Summary
The ability of Pseudomonas species to thrive in all major natural environments (i.e. terrestrial, freshwater and marine) is based on its exceptional capability to adapt to physicochemical changes. Thus, environmental bacteria have to tightly control the maintenance of numerous physiological traits across different conditions. The intracellular pH (pH i) homoeostasis is a particularly important feature, since the pH i influences a large portion of the biochemical processes in the cell. Despite its importance, relatively few reliable, easy‐to‐implement tools have been designed for quantifying in vivo pH i changes in Gram‐negative bacteria with minimal manipulations. Here we describe a convenient, non‐invasive protocol for the quantification of the pH i in bacteria, which is based on the ratiometric fluorescent indicator protein PHP (pH indicator for Pseudomonas). The DNA sequence encoding PHP was thoroughly adapted to guarantee optimal transcription and translation of the indicator in Pseudomonas species. Our PHP‐based quantification method demonstrated that pH i is tightly regulated over a narrow range of pH values not only in Pseudomonas, but also in other Gram‐negative bacterial species such as Escherichia coli. The maintenance of the cytoplasmic pH homoeostasis in vivo could also be observed upon internal (e.g. redirection of glucose consumption pathways in P. putida) and external (e.g. antibiotic exposure in P. aeruginosa) perturbations, and the PHP indicator was also used to follow dynamic changes in the pH i upon external pH shifts. In summary, our work describes a reliable method for measuring pH i in Pseudomonas, allowing for the detailed investigation of bacterial pH i homoeostasis and its regulation.
Introduction
Microbial cells have a remarkable ability to maintain physiological homoeostasis under a wide variety of environmental conditions. Thereby, the tight control of the intracellular pH (pHi, i.e. cytoplasmic pH) is of particular importance (Slonczewski et al., 2009), because pHi influences enzyme activity, protein stability, nucleic acid structure and many other biological processes (Booth, 1985). It is well known that few species of adapted microbes can grow under extreme pH conditions. Bacterial life is possible even at pH values close to 0 (Nordstrom et al., 2000) or above pH = 13 (Roadcap et al., 2006). Nonetheless, the cytoplasmic pH value is remarkably similar for most bacteria, even when exposed to extreme pH environments (Krulwich et al., 2011; Martínez et al., 2012). Generally, bacteria have evolved a series of mechanisms to maintain this tight regulation of pHi homoeostasis. A major strategy to this effect is to regulate the activity of proton (H+) transporters, such as H+‐pumping respiratory chain complexes and H+‐coupled ATPases or cation/H+ antiporters (Krulwich et al., 2011). As H+ ions are transported either way across the cell membrane by these mechanisms, major changes in the transmembrane pH gradient (ΔpH) are also generated. Since both ΔpH and the transmembrane electrical potential (ΔΨ) are directly connected to the H+‐motive force (PMF; given that PMF = ΔΨ – 60 × ΔpH at 25°C), the demand of pH homoeostasis directly influences the overall energetic status of the cell (Deamer and Nichols, 1989).
Several methods have been developed to monitor the pHi in bacteria (Slonczewski et al., 2009; Chen and Lo, 2016). Many of them rely on the measurement of in vivo 31P nuclear magnetic resonance (NMR) spectra (Roberts et al., 1981; Sijbesma et al., 1996) or on the ratiometric fluorescence of specific chemical dyes (Molenaar et al., 1991; Breeuwer et al., 1996; Riondet et al., 1997; Awaji et al., 2001; Hunter and Beveridge, 2005; Rupprecht et al., 2017). While NMR is a laborious and not widely accessible technique, some of the fluorescent chemical probes commercially available are usually expensive, short‐lived and their use for pHi determination requires a number of time‐consuming manipulations. In addition to these drawbacks, some fluorescein‐based fluorophores can rapidly leak out of cells once they are liberated in the cytosol via intracellular hydrolysis (Han and Burgess, 2010). Other chemical dyes also pose problems due to their (i) low permeability (which in turn result in uneven loading of dyes within a cell population), (ii) high chemical instability and (iii) cytotoxicity. Derivatives of fluorescent proteins that respond to changes in pH, including the model green fluorescent protein (GFP), have been described in the past decades (Olsen et al., 2002; Mahon, 2011; Martínez et al., 2012; Rupprecht et al., 2017; Reifenrath and Boles, 2018). The expression of genes encoding fluorescent proteins and the detection of the resulting fluorescence signals are an inexpensive and fast technique, enabling online measurements in vivo, requiring considerably less technical effort than other techniques deployed for pHi determination. The dependence of the fluorescence intensity on the pHi in GFP derivatives is based on the direct protonation of the phenolate moiety of the fluorophore derived from tryptophan, an extremely fast exchange reaction (McAnaney et al., 2005). Thus, fluorescence determinations allow for the assessment of the rate of pHi change in response to rapid shifts in extracellular pH. Additionally, the continuous expression of the protein allows for live‐imaging over a longer timescale than is possible using chemical fluorophores (Grillo‐Hill et al., 2014).
Given the importance of assessing cytoplasmic pH values to understand the principles governing metabolic homoeostasis and energy conservation mechanisms in bacteria, the improvement of non‐invasive methods for calculating the pHi is needed. In the present work, we propose a standard, non‐invasive method for the determination of pHi in Pseudomonas species, as well as other Gram‐negative bacteria such as Escherichia coli. This procedure is based on the enhanced expression of a pH‐sensitive variant of GFP, and the ratiometric assessment of pHi against cells equilibrated to a range of physiologically relevant pH values. The usability of this novel tool for the quantification of pHi in Gram‐negative bacteria is illustrated by investigating the changes in pH homoeostasis brought about by internal metabolic perturbations in the cell factory platform Pseudomonas putida KT2440 and by antibiotic treatment in the opportunistic pathogen P. aeruginosa. The PHP indicator was also used to explore the response of pHi in E. coli upon several perturbations. Moreover, the intrinsic compatibility of our tool with plasmids following the Standard European Vector Architecture (SEVA) (Silva‐Rocha et al., 2013) allows to modulate the replication and expression of PHP in other bacteria. As indicated by the examples below, the PHP indicator constitutes a flexible, non‐invasive and easy‐to‐implement tool to study both endogenous and exogenous pH perturbations in bacteria.
Results and discussion
Construction, benchmarking and calibration of the PHP indicator as a reliable tool to calculate the cytoplasmic pH in Pseudomonas
Our earlier attempts to use previously characterized ratiometric fluorescent proteins as pHi indicators in Pseudomonas species (and, in particular, P. putida and P. aeruginosa) did not yield satisfactory signal‐to‐noise ratios. We attributed this lack of reliable fluorescent signal to two main reasons: (i) deficient (and potentially stochastic) expression of the gene(s) encoding the pH indicator(s) and (ii) some of the currently available ratiometric fluorescent protein indicators have been tailored for eukaryotic systems (Tournu et al., 2017; Reifenrath and Boles, 2018). In order to overcome these issues, we constructed a broad‐host‐range vector for the calibrated expression of a variant of the pH‐sensitive pHluorin2 indicator protein (Mahon, 2011). pHluorin2 is a derivative of GFP that carries 11 point substitutions in its amino acid sequence, including the F64L mutation, known to enhance protein folding (Heim et al., 1995; Mahon, 2011). The sequence encoding pHluorin2 was codon‐optimized for Pseudomonas species, and a synthetic ribosome binding site was added to ensure sufficient expression, as indicated in Experimental Procedures (see also Sequence S1 in the Supporting Information). The synthetic module, which we termed PHP (pH indicator for Pseudomonas), was placed under the transcriptional control of the constitutive EM7 promoter (P EM7) in the pSEVA2513 vector [kanamycin (Km)‐resistant, oriV(RSF1010)]. The resulting plasmid, termed pS2513·PHP, ensures the constitutive production of the PHP fluorescent protein in the bacterial cytosol regardless of the growth stage of the cells.
We first set to compare the performance of the original pHluorin2 protein and the PHP indicator in P. putida KT2440 by comparing the fluorescence of the two proteins in individual bacterial cells by flow cytometry (Fig. 1). To this end, two identical, pSEVA2513‐based plasmid systems were used, in which the expression of either pHluorin2 or PHP is driven by the same constitutive promoter under an equivalent vector copy number (Fig. 1A). These plasmids were individually transformed into P. putida KT2440, and the fluorescence stemming from either pHluorin2 or PHP was measured in exponentially growing cultures in LB medium by means of flow cytometry (Fig. 1B). Both the PHP and pHluorin2 fluorescence could be easily detected with the instrument settings used for GFP and related fluorescent proteins. As indicated by the histogram plots of cell count versus level of fluorescence of the indicators, cells expressing pHluorin2 exhibited a bimodal distribution of fluorescence, with approximately 21% of the cell population displaying fluorescence levels similar to that of a negative control (i.e. P. putida KT2440 transformed with an empty pSEVA2513 vector). Cultures of strain KT2440 transformed with plasmid pS2513·PHP, in contrast, had a clearly unimodal fluorescence distribution, with > 99% of the bacterial population testing positive for the PHP signal. Moreover, the geometric mean value of fluorescence was considerably higher for PHP (fluorescence x mean = 42.5) than for the original pHluorin2 indicator (fluorescence x mean = 22.3). Next, we investigated the signal‐to‐noise ratio, a parameter reflecting the efficiency of fluorescence detection (Giesecke et al., 2017), for the two indicator proteins. The signal‐to‐noise ratio was 40% higher for the PHP indicator than for pHluorin2 (Fig. 1C) – an advantageous property that addresses one of the main problems previously experienced with fluorescent protein‐based pH indicators in Pseudomonas species. The coefficients of variation, a measure of the height versus width of fluorescence histograms and therefore an indication of the dispersion of the signal (Sharrow, 2001), were calculated for the two pH indicators according to the distribution of positive (i.e. pHluorin2+ or PHP+) cells of Fig. 1A. Again, the PHP protein outperformed pHluorin2, with a coefficient of variation which was approximately half of the original pHluorin2 indicator (Fig. 1C). Taken together, these results accredit the value of PHP as a uniformly accumulated fluorescent protein in Pseudomonas, with fluorescence characteristics similar to both GFP and pHluorin2.
Figure 1.
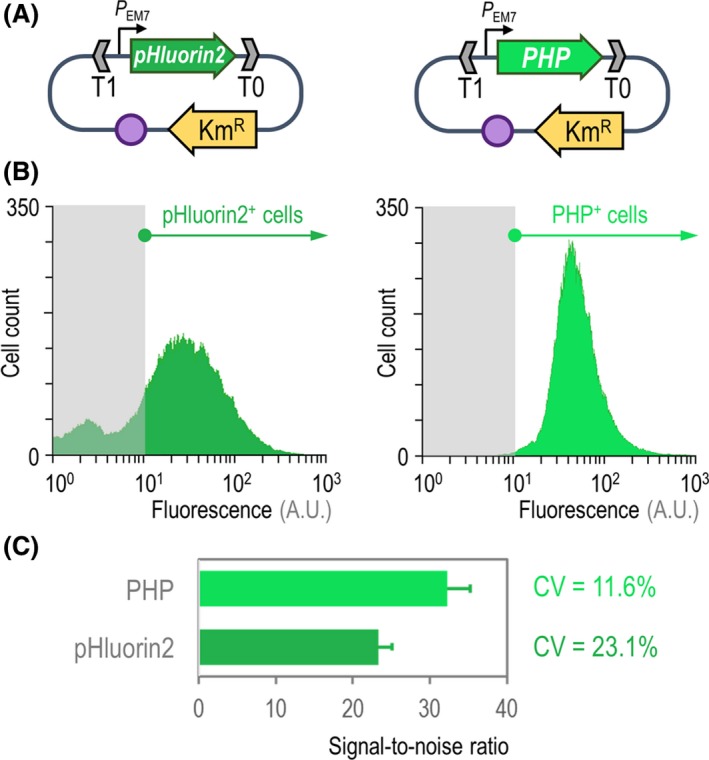
Benchmarking of the PHP indicator for the assessment of intracellular pH.
A. Two separate expression plasmids were constructed for the calibrated expression of the synthetic genes encoding the pHluorin2 or PHP indicators. In both cases, the expression of the genes is driven by the constitutive PEM 7 promoter, and the backbone used for these constructs (vector pSEVA2513) carries the oriV(RSF1010) origin of replication, indicated in the scheme as a purple circle. The kanamycin‐resistance (KmR) determinant and the T1 and T0 transcriptional terminators are likewise highlighted in the plasmid maps.
B. Flow cytometry exploration of the pHluorin2 and PHP indicators. Plasmid pS2513·pHluorin2 or pS2513·PHP was transformed in P. putida KT2440, and the cells were grown in LB medium until the mid‐exponential phase of growth. Cells were processed as indicated in Experimental Procedures , and the histograms show the distribution of fluorescence (in arbitrary units, A.U.) stemming from pHluorin2 or PHP (the bacterial populations positive for the indicators are indicated in each case). The light grey rectangle in each plot identifies the region considered negative for the fluorescence signal, which was assessed with P. putida carrying the empty pSEVA2513 vector and grown under the same conditions.
C. Key performance parameters of the pH indicators. The signal‐to‐noise ratio was calculated from flow cytometry measurements of P. putida KT2440 carrying either plasmid pS2513·pHluorin2 or pS2513·PHP as indicated above. The bars represent the signal‐to‐noise ratio for each sensor and the 95% confidence interval calculated from four independent experiments. The coefficient of variation (CV) of the fluorescence signal was also calculated from these measurements.
Next, we experimentally verified the PHP expression by means of fluorescence microscopy of exponentially growing E. coli DH5α, P. putida KT2440 and P. aeruginosa PA14 carrying plasmid pS2513·PHP in LB medium. As observed in Fig. 2A, a homogeneous distribution of fluorescence was spotted across the individual cells of the three bacterial species examined, both during exponential growth and during the stationary phase – consistent with the results of flow cytometry experiments (Fig. 1). Subsequent tests in LB medium and M9 minimal medium indicated that the presence of plasmid pS2513·PHP did not affect the growth of E. coli or the two Pseudomonas species tested, neither in terms of specific growth rates nor the final cell density achieved in these cultures (data not shown). Moreover, flow cytometry analysis of the corresponding bacterial populations showed that the fluorescence signal followed a unimodal distribution in all three species both in complex and minimal culture media supplemented with glucose as the sole carbon source – indicating that the PHP protein can be used for studying pHi across different culture conditions. The next question is how does the fluorescence stemming from PHP correlates with changes in pHi, as indicated below.
Figure 2.
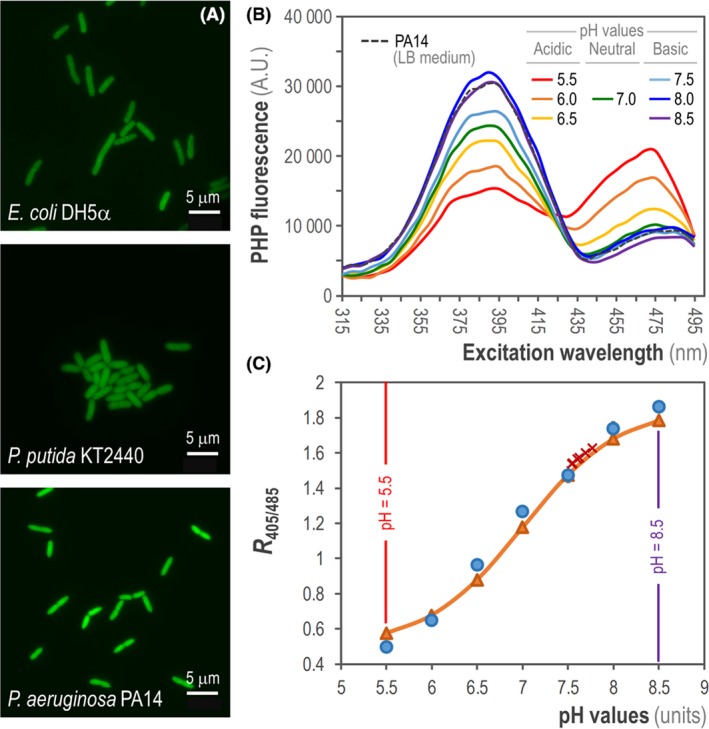
Assessment of intracellular pH (pH i) in different Gram‐negative bacteria.
A. Fluorescent microscopy from exponentially growing cultures of E. coli DH5α, P. putida KT2440 and P. aeruginosa PA14 transformed with plasmid pS2513·PHP and grown in LB medium. Scale bars are indicated in each case.
B. Fluorescence excitation spectrum of the PHP indicator in P. aeruginosa PA14/pS2513·PHP cultured in LB medium. The intracellular pH (pH i) of the cells was collapsed to the extracellular pH as outlined in Experimental Procedures . The mean values obtained from three independent experiments are shown. A.U., arbitrary units.
C. Calibration curve of the PHP indicator in P. aeruginosa PA14/pS2513·PHP cells cultured in LB medium supplemented with 20 μM carbonyl cyanide m‐chlorophenyl hydrazine (CCCP). The data for the calibration curve were obtained as the ratio between the excitation peaks at λexcitation = 405 nm and 485 nm (R 405/485) plotted against the pH values of equilibrated cells (blue circles). These values were fitted using the Boltzmann sigmoid best‐fitting curve to obtain the calibration curve (orange triangles), which was used to calculate the pH i values in P. aeruginosa PA14 during exponential growth (red crosses). The lowest and highest pH values tested (pH = 5.5 and 8.5) are highlighted. Data points are representative of independent triplicates, and the red crosses (corresponding to six individual determinations) are shown in the curve to indicate the narrow dispersion of experimentally calculated pH i values.
All ratiometric derivatives of GFP (e.g. pHluorin and pHluorin2) rely on the bimodality shown in their excitation spectra (Llopis et al., 1998; Esposito et al., 2008), which exhibit two distinct maxima at an excitation wavelength (λexcitation) ~ 395 nm and λexcitation ~ 475 nm (Miesenböck et al., 1998; Mahon, 2011). We first aimed at scanning the excitation spectrum of the engineered PHP protein in P. aeruginosa PA14 cells harbouring plasmid pS2513·PHP. To this end, we recorded the culture's fluorescence emission at an emission wavelength (λemission) = 515 nm in a standard microtiter plate fluorimeter. In order to check the effect of pH on the two excitation peaks of the PHP indicator, cells (previously grown in LB medium) were pelleted and resuspended in M9 minimal medium buffered at different physiologically relevant pH values (ranging from pH = 5.5 to pH = 8.5; see Experimental Procedures for details) in which the pHi was collapsed to equalize the extracellular pH. This effect was achieved by the addition of benzoate (a permeant acid) and methylamine (a permeant base), and this pH collapse was experimentally verified by using the pH‐responsive, ratiometric chemical fluorophore 5(6)‐carboxyfluorescein diacetate succinimidyl ester (data not shown). We observed that upon acidification, the excitation peak of PHP at λexcitation ~ 395 nm decreased with the corresponding increase in the peak at λexcitation ~ 475 nm (Fig. 2B). Conversely, the excitation peak at λexcitation ~ 395 nm increased when pHi alkalized, while the intensity of the peak at λexcitation ~ 475 nm concomitantly decreased. Almost identical spectra were obtained in PHP‐expressing E. coli DH5α and P. putida KT2440 (data not shown), confirming the expected PHP's sensitivity to changes in pH in the range of physiological pH values in different bacterial species. Although the bimodal pattern in both excitation peaks of PHP was essentially the same across the different Gram‐negative bacteria tested herein, we observed that the λexcitation values of 405 nm and 485 nm reported by Martínez et al. (2012) gave rise to the same results regarding calculated pHi values. We thus adopted these experimentally determined λexcitation values (405 and 485 nm) for all subsequent experiments. In summary, our results thus far indicate that the spectral properties of the PHP fluorescent protein are very similar to those of pHluorin2, the parental ratiometric protein from which it derives (Mahon, 2011), across different bacterial species.
The next step was to use this dual excitation attribute of PHP to obtain a calibration curve by plotting the ratio between the fluorescence peaks at λexcitation = 405 nm and λexcitation = 485 nm (R 405/485) against the pH values of equilibrated cells in the set of pH‐modified buffers described above. At first, we harvested exponentially growing P. aeruginosa PA14/pS2513·PHP cells incubated in LB medium. Once again, the pHi of the cells was rapidly collapsed to equalize the pH of the different buffered solutions as detailed above. This allowed us to calculate highly reproducible sigmoid‐shape calibration curves using the Boltzmann best‐fitting equation as indicated in Experimental Procedures (Fig. 2C). The same procedure was applied to obtain calibration curves in cells sampled from exponentially growing cultures of E. coli DH5α/pS2513·PHP and P. putida KT2440/pS2513·PHP (Fig. S1A and S1B in the Supporting Information respectively), and of P. aeruginosa CH2682/pS2513·PHP (data not shown) in LB medium.
We observed some inconsistencies in the R 405/485 values when the pHi of Pseudomonas cells was collapsed in buffers with pH > 7.5. The differences in pH sensitivity of GFP‐borne indicators under alkaline conditions have been reported to be caused by the accessibility of chromophores to H+ ions in the solvent (Benčina, 2013). In order to overcome this problem, we set to supplement all Pseudomonas cultures with a sublethal amount of the hydrazone protonophore carbonyl cyanide m‐chlorophenyl hydrazine (CCCP). At low concentrations, this compound is known to translocate H+ ions across the cell wall, thereby dissipating the H+ gradient (i.e. ΔpH) across the cytoplasmic membrane without significantly compromising viability (Ghoul et al., 1989; Clark et al., 2015). Note that CCCP addition was necessary for Pseudomonas cultures as these species are known to display a very limited membrane permeability (see, for instance, the section below on antibiotic treatment). Protonophores (e.g. CCCP) increase membrane permeability by altering the electrochemical gradient, thus establishing an equilibrium across the cell membrane (Yu et al., 2015).
The addition of CCCP at 20 μM to Pseudomonas cultures tackled the problem observed during incubation of the cells in alkaline‐buffered solutions, and the presence of this protonophore did not impact cell growth across all the conditions tested herein, both in complex and minimal culture media. By applying this strategy, we could obtain highly reproducible sigmoid‐shape calibration curves for all Pseudomonas species harbouring plasmid pS2513·PHP, as shown for P. aeruginosa PA14 (Fig. 2C) and P. putida KT2440 (Fig. S1B in the Supporting Information). Moreover, addition of CCCP to E. coli cultures did not affect the calibration curves obtained for this species (data not shown), indicating that this strategy can be used in other Gram‐negative bacteria to collapse the pHi. With these tools and procedures at hand, we set to calculate pHi in E. coli and in all Pseudomonas species under different culture conditions as indicated below.
Physiological cytoplasmic pH values are conserved across Gram‐negative bacterial species
The calibration curves constructed for wild‐type P. aeruginosa strains (both PA14 and CH2682), P. putida KT2440 and E. coli DH5α were used to calculate the pHi in actively growing cells cultured in LB medium (Table 1). Cell pellets of the four species, promptly harvested from the corresponding cultures, were resuspended in M9 medium salts lacking any permeant acid or base. Under these conditions, cells were able to maintain their pHi homoeostasis. We observed that the pHi values calculated for E. coli DH5α, P. aeruginosa PA14 and P. putida KT2440 were highly similar across all strains tested – with an average value of pHi = 7.65 ± 0.14, close to neutrality. Furthermore, the pHi values obtained correspond to those previously reported for neutrophilic bacteria using other detection methods, including fluorescent pH indicator proteins (Olsen et al., 2002; Sezonov et al., 2007; Krulwich et al., 2011; Martínez et al., 2012; Gao et al., 2016). A slightly higher pHi value was observed in the CH2682 clinical isolate of P. aeruginosa (the cytoplasmic pH of this strain was consistently > 0.2 units above the values observed in the prototypical laboratory strain PA14). Conversely, the lowest pHi value was observed for E. coli DH5α cells. The external pH values determined in the culture supernatants indicated that cells were able to maintain stable pHi values while the external pH suffered changes (Table 1), e.g. due to the secretion of metabolic by‐products by the cells. A case in point is that of E. coli, for which the pHi was approximately 1.5 units above the external pH. E. coli is known to secrete acetate and other low‐molecular‐weight organic acids to the culture medium (Nikel et al., 2008; Bernal et al., 2016), which is probably the reason explaining the acidification of the extracellular medium in these cultures. All the cultures of Pseudomonas species, in contrast, had pH values close to neutrality – displaying differences between pHi and extracellular pH smaller than in E. coli.
Table 1.
Intracellular pH measurements of selected Gram‐negative bacteria growing in LB medium
| Bacterial strain | Relevant characteristics | Specific growth rate (h−1) | Cytoplasmic pH | Extracellular pH |
|---|---|---|---|---|
| E. coli DH5α | Laboratory strain typically used for cloning | 1.46 ± 0.05 | 7.54 ± 0.11 | 6.05 ± 0.02 |
| P. putida KT2440 | Wild‐type strain, platform bacterial host | 1.89 ± 0.06 | 7.77 ± 0.11 | 7.51 ± 0.01 |
| P. aeruginosa PA14 | Wild‐type strain, wide host virulence spectrum | 0.92 ± 0.02 | 7.63 ± 0.08 | 7.12 ± 0.02 |
| P. aeruginosa CH2682 | Wild‐type strain, clinical isolate | 0.57 ± 0.03 | 7.87 ± 0.07 | 7.54 ± 0.02 |
All strains, carrying plasmid pS2513·PHP, were grown in LB medium, and the cytoplasmic pH was determined during exponential growth. The pH of the LB medium was determined to be 7.05 ± 0.02 just prior to inoculation of the cultures. These results represent the mean value of each parameter ± standard deviation of triplicate measurements from at least three independent experiments.
In order to validate these results against a different protocol for determination of pHi, we calculated the pH values of both P. aeruginosa PA14 and P. putida KT2440 by applying the well‐established method of staining the cells with the pH‐responsive fluorophore 5(6)‐carboxyfluorescein diacetate succinimidyl ester (Riondet et al., 1997). Exponentially growing cells of each species cultured in LB medium were harvested and treated with the chemical fluorophore as indicated in Experimental Procedures. The pHi values calculated with this method (pHi P. aeruginosa PA14 = 7.78 ± 0.19 and pHi P. putida KT2440 = 7.59 ± 0.12) had a variation of < 3% with respect to those obtained via the PHP indicator – yet, the dispersion of experimental pH determinations across replicates of the same experiment was, on average, higher for the cells treated with 5(6)‐carboxyfluorescein diacetate succinimidyl ester than for the cells carrying pS2513·PHP. Taken together, these results demonstrate that PHP is a reliable indicator of pHi in Pseudomonas species, indicating that cells tightly maintain a pHi when cultured in favourable conditions (i.e. fast growth in a rich culture medium), and the next question was to evaluate how they respond to internal and external metabolic perturbations in terms of pH homoeostasis.
The cytoplasmic pH of P. putida fluctuates depending on the glucose consumption routes
In the model platform bacterium P. putida KT2440 (Benedetti et al., 2016; Nikel and de Lorenzo, 2018; Calero and Nikel, 2019), the initial steps for the catabolism of glucose occur simultaneously through the phosphorylation of the sugar to glucose‐6‐P by the enzyme glucokinase (Glk), or by its oxidation to gluconate by means of glucose 2‐dehydrogenase (Gcd) (del Castillo et al., 2007; Nikel and de Lorenzo, 2014; Nikel et al., 2015; Nikel and Chavarría, 2016). The oxidation of glucose in the periplasm yields low‐molecular‐weight organic acids, e.g. gluconate and 2‐ketogluconate (Hirshfield et al., 2003; Sánchez‐Pascuala et al., 2019). Hence, we set to study how the use of this peripheral pathway influences the pHi in strain KT2440 and its glycolytic mutant derivatives. We transformed the pS2513·PHP plasmid into two P. putida mutants, each devoid of one of the two aforementioned enzymes, and calculated the pHi in exponential and stationary phase cultures grown in M9 minimal medium containing glucose as the only carbon source (i.e. glycolytic conditions). In a Δglk mutant of strain KT2440, the bulk of hexoses (80–90%) is oxidized into gluconate, whereas in a Δgcd mutant, sugars are exclusively phosphorylated to glucose‐6‐P (Dvořák et al., 2017; Sánchez‐Pascuala et al., 2017, 2019). Once again, cultures of P. putida KT2440 grown in the aforementioned medium supplemented with CCCP were used to calculate the calibration curve for pHi quantification in all strains (Fig. S2 in the Supporting Information).
As indicated in Fig. 3, cytoplasmic pH values calculated for exponentially growing P. putida KT2440 in M9 minimal medium with glucose (pH = 7.66 ± 0.09) were similar to the values calculated when the same bacteria were grown in LB medium (Table 1). Additionally, the cytosolic pH of wild‐type cells grown in the presence of CCCP showed a drop in pHi, decreasing to levels approaching the extracellular culture medium (i.e. around neutrality). This pHi range was observed to be maintained around the same values along the growth curve, even after the cultures reached the stationary phase (after 24 h of cultivation; see Fig. 3). The PHP indicator thus captured the increase in pHi as cells transitioned from exponential growth into the stationary phase, a well‐described phenomenon in Gram‐negative bacteria (Lambert et al., 1997; Kurbatov et al., 2006; Reva et al., 2006).
Figure 3.
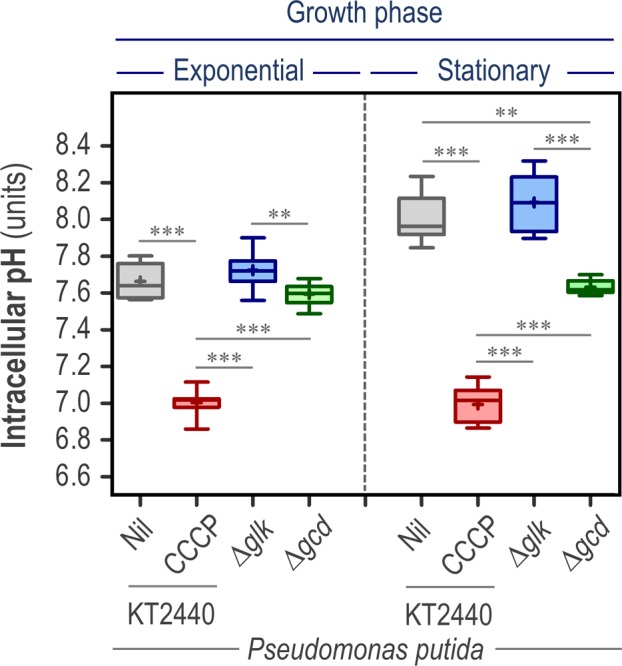
The cytoplasmic pH of P. putida is altered by the pathway used for glucose consumption. Wild‐type P. putida KT2440 and its isogenic mutants in the enzymes glucokinase (Δglk) and glucose 2‐dehydrogenase (Δglk) were transformed with plasmid pS2513·PHP and grown in M9 minimal medium supplemented with glucose at 20 mM as the sole carbon source. The intracellular pH (pH i) was investigated during the exponential and stationary (24 h post‐inoculation) phase of growth. The pH i of wild‐type cells cultured in the presence of 20 μM carbonyl cyanide m‐chlorophenyl hydrazine (CCCP) is also shown in the figure. Independent triplicates of the experiments were carried out, and the level of statistical significance in the pairwise comparisons indicated in the comparisons across experimental conditions is indicated as ** P < 0.01 and *** P < 0.001 (ANOVA).
Interestingly, we observed significant differences in the cytoplasmic pH between the Δglk and Δgcd derivatives of P. putida (Fig. 3). The Δglk strain had a slightly higher (albeit statistically significant) pHi than the Δgcd mutant during exponential growth; this difference became more evident during stationary phase (e.g. pHi = 8.11 for the Δglk strain and pHi = 7.61 for the Δgcd strain after 24 h of cultivation). Forcing the carbon flow through oxidation (in the Glk‐deficient strain) leads to the generation of gluconate (and 2‐ketogluconate, the further oxidized product of this metabolic branch of hexose processing). Gram‐negative bacteria, including Pseudomonas species, are known to respond to slight acidic stress by increasing their pHi (Krulwich et al., 2011), and this general response could partially account for the results. In addition, the ΔpH (and PMF), which is another determining factor of the differences in pHi across the strains tested (Casey et al., 2010), is likewise expected to be affected in these mutants. In turn, the differences in ΔpH result in an alteration of the energetic status of the cells (Fuhrer et al., 2005; Voskuil et al., 2018). Interestingly, the Gcd‐deficient P. putida strain was able to stably maintain pHi values around neutrality during the whole cultivation period. Experimental determination of gluconate and 2‐ketogluconate confirmed that there were no detectable acidic by‐products secreted into culture supernatants, while cultures of both the wild‐type strain and its Δglk derivative reached gluconate and 2‐ketogluconate concentrations of approximately 5 and 1.5 mM and 7.8 and 2.9 mM respectively. This phenomenon could account for the stable pHi values observed in the Δgcd mutant, which would not be subjected to acidic stress caused by the secretion of metabolic by‐products from glucose oxidation. The extracellular pH was assessed in all these cultures (Table S1 in the Supporting Information), providing further support to this notion: cultures of both the wild‐type strain and the Δglk derivative had an acidic external milieu (in particular, for the later strain), whereas the extracellular pH in cultures of the Gcd‐deficient variant was maintained around neutrality values. When cultures of all the three strains were grown in M9 minimal medium containing succinate as the sole carbon source (i.e. under gluconeogenic conditions), no major differences were observed in pHi, neither during exponential growth nor during the stationary phase (in both cases, the average pHi = 7.35 ± 0.18 for all the strains tested) – thus indicating that the differences observed in pHi values in the glucose cultures could be attributed to the use of different metabolic routes deployed for sugar utilization. While endogenous (i.e. metabolic) perturbations have been shown to exert a strong effect on pH homoeostasis (and hence, in the pHi values) in these Pseudomonas strains, we next explored how these parameters are affected by external perturbations, such as a sudden pH shift and treatment with antimicrobial agents, as indicated in the sections below.
The PHP indicator can be used to track time‐dependent changes in cytoplasmic pH upon a shift in extracellular pH
We investigated if the PHP indicator could reflect adaptive changes in pHi upon a sudden shift in extracellular pH values, both acidic and alkaline. To this end, both wild‐type E. coli MG1655 and P. putida KT2440 harbouring plasmid pS2513·PHP were grown in M9 minimal medium containing glucose, and the cell suspensions were challenged by exposure to a buffer at pH = 5.5 or pH = 8.5 (see Experimental Procedures for details). The pHi values were calculated from fluorescence readings taken every 10 sec over 5 min to capture the short‐term dynamics of cytoplasmic pH homoeostasis upon the acidic or alkaline challenge (Fig. 4). Control experiments were run for both bacterial species by adding benzoic acid to the cell suspension in order to collapse pHi – an operation which, as expected, resulted in an almost immediate drop in pHi to match the pH value of the external medium (i.e. pH = 6.95 ± 0.05). The pHi values recorded for E. coli and P. putida did not recover after exposure to sodium benzoate (nor did they recover during 1 h post‐treatment, data not shown). In experiments without any perturbation in extracellular pH, pHi values were stably maintained during the assay within the values previously observed for these bacteria (i.e. in the range of 7.5–7.8 units; see also Table 1). When E. coli was challenged with a buffer at pH = 5.5, the cells did not react with a sudden drop in pHi, in contrast with the response to a pH = 8.5 shift, which entailed a sudden increased in pHi, followed by a slow, steady decrease of the pHi to the values pre‐shift (Fig. 4A). From 9 min onwards, the pHi of base‐challenged cells matched that of untreated control cultures (data not shown). Overall, these observations are similar to the results previously reported by Padan et al. (2005) and Šeputienė et al. (2006), indicating that the cytoplasmic pH of E. coli K‐12 is more sensitive to alkaline challenges than to acidification of the surrounding culture medium. The pattern of pHi values in P. putida KT2440 upon pH shifts was much more rigid in comparison (Fig. 4B). Exposure of the bacterial cells to pH = 5.5 or pH = 8.5 did not result in any major change in pHi over the whole experimental period, and the fluctuations observed in PHP fluorescence (hence, in pHi) were within the range of the experimental error for these determinations. Taken together, the results of these experiments reflect the increased membrane permeability of E. coli as compared to that in P. putida. In fact, the outer membrane of several Pseudomonas species is known to exhibit from 100‐ to 400‐fold lower permeability to hydrophilic compounds than that of E. coli (Yoshimura and Nikaido, 1982; Llamas et al., 2000; Sohlenkamp, 2017). Our results are also in line with the observations reported by Reva et al. (2006), suggesting that P. putida KT2440 displays increased resistance to pH shifts as compared to Gram‐negative Enterobacteria. Since the PHP indicator enabled to follow changes in pHi upon alkalinization or acidification of the external milieu, we also explored if this procedure could be used to capture the dynamic changes in pHi brought about by antibiotic treatment as disclosed below.
Figure 4.
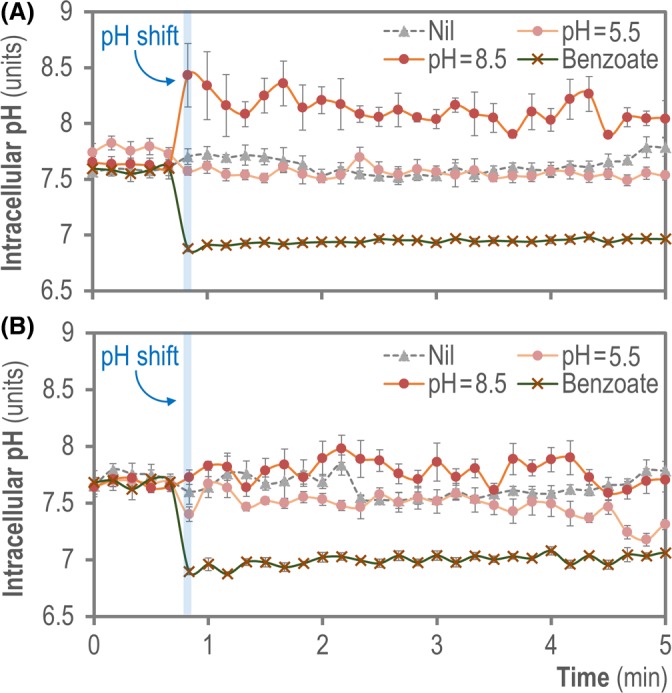
Monitoring the dynamic changes of cytoplasmic pH upon external pH shifts. Exponentially growing cells of (A) E. coli MG1655/pS2513·PHP or (B) P. putida KT2440/pS2513·PHP cultured in M9 minimal medium containing 20 mM glucose were aliquoted into a 96‐well microtiter plate and the cytoplasmic pH was continuously monitored, before and after a shift in the extracellular pH was induced by the addition of the same culture medium buffered at pH = 5.5 or pH = 8.5 as indicated in the Experimental Procedures , or sodium benzoate (permeant acid). Changes in cytoplasmic pH were monitored every 10 s during the first 5 min of incubation. Data shown in each case indicate mean values from independent triplicates ± SD.
Response of the cytoplasmic pH of P. aeruginosa to treatment with different antibiotics
The next phenotype that we addressed was the changes of pHi due to addition of clinically relevant antibiotics often used for the treatment of P. aeruginosa infections. To this end, we first grew P. aeruginosa PA14 harbouring plasmid pS2513·PHP in LB medium to reach the mid‐exponential phase of growth [i.e. optical density at 600 nm (OD600) = 0.4–0.5]. At this point, we treated the cultures with three different antibiotics at one‐fifth (0.2×) or at full (1×) minimal inhibitory concentrations (MIC) of the antibiotics, and we followed the changes in pHi continuously over the course of 4 h (Fig. 5). The antibiotics chosen for this purpose belong to three different functional classes based on their mode of action: ceftazidime (CAZ, a cephalosporin), amikacin (AMK, an aminoglycoside) and ciprofloxacin (CIP, a fluoroquinolone) (Lambert, 2002; Morita et al., 2014; Bassetti et al., 2018).
Figure 5.
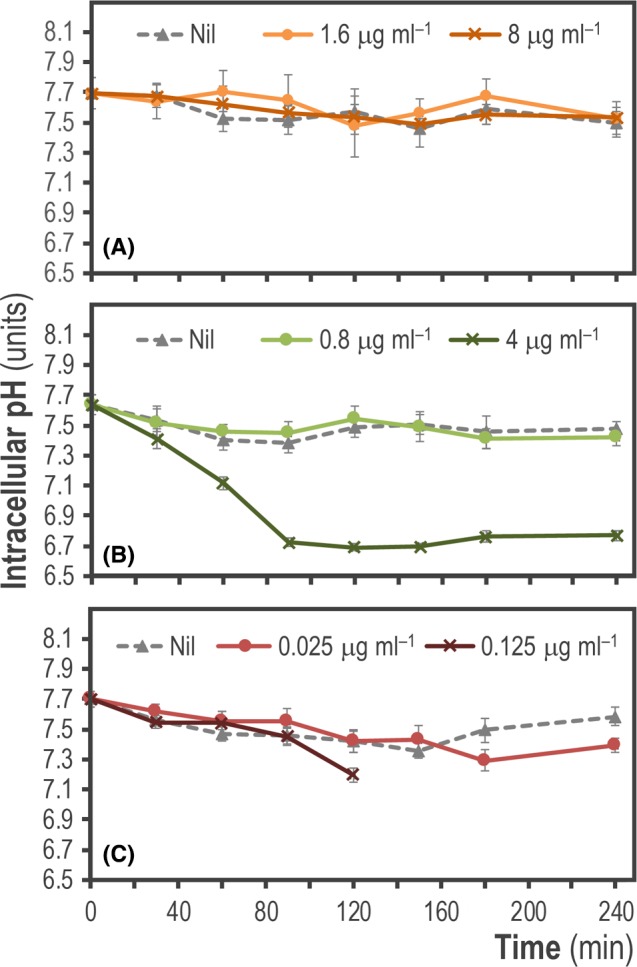
Monitoring the changes of intracellular pH (pH i) in P. aeruginosa PA14 upon treatment with different antibiotics. Exponentially growing cultures of P. aeruginosa PA14 transformed with plasmid pS2513·PHP and grown in LB medium were challenged with one‐fifth (0.2×) and full (1×) minimal inhibitory concentrations of (A) ceftazidime (CAZ), (B) amikacin (AMK) and (C) ciprofloxacin (CIP) as indicated in the graphs. The data shown in each case indicate mean values of pH i from independent triplicates ± SD.
No significant changes in the cytosolic pH were observed after exposure of P. aeruginosa PA14 to 0.2 × (1.6 μg ml−1) and 1 × (8 μg ml−1) MIC concentrations of CAZ when compared to untreated cells (Fig. 5A). However, a clear decrease in the pHi values was observed after the treatment of the same cells with AMK, albeit only when the antibiotic concentration matched the full MIC (4 μg ml−1, Fig. 5B). The drop in pHi could be observed very early on (becoming noticeable just 1 h after treatment) and was retained until the end of the experiment. Remarkably, the pHi in cells exposed to 1 × MIC concentration of AMK dropped to levels close to pH = 6.7. CIP also caused a reduction in the pH, which was evident from 2 h post‐antibiotic treatment with 1 × MIC (0.125 μg ml−1), and after 3 h when cells were challenged with 0.2 × MIC (0.025 μg ml−1, Fig. 5C). This decrease was, however, not as strong as the pH drop observed in AMK‐treated cultures. Interestingly, it was not possible to continue the pHi assessment in cultures amended with 1 × MIC of CIP beyond 2 h after treatment. In this particular case, the integrity of the cells was likely compromised due to the harsh antimicrobial treatment, making it very difficult to obtain consistent cell pellets after centrifugation of the bacterial suspension. This phenomenon has been reported for P. aeruginosa when exposed to different antibiotics, including treatment with CIP (Day et al., 1993; Lee et al., 2017).
The results above shed light on the mechanism of action of antibiotics on P. aeruginosa cells, an area of active research. Bartek et al. (2016) suggested that antibiotic exposure with different types of bactericidal compounds increases the cytosolic pH in Mycobacterium smegmatis. Furthermore, they proposed that there is a general mechanism by which antibiotics kill bacteria through disruption of cellular biosynthetic processes (caused by the primary inhibition of either nucleic acids, proteins or cell wall synthesis) which in turn drives an imbalance in H+ homoeostasis leading to a potentially lethal increase in the pHi. In our experiments, we have used three different types of antibiotics, both at MIC and sub‐MIC concentrations. However, we could not observe an increase in pHi along the 4 h of antibiotic treatment. Conversely, exposure to high concentrations of AMK or CIP caused a drop in the cytosolic pH (Fig. 5). This effect could be caused by the permeabilization of the cell membrane due to a physiological response to the damage caused by the aforementioned antibiotics, a phenomenon previously characterized in Gram‐negative bacteria (Dougherty and Saukkonen, 1985; Davis, 1987; Novo et al., 2000). The differential permeability of the cell membrane of Pseudomonas species to chemicals is a decisive factor that also dictates tolerance or sensitivity to antimicrobials (Novo et al., 2000; Shen et al., 2019). On the other hand, the phenomenon of pH increase upon antibiotic treatment could be affected by the nature of the bacteria tested [e.g. Gram‐positive versus Gram‐negative species (Eichenberger and Thaden, 2019)], and also the experimental conditions used (e.g. addition of antibiotics at the onset of the cultivation or during active growth of the bacterial cells). Taken together, our results suggest that P. aeruginosa may react differently to treatment with diverse types of antibiotics, differing in their mechanism of action, at the level of pHi homoeostasis.
Conclusions
In this work, we described the construction and calibration of a non‐invasive pHi indicator and we have demonstrated its efficacy in E. coli and Pseudomonas species by using a standard protocol that can be easily reproduced in different bacteria (Fig. 6). We have shown the efficacy of this new tool in P. aeruginosa and P. putida by quantifying the alterations in pHi homoeostasis elicited after internal (redirection of glucose consumption pathways) and external (antibiotic treatment) metabolic perturbations, as well as pH shifts for both E. coli and P. putida. Although the vector platform containing the PHP indicator was primarily designed for Pseudomonas species, it can be easily adapted to other Gram‐negative bacteria (unlike other pH ratiometric protein‐bearing plasmids), since our system was constructed following the rules set in the SEVA format (Silva‐Rocha et al., 2013). Plasmids constructed according to this format allow for the easy exchange of different parts at the user's will. Unique oriV modules can be swapped to further broaden the (already sizeable) host range of the plasmids. Typically, different intracellular amounts of PHP will be expected by increasing or decreasing the plasmid copy number. Other manipulations are also possible: different antibiotic markers can be exchanged into the vector depending on the resistance profile of the bacterium under scrutiny. Simple approaches for the online assessment of intracellular properties such as pHi will thus enable both fundamental and applied studies, e.g. biosensor‐based evolution of phenotypic traits (Fernández‐Cabezón et al., 2019).
Figure 6.
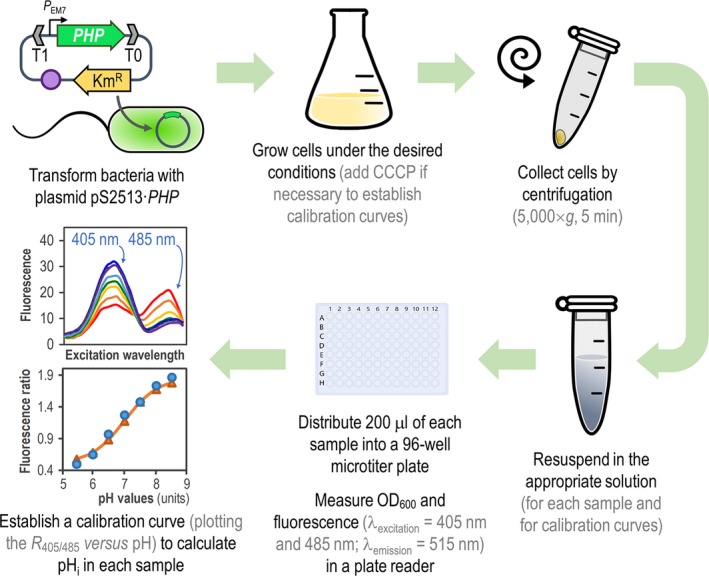
General procedure for in vivo assessment of intracellular pH (pH i) in Pseudomonas species by means of the PHP indicator. Plasmid pS2513·PHP is introduced into the bacteria of interest by chemical transformation, mating or electroporation. The expression of PHP is driven by the constitutive PEM 7 promoter, and vector pSEVA2513 carries the oriV(RSF1010) origin of replication, indicated as a purple circle. The kanamycin‐resistance (KmR) determinant and the T1 and T0 transcriptional terminators are likewise highlighted in the plasmid. Cells are grown in the appropriate culture medium, and a small culture sample is harvested by centrifugation. The cell pellet is resuspended in M9 medium salts to a final optical density at 600 nm (OD 600) = 0.4–0.5. To establish the calibration curve, a second culture is prepared in the same culture medium supplemented with carbonyl cyanide m‐chlorophenyl hydrazine (CCCP). Cells from this culture are also harvested by centrifugation and equilibrated in M9 medium salts adjusted to different pH values (ranging from pH = 5.5–8.5, see Experimental Procedures ) to a final OD 600 = 0.4–0.5. A 200‐μl aliquot of each cell suspension is transferred into a 96‐well microtiter plate, and OD 600 and fluorescence (λexcitation = 405 nm and 485 nm; λemission = 515 nm) are measured in a plate reader. Finally, the R 405/485 values are plotted versus the pH values of pH‐equilibrated cells (calibration curve), and the pH i is calculated using a Boltzmann sigmoid best‐fitting equation.
Experimental procedures
Bacterial strains and culture conditions
All strains used in this work are derivatives of E. coli strains MG1655 (Blattner et al., 1997) and DH5α (Manoil and Beckwith, 1985), P. putida KT2440 (Bagdasarian et al., 1981; Belda et al., 2016), and P. aeruginosa strains PA14 (Friedman and Kolter, 2004) and CH2682 (Hornischer et al., 2018). E. coli DH5α was used both for the propagation and construction of plasmids and for pHi calculations, along with the well‐characterized K‐12 strain MG1655 (Blattner et al., 1997). The P. putida Δglk and Δgcd mutants are derivatives of the parental strain KT2440 with specific mutations in the genes encoding the enzymes that execute the first steps of glucose utilization (Sánchez‐Pascuala et al., 2017, 2019). P. aeruginosa CH2682 is a meropenem‐resistant clinical strain isolated from a patient with a rectal infection (Hornischer et al., 2018).
Unless indicated otherwise, bacteria were cultured in 50‐ml Erlenmeyer flasks containing 10 ml of LB medium (Green and Sambrook, 2012) with agitation at 180 rpm at either 30°C (P. putida strains) or 37°C (E. coli and P. aeruginosa strains). In some experiments, P. putida strains were also grown in 10 ml of M9 minimal medium, containing 8.5 g l−1 Na2HPO4·2H2O, 3 g l−1 KH2PO4, 1 g l−1 NH4Cl, 0.5 g l−1 NaCl, 0.5 g l−1 MgSO4·7H2O and 2.5 ml l−1 of trace element solution (Abril et al., 1989; Nikel and de Lorenzo, 2013) with glucose as the sole carbon source at 3.6 g l−1 (i.e. 20 mM). P. putida strains were also grown under gluconeogenic conditions by using succinate at 20 mM as the sole carbon source. Km was added when needed to ensure plasmid maintenance either at 50 μg ml−1 (for E. coli strains and P. putida KT2440 and its derivatives) or 500 μg ml−1 (for all P. aeruginosa strains). Additionally, the MIC of the antibiotics CAZ, AMK and CIP was determined in P. aeruginosa strains using macrodilutions in Mueller–Hinton broth as described previously (Wiegand et al., 2008).
For growth rate measurements, overnight pre‐cultures of all the strains were grown for 16 h in 3 ml of LB medium supplemented with Km. One millilitre of each pre‐culture was harvested by centrifugation (5000 g, 5 min) and washed twice with 1 ml of fresh LB. The cells were resuspended in fresh LB medium containing Km, diluted to a starting OD600 = 0.02 and distributed (200 μl each well) into a Honeycomb 2 TM plate (Thermo Fisher Scientific, Waltham, MA, USA). The plate was incubated into an automated Bioscreen C MBR™ plate reader (Oy Growth Curves Ab, Helsinki, Finland), and the growth of each sample was constantly monitored by measuring its OD600. The growth rates of each strain were calculated from the exponential fraction of the growth curves (Nikel and Chavarría, 2016). The extracellular pH of culture media was measured in 1‐ml aliquots of culture broth harvested at the times indicated, after separation of the bacterial biomass by centrifugation (5000 g, 5 min) and filtering the supernatant with a 0.2‐μm filter‐membrane (Filtropur S0.2; Sarstedt, Nümbrecht, Germany), with a SevenCompact™ pH meter S210 system (Metler‐Toledo A/S, Glostrup, Denmark). In experiments involving P. putida and its glycolytic mutant derivatives, the concentration of gluconate and 2‐ketogluconate in culture supernatants was determined by high‐pressure liquid chromatography as explained by Nikel et al. (2015).
Construction of a vector for the constitutive expression of the gene encoding the PHP indicator
The DNA sequence of the pHluorin2 gene was codon‐optimized for P. putida, and the resulting coding sequence was ordered as a synthetic DNA fragment (Integrated DNA Technologies, Leuven, Belgium). For the sake of clarity, we refer to the optimized sequence as PHP throughout the text. The entire PHP sequence was PCR‐amplified from the synthetic DNA with primers 5′‐AAA AAG CTT AGG AGG AAA AAC ATA TGA GCA‐3′ and 5′‐TTT ACT AGT TTA CTT GTA GAG TTC ATC CAT ACC‐3′ containing the restriction sites HindIII and SpeI (underlined) and a ribosome binding site (italics) adjacent to the start ATG codon, indicated in boldface in the forward primer (see also Sequence S1 in the Supporting Information). The PCR product and the expression vector pSEVA2513 were double‐digested with the restriction enzymes HindIII and SpeI and column purified (QIAquick™ PCR purification kit; Qiagen, Hilden, Germany). Both DNA fragments were ligated to give rise to plasmid pS2513·PHP, used throughout this study for pHi determinations. The correct plasmid sequence was confirmed by sequencing of plasmid DNA retrieved from selected E. coli DH5α clones. Plasmid pS2513·PHP was later introduced into P. putida and P. aeruginosa strains by electroporation (Choi et al., 2006; Volke et al., 2019; Wirth et al., 2019). Plasmid pS2513·PHP has been deposited in AddGene (https://www.addgene.org/) with catalogue # 122590.
Fluorescence microscopy of bacterial cells expressing PHP
Overnight cultures of bacteria transformed with plasmid pS2513·PHP were grown in LB medium supplemented with Km at the appropriate concentrations (see above). Next, 1 ml of each pre‐culture was pelleted by centrifugation (5000 g, 5 min) and washed twice with 1 ml of fresh LB. Erlenmeyer flasks (50 ml) filled with 10 ml of fresh LB medium (without Km) were inoculated with the washed cells to an OD600 of 0.05 and grown until mid‐exponential phase (OD600 = 0.4–0.5). Cells were fixed by mixing an aliquot of the culture with 5% (v/v) paraformaldehyde in a 1:1 (v/v) ratio, followed by a 30 min incubation at room temperature. Finally, 10 μl of the cell suspensions were placed onto 1.5% (w/v) agarose pads as described by Young et al. (2011) and visualized with a 100 × immersion oil objective in a Nikon Eclipse™Ti microscope (Nikon Instruments, Amsterdam, Netherlands) equipped with a eGFP·ET filter set and a Hamamatsu Orca Flash 4.0 camera (Hamamatsu Photonics K.K., Shizuoka, Japan). Fluorescence images were analysed with the Fiji software package (Schindelin et al., 2012).
In vivo assessment of cytoplasmic pH by flow cytometry and fluorescence spectroscopy using the pHluorin2 or PHP indicators and fluorescent chemical probes
Overnight grown pre‐cultures of each bacterium carrying plasmid pS2513·PHP or plasmid pS2513·pHluorin2 were prepared in the indicated culture media (LB or M9 minimal medium, as specified in each case) added with the necessary antibiotics as described above for the fluorescence microscopy experiments. Unless stated otherwise, 10 ml cultures of each strain were prepared in the same media devoid of antibiotics and grown until mid‐exponential phase (OD600 = 0.4–0.5). Next, 1 ml aliquots were taken from these cultures, and the OD600 of each sample was adjusted (if necessary) to 0.4 before centrifugation (5000 g, 5 min) and cell resuspension in 1 ml of M9 salts devoid of any carbon source, which yielded a cell density of approximately 105 cells ml−1. For flow cytometry experiments, the resulting cell suspension was directly subjected to analysis in a MACSQuant VYB flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany). Both pHluorin2 and PHP were assessed by using the same instrument settings. The fluorescence emission at 525 nm was detected using a 525/40‐nm band pass filter array upon excitation with a diode‐pumped solid‐state laser at 488 nm (Nikel et al., 2016). Size‐related forward scatter signals gathered by the cytometer were used by the FlowJo software v. 9.6.2 (FlowJo LLC, Ashland, OR, USA) to calculate the geometric mean of fluorescence and coefficients of variation of the positive bacterial populations as indicated elsewhere (Benedetti et al., 2016; Akkaya et al., 2018).
For the fluorimetry determinations, 200 μl of each diluted sample were distributed into a 96‐well microtiter plate (Corning & Costar, NY, USA) and incubated for 10 min at 30°C. After incubation, both the OD600 of the cell suspension and the fluorescence emission of PHP (λemission = 515 nm) were recorded at λexcitation of either 405 nm or 485 nm with an EnSpire™ Plate Reader (Perkin‐Elmer, Waltham, MA, USA). In order to generate a calibration curve to calculate pHi, another set of cultures containing 10 ml of either LB medium or M9 minimal medium were inoculated with each bacterial strain. In the case of P. putida and P. aeruginosa strains, CCCP was added to the cultures at 20 μM (in the case of the standard curve for E. coli, the addition of CCCP was not necessary). The cultures were also grown to mid‐exponential phase as detailed before. One millilitre of each cell culture was spun down and resuspended into 1 ml of M9 medium salts containing 50 mM sodium benzoate and 50 mM methylamine HCl, and adjusted to the desired pH with 50 mM of the appropriate buffering agent [2‐(N‐morpholino)ethanesulfonic acid (MES) for pH = 5.5 to 6.5, 3‐(N‐morpholino)propanesulfonic acid (MOPS) for pH = 7.0 and 7.5, and N‐(Tris(hydroxymethyl)methyl)‐3‐aminopropanesulfonic acid (TAPS) for pH = 8.0 and 8.5]. The effect of adding sodium benzoate or methylamine on collapsing pHi was evaluated with the fluorescent probe 5(6)‐carboxyfluorescein diacetate succinimidyl (see section below), and it was found that pHi equalled the pH of the surrounding medium within a ± 0.5 units range. For both the PHP‐ and pHluorin2‐dependent determinations of pHi, culture samples (200 μl) were distributed into a 96‐well microtiter plate and both OD600 and fluorescence were also recorded as described in the preceding section. A Boltzmann sigmoid best‐fitting curve was applied to generate the calibration curves from the λexcitation = 405 nm and λexcitation = 485 nm excitation ratios (R 405/485) obtained by incubating the cells into the buffered solutions, essentially as described by Martínez et al. (2012). The equation used to adjust the best‐fitting curve is as follows:
where R(pH) is the function representing the Boltzmann sigmoid best‐fitting curve; R 1 and R 2 are the lower and upper asymptotes of the curve (representing the minimum and maximum values of R 405/485) respectively; pH is the actual pHi value; pH0 is the half‐value for pHi within the range assessed; and φ is the slope of the fitting curve. The pHi values are expressed as the mean value of two measurements from at least three independent replicates ± SD.
In order to benchmark the PHP indicator against a well‐established protocol, the pHi of the cells was calculated in some experiments by using the ratiometric fluorescent probe 5(6)‐carboxyfluorescein diacetate succinimidyl ester according to the standard protocols of Riondet et al. (1997) and Olivares Pacheco et al. (2017). Briefly, cells, grown as indicated in the text, were collected at an OD600 of 0.4–0.5 and centrifuged at 5000 g for 5 min at room temperature. The bacterial pellets were resuspended at a cell density of approximately 105 cells ml−1 in 50 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES) buffer (pH = 9.0) containing 1 mM EDTA and incubated for 10 min at 30°C with 2.5 μM 5(6)‐carboxyfluorescein diacetate succinimidyl ester (Thermo Fisher Scientific, Waltham, MA, USA). Following this incubation, the samples were washed twice and resuspended in phosphate‐buffered saline buffer (pH = 7.0) containing 1 mM MgCl2. In order to eliminate any possible excess from the non‐conjugated probe, samples were treated with 10 mM glucose in phosphate‐buffered saline buffer. Cells were then centrifuged at 5000 g for 5 min, and bacterial pellets were washed twice and resuspended in a buffer composed of 50 mM Tris, 50 mM MES, 140 mM choline chloride, 1 mM MgCl2, 10 mM KCl, 10 mM NaHCO3 and 0.5 mM CaCl2 (pH = 7.0). One hundred fifty microlitres of each sample was placed on black 96‐well plates (Nunclon Δ Surface; Sigma‐Aldrich Co., St. Louis, MO, USA) and incubated at 30°C in a microtiter plate spectrophotometer (Infinite 200 Pro™; Tecan Trading AG, Männedorf, Switzerland). Fluorescence intensities were measured at excitation wavelengths λexcitation = 490 nm (pH‐sensitive wavelength) and λexcitation = 440 nm (pH‐insensitive wavelength). The emission wavelength used in all of these experiments was λemission = 525 nm. The excitation and emission intensities were used to calculate the corresponding ratios, which were associated to pHi through a linear regression in a standard curve prepared with cells exposed to a set of calibrated buffers following the procedure described by Riondet et al. (1997). The pHi values calculated with the chemical probe are expressed as the mean value of three measurements from at least three independent replicates ± SD.
In vivo tracking of the dynamic changes of cytoplasmic pH upon pH shifts in the extracellular milieu
These experiments were carried out with 10 ml cultures of E. coli MG1655 or P. putida KT2440, each carrying plasmid pS2513·PHP, grown in M9 minimal medium added with 20 mM glucose from overnight pre‐cultures prepared as stated above. Once the cells reached mid‐exponential phase (OD600 = 0.4–0.5), 150‐μl aliquots of each culture were distributed into 96‐well microtiter plates and incubated for 10 min at 30 or 37°C (as appropriate for each strain) with orbital agitation at 180 rpm. The plate was transferred to the EnSpire™ Plate Reader, and the fluorescence emission was recorded continuously every 10 s as described previously. After 1 min incubation, 50 μl of acidic (pH = 5.5; buffered with 200 mM MES) or basic (pH = 8.5; buffered with 200 mM TAPS) M9 minimal medium containing 20 mM glucose was added to each well. Additionally, 50 μl of 250 mM sodium benzoate was added to a set of samples in order to fully collapse the pHi of the cells as indicated above. The recording of the fluorescence emission was resumed immediately after these additions. The pHi for each sample was calculated from the R 405/485 values obtained for the fluorescence emission plotted against the standard curves prepared as described above.
Conflict of interest
None declared.
Supporting information
Fig. S1. Calibration curves for E. coli and P. putida carrying the PHP indicator in a complex culture medium.
Fig. S2. Calibration curve for P. putida carrying the PHP indicator in a minimal culture medium.
Table S1. Intracellular pH assessment of P. putida KT2440 and mutants in glycolytic genes in glucose cultures.
Sequence S1. Synthetic DNA encoding the PHP indicator.
Acknowledgements
The authors are indebted to Prof. Søren Molin and his team (The Novo Nordisk Foundation Center for Biosustainability, Denmark) for sharing research materials. Christopher Kesthely is acknowledged for his critical reading of the manuscript text. The financial support from The Novo Nordisk Foundation (NNF10CC1016517), the European Union's Horizon2020 Research and Innovation ProGramme under grant agreement No. 814418 (SinFonia), and CFT Project No. SRC 017 to P.I.N. is gratefully acknowledged. This work was also partially supported by an ERC Consolidator Grant (COMBAT 724290) from the European Union to S.H.
Microbial Biotechnology (2019) 12(4), 799–813
Funding Information
The financial support from The Novo Nordisk Foundation (NNF10CC1016517), the European Union's Horizon2020 Research and Innovation ProGramme under grant agreement No. 814418 (SinFonia), and CFT Project No. SRC 017 to P.I.N. is gratefully acknowledged. This work was also partially supported by an ERC Consolidator Grant (COMBAT 724290) from the European Union to S.H.
Contributor Information
Alejandro Arce‐Rodríguez, Email: alejandro.arce@helmholtz-hzi.de.
Pablo I. Nikel, Email: pabnik@biosustain.dtu.dk.
References
- Abril, M.A. , Michan, C. , Timmis, K.N. , and Ramos, J.L. (1989) Regulator and enzyme specificities of the TOL plasmid‐encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol 171: 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya, Ö. , Pérez‐Pantoja, D. , Calles, B. , Nikel, P.I. , and de Lorenzo, V. (2018) The metabolic redox regime of Pseudomonas putida tunes its evolvability toward novel xenobiotic substrates. mBio 9: e01512‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awaji, T. , Hirasawa, A. , Shirakawa, H. , Tsujimoto, G. , and Miyazaki, S. (2001) Novel green fluorescent protein‐based ratiometric indicators for monitoring pH in defined intracellular microdomains. Biochem Biophys Res Commun 289: 457–462. [DOI] [PubMed] [Google Scholar]
- Bagdasarian, M. , Lurz, R. , Rückert, B. , Franklin, F.C.H. , Bagdasarian, M.M. , Frey, J. , and Timmis, K.N. (1981) Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010‐derived vectors, and a host‐vector system for gene cloning in Pseudomonas . Gene 16: 237–247. [DOI] [PubMed] [Google Scholar]
- Bartek, I.L. , Reichlen, M.J. , Honaker, R.W. , Leistikow, R.L. , Clambey, E.T. , Scobey, M.S. , et al (2016) Antibiotic bactericidal activity is countered by maintaining pH homeostasis in Mycobacterium smegmatis . mSphere 1: e00176‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti, M. , Vena, A. , Croxatto, A. , Righi, E. , and Guery, B. (2018) How to manage Pseudomonas aeruginosa infections. Drugs Cont 7: 212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda, E. , van Heck, R.G.A. , López‐Sánchez, M.J. , Cruveiller, S. , Barbe, V. , Fraser, C. , et al (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis . Environ Microbiol 18: 3403–3424. [DOI] [PubMed] [Google Scholar]
- Benčina, M. (2013) Illumination of the spatial order of intracellular pH by genetically encoded pH‐sensitive sensors. Sensors 13: 16736–16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, I. , de Lorenzo, V. , and Nikel, P.I. (2016) Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab Eng 33: 109–118. [DOI] [PubMed] [Google Scholar]
- Bernal, V. , Castaño‐Cerezo, S. , and Cánovas, M. (2016) Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl Microbiol Biotechnol 100: 8985–9001. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R. , Plunkett, G. , Bloch, C.A. , Perna, N.T. , Burland, V. , Riley, M. , et al (1997) The complete genome sequence of Escherichia coli K‐12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- Booth, I.R. (1985) Regulation of cytoplasmic pH in bacteria. Microbiol Rev 49: 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer, P. , Drocourt, J. , Rombouts, F.M. , and Abee, T. (1996) A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6‐)‐carboxyfluorescein succinimidyl ester. Appl Environ Microbiol 62: 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero, P. , and Nikel, P.I. (2019) Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non‐traditional microorganisms. Microb Biotechnol 12: 98–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J.R. , Grinstein, S. and Orlowski, J. (2010) Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11: 50–61. [DOI] [PubMed] [Google Scholar]
- del Castillo, T. , Ramos, J.L. , Rodríguez‐Herva, J.J. , Fuhrer, T. , Sauer, U. , and Duque, E. (2007) Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J Bacteriol 189: 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.T. , and Lo, C.J. (2016) Using biophysics to monitor the essential protonmotive force in bacteria. Adv Exp Med Biol 915: 69–79. [DOI] [PubMed] [Google Scholar]
- Choi, K.H. , Kumar, A. , and Schweizer, H.P. (2006) A 10‐min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64: 391–397. [DOI] [PubMed] [Google Scholar]
- Clark, M.W. , Yie, A.M. , Eder, E.K. , Dennis, R.G. , Basting, P.J. , Martinez, K.A. , et al (2015) Periplasmic acid stress increases cell division asymmetry (polar aging) of Escherichia coli . PLoS ONE 10: e0144650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, B.D. (1987) Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C.A. , Marceau‐Day, M.L. , and Day, D.F. (1993) Increased susceptibility of Pseudomonas aeruginosa to ciprofloxacin in the presence of vancomycin. Antimicrob Agents Chem 37: 2506–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer, D.W. , and Nichols, J.W. (1989) Proton flux mechanisms in model and biological‐membranes. J Membr Biol 107: 91–103. [DOI] [PubMed] [Google Scholar]
- Dougherty, T.J. , and Saukkonen, J.J. (1985) Membrane permeability changes associated with DNA gyrase inhibitors in Escherichia coli . Antimicrob Agents Chemother 28: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák, P. , Nikel, P.I. , Damborský, J. , and de Lorenzo, V. (2017) Bioremediation 3.0: engineering pollutant‐removing bacteria in the times of systemic biology. Biotechnol Adv 35: 845–866. [DOI] [PubMed] [Google Scholar]
- Eichenberger, E.M. , and Thaden, J.T. (2019) Epidemiology and mechanisms of resistance of extensively drug resistant Gram‐negative bacteria. Antibiotics 8: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, A. , Gralle, M. , Dani, M.A.C. , Lange, D. , and Wouters, F.S. (2008) pHlameleons: a family of FRET‐based protein sensors for quantitative pH imaging. Biochemistry 47: 13115–13126. [DOI] [PubMed] [Google Scholar]
- Fernández‐Cabezón, L. , Cros, A. and Nikel, P.I. (2019) Evolutionary approaches for engineering industrially‐relevant phenotypes in bacterial cell factories. Biotechnol J, In press. 10.1002/biot.201800439 [DOI] [PubMed] [Google Scholar]
- Friedman, L. , and Kolter, R. (2004) Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51: 675–690. [DOI] [PubMed] [Google Scholar]
- Fuhrer, T. , Fischer, E. , and Sauer, U. (2005) Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187: 1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S.H. , Fan, L. , Peng, L. , Guo, J. , Agulló‐Barceló, M. , Yuan, Z. , and Bond, P.L. (2016) Determining multiple responses of Pseudomonas aeruginosa PAO1 to an antimicrobial agent, free nitrous acid. Environ Sci Technol 50: 5305–5312. [DOI] [PubMed] [Google Scholar]
- Ghoul, M. , Pommepuy, M. , Moillo‐Batt, A. , and Cormier, M. (1989) Effect of carbonyl cyanide m‐chlorophenylhydrazone on Escherichia coli halotolerance. Appl Environ Microbiol 55: 1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke, C. , Feher, K. , von Volkmann, K. , Kirsch, J. , Radbruch, A. , and Kaiser, T. (2017) Determination of background, signal‐to‐noise, and dynamic range of a flow cytometer: a novel practical method for instrument characterization and standardization. Cytometry 91: 1104–1114. [DOI] [PubMed] [Google Scholar]
- Green, M.R. , and Sambrook, J. (2012) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Grillo‐Hill, B.K. , Webb, B.A. , and Barber, D.L. (2014) Ratiometric imaging of pH probes. Methods Cell Biol 123: 429–448. [DOI] [PubMed] [Google Scholar]
- Han, J. , and Burgess, K. (2010) Fluorescent indicators for intracellular pH. Chem Rev 110: 2709–2728. [DOI] [PubMed] [Google Scholar]
- Heim, R. , Cubitt, A.B. , and Tsien, R.Y. (1995) Improved green fluorescence. Nature 373: 663–664. [DOI] [PubMed] [Google Scholar]
- Hirshfield, I.N. , Terzulli, S. , and O'Byrne, C. (2003) Weak organic acids: a panoply of effects on bacteria. Sci Prog 86: 245–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornischer, K. , Khaledi, A. , Pohl, S. , Schniederjans, M. , Pezoldt, L. , Casilag, F. , et al (2018) BACTOME—A reference database to explore the sequence‐ and gene expression‐variation landscape of Pseudomonas aeruginosa clinical isolates. Nucleic Acids Res 47: D716–D720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, R.C. , and Beveridge, T.J. (2005) Application of a pH‐sensitive fluoroprobe (C‐SNARF‐4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71: 2501–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich, T.A. , Sachs, G. , and Padan, E. (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9: 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurbatov, L. , Albrecht, D. , Herrmann, H. , and Petruschka, L. (2006) Analysis of the proteome of Pseudomonas putida KT2440 grown on different sources of carbon and energy. Environ Microbiol 8: 466–478. [DOI] [PubMed] [Google Scholar]
- Lambert, P.A. (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa . J Roy Soc Med 95: 22–26. [PMC free article] [PubMed] [Google Scholar]
- Lambert, L.A. , Abshire, K. , Blankenhorn, D. , and Slonczewski, J.L. (1997) Proteins induced in Escherichia coli by benzoic acid. J Bacteriol 179: 7595–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.M. , Lee, K. , Go, J. , Park, I.H. , Shin, J.S. , Choi, J.Y. , et al (2017) A genetic screen reveals novel targets to render Pseudomonas aeruginosa sensitive to lysozyme and cell wall‐targeting antibiotics. Front Cell Infect Microbiol 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas, M.A. , Ramos, J.L. , and Rodríguez‐Herva, J.J. (2000) Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol 182: 4764–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis, J. , McCaffery, J.M. , Miyawaki, A. , Farquhar, M.G. , and Tsien, R.Y. (1998) Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA 95: 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon, M.J. (2011) pHluorin2: an enhanced, ratiometric, pH‐sensitive green florescent protein. Adv Biosci Biotechnol 2: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil, C. , and Beckwith, J. (1985) TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA 82: 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, K.A. , Kitko, R.D. , Mershon, J.P. , Adcox, H.E. , Malek, K.A. , Berkmen, M.B. , and Slonczewski, J.L. (2012) Cytoplasmic pH response to acid stress in individual cells of Escherichia coli and Bacillus subtilis observed by fluorescence ratio imaging microscopy. Appl Environ Microbiol 78: 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnaney, T.B. , Zeng, W. , Doe, C.F. , Bhanji, N. , Wakelin, S. , Pearson, D.S. , et al (2005) Protonation, photobleaching, and photoactivation of yellow fluorescent protein (YFP 10C): a unifying mechanism. Biochemistry 44: 5510–5524. [DOI] [PubMed] [Google Scholar]
- Miesenböck, G. , de Angelis, D.A. , and Rothman, J.E. (1998) Visualizing secretion and synaptic transmission with pH‐sensitive green fluorescent proteins. Nature 394: 192–195. [DOI] [PubMed] [Google Scholar]
- Molenaar, D. , Abee, T. , and Konings, W.N. (1991) Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim Biophys Acta 1115: 75–83. [DOI] [PubMed] [Google Scholar]
- Morita, Y. , Tomida, J. , and Kawamura, Y. (2014) Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 4: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , and Chavarría, M. (2016) Quantitative physiology approaches to understand and optimize reducing power availability in environmental bacteria In Hydrocarbon and Lipid Microbiology Protocols–Synthetic and Systems Biology–Tools. McGenity T.J., Timmis K.N., and Nogales‐Fernández B. (eds). Heidelberg, Germany: Humana Press, pp. 39–70. [Google Scholar]
- Nikel, P.I. , and de Lorenzo, V. (2013) Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3‐dichloroprop‐1‐ene. Metab Eng 15: 98–112. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , and de Lorenzo, V. (2014) Robustness of Pseudomonas putida KT2440 as a host for ethanol biosynthesis. New Biotechnol 31: 562–571. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , and de Lorenzo, V. (2018) Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans‐metabolism. Metab Eng 50: 142–155. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Pettinari, M.J. , Ramírez, M.C. , Galvagno, M.A. , and Méndez, B.S. (2008) Escherichia coli arcA mutants: metabolic profile characterization of microaerobic cultures using glycerol as a carbon source. J Mol Microbiol Biotechnol 15: 48–54. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarría, M. , Fuhrer, T. , Sauer, U. , and de Lorenzo, V. (2015) Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner‐Doudoroff, Embden‐Meyerhof‐Parnas, and pentose phosphate pathways. J Biol Chem 290: 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Pérez‐Pantoja, D. , and de Lorenzo, V. (2016) Pyridine nucleotide transhydrogenases enable redox balance of Pseudomonas putida during biodegradation of aromatic compounds. Environ Microbiol 18: 3565–3582. [DOI] [PubMed] [Google Scholar]
- Nordstrom, D.K. , Alpers, C.N. , Ptacek, C.J. , and Blowes, D.W. (2000) Negative pH and extremely acidic mine waters from Iron Mountain, California. Environ Sci Technol 34: 254–258. [Google Scholar]
- Novo, D.J. , Perlmutter, N.G. , Hunt, R.H. , and Shapiro, H.M. (2000) Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus . Antimicrob Agents Chemother 44: 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares Pacheco, J. , Alvarez‐Ortega, C. , Alcalde Rico, M. , and Martínez, J.L. (2017) Metabolic compensation of fitness costs is a general outcome for antibiotic‐resistant Pseudomonas aeruginosa mutants overexpressing efflux pumps. mBio 8: e00500‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.N. , Budde, B.B. , Siegumfeldt, H. , Rechinger, K.B. , Jakobsen, M. , and Ingmer, H. (2002) Noninvasive measurement of bacterial intracellular pH on a single‐cell level with green fluorescent protein and fluorescence ratio imaging microscopy. Appl Environ Microbiol 68: 4145–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan, E. , Bibi, E. , Ito, M. , and Krulwich, T.A. (2005) Alkaline pH homeostasis in bacteria: new insights. Biochim Biophys Acta 1717: 67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenrath, M. , and Boles, E. (2018) A superfolder variant of pH‐sensitive pHluorin for in vivo pH measurements in the endoplasmic reticulum. Sci Rep 8: 11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva, O.N. , Weinel, C. , Weinel, M. , Böhm, K. , Stjepandic, D. , Hoheisel, J.D. , and Tümmler, B. (2006) Functional genomics of stress response in Pseudomonas putida KT2440. J Bacteriol 188: 4079–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riondet, C. , Cachon, R. , Waché, Y. , Alcaraz, G. , and Diviès, C. (1997) Measurement of the intracellular pH in Escherichia coli with the internally conjugated fluorescent probe 5‐ (and 6‐)carboxyfluorescein succinimidyl ester. Biotechnol Techniques 11: 735–738. [Google Scholar]
- Roadcap, G.S. , Sanford, R.A. , Jin, Q. , Pardinas, J.R. , and Bethke, C.M. (2006) Extremely alkaline (pH > 12) ground water hosts diverse microbial community. Ground Water 44: 511–517. [DOI] [PubMed] [Google Scholar]
- Roberts, J.K. , Wade‐Jardetzky, N. , and Jardetzky, O. (1981) Intracellular pH measurements by 31P nuclear magnetic resonance: influence of factors other than pH on 31P chemical shifts. Biochemistry 20: 5389–5394. [DOI] [PubMed] [Google Scholar]
- Rupprecht, C. , Wingen, M. , Potzkei, J. , Gensch, T. , Jaeger, K.E. , and Drepper, T. (2017) A novel FbFP‐based biosensor toolbox for sensitive in vivo determination of intracellular pH. J Biotechnol 258: 25–32. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Pascuala, A. , de Lorenzo, V. , and Nikel, P.I. (2017) Refactoring the Embden‐Meyerhof‐Parnas Pathway as a whole of portable Glucobricks for implantation of glycolytic modules in Gram‐negative bacteria. ACS Synth Biol 6: 793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Pascuala, A. , Fernández‐Cabezón, L. , de Lorenzo, V. , and Nikel, P.I. (2019) Functional implementation of a linear glycolysis for sugar catabolism in Pseudomonas putida . Metab Eng 54: 200–211. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šeputienė, V. , Daugelavičius, A. , Sužiedėlis, K. , and Sužiedėlienė, E. (2006) Acid response of exponentially growing Escherichia coli K‐12. Microbiol Res 161: 65–74. [DOI] [PubMed] [Google Scholar]
- Sezonov, G. , Joseleau‐Petit, D. , and D'Ari, R. (2007) Escherichia coli physiology in Luria‐Bertani broth. J Bacteriol 189: 8746–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrow, S.O. (2001) Analysis of flow cytometry data. Curr Protoc Immunol 5: 5.2.1‐5.2.10. [DOI] [PubMed] [Google Scholar]
- Shen, X. , Johnson, N.V. , Kreamer, N.N.K. , Barnes, S.W. , Walker, J.R. , Woods, A.L. , et al (2019) Genetic defects in efflux (oprM), beta‐lactamase (ampC) and lipopolysaccharide transport (lptE) mediate antibiotic hypersusceptibility of Pseudomonas aeruginosa strain Z61. Antimicrob Agents Chemother, In press. 10.1128/aac.00784-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbesma, W.F. , Almeida, J.S. , Reis, M.A. , and Santos, H. (1996) Uncoupling effect of nitrite during denitrification by Pseudomonas fluorescens: an in vivo 31P‐NMR study. Biotechnol Bioeng 52: 176–182. [DOI] [PubMed] [Google Scholar]
- Silva‐Rocha, R. , Martínez‐García, E. , Calles, B. , Chavarría, M. , Arce‐Rodríguez, A. , de las Heras, A. , et al (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41: D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski, J.L. , Fujisawa, M. , Dopson, M. , and Krulwich, T.A. (2009) Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55: 799–79. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp, C. (2017) Membrane homeostasis in bacteria upon pH challenge In Biogenesis of Fatty Acids, Lipids and Membranes. Geiger O. (ed). Cham, Switzerland: Springer, pp. 799–13. [Google Scholar]
- Tournu, H. , Luna‐Tapia, A. , Peters, B.M. and Palmer, G.E. (2017) In vivo indicators of cytoplasmic, vacuolar, and extracellular pH using pHluorin2 in Candida albicans . mSphere 2: e00276‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke, D.C. , Turlin, J. , Mol, V. , and Nikel, P.I. (2019) Physical decoupling of XylS/Pm Regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida . Microb Biotechnol, In press. 10.1111/1751-7915.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil, M.I. , Covey, C.R. , and Walter, N.D. (2018) Antibiotic lethality and membrane bioenergetics. Adv Microb Physiol 73: 77–122. [DOI] [PubMed] [Google Scholar]
- Wiegand, I. , Hilpert, K. , and Hancock, R.E. (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3: 163–175. [DOI] [PubMed] [Google Scholar]
- Wirth, N.T. , Kozaeva, E. and Nikel, P.I. (2019) Accelerated genome engineering of Pseudomonas putida by I‐SceI―mediated recombination and CRISPR‐Cas9 counterselection. Microb Biotechnol, In press. 10.1111/1751-7915.13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, F. , and Nikaido, H. (1982) Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol 152: 636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.W. , Locke, J.C. , Altinok, A. , Rosenfeld, N. , Bacarian, T. , Swain, P.S. , et al (2011) Measuring single‐cell gene expression dynamics in bacteria using fluorescence time‐lapse microscopy. Nat Protoc 7: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z. , Cai, Y. , Qin, W. , Lin, J. , and Qiu, J. (2015) Polymyxin E induces rapid Paenibacillus polymyxa death by damaging cell membrane while Ca2+ can protect cells from damage. PLoS ONE 10: e0135198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Calibration curves for E. coli and P. putida carrying the PHP indicator in a complex culture medium.
Fig. S2. Calibration curve for P. putida carrying the PHP indicator in a minimal culture medium.
Table S1. Intracellular pH assessment of P. putida KT2440 and mutants in glycolytic genes in glucose cultures.
Sequence S1. Synthetic DNA encoding the PHP indicator.


