Abstract
Background: Human N-acetyltransferase 10 (NAT10) plays pivotal roles in cellular biological processes, such as senescence, autophagy and cytokinesis. The expression of NAT10 is dysregulated in colorectal cancer (CRC) and is associated with the prognosis of patients. However, it remains unclear how NAT10 is regulated in CRC.
Methods: The microRNA(miRNA) regulating NAT10 was predicted by bioinformatics analysis and further validated by real-time quantitative PCR(RT-qPCR),Western blot and dual luciferase reporter assays. The expression of the miRNA regulating NAT10 in CRC tissues was examined using RT-qPCR. Cell proliferation, cell apoptosis, cell migration and cell invasion assays were performed after transfection with miRNA mimic and inhibitor.
Results: Here, we report that miR-6716-5p inhibits the expression of NAT10 in CRC. The NAT10 protein level was downregulated by the miR-6716-5p mimic, and was upregulated by the miR-6716-5p inhibitor in CRC cell lines. In addition, miR-6716-5p bound to the 3ʹ-untranslated region of NAT10 mRNA and decreased NAT10 mRNA levels. Significantly, the miR-6716-5p level was higher in the tumor tissues of the CRC patients with liver metastasis than that in the non-metastatic CRC patients. In addition, the miR-6716-5p level was correlated with poor overall survival of CRC patients with liver metastasis. The miR-6716-5p inhibitor inhibited CRC cell migration and invasion. Consistently, the miR-6716-5p mimic significantly promoted cell migration and invasion, and this effect is dependent on NAT10. However, miR-6716-5p had no effect on CRC cell proliferation and apoptosis. We found that miR-6716-5p negatively regulated E-cadherin protein levels. In addition, E-cadherin was upregulated by NAT10 in CRC cells, confirming that miR-6716-5p downregulated E-cadherin levels by inhibiting NAT10 expression.
Conclusion: We demonstrated that miR-6716-5p acts as a crucial regulator of NAT10 to promote cell migration and invasion in CRC by inhibiting NAT10 expression. Our data suggest that miR-6716-5p/NAT10 might act as a potential therapeutic target for CRC treatment.
Keywords: miR-6716-5p, NAT10, colorectal cancer; CRC, liver metastasis
Introduction
Colorectal cancer (CRC), one of the most common malignancies, is the second leading cause of cancer-related death worldwide. It has been estimated that 1.8 million of CRC cases are diagnosed and approximately 860,000 patients die of CRC annually.1 The 5-year survival rate of CRC patients with tumors confined to the colorectum is about 90%, while either lymphatic or distant metastasis decreases the 5-year survival rate of the CRC patients to range from 72% to 13%.2 Tumor metastasis is the major cause of mortality in CRC patients.3 Liver is the most common metastatic site of CRC.4 Approximately 15% of the CRC patients are diagnosed with synchronous hepatic metastasis, and 50% of CRC patients developed metachronous hepatic metastasis after the primary tumor resection.5 The 5-year survival rate of CRC patients with liver metastasis is 25–40% even after resection of the metastatic lesions.6 Therefore, identification of the molecules acting as prognostic marker for CRC with liver metastasis and inhibiting cell invasion will contribute to improving the prognosis of patients.
MicroRNAs (miRNAs) are a class of small endogenous noncoding RNAs. Generally, miRNA is composed of about 22 nucleotides, and regulates gene expression by binding to the 3ʹ-untranslated region (UTR) of its target mRNAs.7 It has been demonstrated that miRNAs play critical roles in cellular processes, such as cell proliferation, mitosis, apoptosis and differentiation.8–10 Numerous evidence have indicated that miRNAs possess tumor-suppressive or oncogenic activities through regulating their downstream targets in human cancers.8,11–13 MiRNAs are abnormally altered during the occurrence and development of CRC, and some of them show potential as diagnostic, prognostic or staging markers of CRC.14–17 For instance, miR-34a-5p has been identified as an independent prognostic factor for CRC recurrence.18 Serum miR-203 could be a promising prognostic and predictive biomarker for CRC metastasis.19 Therefore, the study of the functions of miRNAs in the metastasis or recurrence of CRC would provide more markers for improving the clinical outcome of CRC patients.
NAT10 belongs to the Gcn5-related N-acetyltransferases family and possesses histone acetyltransferase (HAT) activity. It was initially found that NAT10 could upregulate telomerase activity through the transactivation of human telomerase reverse transcriptase promoter.20 NAT10 controls various cellular processes via its acetyltransferase activity. For instance, NAT10 binds to hsSUN1 and regulates the decondensation of chromosomes during mitosis.21 NAT10 also controls cytokinesis through acetylating α-tubulin.22 NAT10 activates RNA polymerase I transcription by binding to and acetylating the upstream binding factor in the nucleolus.23 Recently, it was found that NAT10 functions in the rRNA processing through acetylating rRNA.24 In addition, NAT10 also acetylates tRNA.25 Importantly, NAT10 promotes translation efficiency through acetylating cytidine in mRNA.26 In response to DNA damage, NAT10 activates p53 through the acetylation of p53 K120 to maintain cellular homeostasis.27 Under energy stress, the deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy.28 Recently, it was reported that chemical inhibition of NAT10 acetyltransferase activity could correct defects of laminopathic cells and treat progeria.29 These findings have identified NAT10 as a key regulator of cellular activities. In addition, an increasing number of studies have reported that dysfunction of NAT10 is involved in tumorigenesis and tumor development. It has been reported that NAT10 is related with the patient’s prognosis of CRC and hepatocellular carcinoma.30–32 NAT10 mRNA levels were significantly downregulated in CRC tumor tissues in comparison to the levels in normal tissues.27 We previously demonstrated that the function of NAT10 is regulated by its autoacetylation.33 However, it remains unclear how NAT10 is regulated in CRC.
In this study, we analyzed the miRNAs targeting NAT10 and identified miR-6716-5p as a critical regulator of NAT10 in the liver metastasis of CRC.
Materials and methods
Patients and follow-up
Human CRC tissues were obtained from 80 CRC patients who received surgical resection at Peking University Cancer Hospital and Institute from October 2010 to July 2015. The patients included 54 men and 36 women, with a median age of 57 years. Forty CRC patients only with liver metastasis belonged to stage IV and the remainder belonged to stage (I, II, III) according to the AJCC/UICC (seventh edition) TNM staging system. All patients provided written informed consent for their samples to be used in this study. This study was approved by the ethics committee of Peking University Cancer Hospital and was conducted in accordance with the Declaration of Helsinki. The patients were followed up once every 3 months during the first 2 years, once every 6 months during the third and fourth year and once a year from the fifth year postoperatively. The overall survival time was calculated from the operation date to the death date, and the recurrence or metastasis date or the last censor time were also recorded.
Cell culture and transfections
CRC cell lines (HCT116, LoVo) and HEK293T cell lines were purchased from the National Infrastructure of Cell line Resource (Beijing, People’s Republic of China). Cells were cultured in DMEM with 10% fetal bovine serum. Cells were transfected with plasmid DNA or siRNA RNA duplexes (GenePharma, Shanghai, People’s Republic of China) by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. MiRNA mimics or inhibitors and siRNA sequences are listed in the Supplemental materials.
Luciferase assay
For the luciferase reporter assay, cells were seeded in 24-well plates at a density of 1×105 cells per well. The cells were cotransfected with pmirGLO luciferase reporters and either wild type or mutant NAT10 3ʹUTR plasmid, in combination with miR-6716-5p mimic or control mimic (GenePharma) using Lipofectamine2000 (Invitrogen). After 24 hrs, the firefly and Renilla luciferase activities were measured and analyzed according to the manufacturer’s instructions (Promega, Fitchburg, WI, USA).
Cell viability assay
Cells were transfected with miR-6716-5p (mimic or inhibitor) or a negative control (GenePharma) using Lipofectamine 2000 (Invitrogen). After 48 hrs of transfection, a thousand of cells were seeded into 96-well plates and cultured with 10% serum containing media. Cell viability was analyzed using the MTS assay (Promega) according to the manufacturer’s instructions. Absorbance at 490 nm was measured using a microplate spectrophotometer once a day for 5 consecutive days.
Colony formation assay
After 48 hrs of transfection, a thousand cells were seeded into 6-well plates and cultured with 10% serum containing media for about 12 days. Cells were fixed and then stained with crystal violet, and visible colonies were counted.
Apoptosis assay
Cells transfected with miR-6716-5p or negative control were harvested. The cells were stained with Annexin V-FITC and Propidium Iodide using an Annexin V-FITC Apoptosis detection kit (KeyGEN, BioTECH, Nanjing, People’s Republic of China) according to the manufacturer’s instructions. Then, the cells were analyzed using FACSCalibur (BD Bioscience). A total of 10,000 cells were acquired for each sample, and the data were analyzed using BD CellQuest software.
Transwell assay
Migration and invasion assays were performed according to the instructions of the manufacturer (Merck, Millipore). Briefly, 2×105 cells were plated in serum-free medium in the upper chamber with a non-coated membrane (24 well insert, 8 μm pore size) for the migration assay and with a Matrigel-coated membrane for the invasion assay. The lower chamber was filled with medium containing 10% serum. Then, the cells on the upper surface were removed by cotton swab and the cells on the lower surface were stained with crystal violet and observed under a microscope.
Western blotting analysis
Western blot experiment was carried out as described previously.27 Anti-NAT10 was a gift from Dr B. Zhang. Anti-actin (abclonal, ac004) was purchased from ABclonal Technology. Anti-E-cadherin (BD, 562869) was purchased from BD Bioscience.
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was performed on the ABI 7500 Fast Real-Time PCR systems (Applied Biosystems, Foster City, CA, USA). Total RNA and miRNA were extracted from CRC cells and tissues using MiPure Cell/Tissue miRNA Kit (Vazyme, Nanjing, People’s Republic of China). cDNA was synthesized by Universal cDNA Synthesis Kit (Roche, USA). The primer mix contained a specific stem-loop primer of miR-6716-5p and a RNU6A (U6) reverse primer for cDNA synthesis of miRNA. Random hexamer primer was used to synthesize the cDNA of the mRNAs. RT-qPCR was performed using the FastStart Universal SYBR Green Master (Roche) in the ABI 7500 Fast Real-Time PCR system (Applied Biosystems). GADPH and U6 were used as endogenous controls. A melting curve analysis was performed for the primer sets, and each showed a single peak indicating the specificity of the primers tested. Relative expression levels were computed using the 2−ΔΔCt method. The primer sequences used are listed in the Supplemental Information.
Statistical analysis
Statistical analysis was performed using the SPSS software package (SPSS Standard version 21.0, SPSS Inc, Chicago, IL, USA). Shapiro–Wilk test was performed before using Student’s t test and Mann–Whitney U test. Levene’s test was also performed to evaluate variance homogeneity before using Student’s t test and Mann–Whitney U test. The statistical significance of differences between groups was assessed by Student’s t test or Mann–Whitney U test. The relationship between miR-6716-5p expression and the clinical pathological parameters was determined by the χ2 test. Bonferroni correction was also performed for determination of relations between miR-6716-5p expression and clinical pathological parameters. Kaplan–Meier analysis was used to evaluate CRC patient survival and a log-rank test was used to compare different survival curves. In cases of comparison of relative expression levels, Non-scaled values were used in the data analysis. The correlation between the expression of NAT10 and miR-6716-5p was analyzed by Spearman’s rank correlation coefficient. Data were presented as the mean±SD. P-values <0.05 were considered statistically significant.
Results
NAT10 expression was regulated by miR-6716-5p
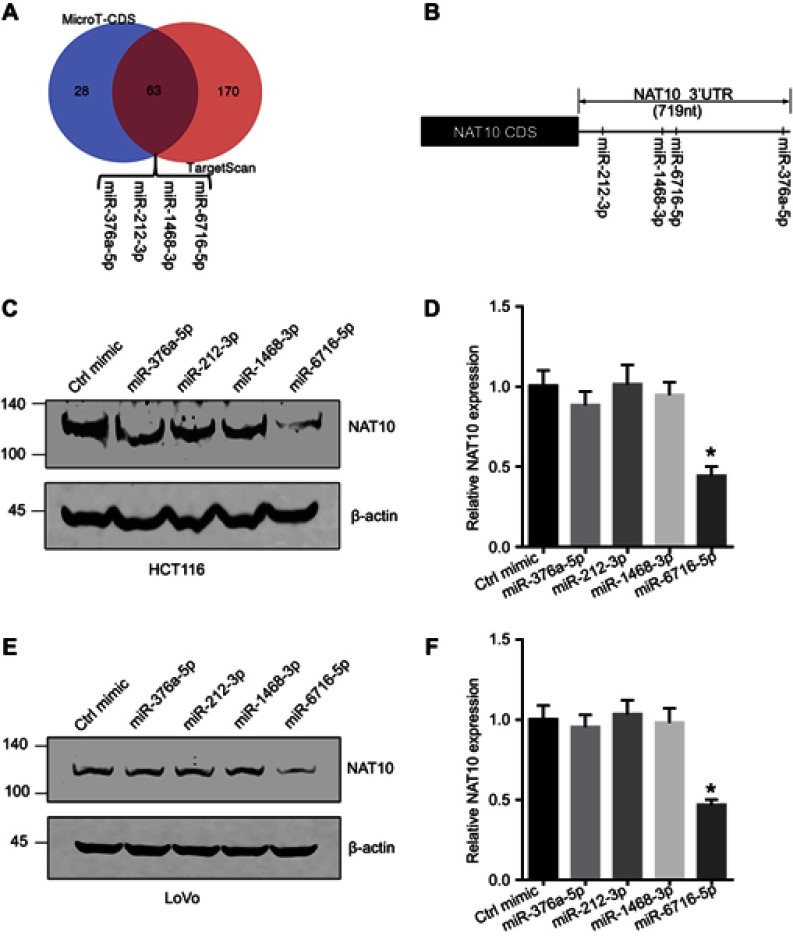
To uncover the potential miRNAs regulating the expression of NAT10, we analyzed the putative miRNAs which might bind 3ʹ-UTR of NAT10 mRNA using two major prediction software programs, TargetScan (release 7.1,http:www.//targetscan.org/vert_71/)34 and DIANA-microT-CDS (5th version, http://www.microrna.gr/microT-CDS, the threshold was set to 0.55) (Figure 1A).35 According to the miRNAs predicted by the above two software programs, we identified the common miRNAs between the two and finally obtained 63 putative miRNAs. According to the scores of the 63 putative miRNAs and the binding sequence in the 3ʹ-UTR of NAT10 mRNA, which were obtained from DIANA-microT-CDS software, miR-376a-5p, miR-212-3p, miR-1468-3p and miR-6716-5p were selected as they might potentially bind to the 3ʹ-UTR of NAT10 mRNA (Figure 1B and Figure S1). To determine if these miRNAs regulate NAT10 expression, we firstly evaluated the NAT10 protein level after transfecting different miRNA mimics in HCT116 cells. We showed that the NAT10 protein level was decreased by the miR-6716-5p mimic but not by the mimics of other miRNAs (Figure 1C and D). Similar results were obtained in LoVo cells (Figure 1E and F). These results indicated that NAT10 expression was regulated specifically by miR-6716-5p.
Figure 1.
NAT10 expression was regulated by miR-6716-5p. (A) NAT10 might be regulated by miR-376a-5p, miR-212-3p, miR-1468-3p and miR-6716-5p according to two prediction software packages. (B) Illustration of binding site between putative miRNAs and the NAT10 3ʹ-UTR was indicated. (C) HCT116 cells were transfected putative miRNAs mimics and harvested. NAT10 protein level was detected by western blotting and NAT10 protein level was quantified by image J software. (D) LoVo cells were transfected putative miRNAs mimics and harvested. NAT10 protein level was detected by western blotting and NAT10 protein level was quantified by image J software. Error bars represent the SD from three independent experiments. P-values were obtained from Student’s t test. *P<0.05.
miR-6716-5p bound to the 3ʹ-UTR of NAT10 mRNA to inhibit the expression of NAT10
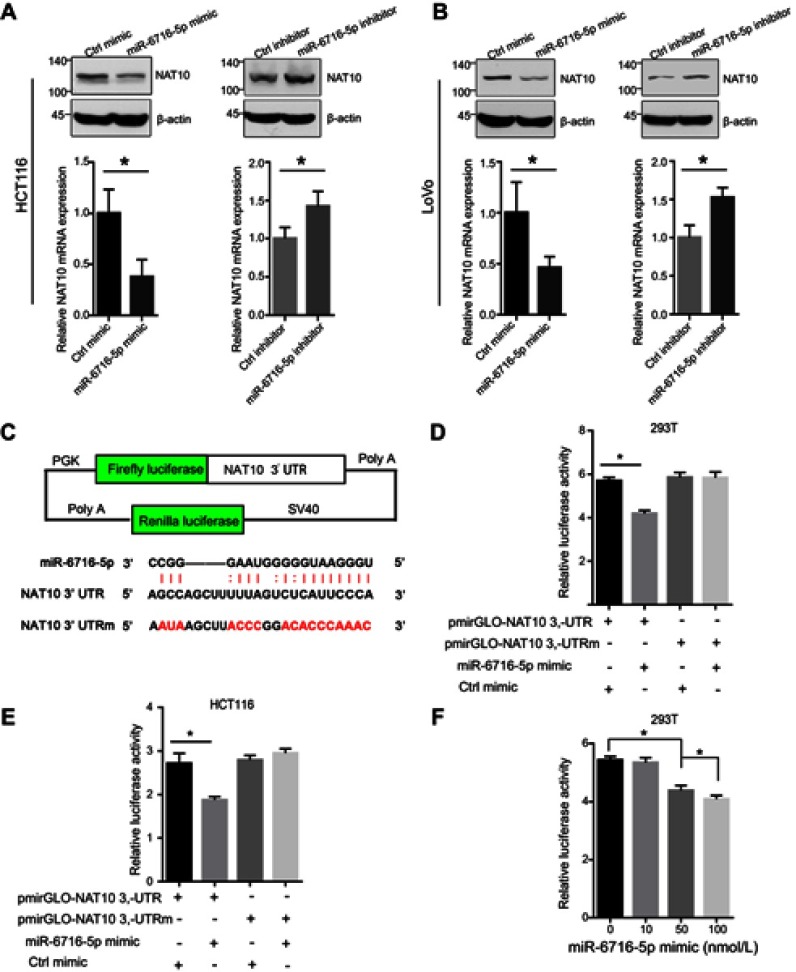
To further confirm that the expression of NAT10 is regulated by miR-6716-5p, the protein and mRNA levels of NAT10 were evaluated when the miR-6716-5p mimic or inhibitor were transfected into HCT116 cells. NAT10 protein and mRNA levels were significantly decreased by the introduction of miR-6716-5p mimic in the HCT116 cells (Figure 2A). Consistently, the miR-6716-5p inhibitor increased the NAT10 protein and mRNA levels in HCT116 cells (Figure 2A). Moreover, similar results were obtained in LoVo cells (Figure 2B). Thereafter, we determined whether miR-6716-5p binds to the 3ʹ-UTR of NAT10 mRNA. The 3ʹ-UTR of NAT10 mRNA or its mutant lacking the miR-6716-5p binding site was cloned into the pmirGLO luciferase reporter vector to generate pmirGLO-NAT10 3ʹ-UTR and pmirGLO-NAT10 3ʹ-UTRm (Figure 2C). The constructed plasmid was transfected with miR-6716-5p mimic into HEK293T cells, respectively. Indeed, the relative luciferase activity of the pmirGLO-NAT10 3ʹ-UTR was dramatically reduced when co-transfected with the miR-6716-5p mimic in HEK293T cells (Figure 2D). Similar results were obtained in HCT116 cells (Figure 2E). These results indicated that miR-6716-5p bound to the 3ʹ-UTR of NAT10 mRNA. To confirm this finding, we further analyzed the luciferase activity of pmirGLO-NAT10 3ʹ-UTRm when co-transfected with the miR-6716-5p mimic. We showed that the miR-6716-5p mimic had no effect on the luciferase reporter activity of pmirGLO-NAT10 3ʹ-UTRm (Figure 2D and E), confirming the binding site of miR-6716-5p in the NAT10 3ʹ-UTR. Moreover, the relative luciferase activity of the pmirGLO-NAT10 3ʹ-UTR was reduced by miR-6716-5p in a dose-dependent pattern in the HEK293T cells (Figure 2F). Taken together, these results indicated that miR-6716-5p directly bound to the 3ʹ-UTR of NAT10 mRNA to inhibit NAT10 expression.
Figure 2.
miR-6716-5p bound to the 3ʹ-UTR of NAT10 mRNA to inhibit expression of NAT10. (A) HCT116 cells were transfected with either miR-6716-5p mimic or inhibitor. Seventy-two hours later, cells were harvested and lysed. Then, NAT10 protein and mRNA expression levels were detected by western blotting and RT-qPCR analysis, respectively. (B) LoVo cells were transfected with either miR-6716-5p mimic or inhibitor. Then, cells were harvested and total proteins were extracted. NAT10 protein and mRNA levels were detected by western blotting and RT-qPCR analysis, respectively. (C) Schematic diagram of pmirGLO luciferase reporter vector was illustrated and the predicted miR-6716-5p binding site in 3ʹ-UTR wild-type sequence of NAT10 (pmirGLO-NAT10-3ʹUTR) and the designed mutant sequence (pmirGLO-NAT10-3ʹUTRm) were indicated. (D) The indicated vectors and miR-6716-5p mimic were transfected into HEK293T cells. Twenty-four hours later, cells were harvested and lysed. Then, the firefly and renilla luciferase activities were measured and analyzed according to the manufacturer’s instructions. Luciferase activities were assessed and normalized. (E) HCT116 cells were transfected with the indicated vectors. Twenty-four hours later, the firefly and renilla luciferase activities were measured and analyzed according to the manufacturer’s instructions. (F) HEK293T cells were transfected as indicated and cultured for 24 hrs. Then, the firefly and renilla luciferase activities were measured and analyzed according to the manufacturer’s instructions. (A, B) Error bars represent the SD from three independent experiments. *P<0.05. (D–F) Data were presented as the mean±SD (n=6, each group). P-values were obtained from Student’s t test. *P<0.05.
The miR-6716-5p level was correlated with liver metastasis and survival in CRC patients
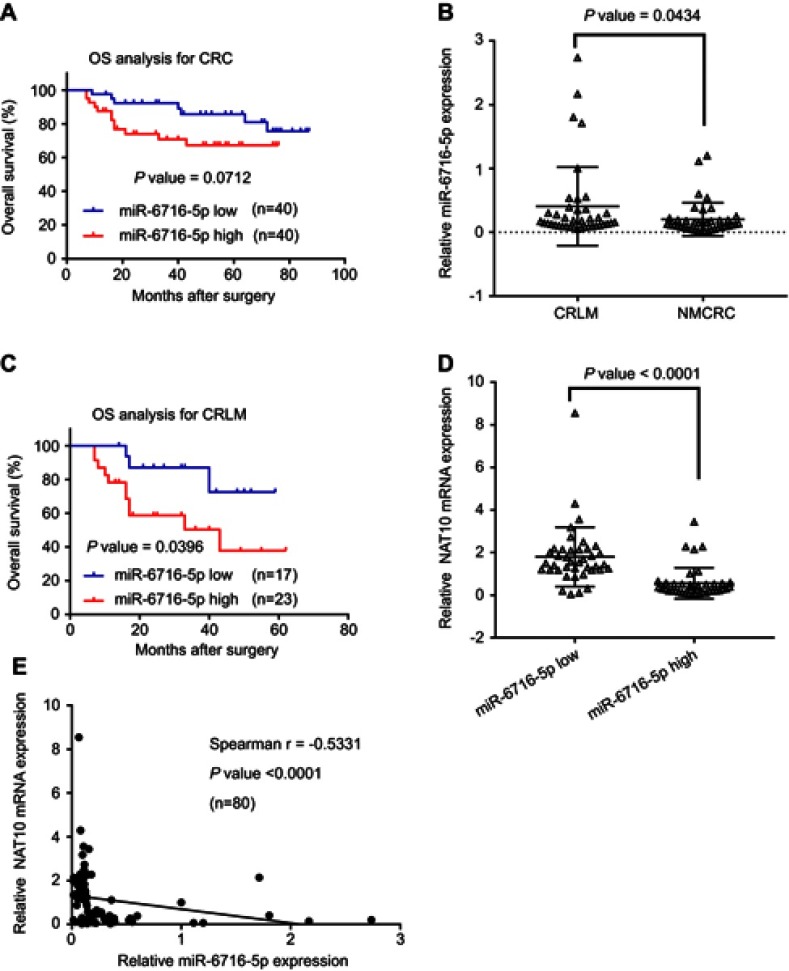
To determine whether miR-6716-5p is related with CRC development, the miR-6716-5p levels were evaluated in human CRC tumor tissues. Firstly, we examined the clinic-pathological significance of miR-6716-5p expression in CRC patients. The median value of the miR-6716-5p levels in 80 CRC tumor tissues was set as the cutoff value. Based on this cutoff value, the CRC patients were divided into a high miR-6716-5p level group and a low miR-6716-5p level group. The characteristics of patients, including gender, age, primary tumor location, vascular invasion, differentiation, TNM stage, and lymph node metastasis, are listed in Table 1. We found that the incidence rate of vascular invasion in the high miR-6716-5p level group was significantly higher than that in the low miR-6716-5p level group (P=0.019), indicating that miR-6716-5p was correlated with vascular invasion (Table 1). The patients with high miR-6716-5p levels had a poor survival rate tendency though there was no significant difference in the overall survival (OS) rate between the CRC patients with high miR-6716-5p levels and those with low miR-6716-5p levels (P=0.0712) (Figure 3A). Since miR-6716-5p was associated with vascular invasion (Table 1), we wanted to know if miR-6716-5p was related with the survival of patients with metastasis. Indeed, the miR-6716-5p levels in the CRC with liver metastasis (CRLM) patients were significantly higher than those in the non-metastatic CRC patients (Figure 3B). Importantly, the OS rate of the CRLM patients with low miR-6716-5p levels was higher than that of the CRLM patients with high miR-6716-5p levels (P=0.0396) (Figure 3C). The characteristics of the CRLM patients are listed in Table 2. It was also found that the incidence rate of vascular invasion in the high miR-6716-5p level group was higher than that in the low miR-6716-5p level group in the CRLM patients (P=0.015). Moreover, the incidence of multiple liver metastases was significantly higher in the CRLM patients with high miR-6716-5p levels than in the CRLM patients with low miR-6716-5p levels (P=0.007). Taken together, these results indicated that high miR-6716-5p levels might be an unfavorable prognostic factor and indicated a poor OS rate in the CRLM patients.
Table 1.
Relationship between miR-6716-5p expression and clinical characteristics in colorectal cancer patients
| Characteristics | No. of patients (%) | miR-6716-5p expression | χ2 | P | |
|---|---|---|---|---|---|
| Low n=40 (%) |
High n=40 (%) |
||||
| Age | 0.457 | 0.499 | |||
| <60 | 45 (56.25) | 21 (52.5) | 24 (60.0) | ||
| ≥60 | 35 (43.75) | 19 (47.5) | 16 (40.0) | ||
| Gender | 0.808 | 0.369 | |||
| Female | 36 (45.0) | 24 (60.0) | 20 (50.0) | ||
| Male | 44 (55.0) | 16 (40.0) | 20 (50.0) | ||
| Location | 0.889 | 0.641 | |||
| Left colon | 25 (31.25) | 11 (27.5) | 14 (35.0) | ||
| Right colon | 38 (47.5) | 19 (47.5) | 19 (47.5) | ||
| Rectum | 17 (21.25) | 10 (25.0) | 7 (17.5) | ||
| TNM stage | 1.800 | 0.180 | |||
| I II III | 40 (50.0) | 23 (57.5) | 17 (42.5) | ||
| IV | 40 (50.0) | 17 (42.5) | 23 (57.5) | ||
| Vascular invasion | 5.495 | 0.019 | |||
| Absent | 52 (65.0) | 31 (77.5) | 21 (52.5) | ||
| Present | 28 (35.0) | 9 (22.5) | 19 (47.5) | ||
| Lymph node metastasis | 0.202 | 0.653 | |||
| N0 | 36 (45.0) | 19 (47.5) | 17 (42.5) | ||
| N1-2 | 44 (55.0) | 21 (52.5) | 23 (57.5) | ||
| Histology differentiation | 0.967 | 0.617 | |||
| Poor | 11 (13.75) | 7 (17.5) | 4 (10.0) | ||
| Moderate | 65 (81.25) | 31 (77.5) | 34 (85.0) | ||
| Well | 4 (5.0) | 2 (5.0) | 2 (5.0) | ||
Figure 3.
miR-6716-5p level was correlated with liver metastasis and survival of CRC patients. (A) Kaplan–Meier analysis for overall survival (OS) rate between miR-6716-5p high-expression group and miR-6716-5p low-expression group in CRC patients. (B) Relative miR-6716-5p level was analyzed between non-metastatic CRC (NMCRC) patients and CRC with liver metastasis (CRLM) patients. (C) Kaplan–Meier analysis for OS rate between miR-6716-5p high group and miR-6716-5p low group in CRLM patients. (D) Relative NAT10 mRNA expression was analyzed between miR-6716-5p high group and miR-6716-5p low group. (E) Correlation between miR-6716-5p level and NAT10 mRNA level was calculated according to Spearman’s rank correlation coefficient in CRC specimen (n=80). CRLM represents CRC with liver metastasis. NMCRC represents non-metastatic CRC. (B, D) The Mann–Whitney U test was used to compare the levels of NAT10 mRNA or miR-6716-5p between groups and horizontal line was the mean value in two groups, respectively.
Table 2.
Relationship between miR-6716-5p expression and clinical characteristics in colorectal cancer with liver metastasis patients
| Characteristics | No. of patients (%) | miR-6716-5p expression | χ2 | P | |
|---|---|---|---|---|---|
| Low n=17 (%) |
High n=23 (%) |
||||
| Age | 0.496 | 0.481 | |||
| <60 | 26 (65.0) | 10 (58.82) | 16 (69.57) | ||
| ≥60 | 14 (35.0) | 7 (41.18) | 7 (30.43) | ||
| Gender | 0.051 | 0.822 | |||
| Female | 22 (55.0) | 9 (52.94) | 13 (56.52) | ||
| Male | 18 (45.0) | 8 (47.06) | 10 (43.48) | ||
| Location | 0.874 | 0.646 | |||
| Left colon | 14 (35.0) | 7 (41.18) | 7 (30.43) | ||
| Right colon | 22 (55.0) | 9 (52.94) | 13 (56.52) | ||
| Rectum | 4 (10.0) | 1 (5.88) | 3 (13.04) | ||
| Vascular invasion | 5.966 | 0.015 | |||
| Absent | 17 (42.5) | 11 (64.71) | 6 (26.09) | ||
| Present | 23 (57.5) | 6 (35.29) | 17 (73.91) | ||
| Lymph node metastasis | 0.349 | 0.555 | |||
| N0 | 11 (45.0) | 6 (35.29) | 5 (21.74) | ||
| N1-2 | 29 (55.0) | 11 (64.71) | 18 (78.26) | ||
| Histology differentiation | 0.644 | 0.725 | |||
| Poor | 4 (13.75) | 1 (5.88) | 3 (13.04) | ||
| Moderate | 32 (81.25) | 14 (82.35) | 18 (78.26) | ||
| Well | 4 (5.0) | 2 (11.76) | 2 (8.70) | ||
| Liver metastasis size | 0.628 | 0.428 | |||
| ≤3(cm) | 23 (57.5) | 11 (64.71) | 12 (52.17) | ||
| >3(cm) | 17 (42.5) | 6 (35.29) | 11 (47.82) | ||
| Number of liver metastases | 7.376 | 0.007 | |||
| 1 | 14 (35.0) | 10 (58.82) | 4 (17.39) | ||
| >1 | 26 (65.0) | 7 (41.18) | 19 (82.61) | ||
Next, we investigated the relationship between miR-6716-5p levels and the NAT10 mRNA levels. Significantly, the NAT10 mRNA levels in the low miR-6716-5p level group were higher than those in the high miR-6716-5p level group (P<0.01) (Figure 3D). We found that there existed negative correlation between NAT10 mRNA and miR-6716-5p (P<0.01) and the correlation coefficient was -0.5331 according to Spearman’s rank correlation coefficient (Figure 3E).
miR-6716-5p promoted cell migration and invasion by downregulating NAT10 expression
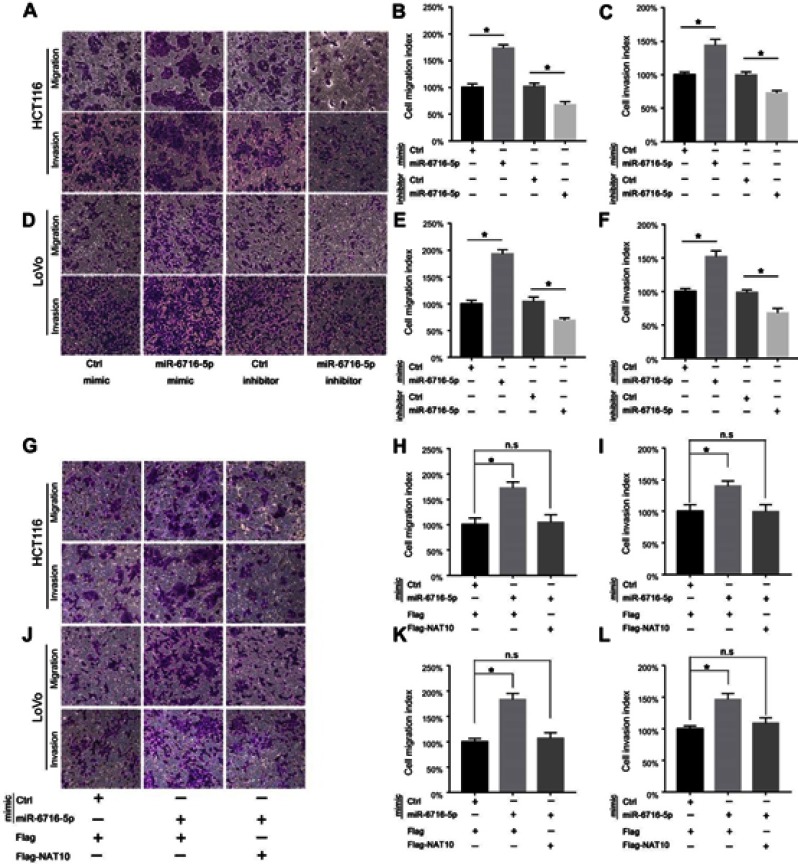
Since miR-6716-5p was correlated with the liver metastasis of CRC, we examined the effect of miR-6716-5p on the migration and invasion of CRC cells. Overexpression of miR-6716-5p significantly increased cell migration and invasion in HCT116 cells (Figure 4A). Meanwhile, inhibitor of miR-6716-5p decreased the cell migration and invasion (Figure 4A, B and C). Similar results were obtained in LoVo cells (Figure 4D, E and F). These results indicated that miR-6716-5p promoted the cell migration and invasion of CRC. To further determine if miR-6716-5p-caused metastasis depends on NAT10 expression, we overexpressed Flag-NAT10 in miR-6716-5p-expressing cells and determined migration and invasion of CRC cells. We showed that overexpression of miR-6716-5p significantly increased the migration and invasion of HCT116 cells, which was alleviated by overexpression of Flag-NAT10 (Figure 4G, H and I). These results indicated that the miR-6716-5p-induced increase of CRC migration and invasion was dependent on NAT10. These results were further confirmed in LoVo cells (Figure 4J, K and L). Together, these findings demonstrated that miR-6716-5p promoted CRC metastasis by downregulating NAT10 expression.
Figure 4.
miR-6716-5p promoted cell migration and invasion by downregulating NAT10 expression. (A) HCT116 cells were transfected as indicated. The ability of cell migration and invasion were assessed by transwell assay. (B) The relative migration index of HCT116 cells was calculated. (C) The relative invasion index of HCT116 cells was calculated. (D) LoVo cells were transfected as indicated. Then, the ability of cell migration and invasion were assessed by transwell assay. (E) The relative migration index of LoVo cells was calculated. (F) The relative invasion index of LoVo cells was calculated. (G) HCT116 cells were transfected as indicated. The ability of cell migration and invasion were assessed by transwell assay. (H) The relative migration index of HCT116 cells was calculated. (I) The relative invasion index of HCT116 cells was calculated. (J) LoVo cells were transfected as indicated. Then, the ability of cell migration and invasion were assessed by transwell assay. (K) The relative migration index of LoVo cells was calculated. (L) The relative invasion index of LoVo cells was calculated. Error bars represent the SD from three independent experiments. P-values were obtained from Student’s t test. *P< 0.05.
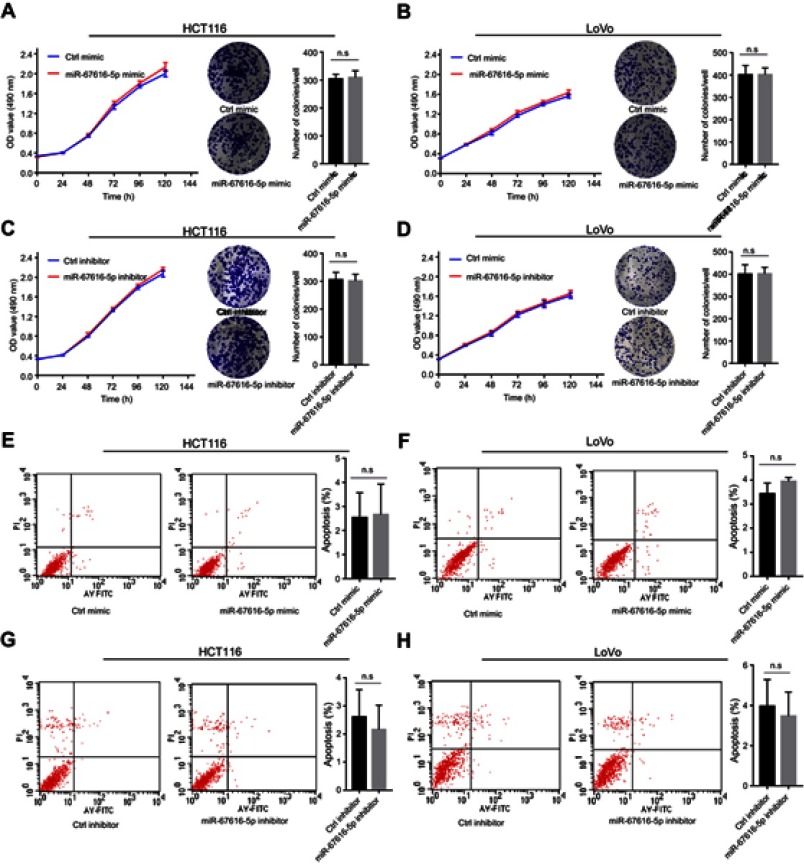
miR-6716-5p had no effect on cell proliferation and apoptosis
It was reported that NAT10 regulated cell proliferation and apoptosis.27,36–38 Thus, we wanted to know whether miR-6716-5p affects cell proliferation and apoptosis. However, overexpression of miR-6716-5p had no influence on the cell proliferation in HCT116 cells (Figure 5A) (P>0.05). Moreover, inhibitor of miR-6716-5p had no effect on cell proliferation (Figure 5C) (P>0.05). The similar results were obtained in LoVo cells (Figure 5B and D) (P>0.05). In addition, the miR-6716-5p mimic and inhibitor had no effect on cell apoptosis in HCT116 cells (Figure 5E and G) (P>0.05). Similar results were observed in LoVo cells (Figure 5F and H) (P>0.05). Thus, miR-6716-5p did not affect cell proliferation and apoptosis in CRC cells.
Figure 5.
miR-6716-5p had no effect on cell proliferation and apoptosis. (A, B) HCT116 or LoVo cells were transfected with either miR-6716-5p mimic or control mimic. Forty-eight hours later, 1,000 cells were seeded into 96-well or 6-well plates and cultured with 10% serum containing media. The ability of cell proliferation ability was assessed by cell viability assay and colony formation assay. (C, D) HCT116 or LoVo cells were transfected with either miR-6716-5p inhibitor or control inhibitor. Forty-eight hours later, 1,000 cells were seeded into 96-well or 6-well plates and cultured with 10% serum containing media. The ability of cell proliferation was assessed by cell viability assay and colony formation assay. (E, F) HCT116 or LoVo cells were transfected with either miR-6716-5p mimic or control mimic. Then, the apoptosis rate of cells was assessed by flow cytometry. (G, H) HCT116 or LoVo cells were transfected with either miR-6716-5p inhibitor or control inhibitor. Then, the apoptosis rate of cells was assessed by flow cytometry. Error bars represent the SD from three independent experiments. P-values were obtained from Student’s t test. n.s. represents no significance.
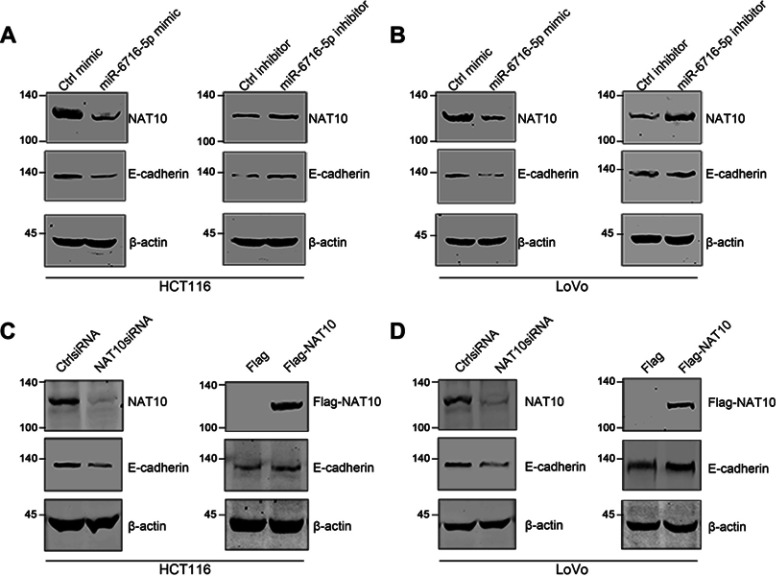
miR-6716-5p might downregulate E-cadherin levels through inhibiting NAT10 expression
It has reported that NAT10 could regulate E-cadherin levels to affect metastasis in HCC and breast cancer (BC) cell lines.37,39 Therefore, we wanted to know if miR-6716-5p regulates E-cadherin levels through inhibiting NAT10 expression. We firstly evaluated E-cadherin levels in HCT116 and LoVo cells when cells were treated with either miR-6716-5p mimic or its inhibitor. Both E-cadherin and NAT10 protein levels were significantly decreased when miR-6716-5p was overexpressed, while the NAT10 and E-cadherin protein levels were increased by the miR-6716-5p inhibitor in HCT116 and LoVo cells (Figure 6A and B). These results suggested that miR-6716-5p may downregulate E-cadherin levels through inhibiting NAT10 expression. To confirm this finding, E-cadherin levels were determined when NAT10 was depleted by siRNA. The depletion of NAT10 by siRNA decreased E-cadherin levels while the ectopic expression of Flag-NAT10 increased E-cadherin levels in CRC cells (Figure 6C and D). Taken together, miR-6716-5p downregulated the E-cadherin level to promote CRC metastasis through downregulating NAT10 expression (Figure 7).
Figure 6.
miR-6716-5p may downregulate E-cadherin levels through inhibiting NAT10 expression. (A) HCT116 cells were transfected with the indicated miRNA mimic or inhibitor. Seventy-two hours later, cells were harvested and lysed. NAT10 and E-cadherin protein levels were detected by western blotting. (B) LoVo cells were transfected as indicated. Then, NAT10 and E-cadherin protein levels were detected by western blotting. (C) HCT116 cells were transfected with siRNAs or plasmid as indicated. Then, cells were harvested and total proteins were extracted. NAT10 and E-cadherin protein expression levels were detected by western blotting. (D) LoVo cells were transfected with siRNAs or plasmid as indicated. Then, NAT10 and E-cadherin protein levels were detected by western blotting.
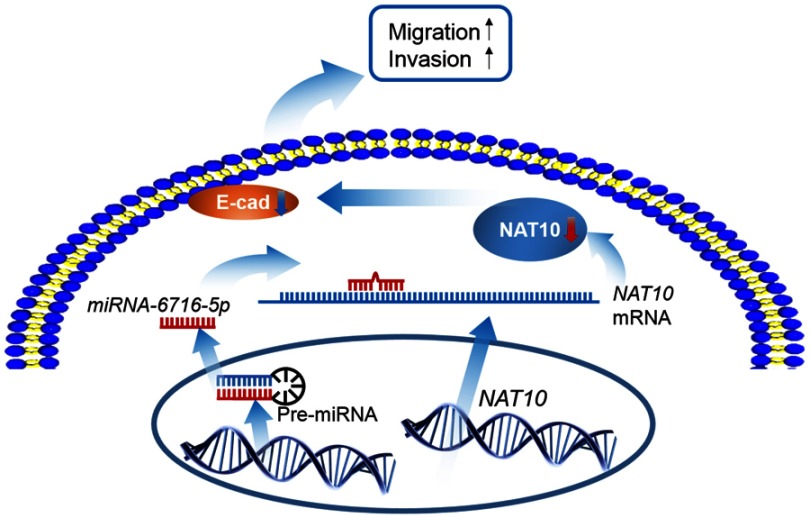
Figure 7.
Working model depicting the role of the miR-6716-5p/NAT10 in CRC progression. MiR-6716-5p downregulates the expression of NAT10, which could downregulate E-cadherin level, and in turn promotes CRC cell migration and invasion.
Discussion
Since NAT10 plays critical roles in tumor development, uncovering the upstream regulators of NAT10 will provide novel insight into the functions of NAT10 in tumor development. In the present study, we identified miR-6716-5p as a critical regulator of NAT10. MiR-6716-5p bound to the 3ʹ-UTR of NAT10 mRNA and inhibited NAT10 expression. Importantly, miR-6716-5p significantly promoted cell migration and invasion in CRC cells and this function was dependent on NAT10. Significantly, high levels of miR-6716-5p in CRLM patients indicated poor OS, suggesting that miR-6716-5p could be a predictive marker for the poor prognosis of CRLM patients.
Tumor metastasis and recurrence are the main causes of poor prognosis in CRC patients.3,40 Therefore, discovering novel regulators of the metastasis or recurrence of CRC would provide a better approach for restraining CRC development. MiRNAs are considered as potential prognostic factors since they play various roles in cellular biological processes by modulating gene expression, and miRNAs are stable and easy to be detected.10 Some miRNAs could be predictive markers for poor prognosis of CRC patients as they act in the metastasis of CRC.41,42 Previous research found that smoking could influence the expression of some mRNAs and miRNAs in CRC patients.43,44 It would make sense to include smoking status into the list of analyzed patient characteristics. However, we have no data about the smoking status of patients in our samples. Thus, miR-6716-5p might be a novel potential clinical biomarker for prognosis of CRC patients.
Previous studies found that the expression stability of some reference genes might be different in different kinds of diseases and under different experimental designs.45–49 Levels of NAT10 and miR-6716-5p were assessed using RT-qPCR with reference gene GAPDH and U6, respectively, which might cause experimental errors induced by reference gene instability. U6 is one of the common reference genes for miRNAs RT-qPCR in CRC research.50,51 Some researchers found that miR-191-5p could be used as reference gene in CRC research48,51 GAPDH and ACTB are commonly used as reference genes for mRNAs RT-qPCR in many research.52,53 To determine the stability of U6 and GAPDH, we have evaluated miR-191-5p and ACTB levels in 18 cases of randomly selected samples. The stabilities of U6, GAPDH, miR-191-5p and ACTB were analyzed using the CT values according to the instruction of the NormFinder and geNorm software.54,55 The results showed that GAPDH and ACTB had the same stability, U6 had the same stability as miR-191-5p (Table S1). In addition, in order to rule out possible experimental errors caused by using U6 and GAPDH as reference genes, we chose miR-191-5p and ACTB as reference genes to analyze the relative levels of miR-6716-5p and NAT10 mRNA, respectively. We showed the tendency of relative miR-6716-5p levels did not change when U6, miR-191-5p, or combination of U6 with miR-191-5p was used as reference gene, respectively (Figure S2A, C and E). Relative NAT10 mRNA levels had similar tendency when GAPDH, ACTB, or combination GAPDH with ACTB was used as reference gene, respectively (Figure S2B, D and F).
It has been reported that high expression of NAT10 correlated with poor prognosis of HCC patients.31,32 Moreover, membranous NAT10 in CRC tissues was associated with a more aggressive CRC phenotype and poor prognosis of CRC patients.30 We demonstrated that simultaneous deletion of residues 68–75 and 989–1018 in NAT10 led to the cytoplasmic and cell membranous localization of NAT10 in HCC cells. We further found that the membranous NAT10 promoted cancer cell invasion and was correlated with poor survival of the HCC patients.56 These results suggested that dysregulation of protein level and cellular location of NAT10 might play critical roles in tumor development and metastasis. Recent research found that NAT10 might act in the tumor metastasis through participating in the epithelial-to-mesenchymal transition process.37,39 It has been reported that inhibition of NAT10 increased E-cadherin expression and repressed cell invasion and migration in HCC and BC cell lines.37,39 In the present study, we found that both miR-6716-5p and the downregulation of NAT10 decreased E-cadherin levels. Consistent with our findings, the database in the GEPIA website (http://gepia.cancer-pku.cn) shows that NAT10 is positively correlated with E-cadherin (P<0.01) in colon adenocarcinoma, stomach adenocarcinoma and pancreas adenocarcinoma (the correlation coefficient was 0.4, 0.4 and 0.32, respectively) (Figure S3A).57 There was no correlation between NAT10 and E-cadherin (P>0.01) in esophageal carcinoma, hepatocellular carcinoma or cholangiocarcinoma (Figure S3B). Thus, the regulation of NAT10-E-cadherin might partially explain why NAT10 functions differently in various tumors. Acetylation of lysine residues in histone H3 is associated with relaxed chromatin structure which facilitates gene expression.58 Liu et al found that treatment with trichostatin A, a histone deacetylase inhibitor, could enhance the acetylation of histone H3K9 and then induce the expression of E-cadherin mRNA in CRC.59 Since NAT10 possesses the HAT activity, we speculate that NAT10 might enhance the acetylation of histones in the promoter region of E-cadherin to activate the transcription of E-cadherin. Chen et al previously reported that there exist three AML1-binding sites, two p300-binding sites and four hepatocyte nuclear factor (HNF3) binding sites in promoter region of E-cadherin.60 They also found that HNF3 could enhance the re-expression of E-cadherin by interacting with p300 and AML1.60 Both NAT10 and p300 belong to the type-A HATs, which might share similar function in transcription regulation.61 However, how NAT10 regulates E-cadherin needs further study in the future.
It has been reported that knockdown of NAT10 mildly promotes cell proliferation in CRC cells27,28 while inhibition of NAT10 suppresses the growth and proliferation of malignant melanoma cells.36 In normal condition, apoptosis was induced by NAT10 suppression in PA-1 cells.38 However, miR-6716-5p had no effect on cell proliferation and apoptosis in CRC cells in the present study. Given that some miRNAs target multiple mRNAs to regulate cellular processes,62 it is likely that miR-6716-5p might target more downstream factors functioning in cell proliferation and apoptosis. The discovery of new miR-6716-5p targets will contribute to completely understanding the functions of miR-6716-5p in cellular processes and tumorigenesis.
A previous study also found that NAT10 was significantly downregulated in CRC tumor samples.27 In this study, we found that decreased level of NAT10 in CRC patients was associated with a poor OS rate (Figure S4A). Consistent with our findings, the Human Protein Atlas website (https://www.proteinatlas.org/) showed that lower NAT10 expression is correlated with a poorer OS rate in rectal cancer patients (Figure S4B).63 These data indicate that NAT10 might act as a prognostic factor in CRC patients.
Conclusion
In the present study, we demonstrated a regulatory mechanism of NAT10 expression by miR-6716-5p in CRC. Disruption of miR-6716-5p-NAT10 regulation may be a potential therapeutic strategy for CRLM patients. Thus, miR-6716-5p inhibitor could be a potential therapeutic drug in the intervention of CRC liver metastasis.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81672735, 81371868 and 81621063), the 973 Program (Grant No. 2013CB837201),Chinese Postdoctoral Science Foundation (2018M641118), and grant from the Innovation Team of Ministry of Education (IRT13001).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 4.Helling TS, Martin M. Cause of death from liver metastases in colorectal cancer. Ann Surg Oncol. 2014;21(2):501–506. doi: 10.1245/s10434-013-3297-7 [DOI] [PubMed] [Google Scholar]
- 5.Hwang M, Jayakrishnan TT, Green DE, et al. Systematic review of outcomes of patients undergoing resection for colorectal liver metastases in the setting of extra hepatic disease. Eur J Cancer (Oxford, England: 1990). 2014;50(10):1747–1757. doi: 10.1016/j.ejca.2014.03.277 [DOI] [PubMed] [Google Scholar]
- 6.Siriwardena AK, Mason JM, Mullamitha S, Hancock HC, Jegatheeswaran S. Management of colorectal cancer presenting with synchronous liver metastases. Nat Rev Clin Oncol. 2014;11(8):446–459. doi: 10.1038/nrclinonc.2014.90 [DOI] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 11.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kida Y, Han YP. MicroRNA expression in colon adenocarcinoma. JAMA. 2008;299(22):2628;author reply 2628–2629. doi: 10.1001/jama.299.22.2628-a [DOI] [PubMed] [Google Scholar]
- 15.Monzo M, Navarro A, Bandres E, et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18(8):823–833. doi: 10.1038/cr.2008.81 [DOI] [PubMed] [Google Scholar]
- 16.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. doi: 10.1136/gut.2008.167817 [DOI] [PubMed] [Google Scholar]
- 17.Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Wang J, Li N, et al. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am J Cancer Res. 2018;8(2):280–290. [PMC free article] [PubMed] [Google Scholar]
- 19.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66(4):654–665. doi: 10.1136/gutjnl-2014-308737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv J, Liu H, Wang Q, Tang Z, Hou L, Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem Biophys Res Commun. 2003;311(2):506–513. [DOI] [PubMed] [Google Scholar]
- 21.Chi YH, Haller K, Peloponese JM Jr., Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem. 2007;282(37):27447–27458. doi: 10.1074/jbc.M703098200 [DOI] [PubMed] [Google Scholar]
- 22.Shen Q, Zheng X, McNutt MA, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315(10):1653–1667. doi: 10.1016/j.yexcr.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 23.Kong R, Zhang L, Hu L, et al. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J Biol Chem. 2011;286(9):7139–7148. doi: 10.1074/jbc.M110.173393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito S, Horikawa S, Suzuki T, et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem. 2014;289(52):35724–35730. doi: 10.1074/jbc.C114.602698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43(4):2242–2258. doi: 10.1093/nar/gkv075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arango D, Sturgill D, Alhusaini N, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018. doi: 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Tan Y, Zhang C, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17(3):349–366. doi: 10.15252/embr.201540505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Cai S, Zhang C, et al. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46(18):9601–9616. doi: 10.1093/nar/gky777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science (New York, NY). 2014;344(6183):527–532. doi: 10.1126/science.1252651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Hou W, Wang HL, et al. GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin Cancer Res. 2014;20(17):4717–4729. doi: 10.1158/1078-0432.CCR-13-3477 [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Liu J, Yan S, Huang K, Bai Y, Zheng S. High expression of N-acetyltransferase 10: a novel independent prognostic marker of worse outcome in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(11):14765–14771. [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Liu X, Jin K, et al. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017;17(1):605. doi: 10.1186/s12885-017-3570-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai S, Liu X, Zhang C, Xing B, Du X. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun. 2017;483(1):624–629. doi: 10.1016/j.bbrc.2016.12.092 [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 35.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–173. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh TI, Lee YM, Lim BO, Lim JH. Inhibition of NAT10 suppresses melanogenesis and melanoma growth by attenuating microphthalmia-associated transcription factor (MITF) expression. Int J Mol Sci. 2017;18:9. doi: 10.3390/ijms18091924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Zhu H, Wu J, Chen W, Guan X. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial-mesenchymal transition in breast cancer. Am J Transl Res. 2018;10(1):256–264. [PMC free article] [PubMed] [Google Scholar]
- 38.Tan TZ, Miow QH, Huang RYJ, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5(7):1051. doi: 10.1002/emmm.201201876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma R, Chen J, Jiang S, Lin S, Zhang X, Liang X. Up regulation of NAT10 promotes metastasis of hepatocellular carcinoma cells through epithelial-to-mesenchymal transition. Am J Transl Res. 2016;8(10):4215–4223. [PMC free article] [PubMed] [Google Scholar]
- 40.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers. 2013;5(2):676–713. doi: 10.3390/cancers5020676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 42.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slattery ML, Pellatt DF, Mullany LE, Wolff RK. Differential gene expression in colon tissue associated with diet, lifestyle, and related oxidative stress. PLoS One. 2015;10(7):e0134406. doi: 10.1371/journal.pone.0134406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullany LE, Herrick JS, Wolff RK, Stevens JR, Slattery ML. Association of cigarette smoking and microRNA expression in rectal cancer: insight into tumor phenotype. Cancer Epidemiol. 2016;45:98–107. doi: 10.1016/j.canep.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caradec J, Sirab N, Keumeugni C, et al. ‘Desperate house genes’: the dramatic example of hypoxia. Br J Cancer. 2010;102(6):1037–1043. doi: 10.1038/sj.bjc.6605573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caradec J, Sirab N, Revaud D, Keumeugni C, Loric S. Is GAPDH a relevant housekeeping gene for normalisation in colorectal cancer experiments? Br J Cancer. 2010;103(9):1475–1476. doi: 10.1038/sj.bjc.6605851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. doi: 10.1186/1471-2407-10-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA (New York, NY). 2008;14(5):844–852. doi: 10.1261/rna.939908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan-Gunn M, Hinch E, Vaughan V, Lewandowski P. Choosing a stable housekeeping gene and protein is essential in generating valid gene and protein expression results. Br J Cancer. 2011;104(6):1055;author reply 1056. doi: 10.1038/bjc.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarzenbach H, Da Silva AM, Calin G, Pantel K. Data normalization strategies for MicroRNA quantification. Clin Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng G, Wang H, Zhang X, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;8(12):e83025. doi: 10.1371/journal.pone.0083025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H, Ma TF, Lin J, et al. Identification of valid reference genes for mRNA and microRNA normalisation in prostate cancer cell lines. Sci Rep. 2018;8(1):1949. doi: 10.1038/s41598-018-19458-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman JR, Waldenstrom J. With reference to reference genes: a systematic review of endogenous controls in gene expression studies. PLoS One. 2015;10(11):e0141853. doi: 10.1371/journal.pone.0141853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):Research0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 56.Tan Y, Zheng J, Liu X, et al. Loss of nucleolar localization of NAT10 promotes cell migration and invasion in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;499(4):1032–1038. doi: 10.1016/j.bbrc.2018.04.047 [DOI] [PubMed] [Google Scholar]
- 57.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407. doi: 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Hong Y, Zhao Y, Ismail TM, Wong Y, Eu KW. Histone H3 (lys-9) deacetylation is associated with transcriptional silencing of E-cadherin in colorectal cancer cell lines. Cancer Invest. 2008;26(6):575–582. doi: 10.1080/07357900701837168 [DOI] [PubMed] [Google Scholar]
- 60.Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005;24(56):8277–8290. doi: 10.1038/sj.onc.1208991 [DOI] [PubMed] [Google Scholar]
- 61.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381. doi: 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genetics. 2014;15(9):599–612. doi: 10.1038/nrg3765 [DOI] [PubMed] [Google Scholar]
- 63.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science (New York, NY). 2017;357:6352. doi: 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]