Abstract
Livestock movements are an important mechanism of infectious disease transmission. Where these are well recorded, network analysis tools have been used to successfully identify system properties, highlight vulnerabilities to transmission, and inform targeted surveillance and control. Here we highlight the main uses of network properties in understanding livestock disease epidemiology and discuss statistical approaches to infer network characteristics from biased or fragmented datasets. We use a ‘hurdle model’ approach that predicts (i) the probability of movement and (ii) the number of livestock moved to generate synthetic ‘complete’ networks of movements between administrative wards, exploiting routinely collected government movement permit data from northern Tanzania. We demonstrate that this model captures a significant amount of the observed variation. Combining the cattle movement network with a spatial between-ward contact layer, we create a multiplex, over which we simulated the spread of ‘fast’ (R0 = 3) and ‘slow’ (R0 = 1.5) pathogens, and assess the effects of random versus targeted disease control interventions (vaccination and movement ban). The targeted interventions substantially outperform those randomly implemented for both fast and slow pathogens. Our findings provide motivation to encourage routine collection and centralization of movement data to construct representative networks.
This article is part of the theme issue ‘Modelling infectious disease outbreaks in humans, animals and plants: epidemic forecasting and control’. This theme issue is linked with the earlier issue ‘Modelling infectious disease outbreaks in humans, animals and plants: approaches and important themes’.
Keywords: livestock networks, network analysis, centrality measures, zoonoses, targeted interventions, Tanzania
1. Introduction
The ‘static network’ concept of a population as a set of ‘individuals’ (nodes) with immutable contacts (links) between them is now well established in infectious disease modelling. The network representation occurs naturally because the ‘individual’ is typically well defined (e.g. a person, animal, city, herd or farm) and the number of potentially infectious contacts per individual is usually few [1–5]. While there are a few studies for human diseases that include comprehensive, explicit network data [6], more frequently, these are either generated indirectly (for example, using mobile phone data or gravity models to predict commuter flow [7–10]) or are explicit but at small geographical scales [11,12]. By contrast, in Great Britain (GB), cattle movement data have been recorded for individuals on a daily basis for almost two decades [13]. This data richness has presented both challenges and opportunities for the application of network analyses in infectious disease epidemiology [4,5]. Similar livestock data now exist in many other countries [14–20]. However, they remain rare in emergent economies where disease burden is often high and zoonotic risk is more pronounced due to the high proportion of people who live and work in close contact with livestock [21]. About one billion of the world's poorest people (earning less than <US$2 per day) depend at least partially on livestock for their livelihoods [22], making the trade of livestock and the freedom to move livestock to access natural resources vital in many impoverished communities [23–25]. In many regions, such as sub-Saharan Africa, there are frequent but poorly recorded cross-border movements [26–28] and, when coupled with poor within-country knowledge of livestock movements, this creates risks for international pathogen transmission.
Though network analyses would be greatly aided by systems for comprehensive routine recording of between-farm and market movement, as occurs in GB and elsewhere, in countries with developing infrastructure collecting these data can be onerous and costly and requires well-evidenced justification. Here, we provide an overview of the role of network analysis in epidemiology, paying particular attention to the challenges of exploiting extensive but fragmented data. These insights are used to analyse livestock movements in northern Tanzania, where there is a high burden of livestock disease including zoonoses [29–35], no formal livestock traceability system implemented at a national level and limited resources for disease control. We demonstrate the utility of our network by identifying nodes to target disease control and surveillance interventions, considering both fast and slowly transmitting pathogens, and interrogate their efficacy through simulation, demonstrating substantial potential benefits in reducing disease spread.
2. Fundamental network concepts applied to livestock diseases
(a). Centrality measures and transmission patterns
Network centrality measures originated in social science [36] and are used to quantify the importance of nodes and links in a network, with obvious applications to identifying disease risks [19,37–41]. Common measures include degree centrality (the number of links associated with a node1), betweenness centrality (the number of times a node or link is traversed by the shortest paths between all other node pairs) and eigenvector centrality (loosely, a measure of how connected a node is to well-connected neighbours).2 Network centrality measures have been used to analyse livestock movement data from many countries, with each using different types of data source [4,17,40–43]. One example showing the relevance of all three of these centrality measures comes from the analysis of the costly [44] 2001 foot-and-mouth disease (FMD) epidemic in GB. First, a small number of ‘cull ewes’ were sold and transported long distance across GB; these were responsible for seeding virus into many otherwise low-risk areas [45]. These seeding movements are a characteristic of ‘small world’ network behaviour [1] with the long-range movements acting as links with high-betweenness centrality [45,46]. Second, Longtown auction market (the largest in GB) played a dominant role in spreading disease [47], demonstrating the importance of high degree centrality. Third, since the epidemic, prohibition of direct market-to-market livestock movements means that some farms now act as ‘middlemen’ between markets, representing a risk that could be effectively targeted to restrict disease spread [4,48]. This role, linking highly connected nodes, is a well-recognized feature of high eigenvector centrality.
(b). Network dynamics
In a static network, the infection pressure from a single individual is reduced over time as each daughter infection ‘uses up’ the link it was infected over [49,50]. Further, the components of the network (groups of nodes which can reach each other) are well defined. In dynamic networks, links can shift between individuals over time (rewiring), nodes can appear or disappear and the components of the network can change in size and composition. Rewiring a link away from an infected individual has the potential to expose another susceptible individual, thus increasing the probability of disease persistence [51,52]. Link dynamics also greatly complicate measures of network structure. For example, for an SIS infection process on a static network, where susceptible individuals (S) can become infected (I) and eventually recover to susceptible again, the eigenvector centrality scores of the nodes of the network contact matrix represent the expected proportion of time those nodes are infected over the long term3. This is the case so long as the probability of recovery before re-infection is high (e.g. if the density of infected nodes is always low, or the recovery time is substantially shorter than the time between infected generations). However, livestock movements vary daily, seasonally and from year-to-year. Contact patterns between farms and therefore eigenvector centrality measures can change dramatically depending on the season and stochastic progression of the epidemic. This influences epidemic spread [4,13,18,51], an effect also seen in human diseases [14,53]. Individual variability in disease progression and severity will also influence disease generation times and therefore what movements are likely to cause infection spread. Thus, predictions of node importance and targeting can depend strongly both on the dynamic properties of the network and the properties of the underlying disease, making the identification of general principles for the targeting of control more challenging (e.g. [54]; also electronic supplementary material).
Livestock movements are also an example where the actual contact occurs episodically. Episodic behaviour is a subject of considerable study in the network literature, especially where there are patterns of concentrated bursts (burstiness) separated by long waiting periods [55–57]. While an infection may itself cause episodic activity, it is most frequently studied as a property of the underlying network. Episodic activity has been shown to slow an epidemic on simulated [58] and real networks [59] but can also increase epidemic speed, for example, due to observed correlations between the topology of the network and the frequency of episodic contacts [60]. Epidemic spread also depends on within-node infection dynamics; in a simulated avian influenza outbreak, patterns of recorded vehicle movement between farms could either slow or accelerate pathogen spread, depending on the disease parameters and detection threshold at the farm level [61].
Infection events themselves can also change the network structure. If the perceived jeopardy is sufficiently high, rumours of pathogen spread may change contact patterns [62,63]. For livestock, farmers may be inclined to sell infected animals due to their condition or may be restricted from selling animals until the farm is officially declared disease-free [64]. In human disease, modelling analyses that included changes in the contact process over the course of the recent West African Ebola epidemic were used to inform changes in policy [65], highlighting the relevance for detailed datasets on contact patterns and their changes over time, both routinely and in response to an outbreak [66].
(c). The role of pathogen sequence data for relating transmission networks to livestock networks
Although livestock movements tell us about potentially infectious contacts, the relationship between these contacts and the transmission network of actual infectious contacts is only partially understood. Duration of contact, heterogeneity in immune response and environmental conditions are some of the factors that could affect which livestock movements transmit infection. The growing availability of high-coverage pathogen sequence data provides an unprecedented opportunity to quantify this relationship [67,68]. A number of tools have been developed to estimate transmission from genetic data [69–78] and new tools continue to be developed [69,73,79]. However, there remain many challenges [80–84]. A key limitation is that pathogen evolution needs to occur on a similar or faster timescale to the disease generation time in order to infer the direction of transmission [80]. Considering larger epidemiological units (e.g. farms rather than animals) can alleviate this problem since the generation time will be concomitantly longer [73,74,77]. Epidemiological information is still required to estimate transmission from genetic data and contact network data are important when trying to identify the most likely transmission events [85,86], but there are few tools to formally integrate these [87]. Phylodynamic approaches that leverage all available data could provide new insights into pathogen transmission and result in more targeted and improved control interventions, but they must overcome the challenge of appropriate weighting of the often biased and/or fragmented data. Nevertheless, even limited genetic data integrated into transmission models can improve epidemiological insights [88] and, in situations where other data are fragmented or sparse, sequence data can greatly strengthen the understanding of transmission and inform control.
3. Exploiting network properties
(a). Evaluating system resilience
Invasion of a livestock network by an infectious pathogen has the capacity to impair or destroy the function of individual nodes, either by the direct impact on livestock or by the restrictions resulting from control efforts. The impact on network structure can be considerable, in extremis resulting in the destruction of the network as a functioning entity. For infectious diseases, interventions such as movement restrictions, culling or prolonged herd testing are all designed to reduce transmission, but will also have varying degrees of impact on livestock movements and potentially impair the nodes' role in the network. Such changes have an economic impact [89,90] and, if sufficiently harmful, can result in node removal and/or substantial long-term harm to the network. Resilience of a network typically focuses on its ability to recover, retain the same structure and adapt to maintain system functionality when exposed to disturbances [91–93]. One approach to eliminate disease, such as during the 2001 FMD epidemic, is to disrupt the network by preventing trade for a period (link removal). These movement restrictions, however, can result in excessive livestock welfare issues, welfare culls, and significant long-term industry damage [94]. Less disruptively, lasting adjustments (link rewiring) can minimize the impact of highly influential nodes, while maintaining overall trade function. An example of this is the implementation of high biosecurity and compartmentalization in some poultry companies to isolate themselves from disease incursion despite close physical proximity to infected farms, allowing operations to continue in the face of national restrictions [95].
Minimizing the number of affected nodes, or protecting particular ones, may be important for resilience. In dynamic networks, slowing the rate at which contacts occur can slow the rate of pathogen spread and maintain communication between nodes [4], improving the network's resilience. Conversely, reducing contact rates can also increase pathogen spread [61]. Additional complications arise when considering multiple layers of a network and multiple diseases that spread on it. Ultimately, targeting control measures that consider the spread of multiple pathogens on a network could be more efficient and robust. Additionally, prior to designing and imposing changes on a network, particularly in economies where livelihoods are heavily dependent on a functioning livestock movement network, the network's resilience to proposed changes should be assessed.
(b). Exploiting network data to improve surveillance
The concepts of network resilience can be used to improve surveillance. Albert et al. showed the extent to which different types of complex network can be resilient to breakdown (which makes disease difficult to control) or vulnerable to breakdown (which makes the disease easier to control) [96]. Nodes (or links) can be removed from a network randomly or using targeted measures such as removing nodes that are highly ranked by one or more centrality measure. In terms of surveillance, random and targeted node removal can be compared to non-targeted and targeted surveillance [4]. Network analysis can thus provide an analytical framework to predict which farms to test in targeted surveillance strategies and estimate net gains in performance. While generic network analysis can be valuable [5], it can be made more robust by an understanding of the characteristics of the real system [97] and the dynamics of the considered pathogen [48]. Network analysis has been used to inform targeted surveillance strategies in many livestock systems [43,97–100], leading to considerable gains in surveillance efficiency [101,102]. Analyses of GB livestock networks have identified highly connected premises with a high risk of both becoming infected with, and spreading, disease [38], and have used simulations to show how targeted surveillance could reduce the size of potential epidemics [4]. For Swedish cattle and pigs, a bespoke metric was identified to consider the timing and sequence of possible incoming and outgoing infection chains [14]. This metric was subsequently expanded to consider the size of the in and out components and then used to analyse the German pig trade movements network to identify high-risk farms [15]. Such data are not typically available in low resource settings; having such network knowledge could enable the use of cost-efficient, network measure-targeted surveillance for disease control but needs justification for the additional cost and effort required.
(c). Multiplexes, multi-layer networks and multi-host pathogen systems
Complex systems are inherently multi-dimensional, with components linked via a complex set of often directed and weighted interactions, giving rise to diverse and unpredictable behaviours [103]. For infectious diseases, these can arise when spread occurs by more than one mechanism (e.g. animal trade, airborne, fomites, sharing a resource or insect vectors), resulting in a multiplex, or where transmission occurs across more than one species, an example of a multi-layer network. Both can compromise disease control [104], especially when there are biases in available data or the ability to exert control [105]. The multiplex representation was first developed in the social sciences to represent different types of interpersonal relationships [106]. It has since been used in a variety of contexts, including ecological systems [107], air transport [108], behavioural biology [109] and epidemiology [110]. In one livestock example, a study of a dairy system in northern Italy explicitly accounted for two independent transmission routes: cattle and veterinarian movements. This study found that at the local-scale veterinarian movements explained the spread of Mycobacterium avium subspecies paratuberculosis better than cattle movements and geographical distance failed to capture the impact of veterinarian visits [111,112]. This highlights a need to identify the potentially multiple transmission routes beyond discrete livestock movements when collecting data to construct a livestock network that is representative of a transmission network.
Many pathogens are multi-host and therefore the network multi-layer. This complication often has severe implications for humans, livestock and wildlife [113]. Unfortunately, most analytical frameworks of resilience are unsuitable for multi-dimensional systems [114], and network resilience can be influenced by interdependence with other networks [115]. Recent work using percolation theory to study the vulnerability of a system of interdependent networks [116] shows that the overlap between network layers can improve network resilience and this makes diseases harder to eradicate [117]. By disentangling system dynamics from system structure, network characteristics can be identified that influence resilience [115]. A well-known exemplar is the transmission of Mycobacterium bovis, the cause of bovine tuberculosis (bTB), between cows and European badgers (Meles meles), where the role of different layers can be quantified by exploiting their spatial patterns (electronic supplementary material, figure S1) [64]. At finer granularities, radio-collar data were used to quantify inter- and intra-species contacts for cattle and badgers [118]; adding a layer of indirect contacts based on badger latrine locations to this network showed better correspondence to badger-to-badger transmission patterns [119].
4. Movement networks where there is a limited resource for explicit traceability
There are many examples where livestock movement data have facilitated the planning of disease control and surveillance [17,19,42,120–122]. Conversely, an absence of movement information can obstruct disease control [45,123]. In settings where comprehensive tracing systems are absent, a variety of methods have been used to quantify livestock movement patterns and construct movement networks. These include the use of GPS collar data to describe mobility patterns of pastoral herds and overlaps with wildlife areas [27,43,124,125], household and market surveys [126], transport vehicle records [127] and international movement permits [28,128].
Movement permits are used in many countries to certify livestock health and/or to regulate movement taxes and have been used to quantify livestock flow and construct movement networks [128,129]. The often ephemeral and patchy nature of these records, due to poor archiving or non-compliance [130], can result in substantial non-random ‘missingness’ that is difficult to quantify. In these cases, movement permits have been used in conjunction with household and/or market survey data to estimate the risk of disease introduction and target surveillance and vaccination campaigns, also illustrating the importance of a regional disease control approach [28,122,131,132]. Such analyses have identified traders as key targets for disease control [130], demonstrated the effects of cattle movement on regional disease transmission [133], identified increased risks of bTB with increased herd introductions [41] and, with serology data, identified the role of between-village cattle movements in transmitting Rift Valley fever virus [134].
Biased network samples can make reconstruction of network characteristics difficult. This was addressed in GB by extrapolating from a small biased network sample via statistical associations between common factors in the network study and a national population survey [135].
Another approach to network construction is to impose an underlying model on observed population densities. Specifically, if census data (populations and locations) are available or can be estimated, gravity [136] and radiation [137] models provide two ways of creating network models of population mobility. While there is ongoing research regarding their relative merits [138], they share the property of describing movement in terms of relative population size and a measure of distance. Gravity models, for example, describe the probability of a movement occurring in inverse proportion to the spatial distance from each hub.
5. Evaluating network-based control strategies for livestock movements in Tanzania
(a). Introduction to the study
Tanzania provides an exemplar of a rapidly developing emerging economy. In northern Tanzania, there is a heavy reliance upon livestock for food, traction power, income, savings and social status. Movements can be over long distances, often on foot, and occasionally over international boundaries with multiple levels of market activity [26,85,139,140]. The pathogen burden is often high, and this impacts productivity, creates herd/flock instability and, in the case of zoonoses, directly affects human health [30,32,33,141–146]. In addition to protecting human health, reducing the burden of endemic livestock pathogens to improve livestock health and productivity is recognized as a route away from poverty and necessary to meet global food demands [23,147–153]. Livestock sales are also a major source of income in rural communities [154–156]. In addition to trade between markets, livestock can be sold privately, borrowed or gifted between households and are regularly moved to access natural resources [41,157,158]. A reduction in endemic livestock disease is therefore paramount to improving livelihoods in such emerging economies.
Historically, there has been no formal, centralized system for identifying and tracing the movement of individual animals in Tanzania; however, a paper movement permit certifying livestock health is officially required whenever animals are traded, recording movements from markets, though not movements to markets. These data are not digitized and the receipt books are stored at administrative Zonal Veterinary Centres in Tanzania. The aims of this study were to quantify cattle and small ruminant movements in a large (97 000 km2) area of northern Tanzania (Arusha, Manyara and Kilimanjaro regions) using archived, routinely collected government movement permit data; infer livestock movement networks; and build this information into livestock disease simulations to inform surveillance and control.
(b). Methods
Summary methods are presented here; for full details see the electronic supplementary material.
(i). Data source and transcription
Access was granted to archived government movement permit receipt books at the Northern Zonal Veterinary Office, Arusha. Movement permit receipt books were selected for analysis from 2009, 2011, 2013 and 2015. Origin, destination, number of each species (cattle, sheep or goat) moved and date were manually entered into spreadsheets from 50% of the available permits (30 946 permits), of which 19 438 (63%) permits yielded complete data. Only cattle movements are analysed here.
(ii). Statistical modelling
Cattle movements were aggregated temporally by month and spatially at the ward level, because origins and destinations often could not be located at a finer scale. A ward is an administrative unit of mean area 243 km2 and mean human population of 12 000 across the 398 wards in the study regions [159]. We aimed to infer the inter-ward cattle movement network within the study area; movements to outside the study area and within wards were excluded (local movements from markets are less likely to generate a movement permit due to non-compliance). The resulting dataset recorded the movement of 86 195 cattle from 98 origin wards to 239 destination wards over the 4 sampled years.
Due to a large number of non-randomly missing permits, it was not possible to use the movement data directly. Instead, the network was inferred by statistical modelling of the observed movements. First, to distinguish true from the artefactual absence of movements (months where an origin ward sent out no cattle), a zero-inflated negative binomial (ZINB) generalized linear model (GLM) was fitted to each origin ward, so that in subsequent modelling steps, movements would be imputed in place of false zeroes. Next, inter-ward livestock movement was modelled using a hurdle model. The movement between each pair of wards in a given month is represented by a two-step process: the binary event of any cattle being moved, modelled by a binomial generalized linear mixed-effects model (GLMM), and the number of animals moved, modelled by a zero-truncated negative binomial (ZTNB) GLMM. Each part of the hurdle model allowed movement to depend multiplicatively on the distance between origin and destination wards and their ‘masses’ (human and cattle population sizes), in addition to other characteristics (electronic supplementary material, table S1). The combined models can therefore be viewed as a gravity model of the livestock movement network. Unexplained spatial and temporal variation was modelled by fitting random effects for the origin and destination ward and for the 48 months.
(iii). Simulated networks
The fitted model was used to simulate monthly movements among the 398 wards for 1 year, with the number of movements inflated twofold to account for using a 50% subsample of the data.
(iv). Network measures
The simulated data were used to create an observed year-aggregated, static, directed, weighted cattle movement network. A spatial contact layer, connecting all adjacent wards, was added to the market movements network as a simplified means of accounting for contacts and movements between wards that are not represented by the movement permit data. Social network analysis was applied to the resulting multiplex network to identify nodes with high in-degree, out-degree, betweenness and eigenvector centrality where disease control interventions could be targeted.
(v). Simulating disease outbreaks and control on the network
The spread of a ‘fast’ (R0 = 3) and ‘slow’ (R0 = 1.5) pathogen was simulated on the multiplex to assess the effects of disease control interventions on the spread of pathogens with varying infectiousness [160]. This was achieved by running a stochastic SIR compartmental model within each ward. The total number of cattle in the susceptible (S), infectious (I) and recovered (R) compartments was updated daily, while cattle were moved monthly between wards. The two sources of simulated cattle movement were long-distance movements via the market network and short-distance movements between adjacent wards to account for unobserved local movements (for a full description, see electronic supplementary material; an animation of a simulated fast epidemic is available in the data repository (http://dx.doi.org/10.5525/gla.researchdata.733)). Two types of intervention were trialled: proactive vaccination of 70% of the cattle in a ward before the start of the epidemic, and a reactive ban on cattle movements one month after the start of the epidemic. Vaccine interventions were applied to all wards, or targeted at 20 (5%) of wards that were selected randomly, based on their total cattle population size or based on their network centrality measures. The network centrality measures used for targeting interventions were betweenness centrality, eigenvector centrality and geometric mean degree. The market movement ban was either implemented in all 111 wards that generated outward cattle movements in the simulations, and were therefore assumed to have a market, or were targeted in a subset of 20 of these wards, the same number as in the targeted vaccination interventions, and based on the same selection criteria.
(c). Results
The two parts of the hurdle model explained a substantial proportion (binomial: 40%; ZTNB: 24%) of the spatial and temporal variation in cattle movement, with movement being more probable over shorter distances and into wards containing a secondary market, and the number of animals moved being most strongly associated with the agro-ecological system of the origin wards and the presence of a primary or secondary market in the origin or destination ward (electronic supplementary material, table S1 and figure S2). All variables were retained in the hurdle model that was used to simulate the monthly cattle market movements.
(i). Network and node measures
The multiplex network is fully strongly connected (all wards can be reached by all other wards) and displays ‘small world’ properties. The spatial network layer connects all adjacent wards and the permit-related movements reduce the network diameter (longest path length between two wards) from 18 on the spatial network to 12 (see electronic supplementary material, table S2 for cattle market, spatial and multiplex networks summary statistics).
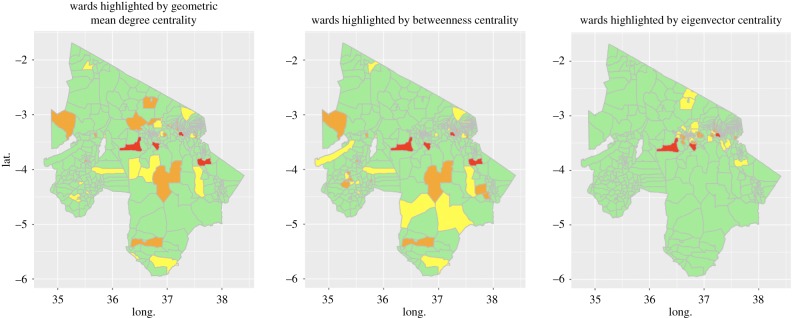
The distributions of the three node centrality measures that were investigated (betweenness, eigenvector and geometric mean degree) were strongly right-skewed. This indicates that the multiplex may be sensitive to targeted disease control interventions at the highly influential nodes. Figure 1 shows the geographical distribution of the top-ranked wards for each centrality measure, showing the potential for substantial differences in the effectiveness of targeting controls based on centrality measures due to their geographical distribution.
Figure 1.
Spatial distribution of wards with highest centrality measures in the northern Tanzania livestock movement network; colour shows position in each centrality measure rank, out of 398: red, top 1%; orange, 1–5%; yellow, 5–10%.
(ii). Simulated movements and pathogen transmission
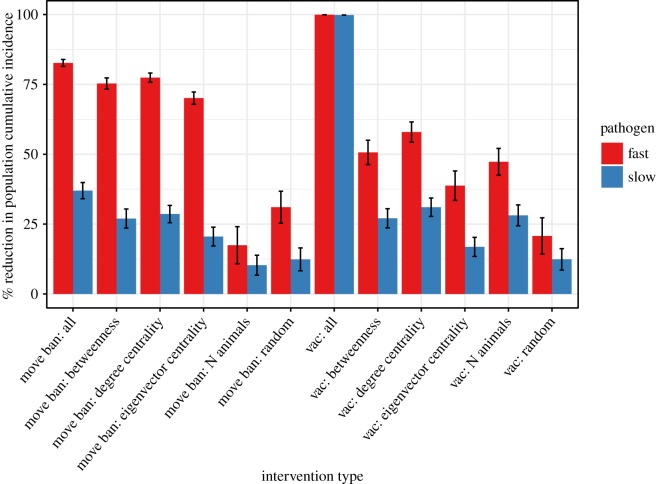
Mean reductions in population cumulative incidence (PCI) after 1 year for the fast and slow pathogens for each intervention scenario are shown in figure 2. Reductions are relative to PCI reached after 1 year with no intervention (fast: 24%; slow: 1.7%). The higher the reduction in PCI, the more effective the intervention. The list of trialled interventions and associated PCI are given in electronic supplementary material, tables S3 and S4. All simulated interventions had a greater reduction in PCI for the fast pathogen example compared to the slow, although the ranking of intervention efficacy was similar for both fast and slow pathogens. The movement ban implemented in all 111 market wards (high economic and logistical costs) performed only slightly better than when targeted in only 20 wards using network measures, and network-based targeting was more effective than selecting wards using population size or randomly, although there was no substantial difference in performance between the network measures. Vaccination applied to all wards achieved a 100% reduction in PCI for both fast and slow pathogens, while the best-performing targeted intervention, degree centrality, achieved reductions in PCI of 58% (fast) and 31% (slow). The ‘common sense’ intervention of targeting using the total number of cattle performed almost as well as degree centrality, and similarly to the second-best network measure, betweenness, but was much less efficient, requiring 3.5× more vaccine doses than degree centrality. Targeting vaccination using eigenvalue centrality performed relatively poorly, particularly against the slow disease, where its performance was comparable to selecting wards randomly.
Figure 2.
Mean (±s.e.) percentage reduction in PCI after 1 year for simulated ‘fast’ and ‘slow’ transmitting pathogens on the northern Tanzania cattle multiplex network for two types of intervention (market movement ban or vaccination at 70% coverage) applied using six strategies: applied to all wards; targeted to 5% (n = 20) of wards using each of three network centrality measures (betweenness, degree and eigenvector centrality); targeted to the 5% of wards with the highest cattle population size; and applied to 5% of wards selected randomly. The greater the reduction in PCI, the more effective the intervention is at reducing the total number of cases. Mean PCI under each scenario is calculated as the geometric mean of 237 simulated epidemics (full data: electronic supplementary material, tables S3 and S4).
6. Discussion
It is well established that the network analysis of livestock movements can be used to better understand and control diseases of commercial and zoonotic importance in higher income countries, where livestock industries tend to be highly structured and movement data are centrally collected and digitized. It is less clear that such approaches are valuable in lower income countries, where movement data are typically unavailable and the cost–benefit ratio less compelling. By exploiting movement permit data collected for health certification and tariff purposes, we have shown that even highly fragmented information about movement patterns can be used to infer network structure. By simulation, we show that the resultant inferred network has the potential to advance strategic understanding. These simulations corroborate that simple network measures can be used to identify good targets for surveillance and disease control that would be appropriate for a range of diseases and reduce the impact of infectious disease at considerably reduced cost and effort. These results could be used to form simple and practical guidelines that could be exploited immediately if, for example, a movement ban was initiated and government needed guidance on where their limited re-enforcement resources should be targeted, although they should not be used for more specific predictions without further data and analysis. They also provide a foundation for deeper research effort, highlighting where the collection of additional empirical data would be useful. For example, the substantial changes in network metrics that result when the spatial spread between wards is incorporated highlight the need to augment movement data with more extensive information about local patterns of contact. The homogeneous mixing assumption used at the within-ward level has previously been shown to be useful for developing strategic understanding, even in highly spatially driven scenarios [161], but more detailed recommendations would require modelling of within-ward heterogeneity supported by higher resolution data. This assumption may be less realistic for small urban wards where cattle are tethered, though in larger pastoral and agro-pastoral wards, shared natural resource points might make homogeneous mixing more appropriate (GL Chaters 2017, unpublished data, and [158]). Similarly, while the assumption that cattle-to-market movements occur from adjacent wards is consistent with two authors' expert knowledge of livestock management practice (O.M.N. and E.S.), verification with further data collection is an important next step. Finally, simulated movements are dynamically generated based on the random variation generated within the stochastic simulation models. We have not investigated in our dataset evidence of dynamic patterns such as changing network patterns over time because the patchy missingness in our data limits the complexity of the movement model. If more complete data became available for analysis, it would be beneficial to assess the evidence for link rewiring throughout the year as this could indicate where control measures should be targeted at specific times. Further potential model deficits include the similar impact of targeting control measures when comparing across centrality measures. This may in part be because of the relative crudeness of the disease model; in a more sophisticated model, where the timescales and frequencies of links were considered in greater detail, more substantial differences might be apparent. Similarly, a more explicit model of spatial spread might also prove discriminatory. Finally, the addition of pathogen sequence data where these are available would provide valuable confirmation of the role of network structure.
7. Conclusion
Despite this demonstration of the value of our inferred network approach, we note that data generation was the result of substantial, time-consuming effort, and the resultant inferred network, while useful, has limitations as noted above. Mobile broadband technology is becoming increasingly accessible and coupled with the availability of inexpensive scanning devices, the adoption of routine, robust digitized data recording should be achievable. In this paper, we have shown the benefits of having these data to be potentially substantial. This will be particularly pertinent in emerging economies such as Tanzania, where changes in industry structure are likely to have unanticipated disease impacts and will require regular monitoring.
Supplementary Material
Acknowledgements
We are grateful to Rigobert Tarimo and Sambeke Kiruswa for movement permit data entry, The Ministry of Livestock and Fisheries, Tanzania for access to the movement permit data, Stefan Widgren for assistance with the SimInf package, and three anonymous reviewers whose comments greatly improved this manuscript.
Endnotes
For directed networks like livestock movements, where transmission is overwhelmingly in the direction of the movement only, the geometric mean of in- and out-degree can be used.
See electronic supplementary material, for a disease-relevant interpretation.
For an irreducible positive definite matrix (e.g. a contact matrix where all nodes belong to a single strong component), the Perron–Frobenius theorem applies and the matrix is guaranteed to have a unique largest eigenvalue (and positive eigenvector). For directed networks, strong connectivity amongst all nodes is required (all nodes can reach each other reciprocally, i.e. are members of the same strong component). Where this is not the case, eigenvector centrality is not well defined, and other network measures need to be considered (for example, by using singular values).
Ethics
Study protocols were approved by the ethical review committees of the Kilimanjaro Christian Medical Centre (KCMC/832) and National Institute of Medical Research (NIMR/2028) in Tanzania and in the UK by the ethics review committee of the College of Medical, Veterinary and Life Sciences, University of Glasgow.
Data accessibility
With the exception of the Tanzanian livestock movement data, which we do not have permission to share publicly, all data and codes used in this manuscript, and an animation of a simulated epidemic, are available in Enlighten Research Data, the University of Glasgow research data repository at: http://researchdata.gla.ac.uk/id/eprint/733.
Competing interests
We declare we have no competing interests.
Funding
The movement permit study was supported by the UK BBSRC Zoonoses and Emerging Livestock Systems (ZELS) Initiative BB/L018926/1.
References
- 1.Watts DJ, Strogatz SH. 1998. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442. ( 10.1038/30918) [DOI] [PubMed] [Google Scholar]
- 2.Keeling MJ. 1999. The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. Lond. B 266, 859–867. ( 10.1098/rspb.1999.0716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liljeros F, Edling CR, Amaral LAN, Stanley HE, Åberg Y. 2001. The web of human sexual contacts. Nature 411, 907–908. ( 10.1038/35082140) [DOI] [PubMed] [Google Scholar]
- 4.Kao RR, Danon L, Green DM, Kiss IZ. 2006. Demographic structure and pathogen dynamics on the network of livestock movements in Great Britain. Proc. R. Soc. B 273, 1999–2007. ( 10.1098/rspb.2006.3505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson SE, Everett MG, Christley RM. 2007. Recent network evolution increases the potential for large epidemics in the British cattle population. J. R. Soc. Interface 4, 669–674. ( 10.1098/rsif.2007.0214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hufnagel L, Brockmann D, Geisel T. 2004. Forecast and control of epidemics in a globalized world. Proc. Natl Acad. Sci. USA 101, 15 124–15 129. ( 10.1073/pnas.0308344101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockmann D, Hufnagel L, Geisel T. 2006. The scaling laws of human travel. Nature 439, 462–465. ( 10.1038/nature04292) [DOI] [PubMed] [Google Scholar]
- 8.Balcan D, Colizza V, Goncalves B, Hu H, Ramasco JJ, Vespignani A. 2009. Multiscale mobility networks and the spatial spreading of infectious diseases. Proc. Natl Acad. Sci. USA 106, 21 484–21 489. ( 10.1073/pnas.0906910106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viboud C, Bjornstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT. 2006. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451. ( 10.1126/science.1125237) [DOI] [PubMed] [Google Scholar]
- 10.Wesolowski A, Buckee CO, Engø-Monsen K, Metcalf CJE. 2016. Connecting mobility to infectious diseases: the promise and limits of mobile phone data. J. Infect. Dis. 214(suppl_4), S414–S420. ( 10.1093/infdis/jiw273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardy JL, et al. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364, 730–739. ( 10.1056/NEJMoa1003176) [DOI] [PubMed] [Google Scholar]
- 12.Meyers LA, Newman ME, Pourbohloul B. 2006. Predicting epidemics on directed contact networks. J. Theor. Biol. 240, 400–418. ( 10.1016/j.jtbi.2005.10.004) [DOI] [PubMed] [Google Scholar]
- 13.Green DM, Kao RR. 2007. Data quality of the cattle tracing system in Great Britain. Vet. Rec. 161, 439–443. ( 10.1136/vr.161.13.439) [DOI] [PubMed] [Google Scholar]
- 14.Noremark M, Håkansson N, Lewerin SS, Lindberg A, Jonsson A. 2011. Network analysis of cattle and pig movements in Sweden: measures relevant for disease control and risk based surveillance. Prev. Vet. Med. 99, 78–90. ( 10.1016/j.prevetmed.2010.12.009) [DOI] [PubMed] [Google Scholar]
- 15.Konschake M, Lentz HHK, Conraths FJ, Hövel P, Selhorst T, Colizza V. 2013. On the robustness of in- and out-components in a temporal network. PLoS ONE 8, e0055223 ( 10.1371/journal.pone.0055223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta BL, Vergu E. 2014. Characteristics of the spatio-temporal network of cattle movements in France over a 5-year period. Prev. Vet. Med. 117, 79–94. ( 10.1016/j.prevetmed.2014.09.005) [DOI] [PubMed] [Google Scholar]
- 17.Natale F, et al. 2009. Network analysis of Italian cattle trade patterns and evaluation of risks for potential disease spread. Prev. Vet. Med. 92, 341–350. ( 10.1016/j.prevetmed.2009.08.026) [DOI] [PubMed] [Google Scholar]
- 18.Bajardi P. et al. 2011. Dynamical patterns of cattle trade movements. PLoS ONE 6, e19869 ( 10.1371/journal.pone.0019869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanderWaal KL, et al. 2016. Network analysis of cattle movements in Uruguay: quantifying heterogeneity for risk-based disease surveillance and control. Prev. Vet. Med. 123, 12–22. ( 10.1016/j.prevetmed.2015.12.003) [DOI] [PubMed] [Google Scholar]
- 20.Leon EA, Stevenson MA, Duffy SJ, Ledesma M, Morris RS. 2006. A description of cattle movements in two departments of Buenos Aires province, Argentina. Prev. Vet. Med. 76, 109–120. ( 10.1016/j.prevetmed.2006.04.010) [DOI] [PubMed] [Google Scholar]
- 21.Klous G, Huss A, Heederik DJJ, Coutinho RA. 2016. Human–livestock contacts and their relationship to transmission of zoonotic pathogens, a systematic review of literature. One Health 2, 65–76. ( 10.1016/j.onehlt.2016.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.FAO. 2009. The state of food and agriculture; livestock in the balance, pp. 32–53. Rome, Italy: FAO. [Google Scholar]
- 23.Perry B, Grace D. 2009. The impacts of livestock diseases and their control on growth and development processes that are pro-poor. Phil. Trans. R. Soc. B 364, 2643–2655. ( 10.1098/rstb.2009.0097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grace D, et al. 2012. Mapping of poverty and likely zoonoses hotspots. London, UK: Department for International Development, https://www.gov.uk/dfid-research-outputs/mapping-of-poverty-and-likely-zoonoses-hotspots. [Google Scholar]
- 25.ILRI. 2018. Why livestock matter 2018 [11/07/2018] See https://www.ilri.org/whylivestockmatter.
- 26.Aklilu Y. 2008. Livestock marketing in Kenya and Ethiopia: a review of policies and practice. Addis Ababa, Ethiopia: Feinstein International Center. [Google Scholar]
- 27.Musemwa L, et al. 2012. The impact of climate change on livestock production amongst the resource-poor farmers of third world countries: a review. Asian J. Agric. Rural Dev. 2, 621–631. [Google Scholar]
- 28.Apolloni A, et al. 2018. Towards the description of livestock mobility in Sahelian Africa: some results from a survey in Mauritania. PLoS ONE 13, e0191565 ( 10.1371/journal.pone.0191565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hummel PH. 1976. Incidence in Tanzania of Cf antibody to Coxiella burnetti in sera from man, cattle, sheep, goats and game. Vet. Rec. 98, 501–505. ( 10.1136/vr.98.25.501) [DOI] [PubMed] [Google Scholar]
- 30.Schoonman L, Swai ES. 2010. Herd- and animal-level risk factors for bovine leptospirosis in Tanga region of Tanzania. Trop. Anim. Health Prod. 42, 1565–1572. ( 10.1007/s11250-010-9607-1) [DOI] [PubMed] [Google Scholar]
- 31.Crump JA, et al. 2013. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. Plos Negl. Trop. Dis. 7, e2324 ( 10.1371/journal.pntd.0002324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assenga JA, Matemba LE, Muller SK, Malakalinga JJ, Kazwala RR. 2015. Epidemiology of Brucella infection in the human, livestock and wildlife interface in the Katavi-Rukwa ecosystem, Tanzania. BMC Vet. Res. 11, 189 ( 10.1186/s12917-015-0504-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumaye RD, Abatih EN, Thiry E, Amuri M, Berkvens D, Geubbels E, Sang RC. 2015. Inter-epidemic acquisition of Rift Valley fever virus in humans in Tanzania. PLoS Negl. Trop. Dis. 9, e0003536 ( 10.1371/journal.pntd.0003536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wensman JJ, Lindahl J, Wachtmeister N, Torsson E, Gwakisa P, Kasanga C, Misinzo G. 2015. A study of Rift Valley fever virus in Morogoro and Arusha regions of Tanzania – serology and farmers' perceptions. Infect. Ecol. Epidemiol. 5, 30025 ( 10.3402/iee.v5.30025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cash-Goldwasser S, et al. 2018. Risk factors for human brucellosis in Northern Tanzania. Am. J. Trop. Med. Hyg. 98, 598–606. ( 10.4269/ajtmh.17-0125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasserman S, Faust K. 1994. Social network analysis: methods and applications. Structural analysis in the social sciences, vol. 8. New York, NY: Cambridge University Press. [Google Scholar]
- 37.Bell D, Atkinson JS, Carlson JW. 1999. Centrality measures for disease transmission networks. Soc. Netw. 21, 1–21. ( 10.1016/S0378-8733(98)00010-0) [DOI] [Google Scholar]
- 38.Christley RM, Pinchbeck GL, Bowers RG, Clancy D, French NP, Bennett R, Turner J. 2005. Infection in social networks: using network analysis to identify high-risk individuals. Am. J. Epidemiol. 162, 1024–1031. ( 10.1093/aje/kwi308) [DOI] [PubMed] [Google Scholar]
- 39.Natale F, Savini L, Giovannini A, Calistri P, Candeloro L, Fiore G. 2011. Evaluation of risk and vulnerability using a disease flow centrality measure in dynamic cattle trade networks. Prev. Vet. Med. 98, 111–118. ( 10.1016/j.prevetmed.2010.11.013) [DOI] [PubMed] [Google Scholar]
- 40.Palisson A, Courcoul A, Durand B. 2016. Role of cattle movements in bovine tuberculosis spread in France between 2005 and 2014. PLoS ONE 11, e0152578 ( 10.1371/journal.pone.0152578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sintayehu DW, Prins HHT, Heitkönig IMA, de Boer WF. 2017. Disease transmission in animal transfer networks. Prev. Vet. Med. 137(Pt A), 36–42. ( 10.1016/j.prevetmed.2016.12.017) [DOI] [PubMed] [Google Scholar]
- 42.Buttner K, Krieter J, Traulsen A, Traulsen I, Moreno Y. 2013. Efficient interruption of infection chains by targeted removal of central holdings in an animal trade network. PLoS ONE 8, e74292 ( 10.1371/journal.pone.0074292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanderWaal K, Enns EA, Picasso C, Alvarez J, Perez A, Fernandez F, Gil A, Craft M, Wells S. 2017. Optimal surveillance strategies for bovine tuberculosis in a low-prevalence country. Sci. Rep. 7, 4140 ( 10.1038/s41598-017-04466-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haydon DT, Kao RR, Kitching RP. 2004. The UK foot-and-mouth disease outbreak—the aftermath. Nat. Rev. Microbiol. 2, 675–681. ( 10.1038/nrmicro960) [DOI] [PubMed] [Google Scholar]
- 45.Gibbens JC, et al. 2001. Descriptive epidemiology of the 2001 foot-and-mouth disease epidemic in Great Britain: the first five months. Vet. Rec. 149, 729. [PubMed] [Google Scholar]
- 46.Shirley MD, Rushton SP. 2005. Where diseases and networks collide: lessons to be learnt from a study of the 2001 foot-and-mouth disease epidemic. Epidemiol. Infect. 133, 1023–1032. ( 10.1017/S095026880500453X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kao RR. 2002. The role of mathematical modelling in the control of the 2001 FMD epidemic in the UK. Trends Microbiol. 10, 279–286. ( 10.1016/S0966-842X(02)02371-5) [DOI] [PubMed] [Google Scholar]
- 48.Kao RR, Green DM, Johnson J, Kiss IZ. 2007. Disease dynamics over very different time-scales: foot-and-mouth disease and scrapie on the network of livestock movements in the UK. J. R. Soc. Interface 4, 907–916. ( 10.1098/rsif.2007.1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeling MJ, Grenfell BT. 2000. Individual-based perspectives on R0. J. Theor. Biol. 203, 51–61. ( 10.1006/jtbi.1999.1064) [DOI] [PubMed] [Google Scholar]
- 50.Green DM, Kiss IZ, Kao RR. 2006. Parameterization of individual-based models: comparisons with deterministic mean-field models. J. Theor. Biol. 239, 289–297. ( 10.1016/j.jtbi.2005.07.018) [DOI] [PubMed] [Google Scholar]
- 51.Enright J, Kao RR. 2018. Epidemics on dynamic networks. Epidemics 24, 88–97. ( 10.1016/j.epidem.2018.04.003) [DOI] [PubMed] [Google Scholar]
- 52.Kao RR. 2010. Networks and models with heterogeneous population structure in epidemiology. In Network science: complexity in nature and technology (eds Estrada E, et al.). Berlin, Germany: Springer. [Google Scholar]
- 53.Takaguchi T, Masuda N, Holme P. 2013. Bursty communication patterns facilitate spreading in a threshold-based epidemic dynamics. PLoS ONE 8, e68629 ( 10.1371/journal.pone.0068629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holme P, Masuda N. 2015. The basic reproduction number as a predictor for epidemic outbreaks in temporal networks. PLoS ONE 10, e0120567 ( 10.1371/journal.pone.0120567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barabasi AL. 2005. The origin of bursts and heavy tails in human dynamics. Nature 435, 207–211. ( 10.1038/nature03459) [DOI] [PubMed] [Google Scholar]
- 56.Vazquez A, Oliveira JG, Dezsö Z, Goh K, Kondor I, Barabási A-L. 2006. Modeling bursts and heavy tails in human dynamics. Phys. Rev. E 73, 036127 ( 10.1103/PhysRevE.73.036127) [DOI] [PubMed] [Google Scholar]
- 57.Oliveira JG, Barabasi AL. 2005. Human dynamics: Darwin and Einstein correspondence patterns. Nature 437, 1251 ( 10.1038/4371251a) [DOI] [PubMed] [Google Scholar]
- 58.Min B, Goh KI, Vazquez A. 2011. Spreading dynamics following bursty human activity patterns. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 83, 036102 ( 10.1103/PhysRevE.83.036102) [DOI] [PubMed] [Google Scholar]
- 59.Iribarren JL, Moro E. 2011. Branching dynamics of viral information spreading. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 84, 046116 ( 10.1103/PhysRevE.84.046116) [DOI] [PubMed] [Google Scholar]
- 60.Karsai M, Kivelä M, Pan RK, Kaski K, Kertész J, Barabási A-L, Saramäki J. 2011. Small but slow world: how network topology and burstiness slow down spreading. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 83, 025102 ( 10.1103/PhysRevE.83.025102) [DOI] [PubMed] [Google Scholar]
- 61.Nickbakhsh S, Matthews L, Dent JE, Innocent GT, Arnold ME, Reid SWJ, Kao RR. 2013. Implications of within-farm transmission for network dynamics: consequences for the spread of avian influenza. Epidemics 5, 67–76. ( 10.1016/j.epidem.2013.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Epstein JM, et al. 2008. Coupled contagion dynamics of fear and disease: mathematical and computational explorations. PLoS ONE 3, e3955 ( 10.1371/journal.pone.0003955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Funk S, Salathe M, Jansen VA. 2010. Modelling the influence of human behaviour on the spread of infectious diseases: a review. J. R. Soc. Interface 7, 1247–1256. ( 10.1098/rsif.2010.0142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green DM, Kiss IZ, Mitchell AP, Kao RR. 2008. Estimates for local and movement-based transmission of bovine tuberculosis in British cattle. Proc. R. Soc. B 275, 1001–1005. ( 10.1098/rspb.2007.1601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drake JM, et al. 2015. Ebola cases and health system demand in Liberia. PLoS Biol. 13, e1002056 ( 10.1371/journal.pbio.1002056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chowell G, Nishiura H. 2015. Characterizing the transmission dynamics and control of Ebola virus disease. PLoS Biol. 13, e1002057 ( 10.1371/journal.pbio.1002057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cottam EM, et al. 2008. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog. 4, e1000050 ( 10.1371/journal.ppat.1000050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kao RR, Haydon DT, Lycett SJ, Murcia PR. 2014. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol. 22, 282–291. ( 10.1016/j.tim.2014.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Maio N, Wu CH, Wilson DJ. 2016. SCOTTI: efficient reconstruction of transmission within outbreaks with the structured coalescent. PLoS Comput. Biol. 12, e1005130 ( 10.1371/journal.pcbi.1005130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall M, Woolhouse M, Rambaut A. 2015. Epidemic reconstruction in a phylogenetics framework: transmission trees as partitions of the node set. PLoS Comput. Biol. 11, e1004613 ( 10.1371/journal.pcbi.1004613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jombart T, Eggo RM, Dodd PJ, Balloux F. 2011. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity 106, 383–390. ( 10.1038/hdy.2010.78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jombart T, . 2009. Spatiotemporal dynamics in the early stages of the 2009 A/H1N1 influenza pandemic. PLoS Curr. 1, RRN1026 ( 10.1371/currents.RRN1026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lau MSY, Marion G, Streftaris G, Gibson G. 2015. A systematic Bayesian integration of epidemiological and genetic data. PLoS Comput. Biol. 11, e1004633 ( 10.1371/journal.pcbi.1004633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morelli MJ, Thébaud G, Chadœuf J, King DP, Haydon DT, Soubeyrand S, Fraser C. 2012. A Bayesian inference framework to reconstruct transmission trees using epidemiological and genetic data. PLoS Comput. Biol. 8, e1002768 ( 10.1371/journal.pcbi.1002768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Numminen E, Chewapreecha C, Siren J, Turner C, Turner P, Bentley SD, Corander J. 2014. Two-phase importance sampling for inference about transmission trees. Proc. R. Soc. B 281, 20141324 ( 10.1098/rspb.2014.1324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Worby CJ, et al. 2016. Reconstructing transmission trees for communicable diseases using densely sampled genetic data. Ann. Appl. Stat. 10, 395–417. ( 10.1214/15-AOAS898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ypma RJ, Bataille AMA, Stegeman A, Koch G, Wallinga J, van Ballegooijen WM. 2012. Unravelling transmission trees of infectious diseases by combining genetic and epidemiological data. Proc. R. Soc. B 279, 444–450. ( 10.1098/rspb.2011.0913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ypma RJ, van Ballegooijen WM, Wallinga J. 2013. Relating phylogenetic trees to transmission trees of infectious disease outbreaks. Genetics 195, 1055–1062. ( 10.1534/genetics.113.154856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pybus OG, Tatem AJ, Lemey P. 2015. Virus evolution and transmission in an ever more connected world. Proc. R. Soc. B 282, 20142878 ( 10.1098/rspb.2014.2878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biek R, Pybus OG, Lloyd-Smith JO, Didelot X. 2015. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 30, 306–313. ( 10.1016/j.tree.2015.03.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frost SDW, Pybus OG, Gog JR, Viboud C, Bonhoeffer S, Bedford T. 2015. Eight challenges in phylodynamic inference. Epidemics 10, 88–92. ( 10.1016/j.epidem.2014.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meehan CJ, et al. 2018. The relationship between. transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine 37 , 410–416. ( ) [DOI]
- 83.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T. 2014. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol. Biol. Evol. 31, 2472–2482. ( 10.1093/molbev/msu179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Worby CJ, Lipsitch M, Hanage WP. 2014. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput. Biol. 10, e1003549 ( 10.1371/journal.pcbi.1003549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Di Nardo A, Knowles NJ, Paton DJ. 2011. Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia. Rev. Sci. Tech. 30, 63–85. [DOI] [PubMed] [Google Scholar]
- 86.VanderWaal KL, Atwill ER, Isbell LA, McCowan B, Altizer S. 2014. Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). J. Anim. Ecol. 83, 406–414. ( 10.1111/1365-2656.12137) [DOI] [PubMed] [Google Scholar]
- 87.Rasmussen DA, Volz EM, Koelle K. 2014. Phylodynamic inference for structured epidemiological models. PLoS Comput. Biol. 10, e1003570 ( 10.1371/journal.pcbi.1003570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viana M, et al. 2016. Integrating serological and genetic data to quantify cross-species transmission: brucellosis as a case study. Parasitology 143, 821–834. ( 10.1017/S0031182016000044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Knight-Jones TJ, Rushton J. 2013. The economic impacts of foot and mouth disease—what are they, how big are they and where do they occur? Prev. Vet. Med. 112, 161–173. ( 10.1016/j.prevetmed.2013.07.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith RL, Tauer LW, Schukken YH, Lu Z, Grohn YT. 2013. Minimization of bovine tuberculosis control costs in US dairy herds. Prev. Vet. Med. 112, 266–275. ( 10.1016/j.prevetmed.2013.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holling CS. 1973. Resilience and stability of ecological systems. Annu. Rev. Ecol. Syst. 4, 1–23. ( 10.1146/annurev.es.04.110173.000245) [DOI] [Google Scholar]
- 92.Holling CS. 1996. Engineering resilience versus ecological resilience. In Engineering within ecological constraints (ed. PE Schulze), pp. 31–44. Washington, DC: National Academy. [Google Scholar]
- 93.Carpenter S, et al. 2001. From metaphor to measurement: resilience of what to what? Ecosystems 4, 765–781. [Google Scholar]
- 94.Anderson I. 2002. Foot and mouth disease 2001: lessons to be learned inquiry. London, UK: The Stationery Office. [Google Scholar]
- 95.Nickbakhsh S, et al. 2014. A metapopulation model for highly pathogenic avian influenza: implications for compartmentalization as a control measure. Epidemiol. Infect. 142, 1813–1825. ( 10.1017/S0950268813002963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Albert R, Jeong H, Barabasi AL. 2000. Error and attack tolerance of complex networks. Nature 406, 378–382. ( 10.1038/35019019) [DOI] [PubMed] [Google Scholar]
- 97.Rossi G, De Leo GA, Pongolini S, Natalini S, Vincenzi S, Bolzoni L. 2015. Epidemiological modelling for the assessment of bovine tuberculosis surveillance in the dairy farm network in Emilia-Romagna (Italy). Epidemics 11, 62–70. ( 10.1016/j.epidem.2015.02.007) [DOI] [PubMed] [Google Scholar]
- 98.Dube C, et al. 2011. Introduction to network analysis and its implications for animal disease modelling. Rev. Sci. Tech. 30, 425–436. ( 10.20506/rst.30.2.2043) [DOI] [PubMed] [Google Scholar]
- 99.Craft ME. 2015. Infectious disease transmission and contact networks in wildlife and livestock. Phil. Trans. R. Soc. B 370, 20140107 ( 10.1098/rstb.2014.0107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dube C, Ribble C, Kelton D, McNab B. 2009. A review of network analysis terminology and its application to foot-and-mouth disease modelling and policy development. Transbound. Emerg. Dis. 56, 73–85. ( 10.1111/j.1865-1682.2008.01064.x) [DOI] [PubMed] [Google Scholar]
- 101.Bessell PR, et al. 2012. Developing a framework for risk-based surveillance of tuberculosis in cattle: a case study of its application in Scotland. Epidemiol. Infect. 141, 314–323. ( 10.1017/S0950268812000635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salvador LCM, Deason M, Enright J, Bessell PR, Kao RR. 2018. Risk-based strategies for surveillance of tuberculosis infection in cattle for low-risk areas in England and Scotland. Epidemiol. Infect. 146, 107–118. ( 10.1017/S0950268817001935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miguel S, et al. 2012. Challenges in complex systems science. Eur. Phys. J. 214, 245–271. ( 10.1140/epjst/e2012-01694-y) [DOI] [Google Scholar]
- 104.Webster JP, Borlase A, Rudge JW. 2017. Who acquires infection from whom and how? Disentangling multi-host and multi-mode transmission dynamics in the ‘elimination’ era. Phil. Trans. R. Soc. B 372, 20160091 ( 10.1098/rstb.2016.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Godfray HCJ, et al. 2013. A restatement of the natural science evidence base relevant to the control of bovine tuberculosis in Great Britain. Phil. Trans. R. Soc. B 280, 20131634 ( 10.1098/rspb.2013.1634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kivelä M, Arenas A, Barthelemy M, Gleeson JP, Moreno Y, Porter MA. 2014. Multilayer networks. J. Comp. Netw. 2, 203–271. ( 10.1093/comnet/cnu016) [DOI] [Google Scholar]
- 107.Pilosof S, Porter MA, Pascual M, Kéfi S. 2017. The multilayer nature of ecological networks. Nat. Ecol. Evol. 1, 0101 ( 10.1038/s41559-017-0101) [DOI] [PubMed] [Google Scholar]
- 108.Cardillo A, Zanin M, Gómez-Gardeñes J, Romance M, García del Amo AJ, Boccaletti S. 2013. Modeling the multi-layer nature of the European Air Transport Network: resilience and passengers re-scheduling under random failures. Eur. Phys. J. 215, 23–33. ( 10.1140/epjst/e2013-01712-8) [DOI] [Google Scholar]
- 109.Barrett L, Henzi SP, Lusseau D. 2012. Taking sociality seriously: the structure of multi-dimensional social networks as a source of information for individuals. Phil. Trans. R. Soc. B 367, 2108–2118. ( 10.1098/rstb.2012.0113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brooks-Pollock E, de Jong MCM, Keeling MJ, Klinkenberg D, Wood JLN. 2015. Eight challenges in modelling infectious livestock diseases. Epidemics 10, 1–5. ( 10.1016/j.epidem.2014.08.005) [DOI] [PubMed] [Google Scholar]
- 111.Rossi G, et al. 2017. The potential role of direct and indirect contacts on infection spread in dairy farm networks. PLoS Comput. Biol. 13, e1005301 ( 10.1371/journal.pone.0075570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rossi G, et al. 2017. Modelling farm-to-farm disease transmission through personnel movements: from visits to contacts, and back. Sci. Rep. 7, 2375 ( 10.1038/s41598-017-02567-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haydon DT, et al. 2002. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 8, 1468–1473. ( 10.3201/eid0812.010317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sole RV, Montoya JM. 2001. Complexity and fragility in ecological networks. Phil. Trans. R. Soc. Lond. B 268, 2039–2045. ( 10.1098/rspb.2001.1767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao J, Barzel B, Barabasi AL. 2016. Erratum: universal resilience patterns in complex networks. Nature 536, 238 ( 10.1038/nature18019) [DOI] [PubMed] [Google Scholar]
- 116.Gao JX, et al. 2013. Percolation of a general network of networks. Phys. Rev. E 88, 062816 ( 10.1103/PhysRevE.88.062816) [DOI] [PubMed] [Google Scholar]
- 117.Cellai D, López E, Zhou J, Gleeson JP, Bianconi G. 2013. Percolation in multiplex networks with overlap. Phys. Rev. E 88, 052811 ( 10.1103/PhysRevE.88.052811) [DOI] [PubMed] [Google Scholar]
- 118.Bohm M, Hutchings MR, White PCL. 2009. Contact networks in a wildlife-livestock host community: identifying high-risk individuals in the transmission of bovine TB among badgers and cattle. PLoS ONE 4, e5016 ( 10.1371/journal.pone.0005016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silk MJ, et al. 2018. Quantifying direct and indirect contacts for the potential transmission of infection between species using a multilayer contact network. Behaviour 155, 731–757. ( 10.1163/1568539X-00003493) [DOI] [Google Scholar]
- 120.Bigras-Poulin M, Thompson RA, Chriel M, Mortensen S, Greiner M. 2006. Network analysis of Danish cattle industry trade patterns as an evaluation of risk potential for disease spread. Prev. Vet. Med. 76, 11–39. ( 10.1016/j.prevetmed.2006.04.004) [DOI] [PubMed] [Google Scholar]
- 121.Kiss IZ, Green DM, Kao RR. 2006. The network of sheep movements within Great Britain: network properties and their implications for infectious disease spread. J. R. Soc. Interface 3, 669–677. ( 10.1098/rsif.2006.0129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Motta P, et al. 2017. Implications of the cattle trade network in Cameroon for regional disease prevention and control. Sci. Rep. 7, 43932 ( 10.1038/srep43932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.New Zealand Government. 2018. National animal identification and tracing. See https://www.mpi.govt.nz/growing-and-harvesting/livestock-and-animal-care/national-animal-identification-and-tracing/.
- 124.Handcock RN, et al. 2009. Monitoring animal behaviour and environmental interactions using wireless sensor networks, GPS collars and satellite remote sensing. Sensors 9, 3586–3603. ( 10.3390/s90503586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Raizman EA, Rasmussen HB, King LE, Ihwagi FW, Douglas-Hamilton I. 2013. Feasibility study on the spatial and temporal movement of Samburu's cattle and wildlife in Kenya using GPS radio-tracking, remote sensing and GIS. Prev. Vet. Med. 111, 76–80. ( 10.1016/j.prevetmed.2013.04.007) [DOI] [PubMed] [Google Scholar]
- 126.Poolkhet C, Chairatanayuth P, Thongratsakul S, Kasemsuwan S, Rukkwamsuk T. 2013. Social network analysis used to assess the relationship between the spread of avian influenza and movement patterns of backyard chickens in Ratchaburi, Thailand. Res. Vet. Sci. 95, 82–86. ( 10.1016/j.rvsc.2013.02.016) [DOI] [PubMed] [Google Scholar]
- 127.Kim Y, et al. 2018. Livestock trade network: potential for disease transmission and implications for risk-based surveillance on the island of Mayotte. Sci. Rep. 8, 11550 ( 10.1038/s41598-018-29999-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lindstrom T, et al. 2013. A Bayesian approach for modeling cattle movements in the United States: scaling up a partially observed network. PLoS ONE 8, e53432 ( 10.1371/journal.pone.0053432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dube C, et al. 2008. Comparing network analysis measures to determine potential epidemic size of highly contagious exotic diseases in fragmented monthly networks of dairy cattle movements in Ontario, Canada. Transbound. Emerg. Dis. 55, 382–392. ( 10.1111/j.1865-1682.2008.01053.x) [DOI] [PubMed] [Google Scholar]
- 130.Poolkhet C, et al. 2016. Social network analysis of cattle movement in Kampong Cham, Kampong Speu and Takeo, Cambodia. Acta Trop. 159, 44–49. ( 10.1016/j.actatropica.2016.03.027) [DOI] [PubMed] [Google Scholar]
- 131.Wongsathapornchai K, Salman MD, Edwards JR, Morley PS, Keefe TJ, Campen HV, Weber S. 2008. Assessment of the likelihood of the introduction of foot-and-mouth disease through importation of live animals into the Malaysia-Thailand-Myanmar peninsula. Am. J. Vet. Res. 69, 252–260. ( 10.2460/ajvr.69.2.252) [DOI] [PubMed] [Google Scholar]
- 132.Selby R, Bardosh K, Picozzi K, Waiswa C, Welburn SC. 2013. Cattle movements and trypanosomes: restocking efforts and the spread of Trypanosoma brucei rhodesiense sleeping sickness in post-conflict Uganda. Parasites Vectors 6, 281 ( 10.1186/1756-3305-6-281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dean AS, et al. 2013. Potential risk of regional disease spread in West Africa through cross-border cattle trade. PLoS ONE 8, e75570 ( 10.1371/journal.pone.0075570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nicolas G, Durand B, Duboz R, Rakotondravao R, Chevalier V. 2013. Description and analysis of the cattle trade network in the Madagascar highlands: potential role in the diffusion of Rift Valley fever virus. Acta Trop. 126, 19–27. ( 10.1016/j.actatropica.2012.12.013) [DOI] [PubMed] [Google Scholar]
- 135.Nickbakhsh S, Matthews L, Bessell PR, Reid SWJ, Kao RR. 2011. Generating social network data using partially described networks: an example informing avian influenza control in the British poultry industry. BMC Vet. Res. 7, 66 ( 10.1186/1746-6148-7-66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xia Y, Bjornstad ON, Grenfell BT. 2004. Measles metapopulation dynamics: a gravity model for epidemiological coupling and dynamics. Am. Nat. 164, 267–281. ( 10.1086/422341) [DOI] [PubMed] [Google Scholar]
- 137.Simini F, González MC, Maritan A, Barabási A-L. 2012. A universal model for mobility and migration patterns. Nature 484, 96–100. ( 10.1038/nature10856) [DOI] [PubMed] [Google Scholar]
- 138.Masucci AP, Serras J, Johansson A, Batty M. 2013. Gravity versus radiation models: on the importance of scale and heterogeneity in commuting flows. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 88, 022812 ( 10.1103/PhysRevE.88.022812) [DOI] [PubMed] [Google Scholar]
- 139.Bouslikhane M. 2015. Cross border movements of animals and animal products and their relevance to the epidemiology of animal diseases in Africa. Paris, France: L'Organisation Mondiale de la Santé Animale (OIE). [Google Scholar]
- 140.Muyunda C. 2009. Hidden value on the hoof: cross-border livestock trade in East Africa. Lusaka, Zambia: Common Market for Eastern and Southern Africa Comprehensive African Agriculture Development Programme. [Google Scholar]
- 141.Biggs HM, Galloway RL, Bui DM, Morrissey AB, Maro VP, Crump JA. 2013. Leptospirosis and human immunodeficiency virus co-infection among febrile inpatients in Northern Tanzania. Vector-Borne Zoonot. Dis. 13, 572–580. ( 10.1089/vbz.2012.1205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Halliday J, et al. 2012. Bringing together emerging and endemic zoonoses surveillance: shared challenges and a common solution. Phil. Trans. R. Soc. B 367, 2872–2880. ( 10.1098/rstb.2011.0362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heinrich N, et al. 2012. High seroprevalence of Rift Valley fever and evidence for endemic circulation in Mbeya region, Tanzania, in a cross-sectional study. PLoS Negl. Trop. Dis. 6, e1557 ( 10.1371/journal.pntd.0001557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karimuribo ED, et al. 2007. Prevalence of brucellosis in crossbred and indigenous cattle in Tanzania. Livest. Res. Rural Dev. 19, 148–152. [Google Scholar]
- 145.Machangu RS, Mgode G, Mpanduji D. 1997. Leptospirosis in animals and humans in selected areas of Tanzania. Belg. J. Zool. 127, 97–104. [Google Scholar]
- 146.Vanderburg S, et al. 2014. Epidemiology of Coxiella burnetii infection in Africa: a OneHealth systematic review. PLoS Negl. Trop. Dis. 8, e2787 ( 10.1371/journal.pntd.0002787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Allen LH. 2003. Interventions for micronutrient deficiency control in developing countries: past, present and future. J. Nutr. 133, 3875S–3878S. ( 10.1093/jn/133.11.3875S) [DOI] [PubMed] [Google Scholar]
- 148.Coker R, et al. 2011. Towards a conceptual framework to support one-health research for policy on emerging zoonoses. Lancet 11, 326–331. ( 10.1016/S1473-3099(10)70312-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kelly AM, Marshak RR. 2007. Veterinary medicine, global health. J. Am. Vet. Med. Assoc. 231, 1806–1808. ( 10.2460/javma.231.12.1806) [DOI] [PubMed] [Google Scholar]
- 150.Muma JB, Mwacalimba KK, Munang'andu HM, Matope G, Jenkins A, Siamudaala V, Mweene AS, Marcotty T. 2014. The contribution of veterinary medicine to public health and poverty reduction in developing countries. Vet. Ital. 50, 117–129. [DOI] [PubMed] [Google Scholar]
- 151.Pradere JP. 2014. Improving animal health and livestock productivity to reduce poverty. Rev. Sci. Tech. 33, 735–744. [PubMed] [Google Scholar]
- 152.Randolph TF, et al. 2007. Invited review: role of livestock in human nutrition and health for poverty reduction in developing countries. J. Anim. Sci. 85, 2788–2800. ( 10.2527/jas.2007-0467) [DOI] [PubMed] [Google Scholar]
- 153.Steinfeld H, et al. 2006. Livestock's long shadow: environmental issues and options. Rome, Italy: FAO.
- 154.Covarrubias K, et al. 2012. Livestock and livelihoods in rural Tanzania: a descriptive analysis of the 2009 National Panel Survey. Washington, DC: World Bank Group.
- 155.Pica-Ciamarra U, et al. 2011. Linking smallholders to livestock markets: combining market and household survey data in Tanzania.
- 156.Williams TO, Spycher BD, Okike I. 2006. Improving livestock marketing and intra-regional trade in West Africa: determining appropriate economic incentives and policy framework. Nairobi, Kenya: ILRI (International Livestock Research Institute). [Google Scholar]
- 157.Coppolillo PB. 2000. The landscape ecology of pastoral herding: spatial analysis of land use and livestock production in East Africa. Hum. Ecol. 28, 527–560. ( 10.1023/A:1026435714109) [DOI] [Google Scholar]
- 158.VanderWaal K, Gilbertson M, Okanga S, Allan BF, Craft ME. 2017. Seasonality and pathogen transmission in pastoral cattle contact networks. R. Soc. open sci. 4, 170808 ( 10.1098/rsos.170808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tanzania National Bureau of Statistics. 2012 Tanzania in figures 2012 See http://www.nbs.go.tz.
- 160.Widgren S, Bauer P, Eriksson R, Engblom S. 2016 SimInf: an R package for data-driven stochastic disease spread simulations. https://arxiv.org/abs/1605.01421v3 . [Google Scholar]
- 161.Keeling MJ, Woolhouse MEJ, May RM, Davies G, Grenfell BT. 2003. Modelling vaccination strategies against foot-and-mouth disease. Nature 421, 136–142. ( 10.1038/nature01343) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
With the exception of the Tanzanian livestock movement data, which we do not have permission to share publicly, all data and codes used in this manuscript, and an animation of a simulated epidemic, are available in Enlighten Research Data, the University of Glasgow research data repository at: http://researchdata.gla.ac.uk/id/eprint/733.