Summary
Cancer immunotherapies have shown substantial clinical activity for a subset of patients with epithelial cancers. Still, technological platforms to study cancer – T cell interactions for individual patients, and understand determinants of responsiveness, are presently lacking. Here, we establish and validate a platform to induce and analyze tumor-specific T cell responses for epithelial cancers in a personalized manner. We demonstrate that co-cultures of autologous tumor organoids and peripheral blood lymphocytes can be used to enrich for tumor-reactive T cells from peripheral blood of patients with mismatch repair deficient colorectal cancer and non-small cell lung cancer. Furthermore, we demonstrate that these T cells can be used to assess the efficiency of killing of matched tumor organoids. This platform provides an unbiased strategy for the isolation of tumor-reactive T cells and provides a means to assess the sensitivity of tumor cells to T cell-mediated attack at the level of the individual patient.
Introduction
The use of antibodies against immune checkpoints, such as CTLA-4 and PD-1/PD-L1, has shown clear clinical benefit for patients with advanced cancer, including melanoma, non-small cell lung cancer (NSCLC), and mismatch repair deficient (dMMR) colorectal cancer (CRC) (Larkin et al., 2015; Garon et al., 2015; Borghaei et al., 2015; Le et al., 2015; Le et al., 2017; Overman et al., 2017; Overman et al., 2018). Furthermore, adoptive transfer of ex vivo-expanded autologous tumor-infiltrating lymphocytes (TIL) has shown impressive clinical responses in melanoma (Rosenberg and Restifo, 2015) and an early clinical signal in cervical cancer (Stevanovic et al., 2015). Despite these encouraging results, a large fraction of patients does not respond to current immunotherapies. Treatment failure may be explained at many different levels that include a low number of immunogenic antigens, defective antigen presentation, and/or the expression of alternative immune checkpoint molecules (Blank et al., 2016; Dijkstra et al., 2016; Pitt et al., 2017; Sharma et al., 2017). Given the large variety in mechanisms of immune evasion by cancers, it is presently challenging to predict whether an individual patient will be sensitive to immunotherapy, what mechanism is likely to underlie resistance and what alternative treatment could potentially overcome such resistance.
Platforms to allow the unbiased and systematic analysis of T cell-mediated tumor recognition on an individual patient basis would greatly contribute to our understanding of the critical factors that determine a successful anti-tumor immune response. Traditionally, ex vivo systems to analyze T cell – tumor interaction have to a very large extent focused on cutaneous melanoma, both because of the availability of robust approaches to expand tumor-infiltrating T cells for this disease (Rosenberg and Restifo, 2015), and because of the relative ease with which melanoma cell lines can be obtained. Importantly however, with the now widespread clinical development and clinical use of immunotherapy for major epithelial cancers, it is critical to develop technology to dissect T cell-mediated tumor recognition in these tumor types. Traditionally, this effort has been limited by both the low success rate of establishing primary tumor cell lines of epithelial cancers such as NSCLC and CRC (success rate of 10% or lower) (Dangles-Marie et al., 2007; Zheng et al., 2011), and the limited feasibility of obtaining matched tumor-reactive T cell populations.
We set out to evaluate the feasibility of an autologous T cell – tumor organoid co-culture platform for individual patients. Tumor organoids are three-dimensional primary tumor cell cultures that retain the histological and mutational features of the original tumor. Organoids can be established from surgical tumor resections (e.g., success rate for CRC of 60-90%) (van de Wetering et al., 2015; Schütte et al., 2017), and from needle biopsies of metastases (e.g. success rate for CRC of ~70%) (Weeber et al., 2015). In this effort, we wished to establish two things: first, whether tumor-reactive T cells can be obtained by co-culture of peripheral blood lymphocytes (PBL) with matched tumor organoids; second, whether such T cells can be used to assess the efficiency of tumor cell killing. The use of peripheral blood as a source of tumor-reactive T cells could provide an easily accessible alternative to TIL. Here we provide proof of concept that co-culture of tumor organoids with PBL forms an unbiased strategy to obtain tumor-reactive T cells from patients with mismatch repair deficient colorectal and non-small cell lung cancer that kill tumor organoids from the same patient. Our findings provide proof of principle for the generation of a novel class of tumor-specific T cell products, derived from peripheral blood, and provide a means to assess the sensitivity of tumor cells to T cell-mediated attack at the level of the individual patient.
Results
Characterization of a panel of mismatch repair deficient (dMMR) CRC organoids
To evaluate whether tumor-reactive T cells can be enriched from PBL by stimulation with autologous tumor organoids, we established a clinical protocol that allowed withdrawing blood and obtaining tumor tissue from patients with CRC. We focused on dMMR CRC given their high mutational load (TCGA, 2012) and frequent response to anti-PD-1 therapy (Le et al., 2015; Le et al., 2017; Overman et al., 2017; Overman et al., 2018). Fresh tumor tissue was obtained from resection specimens or core needle biopsies and used for the establishment of organoids. Mismatch repair deficiency was evaluated by an independent pathologist based on expression of mismatch repair proteins according to standard diagnostic criteria (Table 1 and STAR* Methods). In line with prior data (Weeber et al., 2015; van de Wetering et al., 2015; Schütte et al., 2017), the success rate of growing tumor organoids from these sources was ~60%. Organoids were expanded by weekly to biweekly passaging at 1:2 to 1:5 split ratios and cryopreserved in low passage master biobanks within 2-9 weeks after derivation. Organoids can be recovered from frozen stocks and expanded for several months to large numbers without loss of proliferative capacity.
Table 1. Characteristics of panel of dMMR CRC samples.
MLH1: mutL homolog 1; PMS2: PMS1 homolog 2, mismatch repair system component; MSH2: mutS homolog 2; MSH6: mutS homolog 6. F: female; M: male.
| Sample | Sex | Age | Tumor location | Stage | Primary tumor / metastasis | Biopsy / resection | Absent MMR stains |
|---|---|---|---|---|---|---|---|
| CRC-1 | F | 68 | Colon | I | Primary tumor | Resection | MLH1/PMS2 |
| CRC-2 | M | 65 | Colon | III | Primary tumor | Resection | MSH6 |
| CRC-3 | F | 65 | Colon | II | Primary tumor | Resection | MLH1/PMS2 |
| CRC-4 | F | 75 | Colon | III | Primary tumor | Resection | MLH1/PMS2 |
| CRC-5 | F | 73 | Rectum | I | Primary tumor | Resection | MSH6 |
| CRC-6P | F | 67 | Colon | IV | Primary tumor | Resection | MLH1/PMS2 |
| CRC-6M | F | 67 | Peritoneum | IV | Metastasis | Resection | MLH1/PMS2 |
| CRC-7 | F | 62 | Colon | III | Primary tumor | Resection | MLH1/PMS2 |
| CRC-8P | M | 50 | Colon | IV | Primary tumor | Resection | MLH1/PMS2 |
| CRC-8M | M | 50 | Liver | IV | Metastasis | Resection | MLH1/PMS2 |
| CRC-9 | F | 51 | Lymph node (neck) | IV | Metastasis | Biopsy | MLH1/PMS2 |
| CRC-10 | F | 66 | Liver | IV | Metastasis | Biopsy | MLH1/PMS2 (partial loss of MSH6) |
| CRC-11 | F | 66 | Peritoneum | IV | Metastasis | Biopsy | MLH1/PMS2 |
| CRC-12 | F | 74 | Colon | I | Primary tumor | Resection | MLH1/PMS2 |
| CRC-13 | F | 70 | Colon | II | Primary tumor | Biopsy | MLH1/PMS2 |
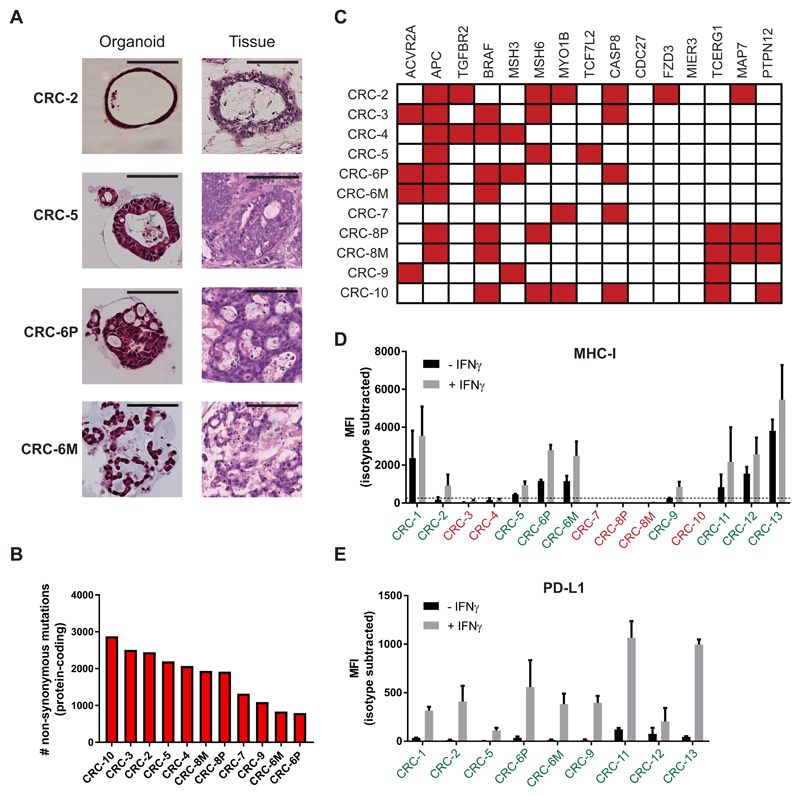
We established a panel of 15 tumor organoids from 13 different patients with dMMR CRC (Table 1). The organoids morphologically reflected the original tumor they were derived from (Figure 1A and Figure S1). Whole-exome sequencing (WES) of tumor organoids showed a high mutational burden (median 1938 non-synonymous mutations per tumor, range 795-2877), well in the range previously reported for hypermutated CRC (Figure 1B) (TCGA, 2012; Vogelstein et al., 2013). Mutations typically associated with hypermutated CRC (TCGA, 2012) were also represented in these organoids (Figure 1C and Table S1). Considering that loss of MHC-I occurs in up to 60% of dMMR CRC (Dierssen et al., 2007), we screened for MHC-I expression after stimulation with IFNγ and identified 9 MHC-I proficient tumor organoids from 8 patients (62% of organoid samples) (Figure 1D and Figure S2). MHC-I expression of tumor organoids correlated with immunohistochemistry data of MHC-I expression of the tumor they were derived from (Spearman r = 0.78; p = 0.01) (Figure S2 and Table S2). Most importantly, tumor organoids classified as MHC-I-deficient were always derived from MHC-I deficient tumors, indicating that loss of MHC-I is not a general feature of organoid culture.
Figure 1. Characterization of a panel of dMMR CRC organoids.
(A) Haematoxylin and eosin (H&E) staining of tumor organoids and original tumor tissue. Organoids show morphology similar to the architecture of the original tumor. CRC-2 organoids demonstrates cystic tubules similar to mucin-filled glands of the primary tumor. CRC-5 organoids and primary tumor are composed of tubular glands with layered epithelium. Organoid CRC-6P is composed of complex cribriform glands, also seen in the primary tumor. Organoid CRC-6M consists of irregular trabecular structures and poorly formed glands, similar to the metastasis. Scale bar = 100 μm.
(B) Mutational load of tumor organoids as determined by the number of non-synonymous mutations per tumor exome.
(C) Mutation status of genes significantly mutated in hypermutated colorectal cancer according to TCGA 2012. Mutated genes are indicated in red.
(D) Cell surface MHC-I expression as determined by flow cytometry. Organoids were stimulated with 200 ng/mL IFNγ for 24 hours or left unstimulated. Bar graphs indicate median fluorescence intensity (MFI) of anti-HLA-A,B,C-PE minus MFI of isotype control. Tumor organoids with MFI < 250 above isotype control (dashed line) were classified as MHC-I deficient (indicated in red). Error bars indicate s.e.m. of at least two independent experiments.
(E) Cell surface PD-L1 expression as determined by flow cytometry. Organoids were stimulated with 200 ng/mL IFNγ for 24 hours or left unstimulated. Bar graphs indicate median fluorescence intensity (MFI) of anti-PD-L1-APC minus MFI of isotype control. Error bars indicate s.e.m. of at least two independent experiments.
See also Figures S1-S2 and Tables S1-S2.
Induction of tumor reactivity in circulating T cells by co-culture with autologous tumor organoids
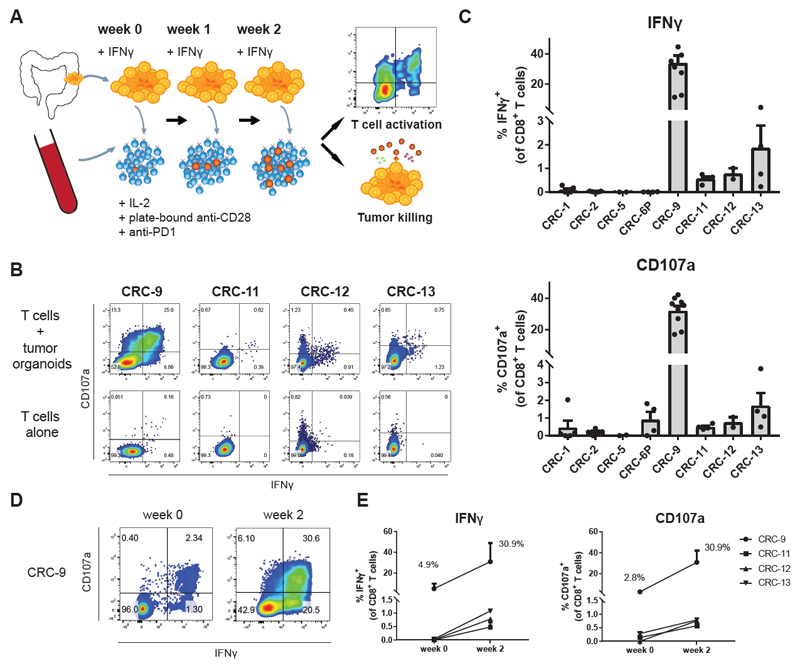
To test whether tumor organoids may be used to obtain tumor reactive T cells, we selected MHC-I proficient tumor organoids and focused on the peripheral blood compartment as a source of T cells with a lower degree of exhaustion. Prior to co-culture with autologous T cells, tumor organoids were pre-stimulated with IFNγ to enhance antigen presentation. IFNγ exposure also led to the induction of PD-L1, a negative regulator of T cell activation (Figure 1E), and to counteract any inhibitory effect of PD-L1 during T cell activation, we added blocking antibodies to PD-1. Plate-bound anti-CD28 and IL-2 were added to provide co-stimulation and to support T cell proliferation, respectively. Peripheral blood mononuclear cells (PBMC) were isolated from patients with dMMR CRC and stimulated weekly with autologous tumor organoids (Figure 2A). Tumor recognition by CD8+ T cells was evaluated at baseline and after two weeks of co-culture, by staining for IFNγ and the degranulation marker CD107a.
Figure 2. Induction of tumor reactivity in circulating T cells by co-culture with autologous tumor organoids.
(A) Experimental workflow. Tumor organoids were established from dMMR CRC (resections or biopsies of primary tumors or metastases) and stimulated with IFNγ for 24 hours prior to co-culture with peripheral blood lymphocytes (PBL) from the same patient. PBL were stimulated weekly with fresh tumor cells. After two weeks of co-culture, T cell effector functions and sensitivity of tumor organoids to T cell-mediated killing were evaluated using flow cytometry.
(B) Representative flow cytometry plots gated on CD8+ T cells tested for reactivity against autologous organoids after two weeks of co-culture with autologous tumor organoids.
(C) Quantification of organoid-induced IFNγ production and CD107a cell surface expression of CD8+ T cells obtained by two week co-culture with autologous tumor organoids. Background (spontaneous IFNγ production or CD107a expression) is subtracted from signal. Error bars indicate s.e.m. of at least 2 biological replicates. Dots indicate biological replicates.
(D) Flow cytometry plots gated on CD8+ T cells of patient CRC-9 tested for reactivity against autologous organoids directly after PBL isolation, or after two weeks of co-culture with autologous tumor organoids.
(E) Quantification of organoid-induced IFNγ production and CD107a cell surface expression of CD8+ T cells directly after PBL isolation, or obtained by two week co-culture with autologous tumor organoids. Background (spontaneous IFNγ production or CD107a expression) is subtracted from signal. Error bars indicate s.e.m of n = 2 biological replicates for CRC-9 and CRC-11. N = 1 for CRC-12 and CRC-13 (low amount of blood available).
In four out of eight (50%) patients with MHC-I+ tumor organoids, stimulation with autologous tumor organoids induced both IFNγ secretion and CD107a upregulation in CD8+ T cells after two weeks of co-culture (Figure 2B-C). Responses were never observed in MHC-I deficient lines (data not shown). The magnitude of the response varied between patients, including small but reproducible responses of 1-3% tumor-reactive CD8+ T cells, as well as a patient of whom ~50% of all CD8+ T cells were tumor-reactive. The high reactivity for CRC-9 suggested the presence of a pre-existing tumor-reactive T cell population. We therefore evaluated tumor reactivity of T cells before co-culture with tumor organoids (i.e. directly after isolation from blood). A substantial proportion of CD8+ T cells from patient CRC-9 was already tumor-reactive, and this population increased approximately 10-fold in frequency upon two weeks of co-culture (Figure 2D-E). In contrast, T cells from patients CRC-11, CRC-12 and CRC-13 did not show any detectable tumor reactivity before organoid co-culture (Figure 2E), indicating that T cell – organoid co-culture systems can also be used to expand previously undetectable tumor-reactive T cell populations.
Induction of tumor reactivity in circulating T cells from patients with NSCLC
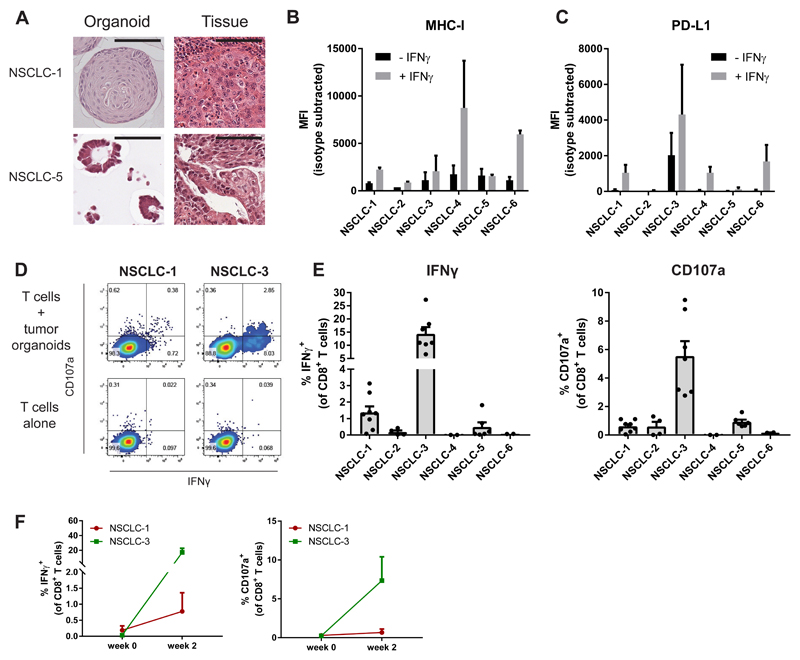
We then assessed whether our strategy to obtain tumor-reactive T cell populations from peripheral blood by organoid co-culture could also be applied to non-small cell lung cancer. As compared to dMMR CRC, mutational burden of NSCLC is approximately 5-fold lower (Vogelstein et al., 2013; Govindan et al., 2012) and response to PD-1/PD-L1 blockade is restricted to approximately 20% of patients (Garon et al., 2015; Borghaei et al., 2015). We generated NSCLC organoids from six patients (Figure 3A, Table S3 and Figure S3), using a method similar to that established for CRC organoids (Sachs et al., 2018). Sample NSCLC-3 contained both normal airway epithelial organoids that grew as cystic thin-walled organoids and tumor organoids that showed a more solid morphology. To select for tumor organoids, we applied the MDM-2 inhibitor Nutlin-3a to selectively grow out p53-mutant cells of this sample, resulting in a pure tumor organoid population (Sachs et al., 2018). All NSCLC organoids were MHC-I proficient and expressed various levels of PD-L1 upon stimulation with IFNγ (Figure 3B-C). In vitro exposure of PBMC to autologous tumor organoids resulted in the expansion of tumor-reactive CD8+ populations in two of these patients after two weeks of co-culture (Figure 3D-E). Notably, none of these responses could be consistently detected before co-culture with organoids (Figure 3F). Collectively, these data demonstrate the feasibility of inducing patient-specific tumor-reactive T cell responses by co-culture of PBMC and autologous tumor organoids in two epithelial tumor types.
Figure 3. Induction of tumor reactivity in circulating T cells from patients with NSCLC.
(A) Haematoxylin and eosin staining of tumor organoids and original tumor tissue. Organoids show morphology similar to the architecture of the original tumor. NSCLC-1 organoids consists of large cells forming a solid mass, similar to the primary tumor. NSCLC-5 organoids are composed of tubular glands similar to the original tumor. Scale bar = 100 μm.
(B-C) Cell surface MHC-I (B) and PD-L1 (C) expression as determined by flow cytometry. Organoids were stimulated with 200 ng/mL IFNγ for 24 hours or left unstimulated. Bar graphs indicate median fluorescence intensity (MFI) of anti-HLA-A,B,C-PE or anti-PD-L1-APC minus MFI of isotype control. Error bars indicate s.e.m. of two independent experiments.
(D) Representative flow cytometry plots gated on CD8+ T cells tested for reactivity against autologous organoids after two weeks of co-culture with autologous tumor organoids.
(E) Quantification of organoid-induced IFNγ production and CD107a cell surface expression of CD8+ T cells obtained by two week co-culture with autologous tumor organoids. Background (spontaneous IFNγ production or CD107a expression) is subtracted from signal. Error bars indicate s.e.m. of at least 2 biological replicates. Dots indicate biological replicates.
(F) Quantification of organoid-induced IFNγ production and CD107a cell surface expression of CD8+ T cells directly after PBL isolation, or obtained by two week co-culture with autologous tumor organoids. Background (spontaneous IFNγ production or CD107a expression) is subtracted from signal. Error bars indicate s.e.m of n = 2 biological replicates.
See also Figure S3.
Specificity of organoid-reactive T cell responses
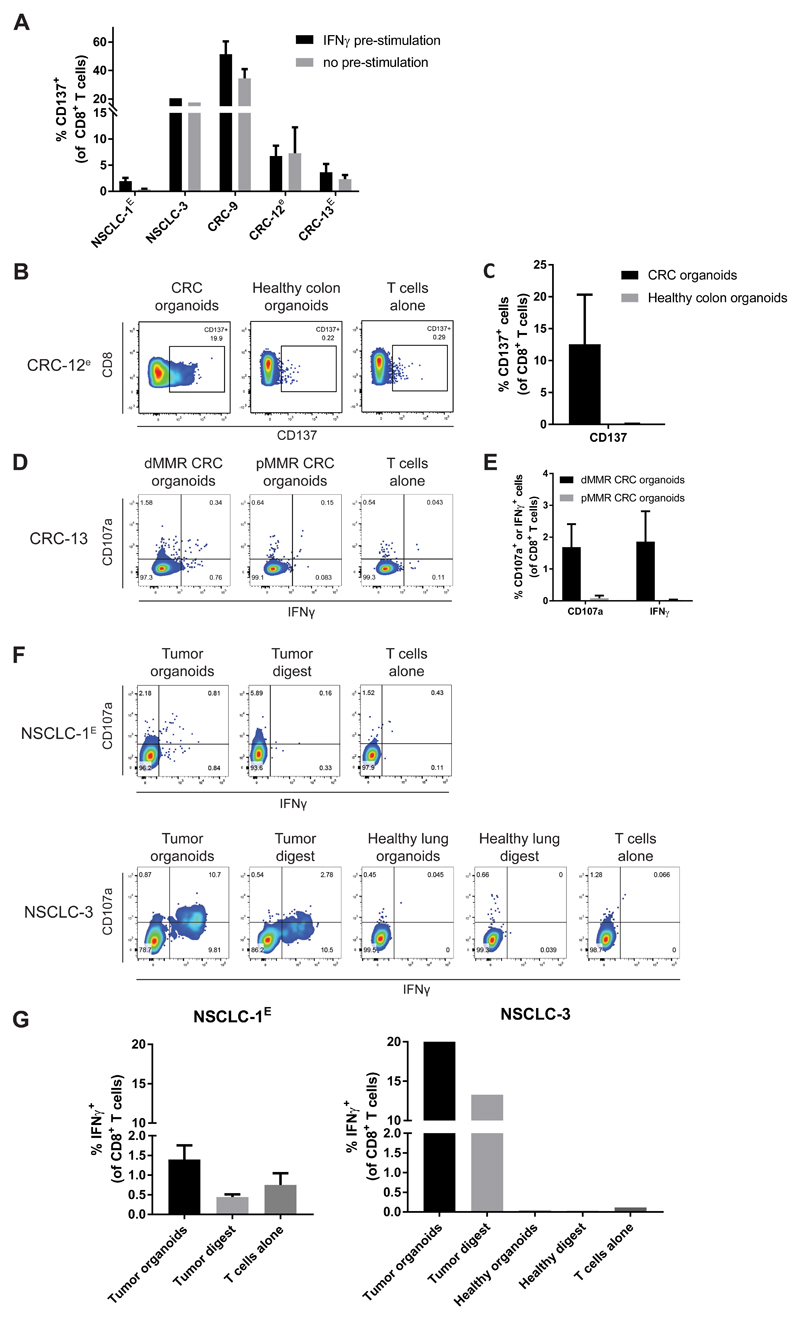
We next wished to evaluate whether the T cell responses induced by organoid co-culture are truly tumor-specific or should be considered artefacts of organoid culture or IFNγ treatment. Specifically, while IFNγ enhances antigen processing and presentation, it also induces expression of a large number of genes, potentially allowing the formation of T cell responses against (self) antigens not expressed in the absence of IFNγ. To evaluate the dependency of tumor organoid-reactive T cell responses on IFNγ, we compared T cell expression of the activation marker CD137 upon stimulation with tumor organoids that were either pre-stimulated with IFNγ or left untreated (Figure 4A). For 4 out of 5 cases tested, similar CD8+ T cell responses were induced toward tumor organoids regardless of IFNγ pre-stimulation. To more directly assess whether the induced T cell responses were specific for tumor antigens, we tested whether T cells also responded to stimulation with organoids of autologous normal tissue, and whether T cells responded to stimulation with autologous tumor digest. To assess restriction of T cell reactivity to the tumor organoid used for T cell induction, we established normal colon or lung organoids from two patients (CRC-12 and NSCLC-3). In addition, we generated organoids of a synchronous mismatch repair proficient (pMMR) CRC from patient CRC-13, from whom we also had obtained a dMMR CRC. In all cases, T cell reactivity was restricted to the tumor organoids to which reactivity had been induced in the two-week co-culture with PBL, i.e. no reactivity was observed against normal tissue or pMMR CRC organoids (Figure 4B-G).
Figure 4. Specificity of organoid-reactive T cell responses.
T cells were obtained by two week co-culture with autologous tumor organoids. To increase the number of T cells available for testing, where indicated (lower case e) tumor-reactive T cells were then expanded using a rapid expansion protocol (Dudley et al. 2013), in some cases preceded by CD137-based enrichment of tumor-reactive cells (indicated by upper case E).
(A) T cells were challenged with organoids pre-stimulated with 200 ng/mL IFNγ for 24 hours or with unstimulated organoids and evaluated for expression of the activation marker CD137. Background (percentage positive cells in T cells alone control) is subtracted from signal. Error bars indicate s.e.m. of at least 2 biological replicates.
(B) Representative flow cytometry plots of CD8+ T cells stimulated with either tumor organoids or healthy colon organoids.
(C) Quantification of CD137 expression by CD8+ T cells stimulated with either tumor organoids or healthy colon organoids. Background (percentage positive cells in T cells alone control) is subtracted from signal. Error bars indicate s.e.m. of 2 biological replicates. One of the replicates for T cells stimulated with tumor organoids is the same as data in (A).
(D) Representative flow cytometry plots of CD8+ T cells obtained by two weeks of co-culture with mismatch repair deficient (dMMR) CRC organoids that were re-stimulated with either dMMR CRC organoids or mismatch repair proficient (pMMR) CRC organoids.
(E) Quantification of organoid-induced IFNγ production and CD107a cell surface expression of CD8+ T cells obtained by two weeks of co-culture with mismatch repair deficient (dMMR) CRC organoids that were re-stimulated with either dMMR CRC organoids or mismatch repair proficient (pMMR) CRC organoids. Background (percentage positive cells in T cells alone control) is subtracted from signal. Error bars indicate s.e.m. of 4 biological replicates. Data for T cells stimulated with dMMR CRC organoids is the same as data in Figure 2A.
(F) Representative flow cytometry plots of CD8+ T cells stimulated with either tumor digest, normal lung digest, NSCLC organoids or healthy lung organoids.
(G) Quantification of IFNγ production of CD8+ T cells stimulated with either tumor digest, normal lung digest, NSCLC organoids or healthy lung organoids. Error bars represent s.d. of technical replicates.
For two patients, single cell digest of autologous tumor tissue was also available to stimulate organoid-reactive T cells (Figure 4F-G). T cells reactive to NSCLC-3 organoids were also reactive to tumor digest but not to normal lung digest. In contrast, T cells reactive to NSCLC-1 organoids did not produce IFNγ upon stimulation with autologous tumor digest. While the relatively low level of reactivity towards NSCLC-1 organoids limits the ability to detect subtle reactivity towards tumor digest, at this stage we cannot rule out that T cell reactivity in this sample is directed against organoid-specific targets. Taken together with the experiments in which we evaluated reactivity toward control organoids, this indicates that for 3 out of 4 tested samples, induced CD8+ T cell responses are tumor-specific. At the same time, it also highlights the value of generating organoids from non-malignant tissue that can serve as controls.
CD4+ T cell reactivity against xenogeneic tissue culture components
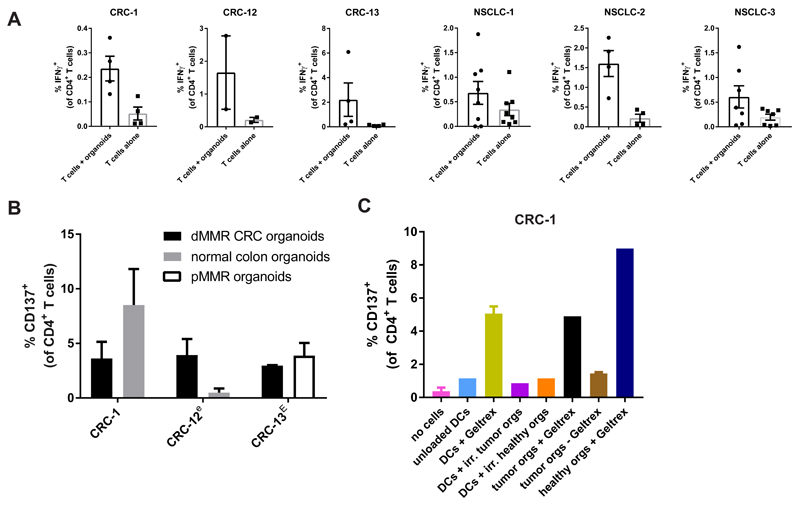
In addition to tumor-reactive CD8+ T cell responses, we occasionally observed variable CD4+ T cell responses upon stimulation with tumor organoids (Figure 5A). Notably, CD4+ T cell reactivity was not restricted to tumor organoids, but was in some cases also directed against control organoids (organoids of normal tissue or of the synchronous pMMR CRC from patient CRC-13) (Figure 5B). Since we only observed cross-reactivity to control organoids for CD4+ T cells and not for CD8+ T cells, we hypothesized this may be directed against foreign antigens derived from the extracellular environment. Organoids are cultured in murine basement membrane matrix (Geltrex), and consequently murine antigens could be presented to T cells. To address this hypothesis, we generated monocyte-derived dendritic cells (moDCs) from PBMC of patient CRC-1 and loaded these with Geltrex, or irradiated cells of tumor or healthy colon organoids. CD4+ T cell reactivity was only induced by stimulation with Geltrex-loaded DCs or organoids grown in Geltrex (Figure 5C). In addition, reactivity was largely abolished when organoids were grown in the absence of Geltrex for three days (Figure 5C and Figure S4). This indicates that co-culture systems involving Geltrex can induce CD4+ T cell reactivity that is not directed against tumor antigens. Of note, strategies to expand organoids in synthetic matrices have been described recently (Gjorevski et al., 2016) and could serve as alternatives to bypass reactivity to animal antigen.
Figure 5. CD4+ T cell reactivity against xenogeneic tissue culture components.
(A) Quantification of spontaneous and organoid-induced IFNγ production by CD4+ T cells obtained by two week co-culture with autologous tumor organoids. Error bars indicate s.e.m. of at least 2 biological replicates.
(B) Quantification of CD137 expression by CD4+ T cells upon re-stimulation with the tumor organoids used for induction, normal colon organoids, or organoids of a mismatch repair proficient (pMMR) CRC from the same patient. T cells were obtained by two week co-culture with autologous tumor organoids and where indicated (lower case e) further expanded using a rapid expansion protocol (Dudley et al. 2013), in some cases preceded by CD137-based enrichment of tumor-reactive cells (indicated by upper case E). Error bars indicate s.e.m. of at least 2 biological replicates.
(C) Quantification of CD137 expression by CD4+ T cells after stimulation with unloaded or Geltrex-loaded monocyte-derived dendritic cells (DCs), irradiated tumor cells, or organoids cultured with or without Geltrex. Error bars indicate s.d. of technical replicates.
See also Figure S4.
Tumor organoids are killed by autologous tumor-reactive T cells
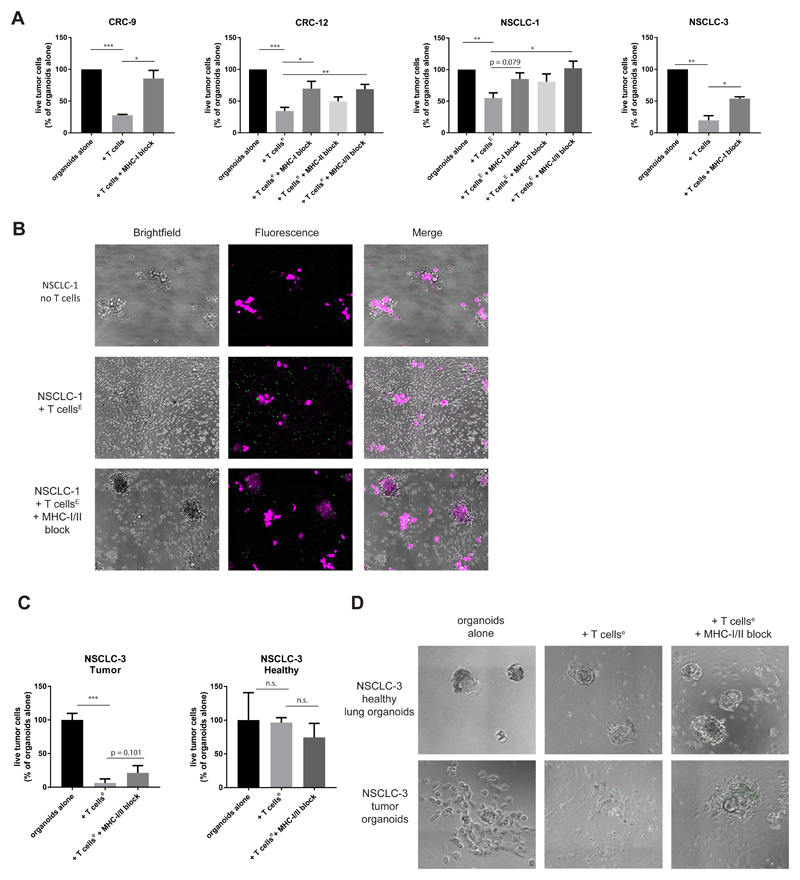
Having established the feasibility of inducing autologous tumor-reactive CD8+ T cell products for a substantial fraction of patients, we next aimed to determine whether organoid systems can be used to assess the efficiency of tumor destruction by these T cells. To determine this, we co-cultured tumor organoids with autologous tumor-reactive T cell populations for three days and quantified the number of live tumor cells by flow cytometry. For all samples tested, exposure of tumor organoids to autologous T cells led to substantially reduced survival (Figure 6A). Addition of MHC-I blocking antibodies rescued tumor cell survival, demonstrating the presence of an antigen-specific CD8+ T cell response. To visualize the cytolytic activity of tumor-reactive T cells, NSCLC-1 organoids were labelled with a tracer dye and imaged in the presence of a green-fluorescent apoptosis probe detecting active caspase 3/7 (“caspase 3/7 probe”) (Figure 6B). Addition of autologous T cells led to reduced organoid size and was accompanied by widespread apoptosis.
Figure 6. Tumor organoids are killed by autologous tumor-reactive T cells.
T cells were obtained by two week co-culture with autologous tumor organoids and where indicated (lower case e) further expanded using a rapid expansion protocol (Dudley et al. 2013), in some cases preceded by CD137-based enrichment of tumor-reactive cells (indicated by upper case E).
(A) Quantification of organoid killing upon T cell co-culture. After 3 days of co-culture, organoids were dissociated into single cells and the number of live cells was quantified using flow cytometry in the presence of counting beads. Where indicated, MHC-I or MHC-II was blocked with W6/32 or Tü39 antibody, respectively. * p < 0.05, ** p < 0.01, *** p < 0.001, Student’s t test. Error bars indicate s.e.m. of at least two biological replicates.
(B) Microphotographs of NSCLC-1 organoids 72 hours after culture with or without T cells in the presence of a green-fluorescent caspase 3/7 probe. Organoids were labelled with CellTrace Yellow (magenta) prior to co-culture. Note the appearance of apoptotic (green) cells upon addition of T cells. When MHC-I and MHC-II are blocked, T cells cluster around organoids but apoptosis is reduced and organoids remain larger in size.
(C) Quantification of killing of matched NSCLC and healthy lung organoids as in (A). Representative of two independent experiments. Error bars indicate s.d. of technical replicates.
(D) Microphotograph images of NSCLC-3 healthy and tumor organoids 72 hours after culture with or without T cells in the presence of a green-fluorescent caspase 3/7 probe.
See also Figure S5.
To provide further evidence for the specificity of tumor cell killing by autologous T cells, we performed parallel cytotoxicity assays with tumor and healthy organoids of NSCLC-3. T cells were first expanded using the rapid expansion protocol previously established to generate TIL products for adoptive cell therapy (Dudley et al., 2003). T cells efficiently killed tumor organoids, but did not affect survival of healthy organoids (Fig. 6C). Live imaging showed strong T-cell mediated killing of tumor organoids when incubated with autologous T cells (Figure 6D, Figure S5 and Video S1). Tumor organoids cultured without T cells (Video S2) or with T cells in the presence of blocking antibodies to MHC-I and MHC-II (Video S3) continued to proliferate. As a further control, healthy organoids continued proliferation regardless of the presence of T cells (Figure 6D, Figure S5 and Videos S4-6). Taken together, these data demonstrate that organoids of epithelial tumors can be used to measure the rate of destruction by autologous T cells. This platform should provide a valuable tool to compare e.g. the relative contribution of different cancer (neo-)antigens to T cell-mediated killing, to assess how the genetic state of tumor cells influences sensitivity to T cell attack, and to test the value of strategies to increase this sensitivity.
Discussion
The potential of patient-specific model systems for the implementation of personalized medicine in the field of targeted therapy is considered substantial (Horvath et al., 2016). We speculate that the value of patient-specific model systems will prove even higher in the field of immuno-oncology, given the inherent diversity in HLA and TCR genes, the private nature of the neo-antigens that are expressed in human cancers, and the multifactorial nature of T cell-mediated tumor destruction. Much of our current understanding of T cell recognition in human cancer has been based on work in melanoma, and robust systems to measure T cell – tumor interactions in a patient-specific manner have to our knowledge not been described for epithelial cancers. Here we provide proof of concept that tumor organoids can be used to establish individualized ex vivo model systems to support T cell-based therapies and to study the interactions between T cells and tumor cells.
We foresee two major applications for the platform that we describe. First, it will allow one to mechanistically dissect the pathways that determine tumor cell sensitivity and resistance to immunotherapy. Second, it opens the possibility to generate patient-specific T cell products in an unbiased manner. With respect to the former application, the ability to establish tumor organoid cultures from very limited amounts of tumor (such as needle biopsies), combined with the potential to expand circulating tumor-reactive T cells from peripheral blood, provides a minimally invasive means to interrogate tumor sensitivity to immunotherapy for individual patients at different time points during treatment. For patients who initially responded to treatment with immune checkpoint inhibitors but eventually relapse, the establishment of co-cultures based on paired (tumor and blood) biopsies before and after relapse should provide a unique assay system for the functional dissection of the underlying cause of relapse. Recent genetic studies on paired biopsies have demonstrated the power of such an approach in a small set of melanoma patients who relapsed while on anti-PD1 therapy, demonstrating the acquisition of JAK1/2 mutations as one cause of resistance (Zaretsky et al., 2016). It will be of interest to determine whether a similar reduced sensitivity to T cell attack by JAK1/2 mutations is selected for in epithelial cancers. In addition to the analysis of natural and therapy-induced variation in tumor cell sensitivity to T cell pressure, there is increasing interest in the use of small molecules and antibodies to enhance tumor cell sensitivity to T cell attack (Patel and Minn, 2018). Parallel tumor organoid – T cell cultures in the presence or absence of drugs of interest should form a straightforward system to identify suitable candidates for combination with immunotherapy.
With respect to the second potential application of this platform, the ability to expand circulating tumor-reactive T cells by co-culture with tumor organoids provides a clinically feasible strategy for the generation of patient-specific T cell products for adoptive T cell transfer. This approach bypasses the need for samples derived from resection specimens to isolate tumor-infiltrating lymphocytes. In our proof of concept study, tumor-reactive CD8+ T cells were expanded from the circulation in four out of thirteen dMMR CRC patients (31%) and two out of six NSCLC patients (33%). When considering only MHC-I proficient dMMR CRC patients, tumor-reactive T cells were induced in 50% of samples, highlighting the need for strategies to revert or bypass MHC-I loss as an immune escape mechanism. Larger sample sizes will be needed to more precisely define the success rate of this approach. Moreover, in this study we focused on tumor types with high mutational load, and it will be important to determine whether the feasibility of inducing tumor-reactive T cells can be extended to poorly immunogenic cancers, such as mismatch repair proficient colorectal cancer. In parallel, efforts to further increase the success rate of this approach could focus on strategies to improve selection of circulating tumor-reactive T cells, for example based on PD-1 expression by T cells (Gros et al., 2016).
STAR* Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human MLH1 | Roche | M1; Cat#6472966001 |
| Mouse anti-human MSH2 | Roche | G219-1129; Cat#5269270001 |
| Mouse anti-human MSH6 | Abcam | EP49; Cat#AC-0047 |
| Mouse anti-human PMS2 | Agilent Technologies | EP51; Cat# M3647 |
| Mouse anti-human CD28 | eBioscience | CD28.2; Cat#16-0289-81 |
| Mouse anti-human HLA-ABC (PE-conjugated) | BD | G46-2.6; Cat#555553 |
| Mouse anti-human CD274 (PD-L1) (APC-conjugated) | eBioscience | MIH1; Cat#17-5983-41 |
| Mouse IgG1 κ isotype control (PE-conjugated) | BD | MOPC-21; Cat#556650 |
| Mouse IgG1 κ isotype control (APC-conjugated) | eBioscience | P3.6.2.8.1; Cat#17-4714-41 |
| Mouse anti-human CD107a (PE-conjugated) | BD | H4A3; Cat#555801 |
| Mouse anti-human CD3 (PerCP-Cy5.5-conjugated) | eBioscience | SK7; Cat#332771 |
| Mouse anti-human CD4 (FITC-conjugated) | BD | RPA-T4; Cat#555346 |
| Mouse anti-human CD8 (BV421-conjugated) | BD | RPA-T8; Cat#562429 |
| Mouse anti-human IFNγ (APC-conjugated) | BD | B27; Cat#554702 |
| Mouse anti-human CD137 (APC-conjugated) | BD | 4B4-1; Cat#550890 |
| Mouse anti-human CD3 | eBioscience | OKT-3; Cat#16-0037-85 |
| Mouse anti-human HLA-ABC | The Netherlands Cancer Institute | W6/32 |
| Mouse anti-human HLA-DR/DP/DQ (sodium azide free) | BD | Tü39; Cat#555556 |
| Mouse anti-human CD3 (Alexa Fluor 700-conjugated) | Invitrogen | UCHT1; Cat#CD0329 |
| Mouse anti-human CD326 (EpCAM) (PE/Cy7-conjugated) | Biolegend | 9C4; Cat#324222 |
| Anti-human PD1 | Merus | N/A |
| Mouse anti-human HLA-ABC | Nordic Mubio | HCA2; Cat#MUB2036P |
| Mouse anti-human HLA-ABC | Nordic Mubio | HC10; Cat#MUB2037P |
| Mouse anti-human p53 | Agilent Technologies | DO-7 |
| Biological Samples | ||
| Patient tumor and normal colon or lung tissue | The Netherlands Cancer Institute | study NL48824.031.14 |
| Patient blood | The Netherlands Cancer Institute | study NL48824.031.14 |
| Healthy donor blood (buffy coat) | Sanquin blood bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| B27 supplement without vitamin A | Gibco | Cat#C12587-010 |
| B27 supplement | Gibco | Cat#17504-044 |
| N-Acetylcysteine | Sigma-Aldrich | Cat#A9165-5G |
| Nicotinamide | Sigma-Aldrich | Cat#N0636 |
| Human recombinant EGF | Peprotech | Cat#AF-100-15 |
| A83-01 | Tocris | Cat#2939 |
| SB202190 | Cayman Chemicals | Cat#10010399 |
| Prostaglandin E2 | Cayman Cehmicals | Cat#14010-1 |
| Y-27632 | Sigma-Aldrich | Cat#Y-0503 |
| Human recombinant FGF-7 | Peprotech | Cat#100-19 |
| Human recombinant FGF-10 | Peprotech | Cat#100-26 |
| Geltrex LDEV-free reduced growth factor basement membrane | Gibco | Cat#A1413202 |
| Collagenase type II | Sigma-Aldrich | Cat#C6885 |
| Hyaluronidase type IV | Sigma-Aldrich | Cat#H3506 |
| Advanced DMEM-F12 | Gibco | Cat#12634-028 |
| Penicillin/streptomycin | Gibco | Cat#15070063 |
| Ultraglutamine type I | Lonza | Cat#BE17-605E |
| HEPES | Gibco | Cat#15630-056 |
| TrypLE Express | Gibco | Cat#12604-013 |
| Recovery Cell Culture Freezing Medium | Gibco | Cat#12648-010 |
| Nutlin-3a | Cayman Chemicals | Cat#10004372 |
| RPMI 1640 | Gibco | Cat#11875093 |
| Human serum, from human male AB plasma | Sigma-Aldrich | Cat#H3667 |
| Benzonase | Merck | Cat#70746-3 |
| Human recombinant interferon gamma | Peprotech | Cat#300-02 |
| GolgiSTOP (Monensin) | BD | Cat#554724 |
| GolgiPLUG (Brefeldin A) | BD | Cat#555029 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#19-144 |
| Ionomycin | Sigma-Aldrich | Cat#I9657 |
| Dispase type II | Sigma-Aldrich | Cat#D4693 |
| Cell Recovery Solution | Corning | Cat#354253 |
| GMP DC medium | CellGenix | Cat#20801-0500 |
| Recombinant human GM-CSF | CellGenix | Cat#1012-050 |
| Recombinant human IL-1β | CellGenix | Cat#1011-050 |
| Recombinant human IL-4 | CellGenix | Cat#1003-050 |
| Recombinant human TNF-α | CellGenix | Cat#1006-050 |
| Recombinant human IL-6 | CellGenix | Cat#1004-060 |
| Interleukin-2 | Proleukin | |
| Prostaglandin E1 | Alprostadil | |
| Critical Commercial Assays | ||
| DNeasy blood and tissue kit | Qiagen | Cat#69504 |
| Fixation/Permeabilization solution kit | BD | Cat#554714 |
| Live/dead fixable near-IR dead cell stain kit | Invitrogen | Cat#L10119 |
| CD137 Microbead kit, human | Miltenyi | Cat#130-093-476 |
| CellTrace Yellow Cell Proliferation Kit | Invitrogen | Cat#C34567 |
| CellTrace Far Red Cell Proliferation Kit | Invitrogen | Cat#C34564 |
| AccuCount blank particles (7.0 – 7.9 μm) | Spherotech | Cat#ACBP-70-10 |
| NucView488 Caspase-3 assay kit | Biotium | Cat#30029 |
| Experimental Models: Cell Lines | ||
| L-Wnt3a cell line | Laboratory of Hans Clevers, Hubrecht Institute | L-Wnt3a |
| R-spondin producer cell line | Laboratory of Calvin Kuo, Stanford | 293T-HA-RspoI-Fc |
| Noggin producer cell line | Laboratory of Hans Clevers, Hubrecht Institute | HEK293-mNoggin-Fc |
| Software and Algorithms | ||
| Optitype | Szolek et al. 2014 | https://github.com/F RED-2/OptiType |
| Somatic Sniper | Larson et al. 2012 | http://gmt.genome.wustl.edu/packages/somatic-sniper/ |
| Somatic Indel Detector (GATK) | McKenna et al. 2010 | https://software.broadinstitute.org/gatk/ |
| snpEff, version 4.3 | Cingolani et al. 2012 | http://snpeff.sourceforge.net |
| FlowJo version 10 | https://www.flowjo.com/ | |
| ImageJ version 1.50i | Schneider et al. 2012 | https://imagej.nih.gov/ij/ |
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Emile E. Voest (e.voest@nki.nl).
Reagents obtained under material transfer agreement (MTA) include producer lines for conditioned medium of Wnt-3a, Noggin (MTA with Hubrecht Institute) and R-spondin-1 (MTA with Calvin Kuo, Stanford).
Patient material, including tissue and organoids, are available for sharing as defined in the signed informed consent (study NL48824.031.14), and as approved by the local Medical Ethical Committee. Deposition of DNA sequencing data in publicly available databases is regulated by the informed consent that participants to this study signed.
Experimental model and subject details
Human subjects
The study (NL48824.031.14) was approved by the Medical Ethical Committee of The Netherlands Cancer Institute – Antoni van Leeuwenhoek hospital and written informed consent was obtained from all patients. Peripheral blood and tumor tissue were obtained from patients with a confirmed diagnosis of colorectal or non-small cell lung cancer. Mismatch repair deficiency was confirmed by immunohistochemical staining for the mismatch repair proteins MSH2, MSH6, MLH1 and PMS2 in routine assessment by a pathologist (see Immunohistochemistry).
Organoid culture
Tumor tissue was obtained either by 18G core needle biopsy or by surgical resection. Tumor tissue was processed for organoid culture within 24 hours. CRC organoids were established essentially as described in van de Wetering et al. 2015. NSCLC organoids were cultured using similar methods; a detailed protocol is accessible at bioRxiv (Sachs et al., 2018). Briefly, tumor tissue derived from needle biopsies was mechanically dissociated into small tumor pieces using needles and embedded in Geltrex (Geltrex LDEV-free reduced growth factor basement membrane extract, Gibco). Tumor tissue derived from surgical resections was cut into small pieces and enzymatically digested using 1.5 mg/mL collagenase II (Sigma-Aldrich), 10 μg/mL hyaluronidase type IV (Sigma-Aldrich) and 10 μM Y-27632 (Sigma-Aldrich) before embedding in Geltrex. After Geltrex solidification for 20 minutes at 37 °C, cells were overlaid with human colorectal cancer (CRC) or non-small cell lung cancer (NSCLC) organoid medium (van de Wetering et al., 2015; Sachs et al., 2018). Human CRC organoids medium is composed of Ad-DF+++ (Advanced DMEM/F12 (Gibco) supplemented with 2 mM Ultraglutamine I (Lonza), 10 mM HEPES (Gibco), and 100/100 U/ml Pencillin/Streptomycin (Gibco)), 10% Noggin-conditioned medium, 20% R-spondin1-conditioned medium, 1x B27 supplement without vitamin A (Gibco), 1.25 mM N-Acetylcysteine (Sigma-Aldrich), 10 mM nicotinamide (Sigma-Aldrich), 50 ng/mL human recombinant EGF (Peprotech), 500 nM A83-01 (Tocris), 3 μM SB202190 (Cayman Chemicals) and 10 nM prostaglandin E2 (Cayman Chemicals). Human wild-type colon organoid medium is identical to CRC medium, except that 50% Wnt3a-conditioned medium is added. Human NSCLC and normal airway organoid medium is composed of Ad-DF+++, 10% Noggin-conditioned medium, 10% R-spondin1-conditioned medium, 1x B27 supplement, 1.25 mM N-Acetylcystein, 10 mM nicotinamide, 25 ng/mL human recombinant FGF-7 (Peprotech), 100 ng/mL human recombinant FGF-10 (Peprotech), 500 nM A83-01, 1 μM SB202190, 5 μM Y-27632. R-spondin1-conditioned medium was produced from 293T-HA-RspoI-Fc producer cell lines (obtained from C. Kuo, Stanford), Wnt3a-conditioned medium from L-Wnt3a cells and Noggin-conditioned medium from HEK293-mNoggin-Fc cell lines (both kind gift from J. den Hertog, Utrecht). In the first two weeks of organoid culture, 1x Primocin (Invivogen) was added to prevent microbial contamination. Organoids were passaged approximately every week by incubating in TrypLE Express (Gibco) for 5-10 minutes at 37 °C to dissociate organoids to single cells and replating in fresh Geltrex. After passaging, 10 μM Y-27632 was added to CRC medium for the first 2-3 days. Organoids were cryopreserved in 10%FCS/DMSO or Recovery Cell Culture Freezing Medium (ThermoFisher) as master and working biobanks. Organoids < passage 30 were used in experiments. To rule out overgrowth by healthy lung organoids for NSCLC organoids derived from intrapulmonal lesions, haematoxylin and eosin stained sections of tumor organoids (see Immunohistochemistry) were evaluated by a pathologist to determine tumor status of organoids. To prevent overgrowth by healthy lung organoids, for patient NSCLC-3, p53-mutant organoids were selected by culturing in the presence of 5 μM Nutlin-3 (Cayman Chemicals). Organoids were regularly checked for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza).
Peripheral blood lymphocytes
The peripheral blood mononuclear cells (PBMC) fraction was isolated from peripheral blood by Ficoll-Paque density gradient separation and cryopreserved until later use.
Organoid line authentication
DNA was isolated from master and working biobanks of organoids and patient-matched peripheral blood using a DNEasy kit (Qiagen). Samples were genotyped using a Taqman-based SNParray targeting 26 SNPs (Hartwig Medical Foundation, Amsterdam). An identity score was calculated as described by (Tanabe et al., 1999) and (Liang-Chu et al., 2015). When comparing two samples, for each locus where both samples had called alleles, the number of distinct alleles in each individual sample, as well as the number of shared alleles was computed. These were summed across all loci and an identity score was computed defined as
A threshold of 0.9 of the summed identity score was defined as a cut-off and organoid lines that did not match to autologous blood were discarded.
Two samples (NSCLC-3 and NSCLC-5), showed identity scores < 0.9 in comparison with autologous blood, but showed exclusively heterozygous to homozygous changes in the tumor, suggesting loss of heterozygosity due to copy number aberrations. We authenticated these lines by performing HLA-typing by PCR (Sanquin, Amsterdam) or based on whole genome sequencing data by Optitype (Szolek et al. 2014).
Method details
Organoid – lymphocyte co-culture
Culture media for PBMC was composed of RPMI 1640 (Gibco), supplemented with 2 mM Ultraglutamine I, 1:100 penicillin/streptomycin and 10% male human AB serum (Sigma-Aldrich) (“T cell medium”). One day before co-culture, PBMC were thawed in pre-warmed (37 °C) T cell medium (human serum was replaced with FCS during thawing) and incubated for 15 minutes with 25 U/mL benzonase (Merck). After washing, cells were resuspended at 2-3*106 cells/mL in T cell medium supplemented with 150 U/mL IL-2 and cultured overnight at 37 °C. Prior to co-culture, tumor organoids were stimulated overnight with 200 ng/mL human recombinant IFNγ (Peprotech). 96-well U-bottom plates were coated with 5 μg/mL anti-CD28 (clone CD28.2, eBioscience) and kept overnight at 4 °C.
The next day, tumor organoids were dissociated to single cells with TrypLE Express and resuspended in T cell medium. Anti-CD28-coated plates were washed twice with PBS and PBMC were seeded at a density of 105 cells/well and stimulated with single cell-dissociated organoids at a 20:1 effector:target ratio. Co-cultures were performed in the presence of 150 U/mL IL-2 and 20 μg/mL anti-PD-1-blocking antibody (kindly donated by Merus, Utrecht). Half of the medium, including IL-2 and anti-PD-1, was refreshed two to three times per week. Every week, PBMC were collected, counted and replated at 105 cells/well, and re-stimulated with fresh tumor organoids.
Flow cytometry
For evaluation of MHC-I and PD-L1 expression by tumor organoids, organoids were dissociated to single cells using TrypLE Express, with or without overnight pre-incubation with 200 ng/mL IFNγ. Tumor cells were washed in FACS buffer (PBS + 5 mM EDTA + 1% bovine serum antigen) and stained with mouse anti-human HLA-A,B,C-PE (BD Bioscience) or anti-CD274-APC (eBioscience) antibodies, or isotype controls (PE mouse IgG1, kappa (BD) and APC mouse IgG1 kappa (eBioscience)) for 30 minutes at 4 °C. Cells were washed twice with FACS buffer and DAPI was added to exclude dead cells prior to recording at a Becton Dickinson Fortessa or LSRII flow cytometer.
For evaluation of tumor reactivity, 105 PBMC were restimulated with tumor organoids at a 2:1 effector: target ratio and seeded in anti-CD28-coated plates in the presence of 20 μg/mL anti-PD-1 and co-cultured for 5 hours. Mouse anti-human CD107a-PE antibodies (BD) were added at the start of co-culture. Golgi-Plug (1:1000, BD) and Golgi-Stop (1:1500, BD) was added after 1 hour and co-culture continued for an additional 4 hours. Cells were washed twice in FACS buffer and stained with the following antibodies: anti-CD3-PerCP-Cy5.5 (BD), anti-CD4-FITC (BD), anti-CD8-BV421 (BD), and near-IR viability dye (Life technologies) for 30 minutes at 4 °C. Cells were washed twice in FACS buffer, fixed and stained for intracellular IFNγ (anti-IFNγ-APC, BD) using the Cytofix/Cytoperm kit (BD), according to manufacturer’s instructions. PBMC stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) and 1 μg/mL ionomycin (Sigma-Aldrich) served as positive controls and PBMC cultured without tumor stimulation as negative controls. In some experiments, PBMC and tumor cells were co-cultured for 24 hours before staining with anti-CD3-PerCP-Cy5.5, anti-CD4-FITC, anti-CD8-PB, anti-CD137-APC (BD) and near-IR viability dye.
Rapid expansion of unselected T cells or tumor-reactive sublines
T cells from patient CRC-12, NSCLC-1 and NSCLC-3 were co-cultured for two weeks with autologous tumor organoids and expanded for two weeks in the presence of irradiated (40 Gy, Gamma Cell-40) pooled PBMC from three healthy donors (1:200 T cell:feeder ratio), 3000 U/mL IL-2 and 30 ng/mL anti-human CD3 (OKT-3, eBioscience). After 5-7 days, medium, including IL-2, was refreshed every 2-3 days. For expansion of tumor-reactive sublines, T cells were sorted on the basis of CD137 expression 24 hours after stimulation with tumor organoids, using a CD137 Microbead Kit (Miltenyi) following manufacturer’s instructions and expanded as described above.
Organoid killing assays
To determine the sensitivity of tumor organoids to T cell-mediated killing, flat-bottom non-tissue culture-treated plates were coated with 5 μg/mL anti-CD28 and kept at 4 °C overnight prior to co-culture. Organoids were stimulated with 200 ng/mL IFNγ for 24h prior to co-culture. The next day, organoids were isolated from Geltrex by density gradient separation. Part of the organoids were dissociated to single cells and counted using a hemocytometer. This was used to infer the number of tumor cells per tumor organoid to allow co-culture of organoids and T cells at a 5:1 effector:target ratio. Next, tumor organoids were resuspended in T cell medium. T cells were collected after two weeks of co-culture with tumor organoids and resuspended in T cell medium. Anti-CD28-coated plates were washed twice with PBS and organoids were seeded in triplicate without T cells or with 5*104 autologous T cells obtained by two weeks of organoid co-culture. To block MHC class I and II, organoids were pre-incubated for 30 minutes with 50 μg/mL pan-MHC-I blocking antibody W6/32, or pan-MHC-II blocking antibody Tü39 (blocking antibody remained present throughout the co-culture). To facilitate visualization, organoids were in some cases stained with 1 μM of CellTrace Yellow (Invitrogen) and T cells with 100 nM CellTrace FarRed (Invitrogen) in PBS for 20 minutes at 37 °C followed by blocking with human serum and washing in PBS. At the start of co-culture, a green-fluorescent caspase 3/7 probe that binds DNA upon cleavage by caspase 3/7 (referred to as “caspase 3/7 probe”) (Biotium) was added at 1:2000 dilution to visualize cells undergoing apoptosis.
After 3 days of co-culture, organoids and PBMC were collected and dissociated into single cells with TrypLE Express, combined with regular resuspension and vortexing. Care was taken to limit the reaction until the moment organoids were fully dissociated. 7.46 μm AccuCount blank counting beads (Spherotech) were added to each well, cells were washed in FACS buffer and stained with anti-CD3-AF700 (Invitrogen) and anti-CD326-PE-Cy7 (Biolegend) for 30 minutes at 4 °C. Organoids or T cells labelled with CellTrace dyes were not stained with antibodies. Cells were washed twice and stained with DAPI to mark dead cells prior to flow cytometric recording.
Immunohistochemistry
Organoids were recovered from Geltrex by density gradient separation (centrifugation at 300g, 5 minutes, 4 °C in cold basal organoid culture medium) when they reached an approximate diameter of 100 μm, fixed in 4% paraformaldehyde for 30 minutes at room temperature or overnight at 4 °C, pelleted, and embedded in paraffin blocks. In some cases, organoids were isolated from Geltrex using 10 mg/mL dispase type II (Sigma) or Cell Recovery Solution (Corning). Immunohistochemistry of samples was performed on a BenchMark Ultra autostainer (Ventana Medical Systems). Briefly, paraffin sections were cut at 3 μm, heated at 75°C for 28 minutes and deparaffinized in the instrument with EZ prep solution (Ventana Medical Systems). Heat-induced antigen retrieval was carried out using Cell Conditioning 1 (CC1, Ventana Medical Systems) for 32 minutes at 95°C. P53 was detected using clone DO-7 (1:7000 dilution, 32 minutes at 37°C, Agilent Technologies), and MHC-I using clone HCA2 (1:5000 dilution, 32 minutes at 37°C, Nordic Mubio) and clone HC10 (1:20000 dilution, 32 minutes at 37°C, Nordic Mubio). For HCA2 signal amplification was applied using the Optiview Amplification Kit (4 minutes, Ventana Medical Systems). Bound antibody was detected using the OptiView DAB Detection Kit (Ventana Medical Systems). Slides were counterstained with Hematoxylin and Bluing Reagent (Ventana Medical Systems). For assessment of mismatch repair status, immunohistochemistry was performed according to standard protocols for the Ventana automated immunostainer. (MLH1, clone M1, Roche; MSH2, clone G219-1129, Roche; MSH6, clone EP49, Abcam; PMS2, clone EP51, Agilent Technologies).
Whole exome DNA sequencing
At least 200 ng of genomic DNA was extracted from CRC organoids and patient-matched PBMC using a DNEasy kit (Qiagen). After exome capture (Integrated DNA Technologies probe set), whole-exome sequencing was performed on an Illumina HiSeq DNA analyzer using 100 bp paired-end reads.
T cell recognition assays of Geltrex
Tumor or healthy organoids were isolated from Geltrex using 10 mg/mL dispase type II (Sigma) and cultured in organoid medium for three days with 10 μM Y-27632 (Cayman Chemicals) in 6-well plates. In some cases, two days later, 100 μL/well Geltrex was added to organoids. After three days, organoids were used in tumor recognition assays as described under flow cytometry.
Monocytes were sorted on the basis of CD14 on a MoFlo Astrios flow cytometer and cultured in GMP DC medium (CellGenix) with 800 U/mL GM-CSF and 400 U/mL IL-4 (CellGenix). Five days after differentiation, DCs were matured with GMC-CSF, IL-4, IL-6 (1 ng/mL), IL-1β (1 ng/mL), TNFα (1 ng/mL) and PGE1 (0.5 μg/mL). DCs were seeded at 2*104 cells / well and loaded with Geltrex or 40 Gy dissociated and irradiated tumor or healthy organoid cells (2*104 cells/well). Two days later, 1*105 T cells were added to each well and cultured for 24h. T cells were assayed for CD137 expression as described under flow cytometry.
Quantification and statistical analysis
Whole exome DNA sequencing data
Reads were aligned to a reference genome (GRCh38). Somatic single nucleotide variations and indels were called using Somatic Sniper (Larson et al., 2012) and SomaticIndelDetector (GATK; McKenna et al., 2010), respectively. Variants were annotated using snpEff (Cingolani et al., 2012), version 4.3. Extra-exonic and synonymous variants were removed from further analyses. Mutational load was defined as the number of non-synonymous mutations per tumor exome. The occurrence of mutations in genes recurrently mutated in hypermutated CRC was based on the top 15 most frequently recurring significantly mutated genes according to TCGA 2012.
Live imaging
During organoid killing assays, cells were co-cultured for 3 days and imaged using a charge coupled device (CCD) camera equipped with a Zeiss AxioCam MRm camera fitted to a Zeiss Axio Observer Z1 inverted microscope. Tile scans were performed every 20 to 30 minutes followed by tile stitching after recording using Zen software. Videos were generated from the stitched tiles or regions of interest after background subtraction using ImageJ software version 1.50i (Schneider et al., 2012).
Flow cytometry
Flow cytometry data was analyzed using FlowJo version 10.
Statistical analysis
Data was analyzed using GraphPad Prism version 7. Group sizes and definition of error bars is indicated in figure legends. In indicated bar graphs, background is subtracted from signal and negative values set to zero. Statistical analysis was performed using a two-tailed Student’s t test. P values <0.05 were considered significant; significance values are indicated as * (P < 0.05), ** (P < 0.01) and *** (P < 0.001).
Data and software availability
Whole exome DNA sequencing data is available in Table S1.
Supplementary Material
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. MHC-I and MHC-II were blocked with W6/32 and Tü39 antibodies, respectively. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids cultured without T cells, in the presence of a green-fluorescent caspase 3/7 probe. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. Note that organoids are unaffected by presence of T cells. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. MHC-I and MHC-II were blocked with W6/32 and Tü39 antibodies, respectively. Note absence of killing and continued proliferation of tumor cells. Video duration: 3 days
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids cultured without T cells in the presence of a green-fluorescent caspase 3/7 probe. Note proliferation of tumor cells that spread out onto the plate toward the end of the assay. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids exposed to autologous T cells obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. Note the destruction of tumor organoids by surrounding T cells and appearance of apoptotic cells visualized by a green-fluorescent caspase 3/7 probe. Video duration: 3 days.
DNA isolated from mismatch repair deficient colorectal cancer organoids and matched PBMCs was analyzed by whole exome sequencing.
Acknowledgements
We thank Suzanne van der Kolk, Judith Westra, Louisa Hoes and Luuk Schipper for help in patient inclusion. We acknowledge Martijn van Baalen, Anita Pfauth and Frank van Diepen for assistance in flow cytometry. We thank Marjolijn Mertz, Lenny Brocks and Bram van den Broek for help in live imaging experiments. We would like to acknowledge Dennis Peters and Ingrid Hofland from the NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI-AVL Biobank material and lab support. DNA sequencing was performed by Marja Nieuwland, Roel Kluin and Ron Kerkhoven. We thank Marc van de Wetering and Hubrecht Organoid Technology (HUB) for advice on organoid culture. We would like to thank Merus for provision of anti-PD-1. Thanks to Marije Marsman, Steven van Houtvin, Hylke Galema, and Koen Verhoef for support in technology transfer. Stef van Lieshout, Ewart de Bruijn and Edwin Cuppen (Hartwig Medical Foundation) performed authentication of organoid lines. Thanks to Salo Ooft, Chelsea McLean, Christina Stangl, Louisa Hoes, Luuk Schipper, Maarten Ligtenberg, David Vredevoogd, Julia Boshuijzen, Wouter Scheper, Joost van den Berg, Renate Groot, Christian Blank and Daniel Peeper for input in the design of experiments.
This work was supported by the NWO gravitation program (NWO 2012-2022) (to E.V. on behalf of CancerGenomics.nl), KWF grant HUBR2014-7006 (to E.V.), the KWF Queen Wilhelmina Award (NKI 2013-6122, to T.S.) and ERC AdG SENSIT (to T.S.).
Footnotes
Author contributions
K.D. designed, performed and analysed experiments and wrote the manuscript. C.C. designed, performed and analysed experiments. F.W. designed the clinical protocol and F.W., M.C., D.V., M.L, A.D., E.S., J.H. and K.H. organized patient inclusion. L.F., S.K. and N.R. designed co-culture experiments. J.V.H., L.F. and M.S. analysed DNA sequencing data. S.K. cultured tumor organoids. M.W. and R.G. provided material for and designed reactivity tests against tumor digest. N.S. and H.C. provided protocols for organoid culture. P.S. and K.M. analysed histology data. T.S. and E.V. supervised the study.
Declaration of interests
H.C. is inventor on several patents related to organoid technology.
N.S. reports grants from The Netherlands Organisation for Scientific Research, during the conduct of the study; other from Vertex Pharmaceuticals Incorporated, outside the submitted work; In addition, N.S. has a patent PCT/EP2015/077990 with royalties paid to Stichting HUB, and a patent PCT/EP2015/077988 with royalties paid to Stichting HUB.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børessen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016;352:658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brea EJ, Oh CY, Manchado E, Budh S, Gejman RS, Mo G, Mondello P, Han JE, Jarvis CA, Ulmert D, et al. Kinase regulation of human MHC class I molecule expression on cancer cells. Cancer Immunol Res. 2016;4:936–947. doi: 10.1158/2326-6066.CIR-16-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Ie L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangles-Marie V, Pocard M, Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N, Validire P, et al. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- Dierssen JW, de Miranda NF, Ferrone S, van Puijenbroek M, Cornelisse CJ, Fleuren GJ, van Wezel T, Morreau H. HNPCC versus sporadic microsatellite-unstable colon cancers follow different routes toward loss of HLA class I expression. BMC Cancer. 2007;7:33–43. doi: 10.1186/1471-2407-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Voabil P, Schumacher TN, Voest EE. Genomics- and transcriptomics-based patient selection for cancer treatment with immune checkpoint inhibitors. JAMA Oncology. 2016;2:1490–1495. doi: 10.1001/jamaoncol.2016.2214. [DOI] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–342. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bagina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, Subrmanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. 2018;48:417–433. doi: 10.1016/j.immuni.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D, Montoya MC, Östling P, Pietiäinen V, Price LS, et al. Screening out irrelevant cell-based models of disease. Nature Rev Drug Discov. 2016;15:751–769. doi: 10.1038/nrd.2016.175. [DOI] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardir ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;28:11–13. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang-Chu MMY, Yu M, Haverty PM, Koeman J, Ziegle J, Lee M, Bourgon R, Neve RM. Human biosample authentication using the high-throughput, cost-effective SNPTrace™ system. PLoS One. 2015;10:e0116218. doi: 10.1371/journal.pone.0116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Noshiko K, Irahara N, Meyerhardt JA, Baba Y, Shima K, Glickman JN, Ferrone CR, Mino-Kenudson M, Tanaka N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (Check Mate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, van Cutsem E, McDermott R, Hill A, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- Pitt J, Vétizou M, Daillére R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, et al. Resistance mechanisms to immune checkpoint blockade in cancer: tumor-intrinsic and –extrinsic factors. Immunity. 2016;44:1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Ritter C, Fan K, Paschen A, Reker Hardrup S, Ferrone S, Nghiem P, Ugurel S, Schrama D, Becker JC. Epigenetic priming restores the HLA class-I antigen presenting machinery expression in Merkel cell carcinoma. Sci Reports. 2017;7:2290–2300. doi: 10.1038/s41598-017-02608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, Zomer-van Ommen DD, Papaspyropoulos A, Heo I, Bottinger L, Klay D, Weeber F, Huelsz-Prince G, Lakobachvili N, Viveen MC, et al. Long-term expanding human airway organoids for disease modeling. bioRxiv. 2018 May 9; doi: 10.1101/318444. 318444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KIW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo J, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic S, Draper LM, Langhan MM, Campbell TE, Kwong ML, Wunderlich JR, Dudley ME, Yang JC, Sherry RM, Kammula US, et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J Clin Oncol. 2015;10:1543–1550. doi: 10.1200/JCO.2014.58.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. Bioinformatics. 2014;1:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–338. [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdegaal EM, Visser M, Ramwadhdoebé TH, van der Minne CE, van Steijn JA, Kapiteijn E, Haanen JB, van der Burg SH, Nortier JW, Osanto S. Successful treatment of metastatic melanoma by adoptive transfer of blood-derived polyclonal tumor-specific CD4+ and CD8+ T cells in combination with low-dose interferon-alpha. Cancer Immunol Immunother. 2011;60:953–963. doi: 10.1007/s00262-011-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van der Velden DL, Peeper DS, Cuppen EP, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad U S A. 2015;112:13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. Prospective derivation of a living biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Sun YH, Ye XL, Chen HQ, Ji HB. Establishment and characterization of primary lung cancer cell lines from Chinese population. Acta Pharmacologica Sinica. 2011;32:385–392. doi: 10.1038/aps.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. MHC-I and MHC-II were blocked with W6/32 and Tü39 antibodies, respectively. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids cultured without T cells, in the presence of a green-fluorescent caspase 3/7 probe. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 healthy lung organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. Note that organoids are unaffected by presence of T cells. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids exposed to autologous T cells, in the presence of a green-fluorescent caspase 3/7 probe. T cells were obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. MHC-I and MHC-II were blocked with W6/32 and Tü39 antibodies, respectively. Note absence of killing and continued proliferation of tumor cells. Video duration: 3 days
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids cultured without T cells in the presence of a green-fluorescent caspase 3/7 probe. Note proliferation of tumor cells that spread out onto the plate toward the end of the assay. Video duration: 3 days.
Time-lapse bright-field and fluorescence microscopy of NSCLC-3 tumor organoids exposed to autologous T cells obtained by two weeks of co-culture with autologous tumor organoids followed by rapid expansion. Note the destruction of tumor organoids by surrounding T cells and appearance of apoptotic cells visualized by a green-fluorescent caspase 3/7 probe. Video duration: 3 days.
DNA isolated from mismatch repair deficient colorectal cancer organoids and matched PBMCs was analyzed by whole exome sequencing.
Data Availability Statement
Whole exome DNA sequencing data is available in Table S1.