Abstract
Interleukin-1 receptor-associated kinase 1 (IRAK1), an essential mediator of innate immunity and inflammatory responses, is constitutively active in multiple cancers. We evaluated the role of IRAK1 in acute myeloid leukemia (AML) and assessed the inhibitory activity of multikinase inhibitor pacritinib on IRAK1 in AML. We demonstrated that IRAK1 is overexpressed in AML and provides a survival signal to AML cells. Genetic knockdown of IRAK1 in primary AML samples and xenograft model showed a significant reduction in leukemia burden. Kinase profiling indicated pacritinib has potent inhibitory activity against IRAK1. Computational modeling combined with site-directed mutagenesis demonstrated high-affinity binding of pacritinib to the IRAK1 kinase domain. Pacritinib exposure reduced IRAK1 phosphorylation in AML cells. A higher percentage of primary AML samples showed robust sensitivity to pacritinib, which inhibits FLT3, JAK2, and IRAK1, relative to FLT3 inhibitor quizartinib or JAK1/2 inhibitor ruxolitinib, demonstrating the importance of IRAK1 inhibition. Pacritinib inhibited the growth of AML cells harboring a variety of genetic abnormalities not limited to FLT3 and JAK2. Pacritinib treatment reduced AML progenitors in vitro and the leukemia burden in AML xenograft model. Overall, IRAK1 contributes to the survival of leukemic cells, and the suppression of IRAK1 may be beneficial among heterogeneous AML subtypes.
Introduction
Acute myeloid leukemia (AML) is a molecularly heterogeneous malignancy with poor outcomes characterized by the clonal expansion of myeloid progenitors [1]. Cytotoxic chemotherapy has remained the mainstay of AML treatment for decades with minimal improvement in outcomes. Significant challenges related to the biological complexity of AML have hindered the development of effective targeted therapies. AML molecular heterogeneity and the rapid emergence of genetically diverse subclones limit the potential effectiveness of a single targeted agent. In addition, prosurvival signals from the bone marrow microenvironment and tumor-intrinsic feedback pathways add further complexities that necessitate characterization of underlying biological mechanisms to identify new therapeutic approaches.
Whole-genome sequencing and gene expression studies have revealed substantial heterogeneity in the molecular abnormalities driving AML [2]. The most commonly mutated gene, FMS-related tyrosine kinase 3 (FLT3), is present in only 25% of AML cases, and FLT-3–targeted therapy has led to rapid emergence of resistance [2]. Other targetable mutations that occur frequently in chronic myeloproliferative disorders, such as those in Janus kinase 2 (JAK2), are rare events in AML [3, 4]. Recurrent activating mutations in these and other kinases have spurred the development of specific inhibitors, including selective agents like quizartinib and ruxolitinib, which inhibit FLT3 and JAK1/2 kinases, respectively. Quizartinib has demonstrated significant activity in clinical studies in patients with FLT3 activating mutations, but secondary mutations and signaling events induced by the microenvironment can counteract FLT3 inhibition and lead to emergence of resistance [5].
The importance of inflammatory pathways in cancer initiation, progression, and therapeutic resistance is now generally accepted [6–9]. We and others recently demonstrated that interleukin-1 (IL-1) contributes to the survival of leukemic cells in AML [7, 10]. Increased secretion of IL-1 in the bone marrow microenvironment leads to activation of IL-1 receptor-associated kinase (IRAK1) and p38MAPK in AML cells. The IRAK protein family consists of four functionally and structurally related members, IRAK1–4. IRAK1 and IRAK4 are active serine/threonine kinases that critical components of the innate immune system and mediate signals downstream of various pathogen-responsive and cytokine-responsive receptors while IRAK2 and IRAK3 are pseudokinases [11, 12]. IRAK1 and IRAK4 have been implicated in hematologic neoplasia [13–15]. IRAK1 acts downstream from IL-1 and lipopolysaccharide through IL-1 receptor (IL1R) and toll-like receptors (TLR), respectively [12]. Activation of IL1R and TLR recruits MYD88, resulting in activation of IRAK4 and IRAK1. Activated IRAK1/4 proteins subsequently activate TRAF6-mediated NF-κB and p38MAPK [16]. In certain B cell lymphomas, activation of the TLR/IRAK pathway occurs often in conjunction with the MYD88L265P gain-of-function mutation. This mechanism occurs in Waldenström’s macroglobulinemia [17, 18], diffuse large B-cell lymphoma (DLBCL) [19], and in primary effusion lymphoma, where IRAK1 gain-of-function mutations lead to constitutive IRAK1 activation [20]. IRAK1 levels are also elevated in a proportion of head and neck squamous-cell carcinoma samples, hepatomas, and triple negative breast cancers [21–23]. Furthermore, MYD88/IRAK signaling plays an indispensable role in the survival of T-cell acute lymphoblastic leukemia (T-ALL) cells [13, 14]. Emerging evidence emphasizes an oncogenic role for IRAK1 in myeloid cancers. Activation and overexpression of IRAK1 has a negative prognostic impact in myelodysplastic syndromes (MDS) [13, 15]. Several studies report that IRAK1 is overexpressed in AML [24–26]. A recent study demonstrated that therapeutic inhibition of IRAK1/4 reduces the growth of mixed lineage leukemia-rearranged leukemic cells [27]. These studies establish IRAK1 and IRAK4 as candidate targets in hematopoietic malignancies and underscore the need for agents that directly inhibit their activity [13–15, 24].
Pacritinib is an ATP-competitive, small-molecule, macrocyclic inhibitor with equipotent activity against JAK2 and FLT3 but not against JAK1. In the previous kinome-wide screen, pacritinib was found to suppress phosphorylation of two other kinases of potential interest in myeloid diseases, specifically IRAK1 (IC50 = 13.6 nM) and CSF1R (IC50 = 46 nM) [28, 29]. Pacritinib is in development as a treatment for myelofibrosis [30, 31]. Clinical studies of pacritinib demonstrate that at relevant peak concentrations (~10 μM), plasma protein binding is 98.8% in human plasma. Based on a free drug fraction of 1.2%, steady-state free drug concentrations of pacritinib are approximately 200–250 nM. Here we provide in vitro and in vivo evidence that IRAK1 is a therapeutic target in AML and pacritinib effectively inhibits IRAK1 activity at clinically relevant concentrations. Our data provide a rationale for evaluation of pacritinib in AML, irrespective of FLT3 mutational status and including patients resistant to FLT3 inhibitors.
Materials and methods
Cell lines and primary samples
All the cell lines were cultured in recommended media and described in supplemental methods. Primary AML samples derived from patients were evaluated according to Institutional Review Board guidelines at Oregon Health & Science University. Mononuclear cells isolated from primary AML samples were cultured in RPMI-1640 supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL penicillin-streptomycin and 10−4 M β-mercaptoethanol. Healthy bone marrow mononuclear cells were purchased from All Cells Inc (Almeda, CA).
shRNA-mediated knockdown
The knockdown was performed as described previously [7, 32] and in supplemental methods.
Transfection, immunoprecipitation, and immunoblotting
Full-length IRAK1 plasmids [33], transfection, immunoprecitipation, and immunoblotting were performed as described previously [7] and in supplemental methods.
Kinase screening assays
In vitro profiling of 439 kinases was performed previously at Reaction Biology Corporation (Malvern, PA,) using the HotSpot assay platform [28] as described in supplemental methods.
Drug sensitivity and apoptosis assays
Cells were seeded into 384-well assay plates at 1000 cells/well for cell lines and 10,000 cells/well for primary mononuclear cells. Pacritinib was provided by CTI BioPharma; quizartinib, and ruxolitinib were purchased from Selleck Chemicals (Houston, TX), IRAK1/4 inhibitor I was purchased from (Sigma Aldrich, ST Louis, MO). Inhibitors were added at a concentration series ranging from 5 or 10 μM to 0.001 μM. After 3 days of culture at 37 °C and 5% CO2, CellTiter96 (Promega) was added, and optical density was measured at 490 nm and used to determine cell viability. All cell lines or primary samples were tested in triplicate for drug sensitivity. The effect of drug treatment on apoptosis was measured by annexin V+ staining (Guava Nexin assay).
Homology alignment and computational docking of pacritinib to IRAK1
A multiple sequence alignment of IRAK1 paralogs was created using Clustal Omega [21988835] and used to generate a homology model of the IRAK1 kinase domain based on the structure of IRAK4 [17312103] (PDB:2OIC). Pacritinib was docked to the homology model using the flexible ring method of AUTODOCK [17585754]; the resulting models were analyzed with UCSF Chimera [15264254].
The gene mutation analysis in primary AML samples and cancer genome atlas (TCGA) somatic mutation data
All the AML samples used in the study were sequenced using institute clinical panel as part of their clinical care (supplemental Table 1). TCGA somatic mutation analysis for IRAK-pathway members was performed as described in supplemental methods.
Xenograft studies
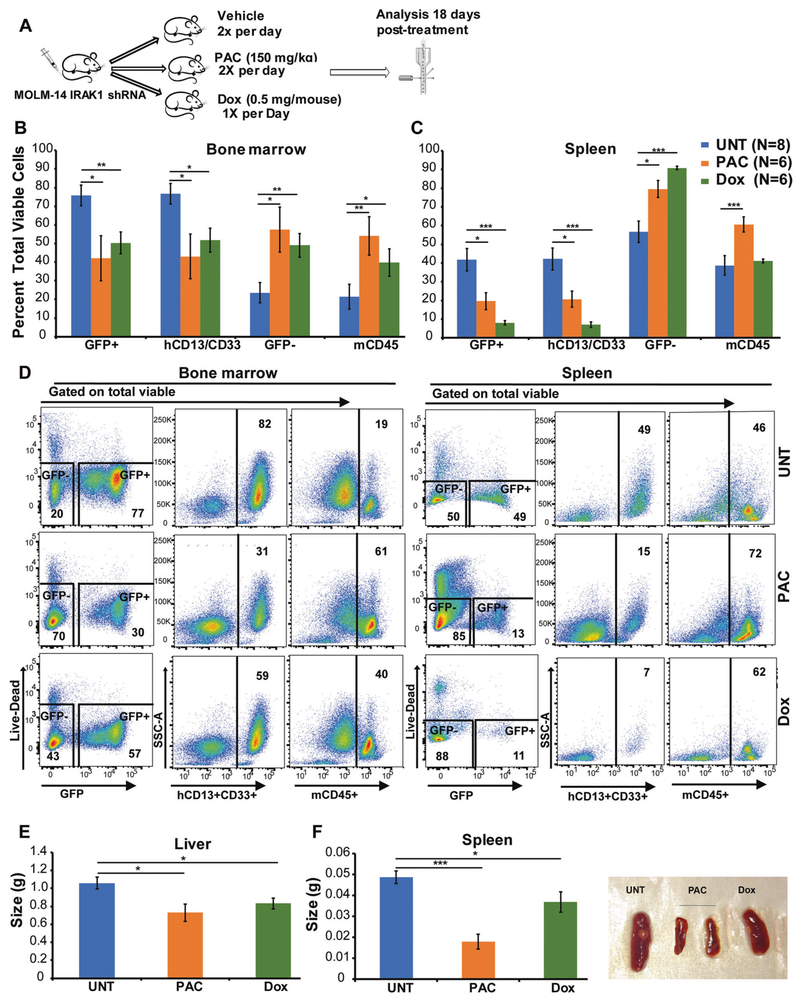
In vivo experiments were performed by xenografting MOLM-14 cells expressing doxycycline-inducible IRAK1 shRNA GFP+ (2 × 105 cell per mouse) into the tail veins of 4-week old NOD-scid IL2Rgnull mice. Pacritinib and doxycycline treatment was started 5 days after the initial injection of cells after confirmation of equal engraftment in peripheral blood (>1%) and as described in supplemental methods.
Phosphoproteomics
Phosphoproteomics analysis was performed as described in supplemental methods using untreated mononuclear cells derived from nine independent AML samples.
Statistical analysis
Fitted probit curves were used to derive three measures of drug effect: IC50, IC90, and area under the curve (AUC). Clinical and genetic features were individually correlated with drug efficacy across the 46 patient samples using the probit-derived IC50 and AUC as described in supplemental methods and were reported in supplemental Table 2. Population curves for all patient samples and FLT3 geno-type subgroups were fitted for inhibitors using a random intercepts and random slopes model with a probit link function; drug-specific curves, IC50 and AUC values were derived using the methods as described for the single-sample curves.
Results
Growth of AML cells is dependent on IRAK1
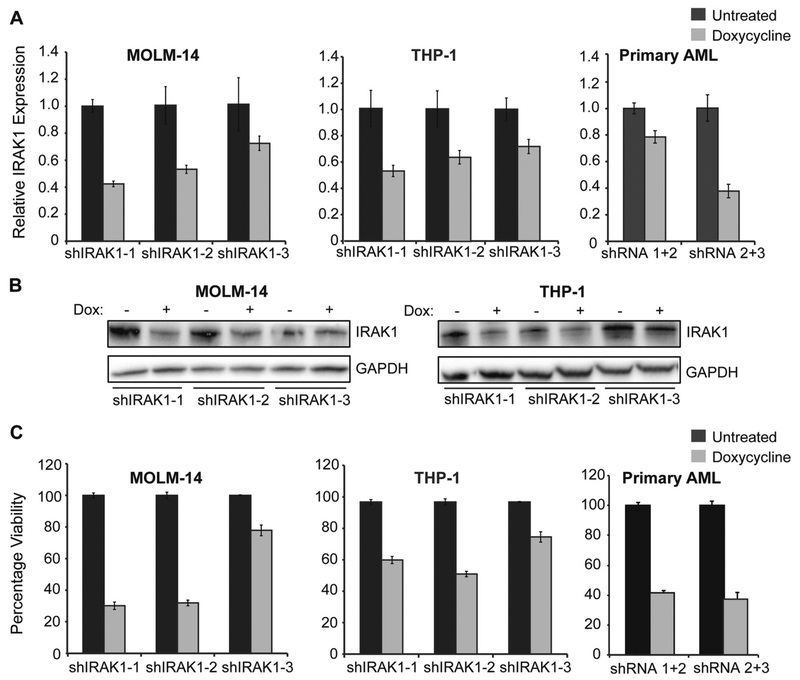
We observed elevated levels of IRAK1 in primary AML cells compared to healthy cells (supplemental Fig. 1). Based on the expression and activity of IRAK1 in AML cells, we evaluated its functional significance in AML. We abrogated IRAK1 expression utilizing doxycycline-inducible shRNAs directed against IRAK1 in the AML cell lines MOLM-14 and THP-1, as well as in primary AML cells. Two of three shRNAs tested produced a 70% reduction in IRAK1 expression in cell lines, as measured by qRT-PCR (Fig. 1a). IRAK1 knockdown was also confirmed by immunoblot analysis (Fig. 1b). Furthermore, the extent of IRAK1 knockdown correlated with a growth impairment in FLT3-ITD-positive (MOLM-14) and FLT3-wild type (THP1) cell lines that ranged from 20 to 80% (Fig. 1c). In primary AML samples, we combined two shRNAs to enhance the knockdown efficiency and observed a growth-inhibitory effect similar to that observed in cell lines (Fig. 1a,c).
Fig. 1.
IRAK1 knockdown by shRNA reduces viability of AML cells. AML cell lines (MOLM-14, THP-1) and primary AML CD34-positive cells were infected with three independent inducible IRAK1 shRNAs (−1, −2, −3), and primary AML CD34-positive cells were infected with a combination of IRAK1 shRNA hairpins. After 48 h of infection, cells were sorted for GFP positivity, and cells stably expressing IRAK1 shRNA were treated with doxycycline for 48 h. The effect of knockdown was determined by measuring change in IRAK1 gene expression by qPCR (a), western blotting (b), and cell growth by MTS assay after 4 days of doxycycline treatment (c). Results shown are representative of three independent experiments.
Pacritinib is a potent and selective IRAK1 inhibitor
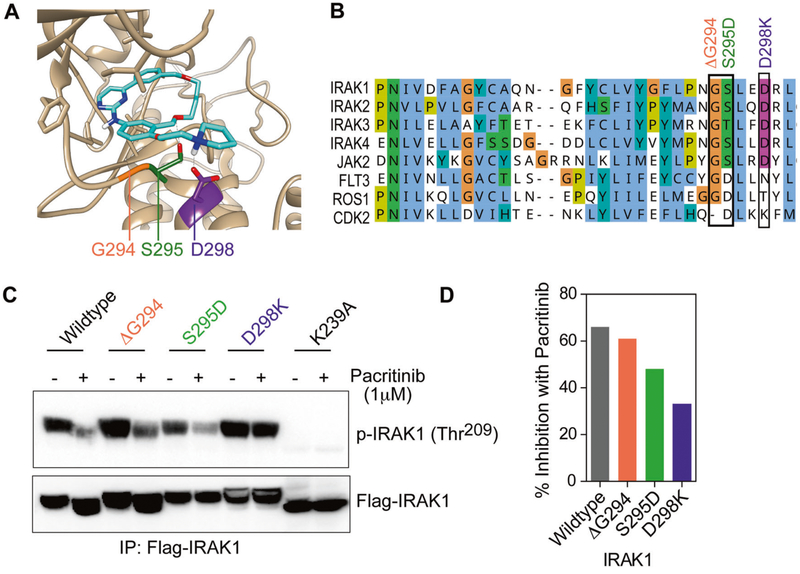
We tested an early-stage development compound IRAK1/4 inhibitor I, which showed a high median IC50 in primary AML samples (data not shown). Previously, a kinome screen of 429 kinases using recombinant proteins revealed that pacritinib potently inhibits IRAK1 (IC50 of < 20 nM) [28]. Pacritinib does not inhibit CDK2 activity [29]; consequently we predicted three residues that are important for pacritinib binding based on amino acid differences between CDK2 and the IRAK family (Fig. 2). In our model (Fig. 2a), serine 295 (S295) forms a hydrogen bond with an oxygen atom in the macrocycle of pacritinib, leading us to predict that a mutation of S295 to aspartate (as observed in CDK2) and a deletion of the adjacent glycine 294 (G294) would disrupt this interaction. Moreover, aspartate 298 (D298) is well-positioned to form a salt-bridge with the tertiary ammonium in pacritinib, leading us to predict that mutation of the negatively charged aspartate to the positively charged residue lysine (D298K), as in CDK2, may form a disfavored interaction.
Fig. 2.
Structural requirements for pacritinib binding in the IRAK1 kinase domain. a Molecular model of pacritinib-bound IRAK1. Ribbon diagram shows crystal structure-based homology model of IRAK1 kinase domain with pacritinib docked in the ATP-binding pocket. Key residues forming favorable interactions with the inhibitor are high-lighted. b Kinase domain alignment of human IRAK family and kinase paralogs. Engineered IRAK1 mutations based on pacritinib-resistant CDK2 (ΔG294, S295D, and D298K) are indicated in the image in orange, green, and purple, respectively. c Representative immunoblot showing phosphorylation of wild-type vs. mutant IRAK1 in heterologous expression system after short-term treatment with pacritinib. d Quantitative assessment of pacritinib inhibition of wild-type and mutant IRAK1. Percent inhibition is based on calculation of ratio of pIRAK1 to total (Flag) IRAK1 after densitometry of immunoblots. Results shown are representative of three independent experiments.
We engineered mutations ΔG294, S295D, and D298K in IRAK1 to resemble CDK2 (Fig. 2b, CDK2 kinase residue numbering is based on IRAK1), and they conferred pacritinib resistance on IRAK1. Mutant IRAK1 exhibited reduced pacritinib sensitivity, with the D298K substitution conferring significant loss of pacritinib sensitivity (Fig. 2c,d). In contrast, the control, kinase-inactive IRAK1 (K239A) mutant, exhibited complete abrogation of auto-phosphorylation.
IRAK1 inhibition by pacritinib suppresses growth of AML cell lines
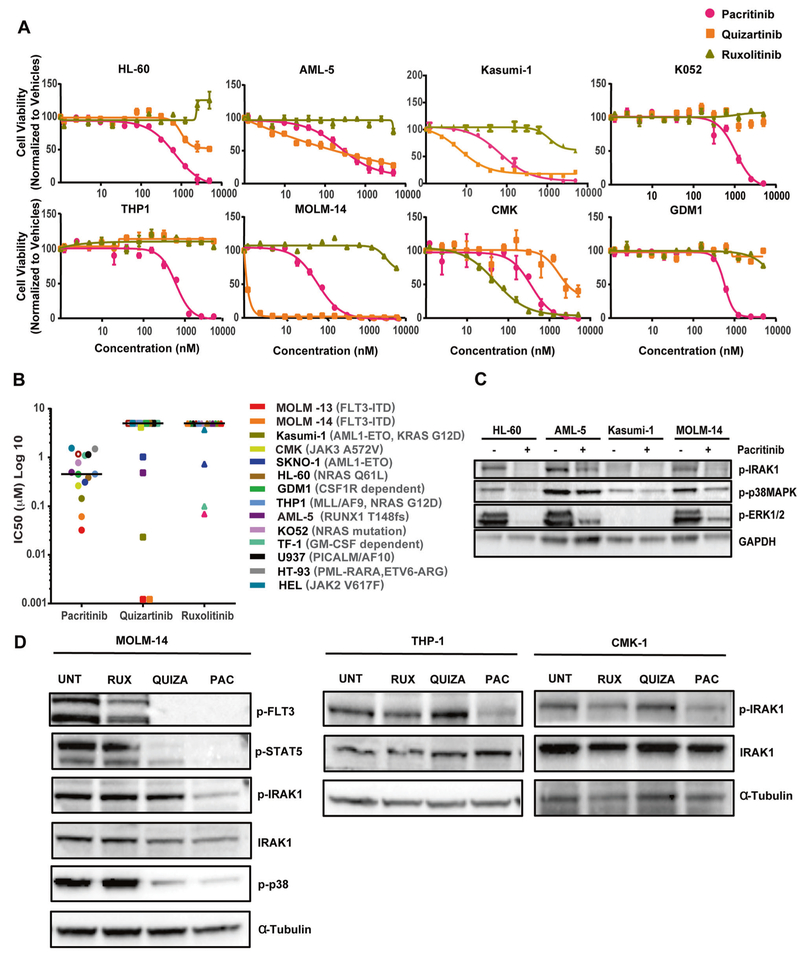
Pacritinib inhibited the growth of FLT3-ITD–positive cells (MOLM-13, MOLM-14) at IC50 values of ~32 nM and 61 nM, respectively, and JAK3 mutation-positive cells (CMK) at an IC50 value of 262 nM. In addition, pacritinib inhibited growth of cell lines harboring various genetic mutations at IC50 values ranging from ~100 to ~500 nM for cell lines Kasumi-1, SKNO-1, OCI-AML5, GDM-1, THP1, and HL-60, and ranging from ~750 to ~1500 nM for cell lines TF-1, HT-93, U937, KO52, and HEL (Fig. 3a,b). In contrast, both quizartinib and ruxolitinib were restricted in their capacity to inhibit growth across this panel. Cell lines OCI-AML5 and Kasumi-1 showed sensitivity to both pacritinib and quizartinib, whereas HL-60 and THP1 showed sensitivity to pacritinib, but not to quizartinib and ruxolitinib. These results indicate pacritinib has broad efficacy against various AML-associated genetic abnormalities (Fig. 3b). Consistent with its effect on cell viability, pacritinib induced apoptosis in AML cells (supplemental Fig. 2). Immunoblot analysis of AML cell lines HL-60, OCI-AML5, Kasumi-1, and MOLM-14, harboring mutant NRAS, mutant RUNX, RUNX translocation, and FLT3-ITD, respectively, showed IRAK1 phosphorylation on residue T209 at variable levels, and treatment with pacritinib inhibited IRAK1 phosphorylation and its downstream effectors (Fig. 3c). Consistent with their sensitivity profiles [34, 35], pacritinib and quizartinib, but not ruxolitinib, reduced phosphorylation of STAT5 and p38 in MOLM-14 cells (Fig. 3d). Further, pacritinib inhibited IRAK1 activation in FLT3-ITD–positive MOLM-14, CSF1R-dependent cells GDM-1, JAK3 mutation-positive CMK-1, and MLL-AF9-positive THP1 cells (Fig. 3d). THP1 and HL-60 cells that are FLT3 and JAK2/3 wild type, showed no sensitivity to quizartinib and ruxolitinib, but showed sensitivity to pacritinib possibly due to IRAK1 inhibition (Fig. 3a,d). We also observed IRAK1 inhibition in PICALM-AF10-dependent U937 cells that are less sensitive to pacritinib, indicating variable sensitivity to pacritinib might be due to differences in the genetic abnormalties (Fig. 3b, supplemental Fig. 3A). Additionally, inhibition of FLT3-ITD–positive cells with quizartinib did not inhibit IRAK1 activity even at 2000 nM, suggesting IRAK1 might not be the direct target of FLT3 (supplemental Fig. 3B).
Fig. 3.
Pacritinib effectively targets AML cell lines harboring a broad range of genomic aberrations. AML cell lines were cultured in their respective culture media for 72 h with graded concentrations of pacritinib, quizartinib, and ruxolitinib from 1.2 to 5000 nM using 2-fold dilution curves. Cell viability was measured by colorimetric MTS-based cell viability assay. a Viability of representative cell lines. Each cell line was tested at least in triplicate, and data are represented as mean ± SEM. b Scatter plot showing cell-based IC50 values for pacritinib, quizartinib, and ruxolitinib determined from cell-based assays with indicated AML cell lines. c Immunoblot analysis of lysates prepared from acritinib-treated AML cell lines as indicated. Phosphorylation status of IRAK1, p38, STAT5, and ERK1/2 are shown before and after pacritinib treatment. GAPDH serves as a loading control. d Immunoblot analysis of lysates prepared from MOLM-14, GDM-1, THP-1, and CMK-1 cells treated with vehicle (UNT) or 1000 nM pacritinib, quizartinib, or ruxolitinib for 16 h. Phosphorylation status of FLT3, IRAK1, p38, and STAT5 before and after inhibitor treatment are shown. α-Tubulin serves as a loading control. Results shown are representative of three independent experiments.
Pacritinib has a potent sensitivity profile across a broad range of genetic subtypes in primary AML patient samples
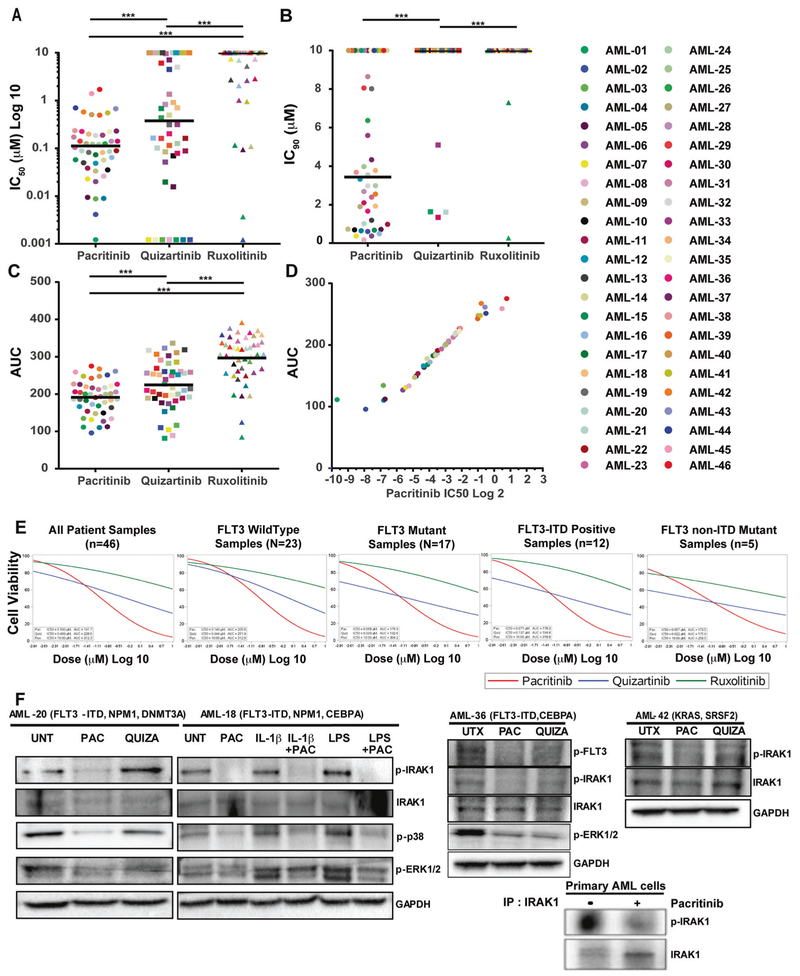
We tested the efficacy of pacritinib in ex vivo assays using primary AML samples with various genetic abnormalities. Pacritinib had a significantly lower median IC50 compared with that for quizartinib (113 nm vs. 376 nM; P < 0.0001) (Fig. 4a). Pacritinib demonstrated a higher response rate (85%; 39/46) than that observed for either quizartinib (54%; 25/46) or ruxolitinib (11%; 5/46), where responsiveness was defined as an IC50 value < 500 nM. Further, 73% (34/46) of primary AML samples showed sensitivity to pacritinib below its free peak drug concentration of 210 nM at steady-state [36, 37]. Notably, comparison of median IC90 values for pacritinib and quizartinib suggested that pacritinib treatment leads to an achievable IC90 (pacritinib IC90 = 3440 nM; range =170–10,000 nM; quizartinib IC90 = 10,000 nM; range = 1340–10,000 nM, P < 0.001) (Fig. 4b). Consistent with lower median IC50 and IC90 values, AML samples treated with pacritinib showed significantly smaller AUC values than samples exposed to either quizartinib or ruxolitinib (Fig. 4c). AUC is significantly correlated with the IC50 values within each tested inhibitor (Fig. 4d). Probit mixed effects population curve modeling on AML samples demonstrated that quizartinib has a lower IC50 estimate for all mutant forms of FLT3, including FLT3-ITD (137 nM, n = 12), FLT3-non-ITD mutant (22 nM, n = 5), and all other FLT3 mutants (93 nM, n = 17) samples, compared with the FLT3 wild-type samples (944 nM, n = 23). In contrast, when a similar analysis was performed for samples plated with pacritinib, sensitivity was apparent for both FLT3 wild type (IC50 = 140 nM) and FLT3 mutation-positive samples (IC50 = 68 nM).
Fig. 4.
Pacritinib treatment showes significant ex vivo efficacy in patient-derived primary AML cells. Freshly processed patient-derived primary AML cells were cultured for 72 h with graded concentrations of pacritinib, quizartinib, and ruxolitinib from 1.2 to 10,000 nM using a 2-fold drug dilution. Cell viability was measured by colorimetric MTS-based assay in triplicate. a Scatter plot showing cell-based median IC50 values. b Scatter plot showing cell-based median IC90 values. c Scatter plot showing AUC values. d Scatter plot showing the correlation between AUC and IC50 values. e Population curves segregated based on FLT3 mutation status and median IC50 and AUC were determined based on mixed probit curve analysis. f Immunoblot analysis of lysates prepared from mononuclear cells derived from four independent patients with AML and treated with vehicle (UNT), 200 nM pacritinib or quizartinib as well as 200 nM pacritinib in the presence and absence of 10 ng/mL IL-1β or LPS for 16 h. Phosphorylation status of IRAK1, p38, and ERK1/2 before and after inhibitor treatment are shown. GAPDH serves as a loading control. Immunoblot analysis of phosphorylated and total IRAK1 from pacritinib-treated primary AML patient-derived mononuclear cells after enrichment of IRAK1 by immunoprecipitation (lower right panel).
In addition, pacritinib showed a significantly lower IC90 value compared with that of quizartinib, and quizartinib was unable to achieve IC90 values at the maximum tested dose in FLT3 mutation-positive patient subgroups (Fig. 4e). Specifically, several AML patient samples (e.g., AML-18) with FLT3-ITD mutations were relatively insensitive to quizartinib (IC50 = ~700 nM) yet sensitive to pacritinib (IC50 = 82 nM). Further, the flow cytometry analysis of primary AML samples revealed that pacritinib treatment significantly reduces CD34+ progenitors after 72 h of treatment supplemental Fig. 4.
We observed no significant association between sensitivity to pacritinib and patient age, sex, sample type, WBC count, blast percentage, karyotype, or prognostic risk was observed (supplemental Fig. 5; supplemental Tables 1 and 2). Pacritinib sensitivity also showed no significant association with gene mutations frequently occurring in AML, with the exception of patients with ASXL1 or CREBBP mutations having reduced sensitivity to this drug (supplemental Fig. 6). Pacritinib reduced IRAK1 and p38 phosphorylation in patient-derived AML cells at basal level as well as in IL-1-stimulated and LPS-stimulated cells (Fig. 4f). Our data suggest that pacritinib is effectively inhibiting IRAK1 activity in less sensitive AML samples such as AML-36 (IC50 = 226 nM) or AML-42 (IC50 = 578 nM), indicating variable sensitivity to pacritinib may be attributed to co-occuring mutations.
Genetic knockdown of IRAK1 or treatment with pacritinib reduces the leukemia burden in AML xenograft model in vivo
We evaluated the efficacy of genetic knockdown of IRAK1 and pacritinib treatment in vivo. We developed xenografts by injecting doxycycline (Dox)-inducible IRAK1 shRNAs expressing AML cells (MOLM-14), used in Fig. 1, into NOD-scid IL2Rgnull (NSG) mice. The post 5 days of injecting cells, the engraftment of GFP+ cells was confirmed in peripheral blood by flow cytometry (>1%) and mice were treated with vehicle or doxycycline to induce IRAK1 knockdown, or pacritinib. Since MOLM-14 cell are GFP+ and express CD13 and CD33, we used GFP positivity and the expression of human CD13/CD33 to quantify the effect of treatment on the leukemia burden. Our data suggested that both IRAK1 knockdown or pacritinib treatment reduced leukemia burden from ~76 to ~50% and ~42%, respectively, in the bone marrow and ~42 to ~8% and ~10%, respectively, in the spleen (Fig. 5b–d). This reduction in leukemia burden is associated with a concomitant increase of the host murine cells in the bone marrow and spleen of NSG mice as shown by GFP− cells or murine CD45+ cells. We also observed a significant reduction in spleen and liver sizes (Fig. 5e,f). We observed slight reduction in white blood counts with no significant differences in platelets, hemoglobin, and hematocrit with IRAK1 knockdown or pacritinib treatment (supplemental Figure 7). These results suggest that IRAK1 knockdown or pacritinib treatment effectively reduces the leukemia burden.
Fig. 5.
Genetic targeting of IRAK1 or pacritinib treatment leads to a significant reduction in leukemia burden. a MOLM-14 cells transduced with the doxycycline (Dox) inducible IRAK1 shRNA were transplanted into NSG mice (2 × 105 cells/mouse) by tail vein injections. Five days postengraftment, after confirmation of equal engraftment in peripheral blood (>1%), the mice were randomly divided in three groups and treated with vehicle control, or 150 mg/kg pacritinib twice daily or 500 μg/mouse doxycycline per day by oral gavage. The doxycycline arm of mice was also maintained at the chow containing 625 mg/kg doxycycline. All the mice were sacrificed 18 days post treatment and MOLM-14 cells engraftment was determined by measuring hCD13/CD33, and GFP-positivity and contribution of murine cells were analyzed by measuring mCD45 and GFP-negetive cells in bone marrow (b) and spleen (c) by flow cytometry. d The representative FCAS plots are shown for all the treatment conditions. e–f The difference in liver and spleen sizes are shown. The data is represented as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Global analysis of primary AML cells reveals phosphoproteomics signature indicative of pacritinib sensitivity
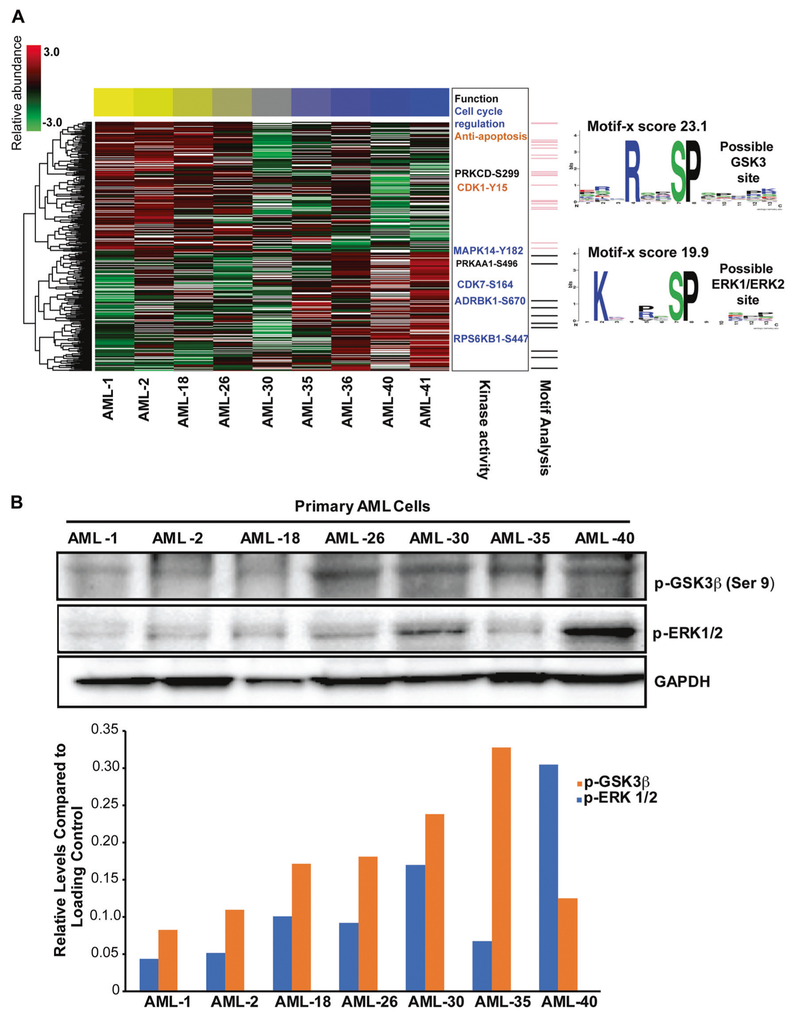
Pacritinib sensitivity in AML samples was not correlated with AML clinical and genetic characteristics in tested samples; consequently, we performed global phosphoproteomics analysis on AML samples with variable pacritinib sensitivity. Immobilized metal affinity chromatography (IMAC) global mass spectrometry identified 2200 phosphopeptides in at least six of nine patients tested prior to pacritinib treatment (supplemental Table 3). Phosphopeptides with a significant correlation or anticorrelation with pacritinib IC50 are presented in Fig. 6 and an interactive correlation biplot: https://activedatabio.github.io/peptideBiPlot/peptideBiPlot.html. We further used known sets targets of kinases to calculate enrichment based on correlation, serving as a proxy of activity. We identified three kinases where known phosphorylation site targets were significantly anticorrelated with pacritinib sensitivity; that is, primary cells with lower IC50 (more sensitive) had higher activity (phosphorylated targets) in these pathways at baseline, prior to drug treatment. The baseline up-regulated pathways in pacritinib-sensitive cells were CDK1, GSK3β, and p38MAPK (Fig. 6; supplemental Table 4); no significantly down-regulated pathways were identified. Phosphopeptides for both CDK1 and p38MAPK were identified in our analysis, and are positioned on sites known to confer activity [38]. GSK3β was not observed directly in our phosphoproteomics data as shown in supplemental Table 3. Additionally, we used an unbiased approach (Motif-X) [39] to search for sequence motifs surrounding phosphorylated sites with significant correlation or anticorrelation with pacritinib IC50 values. We identified two motifs, the [P/R] XpSP motif, which is correlated with pacritinib IC50 is consistent with an ERK1/ERK2 binding site, and the RXXpSP motif, which is anticorrelated with pacritinib IC50 is consistent with a GSK3β binding site (Fig. 6a). Because we predicted GSK3β to be active in pacritinib-sensitive patients based on both substrate enrichment and an unbiased sequence motif analysis, but did not directly observe the kinase in our phosphoproteomic data, we validated GSK3β activity. We used an antibody to GSK3β-pSer9, which inactivates the kinase, and found that this was relatively lower in sensitive patients (consistent with activation). Additionally, we show that ERK1/ERK2 activity is relatively higher in several resistant patients, consistent with our phosphoproteomic data (Fig. 6b).
Fig. 6.
Phosphoproteomics signature associated with pacritinib sensitivity. a Global phosphoproteomics analysis on nine AML samples with variable pacritinib sensitivity used in Fig. 4. Abundance of phosphopeptides correlated or anticorrelated (−0.5 > r > 0.5) with pacritinib IC50 are shown as rows and primary AML cells as columns. Color of cells indicate Z score from mean abundance (3.0 red, −3.0 green). The AML cells are arranged from most sensitive (yellow bar) to most resistant (blue bar) as determined by Probit IC50 in Fig. 4a. Phosphosites known to regulate kinase activity are shown to the right, with an indication of functional roles. Motif analysis identified two partial motifs and peptides matching those motifs are shown in the side bar as pink lines (possible GSK3β motif) and grey lines (possible ERK1/ERK2 motif). The motifs themselves are shown to the right indicating overrepresentation of amino acid residues surrounding the phosphorylated site. b Cell lysates were prepared from the mononuclear cells derived from the available AML samples used in Fig. 6a. Cell lysates were evaluated by immunoblot analysis for phospho-GSK3β (Ser 9) and phospho-ERK1/ERK2. Densitometry analysis was performed to measure p-GSK3β (Ser 9) and p-ERK1/ERK2 levels using GAPDH as the loading control.
Discussion
In AML, due to rapid emergence of resistance, inhibition of any one of the many molecular events driving disease progression, is unlikely to eradicate the disease. For example, patients with mutations in FLT3, one of the more frequently mutated genes in AML, often rapidly, relapse after treatment with an FLT3 inhibitor due to resistance mediated by secondary genetic events or survival signals mediated by the bone marrow microenvironment [5]. This challenge has prompted additional efforts to identify specific mechanisms responsible for AML progression. Here we provide evidence that the IRAK1 pathway is a therapeutic target across a variety of genetic subtypes in AML due to the proximal nature of its signal.
IRAK1 is a central mediator of innate immunity and inflammatory responses. Our data suggest that IRAK1 is constitutively active in AML cells, as shown by constitutive phosphorylation of IRAK1 in different genetic subtypes. Although IRAK1 is overexpressed in AML relative to healthy cells, we observed no obvious difference in expression and activation of IRAK1 across AML subtypes, suggesting the IRAK1 pathway is uniformly active. This is consistent with previous studies showing that IL-1 signaling mediators, including IL-1, IL1RAcP, and IL1R1, are overexpressed in various AML subtypes [7, 40]. Similarly, IRAK1 dysregulation was ubiquitously observed in MDS and T-ALL, irrespective of stage of maturation and/or oncogenic dysregulation [13–15].
Previous studies suggested multiple mechanisms may lead to overexpression or hyperactivity of IRAK1/4 in cancer. One mechanism involves post-transcriptional dysregulation by miRNAs such as miR-146a, which targets IRAK1, as evidenced by low expression levels of miR-146a resulting in the upregulation of IRAK1 in MDS [41]. Further, overexpression of IL1RAcP was observed in patients with MDS [40], suggesting that hyperphosphorylation of IRAK1 in MDS may be the result of aberrant activation of TLR/IL1R-mediated signaling. Activation of IRAK1 can also occur by gain-of-function mutations or aberrant expression of upstream signaling molecules. For example, human lymphomas with oncogenically active MYD88 mutations have constitutive IRAK1 phosphorylation [19]. In Fanconi anemia, IRAK1 exists in a hyperphosphorylated state, potentially as a consequence of aberrant TLR8 signaling [11]. It is noteworthy that mutations in MYD88, IRAKs, TLRs, or IL1R are rare in AML [2], suggesting that alternate molecular changes may activate IRAK1 in AML. Consistent with the MDS study, increased IL-1 and IL-1 receptor (IL1R1, IL1RAcP) levels in AML patients with various genetic abnormalities [7, 40], suggesting that IRAK activation may be prevalent in AML. Accordingly, a retrospective analysis revealed that chronic immune stimulation acts as a trigger to increase the risk for MDS and AML development [42]. Collectively, these studies indicate IRAKs are therapeutic targets in hematological malignancies.
Consistent with previous studies of MDS and T-ALL [14, 25], our data showed that genetic knockdown of IRAK1 significantly reduces the viability of AML cells and reduces in vivo leukemia burden. This prompted us to test the efficacy of IRAK1 inhibitors in AML. Small-molecule inhibitors targeting IRAK1 were originally developed for autoimmune and inflammatory diseases [24, 43]. These inhibitors are effective in vitro and in vivo in hematological malignancies that are dependent on the IRAK1 pathway [14, 26]. However, these IRAK1 inhibitors lack potency; therefore, there is a need for more effective IRAK1 inhibitors for therapeutic use in leukemias.
To inhibit IRAK activity effectively, we used pacritinib, an oral selective inhibitor for JAK2 and FLT3 [29, 44–46]. Kinase profiling showed that pacritinib inhibits the activity of IRAK1 but not IRAK4 [37]. We used computational modeling and biochemical analysis to demonstrate that pacritinib binds to and inhibits IRAK1 activity. Previous studies have reported its pharmacological profile and efficacy in preclinical models of JAK2-driven myeloid and lymphoid malignancies [45, 46]. In addition, pacritinib inhibits FLT3 signaling in AML cell lines with the highest potency against cells harboring FLT3-ITD mutations [44–46]. Two phase III clinical trials in myelofibrosis have demonstrated pacritinib’s efficacy [31], and a clinical trial in late-stage relapsed/refractory lymphoma demonstrated similar pacritinib efficacy [30, 36, 44]. Peak plasma concentrations of free pacritinib are 210 nM, based on a geometric mean Cmax value of 8290 ng/mL at steady state and 98.8% plasma protein binding, and were achieved following administration of 200 mg twice a day [36, 37]. These pacritinib plasma concentrations are substantially above the IC50 values for recombinant kinases, including FLT3, JAK2, IRAK1, and CSF1R [37]. Our data establish the ex vivo and in vivo efficacy of pacritinib to inhibit IRAK1 activity in preclinical models of AML and provide a rationale for clinical trials for this indication.
We showed that pacritinib has potent inhibitory effects on AML cell lines and primary AML samples harboring a wide variety of genetic mutations, whereas the FLT3 inhibitor (quizartinib) and the JAK inhibitor (ruxolitinib) are restricted in their respective efficacy to FLT3 and JAK mutation-positive samples. Consistent with previous studies [47], FLT3 mutation-negative samples in our study exhibited a low level of sensitivity to quizartinib. This may be because quizartinib inhibits wild-type FLT3 and c-KIT with approximately 4-fold and 10-fold higher IC50 values, respectively, than mutant FLT3-ITD, and both FLT3 and c-KIT are frequently overexpressed in AML [47]. Our data suggest that pacritinib is effective in inhibiting the growth of AML cells not only with FLT3 and JAK2 mutations, but also with a variety of other mutations. Our biochemical data, along with ex vivo sensitivity results, suggest that IRAK1 is constitutively active in AML cells harboring a spectrum of mutations, and pacritinib effectively inhibits the activity of IRAK1. Notably, we have not observed significant associations between pacritinib sensitivity and various clinical features, with the exception of AML with ASXL1 or CREBBP mutations, which show less sensitivity to pacritinib. Previous studies have shown that an IRAK1/4 inhibitor alone is not sufficient to kill MDS or T-ALL cells, although it can sensitize these cells to BCL2 inhibitor treatment [13]. In contrast, we found that pacritinib profoundly inhibits the growth of AML cells, reduces primary AML progenitors and decreases the leukemia burden in a murine xenograft model. The broader efficacy may be due to pacritinib targeting of multiple kinases in addition to IRAK1. However, in our in vivo model the genetic targeting of IRAK1 is as effective as pacritinib treatment. Together these results suggest that IRAK1 can serve as a therapeutic target across various genetic subtypes.
In conclusion, we provide evidence that the innate immunity pathway involving IRAK1 is important in AML pathobiology. These results augment previous findings that IRAK1 targeting can suppress an altered TLR/IL1R/NF-κB/p38MAPK pathway and eliminate a leukemic clone [7, 13–15, 26, 27]. Furthermore, we showed that pacritinib is effective in inhibiting the activity of IRAK1.
Therefore, further clinical exploration of IRAK1 as a target for intervention with pacritinib and newer more specific agents is justified in AML and other neoplastic disorders associated with IRAK pathway activation [43].
Supplementary Material
Acknowledgements
This study was supported by National Institutes of Health grant 5R00CA151670–03 (AA), V Foundation Scholar Award (AA), American Cancer Society Research Scholar Award (AA), CTI Biopharma (AA), NIH Build Exito Pilot project (AA) and 1U01CA214116–01 (Rodland/Druker). BJD is a Howard Hughes Medical Institute Investigator and is supported by the Leukemia & Lymphoma Society Beat AML initiative. MM is an NIH Build Exito scholar. The authors thank Peter Kurre, Cristina Tognon, and Pierrette Lo for their critical feedback; Brian Junio for compiling the clinical characteristics data for primary AML samples. Dorian LaTocha and Brianna Garcia for flow cytometry data acquisition and analysis; Marina A. Gritsenko, Therese R. Clauss, Matthew E. Monroe, and Ronald J. Moore for help with phosphoproteomics; and Sarah Bowden for administrative support.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41375-018-0112-2) contains supplementary material, which is available to authorized users.
Conflict of interest JWS is employed by and has equity ownership in CTI BioPharma, Corp. The authors report no other potential conflicts of interest to declare.
References
- 1.Thein MS, Ershler WB, Jemal A, Yates JW, Baer MR. Outcome of older patients with acute myeloid leukemia: an analysis of SEER data over 3 decades. Cancer. 2013;119:2720–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Eng J Med. 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365: 1054–61. [DOI] [PubMed] [Google Scholar]
- 4.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(Pt 8):1281–3. [DOI] [PubMed] [Google Scholar]
- 5.Traer E, Martinez J, Javidi-Sharifi N, Agarwal A, Dunlap J, English I, et al. FGF2 from marrow microenvironment promotes resistance to FLT3 inhibitors in acute myeloid leukemia. Cancer Res. 2016;76:6471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giles FJ, Krawczyk J, O’Dwyer M, Swords R, Freeman C. The role of inflammation in leukaemia. Adv Exp Med Biol. 2014;816:335–60. [DOI] [PubMed] [Google Scholar]
- 7.Carey A, Edwards D, Eide CA, Newell L, Traer E, Medeiros B, et al. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep. 2017;18:p3204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Chu S, Agarwal P, Campbell VL, Hopcroft L, Jorgensen HG, et al. Inhibition of interleukin-1 signaling enhances elimination of tyrosine kinase inhibitor treated CML stem cells. Blood. 2016;128:2671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welner RS, Amabile G, Bararia D, Czibere A, Yang H, Zhang H, et al. Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell. 2015;27:671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrov Z, Kurzrock R, Estey E, Wetzler M, Ferrajoli A, Harris D, et al. Inhibition of acute myelogenous leukemia blast proliferation by interleukin-1 (IL-1) receptor antagonist and soluble IL-1 receptors. Blood. 1992;79:1938–45. [PubMed] [Google Scholar]
- 11.Vanderwerf SM, Svahn J, Olson S, Rathbun RK, Harrington C, Yates J, et al. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood. 2009;114:5290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain A, Kaczanowska S, Davila E. IL-1 receptor-associated kinase signaling and its role in inflammation, cancer progression, and therapy resistance. Front Immunol. 2014;5:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Younger K, Gartenhaus R, Joseph AM, Hu F, Baer MR, et al. Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J Clin Invest. 2015;125:1081–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dussiau C, Trinquand A, Lhermitte L, Latiri M, Simonin M, Cieslak A, et al. Targeting IRAK1 in T-cell acute lymphoblastic leukemia. Oncotarget. 2015;6:18956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhyasen GW, Bolanos L, Fang J, Jerez A, Wunderlich M, Rigolino C, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 2013;24:90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465: 885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez C, Sebastian E, Chillon MC, Giraldo P, Mariano Her-nandez J, Escalante F, et al. MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenstrom’s macro-globulinemia. Leukemia. 2013;27:1722–8. [DOI] [PubMed] [Google Scholar]
- 18.Yang G, Zhou Y, Liu X, Xu L, Cao Y, Manning RJ, et al. A mutation in MYD88 (L265P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenstrom macroglobulinemia. Blood. 2013;122:1222–32. [DOI] [PubMed] [Google Scholar]
- 19.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D, Chen W, Xiong J, Sherrod CJ, Henry DH, Dittmer DP. Interleukin 1 receptor-associated kinase 1 (IRAK1) mutation is a common, essential driver for Kaposi sarcoma herpesvirus lymphoma. Proc Natl Acad Sci USA. 2014;111:E4762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wee ZN, Yatim SM, Kohlbauer VK, Feng M, Goh JY, Bao Y, et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat Commun. 2015; 6:8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams AK, Bolanos LC, Dexheimer PJ, Karns RA, Aronow BJ, Komurov K, et al. IRAK1 is a novel DEK transcriptional target and is essential for head and neck cancer cell survival. Oncotarget. 2015;6:43395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye ZH, Gao L, Wen DY, He Y, Pang YY, Chen G. Diagnostic and prognostic roles of IRAK1 in hepatocellular carcinoma tissues: an analysis of immunohistochemistry and RNA-sequencing data from the cancer genome atlas. Onco Targets Ther. 2017; 10:1711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhyasen GW, Starczynowski DT. IRAK signalling in cancer. Br J Cancer. 2015;112:232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhyasen GW, Bolanos L, Starczynowski DT. Differential IRAK signaling in hematologic malignancies. Exp Hematol. 2013;41:1005–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beverly LJ, Starczynowski DT. IRAK1: oncotarget in MDS and AML. Oncotarget. 2014;5:1699–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang K, Volk AG, Haug JS, Marshall SA, Woodfin AR, Bartom ET, et al. Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell. 2017;168:59–72 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulsen A, William A, Blanchard S, Lee A, Nagaraj H, Wang H, et al. Structure-based design of oxygen-linked macrocyclic kinase inhibitors: discovery of SB1518 and SB1578, potent inhibitors of Janus kinase 2 (JAK2) and Fms-like tyrosine kinase-3 (FLT3). J Comput Aided Mol Des. 2012;26:437–50. [DOI] [PubMed] [Google Scholar]
- 30.Mascarenhas J, Hoffman R, Talpaz M, Gerds AT, Stein B, Gupta V, et al. Results of the persist-2 phase 3 study of pacritinib (PAC) versus best available therapy (BAT), including ruxolitinib (RUX), in patients (pts) with myelofibrosis (MF) and platelet counts 100,000/μl late breaking abstract at 58th American Society of Hematology (ASH) Annual Meeting and Exposition in San Diego; 2016.
- 31.Mesa RA, Vannucchi AM, Mead A, Egyed M, Szoke A, Suvorov A, et al. Pacritinib versus best available therapy for the treatment of myelofibrosis irrespective of baseline cytopenias (PERSIST-1): an international, randomised, phase 3 trial. Lancet Haematol. 2017;4:e225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal A, Mackenzie RJ, Besson A, Jeng S, Carey A, LaTocha DH, et al. BCR-ABL1 promotes leukemia by converting p27 into a cytoplasmic oncoprotein. Blood. 2014; 124:3260–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med. 2005;201:915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desterke C, Bilhou-Nabera C, Guerton B, Martinaud C, Tonetti C, Clay D, et al. FLT3-mediated p38-MAPK activation participates in the control of megakaryopoiesis in primary myelofibrosis. Cancer Res. 2011;71:2901–15. [DOI] [PubMed] [Google Scholar]
- 35.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–4. [DOI] [PubMed] [Google Scholar]
- 36.Komrokji RS, Seymour JF, Roberts AW, Wadleigh M, To LB, Scherber R, et al. Results of a phase 2 study of pacritinib (SB1518), a JAK2/JAK2(V617F) inhibitor, in patients with myelofibrosis. Blood. 2015;125:2649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer JW, Al-Fayoumi S, Ma H, Komrokji RS, Mesa R, Ver-stovsek S. Comprehensive kinase profile of pacritinib, a non-myelosuppressive Janus kinase 2 inhibitor. J Exp Pharmacol. 2016;8:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–8. [DOI] [PubMed] [Google Scholar]
- 40.Barreyro L, Will B, Bartholdy B, Zhou L, Todorova TI, Stanley RF, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120:1290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starczynowski DT, Kuchenbauer F, Wegrzyn J, Rouhi A, Petriv O, Hansen CL, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39:167–78. e164 [DOI] [PubMed] [Google Scholar]
- 42.Kristinsson SY, Bjorkholm M, Hultcrantz M, Derolf AR, Landgren O, Goldin LR. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. J Clin Oncol. 2011;29:2897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KL, Ambler CM, Anderson DR, Boscoe BP, Bree AG, Brodfuehrer JI, et al. Discovery of clinical candidate 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}−7-methoxyisoquinoli ne-6-carboxamide (PF-06650833), a potent, selective inhibitor of interleukin-1 receptor associated kinase 4 (IRAK4), by fragment-based drug design. J Med Chem. 2017; 60: 5521–42. [DOI] [PubMed] [Google Scholar]
- 44.William AD, Lee AC, Blanchard S, Poulsen A, Teo EL, Nagaraj H, et al. Discovery of the macrocycle 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6). 1 (8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene (SB1518), a potent Janus kinase 2/fms-like tyrosine kinase-3 (JAK2/FLT3) inhibitor for the treatment of myelofibrosis and lymphoma. J Med Chem. 2011;54:4638–58. [DOI] [PubMed] [Google Scholar]
- 45.Hart S, Goh KC, Novotny-Diermayr V, Tan YC, Madan B, Amalini C, et al. Pacritinib (SB1518), a JAK2/FLT3 inhibitor for the treatment of acute myeloid leukemia. Blood Cancer J. 2011; 1:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart S, Goh KC, Novotny-Diermayr V, Hu CY, Hentze H, Tan YC, et al. SB1518, a novel macrocyclic pyrimidine-based JAK2 inhibitor for the treatment of myeloid and lymphoid malignancies. Leukemia. 2011;25:1751–9. [DOI] [PubMed] [Google Scholar]
- 47.Cortes JE, Kantarjian H, Foran JM, Ghirdaladze D, Zodelava M, Borthakur G, et al. Phase I study of quizartinib administered daily to patients with relapsed or refractory acute myeloid leukemia irrespective of FMS-like tyrosine kinase 3-internal tandem duplication status. J Clin Oncol. 2013;31:3681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.