Abstract
Inflammatory bowel disease (IBD) is a chronic and heterogeneous intestinal inflammatory disorder. The medical management of IBD aims for long-lasting disease remission to prevent complications and disease progression. Early introduction of immunosuppression forms the mainstay of medical IBD management. Large inter-individual variability in drug responses, in terms of both efficacy and toxicity, leads to high rates of therapeutic failure in the management of IBD. Better patient stratification is needed to maximize patient benefit and minimize the harm caused by adverse events. Pre-treatment pharmacogenetic testing has the potential to optimize drug selection and dose, and to minimize harm caused by adverse drug reactions. In addition, optimizing the use of cheap conventional drugs, and avoiding expensive ineffective drugs, will lead to a significant reduction in costs. Genetic variation in both TPMT and NUDT15, genes involved in thiopurine metabolism, is associated to an increased risk of thiopurine-induced myelosuppression. Moreover, specific HLA haplotypes confer risk to thiopurine-induced pancreatitis and to immunogenicity to tumor necrosis factor-antagonists, respectively. Falling costs and increased availability of genetic tests allow for the incorporation of pre-treatment genetic tests into clinical IBD management guidelines. In this paper, we review clinically useful pharmacogenetic associations for individualized treatment of patients with IBD and discuss the path from identification of a predictive pharmacogenetic marker to implementation into IBD clinical care.
Keywords: Inflammatory bowel disease, Crohn’s disease, Ulcerative colitis, Pharmacogenetics, Personalized medicine
Core tip: In recent years, strong pharmacogenetic associations for drugs used in the management of inflammatory bowel disease (IBD) have been identified. However, the implementation of pre-treatment pharmacogenetic testing into clinical guidelines has been challenging. Particular groups of patients are needlessly exposed to (expensive) drugs that are either ineffective or harmful. Pre-treatment screening for TPMT and NUDT15 genetic variation should be incorporated into clinical IBD management guidelines. Therapeutic recommendations based on HLA genetic variants, conferring risk for thiopurine-induced pancreatitis and immunogenicity to tumor necrosis factor-antagonists, respectively, should be further evaluated.
INTRODUCTION
Inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis, is a chronic inflammatory disorder of the gastrointestinal tract. The therapeutic armamentarium for IBD has rapidly expanded over the last two decades. Beyond conventional therapies such as amino-salicylates, corticosteroids and thiopurines, the development of biological therapies has revolutionized the management of IBD. Although all of these therapies are effective, it is well known that the inter-individual variability in therapy response is high with respect to both efficacy and toxicity. In consequence, particular groups of patients are needlessly exposed to (expensive) drugs that are either ineffective or harmful. This in turn causes a therapeutic delay and is associated with increased morbidity and costs. Ultimately, therapeutic failure may necessitate surgical treatment[1].
Personalized management in patients with IBD
To optimize patient outcomes and better utilize resources, it is crucial to be able to appropriately select the correct initial therapy for each patient; that means we need to determine the predictive factors of beneficial response without adverse events to therapy. In fact, many physicians have been practicing personalized medicine with their IBD patients for many years. It has been demonstrated, for example, that young age, isolated colonic Crohn’s disease and elevated C-reactive protein levels at the initiation of therapy are variables that favor a short-term response to infliximab[2,3]. Conversely, smokers are less likely to respond than nonsmokers, and those with disease duration longer than 2 years are less likely to respond than those with a shorter disease duration[4,5]. These clinical factors, however, explain only a small proportion of inter-individual variability in therapy response.
Pharmacogenetics
Genetic variation can affect individual responses to drugs in terms of both therapeutic effects and adverse effects. In fact, pre-treatment genetic testing has already been implemented in clinical practice outside the management of IBD. In metastatic colorectal cancer patients, for example, pre-treatment screening for mutations in the KRAS gene is recommended to predict the efficacy of anti-EGFR-targeted drugs such as cetuximab[6]. Likewise, the human leukocyte antigen (HLA) class I allele HLA-B*57:01 has been shown to be a major determinant of hypersensitivity to abacavir, a drug used in the management of HIV. In Europe, the United States and Australia, HLA-B*57:01 testing is now mandatory before prescribing abacavir[7].
PHARMACOGENETICS IN THE MANAGEMENT OF IBD
Within the context of IBD, implementation of pharmacogenetic (PGx) testing has the potential to maximize patient benefit by optimizing drug selection and dose and minimize harm caused by adverse events, as several PGx associations have been identified for drugs used in the management of IBD. This will lead to a significant reduction in costs by several means. First of all, the use of relatively safe and cheap conventional therapies can be optimized. Secondly, the prescription of expensive, potentially ineffective or harmful drugs can be avoided. Finally, achieving optimal dosing as early as possible will prevent morbidity during a dose-finding period.
In this opinion article, we focus on robust PGx associations identified in well-characterized cohorts of patients treated for IBD. Unfortunately, many PGx associations proposed in small retrospective candidate-gene studies have failed replication in independent cohorts[8]. Therefore, we will only consider genome-wide significant associations that have been replicated in independent cohorts (Table 1). Next, we will discuss challenges and future perspectives associated with the im-plementation of PGx testing in clinical care of IBD.
Table 1.
Pharmacogenetic associations in inflammatory bowel disease
| Class of drugs | Response | Gene | Clinical utility |
| 5-Aminosalicylates1 | Nephrotoxicity | HLA-DRB1 | No (low incidence, high allele frequency) |
| Thiopurines | Myelosuppression | TPMT NUDT15 | Yes |
| Alopecia | NUDT15 | Yes | |
| Pancreatitis | HLA-DQA1-HLA-DRB1 | Yes | |
| TNF-antagonists | Immunogenicity | HLA-DQA1 | Yes |
No attempt has been made to replicate this association in an independent cohort because of the scarcity of cases. Gene refers to the genes within genetic variants that have been associated to specific drug responses. HLA: Human leukocyte antigen; TPMT: Thiopurine S-methyltransferase; NUDT15: Nudix hydrolase 15.
Aminosalicylates
5-Aminosalicylates (5-ASA), such as mesalazine and olsalazine, are the most frequently prescribed class of drugs to induce and maintain remission in patients with mild to moderately active ulcerative colitis[9]. 5-ASA treatment is considered safe, cheap and effective treatment to achieve long-term sustained steroid-free remission in patients with mild to moderately active ulcerative colitis. Common side effects associated with 5-ASA include flatulence, abdominal pain, nausea, diarrhea, headache, dyspepsia and nasopharyngitis, which may occur in up to 10% of patients using these drugs[9]. Rare adverse events include, among others, pancreatitis (1%) and nephrotoxicity (approximately 0.2%). While 5-ASA induced nephrotoxicity is rare, 70% of these patients will develop irreversible renal injury and 10% require permanent renal replacement therapy[10]. A recent genome-wide association study identified HLA-DRB1*03:01 as a determinant of 5-ASA-induced nephrotoxicity[10]. Carriership of the risk allele is associated with a 3-fold increased risk of renal injury after 5-ASA administration. However, the high frequency of this risk allele and the low frequency of the adverse event limits its clinical utility, and it is therefore currently not recommended to consider it in guiding treatment choice or monitoring intervals.
Thiopurines
Thiopurines, consisting of azathioprine and its analogues 6-mercaptopurine and 6-thioguanine, are the most commonly prescribed immunosuppressive agents used to maintain corticosteroid-free remission and prevent postoperative recurrence in patients with IBD[11]. Although thiopurines are an effective and cheap therapeutic option for maintenance of remission, its use is limited by commonly occurring adverse events. It has been estimated that 17% of European patients with IBD using thiopurines develop adverse events that necessitate drug withdrawal[12]. Thiopurine adverse events can be divided into dose-independent events, like pancreatitis and flu-like illness, and dose-dependent events, such as myelosuppression and hepatotoxicity. Although the standard doses of thiopurines in Asian countries are lower than in Europe, the incidence of dose-dependent adverse events is much higher in Asian populations than in Europeans[13,14]. Genetic polymorphisms, both in the HLA region and in genes encoding enzymes involved in thiopurine metabolism, have been identified as important determinants of adverse events. Large international initiatives to identify genetic variants associated to the other common thiopurine adverse events, such as flu-like illness and hepatotoxicity, are currently ongoing (e.g., the UK IBD PRED4 and Helmsley IBD Exome studies)[15,16].
Thiopurine-induced myelosuppression
Thiopurine-induced myelosuppression (TIM) occurs in 4% of European individuals and in up to 15% of individuals of Asian descent[12,14]. TIM usually occurs within a few weeks of starting the drug but can happen at any time during the treatment course. Most patients are asymptomatic, but serious opportunistic infection may occur and there is an estimated mortality of 1%[17]. There is substantial evidence linking thiopurine S-methyltransferase (TPMT) and nudix hydrolase 15 (NUDT15) enzyme activity to TIM[18,19]. TPMT is a well-known enzyme that inactivates azathioprine and 6-mercaptopurine through methylation, leaving less parent drug available for eventual anabolism to cytotoxic thioguanine metabolites. Genetic variants in the TPMT gene may lead to reduced or absent TPMT enzyme activity levels, which in turn leads to high levels of cytotoxic thioguanine metabolites[20]. NUDT15 catalyzes the conversion of cytotoxic thioguanine metabolites to non-toxic thioguanine metabolites. Genetic variants in the NUDT15 gene lead to deficient NUDT15 enzyme activity levels, which also results in excessive levels of cytotoxic thioguanine[21]. Cytotoxic thioguanine metabolites contribute to the therapeutic effects of thiopurines but may lead to myelosuppression.
TPMT and NUDT15 act independently, and the likelihood of an individual having deficient enzyme activities depends upon allele frequencies within the population. Approximately 10% of individuals of European descent carry TPMT genetic variants. Three TPMT genetic variants account for 90% of the TPMT deficiency in Europeans, which together explain 25% of TIM[22,23]. In contrast, only 3% of Asians carry TPMT genetic variants despite a higher incidence of TIM in Asians populations[14,24]. This difference is largely explained by common genetic variation in NUDT15 in Asian populations, which has been identified as a strong genetic determinant of TIM[25]. The most common NUDT15 genetic variant, R139C, is present in approximately 10% of individuals of Asian descent[25]. More recent studies have identified additional NUDT15 genetic variants predictive for TIM that are also present outside Asian populations[26,27]. Although rare, these variants are associated with very large effect sizes. For example, a recently identified rare NUDT15 genetic variant, present in approximately 2% of individuals of European descent, is associated with a 38-fold increased risk of TIM[27].
Patients with loss-of-function TPMT and NUDT15 genetic variants are at excessive risk of TIM if they receive standard thiopurine dosing. Genetic variation within TPMT or NUDT15 is defined by so-called star (*) alleles. Each star allele is defined by the genotype at one or more loci within the gene, and these star alleles are used for the annotation of enzyme activity levels of TPMT and NUDT15, respectively. These annotations can then be used for dosing recommendations based on pre-treatment genotyping. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has recently published detailed dosing recommendations based on TPMT and NUDT15 genotypes[28]. In short, reduced starting doses (30%-80% of target dose) should be considered for TPMT or NUDT15 intermediate metabolizers, while substantially reduced doses (10% of target dose) or the use of an alternative agent should be used for TPMT or NUDT15 poor metabolizers[28].
Thiopurine-induced pancreatitis
Thiopurine-induced pancreatitis (TIP) complicates thiopurine therapy in about 4% of patients exposed[12]. TIP usually occurs within the first few weeks after initiation of therapy. The pathogenesis of this potentially life-threatening, idiosyncratic adverse event remains poorly understood. Genetic variants in HLA-DQA1-HLA-DRB1 have recently been identified as strong genetic determinants of TIP in European patients with IBD[29,30]. Carriership of a HLA-DQA1*02:01-HLA-DRB1*07:01 haplotype is associated with a 2.5-fold increased risk of TIP[29]. Given the risk allele frequency of 27% in European populations, homozygous patients would be subject to a risk of approximately 17% of developing TIP[29]. Development of TIP is independent of the thiopurine dose administered. To avoid administration of thiopurines to patients with IBD who are homozygous at the HLA-DQA1-HLA-DRB1 haplotype, clinical utility estimates indicate that 76 patients need to be genotyped to prevent one case of TIP[29]. Although these data show the potential of pre-treatment HLA-DQA1-HLA-DRB1 genotyping, it has not yet been incorporated in clinical treatment protocols.
Thiopurine-induced alopecia
Thiopurine-induced alopecia (TIA) is a well-recognized, dose-dependent, adverse event in Asian populations, with an incidence around 1.5%[14,31]. In contrast, this adverse event is rare in individuals of European ancestry[12]. Although not life-threatening, severe alopecia may cause cosmetic problems requiring a long recovery. Small case-control studies reported co-occurrence of severe TIA with severe TIM in patients homozygous for NUDT15 R139C[32,33]. A recent genome-wide association study in Japanese patients with IBD has shown that carrying the NUDT15 R139C risk allele is associated with a 10-fold increased risk of TIA[13]. Avoidance of thiopurines in patients homozygous for NUDT15 R139 to reduce the risk of TIM would similarly reduce the risk of TIA.
TNF-antagonists
Tumor necrosis factor (TNF)-antagonists, mainly infliximab and adalimumab, are the most commonly prescribed biologicals in the management of IBD[34]. Biological therapy has transformed the management of IBD, and has become the largest contribution to IBD healthcare expenditure[35]. Despite their established efficacy, up to a third of patients with IBD will have no response at all to these agents (primary non-response)[36]. An additional third of patients will eventually fail TNF-antagonist therapy after initial response (loss-of-response)[36]. Small hypothesis-driven studies into genetic determinants of primary non-response have reported conflicting data or have been underpowered to identify genetic variants worth translating into clinical practice[8,37].
Immunogenicity of TNF-antagonists
The formation of anti-drug antibodies, referred to as immunogenicity, is the most important cause of the loss-of-response and hypersensitivity reactions that often lead to treatment failure[38]. Although concomitant use of immunosuppressive therapy reduces immunogenicity, up to 65% of patients treated with infliximab and 38% of patients treated with adalimumab will eventually develop anti-drug antibodies[38]. Moreover, many patients are still treated with TNF-antagonist monotherapy because of adverse events, infections and possibly malignancies associated with im-munosuppressive therapy[39,40]. Recently, the HLA-DQA1*05 haplotype was identified as a genetic determinant of immunogenicity to TNF-antagonists. Carriership of the HLA-DQA1*05 haplotype is associated with a 2-fold increased risk of immunogenicity to TNF-antagonists, regardless of the type of drug used [i.e., infliximab (Remicade, CT-P13) or adalimumab][41]. Pre-treatment HLA-DQA1*05 genetic testing has not yet been considered for clinical implementation. TNF-antagonist therapy is a major driver of IBD healthcare expenditure, and im-munogenicity often leads to therapeutic failure and adverse events. Pre-treatment HLA-DQA1*05 genetic testing thus has the potential to personalize TNF-antagonist therapy and allow targeted use of concomitant immunosuppressive therapy to minimize risk and maximize response[41].
From identification to implementation
Patients with IBD need life-long adherence to drugs to maintain disease remission, improve quality of life and reduce the need for surgery. Although the outcomes of medical therapies have greatly improved over the last decades, substantial inter-individual variability remains in terms of both efficacy and toxicity. Many patients with IBD do not achieve disease remission, lose response after initial successful treatment, or develop severe drug-induced adverse events. Indeed, IBD experts will agree that there is no such thing as “one-size-fits-all“ in the management of IBD. However, there is still an unmet need for patient stratification to guide this personalized care.
Several genetic markers have been identified as strong determinants of (adverse) response to drugs used in the management of IBD. However, the uptake of routine pre-treatment genetic testing to better stratify patients with IBD has been slow. For example, the association between TPMT genotypes and severe TIM was established nearly three decades ago[18]. Although the American Gastroenterological Association and the British Society of Gastroenterology now recommend routine TPMT testing (enzymatic activity or genotype) prior to initiating thiopurine therapy, the European Crohn’s and Colitis Organisation guidelines still do not[42-44]. Likewise, TPMT screening is used in IBD clinical practice on a limited scale in many European countries. No other PGx tests are currently implemented in IBD clinical care.
This slow uptake of pre-treatment genetic testing in the management of IBD is a reflection of the challenges associated with the path from identification of a genetic marker to clinical implementation. First, idiosyncratic adverse events are often notoriously difficult to characterize due to the small number of cases available to individual researchers. Recent efforts led by the United Kindom IBD Genetics Consortium have successfully conducted both prospective (e.g., PANTS) and retrospective (e.g., PRED4) PGx studies in the context of IBD management[15,29,41,45] .However, additional large international consortia are needed to facilitate the collection of rigorously characterized cohorts of patients who develop (rare) adverse events. Stringent phenotype definitions should be established to prevent heterogeneity, which significantly reduces the power to find consistent associations[46]. Then, the clinical validity of a PGx test has to be established. Clinical validity refers to the performance of the test, such as the discriminative ability and predictive value[47]. Key to this is replication in independent cohorts. Failure to replicate a PGx association makes it difficult to establish clinical validity estimates. The implementation of a pre-treatment PGx test into clinical care, however, is generally motivated by more than the discriminative ability of a test. Clinical utility takes assessment of the test a step further and focuses on the likelihood that the test concerned will lead to an improved health outcome[48,49]. Therefore, the clinical utility of a given test is dependent upon parameters such as whether or not an effective alternative therapy is available and the relative risk of the predicted outcome (i.e., response or adverse event). The development of novel therapies, although more expensive, provides alternatives for patients with IBD genetically at risk for adverse events. This adds to the clinical utility of pre-treatment PGx testing in IBD care. Finally, the incidence of the predicted outcome and prevalence of the risk allele may differ between different ethnic populations, and both affect the clinical utility of a given test within a specific population. In the context of IBD, for example, the incidence TIM is much higher in Asian populations compared to European populations, which can be largely attributed to trans-ethnic differences in TPMT and NUDT15 risk allele frequencies[24]. This renders the clinical utility of PGx tests before the start of thiopurine treatment higher in Asian populations than in European populations.
Like most diagnostic or therapeutic interventions, clinical implementation of pre-treatment PGx tests has traditionally demanded a randomized controlled trial (RCT) to assess its clinical utility and cost-effectiveness. RCTs are, however, very expensive, require large sample sizes and often fail to produce consistent results. Primary outcomes of RCTs are often averaged across the entire population per study arm. This is, of course, the exact opposite of personalized medicine, where a particular treatment is not chosen based on the “average patient”, but on characteristics of an individual, for example, a genetic profile. A good example in the context of IBD is a recently performed RCT to determine whether pre-treatment TPMT genotyping would affect outcomes of patients with IBD[50]. In this study, pre-treatment TPMT genotyping did not reduce overall incidence of TIM in the intervention group. However, there was a 10-fold reduction in TIM in TPMT variant carriers who received dose reduction, compared with variant carriers who did not. While 609 patients with IBD were enrolled in this study, post-hoc analysis showed that 42,556 patients should have been enrolled to show a reduction in TIM for the entire intervention group. It is unlikely that there will be much interest from industry and academia to conduct (and fund) these large clinical trials for drugs already approved. Indeed, decisions to use PGx tests will probably need to be based on other types of evidence, including case-control studies, cohort studies and retrospective analyses of data[51].
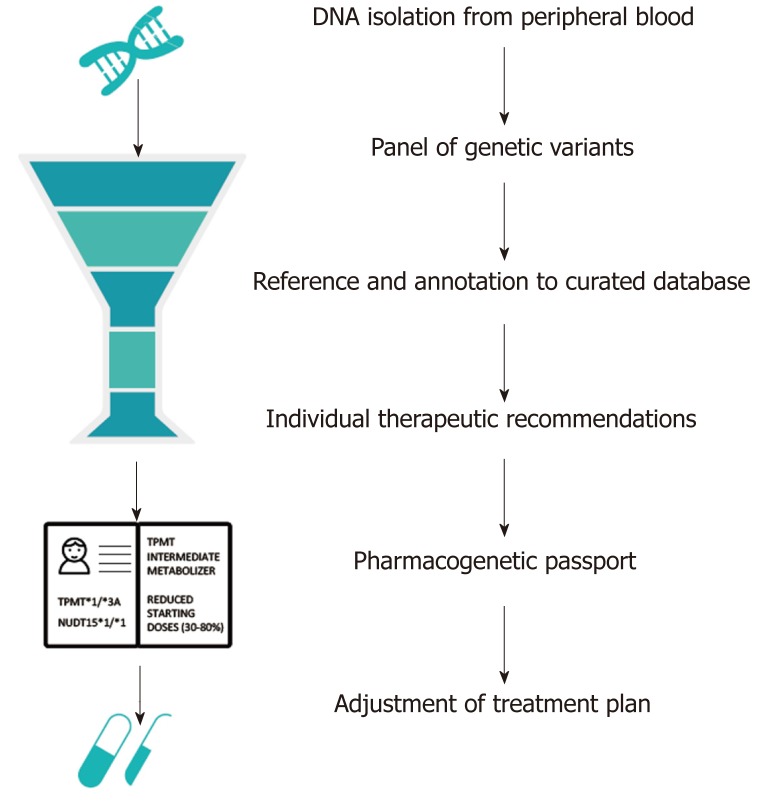
Greater availability and falling costs of genetic testing means that increasingly the question is not whether to genotype, but how to best utilize genetic data. Future studies should focus on the cost-effectiveness of different means of genotyping for different ethnic populations. Targeted sequencing of NUDT15 and TPMT would cost approximately $300 per patient, while genome-wide genotyping arrays are available for less than $40 per patient[52]. Custom genotyping arrays could be designed that include known pharmacogenetic variants, extensive coverage of areas with a high likelihood of pharmacogenetic associations and medium coverage of the rest of the genome. Clinical genetic testing requires accredited pipelines with a genetic assay of high accuracy, consistent quality of data analysis and interpretation, and a short overall turnaround time. These pipelines take genetic data as input, functionally annotate the genetic data via curated databases such as PharmGKB, and translate these data into individual therapeutic recommendations for all well-known gene-drug pairs, including TPMT and NUDT15[53]. These individual reports are referred to as pharmacogenetic passports (Figure 1). Once a patient has been genotyped, novel PGx associations could be automatically added via continuous annotation to curated databases. Worldwide, many PGx implementation programs have already been initiated[54]. The use of a custom genotyping array, in contrast to sequencing, limits opportunities for the identification of novel PGx associations to a degree, but makes the required quality control, quick data interpretation and global registration of known PGx associations more easily achievable.
Figure 1.
Example of an automated computational pipeline that creates an individual pharmacogenetic passport based on an individual’s genotype. In this case, the individual is a heterozygous carrier of the loss-of-function TPMT*3A allele and homozygous carrier of the NUDT15*1 allele. Heterozygous carriers of TPMT*3A are at risk for thiopurine-induced myelosuppression due to intermediate TPMT enzyme activity levels. Hence, a reduced dose (30%-80% of target dose) is strongly recommended. Patients with a NUDT15*1/*1 genotype are considered as NUDT15 normal metabolizers[28]. TPMT: Thiopurine S-methyltransferase; NUDT: Nudix hydrolase.
CONCLUSION
We have discussed several clinically useful genetic determinants of response to IBD drugs. Given the enormous potential it has for the care of patients with IBD, pre-treatment genetic tests should be incorporated into clinical IBD management guidelines. Detailed thiopurine-dosing recommendations based on TPMT and NUDT15 genotypes, provided by CPIC, can be easily adopted. Follow-up studies are required to assess how to translate genetic variation in HLA into therapeutic recommendations regarding the risk of TIP and ITA, respectively. It is likely that future pharmacogenetic studies will identify more predictive genetic variants. For clinical implementation of pre-treatment genetic tests, we suggest a custom genotyping array that can be easily deployed across centers of secondary care around the globe and adjusted if more predictive variants are identified.
ACKNOWLEDGEMENTS
This article was edited for language and formatting by Kate Mc Intyre, Scientific Editor in the Department of Genetics, University Medical Center Groningen.
Footnotes
Conflict-of-interest statement: R.K.W. received unrestricted research grants from Takeda, Tramedico and Ferring. E.A.M.F. received an unrestricted research grant from Takeda. The remaining authors disclose no conflicts.
Peer-review started: January 29, 2019
First decision: March 27, 2019
Article in press: April 20, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: Netherlands
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Massironi S, Ozen H S-Editor: Yan JP L-Editor: A E-Editor: Ma YJ
Contributor Information
Michiel Dirk Voskuil, Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands. m.d.voskuil@umcg.nl; Department of Genetics, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands.
Amber Bangma, Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands; Department of Genetics, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands.
Rinse Karel Weersma, Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands.
Eleonora Anna Margaretha Festen, Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands; Department of Genetics, University of Groningen and University Medical Center Groningen, Groningen 9713 GZ, the Netherlands.
References
- 1.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Louis E, Vermeire S, Rutgeerts P, De Vos M, Van Gossum A, Pescatore P, Fiasse R, Pelckmans P, Reynaert H, D'Haens G, Malaise M, Belaiche J. A positive response to infliximab in Crohn disease: Association with a higher systemic inflammation before treatment but not with -308 TNF gene polymorphism. Scand J Gastroenterol. 2002;37:818–824. [PubMed] [Google Scholar]
- 3.Vermeire S, Louis E, Carbonez A, Van Assche G, Noman M, Belaiche J, De Vos M, Van Gossum A, Pescatore P, Fiasse R, Pelckmans P, Reynaert H, D'Haens G, Rutgeerts P Belgian Group of Infliximab Expanded Access Program in Crohn's Disease. Demographic and clinical parameters influencing the short-term outcome of anti-tumor necrosis factor (infliximab) treatment in Crohn's disease. Am J Gastroenterol. 2002;97:2357–2363. doi: 10.1111/j.1572-0241.2002.05991.x. [DOI] [PubMed] [Google Scholar]
- 4.Parsi MA, Achkar JP, Richardson S, Katz J, Hammel JP, Lashner BA, Brzezinski A. Predictors of response to infliximab in patients with Crohn's disease. Gastroenterology. 2002;123:707–713. doi: 10.1053/gast.2002.35390. [DOI] [PubMed] [Google Scholar]
- 5.Arnott ID, McNeill G, Satsangi J. An analysis of factors influencing short-term and sustained response to infliximab treatment for Crohn's disease. Aliment Pharmacol Ther. 2003;17:1451–1457. doi: 10.1046/j.1365-2036.2003.01574.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Yun YM, Chae HJ, Cho HJ, Ji M, Kim IS, Wee KA, Lee W, Song SH, Woo HI, Lee SY, Chun S. Clinical Pharmacogenetic Testing and Application: Laboratory Medicine Clinical Practice Guidelines. Ann Lab Med. 2017;37:180–193. doi: 10.3343/alm.2017.37.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jägel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A PREDICT-1 Study Team. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 8.Bek S, Nielsen JV, Bojesen AB, Franke A, Bank S, Vogel U, Andersen V. Systematic review: Genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;44:554–567. doi: 10.1111/apt.13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Parker CE, Feagan BG, MacDonald JK. Oral 5-aminosalicylic acid for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2016:CD000544. doi: 10.1002/14651858.CD000544.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heap GA, So K, Weedon M, Edney N, Bewshea C, Singh A, Annese V, Beckly J, Buurman D, Chaudhary R, Cole AT, Cooper SC, Creed T, Cummings F, de Boer NK, D'Inca R, D'Souza R, Daneshmend TK, Delaney M, Dhar A, Direkze N, Dunckley P, Gaya DR, Gearry R, Gore S, Halfvarson J, Hart A, Hawkey CJ, Hoentjen F, Iqbal T, Irving P, Lal S, Lawrence I, Lees CW, Lockett M, Mann S, Mansfield J, Mowat C, Mulgrew CJ, Muller F, Murray C, Oram R, Orchard T, Parkes M, Phillips R, Pollok R, Radford-Smith G, Sebastian S, Sen S, Shirazi T, Silverberg M, Solomon L, Sturniolo GC, Thomas M, Tremelling M, Tsianos EV, Watts D, Weaver S, Weersma RK, Wesley E, Holden A, Ahmad T. Clinical Features and HLA Association of 5-Aminosalicylate (5-ASA)-induced Nephrotoxicity in Inflammatory Bowel Disease. J Crohns Colitis. 2016;10:149–158. doi: 10.1093/ecco-jcc/jjv219. [DOI] [PubMed] [Google Scholar]
- 11.Chande N, Patton PH, Tsoulis DJ, Thomas BS, MacDonald JK. Azathioprine or 6-mercaptopurine for maintenance of remission in Crohn's disease. Cochrane Database Syst Rev. 2015:CD000067. doi: 10.1002/14651858.CD000067.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaparro M, Ordás I, Cabré E, Garcia-Sanchez V, Bastida G, Peñalva M, Gomollón F, García-Planella E, Merino O, Gutiérrez A, Esteve M, Márquez L, Garcia-Sepulcre M, Hinojosa J, Vera I, Muñoz F, Mendoza JL, Cabriada JL, Montoro MA, Barreiro-de Acosta M, Ceña G, Saro C, Aldeguer X, Barrio J, Maté J, Gisbert JP. Safety of thiopurine therapy in inflammatory bowel disease: Long-term follow-up study of 3931 patients. Inflamm Bowel Dis. 2013;19:1404–1410. doi: 10.1097/MIB.0b013e318281f28f. [DOI] [PubMed] [Google Scholar]
- 13.Kakuta Y, Kawai Y, Okamoto D, Takagawa T, Ikeya K, Sakuraba H, Nishida A, Nakagawa S, Miura M, Toyonaga T, Onodera K, Shinozaki M, Ishiguro Y, Mizuno S, Takahara M, Yanai S, Hokari R, Nakagawa T, Araki H, Motoya S, Naito T, Moroi R, Shiga H, Endo K, Kobayashi T, Naganuma M, Hiraoka S, Matsumoto T, Nakamura S, Nakase H, Hisamatsu T, Sasaki M, Hanai H, Andoh A, Nagasaki M, Kinouchi Y, Shimosegawa T, Masamune A, Suzuki Y MENDEL study group. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: A multicenter study. J Gastroenterol. 2018;53:1065–1078. doi: 10.1007/s00535-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Y, Mao R, Zhang SH, Li MY, Guo J, Chen BL, He Y, Zeng ZR, Chen MH. Safety Profile of Thiopurines in Crohn Disease: Analysis of 893 Patient-Years Follow-Up in a Southern China Cohort. Medicine (Baltimore) 2015;94:e1513. doi: 10.1097/MD.0000000000001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Predicting Serious Drug Side Effects in Gastroenterology - PRED4 studies [cited January 20 2019] Available from: https://www.ibdresearch.co.uk/pred4/
- 16.Helmsley IBD Exome Sequencing Pogram [cited January 20 2019] Available from: https://sites.google.com/a/broadinstitute.org/helmsley-ibd-exome-sequencing-program-hiesp?pli=1authuser=2.
- 17.Gisbert JP, Gomollón F. Thiopurine-induced myelotoxicity in patients with inflammatory bowel disease: A review. Am J Gastroenterol. 2008;103:1783–1800. doi: 10.1111/j.1572-0241.2008.01848.x. [DOI] [PubMed] [Google Scholar]
- 18.Lennard L, Van Loon JA, Weinshilboum RM. Pharmacogenetics of acute azathioprine toxicity: Relationship to thiopurine methyltransferase genetic polymorphism. Clin Pharmacol Ther. 1989;46:149–154. doi: 10.1038/clpt.1989.119. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Meng Y, Wang L, Liu Z, Li J, Dong W. Associations between the <i>NUDT15</i> R139C polymorphism and susceptibility to thiopurine-induced leukopenia in Asians: A meta-analysis. Onco Targets Ther. 2018;11:8309–8317. doi: 10.2147/OTT.S177007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: Catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 21.Moriyama T, Nishii R, Perez-Andreu V, Yang W, Klussmann FA, Zhao X, Lin TN, Hoshitsuki K, Nersting J, Kihira K, Hofmann U, Komada Y, Kato M, McCorkle R, Li L, Koh K, Najera CR, Kham SK, Isobe T, Chen Z, Chiew EK, Bhojwani D, Jeffries C, Lu Y, Schwab M, Inaba H, Pui CH, Relling MV, Manabe A, Hori H, Schmiegelow K, Yeoh AE, Evans WE, Yang JJ. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48:367–373. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, Zanger UM, Schwab M. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407–417. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- 23.Yates CR, Krynetski EY, Loennechen T, Fessing MY, Tai HL, Pui CH, Relling MV, Evans WE. Molecular diagnosis of thiopurine S-methyltransferase deficiency: Genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 24.CPIC guidelines for thiopurines and TPMT and NUDT15 [cited January 17 2019] Available from: https://cpicPGx.org/guidelines/guideline-for-thiopurines-and-tpmt.
- 25.Yang SK, Hong M, Baek J, Choi H, Zhao W, Jung Y, Haritunians T, Ye BD, Kim KJ, Park SH, Park SK, Yang DH, Dubinsky M, Lee I, McGovern DP, Liu J, Song K. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46:1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriyama T, Yang YL, Nishii R, Ariffin H, Liu C, Lin TN, Yang W, Lin DT, Yu CH, Kham S, Pui CH, Evans WE, Jeha S, Relling MV, Yeoh AE, Yang JJ. Novel variants in NUDT15 and thiopurine intolerance in children with acute lymphoblastic leukemia from diverse ancestry. Blood. 2017;130:1209–1212. doi: 10.1182/blood-2017-05-782383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker GJ, Harrison JW, Heap GA, Voskuil MD, Andersen V, Anderson CA, Ananthakrishnan AN, Barrett JC, Beaugerie L, Bewshea CM, Cole AT, Cummings FR, Daly MJ, Ellul P, Fedorak RN, Festen EAM, Florin TH, Gaya DR, Halfvarson J, Hart AL, Heerasing NM, Hendy P, Irving PM, Jones SE, Koskela J, Lindsay JO, Mansfield JC, McGovern D, Parkes M, Pollok RCG, Ramakrishnan S, Rampton DS, Rivas MA, Russell RK, Schultz M, Sebastian S, Seksik P, Singh A, So K, Sokol H, Subramaniam K, Todd A, Annese V, Weersma RK, Xavier R, Ward R, Weedon MN, Goodhand JR, Kennedy NA, Ahmad T IBD Pharmacogenetics Study Group. Association of Genetic Variants in NUDT15 With Thiopurine-Induced Myelosuppression in Patients With Inflammatory Bowel Disease. JAMA. 2019;321:773–785. doi: 10.1001/jama.2019.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Relling MV, Schwab M, Whirl-Carrillo M, Suarez-Kurtz G, Pui CH, Stein CM, Moyer AM, Evans WE, Klein TE, Antillon-Klussmann FG, Caudle KE, Kato M, Yeoh AEJ, Schmiegelow K, Yang JJ. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther. 2018 doi: 10.1002/cpt.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heap GA, Weedon MN, Bewshea CM, Singh A, Chen M, Satchwell JB, Vivian JP, So K, Dubois PC, Andrews JM, Annese V, Bampton P, Barnardo M, Bell S, Cole A, Connor SJ, Creed T, Cummings FR, D'Amato M, Daneshmend TK, Fedorak RN, Florin TH, Gaya DR, Greig E, Halfvarson J, Hart A, Irving PM, Jones G, Karban A, Lawrance IC, Lee JC, Lees C, Lev-Tzion R, Lindsay JO, Mansfield J, Mawdsley J, Mazhar Z, Parkes M, Parnell K, Orchard TR, Radford-Smith G, Russell RK, Reffitt D, Satsangi J, Silverberg MS, Sturniolo GC, Tremelling M, Tsianos EV, van Heel DA, Walsh A, Watermeyer G, Weersma RK, Zeissig S, Rossjohn J, Holden AL International Serious Adverse Events Consortium; IBD Pharmacogenetics Study Group, Ahmad T. HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat Genet. 2014;46:1131–1134. doi: 10.1038/ng.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson A, Jansen LE, Rose RV, Gregor JC, Ponich T, Chande N, Khanna R, Yan B, Jairath V, Khanna N, Sey M, Beaton M, McIntosh K, Teft WA, Kim RB. HLA-DQA1-HLA-DRB1 polymorphism is a major predictor of azathioprine-induced pancreatitis in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2018;47:615–620. doi: 10.1111/apt.14483. [DOI] [PubMed] [Google Scholar]
- 31.Takatsu N, Matsui T, Murakami Y, Ishihara H, Hisabe T, Nagahama T, Maki S, Beppu T, Takaki Y, Hirai F, Yao K. Adverse reactions to azathioprine cannot be predicted by thiopurine S-methyltransferase genotype in Japanese patients with inflammatory bowel disease. J Gastroenterol Hepatol. 2009;24:1258–1264. doi: 10.1111/j.1440-1746.2009.05917.x. [DOI] [PubMed] [Google Scholar]
- 32.Kakuta Y, Naito T, Onodera M, Kuroha M, Kimura T, Shiga H, Endo K, Negoro K, Kinouchi Y, Shimosegawa T. NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. Pharmacogenomics J. 2016;16:280–285. doi: 10.1038/tpj.2015.43. [DOI] [PubMed] [Google Scholar]
- 33.Lee YJ, Hwang EH, Park JH, Shin JH, Kang B, Kim SY. NUDT15 variant is the most common variant associated with thiopurine-induced early leukopenia and alopecia in Korean pediatric patients with Crohn's disease. Eur J Gastroenterol Hepatol. 2016;28:475–478. doi: 10.1097/MEG.0000000000000564. [DOI] [PubMed] [Google Scholar]
- 34.Chan HC, Ng SC. Emerging biologics in inflammatory bowel disease. J Gastroenterol. 2017;52:141–150. doi: 10.1007/s00535-016-1283-0. [DOI] [PubMed] [Google Scholar]
- 35.van der Valk ME, Mangen MJ, Leenders M, Dijkstra G, van Bodegraven AA, Fidder HH, de Jong DJ, Pierik M, van der Woude CJ, Romberg-Camps MJ, Clemens CH, Jansen JM, Mahmmod N, van de Meeberg PC, van der Meulen-de Jong AE, Ponsioen CY, Bolwerk CJ, Vermeijden JR, Siersema PD, van Oijen MG, Oldenburg B COIN study group and the Dutch Initiative on Crohn and Colitis. Healthcare costs of inflammatory bowel disease have shifted from hospitalisation and surgery towards anti-TNFα therapy: Results from the COIN study. Gut. 2014;63:72–79. doi: 10.1136/gutjnl-2012-303376. [DOI] [PubMed] [Google Scholar]
- 36.Allez M, Karmiris K, Louis E, Van Assche G, Ben-Horin S, Klein A, Van der Woude J, Baert F, Eliakim R, Katsanos K, Brynskov J, Steinwurz F, Danese S, Vermeire S, Teillaud JL, Lémann M, Chowers Y. Report of the ECCO pathogenesis workshop on anti-TNF therapy failures in inflammatory bowel diseases: Definitions, frequency and pharmacological aspects. J Crohns Colitis. 2010;4:355–366. doi: 10.1016/j.crohns.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland A, Power RJ, Rahman P, O'Rielly DD. Pharmacogenetics and pharmacogenomics in psoriasis treatment: Current challenges and future prospects. Expert Opin Drug Metab Toxicol. 2016;12:923–935. doi: 10.1080/17425255.2016.1194394. [DOI] [PubMed] [Google Scholar]
- 38.Vermeire S, Gils A, Accossato P, Lula S, Marren A. Immunogenicity of biologics in inflammatory bowel disease. Therap Adv Gastroenterol. 2018;11:1756283X17750355. doi: 10.1177/1756283X17750355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lichtenstein GR, Rutgeerts P, Sandborn WJ, Sands BE, Diamond RH, Blank M, Montello J, Tang L, Cornillie F, Colombel JF. A pooled analysis of infections, malignancy, and mortality in infliximab- and immunomodulator-treated adult patients with inflammatory bowel disease. Am J Gastroenterol. 2012;107:1051–1063. doi: 10.1038/ajg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osterman MT, Sandborn WJ, Colombel JF, Robinson AM, Lau W, Huang B, Pollack PF, Thakkar RB, Lewis JD. Increased risk of malignancy with adalimumab combination therapy, compared with monotherapy, for Crohn's disease. Gastroenterology. 2014;146:941–949. doi: 10.1053/j.gastro.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Sazonovs A, Kennedy N, Moutsianas L, Heap GA, Rice DL, Reppell M, Bewshea C, Walker GJ, Perry M, McDonald TJ, Lees C, Cummings F, Parkes M, Mansfield J, Barrett JC, McGovern D, Goodhand J, Anderson CA, Ahmad T PANTS consortium. HLA-DQA1*05 is associated with the development of antibodies to anti-TNF therapy 2018; Preprint Available from: bioRxiv. [Google Scholar]
- 42.Feuerstein JD, Nguyen GC, Kupfer SS, Falck-Ytter Y, Singh S American Gastroenterological Association Institute Clinical Guidelines Committee. American Gastroenterological Association Institute Guideline on Therapeutic Drug Monitoring in Inflammatory Bowel Disease. Gastroenterology. 2017;153:827–834. doi: 10.1053/j.gastro.2017.07.032. [DOI] [PubMed] [Google Scholar]
- 43.Mowat C, Cole A, Windsor A, Ahmad T, Arnott I, Driscoll R, Mitton S, Orchard T, Rutter M, Younge L, Lees C, Ho GT, Satsangi J, Bloom S IBD Section of the British Society of Gastroenterology. Guidelines for the management of inflammatory bowel disease in adults. Gut. 2011;60:571–607. doi: 10.1136/gut.2010.224154. [DOI] [PubMed] [Google Scholar]
- 44.Gomollón F, Dignass A, Annese V, Tilg H, Van Assche G, Lindsay JO, Peyrin-Biroulet L, Cullen GJ, Daperno M, Kucharzik T, Rieder F, Almer S, Armuzzi A, Harbord M, Langhorst J, Sans M, Chowers Y, Fiorino G, Juillerat P, Mantzaris GJ, Rizzello F, Vavricka S, Gionchetti P ECCO. 3rd European Evidence-based Consensus on the Diagnosis and Management of Crohn's Disease 2016: Part 1: Diagnosis and Medical Management. J Crohns Colitis. 2017;11:3–25. doi: 10.1093/ecco-jcc/jjw168. [DOI] [PubMed] [Google Scholar]
- 45.Personalised Anti-TNF Therapy in Crohn's disease - PANTS trial [cited January 20 2019] Available from: https://www.ibdresearch.co.uk/pants/
- 46.Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci U S A. 2008;105:617–622. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burke W. Genetic tests: Clinical validity and clinical utility. Curr Protoc Hum Genet. 2014;81:9.15.1–9.15.8. doi: 10.1002/0471142905.hg0915s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jansen ME, Rigter T, Rodenburg W, Fleur TMC, Houwink EJF, Weda M, Cornel MC. Review of the Reported Measures of Clinical Validity and Clinical Utility as Arguments for the Implementation of Pharmacogenetic Testing: A Case Study of Statin-Induced Muscle Toxicity. Front Pharmacol. 2017;8:555. doi: 10.3389/fphar.2017.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapoor R, Tan-Koi WC, Teo YY. Role of pharmacogenetics in public health and clinical health care: A SWOT analysis. Eur J Hum Genet. 2016;24:1651–1657. doi: 10.1038/ejhg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coenen MJ, de Jong DJ, van Marrewijk CJ, Derijks LJ, Vermeulen SH, Wong DR, Klungel OH, Verbeek AL, Hooymans PM, Peters WH, te Morsche RH, Newman WG, Scheffer H, Guchelaar HJ, Franke B TOPIC Recruitment Team. Identification of Patients With Variants in TPMT and Dose Reduction Reduces Hematologic Events During Thiopurine Treatment of Inflammatory Bowel Disease. Gastroenterology. 2015;149:907–17.e7. doi: 10.1053/j.gastro.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Frueh FW. Back to the future: Why randomized controlled trials cannot be the answer to pharmacogenomics and personalized medicine. Pharmacogenomics. 2009;10:1077–1081. doi: 10.2217/pgs.09.62. [DOI] [PubMed] [Google Scholar]
- 52.Pricing of Illumina Global Screening Array via the HuGe-F consortium [cited January 20 2019] Available from: http://glimdna.org/faq.html#pricing-of-the-array.
- 53.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukerjee G, Huston A, Kabakchiev B, Piquette-Miller M, van Schaik R, Dorfman R. User considerations in assessing pharmacogenomic tests and their clinical support tools. NPJ Genom Med. 2018;3:26. doi: 10.1038/s41525-018-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]