Summary
Listeria monocytogenes has been implicated in a number of outbreaks including the recent largest outbreak in South Africa. Current methods for prevention of foodborne L. monocytogenes infection are inadequate, thus raising a need for an alternative strategy. Probiotic bioengineering is considered a prevailing approach to enhance the efficacy of probiotics for targeted control of pathogens. Here, the ability of Lactobacillus casei expressing the L. monocytogenes invasion proteins Internalins A and B (inlAB) to prevent infection was investigated. The inlAB operon was cloned and surface‐expressed on L. casei resulting in a recombinant strain, LbcInl AB, and subsequently, its ability to inhibit adhesion, invasion and translocation of L. monocytogenes through enterocyte‐like Caco‐2 cells was examined. Cell surface expression of InlAB on the LbcInl AB was confirmed by Western blotting and immunofluorescence staining. The LbcInl AB strain showed significantly higher (P < 0.0001) adherence, invasion and translocation of Caco‐2 cells than the wild‐type L. casei strain (LbcWT), as well as reduced L. monocytogenes adhesion, invasion and transcellular passage through the cell monolayer than LbcWT. Furthermore, pre‐exposure of Caco‐2 cells to LbcInl AB significantly reduced L. monocytogenes‐induced cell cytotoxicity and epithelial barrier dysfunction. These results suggest that InlAB‐expressing L. casei could be a potential practical approach for prevention of listeriosis.
Introduction
Listeria monocytogenes is a Gram‐positive, facultative intracellular foodborne pathogen that persists in the diverse environment within and outside mammalian host cells (Vazquez‐Boland et al., 2001; Czuprynski, 2005). The severity of the disease listeriosis depends on the host immune status. The infection in immunocompetent individuals is commonly self‐limiting febrile gastroenteritis, while it results in meningitis and encephalitis in immunocompromised individuals. In expectant women, it can spread to the uterus, thereby affecting the fetus, consequently causing complications such as spontaneous abortions, stillbirths or premature births (Schuchat et al., 1991; Wolfe et al., 2017). Incidences of listeriosis are much lower than diseases caused by most foodborne pathogens; however, its high case fatality rate (20–30%) has made it a considerable public health concern (Scallan et al., 2011; de Noordhout et al., 2014). The largest listeriosis outbreak ever recorded is the recent one reported in South Africa (2017–2018), linked to consumption of ready‐to‐eat (RTE) sausage called Polony. It resulted in a total of 1060 cases, of which 216 were fatal (Allam et al., 2018) (http://www.nicd.ac.za/index.php/listeriosis-outbreak-situation-report-_4july2018/).
As an intracellular pathogen, L. monocytogenes can invade non‐phagocytic cells and cross the intestinal (Nikitas et al., 2011; Drolia et al., 2018), blood–brain (Ghosh et al., 2018) and feto‐placental (Robbins et al., 2010; Wolfe et al., 2017) barriers. It attaches to and enters into mammalian cells, evades destruction by host phagocytic cells, multiples intracellularly and then spreads to adjacent cells (Radoshevich and Cossart, 2018). Virulence factors responsible for its adhesion include but are not limited to Listeria adhesion protein (LAP), autolysin amidase (AmiA) and the Internalin (Inl) family of proteins (InlA, InlB, InlJ and InlF) (Camejo et al., 2011; Radoshevich and Cossart, 2018). Listeria adhesion protein is an alcohol acetaldehyde dehydrogenase (lmo1634) that promotes adhesion of Listeria during the intestinal infection phase (Pandiripally et al., 1999; Jagadeesan et al., 2010; Bailey et al., 2017). It interacts with the epithelial receptor, heat‐shock protein 60 (Hsp60) (Wampler et al., 2004; Jagadeesan et al., 2011), and activates NF‐κB and myosin light chain kinase (MLCK) resulting in mislocalization of tight junction proteins and opening of the cell–cell junction for bacterial passage into the lamina propria (Drolia et al., 2018).
For host cell invasion, the pathogen uses InlA and InlB (Robbins et al., 2010; Stavru et al., 2011), which binds to the host cell receptor E‐cadherin (Mengaud et al., 1996) and the hepatocyte growth factor receptor c‐Met (Shen et al., 2000) respectively. InlA also aids crossing of the gut epithelial barrier by transcytosis (Nikitas et al., 2011), while InlB facilitates the invasion of human hepatic and M cells (Chiba et al., 2011; Disson and Lecuit, 2013). InlA and InlB are secreted proteins (Trost et al., 2005) and remain covalently attached to the peptidoglycan via LPXTG motif and teichoic acid via GW motif of L. monocytogenes cell wall respectively (Braun et al., 1997; Schubert et al., 2002). The bacterium then employs listeriolysin O (LLO) and phospholipases (PlcA and PlcB) to escape from the vacuoles and actin polymerization protein (ActA) to move from cell to cell (Portnoy et al., 1992; Camejo et al., 2011). There is currently no vaccine for this pathogen. Only precautionary guidance stated by the Centers for Disease Control and Prevention summarizes the importance of hygiene during food preparation and handling, as well as avoidance of certain RTE foods by high‐risk groups.
Probiotics have been used to restore the balance of the gut microbial ecosystem and for control of pathogenic infections. They prevent or control foodborne illnesses through competitive exclusion of pathogens, stimulation of the host immune system and tightening of the gut barrier (Amalaradjou and Bhunia, 2012; Behnsen et al., 2013). Several studies have reported their use to combat L. monocytogenes (Touré et al., 2003; Corr et al., 2007; Aguilar et al., 2011). Despite the proven success of probiotics for control of enteric pathogens, they are not without shortcomings. Their disadvantages are that their action is non‐specific in nature, they sometimes fail to block attachment of some pathogens to their specific receptors, and in certain instances, they induce low levels of an immune response (Bauer et al., 2002; McCarthy et al., 2003; Koo et al., 2012). Novel probiotic strains with enhanced desirable attributes can be designed by considering these limitations of traditional probiotics, as well as the behaviour and disease processes of the pathogens (O'Toole et al., 2017; do Carmo et al., 2018). These novel strains that can prevent pathogenic infections, deliver drugs or vaccines, mimic surface receptors and enhance host immune responses are developed using genetic modification (Steidler, 2003; Buccato et al., 2006; Kajikawa et al., 2007; Wells and Mercenier, 2008; Unnikrishnan et al., 2012; Amalaradjou and Bhunia, 2013; Ryan and Bhunia, 2017).
A recombinant Lactobacillus paracasei strain expressing the LAP of L. monocytogenes (LbpLAP) was previously developed in our laboratory, and it showed enhanced inhibition of L. monocytogenes interaction with Caco‐2 cells when compared to its wild‐type counterpart (Koo et al., 2012). Recently, this same gene was cloned and expressed into L. casei ATCC344 strain and the resultant recombinant strain LbcLAP exhibited a similar anti‐listeria effect (unpublished). Researchers elsewhere also cloned and expressed InlA into Lactococcus lactis for delivering DNA intracellularly (Guimaraes et al., 2005; Innocentin et al., 2009; De Azevedo et al., 2015; Yano et al., 2018). Paradoxically, none of these studies examined whether these InlA‐expressing recombinant strains could prevent L. monocytogenes infection in a model system. Therefore, in the current study, our goal was to simultaneously clone and express both InlA and InlB (since both are required for cell invasion) into L. casei, a well‐studied probiotic strain with proven health beneficial effects (Lenoir et al., 2016; Jacouton et al., 2017), and then investigate the ability of the resultant recombinant strain to inhibit adhesion, invasion and translocation of L. monocytogenes in vitro in a cell culture model.
Results
InlAB was successfully cloned and expressed in Lactobacillus casei
To engineer the probiotic Lactobacillus casei expressing inlAB of L. monocytogenes, the PCR‐amplified inlAB gene product and the plasmid pLP401‐T were both digested with the restriction enzymes NotI and XhoI and then subsequently ligated to produce the recombinant vector designated pLP401‐InlAB (Fig. S1A). This construct was electrotransferred into L. casei ATCC344 (LbcWT), and three selected transformants were confirmed by PCR to contain inlAB operon (Fig. 1A).
Figure 1.
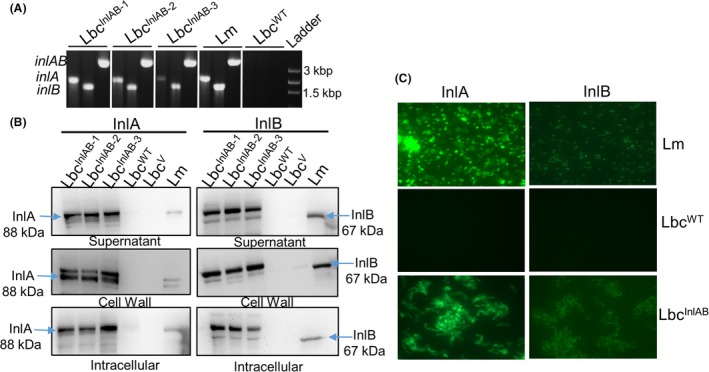
Agarose gel showing (A) PCR‐amplified gene products for inlAB, inlA and inlB of InlAB‐expressing 3 recombinant Lactobacillus casei strains (LbcInl AB −1, LbcInl AB −2, LbcInl AB −3) and L. monocytogenes (Lm) and LbcWT. Lm: L. monocytogenes F4244 (positive control) and LbcWT (negative control).B. Western blot showing expression of Internalins InlA and InlB in the recombinant L. casei strains (LbcInl AB −1, LbcInl AB −2, LbcInl AB −3, LbcWT and LbcV) in the different cellular fractions (supernatant, cell wall and intracellular) and L. monocytogenes F4244 (Lm). C. Immunofluorescence staining of bacteria (magnification 1000 × ) with anti‐InlA mAb‐2D12 and anti‐InlB pAb404. LbcInl AB and Lm (control) cells indicated the presence of InlA (green) and no expression in LbcWT. Anti‐InlB pAb‐404 staining produced weak signal, suggesting this antibody may not be suitable for immunofluorescence staining.
Western blot assay confirmed the expression of both InlA and InlB proteins in the different cellular fractions (supernatant, cell wall and intracellular) of LbcInlAB while absent in LbcWT or LbcV (Lbc carrying only empty pLP401‐T vector) cell fractions (Fig. 1B, Fig. S1B). Immunofluorescence staining also confirmed the surface expression of InlA and InlB in LbcInlAB strain (Fig. 1C). Listeria monocytogenes F4244 (serotype 4b) was used as a positive control (Fig. 1B). These data indicate that both InlA and InlB were successfully expressed in LbcInlAB strain and were associated with the cell wall. Transformant 1 (LbcInlAB−1) was used for the rest of the experiments.
The InlAB expression did not affect the growth rate of LbcInlAB strain
In order to determine whether the expression of InlAB affects the growth of L. casei, we compared growth curves of the LbcWT, LbcV and LbcInlAB. Both optical density (Fig. 2A) and the viable cell count (log CFU ml−1) (Fig. 2B) data showed similar growth profiles for all three strains over time. Furthermore, in phase‐contrast micrographs (Fig. 2C), all three strains LbcWT, LbcV and LbcInlAB maintained a typical elongated curve‐shaped morphology; however, LbcV and LbcInlAB formed slightly longer chains.
Figure 2.
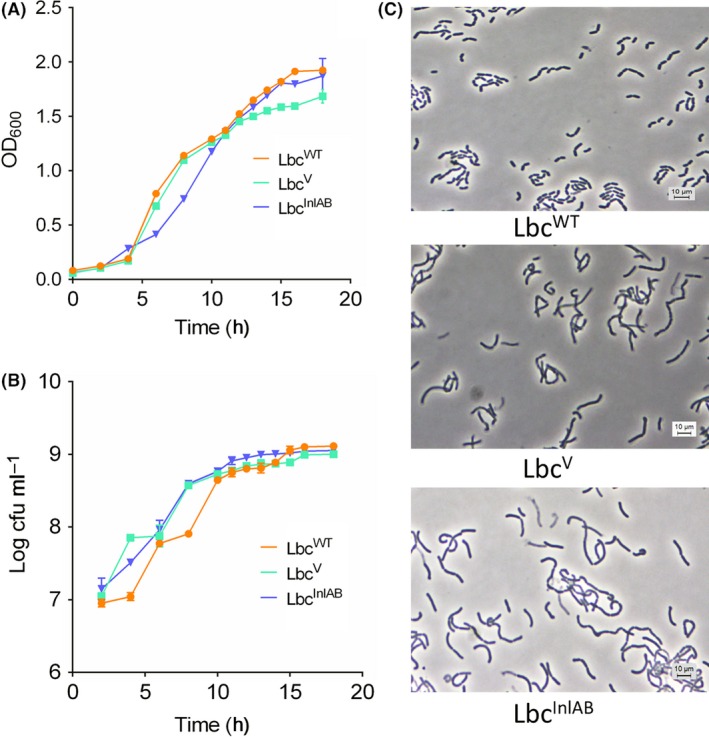
Panel showing L. casei growth curves. (A) Optical density measurement (OD at 600 nm), (B) bacterial counts and (C) phase‐contrast microscopic images of LbcWT, LbcV and LbcInl AB. This experiment was performed twice in triplicates.
Adhesion, invasion and translocation characteristics of recombinant LbcInlAB
We compared the abilities of the L. casei strains (LbcWT, LbcV and LbcInlAB) to adhere to, invade and translocate through or across the Caco‐2 cells versus those of L. monocytogenes and LbcLAP. LbcWT (P = 0.8466) and LbcV (P = 0.9964) showed similar adhesion profiles to Caco‐2 cells when compared to L. monocytogenes (Fig. 3A); however, adhesion of LbcInlAB was significantly higher than that of LbcWT (P = 0.0153). As expected, LbcLAP also showed higher adhesion (17.95%) than LbcWT (11.13%). These data indicate that InlAB expression augmented the ability of LbcInlAB strain to adhere to Caco‐2 cells.
Figure 3.
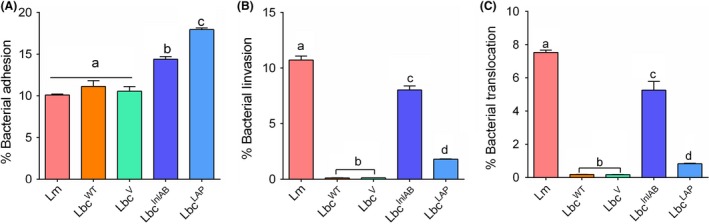
Adhesion, invasion and translocation profiles of Listeria monocytogenes (Lm) and Lactobacillus casei (Lbc) to Caco‐2 cells.A. Adhesion, (B) invasion and (C) translocation of the Caco‐2 cells by L. monocytogenes and L. casei strains (LbcWT, LbcV, LbcInl AB and LbcLAP). Percentages were calculated relative to the inoculums that were added to the Caco‐2 cells. Data are average (SD) of three independent experiments performed in duplicate. For each time point, bars marked with different letters (a, b, c, d) indicate significant difference at P < 0.05.
In Caco‐2 cell invasion assay, LbcInlAB (8.0%) showed a significantly higher invasion (P < 0.05) than the LbcWT (0.18%) or LbcV (0.13%) (Fig. 3B). Listeria monocytogenes as a positive control showed high invasion (10.7%). As anticipated, LbcLAP had a low invasion (0.83%), which was significantly lower (P < 0.0001) than that of LbcinlAB.
Likewise, LbcInlAB also showed a significantly higher (P < 0.0001) transcellular translocation through epithelial (Caco‐2) barrier in a trans‐well set‐up than the LbcWT or LbcV strains (Fig. 3C). L. monocytogenes was able to invade and translocate across the Caco‐2 cells at significantly higher levels (P < 0.0001) than those obtained for all the L. casei strains. Interestingly, LbcLAP showed a significantly lower (P < 0.0001) paracellular translocation than the LbcInlAB strain.
Competitive exclusion of L. monocytogenes by recombinant LbcInlAB
Probiotics inhibit pathogen colonization through the mechanism of competition, either for attachment site or for food. There are different ways by which probiotics can competitively inhibit pathogen adhesion and infection: competitive adhesion, inhibition of adhesion and displacement of adhesion (Fig. 4). Adhesion of L. monocytogenes to Caco‐2 cells in the absence of L. casei strains was recorded as 100% in all the assays and was used to calculate the relative adhesion in the presence of these strains. In the competitive adhesion assay, adhesion of L. monocytogenes was significantly reduced (P < 0.0001) by 24% when co‐inoculated with LbcInlAB for 1 h (Fig. 4A), while it was not reduced when it was added simultaneously with either LbcWT (P = 0.9136) or LbcV (P = 0.9986). Similar results were obtained for inhibition of adhesion assay where L. casei strains were allowed to adhere for 1 h before inoculation with L. monocytogenes for 1 h (Fig. 4B). Conversely, LbcInlAB failed to displace already adhered L. monocytogenes to Caco‐2 cells and showed no statistical differences (P < 0.05) when compared with LbcWT or LbcV (Fig. 4C). Interestingly, LbcLAP showed significantly higher inhibition (P < 0.0007) of L. monocytogenes than LbcInlAB (26% vs. 19%) and was unable to displace attached L. monocytogenes cells (Fig. 4).
Figure 4.
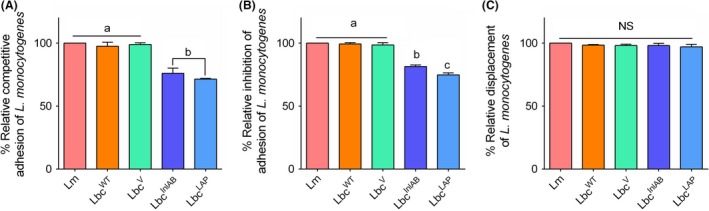
Competitive exclusion of Listeria monocytogenes (Lm) adhesion to Caco‐2 cells by L. casei strains (LbcWT, LbcV, LbcInl AB and LbcLAP), analysed by three different exclusion mechanisms: (A) competitive adhesion – Caco‐2 cells were exposed to L. casei strains with Lm simultaneously; (B) inhibition of adhesion – Caco‐2 cells were pre‐exposed to L. casei strains for 1 h before infection with Lm; and (C) displacement of adhesion – Caco‐2 cells were infected with Lm for 1 h before L. casei treatment (1 h). Adhesion of Lm alone to Caco‐2 cells was presented as 100%, and per cent adhesion was calculated relative to that. For each time point, bars marked with different letters (a, b, c) indicate significant difference at P < 0.05.
Inhibition of L. monocytogenes adhesion, invasion and transcellular migration over time
Next, we compared the inhibitory effect of LbcInlAB pre‐exposed to Caco‐2 cells for 1, 4, 16 and 24 h of duration against L. monocytogenes infection (adhesion, invasion and translocation) for 1 h. Overall, the results indicated that reduction of the interaction between L. monocytogenes and Caco‐2 cells increased with increasing pre‐exposure time of the Caco‐2 monolayer to LbcInlAB, while it was not or was negligibly affected by prolonged exposure to either LbcWT or LbcV (Fig. 5).
Figure 5.
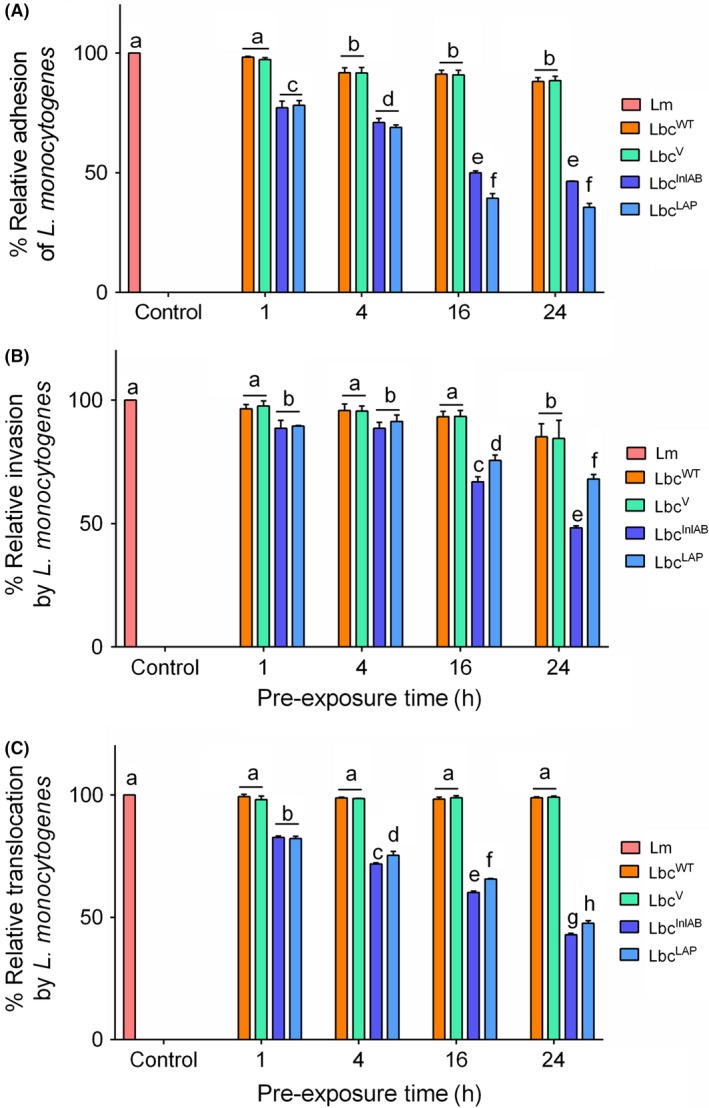
Inhibition of Listeria monocytogenes (Lm) adhesion (A), invasion (B) and transcellular translocation (C) by the L. casei strains (LbcWT, LbcV, LbcInl AB and LbcLAP). Caco‐2 cells were pre‐exposed to L. casei strains for 1, 4, 16 and 24 h before infection with Lm for 1 h for adhesion and invasion and 2 h for translocation. Data are averages of three experiments ran in duplicates. For each time point, bars marked with different letters (a, b, c, d, e, f, g, h) indicate significant difference at P < 0.05.
In the adhesion assay, LbcInlAB reduced L. monocytogenes adhesion by 50–53.6% at 16 and 24 h, while LbcWT and LbcV reduced by only 8% (Fig. 5A). As a positive control, LbcLAP showed about 64.43% reduction in L. monocytogenes adhesion to Caco‐2 cells at 24 h, which is significantly higher (P < 0.0001) than that of the LbcInlAB strain (Fig. 5A).
In the invasion assay, LbcInlAB reduced L. monocytogenes invasion by 51.7% at 24 h, while LbcWT and LbcV reduced invasion by only 15%. As anticipated, LbcLAP showed about a 32% reduction in L. monocytogenes invasion to Caco‐2 cells at 24 h, which is significantly lower than LbcInlAB (Fig. 5B).
In the transcellular translocation assay, LbcInlAB reduced L. monocytogenes translocation by 57.14% at 24 h, while LbcWT and LbcV did not show any reduction at the same pre‐exposure period. As a positive control, LbcLAP showed about 52.46% reduction in L. monocytogenes translocation to Caco‐2 cells at 24 h, similar to LbcInlAB (P = 0.1595) (Fig. 5C). These results collectively indicate that InlAB‐expressing L. casei reduced L. monocytogenes adhesion, invasion and transcellular translocation in the Caco‐2 cell model showing a pronounced inhibitory effect after 16–24 h pre‐exposure.
Inhibition of cytotoxic effects of L. monocytogenes on Caco‐2 cells by L. casei
We investigated the cytotoxic effect by measuring lactate dehydrogenase (LDH) release induced by L. monocytogenes (1 h) from Caco‐2 cells in the presence or absence of L. casei strains (Fig. 6). L. monocytogenes treatment for 1 h induced 64.38% cytotoxicity to Caco‐2 cells in the absence of L. casei, while it induced only 5.93% and 28.7% cytotoxicity after 1 h and 24 h pre‐exposure to LbcInlAB, respectively, and by 57% and 62.3% after 1 and 24 h pre‐exposure to LbcWT respectively (Fig. 6). Interestingly, L. monocytogenes induced only 0.09% and 13.3% cytotoxicity after 1 and 24 h pre‐exposure to LbcLAP respectively. Pre‐treatment of Caco‐2 cells with recombinant L. casei strains resulted in their significant protection (P < 0.0001) against the cytotoxic effect of L. monocytogenes compared to pre‐treatment with LbcWT.
Figure 6.
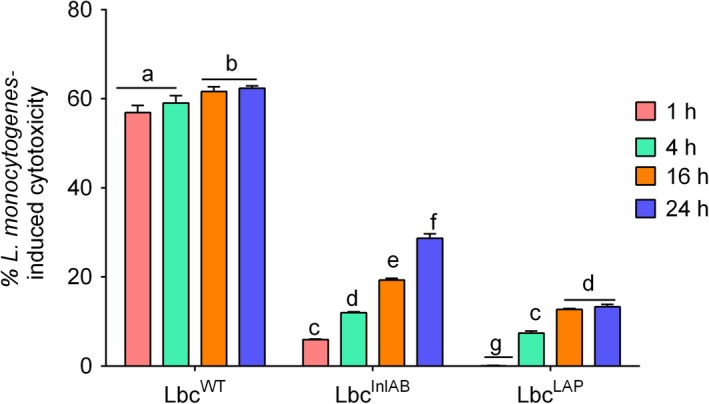
Cytotoxicity of Listeria monocytogenes in Caco‐2 cells pre‐exposed with Lactobacillus casei over time (1, 4, 16, 24 h). Cytotoxicity value for L. monocytogenes treatment (1 h) in the absence of L. casei strains was 64.38%. Data are averages of three experiments ran in duplicates. For each time point, bars marked with different letters (a, b, c, d, e, f, g) indicate significant difference at P < 0.05.
Recombinant LbcInlAB protects epithelial tight junction barrier integrity
We further monitored the effect of recombinant L. casei strains on L. monocytogenes‐mediated tight junction barrier function of Caco‐2 cells by measuring the transepithelial electrical resistance (TEER) and permeability of 4 kDa of dextranFITC (FD4). The TEER value for Caco‐2 cells exposed to L. monocytogenes for 2 h without L. casei pre‐treatment was 16.9%. When Caco‐2 cells were pre‐exposed to L. casei strains, TEER values were between 9.5% and 16.7%, 2.6% and 8.53%, and 1.67% and 6.52% for LbcWT, LbcInlAB and LbcLAP respectively (Fig. 7A). There was a significant (P < 0.0001) protection of epithelial barrier disruption by LbcInlAB and LbcLAP strains compared with LbcWT. However, prolonged pre‐exposure (24 h) to all L. casei strains resulted in a decrease in TEER values for all the treatments.
Figure 7.
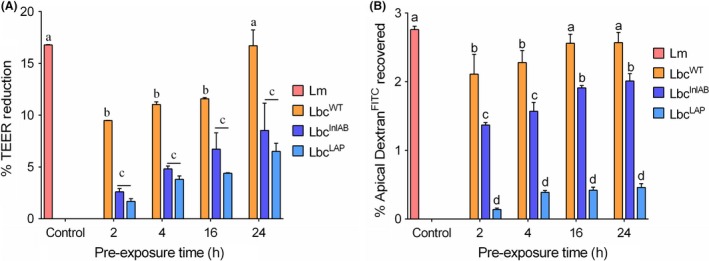
Caco‐2 cell permeability analysis using (A) transepithelial electrical resistance (TEER) and (B) 4 kDa of dextranFITC (FD4) permeability assay. Caco‐2 cell monolayers were grown in trans‐well inserts and treated with L. casei strains (LbcWT, LbcV, LbcInl AB or LbcLAP) for 2, 4, 16 and 24 h, before their infection with L. monocytogenes (Lm) for 2 h. TEER measurements before and after exposure to L. monocytogenes treatment alone were 268.9 ± 2.3 and 224.5 ± 4.7, respectively, with a 16.5% change. Values are averages of two experiments analysed in triplicate. Per cent TEER reduction was calculated as per Koo et al. (Koo et al., 2012) as 1 – TEER after/TEER before ×100.B. FD4 recovery after Lm was 2.76 ± 0.03%. Values are averages of three independent experiments performed in triplicates. For each time point, bars marked with different letters (a, b, c, d) indicate significant difference at P < 0.05.
We also measured the FD4 (paracellular marker) permeability through the epithelial barrier in a trans‐well set‐up to assess the epithelial barrier integrity. When the Caco‐2 cells were only infected with L. monocytogenes for 2 h, 2.76% of the FD4 was recovered at the basal side (Fig. 7B). The FD4 level decreased to 1.3% when Caco‐2 cells were pre‐exposed to LbcInlAB, and 2.1% when pre‐exposed to LbcWT for 1 h. A similar trend was observed at 4, 16 and 24 h. As a positive control, LbcLAP showed the highest protection against L. monocytogenes‐mediated epithelial barrier disruption showing FD4 permeability of only 0.1–0.3%. Nevertheless, these results show that LbcInlAB can prevent epithelial barrier disruption from L. monocytogenes infection much greater than the LbcWT.
Discussion
Most pathogens initiate infection of their host through the interaction of specific receptors using adhesive molecules on their surfaces (Kline et al., 2009). Beneficial bacteria (probiotics) prevent pathogen colonization by virtue of occupying the host cell surface receptors (Ryan and Bhunia, 2017; Jayashree et al., 2018). Therefore, the development of strategies to prevent pathogen interaction with the host provides a logical and effective intervention step. This can be achieved through expression of the virulence genes coding for molecules that bind to host cell receptors, in probiotic bacteria (Steidler, 2003; Paton et al., 2010; Aguilar et al., 2011; Kajikawa et al., 2011; Amalaradjou and Bhunia, 2013; Wolfe et al., 2017).
Listeria monocytogenes is responsible for a fatal infection in immunocompromised population, and pathogenesis depends on its ability to adhere and invade host cells in the gastrointestinal tract (Nikitas et al., 2011; Drolia et al., 2018; Drolia and Bhunia, 2019). Hence, blocking of adhesion and invasion events would be a logical robust option for preventing L. monocytogenes infection through its targeted inactivation (Amalaradjou and Bhunia, 2013; O'Toole et al., 2017). InlA and InlB are considered major invasion proteins required for L. monocytogenes adhesion and invasion into host cells (Radoshevich and Cossart, 2018). In this study, we successfully expressed InlA and InlB into L. casei (LbcInlAB) (Fig. 1) to prevent L. monocytogenes interaction with an intestinal cell line. Molecular weight of InlB in LbcInlAB in the cell wall fraction was found to be slightly higher (~80 kDa) than the actual MW in L. monocytogenes WT (Lm) (Fig. 1, Fig. S1) possibly because of coexpression of InlB (67 kDa) with the PrtP (PII‐type Proteinase) anchor with cell wall (117 aa = 12.87 kDa) (Maassen et al., 1999), while the MW of InlA remained the same in LbcInlAB because it possibly employed its own LPXTG motif to anchor the cell wall peptidoglycan (Bierne and Cossart, 2007). Often, the expression of new genes in a heterologous strain can result in changes in the growth and physiology of the recombinant strain (Ramos et al., 2004; Li et al., 2016). The growth rates of LbcInlAB and LbcWT were similar (Fig. 2), suggesting that expression of the extra genes by the recombinant L. casei did not affect its growth. This is a desirable outcome as it indicates that growth and potential consequent colonization of the recombinant would be comparable to those of the parental strain.
Expression of InlAB in LbcInlAB strain enhanced its ability to adhere, invade and translocate across the epithelial cell barrier. Increased adhesion of LbcInlAB to epithelial cells (Fig. 3) is highly desirable for its optimal functionality (Candela et al., 2008; Duary et al., 2011) and for creating a barrier for pathogen interaction with the host cells (Lee and Puong, 2002; Koo et al., 2012). LbcInlAB strain exhibited higher invasion and paracellular translocation through the epithelial barrier than LbcWT and LbcLAP (Fig. 3). The lower invasion by LbcLAP was expected since LAP is not involved in intracellular invasion (Burkholder and Bhunia, 2010). These findings, in part, corroborate with the previous studies where InlA‐expressing L. lactis was able to invade enterocytes efficiently (Guimaraes et al., 2005; Innocentin et al., 2009; De Azevedo et al., 2015) by transcytosis (Nikitas et al., 2011; Drolia and Bhunia, 2019). The major concern remains if such a strain could cross the epithelial barrier to spread systemically and consequently cause undesirable effects such as bacteraemia or septicaemia (Didari et al., 2014). L. casei is a widely used non‐pathogenic probiotic strain (Galdeano and Perdigon, 2006; Amalaradjou and Bhunia, 2012) lacking other virulence factors required for the systemic spread. It is thus anticipated that its recombinant strain expressing only InlAB will be cleared by immune cells rapidly from lamina propria. Previous studies indicated that some lactobacilli spontaneously translocate across the gut barrier; however, they were cleared within a short period by the host immune system, even when administered in higher dosages (Pavan et al., 2003; Liong, 2008). However, since these studies assessed natural infections using unmodified commensals, the safety of our recombinant strain will have to be tested and confirmed using in vivo experiments.
Others (Gueimonde et al., 2006; Collado et al., 2007) revealed that the degrees of probiotic strain adhesion and of its competitive adhesion, inhibition and/or displacement of the pathogen are not proportional. Therefore, adhesion of the probiotic should always be investigated simultaneously with its ability to reduce the adhesion of the pathogen to the same cells. Lee et al. (2003) reported that when incubated together, lactobacilli were able to compete with eight pathogens for adhesion to Caco‐2 cells. However, Collado et al. (2007) found that co‐incubation of probiotics and pathogens resulted in an increase in the adhesion of some pathogens. In our study, adhesion of L. monocytogenes to Caco‐2 cells was similar in the presence or absence of LbcWT, indicating the limitation of this wild‐type strain to compete with this pathogen for the adhesion site on the cells (Fig. 4). Conversely, we found that both co‐incubation with and pre‐exposure to the recombinant LbcInlAB and LbcLAP significantly decreased L. monocytogenes adhesion, findings similar to previous reports (Lee and Puong, 2002; Jankowska et al., 2008; Koo et al., 2012). Furthermore, all L. casei strains were unable to displace L. monocytogenes already attached to Caco‐2 cell monolayer, similar to previously published studies (Lee et al., 2003; Candela et al., 2008; Koo et al., 2012). Our results suggest that the recombinant L. casei will be effective as a prophylactic rather than a therapeutic intervention.
Next, we examined whether prolonged exposure to L. casei strains would offer higher protection against L. monocytogenes infection. A 16–24 h pre‐exposure to LbcInlAB showed the highest anti‐listeria effect for all three stages of infection modalities: adhesion, invasion and translocation (Fig. 5). The anti‐adhesive and anti‐invasive activities of LbcInlAB can be explained by its preoccupation of E‐cadherin or c‐Met receptors, which prevents L. monocytogenes adhesion and invasion by physical hindrance. Likewise, reduction in L. monocytogenes transcellular translocation is the consequential result of inhibition of its adhesion to the host cell receptor by LbcInlAB. LbcLAP also showed reduced L. monocytogenes translocation, which could be attributed to probiotic‐induced physical hindrance and maintenance of tight junction integrity thus preventing pathogen passage (Pagnini et al., 2010; Bron et al., 2017). Indeed, both LbcInlAB and LbcLAP were able to prevent L. monocytogenes‐mediated epithelial barrier dysfunction and helped maintain epithelial barrier integrity since dextran (paracellular marker) movement was significantly reduced in the Caco‐2 monolayer from apical to the basal compartment in the trans‐well set‐up (Fig. 7).
In conclusion, expression of key virulence genes by probiotic strains offers an alternative strategy with potential for targeted control of L. monocytogenes infection. LAP‐expressing probiotic provided protection against infection in our previous in vitro study (Koo et al., 2012). In this study, we also show that expression of InlAB by L. casei can also provide protection against infection in vitro. Therefore, recombinant Lactobacillus strains expressing different virulence genes of L. monocytogenes can be targeted at different stages of its infection cycle such as adhesion, invasion and translocation. These recombinant strains will be effective as a prophylactic rather than therapeutic intervention for pathogens and for conferring general health beneficial effects.
Although the findings reported in this paper for use of recombinant L. casei strain expressing inlA and inlB for control of L. monocytogenes infection are promising, additional in vivo studies are required to determine its suitability for direct application in humans. Such in vivo trials should determine the persistence of the recombinant strain, the stability of the plasmid and expression of foreign genes in the absence of antibiotic pressure and presence of glucose, demonstrate L. monocytogenes disease reduction and address safety issues relating to its applications.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. L. monocytogenes F4244 (serovar 4b, clinical epidemic strain) was cultured in tryptone soy broth supplemented with 0.6% yeast extract (TSB‐YE) or brain heart infusion (BHI) broth at 37°C for 18 h. The vector pLP401‐T (Pouwels et al., 2001) containing the pAmy promoter was used for the expression of InlAB in L. casei ATCC344. E. coli DH5α with vector was grown in Luria–Bertani (LB) broth supplemented with 50 μg ml−1 ampicillin. Wild‐type L. casei (LbcWT) was grown in de Man Rogosa and Sharpe (MRS) broth, while the L. casei carrying the pLP401T empty vector (LbcV) and recombinant LbcInlAB and LbcLAP (unpublished) strains were grown anaerobically at 37°C for 16 h in MRS broth containing 2 μg ml−1 erythromycin. To induce expression of InlAB and LAP by recombinant L. casei strains, the recombinants were grown in modified MRS broth (1% w/v protease peptone, 0.5% w/v yeast extract, 0.2% w/v meat extract, 0.1% v/v Tween‐80, 37 mM C2H3NaO2, 0.8 mM MgSO4, 0.24 mM MnSO4, 8.8 mM C6H14N2O7 in 0.1 M potassium phosphate buffer, pH 7.0) supplemented with mannitol (1% w/v) (Koo et al., 2012) at 37°C for 16 h.
Table 1.
Bacterial strains and plasmids
| Bacterial/plasmids | Strains | Description | Source |
|---|---|---|---|
| Listeria monocytogenes | F4244 | Wild type, serotype 4b, epidemic strain | Our collection |
| Lactobacillus casei | ATCC344 | Wild type | ATCC |
| Escherichia coli | DH5α | Wild type | Our collection |
| L. casei | AKB904 (LbcLAP) | L. casei expressing Listeria adhesion protein of L. monocytogenes F4244 (EmR 2 μg ml−1) | Our laboratory |
| L. casei | AKB908 (LbcInlAB) | L. casei expressing InlAB of L. monocytogenes F4244 (EmR 2 μg ml−1) | This study |
| L. casei | AKB909 (LbcV) | Vector control; L. casei carrying pLP401T plasmid without an insert (EmR 2 μg ml−1) | This study |
| Plasmids | |||
| pLP401T | Lactobacillus expression vector, (AmR 50 μg ml−1 and EmR 2 μg ml−1) | (Pouwels et al., 2001) | |
| pLP401‐InlAB | pLP401T carrying inlAB of L. monocytogenes F4244 | This study | |
Construction of Lactobacillus casei harbouring Internalin A and B (inlAB) operons
The construction of the recombinant L. casei was done according to the methods described before (Maassen et al., 1999; Koo et al., 2012) with minor modifications. Briefly, chromosomal DNA of L. monocytogenes F4244 was extracted and inlAB operon was amplified with PCR using the primers: InlABExp‐F (NotI): TAGCGGCCGCAACTATTGAAAAAGGAGTGTATATAGTG and InlABExp‐R (XhoI): GTCTCGAGTTTCTGTGCCCTTAAATTAGC (Integrated DNA Technologies, Coralville, IA, USA) with an expected amplicon size of 4371 bp. The plasmid (pLP401T) containing the pAmy promoter was used for expression of inlAB in the probiotic L. casei ATCC344. InlA is anchored into L. casei through its own LPXTG motif while InlB has a GW motif, but it is possibly also fused to the C‐terminal region of PrtP containing an LPXTG motif (Fig. S1). The plasmid and the purified DNA were digested using the restriction enzymes NotI and XhoI (NEB) and subsequently ligated (T4 DNA ligase). The product of ligation was then designated pLP401T‐InlAB, which was used for electroporation into competent cells of E. coli and L. casei (Koo et al., 2012). Electroporated E. coli and L. casei cells were then incubated at 37°C for 1 h and 3 h respectively. Transformants harbouring pLP401T‐inlAB were subsequently selected on LB agar containing 50 μg ml−1 ampicillin and MRS agar containing 2 μg ml−1 erythromycin for E. coli and L. casei respectively. The plates were incubated at 37°C overnight for E. coli and 72 h for L. casei. Confirmation of the identity of inlA and inlB genes in LbcInlAB strain was done using PCR and sequencing.
Analysis of InlAB expression by L. casei
The overnight (18 h) grown cultures of L. monocytogenes, LbcWT, LbcV and LbcInlAB were centrifuged (7000 g, 10 min, 4°C), and proteins were harvested from the supernatant, cell wall and intracellular fractions as before (Burkholder et al., 2009; Koo et al., 2012). Equal amounts of proteins (10 μg) from each fraction were separated using SDS‐PAGE(7.5%). Protein bands were transferred to an Immobilon‐P membrane (Millipore, Billerica, MA, USA) and then immunoprobed with anti‐InlA antibody mAb‐2D12 (1.0 mg ml−1) (Mendonca et al., 2012) or anti‐InlB pAb‐404 (1:1000) (Lathrop et al., 2008) and reacted with horseradish peroxidase‐coupled anti‐mouse or anti‐rabbit secondary antibodies (Jackson Immuno Research, West Grove, PA) at 37°C for 1 h. The membranes were developed with an enhanced chemiluminescence kit (Thermo Fisher, Canoga Park, CA, USA).
Additionally, expression of InlA and InlB in the recombinant L. casei strains was determined by immunofluorescence staining. Overnight cultures were washed twice in PBS and incubated with the anti‐InlA mAb 2D12 and anti‐InlB pAb 404 (diluted 1:500 in PBS) at 37°C for 1 h. Subsequently, cells were treated with Alexa‐conjugated anti‐mouse IgG Fab2 Alexa Flour R555 and anti‐rabbit IgG Fab2 Alexa Flour R488 (Cell Signaling, Danvers, MA, USA) secondary antibodies diluted 1:500 in PBS and incubated in the dark at 37°C for 1 h. Between the treatments, cells were washed at least four times with 0.5% PBS‐Tween‐20. The cells were viewed under a fluorescence microscope (Leica, Wetzlar, Germany) equipped with SPOT Software version 4.6.4.2 (Diagnostic Instruments, Sterling Heights, MI, USA).
Effect of InlAB expression on the growth of L. casei
The growth curve of LbcWT, LbcV and LbcInlAB strains in MRS broth was conducted for 24 h by measuring the cell density (OD600 nm) in a spectrophotometer (Beckman DU80) and by plate counting. At each time point, the OD reading was taken, and culture (1 ml) was used for plating. LbcWT cells were counted on MRS agar, while those of LbcV and recombinant LbcInlAB were counted on MRS agar containing 2 μg ml−1 erythromycin grown anaerobically at 37°C for 48 h. This experiment was performed twice in triplicates. Additionally, the morphologies of overnight L. casei cultures were examined using phase‐contrast micrographs (Leica).
Recombinant L. casei strain adhesion and invasion into Caco‐2 cells
Caco‐2 cell culturing: Human colon carcinoma cell line Caco‐2 (HTB37; American Type Culture Collection, Manassas, VA, USA) was cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose (HyClone™; GE, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA) (D10F). The cells were grown in flasks (Greiner Bio‐One) for up to 10–12 days or until differentiated and then trypsinized (Gaillard and Finlay, 1996). They were then seeded in 12‐well plates at a density of 1 × 105 cells/well and incubated at 37°C in the presence of 7% CO2 for 10–12 days until they were differentiated and reached confluence (106 cells/well).
Adhesion and invasion assays: Overnight (18 h) grown bacterial cultures were washed twice with PBS, adjusted to OD 600 = 1 and were suspended in D10F to a final concentration of 1 × 107 CFU ml−1 to achieve a multiplicity of infection (MOI) or multiplicity of exposure (MOE), 10. The Caco‐2 cell monolayer was washed three times using DMEM, and then exposed separately to the L. casei strains (LbcWT, LbcV, LbcInlAB or LbcLAP) and L. monocytogenes and incubated for 1 h at 37°C in a gas atmosphere with 5% CO2 (Koo et al., 2012). To enumerate bacterial adhesion, the Caco‐2 cell monolayer was first washed thrice using DMEM and then treated with 0.1% Triton X‐100 (37°C, 10 min). For the invasion assay, the monolayers were exposed to L. monocytogenes and L. casei and then washed as performed in the adhesion assay, and treated with gentamicin (50 μg ml−1, 1 h) and with 0.1% Triton X‐100 (37°C, 10 min). The lysed cell suspensions from both adhesion and invasion experiments were serially diluted in PBS before plating on MRS, supplemented with erythromycin (2 μg ml−1) and modified Oxford (MOX) agar for LbcWT, recombinant L. casei and L. monocytogenes respectively. All the plates were incubated at 37°C for 24–48 h before bacterial enumeration.
Determination of L. monocytogenes exclusion mode by the recombinant L. casei strains
The competitive exclusion assay was performed as before (Koo et al., 2012) with minor modifications. Bacterial cultures were prepared as above and were suspended in D10F to a final concentration of 1 × 107 CFU ml−1. For competitive adhesion, L. monocytogenes was co‐inoculated with each of the L. casei strains (LbcWT, LbcV, LbcInlAB or LbcLAP) to Caco‐2 cell monolayer (MOI, 10) and incubated for 1 h. Adherent bacteria were enumerated as above.
In the inhibition of adhesion assay, the Caco‐2 monolayers were first inoculated with each L. casei strain (MOE, 10) and incubated for 1 h, and washed to remove unbound bacteria using DMEM. L. monocytogenes was then added to the wells, and plates were incubated for 1 h, followed by an enumeration of adherent bacteria by plating. For displacement of adhesion, Caco‐2 cells were first inoculated with L. monocytogenes (MOI, 10) and incubated for 1 h, and washed to remove unbound bacteria. L. casei strains were then added to the wells, and plates were incubated for 1 h. Adhered bacteria were released by treatment with 0.1% Triton X‐100 (37°C, 10 min) and plated on MRS, supplemented with 2 μg ml−1 of erythromycin and MOX agar plates for enumeration of LbcWT, recombinant L. casei and L. monocytogenes respectively.
Inhibition of L. monocytogenes adhesion and invasion by L. casei strains
The Caco‐2 cell monolayers were washed and then exposed to the L. casei strains (MOE, 10) for 1, 4, 16 and 24 h at 37°C in the humidified incubator with 5% CO2. Excess medium in the wells containing unbound L. casei was removed and replaced with 500 μl of L. monocytogenes suspended in D10F (MOI, 10), and the plates were incubated for 1 h at 37°C with 5% CO2. The adherent bacteria were enumerated by plating as above.
For inhibition of L. monocytogenes invasion, the Caco‐2 cell monolayers were exposed to each L. casei strain (MOE, 10) for 1, 4, 16 and 24 h at 37°C with 5% CO2. Excess L. casei cells were removed and replaced with 500 μl of L. monocytogenes suspended in D10F (MOI, 10) and then incubated at 37°C with 5% CO2 for 1 h. The cell monolayers were washed, treated with gentamicin (50 μg ml−1) for 1 h and determined for invading bacteria by plating.
Caco‐2 cell cytotoxicity
Caco‐2 cell cytotoxicity induced by L. monocytogenes after pre‐exposure to L. casei strains over time was determined by using the lactate dehydrogenase (LDH) release assay as previously described (Koo et al., 2012).
Transcellular translocation of L. casei strains and subsequent inhibition of L. monocytogenes transepithelial translocation by recombinant L. casei
The Caco‐2 cells were grown in 12‐well trans‐well filter inserts (3 μm pore size) for 20–25 days to reach confluence (Burkholder and Bhunia, 2010; Drolia et al., 2018). Transepithelial electrical resistance of Caco‐2 cells was quantified using the Millicell ERS (Millipore), and a TEER value of more than 200 Ω cm−2 was used for all the experiments. For determining baseline translocation by L. casei strains or L. monocytogenes, the Caco‐2 cells were washed, and then, the bacteria were added (MOI, 10) separately to the apical side of the trans‐well at 37°C with 5% CO2 for 2 h. The liquid from the basal well was collected, serially diluted in PBS and then plated for the enumeration of bacterial cells (CFU ml−1).
For the inhibition of L. monocytogenes translocation, L. casei cells were first added to the apical wells (MOE, 10) and incubated for 1, 4, 16 and 24 h at 37°C with 5% CO2 and L. casei counts in the basal wells were determined by plating on MRS agar. Subsequently, excess L. casei cells were removed from the apical well, and replaced with 500 μl of L. monocytogenes (MOI, 10) and then incubated at 37°C with 5% CO2 for 2 h. L. monocytogenes counts in the basal wells were determined by plating on MOX agar plates.
Epithelial tight junction integrity analysis
Quantification of TEER of Caco‐2 cells before and after the exposure to the bacteria was performed using a Millicell ERS system (Millipore) as described before (Burkholder and Bhunia, 2010). Furthermore, the integrity of the tight junctions between Caco‐2 cells was determined by measuring FD4 permeability in a spectrofluorometer (Burkholder and Bhunia, 2010; Koo et al., 2012).
Statistical analysis
All data were analysed using Prism 7 software (GraphPad Software Inc., San Diego, CA, USA), and significance was assigned at P < 0.05. Where appropriate, Tukey's multiple comparisons test, with P < 0.05 as a significant difference, was used to identify statistically significant differences.
Conflict of interest
None declared.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Supporting information
Figure S1. (A) Plasmid map (14.2 kb) of InlAB expression vector pLP401T (9.8 kb)‐InlAB (4.4 kb) (Pouwels et al., 2001). Ery, erythromycin resistance gene; Amp, ampicillin resistance gene; Ori+ = origin of replication of E. coli, Ori‐ = origin of replication of Lactobacillus; InlAB, Internalin A and B; Pamy, a‐amylase promoter gene; ssAmy, secretion signal (36 aa) and the N‐terminus (26 aa) of a‐amylase gene; Anchor, cell wall anchor region (117 aa) of the prtP (PII‐type Proteinase) gene of L. casei; Tcbh, transcription terminator of the cbh (conjugated bile acid hydrolase) gene; Rep, repA gene. (B) Western blot showing expression of Internalin (InlA) and InlB in the recombinant L. casei strains (LbcInlAB−1, LbcInlAB−2, LbcInlAB−3, LbcWT and LbcV in the different cellular fractions (supernatant, cell wall and intracellular) and L. monocytogenes F4244 (Lm). Molecular weight of InlB in LbcInlAB was slightly higher (~80 kDa) than the actual MW in L. monocytogenes WT (Lm) in the cell wall fraction possibly because of co‐expression of InlB (67 kDa) with the PrtP anchor (117 aa = 12.87 kDa) while the MW of InlA remained the same because it is using LPXTG motif to anchor the cell wall.
Acknowledgements
The research was supported in part by the funds from The National Research Foundation (Grant No. 102107) with the University of Pretoria for providing financial aid to MM to conduct research at Purdue University, the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch accession no. 1016249), and USDA Agricultural Research Service, under Agreement No. 59‐8072‐6‐001. Any opinions, findings, conclusion or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture. Dr. Mike Miller at University of Illinois is acknowledged for kindly providing the Lactobacillus casei ATCC344 strain and Chad Coakley and Rishi Drolia for critical review of the manuscript.
Microbial Biotechnology (2019) 12(4), 715–729
Funding Information
The research was supported in part by the funds from The National Research Foundation (Grant No. 102107) with the University of Pretoria for providing financial aid to MM to conduct research at Purdue University, the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch accession no. 1016249), and USDA Agricultural Research Service, under Agreement No. 59‐8072‐6‐001.
References
- Aguilar, C. , Vanegas, C. , and Klotz, B. (2011) Antagonistic effect of Lactobacillus strains against Escherichia coli and Listeria monocytogenes in milk. J Dairy Res 78: 136–143. [DOI] [PubMed] [Google Scholar]
- Allam, M. , Tau, N. , Smouse, S.L. , Mtshali, P.S. , Mnyameni, F. , Khumalo, Z.T. , et al (2018) Whole‐genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc 6: e00538‐00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalaradjou, M.A.R. , and Bhunia, A.K. (2012) Modern approaches in probiotics research to control foodborne pathogens. Adv Food Nutr Res 67: 185–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalaradjou, M.A.R. , and Bhunia, A.K. (2013) Bioengineered probiotics, a strategic approach to control enteric infections. Bioengineered 4: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.W. , do Nascimento, N.C. and Bhunia, A.K. (2017) Genome sequence of Listeria monocytogenes strain F4244, a 4b serotype. Genome Announc 5, e01324‐01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, T.M. , Fernández, J. , Navasa, M. , Vila, J. , and Rodés, J. (2002) Failure of Lactobacillus spp. to prevent bacterial translocation in a rat model of experimental cirrhosis. J Hepatol 36: 501–506. [DOI] [PubMed] [Google Scholar]
- Behnsen, J. , Deriu, E. , Sassone‐Corsi, M. , and Raffatellu, M. (2013) Probiotics: properties, examples, and specific applications. Cold Spring Harbor Perspect Biol 3: a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, H. , and Cossart, P. (2007) Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev 71: 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, L. , Dramsi, S. , Dehoux, P. , Bierne, H. , Lindahl, G. , and Cossart, P. (1997) InIB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol 25: 285–294. [DOI] [PubMed] [Google Scholar]
- Bron, P.A. , Kleerebezem, M. , Brummer, R.‐J. , Cani, P.D. , Mercenier, A. , MacDonald, T.T. , et al (2017) Can probiotics modulate human disease by impacting intestinal barrier function? Brit J Nutr 117: 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccato, S. , Maione, D. , Rinaudo, C.D. , Volpini, G. , Taddei, A.R. , Rosini, R. , et al (2006) Use of Lactococcus lactis expressing pili from group B Streptococcus as a broad‐coverage vaccine against streptococcal disease. J Infect Dis 194: 331–340. [DOI] [PubMed] [Google Scholar]
- Burkholder, K.M. , and Bhunia, A.K. (2010) Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp 60. Infect Immun 78: 5062–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder, K.M. , Kim, K.‐P. , Mishra, K. , Medina, S. , Hahm, B.‐K. , Kim, H. , and Bhunia, A.K. (2009) Expression of LAP, a SecA2‐dependent secretory protein, is induced under anaerobic environment. Microbes Infect 11: 859–867. [DOI] [PubMed] [Google Scholar]
- Camejo, A. , Carvalho, F. , Reis, O. , Leitao, E. , Sousa, S. , and Cabanes, D. (2011) The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2: 379–394. [DOI] [PubMed] [Google Scholar]
- Candela, M. , Perna, F. , Carnevali, P. , Vitali, B. , Ciati, R. , Gionchetti, P. , et al (2008) Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL‐8 production. Int J Food Microbiol 125: 286–292. [DOI] [PubMed] [Google Scholar]
- do Carmo, F.L.R. , Rabah, H. , De Oliveira Carvalho, R.D. , Gaucher, F. , Cordeiro, B.F. , da Silva, S.H. , et al (2018) Extractable bacterial surface proteins in probiotic–host interaction. Front Microbiol 9, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, S. , Nagai, T. , Hayashi, T. , Baba, Y. , Nagai, S. , and Koyasu, S. (2011) Listerial invasion protein internalin B promotes entry into ileal Peyer's patches in vivo. Microbiol Immunol 55: 123–129. [DOI] [PubMed] [Google Scholar]
- Collado, M. , Meriluoto, J. , and Salminen, S. (2007) Development of new probiotics by strain combinations: is it possible to improve the adhesion to intestinal mucus? J Dairy Sci 90: 2710–2716. [DOI] [PubMed] [Google Scholar]
- Corr, S. , Li, Y. , Riedel, C.U. , O'Toole, P.W. , Hill, C. , and Gahan, C.G.M. (2007) Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Nat Acad Sci USA 104: 7617–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuprynski, C.J. (2005) Listeria monocytogenes: silage, sandwiches and science. Anim Health Res Rev 6: 211–217. [DOI] [PubMed] [Google Scholar]
- De Azevedo, M. , Santos Rocha, C. , Pereira, V. , De Junior, A.D. , De Sousa, C.S. , Azevedo, V. , et al (2015) Prospective uses of recombinant Lactococcus lactis expressing both listeriolysin O and mutated internalin A from Listeria monocytogenes as a tool for DNA vaccination. Genet Mol Res 14: 18485–18493. [DOI] [PubMed] [Google Scholar]
- Didari, T. , Solki, S. , Mozaffari, S. , Nikfar, S. , and Abdollahi, M. (2014) A systematic review of the safety of probiotics. Expert Opin Drug Safety 13: 227–239. [DOI] [PubMed] [Google Scholar]
- Disson, O. , and Lecuit, M. (2013) In vitro and in vivo models to study human listeriosis: mind the gap. Microbes Infect 15: 971–980. [DOI] [PubMed] [Google Scholar]
- Drolia, R. and Bhunia, A.K. (2019) Crossing the intestinal barrier via Listeria adhesion protein and internalin A. Trends Microbiol . [Epub ahead of print]. 10.1016/j.tim.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Drolia, R. , Tenguria, S. , Durkes, A.C. , Turner, J.R. , and Bhunia, A.K. (2018) Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 23: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duary, R.K. , Rajput, Y.S. , Batish, V.K. , and Grover, S. (2011) Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Ind J Med Res 134: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard, J.L. , and Finlay, B.B. (1996) Effect of cell polarization and differentiation on entry of Listeria monocytogenes into the enterocyte‐like Caco‐2 cell line. Infect Immun 64: 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdeano, C.M. , and Perdigon, G. (2006) The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vacc Immunol 13: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, P. , Halvorsen, E.M. , Ammendolia, D.A. , Mor‐Vaknin, N. , O'Riordan, M.X. , Brumell, J.H. , et al (2018) Invasion of the brain by Listeria monocytogenes is mediated by InlF and host cell vimentin. mBio 9: e00160‐00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueimonde, M. , Jalonen, L. , He, F. , Hiramatsu, M. and Salminen, S. (2006) Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res Int 39: 467–471. [Google Scholar]
- Guimaraes, V.D. , Gabriel, J.E. , Lefevre, F. , Cabanes, D. , Gruss, A. , Cossart, P. , et al (2005) Internalin‐expressing Lactococcus lactis is able to invade small intestine of guinea pigs and deliver DNA into mammalian epithelial cells. Microbes Infect 7: 836–844. [DOI] [PubMed] [Google Scholar]
- Innocentin, S. , Guimaraes, V. , Miyoshi, A. , Azevedo, V. , Langella, P. , Chatel, J.M. , and Lefevre, F. (2009) Lactococcus lactis expressing either Staphylococcus aureus fibronectin‐binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl Environ Microbiol 75: 4870–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacouton, E. , Chain, F. , Sokol, H. , Langella, P. and Bermúdez‐Humarán, L.G. (2017) Probiotic strain Lactobacillus casei BL23 prevents colitis‐associated colorectal cancer. Front Immunol 8: 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesan, B. , Koo, O.K. , Kim, K.P. , Burkholder, K.M. , Mishra, K.K. , Aroonnual, A. , and Bhunia, A.K. (2010) LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte‐like Caco‐2 cells only in pathogenic species. Microbiology 156: 2782–2795. [DOI] [PubMed] [Google Scholar]
- Jagadeesan, B. , Fleishman Littlejohn, A.E. , Amalaradjou, M.A.R. , Singh, A.K. , Mishra, K.K. , La, D. , et al (2011) N‐Terminal Gly224 ‐ Gly411 domain in Listeria adhesion protein interacts with host receptor Hsp60. PLoS ONE 6: e20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska, A. , Laubitz, D. , Antushevich, H. , Zabielski, R. and Grzesiuk, E. (2008) Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco‐2 cells. J Biomed Biotechnol 2008: 357964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayashree, S. , Karthikeyan, R. , Nithyalakshmi, S. , Ranjani, J. , Gunasekaran, P. , and Rajendhran, J. (2018) Anti‐adhesion property of the potential probiotic strain Lactobacillus fermentum 8711 against methicillin‐resistant Staphylococcus aureus (MRSA). Front Microbiol 9: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, A. , Satoh, E. , Leer, R.J. , Yamamoto, S. , and Igimi, S. (2007) Intragastric immunization with recombinant Lactobacillus casei expressing flagellar antigen confers antibody‐independent protective immunity against Salmonella enterica serovar Enteritidis. Vaccine 25: 3599–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, A. , Nordone, S.K. , Zhang, L. , Stoeker, L.L. , LaVoy, A.S. , Klaenhammer, T.R. , and Dean, G.A. (2011) Dissimilar properties of two recombinant Lactobacillus acidophilus strains displaying Salmonella FliC with different anchoring motifs. Appl Environ Microbiol 77: 6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, K.A. , Fälker, S. , Dahlberg, S. , Normark, S. , and Henriques‐Normark, B. (2009) Bacterial adhesins in host‐microbe interactions. Cell Host Microbe 5: 580–592. [DOI] [PubMed] [Google Scholar]
- Koo, O.K. , Amalaradjou, M.A.R. , and Bhunia, A.K. (2012) Recombinant probiotic expressing Listeria adhesion protein attenuates Listeria monocytogenes virulence in vitro. PLoS ONE 7: e29277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop, A.A. , Banada, P.P. , and Bhunia, A.K. (2008) Differential expression of InlB and ActA in Listeria monocytogenes in selective and nonselective enrichment broths. J Appl Microbiol 104: 627–639. [DOI] [PubMed] [Google Scholar]
- Lee, Y.K. , and Puong, K.Y. (2002) Competition for adhesion between probiotics and human gastrointestinal pathogens in the presence of carbohydrate. Brit J Nutr 88: S101–S108. [DOI] [PubMed] [Google Scholar]
- Lee, Y.‐K. , Puong, K.‐Y. , Ouwehand, A.C. , and Salminen, S. (2003) Displacement of bacterial pathogens from mucus and Caco‐2 cell surface by lactobacilli. J Med Microbiol 52: 925–930. [DOI] [PubMed] [Google Scholar]
- Lenoir, M. , Del Carmen, S. , Cortes‐Perez, N.G. , Lozano‐Ojalvo, D. , Muñoz‐Provencio, D. , Chain, F. , et al (2016) Lactobacillus casei BL23 regulates Treg and Th17 T‐cell populations and reduces DMH‐associated colorectal cancer. J Gastroenterol 51: 862–873. [DOI] [PubMed] [Google Scholar]
- Li, C. , Sun, J.W. , Zhang, G.F. , and Liu, L.B. (2016) Effect of the absence of the CcpA gene on growth, metabolic production, and stress tolerance in Lactobacillus delbrueckii ssp. bulgaricus . J Dairy Sci 99: 104–111. [DOI] [PubMed] [Google Scholar]
- Liong, M.T. (2008) Safety of probiotics: translocation and infection. Nutr Rev 66: 192–202. [DOI] [PubMed] [Google Scholar]
- Maassen, C.B. , Laman, J.D. , den Bak‐Glashouwer, M.J. , Tielen, F.J. , van Holten‐Neelen, J.C. , Hoogteijling, L. , et al (1999) Instruments for oral disease‐intervention strategies: recombinant Lactobacillus casei expressing tetanus toxin fragment C for vaccination or myelin proteins for oral tolerance induction in multiple sclerosis. Vaccine 17: 2117–2128. [DOI] [PubMed] [Google Scholar]
- McCarthy, J. , O'mahony, L. , O'callaghan, L. , Sheil, B. , Vaughan, E.E. , Fitzsimons, N. , et al (2003) Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut 52: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca, M. , Conrad, N. , Conceicao, F. , Moreira, A. , da Silva, W. , Aleixo, J. and Bhunia, A. (2012) Highly specific fiber optic immunosensor coupled with immunomagnetic separation for detection of low levels of Listeria monocytogenes and L. ivanovii . BMC Microbiol 12: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud, J. , Ohayon, H. , Gounon, P. , Mege, R.M. , and Cossart, P. (1996) E‐cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84: 923–932. [DOI] [PubMed] [Google Scholar]
- Nikitas, G. , Deschamps, C. , Disson, O. , Niault, T. , Cossart, P. , and Lecuit, M. (2011) Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E‐cadherin. J Exp Med 208: 2263–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Noordhout, C.M. , Devleesschauwer, B. , Angulo, F.J. , Verbeke, G. , Haagsma, J. , Kirk, M. , et al (2014) The global burden of listeriosis: a systematic review and meta‐analysis. Lancet Infect Dis 14: 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, P.W. , Marchesi, J.R. , and Hill, C. (2017) Next‐generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol 2: 17057. [DOI] [PubMed] [Google Scholar]
- Pagnini, C. , Saeed, R. , Bamias, G. , Arseneau, K.O. , Pizarro, T.T. , and Cominelli, F. (2010) Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci USA 107: 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiripally, V.K. , Westbrook, D.G. , Sunki, G.R. , and Bhunia, A.K. (1999) Surface protein p104 is involved in adhesion of Listeria monocytogenes to human intestinal cell line, Caco‐2. J Med Microbiol 48: 117–124. [DOI] [PubMed] [Google Scholar]
- Paton, A.W. , Morona, R. , and Paton, J.C. (2010) Bioengineered bugs expressing oligosaccharide receptor mimics: toxin‐binding probiotics for treatment and prevention of enteric infections. Bioengineered Bugs 1: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan, S. , Desreumaux, P. , and Mercenier, A. (2003) Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin Diagn Lab Immunol 10: 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy, D.A. , Chakraborty, T. , Goebel, W. , and Cossart, P. (1992) Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun 60: 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels, P.H. , Vriesema, A. , Martinez, B. , Tielen, F.J. , Seegers, J.F. , Leer, R.J. , et al (2001) Lactobacilli as vehicles for targeting antigens to mucosal tissues by surface exposition of foreign antigens. Methods Enzymol 336: 369–389. [DOI] [PubMed] [Google Scholar]
- Radoshevich, L. , and Cossart, P. (2018) Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 16: 32–46. [DOI] [PubMed] [Google Scholar]
- Ramos, A. , Neves, A.R. , Ventura, R. , Maycock, C. , Lopez, P. , and Santos, H. (2004) Effect of pyruvate kinase overproduction on glucose metabolism of Lactococcus lactis . Microbiology 150: 1103–1111. [DOI] [PubMed] [Google Scholar]
- Robbins, J.R. , Skrzypczynska, K.M. , Zeldovich, V.B. , Kapidzic, M. , and Bakardjiev, A.I. (2010) Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes . PLoS Pathog 6: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, V. and Bhunia, A.K. (2017) Mitigation of foodborne illnesses by probiotics In: Foodborne Pathogens. Gurtler J.B., Doyle M.P., and Kornacki J.L. (eds). Cham, Switzerland: Springer International Publishing AG, pp. 603–634. [Google Scholar]
- Scallan, E. , Hoekstra, R.M. , Angulo, F.J. , Tauxe, R.V. , Widdowson, M.A. , Roy, S.L. , et al (2011) Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, W.D. , Urbanke, C. , Ziehm, T. , Beier, V. , Machner, M.P. , Domann, E. , et al (2002) Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E‐cadherin. Cell 111: 825–836. [DOI] [PubMed] [Google Scholar]
- Schuchat, A. , Swaminathan, B. , and Broome, C.V. (1991) Epidemiology of human listeriosis. Clin Microbiol Rev 4: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Naujokas, M. , Park, M. , and Ireton, K. (2000) InlB‐dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103: 501–510. [DOI] [PubMed] [Google Scholar]
- Stavru, F. , Archambaud, C. , and Cossart, P. (2011) Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol Rev 240: 160–184. [DOI] [PubMed] [Google Scholar]
- Steidler, L. (2003) Genetically engineered probiotics. Best Pract Res Clin Gastroenterol 17: 861–876. [DOI] [PubMed] [Google Scholar]
- Touré, R. , Kheadr, E. , Lacroix, C. , Moroni, O. , and Fliss, I. (2003) Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes . J Appl Microbiol 95: 1058–1069. [DOI] [PubMed] [Google Scholar]
- Trost, M. , Wehmhöner, D. , Kärst, U. , Dieterich, G. , Wehland, J. , and Jänsch, L. (2005) Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5: 1544–1557. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan, M. , Rappuoli, R. , and Serruto, D. (2012) Recombinant bacterial vaccines. Curr Opin Immunol 24: 337–342. [DOI] [PubMed] [Google Scholar]
- Vazquez‐Boland, J.A. , Kuhn, M. , Berche, P. , Chakraborty, T. , Dominguez‐Bernal, G. , Goebel, W. , et al (2001) Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14: 584–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampler, J.L. , Kim, K.P. , Jaradat, Z. , and Bhunia, A.K. (2004) Heat shock protein 60 acts as a receptor for the Listeria adhesion protein in Caco‐2 cells. Infect Immun 72: 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J.M. , and Mercenier, A. (2008) Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol 6: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B. , Wiepz, G.J. , Schotzko, M. , Bondarenko, G.I. , Durning, M. , Simmons, H.A. , et al (2017) Acute fetal demise with first trimester maternal infection resulting from Listeria monocytogenes in a nonhuman primate model. mBio 8: e01938–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, A. , Takahashi, K. , Mori, Y. , Watanabe, S. , Hanamura, Y. , Sugiyama, T. , and Inoue, N. (2018) Peyer's patches as a portal for DNA delivery by Lactococcus lactis in vivo. Biol Pharmaceut Bull 41: 190–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Plasmid map (14.2 kb) of InlAB expression vector pLP401T (9.8 kb)‐InlAB (4.4 kb) (Pouwels et al., 2001). Ery, erythromycin resistance gene; Amp, ampicillin resistance gene; Ori+ = origin of replication of E. coli, Ori‐ = origin of replication of Lactobacillus; InlAB, Internalin A and B; Pamy, a‐amylase promoter gene; ssAmy, secretion signal (36 aa) and the N‐terminus (26 aa) of a‐amylase gene; Anchor, cell wall anchor region (117 aa) of the prtP (PII‐type Proteinase) gene of L. casei; Tcbh, transcription terminator of the cbh (conjugated bile acid hydrolase) gene; Rep, repA gene. (B) Western blot showing expression of Internalin (InlA) and InlB in the recombinant L. casei strains (LbcInlAB−1, LbcInlAB−2, LbcInlAB−3, LbcWT and LbcV in the different cellular fractions (supernatant, cell wall and intracellular) and L. monocytogenes F4244 (Lm). Molecular weight of InlB in LbcInlAB was slightly higher (~80 kDa) than the actual MW in L. monocytogenes WT (Lm) in the cell wall fraction possibly because of co‐expression of InlB (67 kDa) with the PrtP anchor (117 aa = 12.87 kDa) while the MW of InlA remained the same because it is using LPXTG motif to anchor the cell wall.


