Abstract
Background
In China multidrug-resistant bacteria pose a considerable threat to public health. Antimicrobial resistance has weakened the effectiveness of many medicines widely used today. Thus, discovering new antibacterial drugs is paramount in the effort to treat emerging drug-resistant bacteria.
Methods
Eravacycline, tigecycline and other clinical routine antibiotics were tested by reference broth micro-dilution method against 336 different strains collected from 11 teaching hospitals in China between 2012 and 2016. These isolates included Enterobacteriaceae, non-fermentative, Staphylococcus spp., Enterococcus, and a number of fastidious organisms. The strains involved in this study possess the most important drug resistance characteristics currently known in China. Drug resistant bacteria such as those producing extended spectrum β-lactamases (ESBL) and carbapenemases (KPC-2 and NDM-1), and those exhibiting colistin resistance (mcr-1) and tigecycline were included in this study. Additionally, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), β-lactamase positive Haemophilus influenzae, and penicillin resistant Streptococcus pneumoniae (PRSP) were also included.
Results
Eravacycline exhibited good efficacy against all the strains tested, especially for organisms with ESBLs, carbapenemases, and mcr-1 gene compared with tigecycline and other antibiotics tested. The MIC values of eravacycline against carbapenemase producing Enterobacteriaceae and OXA-23-producing A. baumannii were much lower than the MIC values of other antibiotics. MRSA, VRE, β-lactamase positive Haemophilus influenza, and PRSP were sensitive to eravacycline in every strain tested. Furthermore, in most strains tested, the MICs of eravacycline were two to four-fold lower than the MICs of tigecycline.
Conclusions
Eravacycline has shown potent antibacterial activity against common and clinically important antibiotic-resistant pathogens. The MIC distribution of eravacycline was generally lower than that of tigecycline which demonstrates that this new drug is potentially more effective than the existing medications.
Electronic supplementary material
The online version of this article (10.1186/s12879-019-4093-1) contains supplementary material, which is available to authorized users.
Keywords: Eravacycline, Tigecycline, Carbapenem resistant Enterobacteriaceae bacteria, Acinetobacter baumannii, Antibiotic resistance
Background
In China, microbial resistance to presently administered antimicrobial agents is increasing steadily owing to the emergence of novel resistance mechanisms in the microbes [1, 2]. Multidrug-resistant bacterium causes a considerable threat to public health. Antimicrobial resistance weakened the effectiveness of many medicines widely used today [3]. Thus discovering new antibacterial drugs are required to combat the threat of these emerging resistant bacteria. Eravacycline (TP-434 or 7-fluoro-9-pyrrolidinoacetamido-6-demethyl-6-deoxytetracycline) is a novel broad-spectrum synthetic tetracycline antibiotic being developed for the treatment of severe life-threatening infections, including those that are resistant to current broad-spectrum antibiotics [4]. Eravacycline has already been proven effective against some clinically important antibiotic-resistant pathogens, including gram-positive and gram-negative aerobic and anaerobic pathogens [5, 6]. Moreover, eravacycline was found to be safer and more effective than carbapenems in patients with complicated intra-abdominal infection (cIAI) during global phase 3 clinical trials (NCT01844856 and NCT02784704) [5, 7]. Additionally, there is a clinical development plan in place to introduce it into China to address bacterial drug resistance. The targets of eravacycline include complicated intra-abdominal infection (cIAI), complicated urinary tract infection (cUTI), and pulmonary infections caused by other susceptible pathogens. Tigecycline is a relatively new competing drug for eravacycline, imipenem, meropenem, and colistin in the treatment of carbapenem-resistant Enterobacteriaceae. The present study was designed to evaluate the in vitro activities of eravacycline against panels of clinical bacterial pathogens, with or without remarkable resistance factors, which were collected in recent years and were similar to pathogenic bacteria that this drug was designed to treat. This study was designed to prove the in-vitro efficacy of eravacycline (presented by minimum inhibitory concentration, MIC) against major target pathogens in China, which will be used to support further clinical development of eravacycline within China.
Methods
In the present study, a total of 336 different clinical isolates, were routinely collected from 11 teaching hospitals representing the south, north, northwest, east, and middle regions of mainland China between 2012 and 2016, and tested (list of the hospitals can be found in Additional file 1). After re-identification with the typical biochemical reaction of each organism, the strains were stored in a Microbank tube and placed in a refrigerator at − 80 degrees Celsius before test. All organisms and their associated drug resistance factors are detailed in Table 1. MIC measurements were performed via the reference broth microdilution method as described by the Clinical and Laboratory Standards Institute (CLSI) M7-A9 (2012) [8]. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were utilized as quality controls in MIC testing of gram-negative bacteria. Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were utilized as quality controls in MIC testing of gram-positive bacteria. Streptococcus pneumoniae ATCC 49619, Haemophilus influenzae ATCC 49247 and Haemophilus influenzae ATCC 49766 were used as quality controls during MIC testing of the fastidious organisms. Tigecycline, the major comparator for eravacycline, imipenem, meropenem and colistin to treat carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii, were selected in the panel of antibiotics to be tested. We evaluated eravacycline with a gradient concentration of 0.002–16 mg/L against common clinical gram-negative bacilli, gram-positive cocci, and fastidious organisms collected from our previous studies [9–13], including Enterobacteriaceae (Klebsiella pneumoniae, Escherichia coli, Enterobacter cloacae), Acinetobacter baumannii, Stenotrophomonas maltophilia, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Enterococcus faecalis, Enterococcus faecium, Streptococcus pneumoniae and Haemophilus influenzae. Antibiotic solutions for susceptibility testing were freshly prepared according to the manual of CLSI [8]. A scatter plot of eravacycline versus tigecycline was drawn for each species of bacteria, to reveal the relationship between the two antibiotics in different organisms. All the results related to resistant genes were readily available, directly from our previous researches [12–14]. Statistical analyses and data visualization were done with R (version 3.4.4) and ggplot2 package (version 2.2.1).
Table 1.
The strains involved in this study and antibiotic resistance characteristics of the strains
| Group | Identification | Resistance features | Number |
|---|---|---|---|
| Enterobacteriaceae | Klebsiella pneumoniae | ESBL | 10 |
| Tigecycline resistant | 13 | ||
| kpc-2 positive | 9 | ||
| NDM-1 positive | 3 | ||
| mcr-1 positive | 4 | ||
| Sensitive a | 10 | ||
| Escherichia coli | ESBL | 10 | |
| mcr-1, NDM-5 | 5 | ||
| Carbapenem resistant | 10 | ||
| Sensitive a | 10 | ||
| Enterobacter cloacae | ESBL | 6 | |
| Carbapenem resistant | 1 | ||
| Sensitive a | 22 | ||
| Non-fermentive | Acinetobacter baumanii | OXA-23 positive | 21 |
| Tigecycline resistant | 9 | ||
| Sensitive a | 9 | ||
| Stenotrophomonas maltophilia | Sensitive a | 29 | |
| Staphylococcus sp. | Staphylococcus aureus | MRSA | 15 |
| MSSA | 6 | ||
| Staphylococcus epidermidis | MRCoNS | 10 | |
| MSCoNS | 10 | ||
| Staphylococcus haemolyticus | MRCoNS | 8 | |
| MSCoNS | 1 | ||
| Staphylococcus hominis | MRCoNS | 6 | |
| MSCoNS | 4 | ||
| Enterococcus | Enterococcus faecalis | Sensitive a | 10 |
| Enterococcus faecium | VRE | 3 | |
| Sensitive a | 8 | ||
| Fastidious | Haemophilus influenzae | β-lactamase negative | 10 |
| β-lactamase positive | 10 | ||
| Streptococcus pneumoniae | PRSP | 10 | |
| PSSP | 10 |
a: Sensitive strains referred to strains do not have specific resistance characteristics such as ESBL, carbapenem resistance, polymyxin resistance and glycopeptide resistance
Results
In vitro activity of eravacycline was evaluated against 336 strains of clinically significant species, with many exhibiting resistance factors (Table 1). In most of the strains tested, the MIC50 and MIC90 values for eravacycline were lower than that of tigecycline and other comparable antibiotics tested for each organism/phenotypic group. Furthermore, eravacycline was highly effective against all of the organisms tested, regardless of resistance factors.
For Enterobacteriaceae bacteria, the MIC values of eravacycline varied with the resistance characteristics, especially for K. pneumoniae. The MIC50 values of eravacycline against E. cloacae and E. coli were much lower than the values of other comparable drugs, especially in strains with resistance phenotypes (Table 2). For K. pneumoniae, the MIC distribution of eravacycline differed depending on the drug resistance features. K. pneumoniae strains which were ESBL-positive (n = 10), kpc-2-positive (n = 9) and NDM-1-positive (n = 3), had similar MIC distributions. The MIC50 value of eravacycline against strains with the above three resistance mechanisms is 0.5 mg/L, and the MIC90 values were 1 mg/L, 2 mg/L and 1 mg/L respectively.
Table 2.
MIC distribution of Eravacycline and relevant antibiotics against E. coli and E. cloacae of different resistance characteristics
| Organism | Antibiotics | Carbapenem resistant a | ESBL | Sensitive b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | ||
| E.coli | Eravacycline | 0.5 | 1 | 0.064–2 | 0.125 | 0.25 | 0.064–0.25 | 0.064 | 0.125 | 0.064–0.25 |
| Tigecycline | 1 | 2 | 0.25–4 | 0.25 | 0.5 | 0.25–0.5 | 0.25 | 0.25 | 0.125–0.5 | |
| Piperacillin/Tazobactam | 256 | 256 | 2–256 | 2 | 8 | 1–256 | 1 | 2 | 0.5–2 | |
| Cefoxitin | 256 | 256 | 64–256 | 8 | 32 | 4–32 | 2 | 4 | 2–8 | |
| Ceftazidime | 256 | 256 | 0.5–256 | 32 | 64 | 16–128 | 0.064 | 0.25 | 0.064–0.25 | |
| Cefoperazone/Sulbactam | 256 | 256 | 8–256 | 16 | 32 | 8–256 | 0.25 | 1 | 0.064–4 | |
| Ceftriaxone | 256 | 256 | 2–256 | 256 | 256 | 64–256 | 0.032 | 0.064 | 0.016–0.064 | |
| Cefotaxime | 256 | 256 | 4–256 | 256 | 256 | 64–256 | 0.032 | 0.064 | 0.032–0.064 | |
| Cefepime | 64 | 256 | 0.25–256 | 32 | 64 | 8–128 | 0.016 | 0.032 | 0.016–0.064 | |
| Ertapenem | 32 | 32 | 16–32 | 0.125 | 0.25 | 0.016–1 | 0.016 | 0.016 | 0.016–0.016 | |
| Imipenem | 8 | 32 | 8–64 | 0.125 | 0.125 | 0.125–1 | 0.125 | 0.125 | 0.064–0.125 | |
| Meropenem | 8 | 32 | 4–32 | 0.032 | 0.064 | 0.016–0.064 | 0.016 | 0.016 | 0.016–0.016 | |
| Amikacin | 4 | 256 | 0.5–256 | 2 | 4 | 1–8 | 2 | 2 | 1–4 | |
| Minocycline | 8 | 16 | 0.5–16 | 1 | 8 | 0.5–16 | 1 | 2 | 0.5–8 | |
| Ciprofloxacin | 64 | 64 | 0.064–64 | 32 | 64 | 0.25–64 | 4 | 32 | 0.016–32 | |
| Levofloxacin | 16 | 64 | 0.125–128 | 16 | 32 | 0.5–64 | 8 | 8 | 0.032–16 | |
| Moxifloxacin | 16 | 32 | 0.5–64 | 16 | 32 | 0.5–64 | 8 | 16 | 0.032–16 | |
| E.cloacae | Eravacycline | 0.5 | 0.5 | 0.5–0.5 | 0.25 | 0.5 | 0.125–0.5 | 0.5 | 0.5 | 0.125–1 |
| Tigecycline | 2 | 2 | 2–2 | 1 | 1 | 0.125–2 | 0.5 | 2 | 0.5–2 | |
| Piperacillin/Tazobactam | 256 | 256 | 256–256 | 4 | 4 | 2–8 | 2 | 64 | 0.5–256 | |
| Cefoxitin | 256 | 256 | 256–256 | 8 | 32 | 4–256 | 256 | 256 | 64–256 | |
| Ceftazidime | 256 | 256 | 256–256 | 16 | 64 | 16–256 | 0.25 | 64 | 0.064–256 | |
| Cefoperazone/Sulbactam | 32 | 32 | 32–32 | 8 | 16 | 4–32 | 0.125 | 32 | 0.016–256 | |
| Ceftriaxone | 256 | 256 | 256–256 | 64 | 128 | 16–256 | 0.125 | 128 | 0.016–256 | |
| Cefotaxime | 256 | 256 | 256–256 | 64 | 128 | 16–256 | 0.125 | 256 | 0.016–256 | |
| Cefepime | 256 | 256 | 256–256 | 8 | 8 | 1–32 | 0.032 | 8 | 0.016–128 | |
| Ertapenem | 32 | 32 | 32–32 | 0.032 | 0.064 | 0.016–0.125 | 0.032 | 0.5 | 0.016–16 | |
| Imipenem | 32 | 32 | 32–32 | 0.25 | 0.25 | 0.125–0.25 | 0.25 | 1 | 0.125–2 | |
| Meropenem | 32 | 32 | 32–32 | 0.016 | 0.032 | 0.016–0.032 | 0.032 | 0.064 | 0.016–4 | |
| Amikacin | 256 | 256 | 256–256 | 1 | 2 | 1–8 | 1 | 2 | 0.5–256 | |
| Minocycline | 4 | 4 | 4–4 | 4 | 4 | 2–8 | 2 | 4 | 1–64 | |
| Ciprofloxacin | 64 | 64 | 64–64 | 2 | 32 | 0.25–64 | 0.032 | 4 | 0.016–64 | |
| Levofloxacin | 4 | 4 | 4–4 | 1 | 8 | 0.5–16 | 0.064 | 4 | 0.032–16 | |
| Moxifloxacin | 8 | 8 | 8–8 | 2 | 16 | 1–16 | 0.125 | 4 | 0.032–16 | |
a: Of the 15 carbapenem resistant E.coli, 5 strains harbored mcr-1 and NDM-5 simultaneously
b: Sensitive strains referred to strains do not have ESBL and carbapenem resistance
K. pneumoniae strains resistant to tigecycline were susceptible to eravacycline at higher MIC50 values of 8 mg/L, while the MIC90 was equivalent to that of tigecycline at 16 mg/L. For mcr-1 positive strains, the MIC50 of eravacycline was 1 mg/L compared with 16 mg/L for tigecycline, while the MIC90 of eravacycline and tigecycline was equivalent at 16 mg/L. The MIC50 (0.5 mg/L) and MIC90 (2 mg/L) values of eravacycline against carbapenem-resistant K. pneumoniae, were much lower than those of other antibiotics such as imipenem, meropenem, cephalosporins, and fluoroquinolones. The MIC distributions for K. pneumoniae of different resistant phenotypes to eravacycline, tigecycline, and other clinically common antibiotics are presented in Table 3.
Table 3.
MIC distribution of eravacycline and relevant antibiotics against K. pneumoniae of different resistance characteristics
| Antibiotics | Sensitive, n=10 | ESBL, n=10 | kpc-2 positive, n=9 | NDM-1 positive, n=3 | mcr-1 positive, n=4 | Tigecycline resistant, n=13 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Eravacycline | 0.25 | 0.5 | 0.125-0.5 | 0.5 | 1 | 0.125-2 | 0.5 | 2 | 0.25-4 | 0.5 | 1 | 0.5-1 | 1 | 16 | 0.5-16 | 8 | 16 | 2-16 |
| Tigecycline | 0.5 | 1 | 0.5-2 | 1 | 4 | 0.5-4 | 1 | 4 | 0.125 | 1 | 2 | 1--2 | 16 | 16 | 2-16 | 8 | 16 | 8-16 |
| Piperacillin/Tazobactam | 2 | 4 | 2-4 | 4 | 256 | 2-256 | 256 | 256 | 256-256 | 256 | 256 | 256-256 | 4 | 4 | 4-4 | 16 | 32 | 4-32 |
| Cefoxitin | 4 | 8 | 2-16 | 8 | 16 | 2-32 | 256 | 256 | 64-256 | 256 | 256 | 256-256 | 8 | 8 | 2-8 | 32 | 64 | 8-128 |
| Ceftazidime | 0.125 | 0.25 | 0.125-0.25 | 64 | 256 | 16-256 | 64 | 256 | 32-256 | 256 | 256 | 256-256 | 1 | 1 | 0.125-1 | 1 | 64 | 0.5-64 |
| Cefoperazone/Sulbactam | 0.25 | 0.25 | 0.125-0.25 | 16 | 64 | 8-64 | 256 | 256 | 256-256 | 256 | 256 | 256-256 | 1 | 1 | 0.5-1 | 2 | 32 | 1-128 |
| Ceftriaxone | 0.064 | 0.064 | 0.032-0.125 | 256 | 256 | 64-256 | 256 | 256 | 16-256 | 256 | 256 | 256-256 | 0.064 | 0.125 | 0.032-0.125 | 0.25 | 256 | 0.064-256 |
| Cefotaxime | 0.032 | 0.125 | 0.032-0.125 | 256 | 256 | 64-256 | 256 | 256 | 32-256 | 256 | 256 | 256-256 | 0.125 | 0.125 | 0.032-0.125 | 0.5 | 128 | 0.125-256 |
| Cefepime | 0.032 | 0.064 | 0.032-0.064 | 32 | 64 | 4-128 | 64 | 256 | 32-256 | 128 | 256 | 128-256 | 2 | 2 | 0.032-2 | 2 | 64 | 0.125-64 |
| Ertapenem | 0.016 | 0.016 | 0.016-0.016 | 0.25 | 0.5 | 0.032-0.5 | 32 | 32 | 32-32 | 32 | 32 | 32-32 | 0.016 | 0.016 | 0.016-0.016 | 0.032 | 0.25 | 0.016-0.5 |
| Imipenem | 0.125 | 0.25 | 0.125-1 | 0.125 | 0.25 | 0.125-0.25 | 8 | 32 | 8-32 | 8 | 32 | 8-32 | 0.125 | 0.25 | 0.125-0.25 | 0.125 | 0.125 | 0.125-0.5 |
| Meropenem | 0.016 | 0.032 | 0.016-0.032 | 0.032 | 0.064 | 0.032-0.125 | 16 | 32 | 8-32 | 16 | 32 | 8-32 | 0.032 | 0.064 | 0.032-0.064 | 0.032 | 0.064 | 0.016-0.064 |
| Colistin | 0.25 | 0.25 | 0.125-0.25 | 0.25 | 0.25 | 0.125-0.25 | 0.25 | 0.25 | 0.125-0.25 | 0.25 | 0.25 | 0.125-0.25 | 32 | 64 | 16-64 | 0.25 | 32 | 0.125-32 |
| Amikacin | 1 | 1 | 0.5-1 | 1 | 4 | 0.5-32 | 1 | 256 | 0.5-256 | 2 | 2 | 1--2 | 1 | 1 | 1-1 | 1 | 2 | 0.5-256 |
| Minocycline | 2 | 4 | 2-8 | 16 | 32 | 2-32 | 32 | 32 | 4-32 | 32 | 32 | 4-32 | 16 | 32 | 16-32 | 32 | 128 | 16-256 |
| Ciprofloxacin | 0.016 | 0.032 | 0.016-0.25 | 2 | 64 | 0.016-64 | 32 | 64 | 16-64 | 64 | 64 | 64-64 | 32 | 32 | 0.032-32 | 32 | 64 | 0.25-64 |
| Levofloxacin | 0.064 | 0.125 | 0.064-0.5 | 2 | 16 | 0.064-64 | 16 | 64 | 16-64 | 32 | 32 | 16-32 | 16 | 16 | 0.064-16 | 8 | 32 | 0.5-64 |
a: Sensitive strains referred to strains do not have ESBL, carbapenem resistance and polymyxin resistance
MIC distributions for A. baumannii also varied by resistance characteristics. A. baumannii isolates were tigecycline resistant and showed slightly elevated MIC50 and MIC90 for eravacycline at 2 mg/L. OXA-23-producing A. baumannii isolates have a MIC50 of 1 mg/L and MIC90 of 2 mg/L for eravacycline, and these values were much lower than the MIC50 and MIC90 of tigecycline (4 mg/L, 4 mg/L), imipenem (64 mg/L, 64 mg/L), and meropenem (32 mg/L, 64 mg/L). The MIC distributions for A. baumannii with different resistant phenotypes to eravacycline, tigecycline, and other clinically relevant antibiotics such as imipenem, meropenem, and colistin are presented in Table 4.
Table 4.
MIC distribution of Eravacycline and relevant antibiotics against A. baumannii of different resistance characteristics
| Antibiotics | Sensitive a, n = 9 | OXA-23 positive, n = 21 | Tigecycline resistant, n = 9 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Eravacycline | 0.125 | 0.25 | 0.016–0.25 | 1 | 2 | 0.5–2 | 2 | 2 | 2–4 |
| Tigecycline | 0.25 | 0.5 | 0.25–0.5 | 4 | 4 | 4–8 | 8 | 8 | 8–8 |
| Piperacillin/Tazobactam | 2 | 4 | 0.016–8 | 256 | 256 | 256–256 | 256 | 256 | 256–256 |
| Ceftazidime | 2 | 8 | 0.125–32 | 256 | 256 | 64–256 | 256 | 256 | 256–256 |
| Cefepime | 1 | 4 | 0.032–32 | 64 | 256 | 32–256 | 256 | 256 | 128–256 |
| Imipenem | 0.125 | 1 | 0.125–1 | 64 | 64 | 16–64 | 64 | 64 | 64–128 |
| Meropenem | 0.032 | 1 | 0.016–1 | 32 | 64 | 16–64 | 64 | 64 | 32–128 |
| Colistin | 0.125 | 0.25 | 0.125–0.25 | 0.25 | 0.25 | 0.125–0.25 | 0.25 | 0.25 | 0.25–0.25 |
| Amikacin | 4 | 4 | 1–4 | 256 | 256 | 256–256 | 256 | 256 | 256–256 |
| Minocycline | 0.125 | 16 | 0.064–16 | 8 | 16 | 4–16 | 8 | 8 | 8–16 |
| Ciprofloxacin | 0.125 | 0.5 | 0.032–32 | 32 | 32 | 32–32 | 32 | 32 | 32–32 |
| Levofloxacin | 0.125 | 1 | 0.064–32 | 16 | 32 | 8–32 | 16 | 16 | 16–32 |
a: Sensitive strains referred to strains do not have carbapenem resistance and tigecycline resistance
For S. maltophilia there is no breakpoints available for tigecycline, the MIC distributions of tigecycline and eravacycline against S. maltophilia were evaluated. The MIC50 and MIC90 for eravacycline were both 1 mg/L, at the same time the MIC50 and MIC90 for tigecycline were 0.5 mg/L and 1 mg/L.
For Staphylococcus spp., the results indicated that MIC50 and MIC90 of eravacycline were 0.25 mg/L and 0.5 mg/L, respectively, for MRSA (methicillin-resistant S. aureus), for MSSA (methicillin-sensitive S. aureus) the MIC50 of eravacycline was as low as 0.064 mg/L, and MIC90 remained the same as that of MRSA. MIC50 and MIC90 of eravacycline for methicillin-resistant coagulase-negative staphylococci (MRCoNS) were 0.25 mg/L and 1 mg/L, respectively, and for MSCoNS (methicillin-sensitive coagulase-negative staphylococci) the values of eravacycline were lower at 0.016 mg/L and 0.25 mg/L, respectively. For other antibiotics, the values are presented in Table 5.
Table 5.
MIC distribution of Eravacycline and relevant antibiotics against Staphylococcus. spp of different resistance characteristics
| Antibiotics | MRSAa, N = 15 | MSSAb, N = 6 | MRCoNSc, N = 24 | MSCoNSd, N = 15 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | ||
| Eravacycline | 0.25 | 0.5 | 0.032–1 | 0.064 | 0.5 | 0.016–2 | 0.25 | 1 | 0.016–2 | 0.016 | 0.25 | 0.008–0.25 | |
| Tigecycline | 0.25 | 0.5 | 0.125–0.5 | 0.25 | 0.25 | 0.125–0.25 | 0.25 | 0.5 | 0.125–0.5 | 0.125 | 0.25 | 0.064–0.25 | |
| Oxacillin | 64 | 64 | 2–64 | 0.25 | 0.5 | 0.25–0.5 | 2 | 64 | 0.5–256 | 0.125 | 0.25 | 0.125–0.25 | |
| Cefoxitin | 256 | 256 | 32–256 | 4 | 4 | 2–4 | 16 | 256 | 2–256 | 2 | 8 | 1–8 | |
| Vancomycin | 1 | 1 | 0.5–1 | 0.5 | 0.5 | 0.5–0.5 | 1 | 2 | 0.5–2 | 0.5 | 1 | 0.25–1 | |
| Teicoplanin | 2 | 2 | 0.5–2 | 0.5 | 0.5 | 0.5–1 | 2 | 4 | 0.064–8 | 0.5 | 2 | 0.125–2 | |
| Erythromycin | 256 | 256 | 0.25–256 | 256 | 256 | 0.25–256 | 64 | 256 | 0.125–256 | 0.25 | 256 | 0.064–256 | |
| Minocycline | 4 | 16 | 0.064–32 | 0.064 | 0.125 | 0.064–0.125 | 0.25 | 0.5 | 0.064–8 | 0.125 | 0.25 | 0.064–0.5 | |
| Ciprofloxacin | 64 | 64 | 0.25–64 | 0.5 | 0.5 | 0.25–0.5 | 16 | 64 | 0.125–64 | 0.25 | 8 | 0.125–64 | |
| Levofloxacin | 32 | 64 | 0.25–64 | 0.25 | 0.25 | 0.125–0.5 | 4 | 128 | 0.25–128 | 0.25 | 0.5 | 0.125–128 | |
| Moxifloxacin | 8 | 16 | 0.016–32 | 0.032 | 0.064 | 0.016–0.064 | 1 | 16 | 0.064–32 | 0.064 | 1 | 0.032–16 | |
| Trimethoprim/Sulfamethoxazole | 0.125 | 16 | 0.032–16 | 0.032 | 0.064 | 0.032–0.25 | 4 | 32 | 0.064–64 | 0.125 | 4 | 0.016–4 | |
| Chloramphenicol | 8 | 8 | 4–32 | 8 | 8 | 4–64 | 4 | 8 | 2–64 | 4 | 4 | 2–8 | |
| Rifampin | 256 | 256 | 0.004–256 | 0.008 | 0.016 | 0.004–0.016 | 0.008 | 256 | 0.004–256 | 0.008 | 0.016 | 0.004–0.016 | |
| Clindamycin | 128 | 256 | 0.064–256 | 0.064 | 256 | 0.064–256 | 0.125 | 256 | 0.064–256 | 0.064 | 0.125 | 0.064–0.25 | |
| Linezolid | 1 | 2 | 0.5–2 | 1 | 2 | 1–2 | 1 | 1 | 0.5–1 | 1 | 1 | 0.5–2 | |
a Methicillin-resistant Staphylococcus aureus. b Methicillin- sensitive Staphylococcus aureus
c Methicillin-resistant coagulase-negative staphylococci. d Methicillin- sensitive coagulase-negative staphylococci
In the results obtained for Enterococcus spp. it was found that MIC50 and MIC90 of eravacycline for E. faecalis were both 0.032 mg/L. The MIC50 and MIC90 of eravacycline for E. faecium were 0.016 mg/L and 0.032 mg/L. For Vancomycin-Resistant Enterococci (VRE) strains, the MIC50 and MIC90 were identical with that of vancomycin-susceptible E. faecium strains. For other antibiotics, the values are presented in Table 6. In general, for gram-positive bacteria with varying resistance factors, eravacycline demonstrated substantial antibacterial activity.
Table 6.
MIC distribution of Eravacycline and relevant antibiotics against Enterococci. spp of different resistance characteristics
| Antibiotics | E.faecalis, n = 10 | E.faecium, n = 8 | VREa, n = 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Eravacycline | 0.032 | 0.032 | 0.016–0.125 | 0.016 | 0.032 | 0.008–0.064 | 0.016 | 0.032 | 0.008–0.032 |
| Tigecycline | 0.064 | 0.064 | 0.064–0.125 | 0.064 | 0.064 | 0.016–0.125 | 0.125 | 0.25 | 0.125–0.25 |
| Ampicillin | 1 | 8 | 1–8 | 64 | 64 | 4–64 | 64 | 64 | 64–64 |
| Vancomycin | 1 | 2 | 0.5–2 | 0.5 | 1 | 0.25–1 | 128 | 128 | 128–128 |
| Teicoplanin | 0.125 | 0.25 | 0.032–0.25 | 0.25 | 0.25 | 0.064–0.25 | 32 | 64 | 32–64 |
| Erythromycin | 1 | 256 | 0.25–256 | 256 | 256 | 0.016–256 | 0.125 | 256 | 0.125–256 |
| Minocycline | 16 | 16 | 0.064–16 | 0.032 | 16 | 0.032–16 | 0.064 | 16 | 0.064–16 |
| Ciprofloxacin | 2 | 32 | 0.5–64 | 64 | 64 | 4–64 | 64 | 64 | 64–64 |
| Levofloxacin | 2 | 64 | 1–64 | 64 | 128 | 1–128 | 64 | 64 | 64–64 |
| Linezolid | 1 | 2 | 1–2 | 1 | 1 | 0.5–1 | 1 | 1 | 1–1 |
a VRE referred to vancomycin-resistant Enterococci. All of the 3 VRE strains in this study were E.faecium
For fastidious strains, including 20 S. pneumoniae isolates and 20 H. influenzae isolates, eravacycline showed high antimicrobial activities against S. pneumoniae with MIC50 (0.008 mg/L) and MIC90 (0.008 mg/L), there was no difference with eravacycline distribution between PRSP (Penicillin-resistant S. pneumoniae) and PSSP (Penicillin-sensitive S. pneumoniae) strains (Table 7). For H. influenzae the MIC50 and MIC90 were 0.064 mg/L and 0.125 mg/L, and they were the same in both β-lactamase-positive and β-lactamase-negative strains (Table 8).
Table 7.
MIC distribution of Eravacycline and relevant antibiotics against S.pneumoniae of different resistance characteristics
| Antibiotics | PSSPa, n = 10 | PRSPb, n = 10 | ||||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Eravacycline | 0.008 | 0.008 | 0.002–0.016 | 0.008 | 0.008 | 0.004–0.008 |
| Tigecycline | 0.016 | 0.016 | 0.008–0.016 | 0.016 | 0.016 | 0.016–0.016 |
| Penicillin | 0.016 | 0.016 | 0.016–0.032 | 4 | 4 | 4–4 |
| Amoxicillin/Clavulanic acid | 0.016 | 0.064 | 0.008–0.25 | 8 | 8 | 8–8 |
| Cefuroxime | 0.032 | 0.125 | 0.016–0.5 | 16 | 32 | 8–32 |
| Cefaclor | 1 | 2 | 1–4 | 256 | 256 | 128–256 |
| Ceftriaxone | 0.032 | 0.064 | 0.016–0.125 | 2 | 8 | 1–8 |
| Erythromycin | 8 | 32 | 0.5–256 | 256 | 256 | 128–256 |
| Azithromycin | 16 | 32 | 4–256 | 256 | 256 | 256–256 |
| Clindamycin | 0.125 | 128 | 0.032–256 | 256 | 256 | 128–256 |
| Clarithromycin | 2 | 32 | 0.25–256 | 256 | 256 | 256–256 |
| Levofloxacin | 1 | 1 | 0.25–32 | 1 | 1 | 1–1 |
| Moxifloxacin | 0.125 | 0.125 | 0.064–16 | 0.125 | 0.25 | 0.125–0.25 |
| Trimethoprim/Sulfamethoxazole | 4 | 8 | 0.064–8 | 8 | 16 | 4–32 |
| Tetracycline | 32 | 64 | 4–64 | 32 | 32 | 32–32 |
| Chloramphenicol | 4 | 8 | 1–16 | 4 | 4 | 4–4 |
| Vancomycin | 0.25 | 0.25 | 0.125–0.25 | 0.25 | 0.25 | 0.25–0.25 |
a PSSP Penicillin-sensitive Streptococcus pneumoniae
b PRSP Penicillin-resistant Streptococcus pneumoniae
Table 8.
MIC distribution of Eravacycline and relevant antibiotics against H. influenza of different resistance characteristics
| Antibiotics | β-lactamases negative, n = 10 | β-lactamases positive, n = 10 | ||||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Eravacycline | 0.064 | 0.125 | 0.064–0.125 | 0.064 | 0.125 | 0.032–0.125 |
| Tigecycline | 0.25 | 0.5 | 0.125–0.5 | 0.125 | 0.25 | 0.064–0.5 |
| Ampicillin | 0.125 | 0.5 | 0.125–1 | 16 | 64 | 0.064–64 |
| Amoxicillin/Clavulanic acid | 0.125 | 0.5 | 0.125–0.5 | 1 | 1 | 0.5–1 |
| Penicillin | 16 | 32 | 0.032–32 | 16 | 32 | 1–64 |
| Cefaclor | 2 | 8 | 0.5–8 | 4 | 16 | 1–32 |
| Cefuroxime | 1 | 2 | 0.25–4 | 1 | 4 | 0.25–16 |
| Azithromycin | 1 | 4 | 0.064–4 | 2 | 64 | 0.25–64 |
| Clarithromycin | 4 | 16 | 0.5–16 | 4 | 64 | 1–64 |
| Levofloxacin | 0.032 | 1 | 0.016–1 | 0.032 | 0.125 | 0.016–0.5 |
| Moxifloxacin | 0.032 | 1 | 0.016–1 | 0.032 | 0.25 | 0.016–0.5 |
| Trimethoprim/Sulfamethoxazole | 16 | 32 | 0.032–32 | 16 | 32 | 1–64 |
| Tetracycline | 1 | 4 | 0.064–4 | 2 | 64 | 0.25–64 |
| Chloramphenicol | 0.5 | 1 | 0.25–1 | 1 | 8 | 0.5–8 |
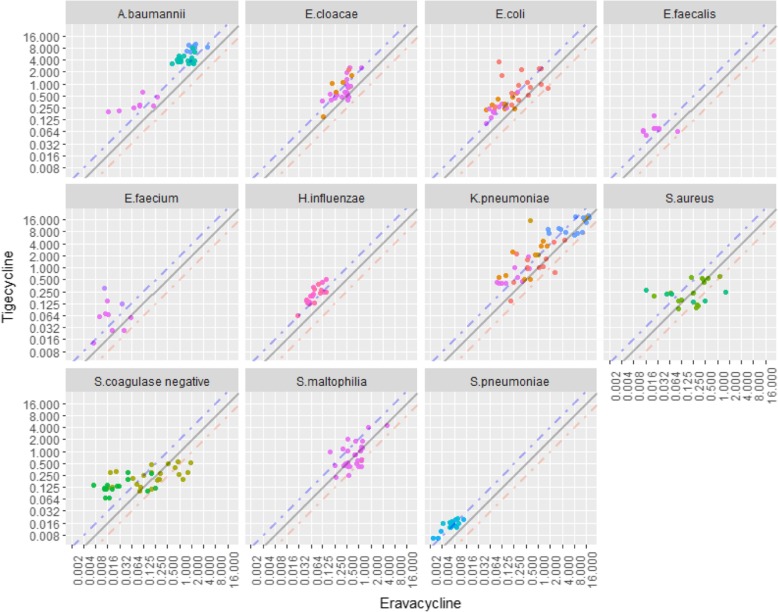
A jittered scatter plot was drawn using the MIC values of eravacycline and tigecycline involving all the strains tested. A clear pattern was found showing that most of the MIC values of tigecycline are higher than the corresponding MIC values of eravacycline (in many cases by 2 to 4 fold). For all of the clinical isolates tested, except for Staphylococcus spp. and S. maltophilia, more points are located above the diagonal y = x line, suggesting that eravacycline has lower MIC distribution than tigecycline (Fig. 1). For Staphylococcus spp. and S. maltophilia the points were distributed on both sides of the diagonal evenly, suggesting a comparable MIC distribution between eravacycline and tigecycline.
Fig. 1.
Scatter plot of MIC values of tigecycline versus MIC value of eravacycline against various bacteria. Note: A tiny displacement was made to the points in this figure in order to avoid overlapping of the strains with the same eravacycline and tigecycline MIC values. This tiny displacement can ensure the actual distribution of all strains visible. The points on the grey solid line indicated the strains shared the identical eravacycline and tigecycline MIC values. The points above the blue dash line indicated that the MIC values of tigecycline were greater than twice than the MIC values of eravacycline. The points below the orange dash line indicated that the MIC values of eravacycline were greater than twice than the MIC values of tigecycline
Legends:  Carbapenem resistant;
Carbapenem resistant;  ESBL;
ESBL;  mcr-1;
mcr-1;  MRCoNS;
MRCoNS;  MRSA;
MRSA;  MSCoNS;
MSCoNS;  MSSA;
MSSA;  OXA-23;
OXA-23;  PRSP;
PRSP;  PSSP;
PSSP;  Tigecycline resistant;
Tigecycline resistant;  VRE;
VRE;  without resistance gene;
without resistance gene;  β-lactamases –;
β-lactamases –;  β-lactamases +.
β-lactamases +.
Discussion
As resistance to antibiotics grows worldwide, it becomes increasingly important to find new treatments for bacterial infections. In the present study, a new antibiotic eravacycline was compared to existing medications. Eravacycline demonstrated high in vitro activity against clinical isolates, including strains with specific resistant factors. Eravacycline was compared to a derivative of tigecycline, and in most cases presented with a lower MIC distribution for the majority of strains tested in this study. Since many years nosocomial pathogens, such as Enterobacteriaceae which are responsible for complicated intra-abdominal infection (cIAI) were increasing in frequency [15]. Moreover, cases of gram-positive cocci such as S. aureus, coagulase-negative staphylococci, and enterococci, the major causative organisms of complicated urinary tract infections (cUTI) were also increasing [16]. The emergence of multiple drug-resistant bacteria, such as Carbapenem resistant Enterobacteriaceae bacteria (CRE), Carbapenem-resistant Acinetobacter baumannii (CRAB) and Methicillin-resistant Staphylococcus aureus (MRSA), has compounded this problem significantly by increasing the difficulty of treatment, the proportion of failures, as well as the mortality rate of patients. Since Tigecycline and eravacycline belong to a different antibiotic class with a mechanism of action distinct from cephalosporins and carbapenem antibiotics, they can evade established resistance mechanisms of Enterobacteriaceae and exhibit higher efficacy against resistant bacteria. In this study, eravacycline showed high antibacterial activity against CRE strains, suggesting that eravacycline could be useful to treat complicated infections caused by CRE. Similarly, CRAB also shows resistance to antibiotics which were commonly used during the clinical practice. CRAB is the most notorious pathogen responsible for nosocomial infections in China at present [17–19]. This study found that the most effective drug for OXA-23 producing A. baumannii was colistin then eravacycline. Eravacycline also demonstrated high potency against OXA-23 producing A. baumannii, with a MIC50 of 1 mg/L which was much lower than other antibiotics, except for colistin. Similar to eravacycline in structure and mechanism, tigecycline has been widely utilized in China for many years, and tigecycline-resistant strains have also emerged with the increase in use of this antibiotic [20, 21]. In the present study, eravacycline also exhibited lower MIC distribution compared with tigecycline in tigecycline-resistant strains, suggesting that the mechanism which leads to tigecycline resistance does not inhibit the activity of eravacycline. Furthermore, high antibiotic potency against CRE and CRAB could make eravacycline a potential option to treat complex infections including respiratory and bloodstream infections. For Staphylococcus spp. the results were entirely different, with tigecycline values much lower than eravacycline. From the scatter plot we observed that the points are evenly distributed on both sides of the diagonal line (line: y = x). This may be either due to the combined effects of different resistance mechanisms, or potentially unknown resistance mechanisms. In addition, the total number of Staphylococcus spp. strains which were tested in this study was relatively small, which may cause random errors in the antibacterial activity of eravacycline. Thus, further validation utilizing different bacterial isolates is required. For fastidious strains, eravacycline demonstrated excellent potency despite resistance characteristics of the strains. From the scatter plot, we can see that although MIC values of eravacycline were generally lower than those of tigecycline, the MIC values of eravacycline were also rising with the MIC values of tigecycline proportionally, thus, we need to be alert to the possible cross-resistance potential of eravacycline and tigecycline, especially in strains with higher MIC values of tigecycline.
Limitation and suggestion
The clinical isolates tested were limited by country as they were exclusively collected in China and within this country, these isolates were only obtained from 11 teaching hospitals. No strains from other hospitals were utilized. Therefore, many different clinical isolates remain untested. Thus, it is important that researchers reproduce our work in other countries with different isolates in order to understand the full spectrum of this new antibiotics’ efficacy. The results of this study show that eravacycline has a positive application potential for the treatment of current drug-resistant bacterial infections. Considering the relatively small number of each organism and limited types of resistant phenotypes, the result of this study only partially represent the resistant phenotype encountered in real clinical practice, and additional studies are needed for a more comprehensive assessment of the antibacterial activity of eravacycline.
Conclusions
The results of this study proved that eravacycline possesses a broad spectrum of activity against a variety of gram-positive and gram-negative bacteria, including multi-drug resistant strains such as A. baumannii and carbapenem-resistant Enterobacteriaceae.
Additional file
The list of committee and the institute to which it belongs for all hospitals that provided Administrative Consent to access or receive samples. This additional file list the committee (and the institute to which it belongs) for all hospitals that provided Administrative Consent to access or receive samples/data (DOCX 13 kb)
Acknowledgements
Not Applicable.
Abbreviations
- CLSI
Clinical and Laboratory Standards Institute
- CRAB
Carbapenem resistant Acinetobacter baumannii
- CRE
Carbapenem resistant Enterobacteriaceae
- cUTI
complicated urinary tract infections
- ESBL
extended-spectrum-lactamases
- MIC
minimum inhibitory concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- MSCoNS
Methicillin- sensitive coagulase-negative staphylococci
- PCR
polymerase chain reaction
- PRSP
penicillin resistant Streptococcus pneumoniae
- VRE
Vancomycin-resistant enterococci
Authors’ contributions
HW, CZ conceived and designed experiments. CZ, XW, YZ, RW, QW and HL performed antibiotic susceptibility testing. HW, CZ wrote the manuscript. CZ performed the data processing and data visualization. All authors read and approved the final manuscript.
Funding
No funding was obtained for this study.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Study protocols were reviewed and granted by the Ethical Committee of Peking University People’s Hospital (No. 2017PHB163). For the hospitals participated, administrative permissions to access the raw samples were granted by the Research Department of the hospitals participated.
Consent for publication
Not applicable as no human subjects.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunjiang Zhao, Email: chunkiang@163.com.
Xiaojuan Wang, Email: curliele@163.com.
Yawei Zhang, Email: z_yw1990@163.com.
Ruobing Wang, Email: ruobing10612@163.com.
Qi Wang, Email: wangqi99887@sina.com.
Henan Li, Email: lhnpku@163.com.
Hui Wang, Phone: +86-10-88326300, Email: whuibj@163.com.
References
- 1.Yang Y, Song W, Lin H, Wang W, Du L, Xing W. Antibiotics and antibiotic resistance genes in global lakes: a review and meta-analysis. Environ Int. 2018;116:60–73. doi: 10.1016/j.envint.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Qiao M, Ying G-G, Singer AC, Zhu Y-G. Review of antibiotic resistance in China and its environment. Environ Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Grossman TH, Murphy TM, Slee AM, Lofland D, Sutcliffe JA. Eravacycline (TP-434) is efficacious in animal models of infection. Antimicrob Agents Chemother. 2015;59:2567–2571. doi: 10.1128/AAC.04354-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomkin JS, Ramesh MK, Cesnauskas G, Novikovs N, Stefanova P, Sutcliffe JA, et al. Phase 2, randomized, double-blind study of the efficacy and safety of two dose regimens of eravacycline versus ertapenem for adult community-acquired complicated intra-abdominal infections. Antimicrob Agents Chemother. 2014;58:1847–1854. doi: 10.1128/AAC.01614-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutcliffe JA, O’Brien W, Fyfe C, Grossman TH. Antibacterial activity of Eravacycline (TP-434), a novel Fluorocycline, against hospital and community pathogens. Antimicrob Agents Chemother. 2013;57:5548–5558. doi: 10.1128/AAC.01288-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomkin J, Evans D, Slepavicius A, Lee P, Marsh A, Tsai L, et al. Assessing the efficacy and safety of Eravacycline vs Ertapenem in complicated intra-abdominal infections in the investigating gram-negative infections treated with Eravacycline (IGNITE 1) trial: a randomized clinical trial. JAMA Surg. 2017;152:224–232. doi: 10.1001/jamasurg.2016.4237. [DOI] [PubMed] [Google Scholar]
- 8.CLSI . Methods for Dilution Antimicrobial Susceptibility Tests f or Bacteria That Grow Aerobically; Approved St andard. 9. Wayne: Clin Lab Stand Inst; 2012. [Google Scholar]
- 9.Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60:6115–6120. doi: 10.1128/AAC.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Chen H, Zhang Y, Wang Q, Zhao C, Li H, et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents. 2015;45:635–640. doi: 10.1016/j.ijantimicag.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Inf Secur. 2015;71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Zhang F, Zhao C, Wang Z, Nichols WW, Testa R, et al. In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob Agents Chemother. 2014;58:1774–1778. doi: 10.1128/AAC.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Xu X, Li Z, Chen H, Wang Q, Yang P, et al. An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb Drug Resist. 2014;20:144–149. doi: 10.1089/mdr.2013.0100. [DOI] [PubMed] [Google Scholar]
- 14.Wang Qi, Wang Xiaojuan, Wang Juan, Ouyang Pengwen, Jin Chunmei, Wang Ruobing, Zhang Yawei, Jin Longyang, Chen Hongbin, Wang Zhanwei, Zhang Feifei, Cao Bin, Xie Liangyi, Liao Kang, Gu Bing, Yang Chunxia, Liu Zhiwu, Ma Xiaobo, Jin Liang, Zhang Xiaoqian, Man Sijin, Li Wei, Pei Fengyan, Xu Xiuli, Jin Yan, Ji Ping, Wang Hui. Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: Data From a Longitudinal Large-scale CRE Study in China (2012–2016) Clinical Infectious Diseases. 2018;67(suppl_2):S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 15.Sartelli M, Catena F, Ansaloni L, Coccolini F, Corbella D, Moore EE, et al. Complicated intra-abdominal infections worldwide: the definitive data of the CIAOW study. World J Emerg Surg. 2014;9:37. doi: 10.1186/1749-7922-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallett A, Hand K. Complicated urinary tract infections: practical solutions for the treatment of multiresistant gram-negative bacteria. J Antimicrob Chemother. 2010;65(Suppl 3):iii25–iii33. doi: 10.1093/jac/dkq298. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Hu J, Harbarth S, Pittet D, Zhou M, Zingg W. Burden of healthcare-associated infections in China: results of the 2015 point prevalence survey in dong Guan City. J Hosp Infect. 2017;96:132–138. doi: 10.1016/j.jhin.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Gong Y, Shen X, Huang G, Zhang C, Luo X, Yin S, et al. Epidemiology and resistance features of Acinetobacter baumannii isolates from the ward environment and patients in the burn ICU of a Chinese hospital. J Microbiol. 2016;54:551–558. doi: 10.1007/s12275-016-6146-0. [DOI] [PubMed] [Google Scholar]
- 19.Yuan X, Liu T, Wu D, Wan Q. Epidemiology, susceptibility, and risk factors for acquisition of MDR/XDR gram-negative bacteria among kidney transplant recipients with urinary tract infections. Infect Drug Resist. 2018;11:707–715. doi: 10.2147/IDR.S163979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng M, Zhu M-H, Li J-J, Bi S, Sheng Z-K, Hu F-S, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014;58:297–303. doi: 10.1128/AAC.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du X, He F, Shi Q, Zhao F, Xu J, Fu Y, et al. The rapid emergence of Tigecycline resistance in blaKPC-2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during Tigecycline treatment. Front Microbiol. 2018;9:648. doi: 10.3389/fmicb.2018.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The list of committee and the institute to which it belongs for all hospitals that provided Administrative Consent to access or receive samples. This additional file list the committee (and the institute to which it belongs) for all hospitals that provided Administrative Consent to access or receive samples/data (DOCX 13 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.