Abstract
Purpose: The emergence of New Delhi metallo-beta-lactamase (NDM) carbapenemase-producing Escherichia coli leaves few therapeutic options. Infections due to NDM-7 carbapenemase-producing E. coli are infrequent. In this study, we report the whole-genome sequence of an NDM-7 carbapenemase-producing E. coli belonging to sequence type (ST) 410 isolated from a patient with a urinary tract infection in China.
Patients and methods: The NDM-7 producing E. coli strain EC25 was isolated from a urine sample of a male patient hospitalized in a tertiary hospital in Zhejiang Province of China. Susceptibility assay of antibiotics was performed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The whole genome of the strain was sequenced, and the blaNDM-7-harboring plasmid was analyzed. The genomic characterization and molecular epidemiology of the strain were further elucidated.
Results: E. coli EC25 was resistant to all antimicrobials tested, except tigecycline and colistin. The whole genome of E. coli EC25 was composed of one chromosomal DNA and five plasmids. Four virulence factors and twenty-five antimicrobial resistance genes, including blaNDM-7, were identified. Resistance genes were all located in an IncF-type plasmid (pEC25-1), except blaNDM-7, which was located in an individual IncX3-type plasmid (pEC25_NDM-7). Twenty-one phylogenetically related strains were identified. The phylogenetically related E. coli ST410 strains exist globally. The closest relative strain of EC25 was a strain isolated from Sichuan province of China in 2016, with a similar IncX3-type plasmid that encoded blaNDM-5.
Conclusion: Our study reports the emergence of an E. coli ST410 strain harboring blaNDM-7 in China. This strain may evolve as a successful epidemic clone of NDM-producing E. coli in China, and the blaNDM gene is prone to mutate during its dissemination.
Keywords: Escherichia coli, blaNDM-7, IncX3 type plasmid, carbapenem resistant, ST410
Introduction
Escherichia coli is a member of Enterobacteriaceae, and it is an important opportunistic pathogen. E. coli is the leading cause of urinary tract infections and the cause of a variety of other infections, including liver abscesses and bacteremia. E. coli that carry extended-spectrum β-lactamase (ESBL) are very common.1 Besides tigecycline and colistin, carbapenems are often the last resort for treating infections due to ESBL-producing E. coli. However, the emergence of carbapenemase has contributed to resistance to all β-lactams including carbapenems. New Delhi metallo-beta-lactamase (NDM) is one of the most important carbapenemases. Since the first report of blaNDM-1 in 2009,2 24 NDM variants (NDM-1 to NDM-24) were identified.3 NDM spread among Enterobacteriaceae and other Gram-negative bacteria, such as Pseudomonas aeruginosa and Acinetobacter baumannii,4,5 and E. coli is the predominant carrier of blaNDM.
NDM-7 differs from NDM-1 by the substitution of two amino acids at positions 130 (Asp→Asn) and 154 (Met→Leu). NDM-7 was first described in an E. coli strain in Germany in 2013, and it exhibited higher carbapenemase activity compared to NDM-1.6 NDM-7 was reported sporadically in many countries.7–13 NDM-7-producing E. coli was first reported in 2016 in China. blaNDM-7 was located on a conjugative IncX3-type plasmid in five clinically isolated E. coli strains of sequence types ST131 and ST650 in the Jiangxi Province.14 Yingying Hao et al recently reported an NDM-7-producing E. coli ST167 strain isolated from a urine sample in the Shandong Province of China, and blaNDM-7 was also located on a conjugative IncX3-type plasmid.15 IncX3-type plasmids likely play an important role in the distribution of blaNDM-7 in China. Considering the global emergence of NDM-7, an epidemiological survey and analysis of blaNDM-7-harboring strains are urgently needed to prevent its future prevalence.
The present study isolated an NDM-7-producing E. coli strain from a male patient hospitalized in a tertiary hospital in the Zhejiang Province of China. The entire genome of the strain was sequenced, and the blaNDM-7-harboring plasmid was analyzed. Genomic characterization and molecular epidemiology of the strain were further elucidated.
Material and methods
Patient and isolate
A 57-year-old male patient diagnosed with pulmonary infection was hospitalized in a tertiary hospital in the Zhejiang Province of China on August 14, 2017. The patient had a pulmonary infection and comorbidities, including hypoxic-ischemic encephalopathy, hypertension, and hypertensive heart disease. During his hospitalization, the patient received an indwelling urinary catheter. Routine urine tests suggested a urinary tract infection, and carbapenem-resistant E. coli was isolated from a urine sample on August 17, 2017. The strain was preliminarily identified using the VITEK MS system (bioMérieux, France) and further confirmed using 16S rRNA gene sequencing.
Antimicrobial susceptibility test
A susceptibility assay to antibiotics was performed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The minimum inhibitory concentrations (MICs) of tigecycline and colistin (Sigma-Aldrich, St. Louis, MO, USA) were determined using standard broth microdilution tests with fresh Mueller-Hinton broth (Cation-adjusted, Oxoid LTD, England). The MICs of other antimicrobial agents were determined using the Etest method. Antimicrobial susceptibility was determined using the breakpoints approved by the CLSI, and the US Food and Drug Administration (FDA) breakpoints were used for tigecycline.
Whole-genome sequencing
Genomic DNA of the isolate was extracted using a QIAamp DNA MiniKit (Qiagen, USA) and subjected to whole-genome sequencing (WGS) using the Illumina HiSeqTM 4000 platform (Illumina, San Diego, CA, USA). The short reads generated were de novo assembled into contigs using SPAdes. To obtain the complete sequence, EC25 was subjected to sequencing using the long-read MinION Sequencer (Nanopore, Oxford, UK). The MinION sequencing libraries were prepared using the rapid barcoding kit and loaded onto a single MinION R9.4 flow cell. The MinION reads were base-called with Albacore v2.0.2 and generated 2.4 Gbp of data. Raw and trimmed Illumina reads were assessed using FastQC v0.11.7, and quality trimming was performed using Trimmomatic v0.32. The MinION reads were demultiplexed and quality-trimmed using Porechop v0.2.3 with the default settings. The de novo hybrid assembly of short Illumina reads and long MinION reads was performed using Unicycler (v0.4.7; parameters: -min_component_size 500-min_dead_end_size 500-verbosity 1-mode conservative) under conservative mode for increased accuracy. Complete circular contigs generated were corrected using Pilon with Illumina reads for several rounds until no change was detected. We obtained one circular contig, which was represented by a complete chromosome with a size of 4,782,653 bp and five plasmids of 227,349 bp, 46,161 bp, 5,167 bp, 4,991 bp, and 3,373 bp. The whole-genome sequence was annotated in the NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) server.
Identification of antimicrobial resistance genes
Acquired antimicrobial resistance genes were identified using ResFinder 3.0 with a 99% threshold for gene identification and a 100% minimum length via depositing of the entire genome sequence into the database.16 The carbapenem-resistant gene blaNDM-7 was confirmed using PCR and Sanger sequencing.
Genomic characterization
Multilocus sequence typing (MLST) of the isolate was analyzed using the BacWGSTdb server of the entire genome sequence.17 Virulence genes and plasmid replicon type were analyzed using VirulenceFinder 1.5 and PlasmidFinder 1.3.16 Further bioinformatics analysis, such as identification of insertion elements (IS), clustered regularly interspaced short palindromic repeat (CRISPR) sequences and secondary metabolite gene clusters, were predicted using applications of ISfinder, CRISPRFinder, and antiSMASH tools, respectively.18,19
Plasmid analysis
The plasmid sequences were annotated in the NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) server. A graphical map of the blaNDM-7-carrying plasmid was converted in the CGView Server and completed with labels and footnotes.20
Phylogenetic relationship analysis
The phylogenetic relationship between EC25 and other E. coli strains was analyzed using the BacWGSTdb server and the entire genome sequence.17 The BacWGSTdb server offers SNP and genome MLST approaches to investigate the phylogenetic relationship of the uploaded genome sequence with sequences deposited in the BacWGSTdb. The database currently contains 10,545 E. coli strains, including 77 strains of ST410. Scheme cgMLST was used with a 200 threshold for phylogenetic analysis.
Nucleotide sequence accession numbers
The complete sequence of strain E. coli EC25 and plasmids were submitted to GenBank under accession number CP035123-CP035128.
Ethical approval
The urine sample and clinical isolate of E. coli EC25 were generated as part of routine hospital laboratory procedures. This study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Zhejiang Provincial People’s Hospital, China. Written informed consent was obtained from the patient, which included publication of the case details.
Results and discussion
The MICs of strain EC25 to different antibiotics are presented in Table 1. The strain was resistant to all antimicrobials tested except tigecycline and colistin.
Table 1.
MICs of the antibiotics tested in E. coli EC25
| Antibiotics | MIC(mg/L) |
|---|---|
| Cefazolina | >256 |
| Ceftriaxonea | >256 |
| Cefotaximea | >256 |
| Cefepimea | >256 |
| Imipenema | >32 |
| Meropenema | >32 |
| Aztreonama | >32 |
| Amoxicillin/clavulanatea | >256 |
| Cefoperazone/sulbactama | >256 |
| Ciprofloxacina | >32 |
| Gentamicina | >256 |
| Tetracyclinea | >256 |
| Tigecyclineb | 0.25 |
| Colistinb | 0.25 |
Notes: aTested by Etest method. bTested by standard broth microdilution tests.
The whole-genome sequence of E. coli EC25 was composed of one chromosomal DNA that comprised 4,782,653 bp and five plasmids of 227,349 bp, 46,161 bp, 5,167 bp, 4,991 bp, and 3,373 bp. The chromosomal DNA contained 88 tRNA genes, 3 rRNA operons, and 4,707 protein-coding sequences, which were identified using the PGAP server. The MLST scheme revealed that EC25 belonged to sequence type ST410. The genome contained two confirmed CRISPRs (CRISPR1 start position and end position: 1,042,955–1,043,471, CRISPR length: 516; CRISPR2 start position and end position: 1,065,855–1,066,433, CRISPR length: 578). Several IS elements were found in the genome, and most belonged to the IS3 and IS5 families. Four secondary metabolite regions, nrps-t1pks, thiopeptide, NRPS, and siderophore, were also identified.
Four virulence factors were found in the genome (Table 2), which were two copies of gad (glutamate decarboxylase), a single copy of iss (increased serum survival) and lpfA (long polar fimbriae). The distribution of the resistance genes in the genome of the strain EC25 is presented in Table 3. We identified the aminoglycoside resistance genes aph(3ʹʹ)-Ib, aph(6)-Id, aac(3)-IId, aadA16, aac(6ʹ)-Ib-cr, and aadA5, the beta-lactam resistance genes blaNDM-7, blaOXA-1, blaTEM-1B, and blaCTX-M-3, the fluoroquinolone resistance genes qnrB2, aac(6ʹ)Ib-cr and qnrS1, the rifampicin resistance gene arr-3, the macrolide, lincosamide and streptogramin B resistance gene mph(A), the sulfonamide resistance genes sul1and sul2, the tetracycline resistance gene tet(A), and the trimethoprim resistance genes dfrA27 and dfrA17. Except for blaNDM-7, all of the resistance genes were located in the IncF-type plasmid pEC25-1, including two copies of aac(6ʹ)Ib-cr and mph(A) and three copies of sul1. The carbapenem-resistant gene blaNDM-7 was located in an individual plasmid, pEC25_NDM-7.
Table 2.
Virulence genes in strain E. coli EC25
| Virulence factor | Identity | Query/template length | Contig | Position in contig | Protein function | Accession number |
|---|---|---|---|---|---|---|
| gad | 100 | 1401/1401 | EC25 | 251,983..253,383 | Glutamate decarboxylase | AP010953 |
| gad | 99.79 | 1401/1401 | EC25 | 2,444,792..2,446,192 | Glutamate decarboxylase | CP002967 |
| iss | 98.64 | 294/294 | EC25 | 3,714,038..3,714,331 | Increased serum survival | CP004009 |
| lpfA | 100 | 573/573 | EC25 | 4,749,714..4,750,286 | Long polar fimbriae | AY646923 |
Table 3.
Distribution of resistance genes in E. coli strain EC25
| Resistance gene | %identity | HSP length/query | Contig | Position in contig | Predicted phenotype | Accession number |
|---|---|---|---|---|---|---|
| Aminoglycoside | ||||||
| aph(3ʹ’)-Ib | 100 | 804/804 | pEC25-1 | 94,244..95,047 | Aminoglycoside resistance | AF321551 |
| aph(6)-Id | 100 | 837/837 | pEC25-1 | 95,047..95,883 | Aminoglycoside resistance | M28829 |
| aac(3)-IId | 99.88 | 861/861 | pEC25-1 | 97,435..98,295 | Aminoglycoside resistance | EU022314 |
| aadA16 | 99.65 | 846/846 | pEC25-1 | 111,488..112,333 | Aminoglycoside resistance | EU675686 |
| aac(6ʹ)-Ib-cr | 100 | 600/600 | pEC25-1 | 113,669..114,268 | Fluoroquinolone and aminoglycoside resistance | DQ303918 |
| aadA5 | 100 | 789/789 | pEC25-1 | 204,055.204,843 | Aminoglycoside resistance | AF137361 |
| aac(6ʹ)-Ib-cr | 100 | 600/600 | pEC25-1 | 208,288.208,887 | Fluoroquinolone and aminoglycoside resistance | DQ303918 |
| Beta-lactam | ||||||
| blaNDM-7 | 100 | 813/813 | pEC25_NDM-7 | 44,052..44,864 | Beta-lactam resistance | JX262694 |
| blaOXA-1 | 100 | 831/831 | pEC25-1 | 209,018..209,848 | Beta-lactam resistance | HQ170510 |
| blaTEM-1B | 100 | 861/861 | pEC25-1 | 214,606.215,466 | Beta-lactam resistance | AY458016 |
| blaCTX-M-3 | 100 | 876/876 | pEC25-1 | 216,248..217,123 | Beta-lactam resistance | Y10278 |
| Fluoroquinolone | ||||||
| qnrB2 | 99.84 | 645/645 | pEC25-1 | 105,800..106,444 | Fluoroquinolone resistance | DQ351242 |
| aac(6ʹ)-Ib-cr | 100 | 600/600 | pEC25-1 | 113,669..114,268 | Fluoroquinolone and aminoglycoside resistance | DQ303918 |
| aac(6ʹ)-Ib-cr | 100 | 600/600 | pEC25-1 | 208,288..208,887 | Fluoroquinolone and aminoglycoside resistance | DQ303918 |
| qnrS1 | 100 | 657/657 | pEC25-1 | 221,088..221,744 | Fluoroquinolone resistance | AB187515 |
| Rifampicin | ||||||
| arr-3 | 100 | 453/453 | pEC25-1 | 113,120..113,572 | Rifampicin resistance | JF806499 |
| MLS - Macrolide, Lincosamide and Streptogramin B | ||||||
| mph(A) | 100 | 906/906 | pEC25-1 | 99,354..100,259 | Macrolide resistance | D16251 |
| mph(A) | 100 | 906/906 | pEC25-1 | 196,453..197,358 | Macrolide resistance | D16251 |
| Sulphonamide | ||||||
| sul2 | 100 | 816/816 | pEC25-1 | 93,368..94,183 | Sulphonamide resistance | HQ840942 |
| sul1 | 100 | 840/840 | pEC25-1 | 104,470..105,309 | Sulphonamide resistance | U12338 |
| sul1 | 100 | 840/840 | pEC25-1 | 110,191..111,030 | Sulphonamide resistance | U12338 |
| sul1 | 100 | 840/840 | pEC25-1 | 202,669..203,508 | Sulphonamide resistance | U12338 |
| Tetracycline | ||||||
| tet(A) | 100 | 1200/1200 | pEC25-1 | 118,110..119,309 | Tetracycline resistance | AJ517790 |
| Trimethoprim | ||||||
| dfrA27 | 100 | 474/474 | pEC25-1 | 112,514..112,987 | Trimethoprim resistance | FJ459817 |
| dfrA17 | 100 | 474/474 | pEC25-1 | 204,974..205,447 | Trimethoprim resistance | FJ460238 |
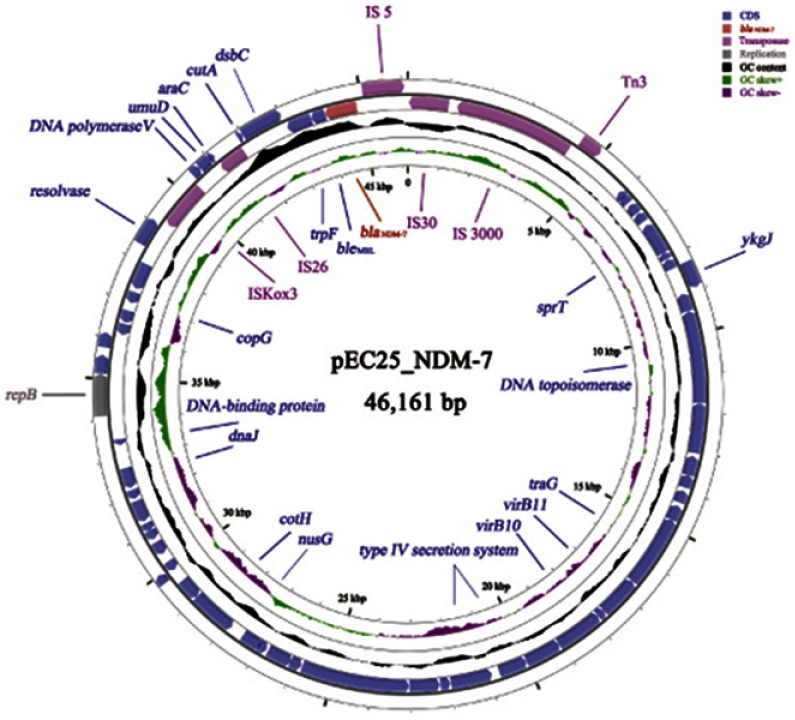
The plasmid profile of pEC25_NDM-7 is presented in Figure 1. It is an IncX3-type plasmid composed of 46,161 bp. The carbapenem-resistant gene blaNDM-7 was preceded by Tn3-IS3000-IS30-IS5 in the upstream region and followed by bleMBL-trpF-dsbC-cutA-IS26 in the downstream region. The similarity of pEC25_NDM-7 to other blaNDM-harboring plasmids was analyzed using Basic Local Alignment Search Tool (BLAST). pEC25_NDM-7 was 99% identical to several previously reported plasmids, namely, pJN05NDM7 (Accession No. MH523639), pAD-19R (Accession No. KX833071), pNDM-20 (Accession No. MF458176), pM216_X3 (Accession No. AP018146), and pKW53T-NDM (Accession No. KX214669). Plasmid pJN05NDM7 (carrying blaNDM-7) was identified in an E. coli ST167 clinical strain isolated from a patient with a urinary tract infection at a teaching hospital in the Shandong Province of China in 2015.15 Plasmid pAD-19R (carrying blaNDM-17) was identified in an E. coli ST48 strain isolated from a chicken at a commercial poultry farm in the Shandong Province of China in 2015.21 Plasmid pNDM-20 (carrying blaNDM-20) was identified in an E. coli ST1114 strain isolated from a swine faecal swab collected from a commercial pig farm in the Shandong province of China in 2016.22 Plasmid pM216_X3 (carrying blaNDM-4) was identified in an E. coli ST101 clinical strain isolated from a urine specimen at a tertiary care hospital in Yangon, Myanmar, in 2015.23 Plasmid pKW53T-NDM (carrying blaNDM-7) was identified in an E. coli ST448 clinical strain isolated from a urine sample in Kuwait in 2012.12 These plasmids encode distinct blaNDM variants, and it is likely that blaNDM mutates from blaNDM-1 to other blaNDM variants via nucleic acid replication in the 46,161 bp IncX3-type plasmid to evolve higher carbapenemase activity. Notably, the 46,161 bp IncX3-type plasmid does not carry antimicrobial resistance genes other than blaNDM, and the plasmid appeared in different ST type of E. coli strain across different countries in humans and animals. It seems the structure of the 46,161 bp plasmid is stable and suitable for the horizontal transfer of blaNDM, which plays an important role in horizontal transmission of NDM carbapenemase worldwide.
Figure 1.
Profile of the blaNDM-7-encoding plasmid pEC25_NDM-7.
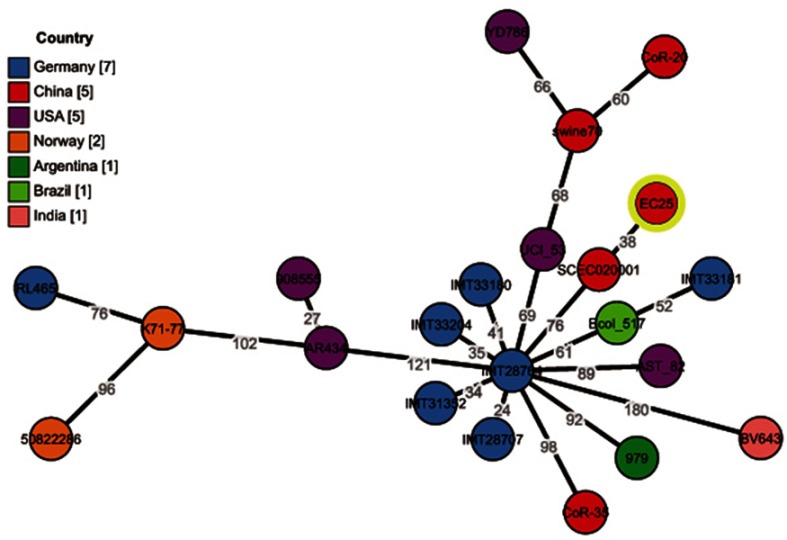
The phylogenetic relationship between E. coli EC25 and other E. coli strains is presented in Table 4 and Figure 2. Twenty-one phylogenetically related strains were identified in the database, all of which belong to ST410. Most of these strains produce ESBL, and only six strains produced carbapenemase, including one KPC-2-producing strain, one VIM-4-producing strain, and four NDM-producing strains. The phylogenetically related E. coli ST410 strains are spread globally. Seven strains were isolated from Germany, five strains from the USA, four strains from China, two strains from Norway, and one strain each from Argentina, Brazil, and India. Of the four strains from China, two of the strains were isolated from the Zhejiang province, one strain was from the Jiangsu province, and one strain was from the Sichuan province. The strain closest to EC25 was E. coli SCEC020001, which was isolated from the Sichuan province of China in 2016. There are only 38 different alleles between E. coli EC25 and E. coli SCEC020001. E. coli SCEC020001 also harbors a 46,161 bp IncX3-type plasmid (pNDM5_020001, Accession No. CP032424) that encodes blaNDM-5. pNDM5_020001 was 99% identical to pEC25_NDM-7. The main difference between the two plasmids was the subtype of blaNDM; pNDM5_020001 encoded blaNDM-5 and pEC25_NDM-7 encoded blaNDM-7. There is a geographic distinction between the Zhejiang province and Sichuan province, and the patient did not have a travel history out of Zhejiang province in recent years, which suggests that the NDM-positive E. coli ST410 strain EC25 was locally acquired, and the blaNDM-7 subtype may evolve during local dissemination.
Table 4.
Information of close isolates (based on cgMLST strategy) to strain E. coli EC25
| Isolate | Accession number | ST | Host | Isolation source | Country state | Collection year | Major resistance genotype | Different alleles | Reference |
|---|---|---|---|---|---|---|---|---|---|
| EC25 | CP035123 | 410 | Homo sapiens | Urine | China: Zhejiang | 2017 | blaNDM-7 | 0 | This study |
| SCEC020001 | CP032426 | 410 | Homo sapiens | N/Aa | China: Sichuan | 2016 | blaNDM-5 | 38 | Unpublished |
| IMT31352 | LJGF01 | 410 | Dog | Feces | Germany | 2013 | ESBL | 96 | 25 |
| IMT28764 | LJGC01 | 410 | Mute swan | Feces | Germany | 2012 | ESBL | 97 | 25 |
| IMT33180 | LJGI01 | 410 | Homo sapiens | Blood | Germany | 2009 | ESBL | 99 | 25 |
| IMT28707 | LJGB01 | 410 | Mute swan | Feces | Germany | 2012 | ESBL | 99 | 25 |
| IMT33204 | LJGK01 | 410 | Homo sapiens | Blood | Germany | 2010 | ESBL | 103 | 25 |
| IMT33181 | LJGJ01 | 410 | Homo sapiens | Blood | Germany | 2009 | ESBL | 118 | 25 |
| Ecol_517 | CP018965 | 410 | Homo sapiens | N/Aa | Brazil: | 2011 | blaKPC-2 | 119 | 26 |
| UCI_53 | JMVS01 | 410 | Homo sapiens | Biliary drain | USA | 2013 | ESBL | 127 | 27 |
| Swine70 | LVOT01 | 410 | Susscrofa | Rectum | China: Jiangsu | 2012 | - b | 135 | Unpublished |
| YD786 | CP013112 | 410 | Homo sapiens | Urine | USA | 2012 | ESBL | 138 | 28 |
| CoR-20 | MKFO01 | 410 | Homo sapiens | Sputum | China: Zhejiang | 2014 | ESBL | 145 | Unpublished |
| AST_82 | LLXC01 | 410 | N/Aa | N/Aa | USA | 2015 | AmpC | 147 | Unpublished |
| 979 | PIJB01 | 410 | Homo sapiens | Urine | Argentina | 2016 | ESBL | 150 | Unpublished |
| CoR-35 | NISA01 | 410 | Homo sapiens | Ascites | China: Zhejiang | 2015 | ESBL | 153 | Unpublished |
| K71-77 | LNGY01 | 410 | Homo sapiens | Blood | Norway | 2010 | blaNDM-1 | 164 | 29 |
| RL465 | FLKU01 | 410 | N/Aa | N/Aa | Germany | N/Aa | ESBL | 174 | 30 |
| AR434 | CP029122 | 410 | N/Aa | N/Aa | USA | N/Aa | - b | 176 | Unpublished |
| 50,822,286 | LNIK01 | 410 | Homo sapiens | Rectal swab | Norway | 2014 | blaVIM-4 | 185 | 31 |
| 908,555 | AXTR01 | 410 | Homo sapiens | N/Aa | USA | N/Aa | - b | 193 | Unpublished |
| BV643 | PVPJ01 | 410 | Homo sapiens | Blood | India | 2016 | blaNDM-5 | 194 | Unpublished |
Notes: aData not available. bNo carbapenemase, ESBL, and AmpC-type ß-Lactamases can be found in the genome.
Figure 2.
Phylogenetic relationship between E. coli ST410 strain EC25 and other E. coli strains. Twenty-one phylogenetically related strains were identified in the BacWGSTdb server. All of these strains belonged to ST410. Seven strains were isolated from Germany, five from the USA, four from China, two from Norway, and one each from Argentina, Brazil, and India.
A variety of STs were found in NDM-positive E. coli strains worldwide, and there are no predominant STs. The number of large-scale studies examining the clonal background of NDM-positive E. coli strains is limited in China. Zhang R et al suggested that ST167 and ST410 were two of the most common sequence types of NDM-positive E. coli strains in China.24 We report the whole-genome sequence of a clinically isolated E. coli ST410 strain harboring blaNDM-7. Our study is the first report on fully sequenced E. coli ST410 strain carrying blaNDM-7 isolated from China because blaNDM-7 is rare. A similar NDM-positive E. coli ST410 strain was found in the BacWGSTdb server in other provinces of China. ST410 is more likely to emerge as a successful epidemic clone of NDM-producing E. coli in China, and the blaNDM gene is prone to mutate during its dissemination. Therefore, more studies are required to illuminate the epidemic clones of NDM-positive E. coli in China.
Conclusion
In summary, our study reports the first identification of a clinical carbapenem-resistant E. coli ST410 strain carrying blaNDM-7 recovered from a urinary tract infection in China. Our study highlights the potential transmission opportunity of carbapenem-resistant plasmid carriage in E. coli. Further studies involving more NDM-producing isolates are warranted to identify reservoirs and monitor the transmission dynamics of blaNDM genes in China.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81702042), Natural Science Foundation of Zhejiang Province (LQ19H200003) and the Zhejiang Provincial Medical and Health Science and Technology Plan (2018KY344 and 2019KY311).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Quan J, Zhao D, Liu L, et al. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J Antimicrob Chemother. 2017;72(1):273–280. doi: 10.1093/jac/dkw372 [DOI] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. ndm metallo-beta-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2):e00115–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jovcic B, Lepsanovic Z, Suljagic V, et al. Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011;55(8):3929–3931. doi: 10.1128/AAC.00226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bontron S, Nordmann P, Poirel L. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(12):7245–7251. doi: 10.1128/AAC.01755-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68(8):1737–1740. doi: 10.1093/jac/dkt088 [DOI] [PubMed] [Google Scholar]
- 7.Cuzon G, Bonnin RA, Nordmann P. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One. 2013;8(4):e61322. doi: 10.1371/journal.pone.0061322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuno Y, Yamaguchi T, Matsumoto T. A first case of New Delhi metallo-beta-lactamase-7 in an Escherichia coli ST648 isolate in Japan. J Infect Chemother. 2014;20(12):814–816. doi: 10.1016/j.jiac.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 9.Lee CS, Vasoo S, Hu F, Patel R, Doi Y. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol. 2014;52(11):4109–4110. doi: 10.1128/JCM.01404-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Peirano G, Lynch T, et al. Molecular characterization by using next-generation sequencing of plasmids containing blaNDM-7 in enterobacteriaceae from Calgary, Canada. Antimicrob Agents Chemother. 2015;60(3):1258–1263. doi: 10.1128/AAC.02661-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Gajendiran R, Daniel JL, Walia K, Veeraraghavan B. First Indian report of IncX3 plasmid carrying blaNDM-7 in Escherichia coli from bloodstream infection: potential for rapid dissemination. New Microbes New Infect. 2017;17:65–68. doi: 10.1016/j.nmni.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal T, Ghazawi A, Darwish D, et al. Characterization of NDM-7 carbapenemase-producing Escherichia coli isolates in the Arabian Peninsula. Microb Drug Resist. 2017;23(7):871–878. doi: 10.1089/mdr.2016.0216 [DOI] [PubMed] [Google Scholar]
- 13.Espinal P, Miro E, Segura C, et al. First description of blaNDM-7 carried on an IncX4 plasmid in Escherichia coli ST679 isolated in Spain. Microb Drug Resist. 2018;24(2):113–119. doi: 10.1089/mdr.2017.0039 [DOI] [PubMed] [Google Scholar]
- 14.Wang LH, Liu PP, Wei DD, et al. Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-beta-lactamase in China. Int J Antimicrob Agents. 2016;48(1):41–45. doi: 10.1016/j.ijantimicag.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 15.Hao Y, Shao C, Bai Y, Jin Y. Genotypic and phenotypic characterization of IncX3 plasmid carrying bla NDM-7 in Escherichia coli sequence type 167 isolated from a patient with urinary tract infection. Front Microbiol. 2018;9:2468. doi: 10.3389/fmicb.2018.02468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan Z, Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44(D1):D682–D687. doi: 10.1093/nar/gkv1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–W57. doi: 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(suppl_1):D32–D36. doi: 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant JR, Stothard P. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(WebServer issue):W181–W184. doi: 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Wang Y, Walsh TR, et al. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother. 2017;61:5. doi: 10.1128/AAC.02233-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Li J, Wang X, et al. Novel variant of New Delhi metallo-beta-lactamase, NDM-20, in Escherichia coli. Front Microbiol. 2018;9:248. doi: 10.3389/fmicb.2018.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugawara Y, Akeda Y, Sakamoto N, et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One. 2017;12(9):e0184720. doi: 10.1371/journal.pone.0184720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaufler K, Semmler T, Wieler LH, et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410–another successful pandemic clone? FEMS microbiol Ecol. 2016;92(1):fiv155. [DOI] [PubMed] [Google Scholar]
- 26.Stoesser N, Sheppard AE, Peirano G, et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep. 2017;7(1):5917. doi: 10.1038/s41598-017-06256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerqueira GC, Earl AM, Ernst CM, et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A. 2017;114(5):1135–1140. doi: 10.1073/pnas.1616248114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Q, Ding B, Jove T, et al. Characterization of a novel IncHI2 plasmid carrying tandem copies of blaCTX-M-2 in a fosA6-harboring Escherichia coli sequence type 410 strain. Antimicrob Agents Chemother. 2016;60(11):6742–6747. doi: 10.1128/AAC.01173-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuelsen O, Thilesen CM, Heggelund L, Vada AN, Kummel A, Sundsfjord A. Identification of NDM-1-producing Enterobacteriaceae in Norway. J Antimicrob Chemother. 2011;66(3):670–672. doi: 10.1093/jac/dkq483 [DOI] [PubMed] [Google Scholar]
- 30.Falgenhauer L, Waezsada SE, Gwozdzinski K, et al. Chromosomal locations of mcr-1 and blaCTX-M-15 in fluoroquinolone-resistant Escherichia coli ST410. Emerg Infect Dis. 2016;22(9):1689–1691. doi: 10.3201/eid2209.160692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuelsen O, Overballe-Petersen S, Bjornholt JV, et al. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS One. 2017;12(11):e0187832. doi: 10.1371/journal.pone.0187832 [DOI] [PMC free article] [PubMed] [Google Scholar]