Summary
Bacterial nanocellulose (BNC) produced by aerobic bacteria is a biopolymer with sophisticated technical properties. Although the potential for economically relevant applications is huge, the cost of BNC still limits its application to a few biomedical devices and the edible product Nata de Coco, made available by traditional fermentation methods in Asian countries. Thus, a wider economic relevance of BNC is still dependent on breakthrough developments on the production technology. On the other hand, the development of modified strains able to overproduce BNC with new properties – e.g. porosity, density of fibres crosslinking, mechanical properties, etc. – will certainly allow to overcome investment and cost production issues and enlarge the scope of BNC applications. This review discusses current knowledge about the molecular basis of BNC biosynthesis, its regulations and, finally, presents a perspective on the genetic modification of BNC producers made possible by the new tools available for genetic engineering.
Introduction
Bacterial nanocellulose (BNC) is a linear polysaccharide composed of β‐d‐glucopyranose monomers linked by β‐1,4‐glycosidic linkages. The repeating unit is the disaccharide cellobiose (McNamara et al., 2015). Bacterial nanocellulose was first described in 1886 by A. J. Brown, who observed the production of cellulose by Acetobacter xylinum cells in the presence of oxygen and glucose (Brown, 1886). Meanwhile, classification of acetic acid bacteria has changed, Acetobacter being reclassified as Gluconacetobacter, which has recently further moved to a new type of Komagataeibacter. The current generic name as proposed in 2012 by Yuzo Yamada (Yamada et al., 2012) comes from the name of the Japanese microbiologist Kazuo Komagata.
Among the many BNC producers, we can distinguish nitrogen‐binding bacteria (Rhizobium leguminosarum), plant pathogens (Dickeya dadantii), Agrobacterium tumefaciens, Escherichia coli, Salmonella enterica, Pseudomonas putida and bacteria of the genus Komagataeibacter, currently regarded as one of the most efficient producers of this exopolysaccharide (Römling and Galperin, 2015). Current literature indicates a particular interest in bacterial strains of the species Komagataeibacter hansenii, which, due to an interesting phenotype and high cellulose yield, became a model organism used for genetic, molecular and biochemical studies (Florea, et al., 2016b). The number of reports of bacteria producing BNC continues to grow, as well as the annotation of the putative operons of cellulose synthase in whole bacterial genome sequences, suggesting that more and wider groups of bacteria may be capable of producing cellulose (Solano et al., 2002; Chawla et al., 2009; Esa et al., 2014; Matsutani et al., 2015; Keshk and El‐Kott, 2016; Moniri et al., 2017; Reiniati et al., 2017).
Of particular interest in Komagataeibacter strains is the study of molecular aspects of BNC biosynthesis. Genetic engineering is a powerful tool being used to produce valuable strains to increase the efficiency of BNC biosynthesis, as will be further presented below.
Molecular mechanisms of BNC biosynthesis by Komagataeibacter genus
The biochemical reactions of BNC biosynthesis by K. xylinus are very well characterized (Brown, 1987; Delmer and Amor, 1995). This is a precise and specifically regulated multi‐step process, involving numerous individual enzymes and complexes of catalytic and regulatory proteins. However, their supramolecular structure has not yet been fully defined. The pathways and mechanisms of the synthesis of uridine diphosphoglucose (UDP‐Glc) are relatively well known, while the molecular mechanisms of glucose polymerization in long and unbranched chains still need to be further investigated.
Bacterial nanocellulose is synthesized in two stages, the first being the production of β‐1,4‐glucan chains and the second the crystallization of cellulose (Brown and Saxena, 2000). The conversion of glucose to cellulose requires four enzymatic steps (Fig. 1): phosphorylation of glucose by glucokinase to glucose‐6‐phosphate (G6P); isomerization of glucose‐6‐phosphate to glucose‐1‐phosphate (G1P) by phosphoglucomutase (PGM); conversion of glucose‐1‐phosphate to uridine diphosphate glucose (UDP‐glucose) by UDP‐glucose pyrophosphorylase; and finally, the synthesis of cellulose from UDP‐glucose by cellulose synthase (Bcs), a complex of four subunits, BcsA, BcsB, BcsC and BcsD, which are encoded by three (bcsAB, bcsC and bcsD) or four (bcsA, bcsB, bcsC and bcsD) genes (Ross et al., 1991; Jedrzejczak‐Krzepkowska et al., 2016). UDP‐glucose pyrophosphorylase is a key enzyme in this process, since its activity in cellulose‐producing bacteria is one hundred times higher than in the non‐producing counterparts (Valla et al., 1989).
Figure 1.
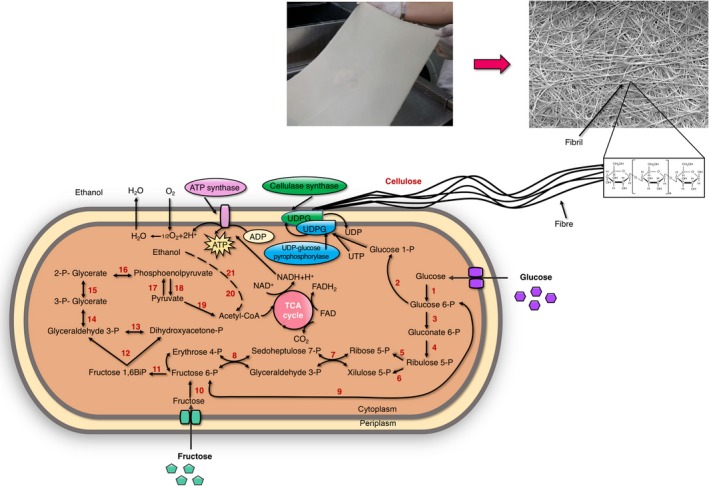
Pathways for the biosynthesis of BNC by K. xylinus and assembly of cellulose molecules into nanofibrils: (1) Glucokinase‐ATP, (2) Phosphoglucomutase, (3) Glucose‐6‐phosphate dehydrogenase, (4) 6‐phosphogluconate dehydrogenase, (5) Phosphorribulose isomerase, (6) Phosphorribulose epimeraase, (7) Transaketolase, (8) Transaldolase, (9) Phosphoglucoisomerase, (10) Fructokinase, (11) Fructokinase ATP, (12) Aldolase, (13) Triosephosphate isomerase, (14) Glyceraldehyde 3‐phosphate dehydrogenase, (15) Phosphoglycerate mutase, (16) Enolase, (17) Pyruvate kinase (18) Pyruvate biphosphate kinase, (19) Pyruvate dehydrogenase, (20) Alcohol dehydrogenase and (21) Aldehyde dehydrogenase.
As shown in Fig. 1, the biosynthesis of BNC is closely related to many metabolic pathways, such as the pentose‐phosphate (PP) pathway, Embden–Meyerhof–Parnas (EMP) pathway, the Krebs cycle (TCA) and gluconeogenesis (GNG) (Lee et al., 2014; Nagashima et al., 2016). Many different compounds, such as hexoses, glycerol, dihydroxyacetone, pyruvate or dicarboxylic acids, can be converted to cellulose. Pyruvate, like glycerol, dihydroxyacetone and intermediates of the pentose monophosphate cycle can be converted into glucose‐6‐phosphate by gluconeogenesis (Krystynowicz et al., 2005). On the other hand, disaccharides, such as sucrose or maltose, are first hydrolysed to monosaccharides and then also converted to glucose‐6‐phosphate (Lee et al., 2014).
Due to the lack or very low phosphofructokinase (pfk) activity, certain cellulose‐producing bacteria, such as K. xylinus, are unable to use EMP pathway for pyruvate synthesis from glucose. Alternatively, pyruvate is obtained from acetate and is used to synthesize glucose through the GNG pathway (Velasco‐Bedrán and López‐Isunza, 2007; Sarkar et al., 2010; Zhong et al., 2013). The PP cycle involves the oxidation of carbohydrates, and the TCA cycle involves the oxidation of acetate‐derived carbohydrates, fat and proteins, such as oxalosuccinate and α‐ketoglutarate. Nevertheless, K. xylinus is not able to metabolize glucose anaerobically since it lacks phosphofructose kinase, which is required for glycolysis (Lee et al., 2014).
The model of Komagataeibacter metabolism, proposed by Velasco‐Bedrán and López‐Isunza, indicates the connection between catabolic pathways of ethanol, glucose and fructose (Velasco‐Bedrán and López‐Isunza, 2007). The G6P can be metabolized into acetate via phospho‐ketolase pathway, linking glucose and ethanol catabolic pathways. However, ethanol dissimilation feeds the TCA cycle and the production of pyruvate feeds gluconeogenesis through phospho enol pyruvate, connecting ethanol to the glucose pathways. In consequence, all four metabolic products (acetic acid, biomass, acetan and cellulose) may be produced from either of the carbon sources or from a mixture of the two, although the energy balance is different in either case (Velasco‐Bedrán and López‐Isunza, 2007).
Recently Zhong et al. (2013, 2014) proposed a similar model of metabolism, but excluded the catabolism of ethanol. In the first study, they clarified as to the higher cellulose yield in K. xylinus (CGMCC no. 2955) from glycerol, glucose and fructose by the increased metabolic flux of carbon to BNC. The analysis of the central carbon metabolic flux showed that about 47.96% of glycerol was conveyed into BNC, a value that drops to only 19.05% for glucose and 24.78% for fructose. Whereas, when glucose was used as the carbon source, 40.03% of glucose was turned into the by‐product gluconic acid (Zhong et al., 2013). Furthermore, the same authors demonstrated that formation of gluconic acid determines BNC productivity (Zhong et al., 2014). These studies indicate that both carbon and energy metabolism influence BNC yields.
The metabolic flux analysis conducted by Li et al. (2016) showed that EMP pathway in K. hansenii was fully active. Moreover, the low phosphofructokinase activity was detected in K. oboediens and this activity was significantly increased along with the gluconate feed concentration (Sarkar et al., 2010).
Characterization of proteins involved in the BNC biosynthesis
The essential enzyme machinery involved in BNC biosynthesis includes cellulose synthase and endo‐1,4‐glucanase (CMCax) flanking genes, a complementary cellulose factor (CcpAx) and β‐glucosidase (BglAx). Cellulose synthase is an enzymatic complex containing three (bcsAB, bcsC, bcsD) or four (bcsA, bcsB, bcsC, bcsD) subunits encoded by genes found in the bcs (bacterial cellulose synthase) operon (Fig. 2). In 1994, the cellulose synthase complex bcsABCD was discovered in K. xylinus (Saxena et al., 1994). The protein subunits included in the cellulose synthase complex and the accessory proteins fulfil various functions identified in Table 1.
Figure 2.
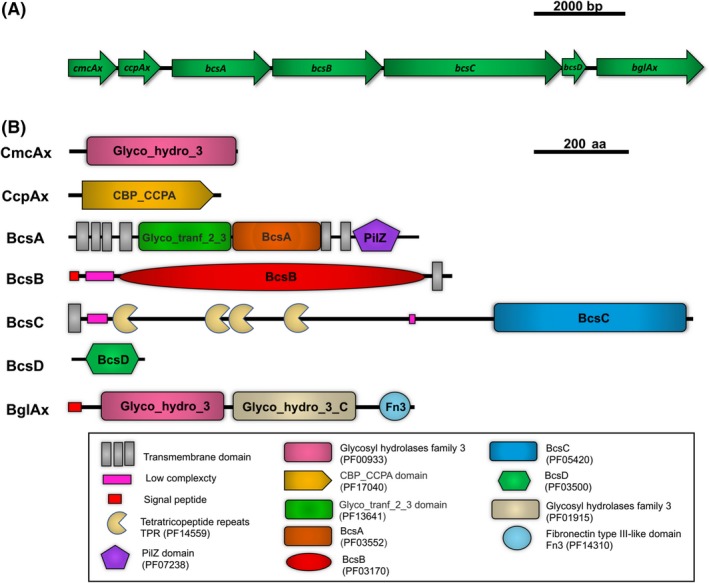
A. Organization of cellulose synthase operon and its flanking regions in Komagataeibacter xylinus E25 Accession no. CP004360 (Ia, locus tags H845_449 → H845_455).B. A cartoon showing the domain organisation of cellulose synthase operon and its flanking regions in Komagataeibacter xylinus E25. Domains were identified by a combined use of Blast (Altschul et al., 1997); HMMER/Pfam (Bateman et al., 1999); and SMART (Schultz, 2000).
Table 1.
Summary of the function of cellulose synthase subunits and proteins involved in the BNC biosynthesis in bacteria of the genus Komagataeibacter
| Protein | Function | References |
|---|---|---|
| BcsA | Cellulose synthase catalytic subunit | McNamara et al. (2015); Römling and Galperin (2015); McManus et al. (2016) |
| Shows the activity of β‐1,4‐glycosyltransferase | ||
| Catalysing the polymerization reaction of UDP‐Glucose monomers to β‐1,4‐glucan chains (cellulose precursor) | ||
| Forms the core of the cellulose synthase complex | ||
| BcsB | Forms the core of the cellulose synthase complex | McNamara et al. (2015); Römling and Galperin (2015); McManus et al. (2016) |
| Takes part in the transport of the newly synthesized β‐1,4‐glucan chain from the cytoplasm through the periplasmic space | ||
| BcsC | Probably creating pores in the outer cell membrane and is involved in the export of the synthesized polysaccharide outside the cell | McNamara et al. (2015); Römling and Galperin (2015); McManus et al. (2016) |
| BcsD | It is probably responsible for the formation of crystalline regions of the cellulose chain by facilitating hydrogen formation bonds between the four newly established chains of β‐1,4‐glucan | McNamara et al. (2015); Römling and Galperin (2015); McManus et al. (2016) |
| CMCax | It exhibits β‐endo‐1,4‐glucanase activity | Römling and Galperin (2015); Castiblanco and Sundin (2016); McManus et al. (2016) |
| Takes part in the regulation of packing cellulose fibrils | ||
| Literature reports indicate that the BcsZ protein is an active participant in the activation of cellulose biosynthesis by c‐di‐GMP | ||
| CcpAx | A protein specific to acetic bacteria | McManus et al. (2016) |
| It probably interacts with the BcsD subunit | ||
| The effect of this protein on the activity of cellulose biosynthesis in K. xylinus and K. hansenii was noted | ||
| BglxA | An enzyme with β‐glucosidase activity | McNamara et al. (2015); Römling and Galperin (2015) |
| BcsX | It probably has cellulose deacetylase activity | McNamara et al. (2015) |
| BcsY | Possible cellulose transacylase | McNamara et al. (2015) |
| Creating modified polysaccharides, e.g. acetylocellulose |
The crystallographic structure of the BcsA and BcsB subunits of the cellulose synthase complex from Rhodobacter sphaeroides was resolved for the first time in 2013 (Morgan et al., 2013). The BcsA and BcsB subunits form the core of the cellulose synthase complex, anchored in the inner cell membrane. This core is extremely important for the production of BNC, both in vivo and in vitro (Römling and Galperin, 2015). The BcsA subunit is associated with the cytoplasmic membrane through 8 TM helices flanked by three amphipathic helices that run parallel to the membrane at the cytosolic water–lipid interface (Kimura et al., 2001; Morgan et al., 2013; McNamara et al., 2015). The cytoplasmic region of the BcsA subunit consists of a catalytic domain with glucosyltransferase (GT) activity using UDP‐Glc as a precursor of the β‐1,4‐glucan chain and the PilZ regulatory domain, that binds a specific allosteric activator to the c‐di‐GMP molecule (Fig. 3). The GT domain of BcsA contains the conserved signature D,D,D,Q (Q/R)XRW which is present in each glycosyltransferases that use a nucleotide‐sugar as a glycosyl donor (Morgan et al., 2013). The first two conserved Asp residues (D‐D) coordinate UDP, while the third D residue is presumably important for catalysis (Jedrzejczak‐Krzepkowska et al., 2016). The Q(Q/R)XRW sequence belonging to IF2 is a part of the cytoplasmic entry to the glucan channel and together with an equally conserved FFCGS sequence, forms a binding site for the terminal disaccharide of the glucan, the acceptor (Morgan et al., 2013).
Figure 3.
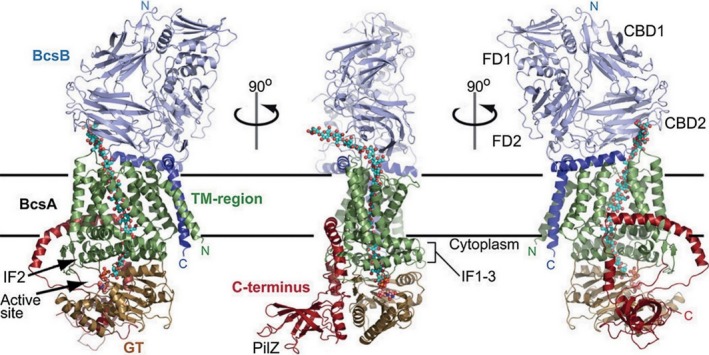
Crystal structure of the BcsA–BcsB complex (Morgan et al., 2013).
The BcsA subunit is associated with the cytoplasmic membrane through 8 TM helices flanked by three amphipathic helices that run parallel to the membrane at the cytosolic water–lipid interface (Kimura et al., 2001; Morgan et al., 2013; McNamara et al., 2015).
The BcsB subunit is a periplasmic protein bound to the cytoplasmic membrane via the C‐terminal end of a single transmembrane helix. The periplasmic fragment contains two copies of the repeating carbohydrate‐binding domain (CBD) fused to the domain of flagroxin (FD) (McNamara et al., 2015). The CBD domains together with the transmembrane helices 3–8 belonging to the BcsA subunit form the export channel of the resulting polysaccharide from the cytoplasmic space (McNamara et al., 2015).
The BcsC subunit, the periplasmic protein, is probably responsible for exporting cellulose out of the cell. It contains an N‐terminal α‐helical motif consisting of several tetratricopeptide (TPR) repeats and a C‐end fragment structurally similar to the β‐barrel characteristic of proteins located in the outer cell membrane (Römling and Galperin, 2015).
The BcsD subunit is a homo‐perceptic periplasmic protein probably involved in the formation of crystalline regions in the newly formed cellulose chain. The functional protein consists of four dimers: AB, CD, EF and GF, forming a cylindrical structure with an internal channel able to host four β‐1,4‐glucan chains. These chains, due to mutual interaction, undergo rearrangement, creating a crystalline region with a strictly defined structure (Hu et al., 2010).
The cellulose synthase operon contains two genes in the upper region, cmcax and ccpAx, and one in the lower region, bglAx. The cmcax gene encodes endo‐β‐1,4‐glucanase, a globular protein belonging to the cluster I in the family of 8 glycoside hydrolases. The CMCax structure consists of eleven α‐helixes and seven β‐strands (Yasutake et al., 2006). CMCax exhibits hydrolytic activity with respect to cellulose and affects the regulation of biosynthesis (Kawano, et al., 2002a; Kawano, et al., 2002b). It has been shown that in small amounts, exogenous CmcAx enhances BNC production of K. xylinus, while endogenous overexpression of cmcAx also enhanced the yield of BNC production (Augimeri et al., 2015).
The second protein found in the upper region of the bcs operon is CcpAx, (Cellulose complementing protein Acetobacter xylinum) also called Ccp (Cellulose complementing factor) (Deng et al., 2013). Research conducted by Standal et al. (1994) showed that the ccpAx gene plays an important role, because mutants with disruption of the ccpAx gene did not produce cellulose in vivo. In turn, Sunagawa et al. (2013) demonstrated, by means of fluorescence microscopy, that CcpAx is colocalized with BcsD along one side of the cell, and furthermore, using pulldown analysis and the isothermal method of calorimetric titration, that CcpAx and BcsD interact with each other. Although these results indicate that CcpAx may act as a mediator of protein‐protein interactions, its exact role in cellulose biosynthesis is still unknown.
In the lower region of the BNC synthase operon, there is a bglAx gene coding for β‐glucosidase, which belongs to the family of three glycoside hydrolases (GH3). BglxA is secreted and has the ability to hydrolyse oligosaccharides larger than three residues to single β‐D‐glucose units (Tahara et al., 1998; Tajima et al., 2001). It was also shown that BglAx from K. xylinus BPR2001 has exo‐1,4‐β ‐glucosidase activity and probably also glucosyltransferase activity (Tahara et al., 1998). Although BglAx is not essential for the production of BNC, the bglAx disruption mutant of K. hansenii ATCC 23769 produced less cellulose than the wild‐type strain. The expression of the bglAx gene has been shown to be regulated transcriptionally by the cyclic‐AMP/fumarate nitrate reductase (CRP/FNRKh) receptor. In contrast, the transposon insertion into crp/fnrKh completely abolished the production of BNC and BglAx, proving that CRP/FNRKh controls the biosynthesis at the transcription level (Deng et al., 2013).
Regulation of the biosynthesis of BNC
The BNC biosynthesis is regulated at both the transcriptional and post‐translational levels (Zogaj et al., 2001). A well‐known regulatory mechanism for this process is the allosteric activation of BcsA by cyclic diguanylate (c‐di‐GMP), a universal bacterial second messenger discovered in K. xylinus (Ross et al., 1990; Römling, 2012). In Komagataeibacter, the cellulose biosynthesis is modulated by the opposing action of two enzymes, diguanylate cyclase (DGC) and c‐di‐GMP diesterase (PDEA), controlling cellular levels of c‐di‐GMP (Ross et al., 1987; Tal et al., 1998). Cyclic‐di‐GMP is produced from two GTP molecules by DGC, whose activity is associated with the conserved (GGDEF) domain (Ausmees et al., 2001; Chang et al., 2001). Specific phosphodiesterases (PDEs) that contain EAL or HD‐GYP domains hydrolyse c‐di‐GMP into 5′‐phosphoguanylyl‐(3′‐5′)‐guanosine (pGpG) or GMP, respectively. The oligoribonuclease Orn, which is a ribonuclease that hydrolyses RNAs that are 2–5 nucleotides in length, is the primary enzyme that is capable of degrading pGpG to GMP (Cohen et al., 2015; Orr et al., 2015). The HD‐GYP domain is a subset of a larger HD family which members have hydrolytic activity for various substrates (Jenal et al., 2017).
Cyclic‐di‐GMP exerts its activity by signalling pathways regulating e.g. biofilm formation, flagella biosynthesis, motility, virulence, the cell cycle, bacterial cell differentiation and other fundamental physiological processes in several bacterial species (Castiblanco and Sundin, 2016; Valentini and Filloux, 2016). It plays a key role in the pathways responsible for the conversion from a planktonic to a sessile lifestyle. High intracellular c‐di‐GMP content enhances biofilm formation via the reduction of motility and production of biofilm matrix, whereas low intracellular c‐di‐GMP content leads to increased motility, biofilm dispersal and the return to planktonic phase (Dow et al., 2007; Schirmer, 2016; Lin Chua et al., 2017). As a bacterial second messenger, it mediates signals coming from the environment onto regulation of many cellular processes (Fig. 4) (Fu et al., 2018; Hall and Lee, 2018). Intracellular c‐di‐GMP levels are balanced by the antagonistic activities of DGCs and PDEs, which are frequently found in multiple copies, and which are related within amino‐terminal sensor domains, such as PER–ARNT–SIM (PAS) for sensing gaseous ligands, and blue light using flavin (BLUF) for sensing light (Fig. 4) (Tarutina et al., 2006; Tschowri et al., 2009).
Figure 4.
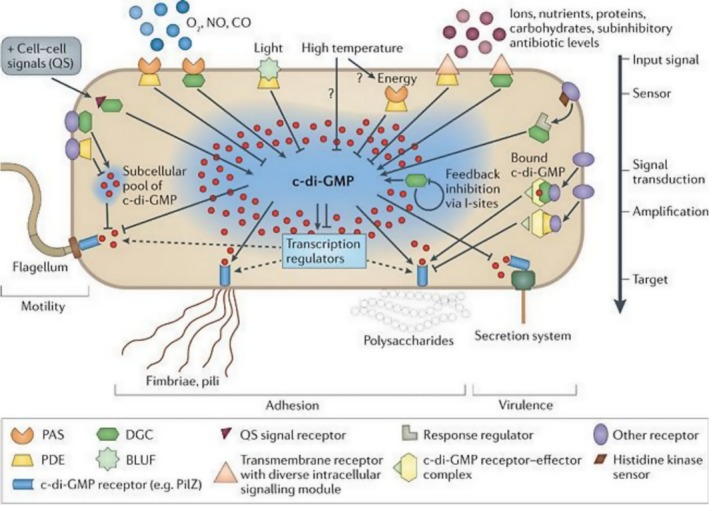
The influence of environmental factors on the biological functions of the bacterial cell (McDougald et al., 2012).
The PAS domain is one of the most common and well‐known sensor domains, which receives environmental signals such as: light, redox potential and oxygen concentration (Vogt and Schippers, 2015). It has been identified in numerous signalling proteins located in the cytoplasm. Depending on the enzyme PAS is associated with, it may bind to different cofactors, e.g. oxygen or the oxidized form of FAD (Dow et al., 2007). Oxygen level sensing was first reported in K. xylinus for the PDEA1 haem‐binding PAS domain (Chang et al., 2001), where it regulates c‐di‐GMP hydrolysis by reversibly binding O2 and through conformational changes in the EAL catalytic domain. These findings provide additional evidence of the significance of oxygen availability for appropriate c‐di‐GMP level regulation and, in consequence, cellulose biosynthesis activation in K. xylinus. Further research revealed that K. xylinus diguanylate cyclase 2 (DGC2) contains the PAS domain that binds a flavin adenine dinucleotide (FAD) cofactor noncovalently (Qi et al., 2009). Binding of the oxidized form of flavin nucleotide regulates the activity of the GGDEF domain and therefore the c‐di‐GMP synthesis. Together with the regulation of PDEA1 by O2, these findings also underline the regulation of c‐di‐GMP concentration and cellulose biosynthesis through both heme‐ and flavin‐containing PAS domains and O2 sensing in K. xylinus (Fig. 5) (Chang et al., 2001; Qi et al., 2009). These studies provide preliminary molecular elucidation of the role of O2 concentration in the regulation of cellulose synthase activity by c‐di‐GMP in K. xylinus, although direct evidence for this correlation is still missing.
Figure 5.
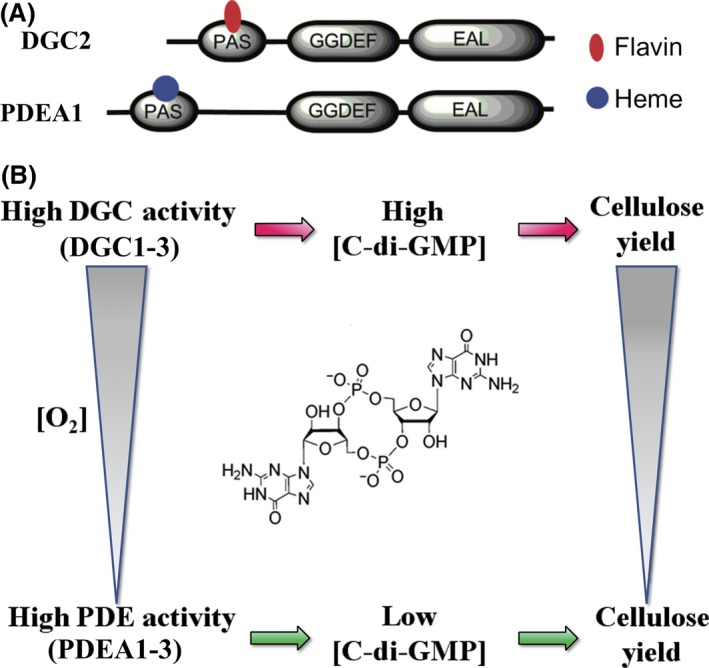
A. Comparison of the domain organization of the PAS domain‐containing proteins in DGC2 and PDEA1 from K. xylinus.B. The connection among oxygen level, cellular c‐di‐GMP concentration and cellulose yield in K. xylinus based on Qi et al. (2009).
Due to the pioneer work of Professor Moshe Benziman, the isolation of the cdg1, cdg2 and cdg3 operons derived from K. xylinus, which encode homologous isoforms of DGC and PDEA, was performed (Ross et al., 1986, 1987). It has been shown that each operon is organized into the pdea gene located above the dgc gene. Genetic analyses among dgc operons indicate that they contribute in different ways to the PDEA and DGC enzymatic activity (cdg1 is responsible for 80% of each activity, while cdg2 and cdg3 account respectively for 15% and 5%). The discovery of the three cdg operons reveals the unusual genetic organization of bacteria. In the genomes of K. hansenii ATCC 23769 and K. hansenii, ATCC 53582 two Cdg operons (cdg1 and cdg2) containing a diguanylate cyclase gene (dgc1, 2) and phosphodiesterase A (pdeA1, 2) as well as four standalone c‐di‐GMP phosphodiesterases (pdeA3–6) were found. Interestingly in K. xylinus, there are three operons encoding enzymes regulating the level of c‐di‐GMP (Tal et al., 1998; Deng et al., 2013; Florea et al., 2016b). The arrangement of genes encoding enzymes with opposite actions within the same genetic unit is rare. In addition, the proteins encoded by the dgc and pdeA genes show a high degree of identity within each isoenzyme package and significant structural conservation. The N‐terminus of all six isoenzymes contains domains similar to those found in various oxygen‐sensing proteins. Furthermore, the DGC and PDEA sequences share a long motif, consisting of two adjacent areas defined by GGDEF and EAL. Coordinated expression of pdeA and dgc provides the necessary balance of PDEA and DGC to achieve optimal c‐di‐GMP concentration, which is essential for the rate of cellulose biosynthesis in accordance with environmental conditions (Tal et al., 1998).
The mechanism of activation of BNC production is relatively simple and consists in post‐translational modification of the BcsA subunit. BcsA without the associated allosteric agent is catalytically inactive and unable to conduct the polymerization reaction (Morgan et al., 2014). In the free state, the gating loop blocks the access of the substrate (UDP‐Glc) to the glucosyltransferase domain (GT) (Fig. 6). The binding of c‐di‐GMP through the PilZ regulatory domain results in a number of conformational changes leading to the exposure of the active site.
Figure 6.
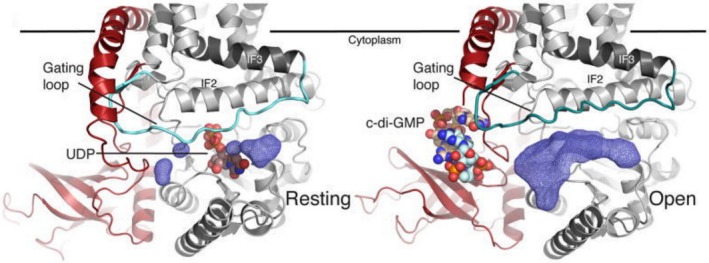
Gating loop positions in the absence and presence of c‐di‐GMP (Morgan et al., 2014).
The C‐terminal PilZ domain contains two motifs: an ‘RXXXR’ with conserved arginine residues surrounding one guanine base of c‐di‐GMP, and a ‘DxSxxG’ motif that surrounds the other guanine of the molecule (Whitney et al., 2015; Chou and Galperin, 2016). The binding of c‐di‐GMP dimer is necessary for the allosteric activation of the BcsA subunit (Castiblanco and Sundin, 2016). The first molecule (c‐di‐GMP‐A) interacts with the ‘DxSxxG’ motif, while the second one (c‐di‐GMP‐B) is stabilized by π–π stacking interactions with c‐di‐GMP‐A (Morgan et al., 2014). Binding of two molecules of the cyclic di‐GMP results in a change in the position of the gating loop and the exposure of the UDP‐Glc binding site in the GT domain, consequently activating the cellulose synthase (Morgan et al., 2014).
It was shown that the GGDEF domain is responsible for the synthesis of c‐di‐GMP thanks to its diguanylate cyclase activity, whereas the EAL domain is responsible for its degradation through its phosphodiesterase activity (Paul et al., 2004). In some cases, only one of those domains is active in GGDEF‐EAL proteins, as it seems to be the case for the three K. xylinus proteins with conserved GGDEF domains, but distinct EAL domains from the active phosphodiesterase A of the same strain (Römling et al., 2005; Schmidt et al., 2005). However, some in vivo observations suggest that some GGDEF‐EAL proteins may have both DGC and PDE activity. For example, the disruption of diguanylate cyclase 1 (dgc‐1), which is responsible for 80% of the c‐di‐GMP production in K. xylinus, caused a reduction in BNC production under certain growth conditions, but a surprisingly increased production under different conditions (Tal et al., 1998; Bae et al., 2004). Therefore, additional work is needed to determine the full enzymatic capacity of proteins containing both the GGDEF and EAL domains.
The availability of high‐resolution crystal structures of the domains GGDEF, EAL and HD‐GYP combined with site‐directed mutagenesis studies allows for the formulation of general principles of differentiation domains. In the GGDEF domain, the active site (A site) contains catalytic Asp/Glu residues surrounded on each side by two highly conserved residues that together form 79GG (D/E) EF83 (amino acid residue numbering is derived from the GGDDEF domain of the NCBI Conserved Domain Database). The first two glycines are involved in GTP binding, while the fourth Glu is involved in the coordination of metal ions. In addition, the active site includes an Asp38 residue that binds Mg2+, as well as Asn46 and Asp55, which binds guanine. Indeed, the RYGGEEF active site motif found in such proteins as PleD, WspR and HmsT requires the presence of conserved residues surrounding the catalytic Glu81. The mutated HmSt form from Y. pestis with the changed motif in the RYAGEEF active centre is not active. In contrast, the motif ‘RxGGDEF’ with Asp81 catalytic residue can contain a variety of hydrophobic residues at position ‘x.’ In addition, DGC activity also remains when the first Glu is replaced with Ala or Ser. In the position of five amino acid residues above the GG (D/E) EF domain, allosteric inhibitory place of the so‐called I site consists of four residues RxxD, where x stands for each residue. It has been demonstrated that the allosteric c‐di‐GMP binding site (I site) is responsible for the non‐competitive inhibition of the DGC product. It has been observed that place I is conserved in the majority of known and potential DGC proteins (Christen et al., 2006; Romling et al., 2013).
It was also reported that the quorum‐sensing (QS) system positively regulate phosphodiesterase, which decompose c‐di‐GMP (Liu et al., 2018). Quorum‐sensing is a cell‐to‐cell signalling system that controls bacterial social behaviours, such as biofilm formation, virulence and motility (An et al., 2014). Quorum sensing controls density‐dependent gene expression by the secretion and detection of chemical signals called QS autoinducers to sense the local population density (Ng and Bassler, 2009). Many gram‐negative bacteria were reported to use a different class of autoinducers: the acyl‐homoserine lactones (AHLs) (Srivastava and Waters, 2012). In Komagataeibacter intermedius, pde expression was shown to be positively regulated by the QS N‐AHL‐dependent system termed GinI/GinR (Iida et al., 2008, 2009). Recently, AHLs were identified in K. xylinus CGMCC 2955, where they are synthesised by the LuxR–LuxI system (Liu et al., 2018). When the population density reaches the ‘quorum,’ AHLs exceed the threshold concentration and bind LuxR, a complex that activates the transcription of a specific operon. Since luxI and luxR are homologs of ginI and ginR, and c‐di‐GMP is an activator of the BcsA–BcsB subunit, Liu et al. (2018) hypothesized that BNC biosynthesis may be regulated by QS by controlling c‐di‐GMP levels.
Recently, a novel cellulose biosynthesis regulator, the quorum‐quenching protein GqqA, has been described. The addition of recombinant GqqA protein to growing cultures of the Komagataeibacter europaeus CECT8546 exerted a strong impact on cellulose production (Valera et al., 2016). The density of the culture increased overtime in the presence of the GqqA protein, whereas the control cultures did not display any altered behaviour. (Valera et al. (2016) suggested that this effect is attributable to alterations in the produced AHL molecules.
Another group of BNC biosynthesis regulator is the CRP/FNR family transcription factors. CRP/FNRKh regulates BNC biosynthesis in K. hansenii ATCC 23769 through positive regulation of bglAx expression (Deng et al., 2013). Interestingly, a Crp/Fnr protein in Burkholderia cenocepacia, named Bcam1349, binds c‐di‐GMP and regulates biofilm formation by enhancing the production of BNC and curli fimbriae (Fazli et al., 2011). Binding of c‐di‐GMP in turn enhanced the ability of Bcam1349 to bind the promoter region and increase the expression of cellulose synthase operon genes, along with the Bcam1330‐Bcam1341 gene cluster involved in exopolysaccharide biosynthesis (Fazli et al., 2011). In recent years, Augimeri and Strap (2015) identified a novel phytohormone‐regulated CRP/FNRKx transcription factor that directly regulates BNC biosynthesis in K. hansenii ATCC 53582 at a transcriptional level, similarly to CRP/FNRKh in K. hansenii. Furthermore, they reported the ethylene up‐regulated crp/fnr Kx expression and enhanced BNC production in K. hansenii, directly by up‐regulating the expression of bcsA and bcsB, and indirectly though the up‐regulation of cmcAx, ccpAx and bglAx (Augimeri and Strap, 2015).
Genetic instability
An important obstacle in the implementation of the BNC biosynthesis process to the industry is the huge diversity between the Komagataeibacter strains in terms of nutritional requirements and the production efficiency. In addition, for some strains, the formation of spontaneous mutants Cel− (that do not produce cellulose) is observed, especially in submerged cultures (Fig. 7). The occurrence of Cel‐mutants in shaken cultures was first observed by Hestrin and Schramm in 1954. They isolated and described three different types of K. xylinus cells, according to the morphology and performance of BNC biosynthesis (Schramm and Hestrin, 1954). Group I corresponds to wild‐type cells producing BNC. Group II corresponds to cells that do not produce cellulose (Cel−), but are capable of reverting over culture passages, while Group III includes irreversible morphotypes of Cel− (Schramm and Hestrin, 1954).
Figure 7.
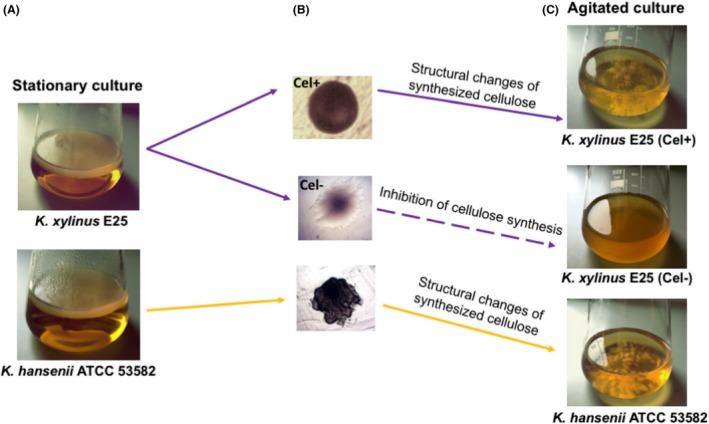
Stable strain of K. hansenii ATCC 53582 and unstable strain K. xylinus E25 generating Cel− and Cel+ forms.A. Stationary culture – thin homogenous BNC membrane formed on the SH medium surface.B. Microscopic pictures of colonies formed by the K. xylinus E25 Cel− and Cel+ forms and K. hansenii ATCC53582.C. Agitated culture – cellulose biosynthesis in the form of small beads (K. hanseii ATCC 53582 and K. xylinus E25 Cel+) or lack of BNC biosynthesis (K. xylinus E25 Cel−).
It was observed that the Cel+ and Cel− morphotypes are characterized by a different morphology. Cel+ colonies are jelly‐like and convex, with smooth edges, while Cel− colonies are large, flat and with wavy edges (Wong et al., 1990) (Fig. 7).
The transformation frequency from Cel+ to Cel− depends on the culture conditions. Cel+ cells dominate in stationary cultures, producing a BNC membrane at the liquid–air interface, where cells find a more oxygen rich environment. It has been shown that homogeneous aeration of liquid culture under agitated conditions favours the spontaneous appearance of Cel−, which become dominant over time. Despite the undertaken research on the emergence of Cel−, this phenomenon has not yet been explained at the molecular level. The search for optimal culture conditions limiting Cel− formation allowed the identification of strategies such as replacing glucose with fructose or enriching the glucose medium with ethanol (2%, v/v), as well as properly selecting the agitation speed (Krystynowicz et al., 2002; Jae et al., 2005).
It was thought that K. xylinus plasmids could have a role in the BNC biosynthesis since these strains usually bear several plasmids of various sizes (from 16 to 300 kb). However, although Cel+ and Cel− cells usually exhibit different plasmid profiles, some BNC‐synthesizing strains devoid of plasmids have been found (Valla and Coucheron, 1983).
It was also reported that insertion elements IS1031 could be involved in an unstable cellulose production by K. xylinus (Coucheron, 1991). When IS elements appear in the middle of genes, they interrupt the coding sequence and inactivate the expression of that gene (Siguier et al., 2014). A correlation has been found between the presence of the IS1031 insertion element and the Cel− morphotype (Coucheron, 1991). Analysis of the protein profile (2D electrophoresis) of the strain K. xylinus E25 shows that the Cel− morphotype does not have two key enzymes in the cellulose biosynthetic pathway: phosphoglucomutase and glucose‐1‐phosphate urydilotransferase (Krystynowicz et al., 2005). On the other hand, other studies have shown that K. hansenii, ATCC 23769 Cel− spontaneous mutant, has a local transposition element inserted in ccpAx (Deng et al., 2013).
In Komagataeibacter hansenii ATCC 23769, the IS1031 A insertion sequence and other isoforms, namely IS1031 B, IS1031 C and IS1031 D, were located. Genetic analysis of the genomes of nine K. xylinus strains showed that all of them contain insertion sequences belonging to the IS1031 group, present in variable number of copies (Coucheron, 1993). The element with slightly lower homology IS 1032 is responsible for the inactivation of genes involved in the synthesis of acetan (Iversen et al., 1994). Nevertheless, the reasons for the appearance of Cel− spontaneous mutants are still unclear.
Genetic modification of Komagataeibacter genus
Despite the long‐term and intensive research involving modifications of culturing conditions, it is still not possible to fully control the biosynthesis and the properties of cellulose produced by Komagataeibacter species. To achieve this goal, extensive genetic testing is necessary, which would uncover the molecular relationships between proteins involved directly and indirectly in BNC production.
So far, genetic engineering of Komagataeibacter strains concerned attempts to increase the efficiency of BNC biosynthesis or to achieve structural changes in the BNC network, hence giving this nanomaterial new properties. These genetic modifications included the expression of a foreign gene or the gene disruption (Edwards et al., 1999; Chien et al., 2006; Yadav et al., 2010).
The most commonly used method of transforming Komagataeibacter is by electroporation, which consists in the formation of unstable pores in the cell membrane under the influence of an electric field. The presence of pores allows the penetration of macromolecules present in the intercellular space into the cells (Wong et al., 1990; Edwards et al., 1999; Chien et al., 2006; Yadav et al., 2010). Another method of transformation described for K. xylinus is the conjugation with E. coli cells (Battad‐Bernardo et al., 2004). For the expression or overexpression of the gene, replicating plasmids in K. xylinus cells such as pLBT, pBBR122, pTI99, pBAV1C and the pSA19 shuttle plasmid are used as vectors (Nakai et al., 1999; Chien et al., 2006; Fang et al., 2015; Mangayil et al., 2017). Nevertheless, the case of using the transposon mutagenesis using the mini‐Tn10 transposon has also been described (Battad‐Bernardo et al., 2004). When the transformation goal is to disrupt one of the genes found in the bacterial chromosome, the corresponding sequence is introduced using a non‐replicating plasmid in K. xylinus cells, e.g. pACYC184, pKE23, BPR2001, pET‐14b (Saxena et al., 1994; Ishida et al., 2002; Shigematsu et al., 2005; Deng et al., 2013, 2015; Kuo et al., 2015). The introduced sequence includes the gene of interest interrupted by, e.g., an antibiotic resistance gene. Since the introduced plasmid does not replicate in K. xylinus cells, antibiotic resistant cells must arise from homologous recombination and the conversion of the chromosomal gene sequence into the one carried by the plasmid. Gene expression is thus suppressed in these cells due to its interruption (Edwards et al., 1999; Yadav et al., 2010).
Inhibition of gene expression
The abolishment of gene expression through disruption was used for the first time in 1990 during the research on the cellulose synthase operon and the function of its genes. For this purpose, the plasmid pACYC184 was used, into which the bcsD gene sequence was cloned, and then interrupted using the ampicillin resistance gene sequence. Obtained mutants showed reduced cellulose synthesis efficiency by about 40%, hence being concluded that the bcsD gene is required for maximum efficiency of biopolymer synthesis in vivo (Wong et al., 1990).
However, further research carried out by Saxena et al. (1994) showed that the disrupted bcsD mutant produced reduced amounts of two cellulose alomorphs (cellulose I and II), suggesting that the bcsD gene is involved in the cellulose crystallization. It has been proposed that the BcsD protein is included in the pore complex or involved in the organization of pores of the linear terminal complex (TC). Additional TEM observations of the mutant cells revealed abnormalities in the orientation of the linear TCs on the BcsD mutant cells, while the components within each liner TC appeared to be aligned normally and functioned similar to that of the wild type (Mehta et al., 2015). Furthermore, results obtained by Mehta et al. (2015) strongly suggest that BcsD aids in the proper orientation of the linear terminal complexes along the longitudinal axis of the cell, indicating a role of this protein in the final level of the hierarchical assembly of cellulose resulting in highly efficient cellulose biosynthesis.
Bae et al. (2004) genetically modified the K. xylinus BPR2001 strain to compare the performance and structural characteristics of BNC produced by mutants with dgc1 gene disruption with the one obtained from the wild‐type strain. Since the dgc1 gene plays an important role in the activation of BNC biosynthesis through c‐di‐GMP, its disruption should reduce the production. However, it was found that the disruption of the dgc1 gene had no apparent effect on BNC production but had a strong impact on its structure. The BNC membranes produced by dgc1 mutant were characterized by smaller and more dispersed fibres that do not form a characteristic compact, hydrated membrane (Bae et al., 2004).
Another modification concerns the disruption of the aceP gene encoding the glycosyltransferase involved in the synthesis of the extracellular branched heteropolysaccharide acetan. This is a water‐soluble polysaccharide produced by Komagataeibacter xylinus from UDP‐Glucose. The K. xylinus CHE5 disrupting mutant synthesized acetan with altered structure and also showed unchanged yield of its production (Edwards et al., 1999). In the following years, Ishida et al. (2002) created a mutant that did not produce acetan (EP1) by disrupting the aceA gene in K. xylinus BPR2001, a gene encodes the β‐glucosyltransferase responsible for the first step of the acetan biosynthetic pathway. Although inhibition of acetan production was expected to increase the concentration of UDP‐Glucose, and thus the yield of BNC, the EP1 mutant produced less BNC under shaking conditions, than the wild‐type strain. After 2 days, the culture medium with the EP1 mutant became a heterogeneous suspension containing large flocs formed by cell aggregates and BNC. Furthermore, the addition of water‐soluble polysaccharides, such as acetan or agar, improved the dispersion of the culture medium and the number of free cells. The authors suggested that the lack of acetan reduces the viscosity of the culture medium and increases the agglomeration of cells and BNC, which in turn led to a reduction in BNC production (Ishida et al., 2002).
A further example is the disruption of the ghd gene sequence coding for glucose dehydrogenase (GDH) with the chloramphenicol resistance gene in K. xylinus BPR2001 cells. The GDH activity is responsible for the extracellular conversion of glucose to gluconic acid, which thus is no longer available as a substrate for the production of BNC in the mutant strain. The GDH coding gene was inserted into the pT7‐Blue T‐Vector plasmid vector and then discontinued with the cat‐1 chloramphenicol acetyltransferase gene sequence. The obtained mutant did not show GDH activity; moreover, it was characterized by a higher efficiency of cellulose biosynthesis (Shigematsu et al., 2005). In addition, Kuo et al. (2015) showed that the mutant (GDH‐KO) K. xylinus BCRC 12334 with the disruption of the gdh gene produced more BNC as compared to the wild‐type strain (40% under stationary conditions and 230% under shaken conditions). The authors suggested that such a significant increase in the production of BNC in shaking culture is the result of a better mass transfer of O2 and nutrients. Kuo et al. (2015) further demonstrated that the GDH‐KO mutant can use glucose to produce BNC without producing gluconic acid as a by‐product.
Gene overexpression
Recently, Mangayil et al. (2017) successfully overexpressed the bcsA genes, bcsAB and a complete cellulose synthase operon (bcsABCD) in K. xylinus DSM 2325. Although there were no significant differences between the growth of mutants and the wild‐type strain, mutants showed faster production, generating twofold to fourfold more BNC. The highest efficiency was observed in the case of the bcsABCD overexpression mutant.
Kawano et al. (2002a) demonstrated that overexpression of the cmcax gene in K. hansenii ATCC 23769 increases the yield of BNC; furthermore, the addition of CMCax protein to the culture medium also promotes the production of cellulose. These authors revealed that the production of BNC may be controlled by regulating the expression of the cmcax gene. Another evidence of the impact of CMCax on BNC biosynthesis was studied by Nakai et al. (2013) The K. xylinus BPR 2001 mutant with disruption of the cmcax gene produced less BNC than the wild‐type strain. Also, these authors observed that the mutant produced mainly cellulose II, thus indicating that CMCax affects the crystallinity of cellulose. Electron microscopy results of Kawano et al. (2002a) showed that CMCax can affect the cellulose fibre structure. Furthermore, cellulose fibres secreted from the K. hansenii strain of ATCC 23769 overexpressing the cmcax gene produced a relaxed structure compared to wild‐type strains.
Expression of foreign genes in cells of bacteria of the genus Komagataeibacter
A successful example concerns the introduction of a mutant sucrose synthase gene derived from Mung bean into the cells of K. xylinus strain BPR2001. Sucrose synthase is an enzyme found in plants that catalyses the reversible reaction of sucrose synthesis with UDP‐glucose and fructose. The gene – under the control of the lac promoter – was introduced into bacterial cells using the pSA19 shuttle plasmid. A gene mutation, involving the replacement of serine at position 11 with glutamic acid, was expected to result in enzyme activation for the synthesis of UDP‐glucose from sucrose and UDP. Indeed, the obtained transformants showed an increase in the content of UDP‐glucose and a twofold and even threefold increase in the synthesis of BNC yield (Nakai et al., 1999).
Another genetic modification of K. xylinus described in the literature concerned the preparation of a strain able to grow on a medium containing lactose as a carbon source, therefore allowing the conversion of cheese whey into BNC. Thus, the promoter‐free lacZ gene coding for β‐galactosidase, one of the genes of the lactose operon, was introduced into the chromosomal DNA of the wild‐type K. xylinus ITDI 2.1 strain. For this purpose, the plasmid pLBT containing the transposon mini‐Tn10 and the transposase gene outside the tansposon sequence was used. The mini‐Tn10 sequence includes the lacZ gene and the gene coding for kanamycin resistance flanked by repeated sequences recognized by transposase (Battad‐Bernardo et al., 2004; Vizváryová and Valková, 2004). The mutagenization process was performed by conjugation of the E. coli S17.1 strain, acting as a vector donor, and the wild strain K. xylinus. Among the obtained mutants, the one showing the highest efficiency of cellulose synthesis was selected. The obtained K. xylinus ITz3 strain showed the stability of mini‐Tn10 transposon insertion into the chromosomal DNA and the ability to synthesize the biopolymer on lactose containing medium, as well as on the whey substrate itself. The β‐galactosidase gene was constitutively expressed, resulting in high enzyme activity (Battad‐Bernardo et al., 2004).
Another interesting modification regards the introduction of the vgb gene present in bacteria of the genus Vitreoscilla, which encodes bacterial haemoglobin (Vhb), into K. xylinus BCRC 12334 cells. In Vitreoscilla cells, VHb synthesis increases with the decrease in extracellular oxygen concentration. This protein binds molecular oxygen and transports it to cytochrome oxidases that are the final enzyme of the respiratory chain. In this way, bacteria have the ability to survive hypoxic environment (Chien et al., 2006). In the case of the introduction of the vgb gene into cells of other bacteria, intensification of growth and synthesis of metabolites is very often observed (Ramandeep et al., 2001). The vgb gene was introduced into K. xylinus BCRC12334 cells by electroporation using the modified plasmid pBBR122. The constitutive expression of the gene and the presence of active protein in the transformants were confirmed, as was the intensification of the growth of microorganisms, probably due to the increased oxygen uptake. Faster bacterial growth resulted in an increase in the amount of synthesized cellulose, with the yield being about 50% higher in the case of micro‐aerobic cultures than in aerobic conditions (Chien et al., 2006).
Another recombinant K. xylinus BCRC12334 was conceived as a sophisticated strategy to express a protein of interest, supporting its stable expression and allowing its self‐immobilization (in the bacteria) on BNC fibres. As a model enzyme expressed in recombinant K. xylinus, d‐Amino oxidase (DAAO; EC1.4.3.3) from Rhodosporidium toruloides was used. The constructed K. xylinus mutant not only successfully produced DAAO activity but was also immobilized by cellulose nanofibres both in stationary and shaken cultures. Although self‐immobilized cells exhibited only 10% of the DAAO activity available in the crude cell extract, they provided several advantages in terms of better thermal stability, stability of operation and easy recovery for repeated use (Setyawati et al., 2009).
Fang et al. (2015) introduced the Curdlan synthase gene, crdS, from Agrobacterium sp. ATCC31749, in K. hansenii AY201, attempting to combine the BNC and Curdlan synthesis metabolic pathways. A significant difference in the morphology of the bionanocomposite surface was noticed, because the BNC pore structure was covered with Curdlan, which also reduced the water permeability. Although Curdlan's secretion significantly changed the surface morphology of the membranes, it had little impact on the crystalline structure, indicating that Curdlan secretion may be of lower priority than BNC nanofibre production (Fang et al., 2015).
Due to the resistance of BNC to in vivo degradation, its use in tissue reconstruction is limited. In order to obtain a polysaccharide susceptible to in vivo degradation, an attempt was made to introduce into K. xylinus 10245 cells an operon containing three genes (NAG5, AGM1 and UAP1) responsible for the synthesis of UDP‐N‐acetylglucosamine (UDP‐GlcNAc). The operon sequence originated from yeast cells Candida albicans. It was assumed that the availability of UDP‐GlcNAc for cellulose synthase would allow the incorporation of GlcNAc into the biopolymer structure. The operon sequence was introduced into K. xylinus cells by electroporation using the modified plasmid pBBR122. Transformants able to synthesize modified cellulose containing GlcNAc were obtained, when N‐acetylglucosamine was present in the culture medium. Transformants showed a significantly lower BNC yield; however, the polysaccharide obtained was characterized by a high content of GlcNAc, and thus lower degree of crystallinity due to the weak interaction between the fibrils. As part of the research, an analysis of its susceptibility to degradation in vivo was also carried out by implanting BNC and modified BNC into mice. It turned out that the polysaccharide containing GlcNAc was completely degraded after only 3 weeks, in contrast to the implant made of native cellulose (Yadav et al., 2010).
Modification of BNC structure has mainly been achieved by chemical or physical modifications of the BNC pellicles or changing culturing conditions (Chanliaud and Gidley, 1999; Luo et al., 2008; Hu et al., 2014). Although genetic modifications of the Komagataeibacter strains were aimed mainly at increasing the productivity, the group from Lodz University of Technology recently obtained two disruption mutants producing stiffer membranes with a more‐packed structure and three overexpression mutants producing membranes with a porous structure (Jacek et al. 2018) demonstrated that by manipulating genes responsible for motility and cell divisions it is possible to obtain mutants producing BNC membranes with desirable properties. These studies initiate a new promising trend in obtaining BNC membranes with changed structure.
Perspectives
Bacterial nanocellulose has been gaining significant attention from scientists and engineers in various research fields due to its unique properties. Nevertheless, the high cost of manufacturing and genetic instability of some Komagataeibacter strains are major obstacle to its wide application. Genetic engineering of bacterial nanocellulose producers is believed to make this process stable, more efficient and less expensive than today. One of the most impressive examples of genetic engineering is the K. xylinus mutant which uses lactose from whey as a carbon source, producing 28‐fold more cellulose from lactose than the wild‐type strain (Battad‐Bernardo et al., 2004).
The growing number of available genome sequences from the genus Komagataeibacter and genetic tools should allow to better understand the molecular aspects of BNC biosynthesis and rationally engineered mutant strains (Florea, et al., 2016a; Pfeffer et al., 2016a,2016b; Zhang et al., 2017). In recent years, comparative genomic and transcriptomic analyses are gaining attention in the scientific community. Recently, it has been shown that high complexity of regulatory mechanisms, directly or indirectly involved in the cellulose biosynthesis in Komagataeibacter strains, could be predicted by comparative genomic analysis (Ryngajłło et al., 2018). Transcriptome analysis‐based RNA‐seq is an important technique to identify differentially expressed genes under distinct experimental conditions in bacteria. Compared with genome analyses, transcriptome analyses only evaluate transcribed genes; thereby, they have a smaller research scope, which may lead to more accurate results. Therefore, it is necessary to study the phenotype of Komagataeibacter new mutants using RNA‐seq technique because this will reveal new dependencies and predict new genes for further research.
Depending on the application, BNC with varying stiffness, tensile strength, porous or denser structure is required. Modification of BNC structure has mainly been achieved by changing the type of bacterial strains, additional supplements to the medium, and differentiation of their growing conditions. Novel approach to change the properties of bacterial nanocellulose is to genetically modify the bacteria. Moreover, genetic engineering may allow a greater range of biomaterials to be produced, by providing precise control over cellulose synthesis genes and production BNC membranes with desired properties. Thus, the fine‐tuning of the properties of BNC through genetic engineering is being successfully achieved, furthering the potential of BNC for application in biomedicine and other fields. Nevertheless, BNC received from genetically modified strains may face regulatory restrictions in medical and food industry.
Conflict of interest
None declared.
Acknowledgements
The authors thank Dr Małgorzata Ryngajłło for reviewing the manuscript.
Microbial Biotechnology (2019) 12(4), 633–649
Funding Information
No funding information provided.
References
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J.H. , Goo, E. , Kim, H. , Seo, Y.‐S. and Hwang, I. (2014) Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci USA 111: 14912–14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augimeri, R.V. and Strap, J.L. (2015) The phytohormone ethylene enhances cellulose production, regulates CRP/FNRKxtranscription and causes differential gene expression within the bacterial cellulose synthesis operon of Komagataeibacter (Gluconacetobacter) xylinus ATCC 53582. Front Microbiol 6: 633–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augimeri, R.V. , Varley, A.J. and Strap, J.L. (2015) Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm‐producing proteobacteria. Front Microbiol 6: 1282–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees, N. , Mayer, R. , Weinhouse, H. , Volman, G. , Amikam, D. , Benziman, M. and Lindberg, M. (2001) Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett 204: 163–167. [DOI] [PubMed] [Google Scholar]
- Bae, S.O. , Sugano, Y. , Ohi, K. , and Shoda, M. (2004) Features of bacterial cellulose synthesis in a mutant generated by disruption of the diguanylate cyclase 1 gene of Acetobacter xylinum BPR 2001. Appl Microbiol Biotechnol 65: 315–322. [DOI] [PubMed] [Google Scholar]
- Bateman, A. , Birney, E. , Durbin, R. , Eddy, S.R. , Finn, R.D. , and Sonnhammer, E.L.L. (1999) Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res 27: 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battad‐Bernardo, E. , McCrindle, S.L. , Couperwhite, I. , and Neilan, B.A. (2004) Insertion of an E. coli lacZ gene in Acetobacter xylinus for the production of cellulose in whey. FEMS Microbiol Lett 231: 253–260. [DOI] [PubMed] [Google Scholar]
- Brown, A.J. (1886) XLIII. – On an acetic ferment which forms cellulose. J Chem Soc Trans 49: 432–439. [Google Scholar]
- Brown, R.M. (1987) The biosynthesis of cellulose. Top Catal 1: 345–351. [Google Scholar]
- Brown, R.M. , and Saxena, I.M. (2000) Cellulose biosynthesis: a model for understanding the assembly of biopolymers. Plant Physiol Biochem 38: 57–67. [Google Scholar]
- Castiblanco, L.F. and Sundin, G.W. (2016) Cellulose production, activated by cyclic di‐GMP through BcsA and BcsZ, is a virulence factor and an essential determinant of the three‐dimensional architectures of biofilms formed by Erwinia amylovora Ea1189. Mol Plant Pathol 19: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A.L. , Tuckerman, J.R. , Gonzalez, G. , Mayer, R. , Weinhouse, H. , Volman, G. , et al (2001) Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme‐based sensor. Biochemistry 40: 3420–3426. [DOI] [PubMed] [Google Scholar]
- Chanliaud, E. , and Gidley, M.J. (1999) In vitro synthesis and properties of pectin/Acetobacter xylinus cellulose composites. Plant J 20: 25–35. [DOI] [PubMed] [Google Scholar]
- Chawla, P.R. , Bajaj, I.B. , Survase, S.a. and Singhal, R.S. (2009) Microbial cellulose: fermentative production and applications (Review). Food Technol Biotechnol 47: 107–124. [Google Scholar]
- Chien, L.J. , Chen, H.T. , Yang, P.F. , and Lee, C.K. (2006) Enhancement of cellulose pellicle production by constitutively expressing Vitreoscilla hemoglobin in Acetobacter xylinum . Biotechnol Prog 22: 1598–1603. [DOI] [PubMed] [Google Scholar]
- Chou, S.H. , and Galperin, M.Y. (2016) Diversity of cyclic di‐GMP‐binding proteins and mechanisms. J Bacteriol 198: 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen, B. , Christen, M. , Paul, R. , Schmid, F. , Folcher, M. , Jenoe, P. , et al (2006) Allosteric control of cyclic di‐GMP signaling. J Biol Chem 281: 32015–32024. [DOI] [PubMed] [Google Scholar]
- Cohen, D. , Mechold, U. , Nevenzal, H. , Yarmiyhu, Y. , Randall, T.E. , Bay, D.C. , et al (2015) Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 112: 11359–11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheron, D.H. (1991) An Acetobacter xylinum insertion sequence element associated with inactivation of cellulose production. J Bacteriol 173: 5723–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucheron, D.H. (1993) A family of IS1031 elements in the genome of Acetobacter xylinum: nucleotide sequences and strain distribution. Mol Microbiol 9: 211–218. [DOI] [PubMed] [Google Scholar]
- Delmer, D.P. , and Amor, Y. (1995) Cellulose biosynthesis. Am Soc Plant Physiol 7: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Nagachar, N. , Xiao, C. , Tien, M. , and Kao, T.H. (2013) Identification and characterization of non‐cellulose‐producing mutants of Gluconacetobacter hansenii generated by Tn5 transposon mutagenesis. J Bacteriol 195: 5072–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Nagachar, N. , Fang, L. , Luan, X. , Catchmark, J.M. , Tien, M. and Kao, T.H. (2015) Isolation and characterization of two cellulose morphology mutants of Gluconacetobacter hansenii ATCC23769 producing cellulose with lower crystallinity. PLoS ONE 10: e0119504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, J.M. , Fouhy, Y. , Lucey, J. and Ryan, R.P. (2007) Cyclic di‐GMP as an intracellular signal regulating bacterial biofilm formation. In The Biofilm Mode of Life, Mechanisms and Adaptations. Kjelleberg S. and Givskov M. (eds). Norwich, UK: Horizon Scientific Press, pp. 71–94. [Google Scholar]
- Edwards, K.J. , Jay, A.J. , Colquhoun, I.J. , Morris, V.J. , Gasson, M.J. , and Griffin, A.M. (1999) Generation of a novel polysaccharide by inactivation of the aceP gene from the acetan biosynthetic pathway in Acetobacter xylinum . Microbiology 145: 1499–1506. [DOI] [PubMed] [Google Scholar]
- Esa, F. , Tasirin, S.M. , and Rahman, N.A. (2014) Overview of bacterial cellulose production and application. Ital Oral Surg 2: 113–119. [Google Scholar]
- Fang, J. , Kawano, S. , Tajima, K. , and Kondo, T. (2015) In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus . Biomacromol 16: 3154–3160. [DOI] [PubMed] [Google Scholar]
- Fazli, M. , O'Connell, A. , Nilsson, M. , Niehaus, K. , Dow, J.M. , Givskov, M. , et al (2011) The CRP/FNR family protein Bcam1349 is a c‐di‐GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia . Mol Microbiol 82: 327–341. [DOI] [PubMed] [Google Scholar]
- Florea, M. , Hagemann, H. , Santosa, G. , Abbott, J. , Micklem, C.N. , Spencer‐Milnes, X. , et al (2016a) Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose‐producing strain. Proc Natl Acad Sci USA 113: E3431–E3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea, M. , Reeve, B. , Abbott, J. , Freemont, P.S. and Ellis, T. (2016b) Genome sequence and plasmid transformation of the model high‐yield bacterial cellulose producer Gluconacetobacter hansenii ATCC 53582. Sci Rep 6: 23635–23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Yu, Z. , Liu, S. , Chen, B. , Zhu, L. , Li, Z. , et al (2018) c‐di‐GMP regulates various phenotypes and insecticidal activity of gram‐positive Bacillus thuringiensis . Front Microbiol 9: 633–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, C.L. and Lee, V.T. (2018) Cyclic‐di‐GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev RNA 9: 633–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S.‐Q. , Gao, Y.‐G. , Tajima, K. , Sunagawa, N. , Zhou, Y. , Kawano, S. , et al (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci USA 107: 17957–17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W. , Chen, S. , Yang, J. , Li, Z. , and Wang, H. (2014) Functionalized bacterial cellulose derivatives and nanocomposites. Carbohydr Polym 101: 1043–1060. [DOI] [PubMed] [Google Scholar]
- Hwang, K.W. , Raje, M. , Kim, K.J. , Stark, B.C. , Dikshit, K.L. , and Webster, D.A. (2001) Vitreoscilla hemoglobin: intracellular localization and binding to membranes. J Biol Chem 276: 24781–24789. [DOI] [PubMed] [Google Scholar]
- Iida, A. , Ohnishi, Y. and Horinouchi, S. (2008) Control of acetic acid fermentation by quorum sensing via N‐acylhomoserine lactones in Gluconacetobacter intermedius . J Bacteriol 190: 2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, A. , Ohnishi, Y. , and Horinouchi, S. (2009) Identification and characterization of target genes of the GinI/GinR quorum‐sensing system in Gluconacetobacter intermedius . Microbiology 155: 3021–3032. [DOI] [PubMed] [Google Scholar]
- Ishida, T. , Sugano, Y. , Nakai, T. , and Shoda, M. (2002) Effects of acetan on production of bacterial cellulose by Acetobacter xylinum . Biosci Biotechnol Biochem 66: 1677–1681. [DOI] [PubMed] [Google Scholar]
- Iversen, T.G. , Standal, R. , Pedersen, T. , and Coucheron, D.H. (1994) Is1032 from Acetobacter xylinum, a new mobile insertion sequence. Plasmid 32: 46–54. [DOI] [PubMed] [Google Scholar]
- Jacek, P. , Szustak, M. , Kubiak, K. , Gendaszewska‐Darmach, E. , Ludwicka, K. , and Bielecki, S. (2018) Scaffolds for chondrogenic cells cultivation prepared from bacterial cellulose with relaxed fibers structure induced genetically. Nanomaterials 8: 1066–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae, Y.J. , Joong, K.P. , and Ho, N.C. (2005) Bacterial cellulose production by Gluconacetobacter hansenii in an agitated culture without living non‐cellulose producing cells. Enzyme Microb Technol 37: 347–354. [Google Scholar]
- Jedrzejczak‐Krzepkowska, M. , Kubiak, K. and Ludwicka, K. (2016) Chapter 2 – bacterial nanocellulose synthesis, recent findings In Bacterial nanocellulose: From Biotechnology to Bio?Economy. Gama M., Dourado F., and Bielecki S. (eds). New York, NY, USA: Elsevier, pp. 19–46. [Google Scholar]
- Jenal, U. , Reinders, A. , and Lori, C. (2017) Cyclic di‐GMP: second messenger extraordinaire. Nat Rev Microbiol 15: 271–284. [DOI] [PubMed] [Google Scholar]
- Kawano, S. , Tajima, K. , Kono, H. , Erata, T. , Munekata, M. , and Takai, M. (2002a) Effects of endogenous endo‐beta‐1,4‐glucanase on cellulose biosynthesis in Acetobacter xylinum ATCC23769. J Biosci Bioeng 94: 275–281. [DOI] [PubMed] [Google Scholar]
- Kawano, S. , Tajima, K. , Uemori, Y. , Yamashita, H. , Erata, T. , Munekata, M. , and Takai, M. (2002b) Cloning of cellulose synthesis related genes from Acetobacter xylinum ATCC23769 and ATCC53582: comparison of cellulose synthetic ability between strains. DNA Res 9: 149–156. [DOI] [PubMed] [Google Scholar]
- Keshk, S.M.A.S. and El‐Kott, A.F. (2016) Natural bacterial biodegradable medical polymers: Bacterial cellulose In Science and Principles of Biodegradable and Bioresorbable Medical Polymers. Zhang X.C. (ed.). Cambridge, UK: Woodhead Publishing, pp. 295–319. [Google Scholar]
- Kimura, S. , Chen, H.P. , Saxena, I.M. , Brown, J. , and Itoh, T. (2001) Localization of c‐di‐GMP‐binding protein with the linear terminal complexes of Acetobacter xylinum . J Bacteriol 183: 5668–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystynowicz, A. , Czaja, W. , Wiktorowska‐Jezierska, A. , Gonc, M.‐M. , Turkiewicz, M. and Bielecki, S. (2002) Factors affecting the yield and properties of bacterial cellulose. J Ind Microbiol Biotechnol 29: 189–195. [DOI] [PubMed] [Google Scholar]
- Krystynowicz, A. , Koziołkiewicz, M. , Wiktorowska‐Jezierska, A. , Bielecki, S. , Klemenska, E. , Masny, A. and Płucienniczak, A. (2005) Molecular basis of cellulose biosynthesis disappearance in submerged culture of Acetobacter xylinum Acta Biochimica Polonica 52: 691–698. [PubMed] [Google Scholar]
- Kuo, C.H. , Teng, H.Y. , and Lee, C.K. (2015) Knock‐out of glucose dehydrogenase gene in Gluconacetobacter xylinus for bacterial cellulose production enhancement. Biotechnol Bioprocess Eng 20: 18–25. [Google Scholar]
- Lee, K.Y. , Buldum, G. , Mantalaris, A. , and Bismarck, A. (2014) More than meets the eye in bacterial cellulose: biosynthesis, bioprocessing, and applications in advanced fiber composites. Macromol Biosci 14: 10–32. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Tian, J. , Tian, H. , Chen, X. , Ping, W. , Tian, C. , and Lei, H. (2016) Mutation‐based selection and analysis of Komagataeibacter hansenii HDM1‐3 for improvement in bacterial cellulose production. J Appl Microbiol 121: 1323–1334. [DOI] [PubMed] [Google Scholar]
- Lin Chua, S. , Liu, Y. , Li, Y. , Jun Ting, H. , Kohli, G.S. , Cai, Z. , et al (2017) Reduced intracellular c‐di‐GMP content increases expression of quorum sensing‐regulated genes in Pseudomonas aeruginosa. Microbiol Front Cell Infect: 7: 633–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. , Liu, L. , Jia, S. , Li, S. , Zou, Y. and Zhong, C. (2018) Complete genome analysis of Gluconacetobacter xylinus CGMCC 2955 for elucidating bacterial cellulose biosynthesis and metabolic regulation. Sci Rep 8: 6266–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , Xiong, G. , Huang, Y. , He, F. , Wang, Y. , and Wan, Y. (2008) Preparation and characterization of a novel COL/BC composite for potential tissue engineering scaffolds. Mater Chem Phys 110: 193–196. [Google Scholar]
- Mangayil, R. , Rajala, S. , Pammo, A. , Sarlin, E. , Luo, J. , Santala, V. , et al (2017) Engineering and characterization of bacterial nanocellulose films as low cost and flexible sensor material. ACS Appl Mater Interfaces 9: 19048–19056. [DOI] [PubMed] [Google Scholar]
- Matsutani, M. , Ito, K. , Azuma, Y. , Ogino, H. , Shirai, M. , Yakushi, T. , and Matsushita, K. (2015) Adaptive mutation related to cellulose producibility in Komagataeibacter medellinensis (Gluconacetobacter xylinus) NBRC 3288. Appl Microbiol Biotechnol 99: 7229–7240. [DOI] [PubMed] [Google Scholar]
- McDougald, D. , Rice, S.A. , Barraud, N. , Steinberg, P.D. , and Kjelleberg, S. (2012) Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10: 39–50. [DOI] [PubMed] [Google Scholar]
- McManus, J.B. , Deng, Y. , Nagachar, N. , Kao, T.H. , and Tien, M. (2016) AcsA‐AcsB: the core of the cellulose synthase complex from Gluconacetobacter hansenii ATCC23769. Enzyme Microb Technol 82: 58–65. [DOI] [PubMed] [Google Scholar]
- McNamara, J.T. , Morgan, J.L.W. , and Zimmer, J. (2015) A molecular description of cellulose biosynthesis. Annu Rev Biochem 84: 895–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, K. , Pfeffer, S. , and Brown, R.M. (2015) Characterization of an acsD disruption mutant provides additional evidence for the hierarchical cell‐directed self‐assembly of cellulose in Gluconacetobacter xylinus . Cellulose 22: 119–137. [Google Scholar]
- Moniri, M. , Boroumand Moghaddam, A. , Azizi, S. , Abdul Rahim, R. , Bin Ariff, A. , Zuhainis Saad, W. , et al (2017) Production and status of bacterial cellulose in biomedical engineering. Nanomaterials 7: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, J.L.W. , Strumillo, J. , and Zimmer, J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, J.L.W. , McNamara, J.T. , and Zimmer, J. (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di‐GMP. Nat Struct Mol Biol 21: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima, A. , Tsuji, T. , and Kondo, T. (2016) A uniaxially oriented nanofibrous cellulose scaffold from pellicles produced by Gluconacetobacter xylinus in dissolved oxygen culture. Carbohydr Polym 135: 215–224. [DOI] [PubMed] [Google Scholar]
- Nakai, T. , Tonouchi, N. , Konishi, T. , Kojima, Y. , Tsuchida, T. , Yoshinaga, F. , et al (1999) Enhancement of cellulose production by expression of sucrose synthase in Acetobacter xylinum . Proc Natl Acad Sci USA 96: 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, T. , Sugano, Y. , Shoda, M. , Sakakibara, H. , Oiwa, K. , Tuzi, S. , et al (2013) Formation of highly twisted ribbons in a carboxymethylcellulase gene‐disrupted strain of a cellulose‐producing bacterium. J Bacteriol 195: 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.‐L. and Bassler, B.L. (2009) Bacterial quorum‐sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, M.W. , Donaldson, G.P. , Severin, G.B. , Wang, J. , Sintim, H.O. , Waters, C.M. , and Lee, V.T. (2015) Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic‐di‐GMP turnover. Proc Natl Acad Sci USA 112: E5048–E5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, R. , Weiser, S. , Amiot, N.C. , Chan, C. , Schirmer, T. , Giese, B. , and Jenal, U. (2004) Cell cycle‐dependent dynamic localization of a bacterial response regulator with a novel di‐guanylate cyclase output domain. Genes Dev 18: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. , Mehta, K. , and Brown, R.M. (2016a) Complete genome sequence of a Gluconacetobacter hansenii ATCC 23769 isolate, AY201, producer of bacterial cellulose and important model organism for the study of cellulose biosynthesis. Genome Announc 4: e00808‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. , Mehta, K. , and Brown, R.M. (2016b) Complete genome sequence of Gluconacetobacter hansenii strain NQ5 (ATCC 53582), an efficient producer of bacterial cellulose. Genome Announc 4: e00785‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y. , Rao, F. , Luo, Z. , and Liang, Z.X. (2009) A flavin cofactor‐binding PAS domain regulates c‐di‐GMP synthesis in AxDGC2 from Acetobacter xylinum . Biochemistry 48: 10275–10285. [DOI] [PubMed] [Google Scholar]
- Reiniati, I. , Hrymak, A.N. , and Margaritis, A. (2017) Recent developments in the production and applications of bacterial cellulose fibers and nanocrystals. Crit Rev Biotechnol 37: 510–524. [DOI] [PubMed] [Google Scholar]
- Römling, U. (2012) Minireview Cyclic di‐GMP, an established secondary messenger still speeding up. Environ Microbiol 14: 1817–1829. [DOI] [PubMed] [Google Scholar]
- Römling, U. , and Galperin, M.Y. (2015) Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol 23: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling, U. , Galperin, M.Y. , and Gomelsky, M. (2013) Cyclic di‐GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 633–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U. , Gomelsky, M. , and Galperin, M.Y. (2005) C‐di‐GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57: 629–639. [DOI] [PubMed] [Google Scholar]
- Ross, P. , Aloni, Y. , Weinhouse, H. , Michaeli, D. , Weinberger‐Ohana, P. , Mayer, R. , and Benziman, M. (1986) Control of cellulose synthesis Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr Res 149: 101–117. [Google Scholar]
- Ross, P. , Weinhouse, H. , Aloni, Y. , Michaeli, D. , Weinberger‐Ohana, P. , Mayer, R. , et al (1987) Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281. [DOI] [PubMed] [Google Scholar]
- Ross, P. , Mayer, R. , Weinhouse, H. , Amikam, D. , Huggirat, Y. , Benziman, M. , et al (1990) The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J Biol Chem 265: 18933–18943. [PubMed] [Google Scholar]
- Ross, P. , Mayer, R. , and Benziman, M. (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55: 35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryngajłło, M. , Kubiak, K. , Jędrzejczak‐Krzepkowska, M. , Jacek, P. and Bielecki, S. (2018) Comparative genomics of the Komagataeibacter strains — Efficient bionanocellulose producers. Microbiologyopen 731: 633–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, D. , Yabusaki, M. , Hasebe, Y. , Ho, P.Y. , Kohmoto, S. , Kaga, T. , and Shimizu, K. (2010) Fermentation and metabolic characteristics of Gluconacetobacter oboediens for different carbon sources. Appl Microbiol Biotechnol 87: 127–136. [DOI] [PubMed] [Google Scholar]
- Saxena, I.M. , Kudlicka, K. , Okuda, K. , and Brown, R.M. (1994) Characterization of genes in the cellulose‐synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 176: 5735–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, T. (2016) C‐di‐GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol 428: 3683–3701. [DOI] [PubMed] [Google Scholar]
- Schmidt, A.J. , Ryjenkov, D.A. and Gomelsky, M. (2005) The ubiquitous protein domain EAL is a cyclic diguanylate‐specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187: 4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, M. , and Hestrin, S. (1954) Factors affecting production of cellulose at the air/liquid interface of a culture of Acetobacter xylinum . J Gen Microbiol 11: 123–129. [DOI] [PubMed] [Google Scholar]
- Schultz, J. (2000) SMART: a web‐based tool for the study of genetically mobile domains. Nucleic Acids Res 28: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyawati, M.I. , Chien, L.J. , and Lee, C.K. (2009) Self‐immobilized recombinant Acetobacter xylinum for biotransformation. Biochem Eng J 43: 78–84. [Google Scholar]
- Shigematsu, T. , Takamine, K. , Kitazato, M. , Morita, T. , Naritomi, T. , Morimura, S. , and Kida, K. (2005) Cellulose production from glucose using a glucose dehydrogenase gene (gdh)‐deficient mutant of Gluconacetobacter xylinus and its use for bioconversion of sweet potato pulp. J Biosci Bioeng 99: 415–422. [DOI] [PubMed] [Google Scholar]
- Siguier, P. , Gourbeyre, E. , and Chandler, M. (2014) Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38: 865–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, C. , García, B. , Valle, J. , Berasain, C. , Ghigo, J.M. , Gamazo, C. , and Lasa, I. (2002) Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol 43: 793–808. [DOI] [PubMed] [Google Scholar]
- Srivastava, D. and Waters, C.M. (2012) A tangled web: regulatory connections between quorum sensing and cyclic Di‐GMP. J Bacteriol 197: 4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standal, R. , Iversen, T.G. , Coucheron, D.H. , Fjaervik, E. , Blatny, J.M. , and Valla, S. (1994) A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J Bacteriol 176: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa, N. , Fujiwara, T. , Yoda, T. , Kawano, S. , Satoh, Y. , Yao, M. , et al (2013) Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum . J Biosci Bioeng 115: 607–612. [DOI] [PubMed] [Google Scholar]
- Tahara, N. , Tonouchi, N. , Yano, H. , and Yoshinaga, F. (1998) Purification and characterization of exo‐1,4‐β‐glucosidase from Acetobacter xylinum BPR2001. J Ferment Bioeng 85: 589–594. [Google Scholar]
- Tajima, K. , Nakajima, K. , Yamashita, H. , Shiba, T. , Munekata, M. , and Takai, M. (2001) Cloning and sequencing of the beta‐glucosidase gene from Acetobacter xylinum ATCC 23769. DNA Res 8: 263–269. [DOI] [PubMed] [Google Scholar]
- Tal, R. , Wong, H.C. , Calhoon, R. , Gelfand, D. , Fear, A.L. , Volman, G. , et al (1998) Three cdg operons control cellular turnover of cyclic di‐GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180: 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutina, M. , Ryjenkov, D.A. , and Gomelsky, M. (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c‐di‐GMP. J Biol Chem 281: 34751–34758. [DOI] [PubMed] [Google Scholar]
- Tschowri, N. , Busse, S. , and Hengge, R. (2009) The BLUF‐EAL protein YcgF acts as a direct anti‐repressor in a blue‐light response of Escherichia coli . Genes Dev 23: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini, M. , and Filloux, A. (2016) Biofilms and Cyclic di‐GMP (c‐di‐GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291: 12547–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera, M.J. , Mas, A. , Streit, W.R. and Mateo, E. (2016) GqqA, a novel protein in Komagataeibacter europaeus involved in bacterial quorum quenching and cellulose formation. Microb Cell Fact 15: 633–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valla, S. and Coucheron, D.H. (1983) Acetobacter xylinum contains several plasmids: evidence for their involvement in cellulose formation. Arch Microbiol 134: 9–11. [Google Scholar]
- Valla, S. , Coucheron, D.H. , Fjærvik, E. , Kjosbakken, J. , Weinhouse, H. , Ross, P. , et al (1989) Cloning of a gene involved in cellulose biosynthesis in Acetobacter xylinum: complementation of cellulose‐negative mutants by the UDPG pyrophosphorylase structural gene. MGG Mol Gen Genet 217: 26–30. [DOI] [PubMed] [Google Scholar]
- Velasco‐Bedrán, H. , and López‐Isunza, F. (2007) The unified metabolism of Gluconacetobacter entanii in continuous and batch processes. Process Biochem 42: 1180–1190. [Google Scholar]
- Vizváryová, M. , and Valková, D. (2004) Transposons – the useful genetic tools. Biol Sect Cell Mol Biol 59: 309–318. [Google Scholar]
- Vogt, J.H.M. and Schippers, J.H.M. (2015) Setting the PAS, the role of circadian PAS domain proteins during environmental adaptation in plants. Front Plant Sci 6: 633–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J.C. , Whitfield, G.B. , Marmont, L.S. , Yip, P. , Neculai, A.M. , Lobsanov, Y.D. , et al (2015) Dimeric c‐di‐GMP is required for post‐translational regulation of alginate production in Pseudomonas aeruginosa . J Biol Chem 290: 12451–12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.C. , Fear, A.L. , Calhoon, R.D. , Eichinger, G.H. , Mayer, R. , Amikam, D. , et al (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum . Proc Natl Acad Sci USA 87: 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, V. , Paniliatis, B.J. , Shi, H. , Lee, K. , Cebe, P. , and Kaplan, D.L. (2010) Novel in vivo‐degradable cellulose‐chitin copolymer from metabolically engineered gluconacetobacter xylinus. Appl Environ Microbiol 76: 6257–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, Y. , Yukphan, P. , Vu, H.T.L. , Muramatsu, Y. , Ochaikul, D. , and Nakagawa, Y. (2012) Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: the proposal of Komagatabacter gen. nov., for strains accommodated to the Gluconacetobacter xylinus group in the α‐Proteobacteria. Ann Microbiol 62: 849–859. [Google Scholar]
- Yasutake, Y. , Kawano, S. , Tajima, K. , Yao, M. , Satoh, Y. , Munekata, M. , and Tanaka, I. (2006) Structural characterization of the Acetobacter xylinum endo‐β‐1,4‐ glucanase CMCax required for cellulose biosynthesis. Proteins Struct Funct Genet 64: 1069–1077. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Xu, X. , Chen, X. , Yuan, F. , Sun, B. , Xu, Y. , et al (2017) Complete genome sequence of the cellulose‐producing strain Komagataeibacter nataicola RZS01. Sci Rep 7: 633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. , Zhang, G.C. , Liu, M. , Zheng, X.T. , Han, P.P. , and Jia, S.R. (2013) Metabolic flux analysis of Gluconacetobacter xylinus for bacterial cellulose production. Appl Microbiol Biotechnol 97: 6189–6199. [DOI] [PubMed] [Google Scholar]
- Zhong, C. , Li, F. , Liu, M. , Yang, X.N. , Zhu, H.X. , Jia, Y.Y. , et al (2014) Revealing differences in metabolic flux distributions between a mutant strain and its parent strain Gluconacetobacter xylinus CGMCC 2955. PLoS ONE 9: 98772–98780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj, X. , Nimtz, M. , Rohde, M. , Bokranz, W. , and Römling, U. (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39: 1452–1463. [DOI] [PubMed] [Google Scholar]


