Abstract
Background
This study aimed to investigate the role of the long noncoding RNA (lncRNA), LINC01555, on the migration and invasion of colorectal cancer (CRC) cells, its expression in CRC tissue, and its interaction with the neuropeptide, neuromedin U (NmU).
Material/Methods
LINC01555 expression in SW620 and HCT116 CRC cells, and NCM460 normal colorectal cells, and 48 resection specimens containing CRC and adjacent normal tissue, was detected by quantitative real-time polymerase chain reaction (qRT-PCR). Cox regression analysis was used to assess the relationship between LINC01555 expression and patient survival. The effects of LINC01555 expression on CRC cell proliferation, migration, and invasion were assessed using the cell counting kit-8 (CCK-8) assay, the colony formation assay, and the transwell assay. Functional studies determined the interaction between LINC01555 and NmU in the development of CRC.
Results
The Cancer Genome Atlas (TCGA) dataset showed that LINC01555 was highly expressed in CRC tissue when compared with adjacent normal colorectal tissue. LINC01555 expression was positively correlated with tumor stage, but negatively correlated with disease-free survival (DFS) and overall survival (OS) and was an independent risk factor for CRC. The receiver operating characteristic (ROC) curve analysis showed the diagnostic specificity of LINC01555 in CRC. Knockdown of LINC01555 inhibited cell proliferation, migration, and invasion of CRC cells. Functional studies showed that knockdown of NmU reduced cell migration and invasion of CRC cells that overexpressed LINC01555.
Conclusions
Increased expression of LINC01555 was found in CRC tissues and promoted the invasion of CRC cells by upregulating the expression of NmU.
MeSH Keywords: Colorectal Neoplasms; Neoplasm Invasiveness; Neoplasm Metastasis; RNA, Long Noncoding
Background
Patients with colorectal cancer (CRC) are managed with a multidisciplinary team (MDT), with the first-line treatment being surgery. Recent developments in targeted therapy using monoclonal antibodies and anti-angiogenic drugs has improved the efficacy of treatment for patients with CRC. However, patients with advanced-stage CRC, postoperative recurrence and chemotherapy resistance remain as factors that reduce patient prognosis [1–3]. CRC seriously affects both health and quality of life and is a global health problem that requires further research. The pathogenesis of CRC involves long-term exposure to environmental factors and genetic factors. Studies have shown that mutations and epigenetic modifications of tumor-suppressor genes, including p53, signaling pathway abnormalities, and multidimensional regulation of noncoding RNA on protein-coding genes are closely associated with the development of CRC [4–6].
A long noncoding RNA (lncRNA) contains more than 200 bases, does not have an open reading frame, and rarely encodes protein. LncRNA was considered initially to represent irrelevant noise from genomic transcription, and to be a byproduct of the transcription process without any biological function [7,8]. Recent studies have identified the physiological functions of lncRNA in regulating protein-encoding genes at the epigenetic, transcriptional, and post-transcriptional levels. LncRNA is capable of altering protein structure, influencing protein distribution, function, and activity [9]. During the process of lncRNA-induced gene encoding, lncRNA participates in chromatin modification, transcriptional activation, transcriptional interference, selective splicing, nuclear transport, and regulation of proto-oncogene activation. The regulatory effects of lncRNA are also found in malignant tumors, mediating the occurrence, development, invasion, and metastasis of tumor cells [10,11].
Therefore, this study aimed to investigate the role of the lncRNA, LINC01555, on the migration and invasion of CRC cells, its expression in CRC tissue, and its interaction with the neuropeptide, neuromedin U (NmU). RNA sequencing data from CRC tissue and adjacent normal tissue were downloaded from The Cancer Genome Atlas (TCGA) to characterize aberrantly expressed lncRNAs,
Material and Methods
Patients and ethics
Forty-eight pairs of surgically resected colorectal cancer (CRC) and adjacent normal colorectal tissues were obtained from patients undergoing surgical resection for CRC in Taizhou Peoples’ Hospital between 2015 to 2016. None of the patients were treated with pre-operative radiotherapy or chemotherapy. The patients included in the study did not have other types of malignancy, autoimmune disease, or severe chronic disease. Tissue samples were immediately preserved in liquid nitrogen after surgical resection. The study subjects and their families performed written informed consent for tissue sampling. This experiment was approved by the Medical Ethics Committee of the Taizhou Peoples’ Hospital.
Cell culture
CRC cell lines, SW620 and HCT116, and the normal colorectal cell line, NCM460, were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermofisher Scientific, Rockville, MD, USA) containing 10% fetal bovine serum (FBS) (Hyclone, South Logan, UT, USA), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA), and cultured in an incubator at 37°C in a 5% CO2 atmosphere.
Cell transfection
Cells in the logarithmic growth phase were selected and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). When cell confluence reached 40–60%, short-interfering (si)-LINC01555, si-NMU, pcDNA-LINC01555, or the negative control (NC) were transfected. Six hours later, serum-free medium was replaced by DMEM containing 10% FBS. Construction of the pcDNA-LINC01555 plasmid was performed by OBiO Technology (Shanghai, China). Primers of si-LINC01555 and si-NMU were constructed by GenePharma (Shanghai, China).
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Tissue and cellular RNAs were extracted by TRIzol (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The RNA concentration was measured, and RNA samples were stored at −80°C. Reverse transcription was performed according to the instructions of the TaKaRa OneStep PrimeScript® miRNA cDNA Synthesis Kit (TaKaRa, Tokyo, Japan). PCR detection was performed using the SYBR Green I fluorescent dye method. Amplification conditions were 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the loading control. Primers were as follows:
Forward, LINC01555: CTCAGGCATCCAACGCACT;
Reverse, LINC01555: GACTTGCCCAACTTCTGTGTC;
Forward, NMU: GTCAAATGTTGTGTCGTCAGTTG;
Reverse, NMU: CCATTCCGTGGCCTGAATAAA.
Cell counting kit-8 (CCK-8) assay
The CCK-8 assay (Beyotime, Shanghai, China) was performed to evaluate cell proliferation following the manufacturer’s recommendations. Cells were cultured in 96-well plates with 3×103 cells per well for 24 hours and incubated with CCK-8 reagent for 1 hour. The absorbance at a wavelength of 490 nm was measured using a TECAN Infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium).
Colony formation assay
CRC cells were seeded in a 6-well plate with 1×103 cells per well and cultured at 37°C in an atmosphere of 5% CO2 for 12 days. Cells were washed twice with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 40 min, and stained with 0.1% crystal violet for 20 min at room temperature. Colonies were imaged under a microscope at a magnification of ×10) and imaged using Image-Pro Plus 6.0 Software (Media Cybernetics, Silver Springs, MD, USA).
Cell migration and invasion assays
Cell migration was measured using the 8-μm transwell chamber (Millipore Corporation, Billerica, MA, USA), with 2×105 cells suspended in 200 μL and the addition of serum-free medium to the upper chamber that was precoated with diluted Matrigel. Then, 600 μL of medium containing 10% FBS was added to the lower chamber. After incubation for 48 hours, non-migratory cells on the upper side were carefully wiped off. The remaining cells were fixed and stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 15 min and then photographed using a microscope at a magnification of ×20. The invasion assay was the same as that described above, except for the Matrigel precoating.
Western blot
Proteins were extracted by radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China). An equal amount of protein sample was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Membranes were blocked with dried skimmed milk powder, and incubated with primary antibody (Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. The following day, the membranes were incubated with secondary antibody for 2–3 h at room temperature. Finally, an image of the protein band was captured by using the electrochemiluminescence (ECL) reagent (Thermofisher Scientific, Waltham, MA, USA). GAPDH was used as the loading control.
Statistical analysis
Data were analyzed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Data were presented as the mean ± standard deviation (SD). Differences between two groups were compared using Student’s t-test. The Cox regression model for univariate and multivariate survival analysis was performed to explore the correlation between LINC01555 expression and patient survival, including the disease-free survival (DFS) or overall survival (OS) of patients with CRC. The receiver operating characteristic (ROC) curve was used to evaluate the diagnostic and prognostic value of LINC01555 in CRC. A P-value <0.05 was considered to be statistically significant.
Results
Expression of the long noncoding RNA (lncRNA), LINC01555, was increased in colorectal cancer (CRC)
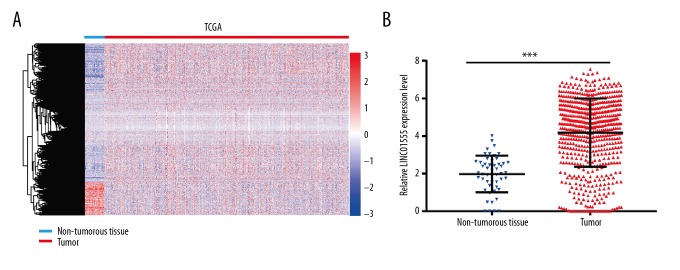
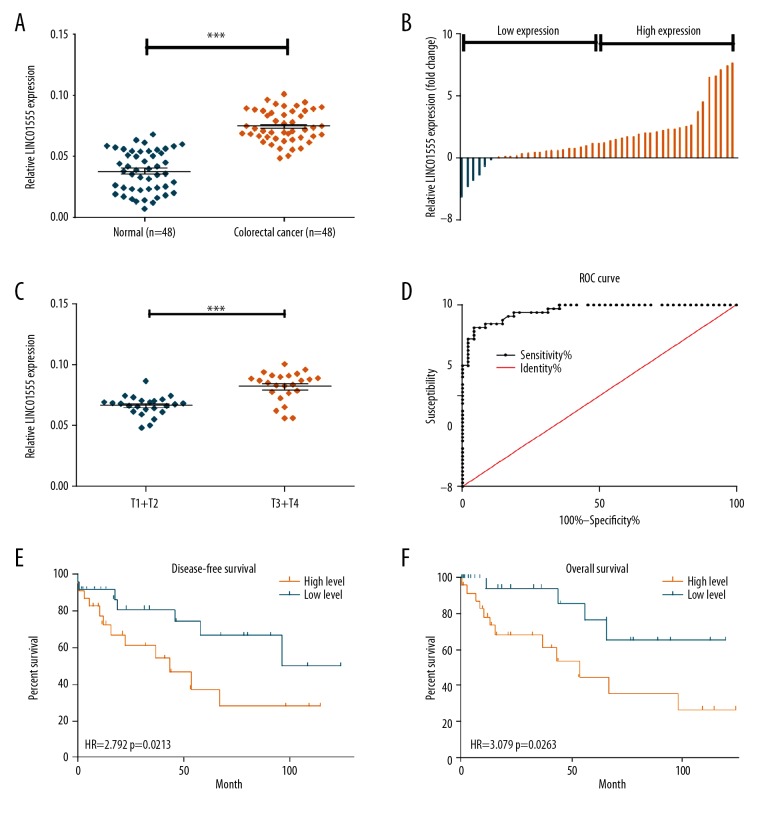
The Cancer Genome Atlas (TCGA) dataset showed that LINC01555 was highly expressed in CRC tissue when compared with adjacent normal colorectal tissue (Figure 1A, 1B). Also, quantitative real-time polymerase chain reaction (qRT-PCR) was performed to detect LINC01555 expression in 48 paired CRC tissue samples and adjacent normal tissues, collected in the Taizhou People’s Hospital. LINC01555 was highly expressed in CRC tissues (Figure 2A, 2B). Increased expression of LINC01555 was found in patients with CRC with stage T3 and T4 tumors compared with stage T1 and T2 tumors (Figure 2C). The receiver operating characteristic (ROC) curve analysis demonstrated the capacity of LINC01555 to distinguish between CRC tissues and normal colorectal tissues (Figure 2D). By analyzing follow-up data, Kaplan-Meier analysis found lower disease-free survival (DFS) in patients with CRC with increased expression of LINC01555 compared with those with lower expression of LINC01555 (Figure 2E). Meanwhile, lower overall survival (OS) was observed in patients with CRC with increased expression of LINC01555 compared with those with lower expression of LINC01555 (Figure 2F).
Figure 1.
The Cancer Genome Atlas (TCGA) dataset showed that the long noncoding RNA (lncRNA), LINC01555, was highly expressed in colorectal cancer (CRC) tissue when compared with adjacent normal colorectal tissue. The expression level of LINC01555 in The Cancer Genome Atlas (TCGA) is significantly upregulated. (A) Heatmap of differentially expressed lncRNAs in the TCGA dataset. (B) LINC01555 expression is significantly upregulated.
Figure 2.
The long noncoding RNA (lncRNA), LINC01555, was highly expressed in colorectal cancer (CRC). (A, B) Quantitative real-time polymerase chain reaction (qRT-PCR) showed that the expression level of LINC01555 in 48 colorectal cancer tissues was significantly higher than that in the normal adjacent control tissue. (C) The expression level of LINC01555 in patients with stage T3 and T4 CRC was significantly higher than those with stage T1 and T2 CRC detected by qRT-PCR. (D) The receiver operating characteristic (ROC) curve shows the diagnostic sensitivity of the expression level of LINC01555 in CRC. (E) The disease-free survival (DFS) rate in patients with CRC with higher expression of LINC01555 was significantly reduced when compared with those with lower expression of LINC01555. (F) The overall survival (OS) rate in patients with CRC with higher expression of LINC01555 was significantly reduced when compared with those with lower expression of LINC01555.
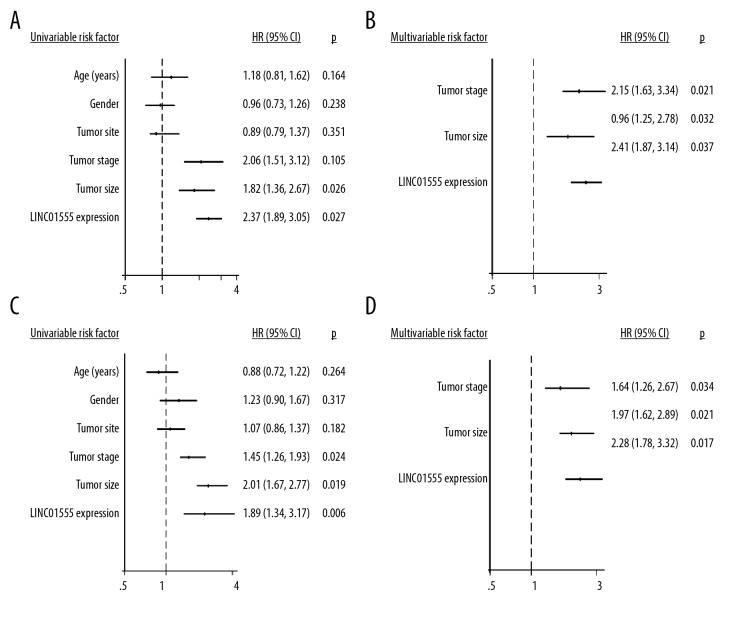
The Cox regression model for univariate survival analysis showed that LINC01555 expression was correlated to tumor size and survival of patients with CRC, but was not correlated with gender, age, tumor location, and tumor stage of CRC (Figure 3A). After correction by multivariate survival analysis, LINC01555 expression, tumor stage, and tumor size were expected to be independent risk factors for the survival and prognosis of patients with CRC (Figure 3B). Also, the Cox regression model for univariate survival analysis indicated that LINC01555 expression, TNM stage, and tumor size were correlated with overall survival (OS) in patients with CRC, rather than gender, age and tumor location (Figure 3C). Multivariate survival analysis showed that the independent risk factors for the survival and prognosis in CRC were LINC01555 expression, TNM stage, and tumor size (Figure 3D).
Figure 3.
The long noncoding RNA (lncRNA), LINC01555, is an important factor in the development of colorectal cancer (CRC). (A) The Cox regression model for univariate survival analysis shows the correlation between LINC01555 expression and disease-free survival (DFS) in patients with colorectal cancer (CRC). (B) The Cox regression model for multivariate survival analysis shows the correlation between LINC01555 expression and DFS in patients with CRC. (C) The Cox regression model for univariate survival analysis showed the correlation between LINC01555 expression and overall survival (OS) in patients with CRC. (D) The Cox regression model for multivariate survival analysis showed the correlation between LINC01555 expression and OS in patients with CRC.
LINC01555 knockdown inhibited the proliferative rate of CRC cells
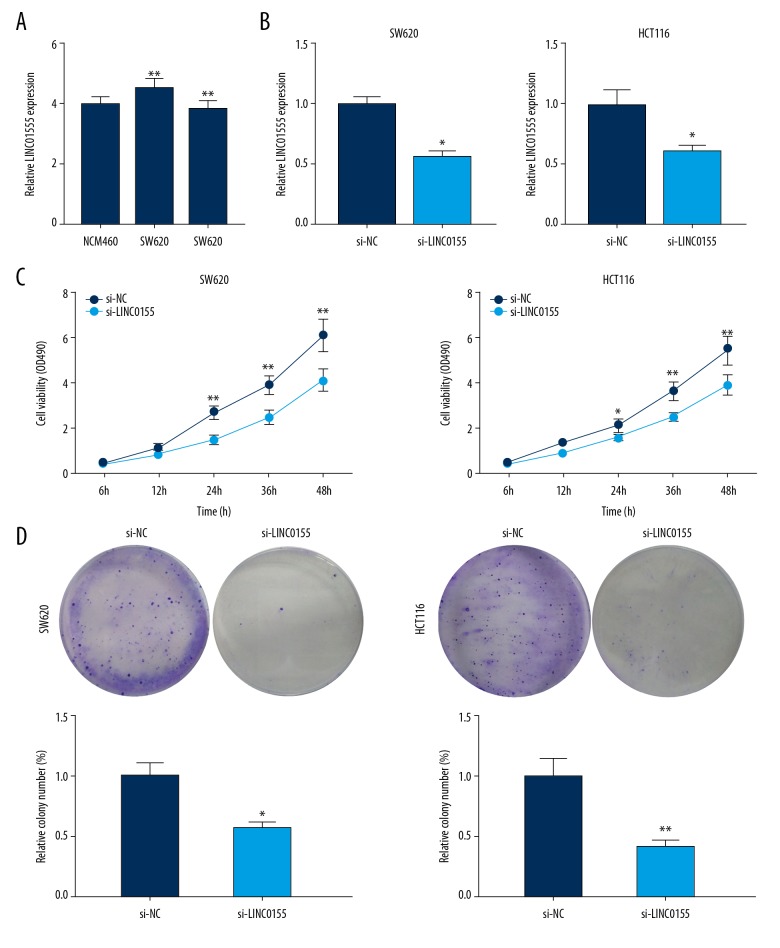
To explore the biological function of LINC01555, we first detected its expression in CRC cell lines. Compared with the normal colorectal cell line, NCM460, LINC01555 was highly expressed in CRC cell lines SW620 and HCT116 (Figure 4A). By exogenous transfection of si-LINC01555, intracellular expression of LINC01555 was downregulated (Figure 4B). Subsequently, the cell counting kit-8 (CCK-8) and colony formation assay were performed to examine the regulatory effects of LINC01555 on the proliferative ability of CRC cells. Decreased proliferative rates of SW620 and HCT116 cells were seen after LINC01555 knockdown (Figure 4C, 4D).
Figure 4.
LINC01555 promoted the proliferation of colorectal cancer (CRC) cells. (A) LINC01555 expression in colorectal cancer (CRC) cell lines SW620 and HCT116 were significantly higher than that in normal colorectal cell line NCM460. LINC01555 was knocked down in SW620 and HCT116 cells. (B) Quantitative real-time polymerase chain reaction (qRT-PCR) showed that LINC01555C expression was significantly downregulated. (C) The cell counting kit-8 (CCK-8) assay showed decreased cell viability. (D) The colony formation assay showed decreased colony formation.
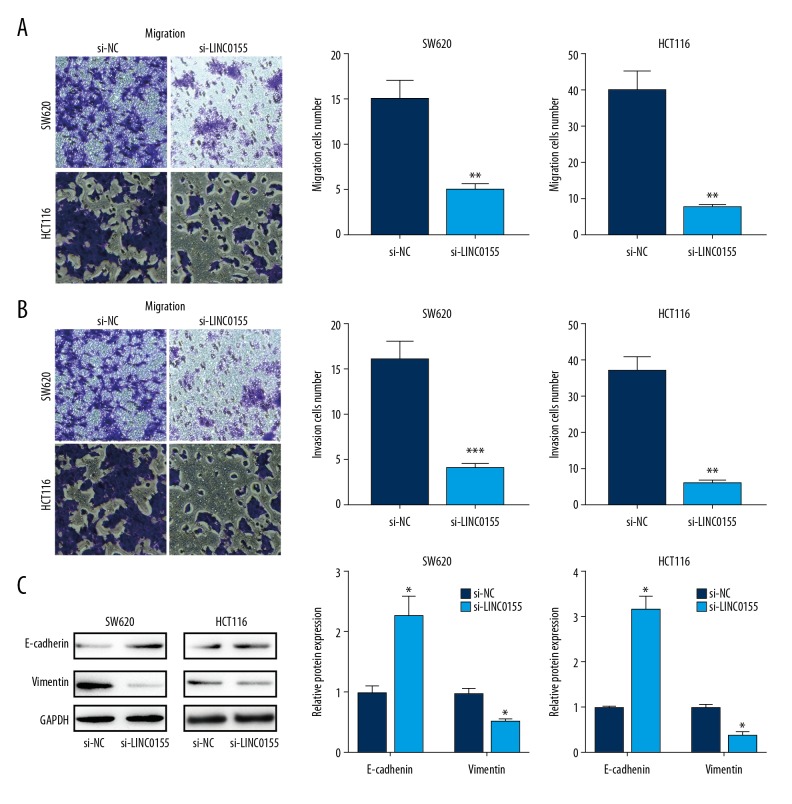
LINC01555 knockdown inhibited the migratory and invasive capacities of CRC cells
Migration and invasion of tumor cells contribute to the development of metastasis. We further evaluated the regulatory effects of LINC01555 on the migratory and invasive capacities of CRC cells. LINC01555 knockdown markedly inhibited the migratory ability of CRC cells (Figure 5A). Similarly, the invasive ability of CRC cells was also suppressed by LINC01555 knockdown (Figure 5B). Epithelial-mesenchymal transition (EMT) has a major role in tumor cell migration and invasion. In this study, the expression of EMT-related genes in CRC cells was examined. Knockdown of LINC01555 significantly upregulated the expression of E-cadherin but downregulated the expression of vimentin expression, suggesting that LINC01555 may be involved in inducing EMT in CRC (Figure 5C).
Figure 5.
LINC01555 promoted the migration and invasion of colorectal cancer (CRC) cells. LINC01555 was knocked down in SW620 and HCT116 cells. (A) The transwell assay showed decreased migratory ability. (B) The transwell assay showed decreased cell invasion. (C) Western blot showed that the protein levels of E-cadherin and vimentin were upregulated.
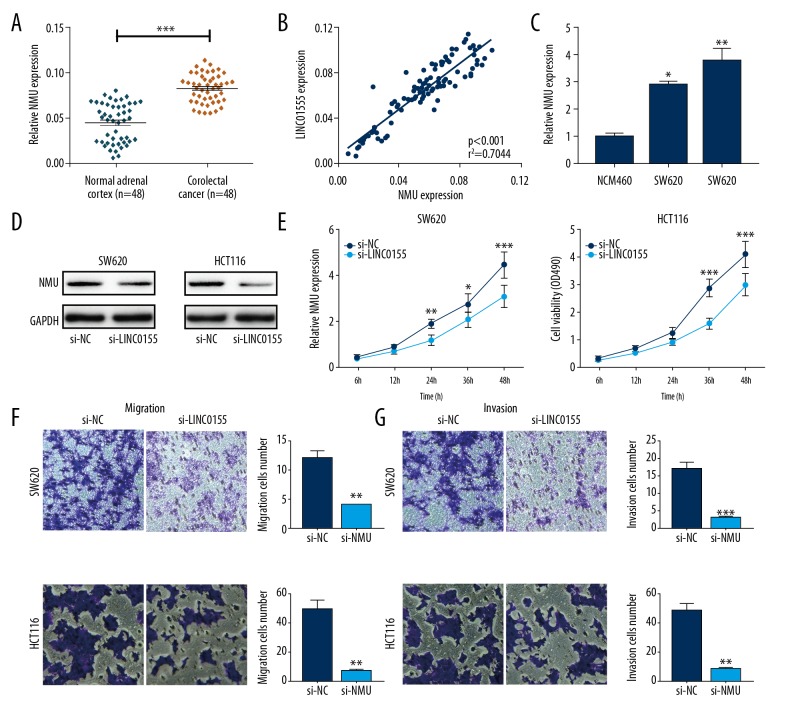
Neuromedin U (NmU) was highly expressed in CRC tissues and promoted CRC cell migration and invasion
NmU was highly expressed in CRC tissues (Figure 6A), and its expression was positively correlated with LINC01555 expression (Figure 6B). NmU was upregulated in CRC cell lines (Figure 6C). Exogenous transfection of si-NMU efficiently reduced NmU expression in CRC cells (Figure 6D). NMU knockdown significantly inhibited the proliferative rate of CRC cells, at 24 hours (Figure 6E). Also, cell migration and invasion of CRC cells were suppressed by NMU knockdown (Figure 6F, 6G).
Figure 6.
Neuromedin U (NmU) was highly expressed in colorectal cancer (CRC) and promoted migration and invasion of CRC cells. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) shows neuromedin U (NmU) expression in 48 colorectal cancer (CRC) tissues was significantly higher than in normal controls. (B) The expression level of LINC01555 was highly correlated with NMU expression in colorectal cancer (CRC) tissues. (C) Protein expression of NmU was downregulated after LINC01555 knockdown in SW620 and HCT116 cells. (D) NmU expression was significantly inhibited, as detected by qRT-PCR. (E) The cell counting kit-8 (CCK-8) assay shows decreased cell viability. (F) The transwell assay shows decreased migratory ability. (G) The transwell assay shows decreased invasive ability. NMU: neuromedin U.
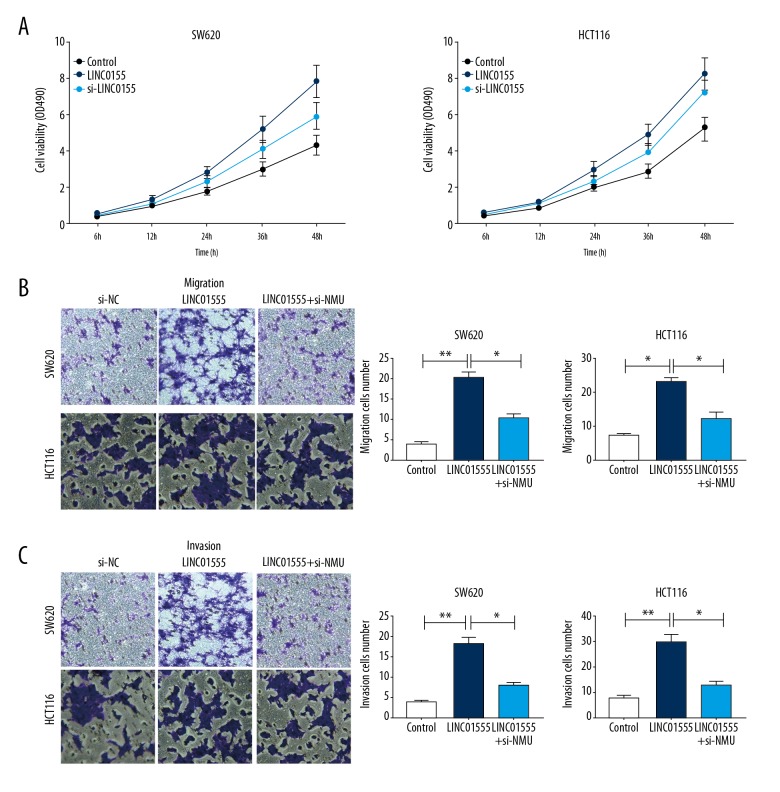
Functional studies were conducted to determine whether LINC01555 could regulate CRC development by targeting the NMU gene. CRC cells co-transfected with pcDNA-LINC01555 and si-NMU showed a decreased proliferative rate compared with those only transfected with pcDNA-LINC01555 (Figure 7A). Similarly, NMU knockdown reversed the increased migratory and invasive capacities of CRC cells that overexpressed LINC01555 (Figure 7B, 7C).
Figure 7.
LINC01555 promoted cell migration and invasion by activating neuromedin U (NmU). Cells were divided into the control group, the LINC01555 overexpression group, and the LINC01555 overexpression + NMU knockdown group. (A) The cell counting kit-8 (CCK-8) assay shows LINC01555 overexpression. NMU knockdown could reverse the increased cell viability induced by LINC01555 overexpression. (B) The transwell assay shows LINC01555 overexpression. NMU knockdown could reverse the increased cell migration induced by LINC01555 overexpression. (C) The transwell assay shows LINC01555 overexpression. NMU knockdown could reverse the increased cell invasion induced by LINC01555 overexpression.
Discussion
Several important long noncoding RNAs (lncRNAs) are associated with the development and progression of colorectal cancer (CRC). For example, the HOTAIR gene has been found to accelerate epithelial-mesenchymal transition (EMT) and to promote invasion and metastasis of colon cancer [12]. High expression levels of the MALAT1 gene promotes the growth, invasion, and metastasis of colon cancer cells, and its deficiency can inhibit the growth and metastasis of colon cancer cells [13]. The CCAT1 gene is located adjacent to the proto-oncogene c-Myc, containing an E-homeobox regulatory element that specifically binds to c-Myc. A chromatin loop is then formed that positively regulates the transcriptional and chromatin conformation of c-Myc, which can increase CCAT1 promoter activity and expression levels to accelerate the proliferative and metastatic abilities of cells [14], thereby stimulating the occurrence and development of CRC [15]. LincRNA p21 is expressed at low levels in CRC tumor tissues when compared adjacent normal tissues. Also, lncRNA p21 expression in rectal cancer is higher than that in colon cancer, and its expression increases with the depth of tumor invasion and with vascular invasion [16]. Increased expression of lncRNA p21 is observed after radiotherapy, suggesting that overexpression of lncRNA p21 improves the radiotherapy sensitivity and tumor cell apoptosis [17]. The GAS5 gene is expressed at low levels in CRC, and its expression is negatively correlated with tumor size, tumor grade, and TNM stage. GAS5 is an independent prognostic factor for CRC and overexpression of GAS5 significantly inhibits the proliferation rate of colon cancer cells [18]. BANCR is highly expressed in CRC, and its expression is positively correlated with the depth of tumor invasion and lymph node metastasis. As an independent prognostic factor, increased expression of BANCR is associated with worse prognosis in patients with CRC [19]. Therefore, lncRNAs are closely related to the clinicopathological features of CRC, serving as crucial diagnostic, therapeutic, and prognostic indicators.
In the present study, upregulation of LINC01555 in tumor tissues was a risk factor for poor prognosis of patients with CRC. LINC01555 knockdown markedly inhibited cell proliferation, cell migration, and cell invasion of CRC cells. The expression of epithelial-mesenchymal transition (EMT)-associated genes in CRC cells were inhibited by LINC01555 knockdown, which was associated with upregulation of E-cadherin and downregulation of vimentin. Therefore, it is possible to hypothesize that LINC01555 exerted its biological role in CRC by targeting neuromedin U (NmU). NmU is a neuropeptide that is associated with smooth muscle contraction and is widely distributed in peripheral organs and the central nervous system. NmU in different species has been highly conserved during evolution [20]. Studies have shown that NmU has crucial physiological functions in regulating smooth muscle contraction, stress, energy metabolism, cancer, and immunity [21,22]. NmU is downregulated in oral cancer cells [22]. Studies have shown that the NmU promoter region is highly methylated in esophageal squamous cell carcinoma (ESCC) cells, which is accompanied by a lack of mRNA expression [23]. Also, 20% of the genes in the NmU promoter region are highly methylated in primary head and neck squamous cell carcinoma (HNSCC) cells [24]. Recombinant expression of NMU can effectively inhibit the growth of ESCC cells, suggesting its tumor-suppressor role [23]. Previous studies showed low expression of c-Myc in tamoxifen-treated K562-MERT cells, and NmU expression was also downregulated [25]. Exogenous NmU could reverse the growth inhibition of K562-MERT cells, trigger the production of calcium flux, and stimulate the proliferation of primary acute myeloid leukemia (AML) cells. It is possible that NmU is closely related to c-Myc, as a previously unknown growth-promoting autocrine loop of the NMU/NMU1R axis in myeloid leukemia cells may exist and be of significance.
The findings from this study showed a positive correlation between LINC01555 and NmU in CRC tissues, and LINC01555 knockdown inhibited NmU expression in CRC cells. Also, knockdown of NmU was shown to inhibit the cell proliferative, cell invasion, and cell migration rates of CRC cells. Functional studies showed that knockdown of NMU reversed the proliferative and migratory capacities of CRC cells that overexpressed LINC01555. Future in vivo experiments are needed to validate the findings of the present study. Also, the specific mechanism by which LINC01555 regulates the expression of NmU expression also requires further investigation.
Conclusions
Increased expression of LINC01555 was found in colorectal cancer (CRC) tissues and promoted the invasion of CRC cells by upregulating the expression of the NMU gene.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Park CH, Eun CS, Han DS. Intestinal microbiota, chronic inflammation, and colorectal cancer. Intest Res. 2018;16:338–45. doi: 10.5217/ir.2018.16.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L, Xia J, Yang J, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON. 2018;23:1343–49. [PubMed] [Google Scholar]
- 3.Tan HY, Zheng YB, Liu J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:8657–63. doi: 10.26355/eurrev_201812_16630. [DOI] [PubMed] [Google Scholar]
- 4.Vassos N, Piso P. Metastatic colorectal cancer to the peritoneum: Current treatment options. Curr Treat Options Oncol. 2018;19:49. doi: 10.1007/s11864-018-0563-8. [DOI] [PubMed] [Google Scholar]
- 5.Veenstra CM, Krauss JC. Emerging systemic therapies for colorectal cancer. Clin Colon Rectal Surg. 2018;31:179–91. doi: 10.1055/s-0037-1602238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itatani Y, Kawada K, Sakai Y. Treatment of elderly patients with colorectal cancer. Biomed Res Int. 2018;2018 doi: 10.1155/2018/2176056. 2176056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng L, Ye L, Zhang D, et al. New insights into the long non-coding RNA SRA: Physiological functions and mechanisms of action. Front Med. 2018;5:244. doi: 10.3389/fmed.2018.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong D, Mu Z, Zhao C, Sun M. ZFAS1: A novel tumor-related long non-coding RNA. Cancer Cell Int. 2018;18:125. doi: 10.1186/s12935-018-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji D, Zhong X, Jiang X, et al. The role of long non-coding RNA AFAP1-AS1 in human malignant tumors. Pathol Res Pract. 2018;214:1524–31. doi: 10.1016/j.prp.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Martens-Uzunova ES, Bottcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Virgilio E, Giarnieri E, Giovagnoli MR, et al. Long non-coding RNAs in the gastric juice of gastric cancer patients. Pathol Res Pract. 2018;214:1239–46. doi: 10.1016/j.prp.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–26. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 13.Yang MH, Hu ZY, Xu C, et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–74. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alaiyan B, Ilyayev N, Stojadinovic A, et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196. doi: 10.1186/1471-2407-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–31. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai H, Fesler A, Schee K, et al. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer. 2013;12:261–66. doi: 10.1016/j.clcc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Li Z, Zhao Q, et al. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol Rep. 2014;31:1839–45. doi: 10.3892/or.2014.3047. [DOI] [PubMed] [Google Scholar]
- 18.Yin D, He X, Zhang E, et al. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:253. doi: 10.1007/s12032-014-0253-8. [DOI] [PubMed] [Google Scholar]
- 19.Guo Q, Zhao Y, Chen J, et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–75. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez VG, O’Driscoll L. Neuromedin U: A multifunctional neuropeptide with pleiotropic roles. Clin Chem. 2015;61:471–82. doi: 10.1373/clinchem.2014.231753. [DOI] [PubMed] [Google Scholar]
- 21.Dalboge LS, Pedersen SL, Secher T, et al. Neuromedin U inhibits food intake partly by inhibiting gastric emptying. Peptides. 2015;69:56–65. doi: 10.1016/j.peptides.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Alevizos I, Mahadevappa M, Zhang X, et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–95. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 24.Tokumaru Y, Yamashita K, Osada M, et al. Inverse correlation between cyclin A1 hypermethylation and p53 mutation in head and neck cancer identified by reversal of epigenetic silencing. Cancer Res. 2004;64:5982–87. doi: 10.1158/0008-5472.CAN-04-0993. [DOI] [PubMed] [Google Scholar]
- 25.Shetzline SE, Rallapalli R, Dowd KJ, et al. Neuromedin U: A Myb-regulated autocrine growth factor for human myeloid leukemias. Blood. 2004;104:1833–40. doi: 10.1182/blood-2003-10-3577. [DOI] [PubMed] [Google Scholar]