Abstract
Importance
Mitochondrial disorders have emerged as a common cause of inherited disease, but their diagnosis remains challenging. Patients with multiple respiratory chain complex defects are particularly difficult to diagnose at the molecular level because of the massive number of nuclear genes potentially involved in intra-mitochondrial protein synthesis, with many not yet linked to human disease.
Objective
To determine the molecular basis of multiple respiratory chain complex deficiencies.
Design
We studied 53 patients referred to 2 national centers in the United Kingdom and Germany between 2005-2012. All had biochemical evidence of multiple respiratory chain complex defects, and no primary pathogenic mitochondrial DNA mutation. Whole exome sequencing was performed using 62Mb exome enrichment, followed by variant prioritization using bioinformatic prediction tools, variant validation by Sanger sequencing, and segregation of the variant with the disease phenotype in the family.
Participants
53 patients with biochemical evidence of multiple respiratory chain complex defects but no primary pathogenic mitochondrial DNA mutation.
Results
Presumptive causal variants were identified in 28 patients (53%, 95% CI=39-67), and possible causal variants were identified in 4 (8%, 95% CI=2-18). Together these account for 32 patients (60%, 95% CI=46-74), and involved 18 different genes. These included recurrent mutations in RMND1, AARS2 and MTO1, each on a haplotype background consistent with a shared founder allele, and potential novel mutations in 4 possible mitochondrial disease genes (VARS2, GARS, FLAD1 and PTCD1). Distinguishing clinical features included deafness and renal involvement associated with RMND1, and cardiomyopathy with AARS2, and MTO1. However, “classical” features were absent in some patients, including normal liver function and Leigh syndrome (subacute necrotizing encephalomyelopathy) seen in association with TRMU mutations, and no cardiomyopathy with founder SCO2 mutations. It was not possible to confidently identify the underlying genetic basis in 21 patients (40%, 95% CI=26-54).
Conclusions and Relevance
Exome sequencing enhances the ability to identify potential nuclear gene mutations in patients with biochemically-defined defects affecting multiple mitochondrial respiratory chain complexes. Additional study will be required in independent patient populations to determine the utility of this approach in comparison to traditional diagnostic methods.
Introduction
Defects of the mitochondrial respiratory chain have emerged as the most common cause of childhood and adult neurometabolic disease, with an estimated prevalence of 1 in 5000 live births.1 Clinically they can present at any time of life, are often seen in association with neurological impairment, and cause chronic disability and premature death.2 Major advances in understanding the molecular basis of mitochondrial disease have been mirrored by a complex, expanding phenotypic spectrum. Although some genetic defects appear to be seen in association with specific clinical features, this is not usually the case, and a systematic multidisciplinary approach is required to make a diagnosis.3 Biochemical and molecular genetic investigations are time consuming, expensive, and highly specialized, often involving a biopsy of an affected tissue or organ. With a growing list of mitochondrial diseases caused by different nuclear gene defects,4 achieving a comprehensive molecular diagnosis is now more labor intensive than ever. This can compromise clinical management through protracted and often repeated investigations, impeding reliable genetic counseling and prenatal diagnosis.
Approximately one third of patients with mitochondrial disease have a biochemical defect involving multiple respiratory chain complexes suggesting a defect of intra-mitochondrial protein synthesis5. With only a minority having a primary defect involving mitochondrial DNA (mtDNA), the remainder presents a particular challenge. The molecular mechanism potentially involves many different gene products affecting mtDNA replication and expression, including ribosomal structural and assembly proteins, aminoacyl-tRNA synthetases, tRNA modifying and methylating enzymes, and several initiation, elongation and termination factors of mitochondrial translation.6 Recent studies have shown that apparently unique genetic defects are common in this group, often involving proteins not previously thought to influence mitochondrial function, nor with clear mitochondrial localization. The objective of this study was to determine whether whole exome sequencing approach could be used to define the molecular basis in these patients.
Methods
Patients
Patients suspected of having mitochondrial disease referred to two nationally-accredited diagnostic laboratories (Newcastle upon Tyne Highly Specialised Service Mitochondrial Diagnostic Laboratory, UK and the Medical Genetics Centre, Munich, Germany) between 2005 and 2012 and meeting the inclusion criteria were included in this study. The inclusion criteria were: (1) histochemical and/or biochemical diagnosis of mitochondrial disease in a clinically-affected tissue (skeletal muscle, liver or heart), confirming decreased activities of multiple respiratory chain complexes based on published criteria (Table 1);7 (2) The absence of large-scale mtDNA rearrangements, mtDNA depletion and mtDNA point mutations,8 with the exception of patients P20, P21, P25, P43 and P45, where decreased levels of mtDNA were confirmed in muscle (mtDNA depletion); (3) The exclusion of major nuclear gene rearrangements by comparative genomic hybridisation arrays in patients with congenital structural abnormalities. Standardized clinical assessments were performed by the study authors. Clinical phenotypes were defined using local reference ranges for cardiomyopathy on echocardiography, abnormal renal and liver function tests, severe lactic acidosis as a blood level >5mM/L, and clinical neurophysiology for peripheral neuropathy. Informed consent was obtained from all participants in accordance with protocols approved by local institutions and research ethics committees.
Table 1. Clinical and molecular genetic characteristics of 53 patients with multiple respiratory chain complex defects.
Patients were categorized into four groups based on the molecular genetic results defined a priori (see Methods). The complete data set is show in eTable 1, including the results of biochemical analyses. Abbreviations: M: male, F: female; *: included in reference,5 C: consanguinity, N: no family history, 1S or 2S: number of other affected sibs, Clinical Presentation, Mu: muscle, C: central nervous system, H: heart, L: liver, Hom: homozygous. + = present. - = absent. Current age = if alive, age at last follow up.
| A) Presumptive | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient Information | Clinical Presentation | Genetic Analysis | |||||||
| ID/Sex | Country of Origin | Family History | Onset/Death(†) or current age | Mu | C | H | L | Gene | Variant |
| 1/M | British Pakistani | C | 6m/4y | + | - | - | - | RMND1 | Hom. c.1349G>C: p.*450Serext*32 |
| 2/M* | British Pakistani | C | 3m/1y† | + | + | - | - | RMND1 | Hom. c.1349G>C: p.*450Serext*32 |
| 3/F | British Pakistani | C | 18m/5y | + | + | + | - | RMND1 | Hom. c.1349G>C: p.*450Serext*32 |
| 4/F* | Turkish | C | 6m/10y† | + | - | + | - | RMND1 | Hom. c.1349G>C: p.*450Serext*32 |
| 5/F | British Pakistani | C | <1m/18m | + | - | - | - | RMND1 | Hom. c.1349G>C: p.*450Serext*32 |
| 6/M | British | N | 18m/5y† | + | - | - | - | RMND1 | c.713A>G:p.Asn238Ser c.829_830+2delGAGT:p.Glu277Glyfs*2 |
| 7/M* | British | N | birth/6w† | + | + | + | - | AARS2 | c.1774C>T: p.Arg592Trp c.2882C>T: p.Ala961Val |
| 8/M | German | N | birth/1m† | + | + | + | - | AARS2 | c.1616A>G: p.Tyr539Cys c.1774C>T: p.Arg592Trp |
| 9/F* | German | C | 3w/2m† | + | - | + | - | AARS2 | Hom. c.1774C>T: p.Arg592Trp |
| 10/F | British | N | birth/3m† | - | - | + | - | AARS2 | c.647_648insG: p.Cys218Leufs*6 c.1774C>T: p.Arg592Trp |
| 11/F | British | N | 6m/11m† | - | - | + | - | AARS2 | Hom. c.1774C>T: p.Arg592Trp |
| 12/F* | Croatian | 1S | birth/1m | + | + | + | - | MTO1 | c.631_631delG: p.Gly211Aspfs*3 c.1282G>A: p.Ala428Thr |
| 13/M* | British Pakistani | C | birth/1y† | + | - | + | - | MTO1 | Hom. c.1232C>T: p.Thr411Ile |
| 14/M* | British Pakistani | C | 1y/3y† | + | + | + | + | MTO1 | Hom. c.1232C>T: p.Thr411Ile |
| 15/M | British | N | <1y/2y | + | - | - | - | MTO1 | c.122T>G: p.Val41Gly c.767A>G: p.His256Arg |
| 16/M* | Turkish | N | birth/3m† | + | - | - | + | EARS2 | Hom. c.193A>G: p.Lys65Glu |
| 17/M* | British | N | 2m/6m† | + | + | - | + | EARS2 | c.322C>T: p.Arg108Trp c.814G>A: p.Ala272Thr |
| 18/F* | German | 1S | 3y/16y | + | - | - | - | MTFMT | c.452C>T: p.Pro151Leu c.994C>T: p.Arg332* |
| 19/F | British | N | birth/20y | + | + | + | - | MTFMT | c.626C>T: p.Ser209Leu c.1100_1101delTT: p.Phe367Serfs*22 |
| 20/M | British | N | 2y/2.5y† | - | - | + | - | MGME1 | c.532C>T: p.Arg178Trp c.794C>T: p.Thr265Ile |
| 21/F | Irish | 1S | 2.5y/13 | + | + | - | - | C12orf65 | Hom. c.96_99dupATCC: p.Pro34Ilefs*25 |
| 22/M* | Lebanese | C | 14y/37y† | + | - | - | - | YARS2 | Hom. c.137G>A: p.Gly46Asp |
| 23/F | Turkish | C | 4y/17y | + | + | - | - | PUS1 | Hom. c. 426C>A: p.Cys142* |
| 24/M | British Pakistani | C | birth/1m† | + | + | + | - | TRMU | Hom. c.287A>G: p.Asn96Ser |
| 25/F | British | C | birth/<1m† | + | + | - | - | TK2 | Hom. c.1A>G: p.Met1Val |
| 26/F | Polish | N | 7m/18m† | + | + | - | - | SCO2 | Hom. c.418G>A: p.Glu140Lys |
| 27/F* | German | N | birth/3w† | + | - | + | - | ELAC2 | c.1478C>T: p.Pro493Leu c.1621G>A: p.Ala541Thr |
| 28/M* | Turkish | 1S | 4y/7y | + | + | - | - | ETHE1 | Hom. c.3G>T: p.Met1Ile |
| B) Possible | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient Information | Clinical Presentation | Genetic Analysis | |||||||
| ID/Sex | Country of Origin | Family History | Onset/Death(†) or current age | Mu | C | H | L | Gene | Variant |
| 29/M | British | N | <1/10y | + | + | - | - | VARS2 | c.1135G>A: p.Ala379Thr c.1877C>A: p.Ala626Asp |
| 30/M* | Turkish | C | 4m/8m† | + | - | - | - | FLAD1 | Hom. c.397_400 delTTCT: p.Phe134Cysfs*8 |
| 31/F* | Turkish | C 2S |
birth/1m† | + | - | + | - | GARS | Hom. c.2065C>T: p.Arg689Cys |
| 32/F* | British | N | 4m/8m† | - | - | + | - | PTCD1 | c.337C>T: p.Arg113Trp c.388C>T: p.Arg130* c.550G>A: p.Gly184Arg |
| C) Variants of unknown significance | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient Information | Clinical Presentation | Genetic Analysis | |||||||
| ID/Sex | Country of Origin | Family History | Onset/Death(†)or Current age | Mu | C | H | L | Gene | Variant |
| 33/F* | Turkish | C | 2y/6y | + | + | - | - |
SLC25A12 METAP1D |
Hom. c.1333G>A: p.Ala445Thr Hom. c.497+2T>A |
| 34/M | British | N | 17y/20y | - | + | - | - | ACSM5 | c.1157A>C: p.Lys386Thr c.1273C>A: p.Pro425Thr |
| 35/F | Georgian | C | 7y/10y | + | + | - | - |
PERP MEF2A ACSM5 |
Hom. c.206T>C: p.Met69Thr c.1262A>C: p.Gln421Pro c.1265A>C: p.Gln422Pro c.68A>G: p.His23Arg c.73A>C:p.Lys25Gln |
| 36/M | German | C | 10y/14y | + | - | - | - |
HKDC1 ETFA IREB2 SMCR7 |
Hom. c.1276C>T: p.Arg426Cys Hom. c.20C>T: p.Pro7Leu Hom. c.2393C>T: p.Thr798Ile Hom. c.241C>T: p.Gln81* |
| 37/F | British | N | <1m/5m | + | + | - | - | PC | c.1876C>T: p.Arg626Trp c.1892G>A: p.Arg631Gln |
| 38/M | Turkish | C | 4y/6y | + | + | - | + | TPO | Hom. c.443C>T: p.Ala148Val |
| 39/M | British | N | <1y/5y | + | + | - | - | HERC2 | c.6448C>G: p.Leu2150Val c.9979G>A: p.Val3327Met |
| 40/M* | Turkish | C | 2w/3w† | + | - | - | - |
MAGI1 NDRG3 TPX2 TAF9 |
Hom. c.2290A>C: p.Thr764Pro Hom. c.469G>A: p.Gly157Ser Hom. c.505C>T: p.Pro169Ser Hom. c.406G>C: p.Glu136Gln |
| 41/M* | Croatian | N | birth/9m† | + | + | + | - |
SLC25A43 FAAH2 |
c.493C>T: p.Arg165* (X-linked) c.368T>C: p.Phe123Ser (X-linked) |
| 42/M | Hungarian | N | 6y/14y | + | - | - | - |
DLAT SDHD POLRMT ARHGEF5 |
c.55G>C: p.Glu19Gln c.626A>G: p.Gln209Arg c.34G>A: p.Gly12Ser c.386T>C: p.Leu129Ser c.112C>T:p.Pro38Ser c.232G>A:p.Val78Met c.1738G>T:p.Gly580Cys c.4066A>G:p.Asn1356Asp |
| 43/F | British | N | birth/<1m† | + | - | - | - |
TYMP ACSM2A LRPPRC HTRA2 ALDH1L1 BCKDHB SLC25A4 |
c.242G>A: p.Arg81Gln c.1003G>A: p.Val335Ile c.4132A>G: p.Ser1378Gly c.1210C>T: p.Arg404Trp c.2143G>C:p.Glu715Gln c.23C>T:p.Ala8Val c.239G>A:p.Arg80His |
| 44/M* | Turkish | 1S | 18m/2y | + | - | - | + |
PPL SLC5A10 |
c.263A>G: p.Asp88Gly c.1003C>A: p.Leu335Met c.674A>G: p.Glu225Gly c.1799T>C: p.Leu600Pro |
| 45/F | German | Healthy Dizygotic twin |
1m/6m† | + | + | - | + |
FASN FNDC1 |
c.1850C>T: p.Pro617Leu c.2657T>C: p.Phe886Ser c.4429A>G: p.Thr1477Ala c.4547C>A: p.Thr1516Asn |
| 46/M | German | N | 6y/21y | + | - | - | - |
PDPR LONP1 |
c.616A>G: p.Ile206Val c.1774A>G: p.Thr592Ala c.79G>C: p.Ala27Pro c.2485G>A: p.Ala829Thr |
| 47/M | Egyptian | C | 2m/2y† | + | + | - | + |
MIPEP STARD13 |
Hom. c.671A>G: p.Asn224Ser Hom. c.1186C>T: p.His396Tyr |
| D) Unresolved | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient Information | Clinical Presentation | Genetic Analysis | ||||||
| ID/Sex | Country of Origin | Family History | Onset/Death(†) or Current age | Mu | C | H | L | |
| 48/M | Turkish | C | 4m/1y | + | + | + | + | No candidate variants detected |
| 49/F* | German | N | 1y/3y | + | + | - | - | |
| 50/F* | British Pakistani | C | birth/4d† | - | - | + | - | |
| 51/F | British Pakistani | C | adult-onset | + | - | - | - | |
| 52/M | German | N | 2y/5y† | + | + | - | + | |
| 53/F | German | N | birth/4y | + | + | - | - | |
Molecular genetics and bioinformatics
Exome sequencing, bioinformatic analysis, variant confirmation, and segregation analysis were performed in Newcastle upon Tyne. Genomic DNA was isolated from primary cell lines, muscle or circulating lymphocytes (DNeasy®, Qiagen, Valencia, CA), fragmented and enriched by Illumina TruSeq™ 62 Mb exome capture, and sequenced (Illumina HiSeq 2000, 100 bp paired-end reads). The in-house bioinformatics pipeline involved the following steps: alignment to the human reference genome (UCSC hg19),9 removal of duplicate sequence reads (Picard v1.85 http://picard.sourceforge.net) and variant detection (Varscan v2.210, Dindel v1.0111). On-target variant filtering excluded those with minor allele frequency greater >0.01 in several databases: dbSNP135, 1000 genomes (February 2012 data release), the National Heart, Lung and Blood Institute (NHLBI, NIH, Bethesda, MD) Exome Sequencing Project (ESP) 6500 exomes, and 238 unrelated in-house controls. We used published and experimentally-validated bioinformatic tools to predict mitochondrial localization and probably impact on mitochondrial function.12,13 Rare homozygous and compound heterozygous variants were defined, and protein altering and/or putative ‘disease causing’ mutations, along with their functional annotation, were identified using ANNOVAR.14 Candidate genes were filtered against a list of bioinformatically predicted mitochondrial proteins,12,13 as well as genes which matched a Gene-Ontology (GO)-term of ‘mitoch’ and prioritized if previously seen in association with a disease phenotype (see eTable 3 for detailed methods). Putative pathogenic variants were confirmed by Sanger sequencing using custom-designed primers (http://frodo.wi.mit.edu) on an ABI 3130XL (BigDye, Applied Biosystems, Carlsbad, CA) and compared to transcripts available at the Nucleotide database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nuccore) allowing segregation analyses where possible.
Variants were classified into four groups, defined a priori: (1) Presumptive pathogenic - homozygous or compound heterozygous mutations in genes previously shown to cause multiple respiratory chain complex deficiencies; (2) Possible pathogenic - homozygous or compound heterozygous mutations in novel genes predicted to cause a mitochondrial translation defect based on their proposed function and similarity to known disease genes; (3) Variants of unknown significance - homozygous or compound heterozygous mutations in novel or known disease genes not known to be associated with mitochondrial pathology; (4) Unresolved - patients where a single plausible genetic cause could not be identified.
Binomial confidence intervals were calculated using the Clopper-Pearson method.
Results
Clinical presentation
The clinical presentation and laboratory findings of the 53 unrelated patients are summarized in Table 1 and eTable 1. The majority (51/53, 96%, 95% CI=87-99) of the patients presented in childhood (<15 years old) and most (35/53, 66%, 95% CI=52-78) developed symptoms within the first year of life. Parental consanguinity was apparent in 24 cases, and 3 cases had an additional affected sibling. The most frequent clinical feature was muscle weakness with hypotonia (47/53, 89%, 95% CI=77-96), followed by clinical or imaging features of central neurological disease (28/53, 53%, 95% CI=39-67), cardiomyopathy (19/53, 36%, 95% CI=23-50) or abnormal liver function (9/53, 17%, 95% CI=8-30), and a combination of these symptoms were present in most cases (34/53, 64%, 95% CI=50-77). Severe lactic acidosis was observed in half of the patients (27/53, 51%, 95% CI=37-65). The presence of deafness (8/53, 15%, 95% CI=7-28), ptosis or progressive external ophthalmoplegia (PEO, 4/53, 8%, 95% CI=2-18), renal impairment (5/53, 9%, 95% CI=3-21), axonal neuropathy (3/53, 6%, 95% CI=1-16), sideroblastic anaemia (2/53, 4%, 95% CI=0-13), immune deficiency (1/53, 2%, 95% CI=0-10) and optic atrophy (2/53, 4%, 95% CI=0-13) were also noted.
DNA analysis, whole exome sequencing
The mean per base depth of coverage for the exome consensus coding sequence was 79-fold, with 88.6% of bases covered >20-fold. Coverage and depth statistics for each patient are shown in eTable 2. The results of the bioinformatic analysis are shown in eTable 3, which also includes the allele frequency data. Confirmed variants are shown in Table 1. Presumptive pathogenic variants were found in 28 of the patients (53%, 95% CI=39-67), and possible pathogenic variants in 4 patients (8%, 95% CI=2-18), giving a combined result of 32 out of 53 (60%, 95% CI=46-74). Variants of uncertain significance were found in 15 patients (28%, 95% CI=17-42), and six cases remained unresolved (11%, 95% CI=11-34).
(1). Presumptive pathogenic group
A single, novel homozygous c.1349G>C (p.*450Serext*32) RMND1 (NM_017909.3)15,16 mutation introducing a premature stop codon was identified in 5 independent patients. In each patient, the phenotype was severe, affecting different organs but including myopathy, profound deafness and renal involvement. The 5 homozygotes were from consanguineous families of Pakistani origin. A founder effect was supported by the presence of a homogeneous haplotype flanking the mutation (Figure 1a). One other patient had different compound heterozygous mutations in RMND1. Mutations in AARS2 (NM_020745.3)17 were the second most frequently identified defect (5 patients). All presented with severe infantile cardiomyopathy, with additional muscle (3 patients) and central neurological features (2 patients) in a subgroup. Interestingly, despite being from different European ethnic backgrounds, all carried the previously reported c.1774C>T (p.Arg592Trp) mutation on at least one allele (Figure 1b).17 Mutations in MTO1 (NM_012123.3)18 were identified in 4 patients. All had muscle weakness on presentation with lactic acidosis, two had central neurological features, and unlike a previous report,18 one did not have a cardiomyopathy; two patients were homozygous for a p.Thr411Ile MTO1 mutation recently shown to cause a severe respiratory phenotype in a yeast model19 (Figure 1c). Homozygous or compound heterozygous mutations were detected in previously characterised mitochondrial translation genes, including 2 patients with EARS2 (NM_001083614.1)20 mutations (one having a leukoencephalopathy and no corpus callosum),21 2 patients with MTFMT (NM_139242.3) mutations,22 and one case with C12orf65 (NM_152269) mutations.23 Single patients with a clinical presentation resembling previously described cases carried homozygous or compound heterozygous mutations in the YARS2 (NM_001040436.2),24, PUS1 (NM_025215.5),25 MGME1 (NM_052865.2),26 ETHE1 (NM_014297.3),27 ELAC2 (NM_018127.6),28 and TK2 (NM_004614.3)29 the latter case seen in association with severe loss of mtDNA copy number due to mutation (c.1A>G, p.Met1Val) of the initiating methionine codon. Atypical presentations included a patient with a homozygous TRMU (NM_018006.4)30 mutation seen in association with heart, CNS and muscle involvement, but no liver involvement, and only a subclinical, mild anaemia in patient 23 carrying a homozygous nonsense mutation in PUS1. In addition, patient 26 had typical features of Leigh syndrome and multiple respiratory chain complex defects at the time of biopsy, but was homozygous for the p.Glu140Lys SCO2 (NM_001169111.1)31 founder mutation which is usually seen in association with an isolated complex IV defect and cardiomyopathy, features not present in this patient.
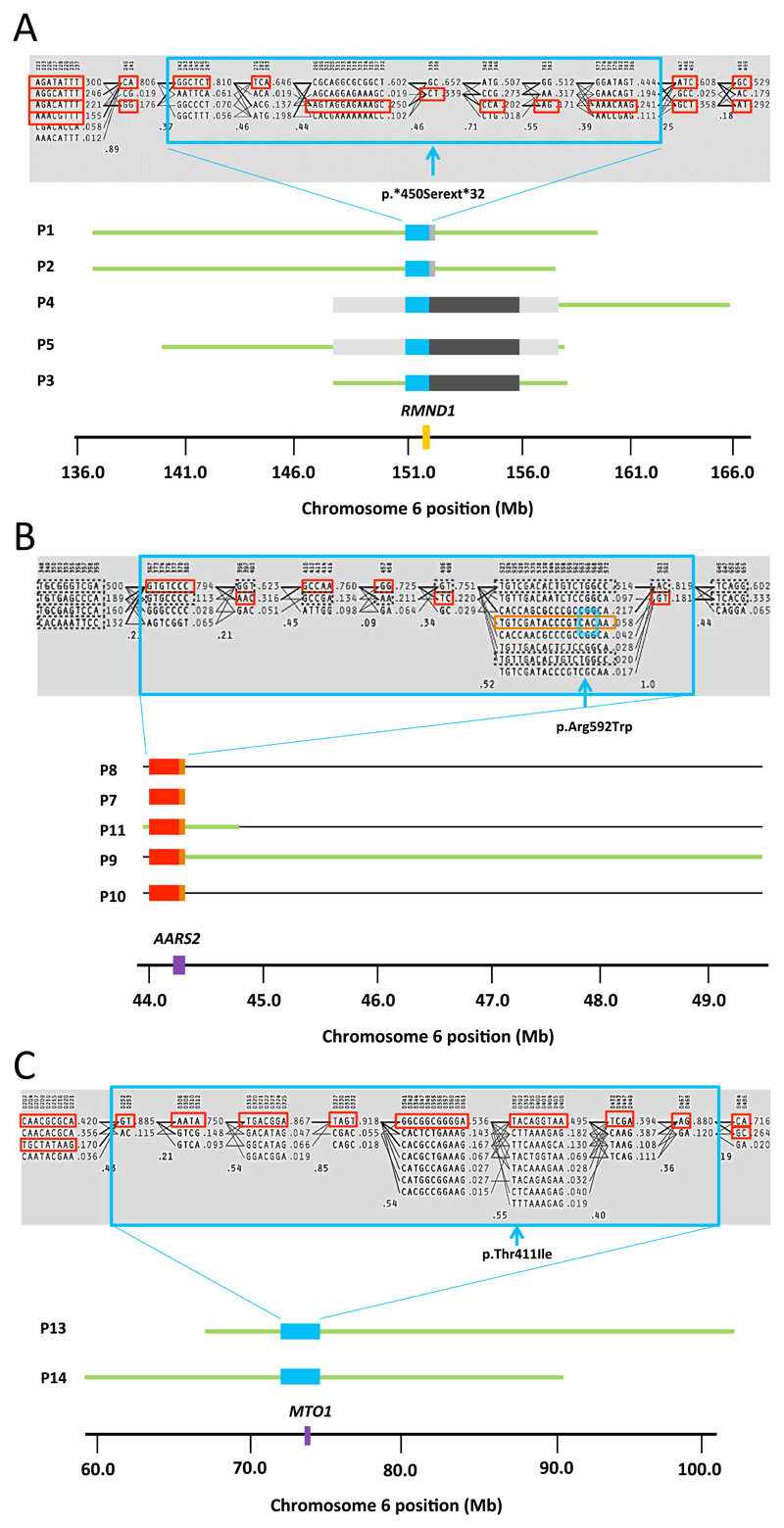
Figure 1. Molecular haplotypes flanking RMND1, AARS2 and MTO1 in the patients studied here.
The grey highlighted box in (A), (B) and (C) shows haplotype blocks generated from the selected markers using 62 in-house control exomes and the patients found to harbor mutations in RMND1, AARS2 and MTO1 in this study. Population frequencies are shown next to each haplotype and lines show the most common crossings from one block to the next. The thicker lines show more common crossings than thinner lines with multilocus D’ below, which is a measure of the linkage disequilibrium between two blocks. The closer the value is to zero, the greater the amount of historical recombination between the two blocks. The large blue boxes group together the single nucleotide variants (SNV), which make up the shared RMND1, AARS2 and MTO1 haplotypes. The red and orange boxes show the alleles at each SNV that make up the shared haplotype. (A) Molecular haplotypes flanking RMND1 in P1, P2, P3, P4 & P5 (blue box). The red boxes show the alleles at each SNV that make up the shared haplotype. The light grey solid boxes represent the two different haplotypes shared between P1/P2, and P4/P5, which are situated 3’ to the RMND1 gene. The dark grey boxes represent the haplotype shared by P3, P4 and P5. Regions of extended homozygosity surrounding the RMND1 gene are represented by green lines. The RMND1 mutation, c.1349G>C (p.*450Serext*32) (between Haploview markers 335 and 338, shown by an asterisks *), violates Hardy-Weinberg equilibrium because there are no heterozygotes and so is not included by Haploview in the blocks. The position of the RMND1 gene is highlighted by a solid orange box in chromosome 6. (B) Molecular haplotype flanking the AARS2 in P7, P8, P8, P10 and P11 (0.3Mb, blue box) including the c.1774C>T (p.Arg592Trp) mutation. A haplotype spanning exons 10-22 is shown by orange boxes. The red boxes represent the further six haplotype blocks which appear to be shared between p.Arg592Trp AARS2 mutation carriers, however, for the carriers of the discrete heterozygous AARS2 mutations (P7, P8 and P10), it was not possible to resolve the phase of these blocks. Regions of extended homozygosity surrounding the AARS2 gene are represented by green lines in patients P9 and P11. The dashed boxes represent the alternative haplotype blocks represent the alternative haplotype blocks in the patients where the mutation was heterozygous (P7, P8 and P10). The AARS2 gene is represented in chromosome 6 by a solid purple box. (C) Molecular haplotype flanking the MTO1 gene in 2 of the 4 affected individuals (P13 and P14, blue box) showing the shared haplotype defining a founder allele. Regions of extended homozygosity surrounding the MTO1 gene are represented by green lines. The homozygous MTO1 mutation, c.1232C>T:p.Thr411Ile is located between Haploview markers 382 and 392 and is shown by a purple asterisks (*). The MTO1 gene is represented in the chromosome by a solid purple box.
(2). Possible pathogenic
Probable disease-causing variants were identified in novel mitochondrial disease genes in 4 patients, each predicted to affect mitochondrial protein synthesis. VARS2 (NM_001167734.1) and GARS (NM_002047.2) encode mitochondrial aminoacyl-tRNA synthetase genes. FLAD1 (NM_025207.4) encodes a key factor of the riboflavin metabolism, and PTCD1 (NM_015545.3) is a gene encoding a mitochondrially-targeted pentatricopeptide reported to be involved in mitochondrial RNA metabolism.32 In silico predictions supported a pathogenic role in each case, but given that they were only identified in single patients, further evidence is required before these variants can be considered definitively pathogenic; where familial samples were available, identified mutations were shown to segregate with disease (Table 1).
(3). Variants of unknown significance
In 15 patients we identified one or more variants in genes predicted to encode mitochondrial proteins (Table 1), where there were several plausible candidate disease genes. Most of the gene defects were detected in single patients only (except for ACSM5 (NM_017888.2) mutations in patients 34 and 36). The current lack of functional data directly linking these genes to multiple respiratory chain complex defects led to their classification as possible and not probable causative variants (Figure 2). Identification of further patients with mutations in these genes and/or functional work is required to validate these findings.
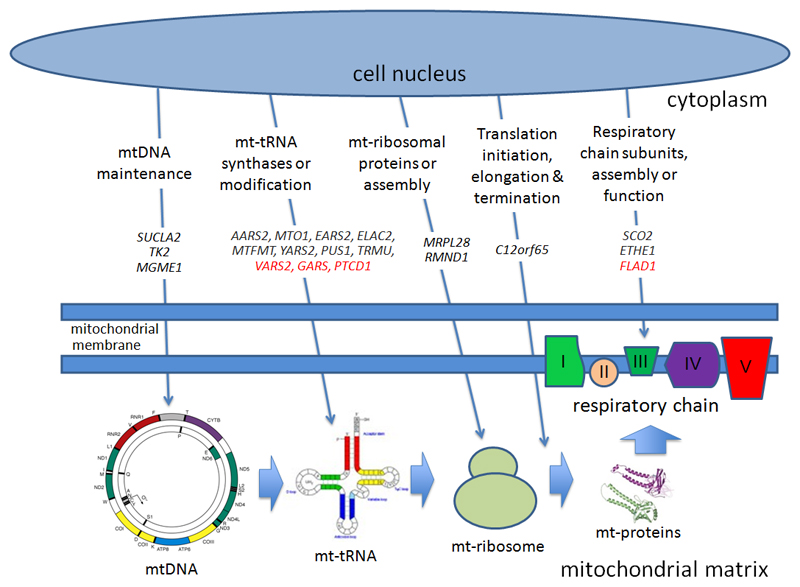
Figure 2. Intra-mitochondrial protein synthesis. Genes present within the cell nucleus encode proteins critical for intra-mitochondrial protein translation.
These are synthesised in the cytoplasm (upper half), and transported through the double mitochondrial membrane into the mitochondrial matrix. Here they regulate (from left to right) mtDNA maintenance; mitochondrial tRNA synthases or tRNA modifications; mitochondrial ribosomal protein components or ribosomal assembly; the initiation, elongation or termination of protein translation; or the protein subunits themselves and their assembly factors. Genes known to cause defects of intra-mitochondrial protein translation are shown in capitals and italics. Genes in red are the novel predicted mitochondrial disease genes described here. mtDNA = mitochondrial DNA, mt = mitochondrial, tRNA = transfer RNA.
(4). Unresolved
Exome sequencing did not identify any candidate pathogenic variants in 6 patients.
Discussion
In the pre-exome era, the systematic biochemical characterisation of 53 patients with multiple respiratory chain complex defects led to the underlying genetic basis in only one patient.5 The work presented here demonstrates the effect of whole exome sequencing in this context, which has defined the genetic aetiology in 32 of 53 patients (60%, including 28 presumptive and 4 probable causative mutations) with a confirmed biochemical defect consistent with a generalised decrease in mitochondrial translation. The detection rate was even higher in children with onset < 1 year of age (24/35, 69%). In 20 patients with prominent cardiac disease we detected the causative mutation in 15 (80%), while the detection rate was much lower in patients with liver disease (3/9, 33%). Our findings contrast with large-scale candidate gene analysis using conventional,33 and next-generation sequencing approaches,34 both of which had a lower diagnostic yield (10-13%) and by definition did not discover new potential disease genes. A more ambitious approach involving exon capture and sequencing of all predicted mitochondrial genes (the “Mitoexome”) has delivered a greater diagnostic yield (22-28%)12,35, although in cohorts with predominantly isolated respiratory chain complex defects which are less challenging to define at the genetic level.36
There are a number of reasons why our approach had a >2-fold greater diagnostic yield than previous studies, despite the known difficulty in making a molecular diagnosis in patients with multiple respiratory chain complex deficiencies. First, not being based on any prior assumptions about known candidate disease genes, the whole exome approach detected four new genes which may to be responsible for the underlying mitochondrial disorder. Further arguments supporting the pathogenic role of mutations in these new genes come from similarities in function to known mitochondrial genes. For example VARS2 and GARS encode tRNA synthase genes with products known to enter mitochondria. Recessive mutations in other mitochondrial tRNA synthetase genes responsible for charging tRNAs with different specific cognate amino acids during protein synthesis have been shown to cause identical biochemical defects and similar clinical phenotypes.17,20,24 Compound heterozygous mutations in GARS have recently been reported in a single family with a multi-systemic mitochondrial disease with cardiomyopathy.37 These findings endorse our approach, and support the pathogenic role of the GARS mutations in our patient who developed a fatal cardiomyopathy, but the lack of functional data mean that mutations in this gene should remain in the “possible pathogenic” group (Table 1). Like GARS, FLAD1 was not originally considered to encode a mitochondrial protein, but ultimately were found to encode alternatively spliced cytoplasmic and mitochondrial transcripts, and thus are plausible candidate genes.38 Although we have not shown proof of the causal link, in silico predictions of the deleterious effect of the mutations in genes showing strong functional similarities with known disease genes, supports a causal association with the mutations. Secondly, the unbiased exome approach has the potential to reveal unexpected results. This was the case for three additional unreported patients who also met the selection criteria for this study. One patient presenting with neonatal myopathy, encephalopathy and lactic acidosis, with a complex I defect and a less prominent complex IV defect, and was found to have novel homozygous mutations in the complex I subunit gene NDUFS6 (c.317_320delAAAC:p.Glu106fs*21). In this context, the complex IV defect was presumably secondary, perhaps mediated through the disruption of respiratory chain super-complexes. Similarly, one child carrying the homozygous common SCO2 mutation also presented with severe combined complex I and IV defects. These results also illustrate the difficulties in interpreting respiratory chain enzyme analysis, which even in skilled hands, can misdirect a candidate gene approach. The higher diagnostic yield could, in part, reflect the growing inventory of genes known to cause mitochondrial diseases, and the relatively high proportion of familial and consanguineous cases in our study cohort.
Our findings implicate 18 different genes and further 33 possible candidates in 53 patients, underscoring the genetic heterogeneity of this group of mitochondrial disorders (Figure 2). Given this complexity, how should we approach the diagnosis in a clinical setting? Despite the relatively small number of individuals with any single mutation, our observations show the phenotypic diversity in patients with multiple respiratory chain complex defects, emerging clinical subgroups do appear to be seen in association with specific genetic defects. For example, AARS2 and MTO1 mutations were preferentially seen in association with cardiomyopathy, mutations in TRMU present with liver failure that can improve spontaneously, YARS2 and PUS1 with sideroblastic anemia and myopathy, and RMND1 with deafness, myopathy, renal involvement and a severe biochemical defect (Table 1). However, not all patients fitted neatly into these subgroups – including a patient with a TRMU mutation and normal liver function, one with a PUS1 mutation and only subclinical anaemia, and a SCO2 mutation with no cardiomyopathy. This heterogeneity is typical for mitochondrial diseases, and supports the use of next-generation sequencing early in the diagnostic approach. Although a molecular diagnosis is unlikely to lead to specific drug treatments at present, defining the genetic etiology may enable accurate genetic counselling and prenatal diagnosis, and personalized disease surveillance for genotype-specific complications.
Given that our case ascertainment was determined by clinical referral and not influenced or biased by the study team, our findings may have broader relevance. However, our study has several limitations. First, we could not identify a potential candidate mutation in only 6 patients (11%), possibly because the causative mutations lay in deep intronic regions, or may involve deletions or duplications (copy number variations), missed by the exome capture and bioinformatic analysis. Second, our work was carried out on carefully phenotyped patients defined by a biochemical defect measured in two specialist centers, and a high proportion of patients were from consanguineous families. It will be important to replicate our findings in similar patient cohorts investigated in other centers from other parts of the world, where the spectrum of nuclear gene defects may be different. The 53 patients we studied account for ~20% of the referrals to our centres with any form of biochemical defect of the respiratory chain determined by a spectrophotometric assay of enzyme activity, and the patients with multiple respiratory chain complex defects account for ~60% of patients with a biochemical defect with no known molecular diagnosis. Therefore, the role of exome sequencing in unselected patients with a clinical diagnosis of suspected multiple respiratory chain complex defects remains to be determined, and the effect of exome sequencing in patients with a general diagnosis of suspected mitochondrial disease is not clear. However, applying a whole-exome approach to a group of patients with multiple respiratory chain complex defects that are difficult to diagnose has delivered a high diagnostic yield. Curiously, most of the genes appear to be involved in intra-mitochondrial gene translation. This explains the phenotypic and biochemical overlap, but it is not at all clear why apparently subtle differences in function should lead to discrete clinical phenotypes. These discrete phenotypes may point to a few candidate genes, but in our large cohort, this only became obvious after performing this study. It is possible that next-generation sequencing will revolutionise the investigation of mitochondrial diseases, and its early application may provide a rapid diagnosis at a relatively low cost, particularly in patients with multiple respiratory chain complex defects.
Conclusions
Exome sequencing enhances the ability to identify the underlying nuclear gene mutations in patients with multiple mitochondrial respiratory chain complex defects. Additional study will be required to determine the utility of this approach compared to traditional diagnostic methods in independent patient populations.
Supplementary Material
Funding/Support
Chinnery, Taylor and Turnbull receive support from the Wellcome Trust Centre for Mitochondrial Research (096919Z/11/Z). Chinnery, Taylor, Horvath, McFarland, Gorman and Turnbull receive support from the Medical Research Council (UK) Centre for Translational Muscle Diseases (G0601943) and Mitochondrial Disease Patient Cohort (G0800674). Chinnery is a Wellcome Trust Senior Fellow in Clinical Science (101876/Z/13/Z), a UK NIHR Senior Investigator, and receives additional support from EU FP7 TIRCON, and the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre based at Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Horvath was supported by the Medical Research Council (UK) (G1000848) and the European Research Council (309548).
Role of the Sponsor
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. Drs Chinnery, Taylor and Horvath had full access to all the data and had final responsibility for the decision to submit for publication.
Footnotes
Author contributions: Drs Chinnery and Taylor had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Equal contributions: Drs Taylor, Pyle, Horvath and Chinnery contributed equally to this work.
Study concept and design: Chinnery, Taylor, Horvath
Acquisition, analysis, or interpretation of the data: All authors
Bioinformatic analysis: Griffin, Santibanez-Koref
Drafting of the manuscript: Chinnery, Horvath, Pyle, Taylor
Critical revision of the manuscript for important intellectual content: All authors
Obtained funding: Chinnery, Taylor, Horvath.
Study supervision: Chinnery, Taylor, Horvath.
Additional contributions
We thank the clinicians who referred the patients for further investigation.
Conflicts of Interest disclosures
The authors report no conflicts of interest
References
- 1.Skladal D, Bernier FP, Halliday JL, Thorburn DR. Birth prevalence of mitochondrial respiratory chain defects in children. J Inher Metab Dis. 2000;23:138. [Google Scholar]
- 2.Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Ann Med. 2012;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- 3.McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurol. 2010;9:829–40. doi: 10.1016/S1474-4422(10)70116-2. [DOI] [PubMed] [Google Scholar]
- 4.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–83. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 5.Kemp JP, Smith PM, Pyle A, et al. Nuclear factors involved in mitochondrial translation cause a subgroup of combined respiratory chain deficiency. Brain. 2011;134:183–95. doi: 10.1093/brain/awq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotig A. Genetic bases of mitochondrial respiratory chain disorders. Diabetes Metab. 2010;36:97–107. doi: 10.1016/j.diabet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods in cell biology. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- 8.Taylor RW, Schaefer AM, Barron MJ, McFarland R, Turnbull DM. The diagnosis of mitochondrial muscle disease. Neuromuscul Disord. 2004;14:237–45. doi: 10.1016/j.nmd.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–5. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–73. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvo SE, Compton AG, Hershman SG, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Science translational medicine. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366:1132–41. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janer A, Antonicka H, Lalonde E, et al. An RMND1 Mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am J Hum Genet. 2012;91:737–43. doi: 10.1016/j.ajhg.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Diaz B, Barros MH, Sanna-Cherchi S, et al. Infantile encephaloneuromyopathy and defective mitochondrial translation are due to a homozygous RMND1 mutation. Am J Hum Genet. 2012;91:729–36. doi: 10.1016/j.ajhg.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotz A, Tyynismaa H, Euro L, et al. Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet. 2011;88:635–42. doi: 10.1016/j.ajhg.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghezzi D, Baruffini E, Haack TB, et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am J Hum Genet. 2012;90:1079–87. doi: 10.1016/j.ajhg.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baruffini E, Dallabona C, Invernizzi F, et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum Mutat. 2013 doi: 10.1002/humu.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steenweg ME, Vanderver A, Ceulemans B, et al. Novel infantile-onset leukoencephalopathy with high lactate level and slow improvement. Arch Neurol. 2012;69:718–22. doi: 10.1001/archneurol.2011.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steenweg ME, Ghezzi D, Haack T, et al. Leukoencephalopathy with thalamus and brainstem involvement and high lactate 'LTBL' caused by EARS2 mutations. Brain. 2012;135:1387–94. doi: 10.1093/brain/aws070. [DOI] [PubMed] [Google Scholar]
- 22.Tucker EJ, Hershman SG, Kohrer C, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell metabolism. 2011;14:428–34. doi: 10.1016/j.cmet.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonicka H, Ostergaard E, Sasarman F, et al. Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am J Hum Genet. 2010;87:115–22. doi: 10.1016/j.ajhg.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley LG, Cooper S, Hickey P, et al. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am J Hum Genet. 2010;87:52–9. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am J Hum Genet. 2004;74:1303–8. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornblum C, Nicholls TJ, Haack TB, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–9. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiranti V, D'Adamo P, Briem E, et al. Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am J Hum Genet. 2004;74:239–52. doi: 10.1086/381653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haack TB, Kopajtich R, Freisinger P, et al. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am J Hum Genet. 2013;93:211–23. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29:342–4. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 30.Zeharia A, Shaag A, Pappo O, et al. Acute infantile liver failure due to mutations in the TRMU gene. Am J Hum Genet. 2009;85:401–7. doi: 10.1016/j.ajhg.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadopoulou LC, Sue CM, Davidson MM, et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet. 1999;23:333–7. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 32.Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 2009;37:5859–67. doi: 10.1093/nar/gkp627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haack TB, Madignier F, Herzer M, et al. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9. J Med Genet. 2012;49:83–9. doi: 10.1136/jmedgenet-2011-100577. [DOI] [PubMed] [Google Scholar]
- 34.Calvo SE, Tucker EJ, Compton AG, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 2010;42:851–8. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieber DS, Calvo SE, Shanahan K, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 2013;80:1762–70. doi: 10.1212/WNL.0b013e3182918c40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haack TB, Haberberger B, Frisch EM, et al. Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J Med Genet. 2012;49:277–83. doi: 10.1136/jmedgenet-2012-100846. [DOI] [PubMed] [Google Scholar]
- 37.McMillan HJ, Schwartzentruber J, Smith A, et al. Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. BMC medical genetics. 2014;15:36. doi: 10.1186/1471-2350-15-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brizio C, Galluccio M, Wait R, et al. Over-expression in Escherichia coli and characterization of two recombinant isoforms of human FAD synthetase. Biochem Biophys Res Commun. 2006;344:1008–16. doi: 10.1016/j.bbrc.2006.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.