instead of scoring all TTNtv equally and assuming an underlying LoF mechanism, we only score TTNtv highly if they are in constitutive exons (proportion spliced in (PSI) > 0.9), and we reduce the strength of evidence by one level from very strong to strong (coded as PVS1_strong).
We also extended PS1 and PM5 to utilise known disease-causing variants in related genes/proteins (paralogues) to identify residues intolerant to variation16 (Figure 2c). Where nothing is known about variants at the equivalent residue of the same gene, we use high confidence variants (i.e. same reference allele and M-coffee mapping score >3) as evidence if they affect the equivalent residue in a paralogue (with the same reference allele), either with the same substitution (rule PS1_moderate - Equivalent amino acid change as an established pathogenic variant in a paralogous gene), or a different substitution (rule PM5_supporting - Missense change at an amino acid residue where a pathogenic missense change has been seen in the equivalent residue of a paralogous gene). This analysis is currently restricted to the families of predominantly ion channel proteins associated with inherited arrhythmia syndromes for which this method has been previously validated16,17.
We have previously shown paralogue annotation to be informative for over one third of novel SNVs17, and independent validation has shown a high specificity and PPV compared with other sources of evidence18,19. To determine the effect of these criteria on variant classification (before inclusion of any case-level or functional data that cannot be computationally predicted) we used 48 clinically curated (i.e. not literature only or research) missense variants from ClinVar identified as ‘Pathogenic’ or ‘Likely Pathogenic’ for LQTS from one or more submitter with at least one review status star, and compared CardioClassifier interpretations with and without paralogue data. Paralogue data were available for 11/48 (22.9%) variants and resulted in a potential change of class from variant of uncertain significance (VUS) to Likely Pathogenic for 63.6% (7/11) of these (Supplementary Table 2).
Code and implementation
CardioClassifier is implemented server-side in perl and PHP. Uploaded variant data is annotated by the Ensembl variant effect predictor (VEP)10 and converted to a table using the tableize_vcf.py script within LOFTEE (https://github.com/konradjk/loftee). Protein altering and splice site variants (coding ±8bps) are analysed for a set of 40 genes associated with inherited cardiac conditions (Table 1). We look to continuously expand this list, focusing on curated genes robustly implicated in disease, emerging from community efforts such as ClinGen4.
Table 1. Details of gene-disease pairs currently analysed by CardioClassifier.
The disease class column details the larger sub-panels relating to broad disorder types that each disease and gene set are within.
| Disease | Disease Class | Genes | Total Genes |
|---|---|---|---|
| DCM | Cardiomyopathy | LMNA, TNNT2, SCN5A, TTN, TCAP, MYH7, VCL, TPM1, TNNC1, RBM20, DSP, BAG3 | 12 |
| HCM | Cardiomyopathy | MYH7, TNNT2, TPM1, MYBPC3, PRKAG2, TNNI3, MYL3, MYL2, ACTC1, CSRP3, PLN, TNNC1, GLA, FHL1, LAMP2, GAA | 16 |
| ARVD/C | Cardiomyopathy | DSP, PKP2, DSG2, DSC2, JUP | 5 |
| RCM | Cardiomyopathy | TNNI3 | 1 |
| ncCM | Cardiomyopathy | MYBPC3, MYH7 | 2 |
| Noonan syndrome | Cardiomyopathy | RAF1, SOS1, PTPN11, KRAS | 4 |
| Long QT syndrome | Arrhythmia | KCNQ1, KCNH2, SCN5A, KCNE1, KCNE2 | 5 |
| Brugada syndrome | Arrhythmia | SCN5A | 1 |
| CPVT | Arrhythmia | RYR2 | 1 |
| Marfan syndrome | Aortopathy | FBN1 | 1 |
| FH | - | LDLR | 1 |
The classifier automatically assesses each variant for 17 rules across three distinct data categories, as defined by the ACMG/AMP guidelines2. It also consults an internal knowledge base of additional evidence, grouped by ACMG rule, either derived from community curation efforts or manually curated internally. The output is displayed on a PHP webpage that allows the user to interact and add (or remove) additional levels of evidence.
Benchmarking
Datasets
In order to test CardioClassifier extensively we used data from the following sources:
ClinVar – all variants identified as ‘Pathogenic’ or ‘Likely Pathogenic’ by multiple submitters with no conflicting data (i.e. no reports of ‘Benign’, ‘Likely Benign’ or ‘Uncertain Significance’) for hypertrophic cardiomyopathy (HCM; n=158), dilated cardiomyopathy (DCM; n=16), long QT syndrome (LQTS; n=18), catecholaminergic polymorphic ventricular tachycardia (CPVT; n=1), Brugada syndrome (Brs; n=4) or arrhythmogenic right ventricular cardiomyopathy (ARVC; n=22) were extracted from the 20161201 release of ClinVar11 using publically available scripts12.
57 protein-altering variants in MYH7 that have been expertly curated by the ClinGen Inherited Cardiomyopathy expert panel (https://www.ncbi.nlm.nih.gov/clinvar/submitters/506161/).
A prospective dataset of 327 HCM cases and 625 healthy volunteers recruited to the NIHR Royal Brompton cardiovascular BRU, all phenotypically characterised using cardiac MRI. Samples were sequenced using the IlluminaTruSight Cardio Sequencing Kit1 on the Illumina NextSeq platform. This study had ethical approval (REC: 09/H0504/104+5) and informed consent was obtained for all subjects.
Comparison with existing resources
We compared the performance of CardioClassifier against the generic tool InterVar7, to assess the importance of our disease-specific annotations. We used the ClinVar dataset of 219 variants described above as a test dataset.
InterVar scripts were downloaded from GitHub (https://github.com/WGLab/InterVar) and individually run for each disease using an engineered VCF file. To ensure a fair comparison, we edited the ‘disorder_cutoff’ to be equivalent to the thresholds used to activate BS1 in CardioClassifier. All other settings were left as default and no additional evidence was uploaded. We compared both the final classifications and the individual rules that were activated by each tool.
Code and tool availability
CardioClassifier is available at www.cardioclassifier.org, with a free license for non-commercial use. The code and data used to produce this manuscript are available at: https://github.com/ImperialCardioGenetics/CardioClassifierManuscript.
Results
Semi-automation leads to high quality and reproducible variant interpretation
CardioClassifier provides a simple-to-use web interface that takes as input either individual variant details or a single sample VCF (Supplementary Figure 1). Users select one of 11 cardiac disorders, and this determines which pre-specified validated disease genes are analysed. Where a diagnosis is uncertain (e.g. sudden cardiac death or complex cardiomyopathy), a wider analysis can be performed for genes associated with a broader phenotype (e.g. all cardiomyopathies, or all arrhythmia syndromes; Table 1), or for all 40 ICC genes parameterised. Details of the key features of CardioClassifier can be found in Table 2.
Table 2. Key features of CardioClassifier.
Included are details of each key feature and which of three currently available tools (Alamut, InterVar and the ClinGen pathogenicity calculator) also includes each feature.
| Feature | Description | CardioClassifier | Alamut | InterVar | ClinGen Pathogenicity Calculator |
|---|---|---|---|---|---|
| Collates data from multiple sources | CardioClassifier retrieves data from multiple databases/resources including ExAC, ClinVar, ACGV and dbNSFP as well as internally derived data | ✓ | ✓ | ✓ | - |
| Takes a standard VCF or variant details as input and annotates with effect on sequence and protein | The Ensembl Variant Effect Predictor is used to annotate all variants according to protein consequence | ✓ | ✓ | ✓ | - |
| ACMG/AMP rules parameterised through expert curation according to specific gene and disease | We have developed expertly-curated gene and disease specific thresholds for 14 computational ACMG/AMP criteria in addition to 3 specifically created ICC specific rules. | ✓ | - | - | - |
| Computational data used to activate ACMG/AMP rules | Each variant is automatically assessed against 17 computational criteria | ✓ | - | ✓ | - |
| Interactive refinement of rules and addition of case-level data | Users can interactively add or remove evidence pertaining to any of the ACMG/AMP rules | ✓ | - | ✓ | ✓ |
| Integration of automated annotations and case-level interactive additions to calculate a classification according to the ACMG logic | The logic from the ACMG/AMP guidelines is used to provide a final classification | ✓ | - | ✓ | ✓ |
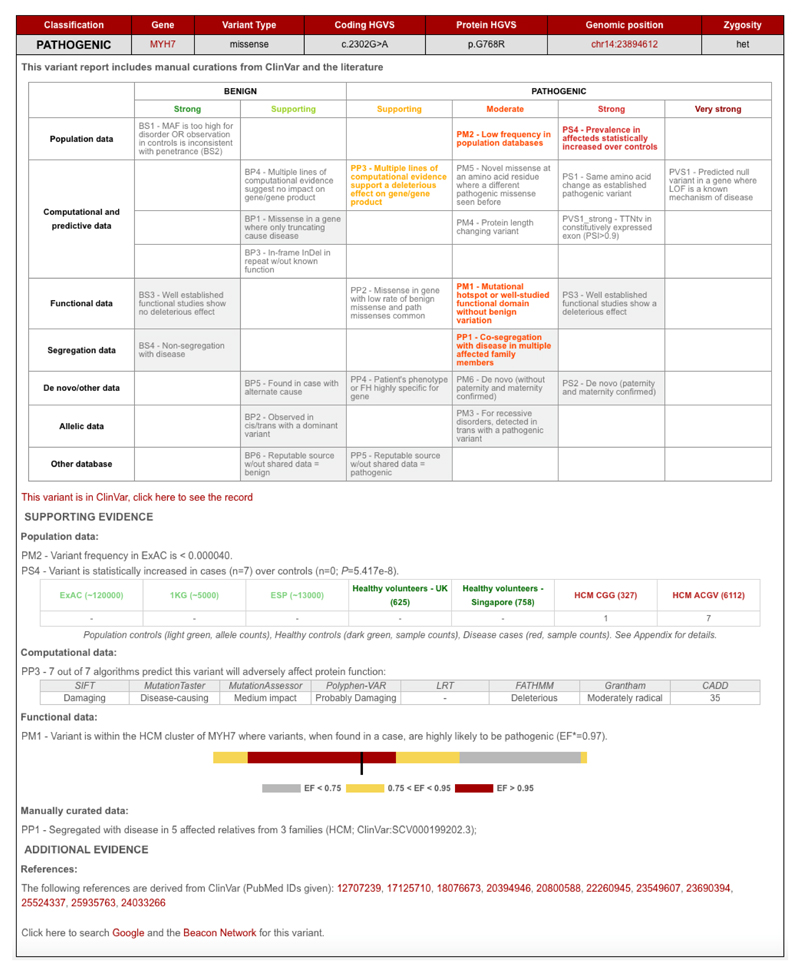
Each variant is annotated for up to 17 computational criteria, with results output to a grid representing the ACMG/AMP framework (Figure 1). The variant report is interactive, allowing a user to add additional case-level evidence to generate and refine a final classification (Supplementary Figure 2). The report is transparent, with all supporting evidence displayed along with links out to eight external resources that are commonly used for interpretation of ICC variants: the ExAC browser14, Ensembl, the UCSC genome browser, ClinVar, PubMed, Google, the Beacon Network (https://beacon-network.org) and the Atlas of Cardiac Genetic Variation (ACGV)9.
Figure 1. Example variant report output by CardioClassifier.
A grid is output for each individual variant. Rules highlighted in colour are activated for the variant and rules in grey on a white background are assessed but not activated. A user can click on a rule to manually add or remove a piece of evidence. All evidence used to assess the variant is displayed under the grid along with links out to external resources. An overall classification for the variant using the ACMG/AMP logic is displayed in the top left corner. *EF - etiological fraction; the prior probability that a variant, identified in a case, is Pathogenic9.
Highly curated datasets of disease cases and healthy controls aid annotation and filtering
As well as publically available data for both cases and population controls, CardioClassifier incorporates data from three highly-curated in-house datasets sequenced with the Illumina TruSight Cardio sequencing panel1. Counts from 877 DCM, 327 HCM cases, and 1383 healthy volunteers, all rigorously phenotyped using cardiac MRI, are used to annotate variants in genes associated with these disorders.
Some genomic regions, especially those that are repetitive or with high GC content, are not fully covered by standard exome sequencing used by major reference datasets. Specifically, 12.5% of sample bases across our 40 ICC genes are covered at <20x (Supplementary Figure 3) in the ExAC dataset. In contrast, our control set has 99.9% of sample bases covered at >20x, allowing accurate identification of common and low-frequency variants and platform specific errors, across all regions of interest (rule BS1). As this dataset is derived from the Illumina TruSight Cardio sequencing panel, users uploading variants derived from different sequencing panels should consider comparison with a local dataset to identify platform specific errors.
In addition to these in-house data, we display counts from published clinical cohorts for HCM9,20, DCM9,21, LQTS22 and Brugada syndrome23. These data are also used to assess individual variants for enrichment in cases over controls (rule PS4).
Results show high concordance with manually curated and gold-standard data
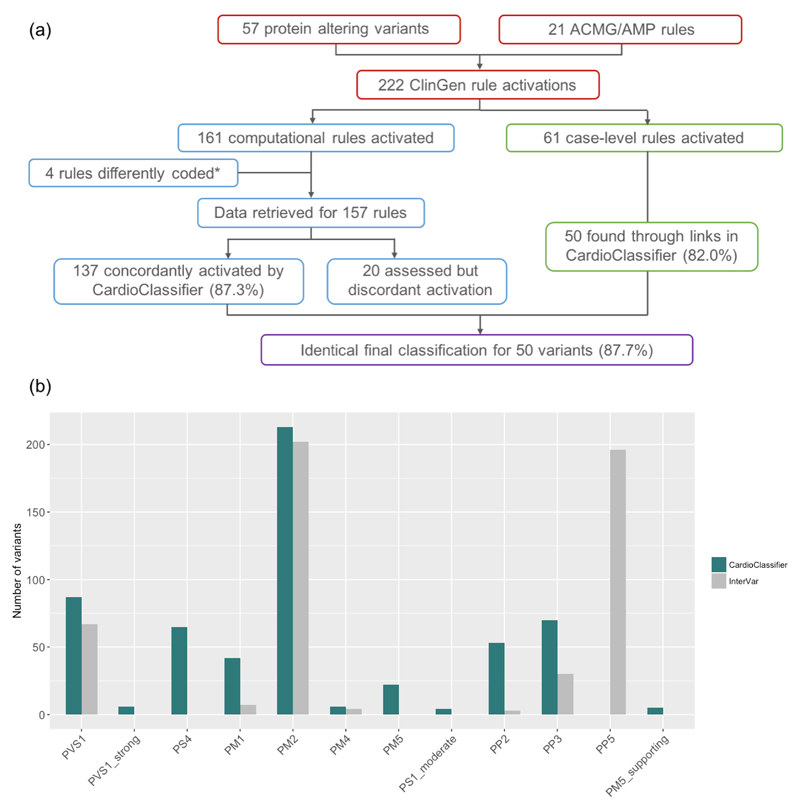
We compared CardioClassifier to 57 gold-standard, manually curated protein-altering variants in MYH7 that have been expertly curated by the ClinGen Inherited Cardiomyopathy expert panel24. Of 222 rules activated by ClinGen for these 57 variants, 157 represented computationally accessible data (from 9 ACMG/AMP rules) that were fully retrieved by CardioClassifier. CardioClassifier concordantly activated 137/157 rules (87.3%; Figure 3; Supplementary Table 3). The discrepancies fall across 3 rules; PP3 (in silico prediction algorithms; n=12), PS4 (prevalence in affected individuals statistically increased over controls; n=7) and PM5 (same amino acid residue as known Pathogenic variant; n=1). CardioClassifier imposes a more stringent threshold on PP3 (allowing only one of eight in silico prediction algorithms to be discordant), and differences in PS4 and PM5 are due to the increased availability of proband data to the ClinGen team (not available from public repositories). In all cases, CardioClassifier successfully returned all available data.
Figure 3. Validation of CardioClassifier.
(a) Comparing CardioClassifier to a set of 57 MYH7 expert panel curated variants. Rules were split into those that can be computationally annotated and those that are 'case-level' and require manual input. CardioClassiifer was run using an 'All Cardiomyopathy' test to reflect the spectrum of phenotypes caused by variants in MYH7. *Of the computational rules, 3 were removed from the comparison as they represent draft modifications to the ACMG framework by the ClinGen Cardiovascular domain working group that were not published at the time of this work, and not yet implemented in CardioClassifier. Specifically, truncating variants in MYH7 activate a new rule PVS1_moderate. Additionally, for variants classified as Benign by frequency alone (BA1) CardioClassifier does not assess any further rules, leading us to remove an additional data point from the comparison as we would not expect it to be retrieved. (b) Counts of individual rules activated by CardioClassifier and InterVar for 219 variants identified as Pathogenic or Likely Pathogenic in ClinVar. Only pathogenic evidence rules and rules activated by one of the tools at least once are shown.
We then tested the ability of the links within the CardioClassifier report to inform activation of the 61 case-level data points activated by the ClinGen team. These links allowed us to manually collate 50/61 (82.0%) individual data points (Supplementary Table 3) with differences again in the availability of proband data (6 PS4_supporting, 1 PS4_moderate, 1 PS2, 1 BS4 and 2 PP1_moderate). After addition of this clinical data, we reached an identical classification to the ClinGen team for 50/57 (87.7%) variants (Figure 3a).
CardioClassifier has higher sensitivity and specificity than non-specific interpretation tools
In February 2017 InterVar, and its companion web-server winterVar, became the first tools to automatically populate criteria from the ACMG/AMP guidelines7. Whilst these tools were crucial steps forward in application of the framework, they aim to support interpretation across the full spectrum of human genes and disorders.
To determine the added value of the disease- and gene-specific annotations included in CardioClassifier, we compared CardioClassifier to InterVar using a set of 219 variants identified as ‘Pathogenic’ or ‘Likely Pathogenic’ on ClinVar, with high confidence, across six ICCs. Based on automatically-retrieved data only, InterVar identified 64/219 (29.2%) variants as Likely Pathogenic or Pathogenic, while CardioClassifier identified over double this number as clinically actionable (156/219) with a sensitivity of 71.2% (Supplementary Table 4). For both tools, sensitivity would be increased further through user addition of clinical and functional data.
Despite the lower sensitivity of InterVar, there are occasions where the tool activates rules inappropriately in the absence of gene-specific knowledge. Firstly, InterVar activates PVS1 in the TTN gene, regardless of protein location, when it is recognised that truncating variants in exons not constitutively expressed in the heart are not associated with DCM, and are commonly found in demonstrably healthy controls15. Consequently, InterVar will categorise rare variants in these regions as ‘Likely Pathogenic’ when they are highly unlikely to be disease causing.
Secondly, InterVar activates rule PP5 (reputable source identifies the variant as Pathogenic) for 89.5% of the variants as they are reported as ‘Pathogenic’ in ClinVar. The ACMG guidelines state that this rule should only be activated when the evidence supporting the classification is unavailable, yet this evidence is often contained within the appropriate ClinVar submission. Full details of the rules activated by both tools are shown in Figure 3b.
To ensure the higher sensitivity of CardioClassifier was not due to over-activating rules, we also tested a set of 67 ‘Benign’ and ‘Likely Benign’ variants from ClinVar across the same six ICCs. CardioClassifier identified 61/67 (91.0%) of these as Benign and the remaining 6 as VUS. Conversely, InterVar identified 41/67 (61.2%) as Benign with 22 as Likely Benign and 4 as VUS. Here InterVar activates BS2 when a variant is seen in the 1000 genomes dataset, which we believe is inappropriate for ICCs which do not fit the important caveat of ‘full penetrance expected at an early age’. We do acknowledge, however, that InterVar was developed for severe congenital and very-early onset developmental disorders with nearly 100% penetrance.
Diagnostic yield in HCM cases matches previous reports
To investigate the clinical utility of CardioClassifier we used a dataset of 327 HCM cases. In 66 cases (20.2%) we identified a Pathogenic (n=11) or Likely Pathogenic (n=55) variant, with a further 76 cases (23.2%) harbouring a VUS. To determine the proportion of these VUSs likely to become clinically actionable after the addition of case-level data, we calculated the excess of VUSs in cases over the background level of rare and presumably benign VUSs in 625 healthy volunteers (HVOLs). Based on a background level of 9.7%, we calculate a case excess of VUSs of 13.5%. Combining this with the 20.2% of cases with a Pathogenic or Likely Pathogenic variant, overall, 33.7% of cases have a potentially clinically relevant variant (Supplementary Figure 4a), comparable to previous reports20.
Manual curation of known variants
In addition to automatic retrieval of computational data, CardioClassifier will store curated case-level data entered by users, or pre-populated by active curation. We have primed this ‘knowledge base’ with data from 120 fully curated cardiomyopathy variants, comprising the 57 expert panel curated MYH7 variants and the most commonly observed variants for the major cardiomyopathies; HCM, DCM and ARVC, defined as those occurring six or more times in the ACGV resource (reflecting a HCM case frequency of ~1/1000)9. There were 84 such recurrent variants in ACGV, together representing 39.5% (1,258/3,186) of all identified variants. We curated 63 that had not already been assessed by the expert panel.
After manual curation of the literature and ClinVar for reports of segregation, de novo occurrence and functional characterisation, 34 variants were classified as Pathogenic, 13 as Likely Pathogenic and 7 as VUS (Supplementary Table 5; Supplementary Figure 4b). The annotations for these 120 variants, accounting for at least 40% of variants identified in Caucasian cardiomyopathy cases, are stored in CardioClassifier, ensuring these variants are correctly classified without further user-input.
Discussion
We describe CardioClassifier, an automated and interactive web-tool to aid clinical variant interpretation across a wide range of ICCs. To the best of our knowledge this represents a unique disease-specific solution that automates data retrieval, incorporates gene- and disease-specific knowledge to refine rule application, is pre-loaded with curated data on prior observations (in health or disease), and integrates evidence according to the widely-adopted framework from ACMG/AMP. The tool is transparent, with all the information incorporated into interpreting each variant displayed along with the final classification. It is also flexible, and designed to be fully interactive, with the user able to add and remove evidence specific to the patient/family of interest.
The strength of CardioClassifier is its disease-specificity. The ACMG/AMP rules are intentionally non-specific to allow adoption in any disease domain. To harness the full power of this framework, the rules need to be applied in a disease- and gene-specific manner25. We have defined criteria and thresholds for each ACMG/AMP rule that are specific to the disorder of interest, and demonstrate the power and effectiveness of this approach over a recently released genome-wide interpretation interface. Incorporation of disease-specific knowledge is limited by current data, and the power of this tool will increase over time as new data become available.
On-going community initiatives, such as the Clinical Genome Resource (ClinGen), are defining consensus disease and gene-specific standards for modifications to the ACMG/AMP guidelines, and it is our intention to continue to develop CardioClassifier to utilise these standards as they become available.
We believe the main limitation to the effectiveness of any computational solution is the retrieval of clinical and patient-specific data that is seldom available as fully structured data for programmatic retrieval. CardioClassifier combines pre-populated computational data with interactive addition of case and variant specific evidence in a structured format to overcome this hurdle. Our growing variant knowledge base will add to available structured representation of this crucial case-level data. Future development of CardioClassifier will streamline data-sharing, expanding our knowledge base and sharing it with the community via submission to the ClinVar database. This increasing knowledge base relies on researchers and clinicians in the field supporting data-sharing initiatives, and facilitating direct ClinVar submission from CardioClassifier for the benefit of the ICC community is a development priority.
A further limitation to CardioClassifier in its current form is the restricted prediction of impact on splicing. This arises for two main reasons; firstly, CardioClassifier uses the Ensembl variant effect predictor to annotate variants, which annotates bases within 8 base-pairs of the exon/intron boundary as splice-site, but will miss more distal bone-fide splice site variants. Secondly, we currently have not incorporated any in silico splice-site prediction algorithms, due to limitations around availability, licensing and accuracy. These issues will be addressed in a future release.
CardioClassifier is designed to work seamlessly with data from any sequencing platform in standard VCF format, whether targeted sequencing (e.g. Illumina TruSight Cardio1), or targeted analysis of exome/genome-wide data. This is a crucial step in broadening the availability of genetic testing for ICCs, and standardising variant interpretation in this field. Furthermore, we hope that in demonstrating the clinical utility of our disease-specific approach, we will encourage others to develop similar tools across other disease specialties.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust (107469/Z/15/Z); Medical Research Council (UK), NIHR Cardiovascular Biomedical Research Unit at Royal Brompton and Harefield NHS Foundation Trust and Imperial College London; the Royal Brompton & Harefield Cardiovascular Research Centre Biobank; NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London; Fondation Leducq (11 CVD-01), the British Heart Foundation (SP/10/10/28431, FS/15/81/31817); and a Health Innovation Challenge Fund award from the Wellcome Trust and Department of Health, UK (HICF-R6-373).
This publication includes independent research commissioned by the Health Innovation Challenge Fund (HICF), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this work are those of the authors and not necessarily those of the Department of Health or Wellcome Trust.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Pua CJ, Bhalshankar J, Miao K, et al. Development of a comprehensive sequencing assay for inherited cardiac condition genes. Journal of Cardiovascular Translational Research. 2016;9(1):3–11. doi: 10.1007/s12265-016-9673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genetics in Medicine. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SM, Dolinsky JS, Johnson AEK, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genetics in Medicine. 2017 Mar; doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehm HL, Berg JS, Brooks LD, et al. ClinGen the clinical genome resource. New England Journal of Medicine. 2015;372(23):2235–2242. doi: 10.1056/nejmsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel RY, Shah Neethu, Jackson AR, et al. ClinGen pathogenicity calculator: A configurable system for assessing pathogenicity of genetic variants. Genome Medicine. 2017;9(1) doi: 10.1186/s13073-016-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinberger J, Maloney KA, Pollin TI, Jeng LJB. An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genetics in Medicine. 2016;18(11):1165–1165. doi: 10.1038/gim.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. The American Journal of Human Genetics. 2017;100(2):267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiffin N, Minikel E, Walsh R, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. GENETICS in MEDICINE. 2017 May; doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh R, Thomson KL, Ware JS, et al. Reassessment of mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in Medicine. 2016;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biology. 2016;17(1) doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research. 2013;42(D1):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Minikel EV, O-Luria AH, MacArthur DG, Ware JS, Weisburd B. ClinVar data parsing. Wellcome Open Research. 2017;2:33. doi: 10.12688/wellcomeopenres.11640.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karczewski KJ, Weisburd B, Thomas B, et al. The ExAC browser: Displaying reference data information from over 60 000 exomes. Nucleic Acids Research. 2016;45(D1):D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts AM, Ware JS, Herman DS, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Science Translational Medicine. 2015;7(270):270ra6–270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ware JS, Walsh R, Cunningham F, Birney E, Cook SA. Paralogous annotation of disease-causing variants in long QT syndrome genes. Human Mutation. 2012;33(8):1188–1191. doi: 10.1002/humu.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh R, Peters NS, Cook SA, Ware JS. Paralogue annotation identifies novel pathogenic variants in patients with brugada syndrome and catecholaminergic polymorphic ventricular tachycardia. Journal of Medical Genetics. 2013;51(1):35–44. doi: 10.1136/jmedgenet-2013-101917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapplinger JD, Tseng AS, Salisbury BA, et al. Enhancing the predictive power of mutations in the c-terminus of the KCNQ1-encoded kv7.1 voltage-gated potassium channel. Journal of Cardiovascular Translational Research. 2015;8(3):187–197. doi: 10.1007/s12265-015-9622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapplinger JD, Giudicessi JR, Ye D, et al. Enhanced classification of brugada syndromeAssociated and long-QT syndromeAssociated genetic variants in theSCN5A-encoded nav1.5 cardiac sodium ChannelCLINICAL PERSPECTIVE. Circulation: Cardiovascular Genetics. 2015;8(4):582–595. doi: 10.1161/circgenetics.114.000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genetics in Medicine. 2015;17(11):880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 21.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genetics in Medicine. 2014;16(8):601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 22.Kapplinger JD, Tester DJ, Salisbury BA, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6(9):1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for brugada syndrome genetic testing. Heart Rhythm. 2010;7(1):33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly MA, Caleshu C, Morales A, et al. Adaptation and Validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen's Inherited Cardiomyopathy Expert Panel. Genetics in Medicine. doi: 10.1038/gim.2017.218. In press (GIM-D-17-00417R2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. The American Journal of Human Genetics. 2016;99(1):247. doi: 10.1016/j.ajhg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.