Abstract
Expression of fat mass and obesity-associated gene (FTO) and ADP-ribosylation factor-like 15 (ARL15) in human islets is inversely correlated with HbA1c. However, their impact on insulin secretion is still ambiguous. Here in, we investigated the role of FTO and ARL15 using GRINCH (Glucose-Responsive Insulin-secreting C-peptide-modified Human proinsulin) clonal rat β-cells. GRINCH cells have inserted GFP into the human C-peptide insulin gene. Hence, secreted CpepGFP served to monitor insulin secretion. mRNA silencing of FTO in GRINCH cells showed a significant reduction in glucose but not depolarization-stimulated insulin secretion, whereas ARL15 silencing had no effect. A significant down-regulation of insulin mRNA was observed in FTO knockdown cells. Type-2 Diabetic islets revealed a reduced expression of FTO mRNA. In conclusion, our data suggest that fluorescent CpepGFP released from GRINCH cells may serve as a convenient marker for insulin secretion. Silencing of FTO expression, but not ARL15,inhibits insulin secretion by affecting metabolic signaling.
1. Introduction
The FTO gene has been identified as an obesity-susceptibility gene in multiple populations (Frayling et al., 2007; Dina et al., 2007; Scuteri et al., 2007; Scott et al., 2007; Peeters et al., 2008). The gene was first discovered in mice where a deletion of 1.6 Mbp in chromosome 8 was reported. Mice heterozygous for the deletion displayed a malformation of forelimbs (fused toes) (Peters et al., 1999), while homozygous mice died during mid-gestation of malformations of the head and face, with defect in the central nervous system (CNS) development and growth retardation. FTO acts as a 2-oxoglutarate (2OG)-dependent nucleic acid demethylase (Gerken et al., 2007), therefore, it is suggested that FTO might down-regulate genes involved in metabolism and lead to obesity.
FTO has been shown to play a vital role in obesity, appetiteand energy homeostasis (Church et al., 2009). The gene is ubiquitously expressed in the brain, pancreatic endocrine cells (β and α) and adipose tissue (Frayling et al., 2007; Gao et al., 2010; Stratigopoulos et al., 2008; Taneera et al., 2012). Its expression is highest in the hypothalamic region that regulates energy intake and expenditure (Poritsanos et al., 2011). Fasted mice exhibit a significant reduction of FTOexpression in the hypothalamic regions (Poritsanos et al., 2011). These data suggest that variation in FTO resulting in decreased expression or activity might provide a signal that promotes feeding and obesity. Moreover, inactivation of the FTO gene in mice was shown to protect from obesity, substantiating a central role of FTO in the control of energy expenditure (Fischer et al., 2009; Merkestein et al., 2014).
In humans, several studies have demonstrated single nucleotide polymorphisms (SNPs) in the FTO gene that are associated with increased body mass index (BMI) (Frayling et al., 2007; Scuteri et al., 2007). Many studies have shown a vital role for FTO in the development of obesity, regulation of appetite and energy homeostasis (Church et al., 2009). Moreover, Genome-wide association studies (GWAS) have identified genetic variants in the FTO gene that are associated with increased risk of type 2 diabetes (T2D) (Frayling et al., 2007; Yajnik et al., 2009). Furthermore, the FTO protein was reported to have a rapid turnover in clonal pancreatic β cells and its overexpression enhanced insulin secretion, suggesting that FTO could play an important role in β-cell function (Russell and Morgan, 2011).
ARL15 belongs to the ADP-ribosylation factor (ARF) family. It has been shown that ARL15 regulates membrane trafficking, lipid composition and the insulin signaling pathway(Donaldson and Jackson, 2011). Several studies reported a strong association of ARL15 with increased risk of T2D, fasting insulin level and even β cell proliferation (Scott et al., 2012; Thomsen et al., 2016). Also, it has been reported that ARL15 acts as insulin-sensitizing protein to phosphorylate the insulin receptor, (IR), the insulin receptor substrate-1 (IRS1) and AKT kinase in the insulin pathway (Zhao et al., 2017). Recently, using RNA-sequencing gene expression from human islets we reported that both FTO and ARL15 are inversely correlated with HbA1c levels (Fadista et al., 2014a). Still, the functional role of FTO and ARL15 in pancreatic β cells remains unclear.
Pancreatic β-cells are specialized cells that regulate glucose homeostasis. Defects of insulin secretion in β-cells represent the culprit of T2D, a disease that is expected to affect more than 640 million people by 2040 (http://www.diabetesatlas.org). Therefore, there is an urgent need to understand mechanisms of dysregulated insulin secretion in humans. One way to achieve this is to develop experimental models that mimic real human pancreatic β-cells. Here we used the stable β-cell line called GRINCH, derived from INS-1 cells but additionally expressing a human proinsulin, in which the superfolder GFP is contained within the C-peptide (Haataja et al., 2013).
In this study, we harness GRINCH cells to investigate the influence of expression silencing of FTO and ARL15 on glucose-stimulated insulin secretion. Our data indicate that FTO expression is reduced in islets from T2D organ donorsand that its silencing attenuates glucose-stimulated insulin secretion. In contrast, knockdown of ARL15 did not alter insulin secretion.
2. Materials and methods
2.1. GRINCH cells
The establishment of GRINCH cells has been described elsewhere (Haataja et al., 2013). Briefly, cDNA encoding superfolder GFP (sfGFP) was amplified and ligated into the ApaI site located at the C-peptide encoding sequence to make hPro-CpepSfGFP. Cells were cultured at 37 C in a humidified atmosphere of 95% air and 5% CO2 in RPMI 1640 culture medium containing 11 mM D-glucose (Gibco, Life Technologies) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/mL streptomycin, 10 mM glutamine, 1 mM sodium pyruvate, 10 mm HEPES, 50 μM β-mercaptoethanol (all from Sigma) and 0.1 mg/mL G418 (CalBiochem, Germany). INS-1 (832/13) cells were cultured under the same conditions but without G418.
2.2. Cell culture and insulin secretion assay
Confluent plates containing (GRINCH or INS-1 (832/13) cells were washed with 1 mL pre-warmed Secretion Assay Buffer (SAB), pH 7.2 (114 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.16 mM MgSO4, 20 mM HEPES, 2.5 mM CaCl2, 25.5 mM NaHCO3 and 0.2% Bovine Serum Albumin) containing 2.8 mM glucose. The cells were then pre-incubated for 2 h in fresh 2 mL SAB with 2.8 mM glucose. Thereafter, separate wells were incubated for 1 h in 1 mL SAB containing either 2.8 mM glucose plus secretagogue (16.7 mM glucose, 1 mM 3-isobutylmethylxanthine (IBMX) and 10 μM Forskolin). Secreted insulin was measured from the supernatant using Coat-a-Count Insulin radioimmunoassay kit (RIA) (Siemens), High Range Rat Insulin ELISA (Mercodia, Uppsala, Sweden) and the GFP signals using TECAN plate-reader (Infinite M200, Switzerland). The fluorescence intensity in the plate-reader was adjusted for excitation wavelength 489 nm and emission wavelength 527 nm.
2.3. RNA interference
GRINCH (passage nr 14) or INS-832/13 (passage nr 59) cells were cultured as previously described (Taneera et al., 2012) and transfected using Lipofectamine RNAiMAX transfectionreagent® (Invitrogen). siRNA sequences against CHL1 (S 44135 and S 44134), FTO (S 146052 and S146053) and ARL15(s187063) were purchased from Thermo-Fisher. For control purposes, a previously described control sequence Silencer®Negative Control #2 from Ambion was employed. Cells were cultured in RPMI 1640 medium for 72 h at 37 °C in a humidified atmosphere containing 95% air and 5% CO2 in the presence of 40 nM siRNA in 24-well cell culturemicroplates (250 000 cells/well). Three days after transfection, insulin secretion measurements were performed by RIA or ELISA and by TECAN plate-reader.
2.4. RT-qPCR
Total RNA was isolated with RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). RNA quality and concentration were measured using an Agilent 2100 bioanalyzer (Bio-Rad, Hercules, CA, USA) and Nanodrop ND-1000 equipment (NanoDrop Technologies, Wilmington, DE, USA). cDNA was synthesized using RevertAid H Minus First Strand cDNA synthesis Kit (Fermentas, Life Sciences). Analysis of gene expression after knockdown was performed using the TaqMan qPCR with an ABI Prism 7900 HT System (Applied Biosystems, USA) using gene-specific primer probes for CHL1 (Rn01420997_m1), FTO (Rn01538186_m1), ARL15(Rn01494252_m1), Ins1 (Rn02121433_g1), Ins2(Rn01774648_g1) and human Ins (Hs02741908_m1). (Applied Biosystems, USA) in triplicates on 384-well plate. 1.2 ng cDNA was used per well in 10-μL reaction volume containing TaqMan master mix (Applied Biosystems, USA). The rat HPRT (Applied Biosystem) was used to normalize gene expression by the ΔΔCt method, where the final normalized quantity was expressed as 2ˆ-(Ct target - Ct control).
2.5. RNA sequencing and human pancreatic islets
Human Islets from 204 cadaver donors (162 nondiabetic and 29 diabetic) of European ancestry were provided by the Nordic Islet Transplantation Program and processed as previously described (Taneera et al., 2015). Islets were obtained from 162 nondiabetic donors (65 females, 97 males, age 57.9 ± 10, BMI 26.2 ± 3.8, HbA1c 5.7 ± 0.5) and 29 T2D donors (10 females, 19 males, age 61.1 ± 10, BMI 27.6 ± 4.5, HbA1c 6.8 ± 0.8). Purity of islets was assessed by dithizonestaining and was 70% ± 17% in the nondiabetic and 67% ± 20% in the diabetic islets (p = 0.45). RNA sequencingwas done using Illumina’s TruSeq RNA Sample Preparation Kit. Detailed materials and methods for RNA-seq were previously described (Fadista et al., 2014b). All procedures were approved by the ethics committees at Uppsala and Lund University.
2.6. Apoptosis analysis
For apoptosis analysis, 1 × 105 cells (72-h post-transfection) were re-suspended in 500 μl of Annexin-V (1X) Binding Buffer (BD, USA). A 5 μl of Annexin V-FITC and 5 μl Propidium Iodide was added and incubated at room temperature for 10 min in the dark. Analysis with Flowcytometer (FACSAria III, BD) was performed using FITCchannel (515 nm) and PI at PE detector (585 nm), 30000 events are collected for the analysis. Compensation was performed by using the single color controls with Annexin FITC and PI.
2.7. Western blot analysis
Cells were lysed in ice-cold NP-40 (1.0% NP-40, 150 mM of NaCl, 50 mM of Tris-Cl, pH 8.0) lysis buffer containing protease cocktail inhibitor tablets (Sigma) 72 h post-transfection. Protein concentrations were quantified using the standard Bradford method (Bio-Rad, CA, USA). A 30 μg sample of protein lysate was separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane (Bio-Rad) and then blocked by 5% skimmed milk powder for 1 h at room temperature, washed with (TBST), and the blot was probed with FTO (1:1000; #Ab92821 Abcam, UK) antibody overnight at 4 °C. The secondary (anti-mouse and anti-rabbit) antibodies were then reacted with the membrane at 1:1000 dilutions for 1 h at room temperature. Chemiluminescence was detected using the ECL kit (Thermo Fisher). Protein band quantification was carried out using the Bio-Rad Image Lab software (ChemiDoc™ Touch Gel and Western Blot Imaging System; Bio-Rad). β-actin was used as a normalization control.
2.8. Statistical analysis
Data are presented as means ± S.D. Differences in expression levels were analyzed by student’s t-test or nonparametric Mann-Whitney tests. P < 0.05 was considered statistically significant.
3. Results
3.1. GFP signal intensity in GRINCH cells
To measure CpepGFP signal intensity of GRINCH cells, we prepared lysate dilutions from GRINCH cells, with regular INS-1 (832/13) cells used as a negative control. Lysates from GRINCH cells exhibited high GFP intensity, which is gradually reduced with higher dilution (Fig. 1-A). We were able to detect CpepGFP signals from the lysate preparation even at dilution factor 1:40 compared to INS-1 (832/13) cell lysate, which showed no GFP signal intensity.
Fig 1. GFP signal intensity, Ins genes expression and insulin secretion in GRINCH cells.
(A) Sonicated lysates from GRINCH and regular INS-832/13 cells were diluted in H2O and CpepGFP signals were measured using the TECAN platereader.(B) Ct values of qRT-PCR demonstrate that GRINCH cells express both rat (Ins1 and Ins2) and human insulin genes. HPRT1 was used as a house keeping gene.(C, D) GRINCH cells were stimulated with 2.8, 6, 10, 16.7 mM glucose, 16.7 mM glucose + 1 mM IBMX or 10 μM Forskolin for 1 hr. Supernatants containing secreted insulin were measured by RIA (C) and CpepGFP signals using TECAN plate-reader method (D).
3.2. GRINCH cells expressed a human and rat insulin gene cDNA along with endogenous rat insulin
As previously described, INS-1 (832/13) cells were derived from INS-1 cells and engineered to stably express human insulin cDNA (Asfari et al., 1992). Moreover, the human insulin content of GRINCH and INS-1 (832/13) cells was similar and both exhibited stimulus-dependent secretion (Haataja et al., 2013). To confirm expression of human and rat insulin mRNA in GRINCH cells, RT-qPCR was performed using distinct sets of amplifying primers. As expected, both human and rat insulin mRNA was expressed in GRINCH cells. The Ct values for the rat Ins1 was 13, rat Ins2 19 and for the human Ins gene was 25 (Fig. 1-B).
3.3. Insulin secretion in GRINCH cells
To test secretegogue-stimulated insulin secretion in GRINCH cells, cells were seeded (700 000 cell/well) and grown overnight. The next day, cells were stimulated with 2.8, 6, 10, 16.7 mM glucose, 1 mM IBMX or 10 μM Forskolinfor 1 h. The supernatant containing secreted insulin was analyzed using RIA and by measuring the GFP signals in a TECAN plate-reader. RIA measurements showed a 3-fold increase between the basal level (2.8 mM Glucose) and secretion level (16.7 mM Glucose) and almost 6-fold between the 16.7 mM glucose and 16.7 mM glucose with IBMX or Forskolin (Fig. 1 -C). Using the GFP signal as a surrogate marker to measure insulin secretion, we observed a 2-fold increase between the basal and stimulating section level (16.7 mM) and an almost 5-fold increase between 16.7 mM glucose and IBMX or Forskolin (Fig. 1-D). Insulin secretion measured by RIA and plate-reader were highly correlated (R2 = 0.99) (data not shown). The percentage of insulin release measured as a fraction of insulin content was 1.8% using CpepGFP signal assay, while it was 2% using the ELISA assay after 16.7 mM glucose stimulation.
3.4. GRINCH cells as a tool to study pancreatic β cell function
The CHL1 gene encodes for a neural adhesion molecule, it has been suggested that CHL1 might be involved in the development of islet structure and determine the distribution of β, α, and δ cells in pancreatic islets. Previously, we reported that expression of CHL1 in human islets is strongly related to HbA1c and insulin secretion and decreased in islets and β cells from patients with T2D (Taneera et al., 2012). Importantly, we showed that siRNAgene silencing of CHL1 in INS-1 (832/13) cells significantly reduced insulin secretion.
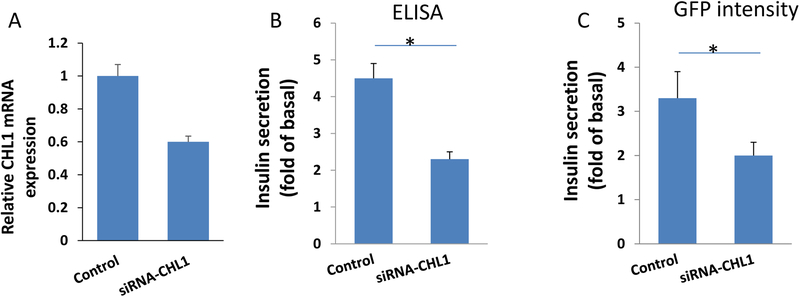
Thus, to validate GRINCH cells as a tool for functional studies of pancreatic β cells, we silenced the expression of CHL1 in GRINCH cells. Knockdown efficacy measured by RT-qPCR after 48 h revealed almost 50% decreased expression of CHL1 (Fig. 2-A. The supernatants containing secreted insulin from transfected cells were analyzed after glucose stimulation using ELISA assay and TECAN plate-reader for CpepGFP signal intensity. We noticed a significant reduction in insulin secretion (45%, p < 0.05) in silenced cells compared to negative control siRNA using ELISA assay (Fig. 2-B), whereas a 37% reduction in insulin secretion was observed using the CpepGFP signal measurement (p < 0.05) (Fig. 3-C). These results further validate the use of GRINCH cells for screening of altered insulin secretion by monitoring fluorescence.
Fig 2. CHL1 knockdown in GRINCH cells.
(A) Knockdown efficiency of siRNA of CHL1 in GRINCH cells as determined by RT-qPCR. (B) Insulin secretion as assessed by RIA in response to 2.8 mM and 16.7 mM glucose at 72 hours post siRNA transfection during 1-hour static incubation. Insulin secretion expressed as a fold increase (insulin secreted at 16.7 mM at 2.8 mM glucose). (C) Insulin secretion measured as GFP intensity by plate-reader. Bars represent mean ± SD. *p < 0.05.
Fig 3. FTO and ARL15 knockdown in GRINCH cells.
(A) knockdown efficiency of siRNA for ARL15 (s187063), FTO-1 (s146052) and FTO-2 (s146053) in INS-1 (832/13) cells. (B) Western blot bands of FTO protein expression relative to the endogenous control protein β-actin in FTO knockdown cells and cells transfected with negative siRNA control. (C) Insulin secretion in response to 2.8 mM and 16.7 mM glucose 72 hours post siRNA transfection measured during 1-hour static incubation. Cells transfected with siRNA against FTO showed almost a 50% reduction in insulin secretion in both siRNAs, whereas those with ARL15 showed no effect compared to siRNA negative control (NC). (D) Insulin secretion measured by Cpep-GFP intensity by plate-reader showed a 40% reduction with FTO transfected cells but not ARL15. (E & F) Transfected cells with siRNA against FTO-2 were stimulated by a depolarizing agent (35 mM KCL) and measured by ELISA and GFP intensity. No significant differences between transfected and control cells were observed. Insulin secretion was normalized for cell count. Data are shown from three independent experiments for each siRNA. Bars represent mean ±SD. * < 0.05, ** < 0.01.
3.5. Silencing of T2D candidate genes
Fadista et al. have identified a list of 17 established T2D and glycemic associated loci (PCSK1, SLC30A8, G6PC2, IKBKAP, FTO, VPS13C, PDX1, ARL15, ADAMTS9, JAZF1, RASGRP1, PROX1, SLC2A2, GRB14, AMT, PTPRD and IRS1) whose gene expression proxies in human islets using RNA-sequencing are associated with HbA1c levels (Fadista et al., 2014b). Of the 17 genes, 2 genes (ARL15 (ADP-ribosylation factor-like 15) and FTO were selected for further investigation in GRINCH cells.
Specifically, siRNA against FTO and ARL15 were used to silence the expression of each gene in GRINCH cells. Transfection efficacy measured by RT-qPCR after 48 h resulted in almost a 90% decreased expression of FTO and 70% for ARL15 (Fig. 3-A). Similar reduction of FTO protein expression was observed by western blot analysis. (Fig. 3-B). The supernatants containing secreted insulin from transfected cells were analyzed after glucose stimulation using ELISA and TECAN plate-reader for GFP signal intensity. Insulin secretion measurement after 72 h in FTO knockdown cells (measured by ELISA) in response to 16.7 mM glucose showed almost 50% reduction (p < 0.5) and 40% reduction (p < 0.05) as measured by CpepGFP compared to negative control cells (Fig. 3-B and C). As the first siRNA silencing of FTO (siFTO-1) revealed a decrease in insulin secretion, we further confirmed the finding using another siRNA (siFTO-2) (Fig. 3-B and C). Moreover, FTO knockdown reduced the first phase of insulin secretion as measured at 10 min after glucose stimulation compared to negative control cells (Supplementary Fig. 1).
Silencing of ARL15 showed no impact on insulin secretion compared to siRNA negative control measured by ELISA or GFP intensity (Fig. 3-B and C).
To elucidate how siRNA gene silencing of FTO contributes to pancreatic β-cell failure, we performed several functional studies to identify the possible mechanism.
First, to explore whether the exocytosis machinery is defective upon silencing of FTO, we stimulated insulin exocytosis with depolarizing (35 mM) KCl, and this showed no difference between transfected cells and controls (Fig. 3-D and E).
Next, we stained transfected cells with Annexin-V/PI to investigate the cell death of INS-1 (832/13) cells following siRNA silencing. Since Annexin-V is monitored using the FITC channel we could not use the GRINCH cells because of fluorescence overlap. As shown in Fig. 4-A, the ratio of the apoptotic cells between FTO knockdown cells and siRNA negative control were similar. The percentage of early apoptotic cells (0.2% vs. 0.3%) or late apoptotic cells (0.1% vs. 0.2%) between FTO knockdown cells and control siRNA (Fig. 4-A).
Figure 4. Assessment of apoptosis in INS-1 cells.
(A) Cell transfected with siRNA negative control or siRNA against FTO were stained with Annexin V-FITC and PI and analyzed by flow cytometry. (B) qPCR expression analysis of Ins1 and Ins2 genes in GRINCH cells transfected with negative control or siRNA against FTO. Cell lysate from transfected cells (with negative control or FTO) were used to measure GFP intensity as indicator for insulin content. Bars represent mean ±SD. * < 0.05.
Finally, we analyzed the mRNA expression level of Ins1 and Ins2 genes in transfected GRINCH cells. qRT-PCR analysis showed a significant decrease of both Ins1 (p = 0.01 and Ins2(p = 0.02) mRNA expression in transfected cells compared to negative siRNA control (Fig. 4-B). This was further confirmed by measuring the GFP signal from cell lysates which showed almost 30% decrease in FTO transfected cells (Fig. 4-C).
3.6. Expression of FTO in human islets
Using RNA-sequencing data from human pancreatic islets, we investigated the expression of FTO in diabetic (n = 30) and nondiabetic donors (n = 172). FTO expression was found to be lower in T2D islets (p = 0.058, adjusted for age, sex and BMI) than in non-diabetic islets (Fig. 5-A). Moreover, stratifying the donors into normal glucose tolerant (NGT; HbA1c < 6.0) and T2D (HbA1c < 6.5) showed that mean expression of FTOwas significantly lower in T2D donors (p = 0.04; adjusted to age, sex and BMI) (Fig. 5-B).
Fig 5. FTO gene expression in human pancreatic islets.
FTO mRNA expression revealed lower level in pancreatic islets from T2D donors compared to those from non-diabetic controls (B), the reduced expression was significant when donors were stratified for NGT vs T2D.
4. Discussion
Several studies have suggested a potential role for FTO in insulin secretion (Russell and Morgan, 2011; Kirkpatrick et al., 2010). Kirkpatrick et al., reported that FTO mRNA level was reduced in purified human β-cells from a small number of T2D patients compared with non-diabetic controls (Kirkpatrick et al., 2010). Fadista et al. found FTO to be suppressed in islets of T2D patients (21), which we confirm in a larger number of organ donor islets. It is likely that the reported increase in DNA methylation of the FTO gene contributes to the lower mRNA expression (Dayeh et al., 2014).
However, the role of FTO in insulin secretion is controversial. Here, we demonstrate that siRNA silencing of FTO results in reduced glucose-stimulated insulin secretion without affecting the stimulant action of depolarizing potassium concentrations. This indicates that the exocytotic machinery is not altered by FTO silencing. In agreement with our finding, Russell et al. showed that conditional expression of the FTO gene product in INS-1 cells enhanced first phase insulin secretion stimulated with 20 mM glucose (Russell and Morgan, 2011). However, the increase in insulin secretion in this case was attributed to the exocytosismachinery rather than upregulated insulin biosynthesis.
Inconsistent with our findings and those of Russel and Morgan (Russell and Morgan, 2011) was the observation that FTO overexpression in MIN6 clonal mouse β-cells by lentivirus resulted in a significant reduction in insulin secretion in the presence of 20 mM glucose or KCl (Fan et al., 2015). Interestingly, silencing of FTO in MIN6 cells revealed no effect on insulin secretion. A possible explanation for the discrepancy could relate to the different approach for FTOsilencing by siRNA, vs. overexpression by lentivirus. INS-1 cells and GRINCH cells were derived from a rat insulinoma, while MIN6 cells are of murine origin. Therefore, species-related variations could underlie the discrepant results. Species-specific results regarding the role of FTO have indeed been reported. For example, while fasted mice exhibit a significant decrease in hypothalamic FTOexpression (independently of leptin levels) as compared to fed controls (Gerken et al., 2007), fasted rats show increased FTO expression in the hypothalamus (Fredriksson et al., 2008). Moreover, loss of FTO function in mice reduces fat mass through increased energy expenditure but not decreased energy intake (Church et al., 2009). It has been suggested that FTO impacts feeding behavior by affecting neuropeptide Y expression in the hypothalamus in rodents (Alkan et al., 2017). In man, FTO-SNPs are associated with appetite and food intake but not energy expenditure (Fischer et al., 2009; Do et al., 2008). Thus, it is likely that the discrepancy between species reflects differences of FTO actions.
GWAS have implicated ARL15 in several metabolic traits (Scott et al., 2012; Thomsen et al., 2016). Despite the fact that ARL15 expression was correlated with HbA1c levels, our results showed that gene silencing of ARL15 in INS-1 cells had no effect on insulin secretion. During the preparation of this study, Rocha et al. reported studies on ARL15 in insulin signal transduction and adipocytes differentiation in 3T3-L1preadipocytes (Rocha et al., 2017). In ARL15 knockdown cells, there was no effect on insulin-stimulated phosphorylation of AKT or ERK nor on insulin-stimulated glucose uptake. However, significant impairment of adiponectin secretion was observed.
Here we report the use of CpepGFP signal intensity from GRINCH cells as a new tool to assay pancreatic β-cell function. GRINCH cells were developed to study proinsulintrafficking, processing and secretion from pancreatic β-cells. The superfolder GFP inserted in human proinsulin C-peptide showed excellent glucose-stimulated insulin secretion upon exposure to high glucose doses and unique feature based on the ability to exhibit a recoverable CpepGFP signal (Haataja et al., 2013). This feature of GRINCH cells allowed us to exploit the CpepGFP signal in GRINCH cells as a marker of β-cell function. We noticed that secretion data from stimulated GRINCH cells measured by GFP-plate reader assay were overall similar to results obtained with RIA or ELISA, except for a marginally smaller fold-stimulation. However, such a difference between the assays was not statistically significant (p > 0.05). Herein, we have used relatively early passage number of GRINCH cells, stability of the CpepGFP over multiple passages need to be tested.
Several reports have described surrogate markers for insulin secretion in pancreatic β-cell lines by fused GFP or luciferase (Burns et al., 2015; Pouli et al., 1998). Recently, Bruns et al. reported a luminescent insulin secretion assay that enables large-scale investigation of β-cell function (Burns et al., 2015). Specifically, Bruns et al. inserted a Gaussia luciferase into the C-peptide portion of proinsulin, and they found that upon stimulation with secretagogues, β-cells expressing this reporter secreted luciferase in close correlation with insulin. In the case of GRINCH cells, insulin secretion measurement is performed directly by plate-reader assay of the supernatants after secretagogues simulations while the luciferase activity requires adding coelenterazine substrate to the supernatant before read-out.
Out of the 17 established T2D genes whose gene expression proxies are associated with HbA1c, eight genes (SLC30A8(Wijesekara et al., 2010), G6PC2 (Pound et al., 2013), PCSK1(Ravassard et al., 2011), PDX1 (Stoffers et al., 1997), VPS13C (Grarup et al., 2011), RASGRP1 (Taneera et al., 2012), SLC2A2(Guillam et al., 1997), and IRS1 (Meur et al., 2010)) were excluded as they have already been implicated for a role in insulin secretion. Additionally, previously we showed that gene silencing of ADAMTS9 and JAZF1 has no impact on insulin secretion (Taneera et al., 2015). For the other 7 genes, the selection criteria included expression in β-cells based upon previously published RNA-sequencing expression data from sorted human pancreatic endocrine cells (Blodgett et al., 2015). The FTO, IKBKAP and ARL15 genes were found to have the highest expression in sorted β-cells. ARL15 is a GTP-binding protein that has been shown to influence adiponectin levels (Richards et al., 2009). IKBKAP encodes a scaffold protein and a regulator for 3 different kinasesinvolved in pro-inflammatory signaling. Mutations in this gene have been associated with familial dysautonomia (Slaugenhaupt et al., 2001). Based on gene function, only FTO and ARL15 were selected for functional studies in GRINCH cells.
Conclusions
We demonstrate the validity of GRINCH cells as a new tool to study pancreatic β-cell secretion. The cells retain insulin secretion responses to glucose and exhibit similar physiologic characteristics comparable to those of INS-1 (832/13) cells. This tool may serve as a rapid, and inexpensive method for screening of compounds, in order to investigate molecular mechanisms influencing insulin secretion. Knockdown of FTO expression in GRINCH cells inhibits insulin secretion, whereas no effect for ARL15 was observed. The reduced insulin secretion in FTO knockdown cells was associated with decreased insulin mRNA levels while cell viability was preserved. Additional work should pin-point the exact role of the FTO gene as a regulator of pancreatic β-cell function.
Supplementary Material
Highlights.
GRINCH cells as a new tool to study pancreatic β-cell secretion facilitating drug screening.
Knockdown of FTO expression inhibits glucose-but not potassium-stimulated insulin secretion.
Knockdown of ARL15 expression has no impact on insulin secretion.
FTO expression is lower in islets from T2D patients.
Acknowledgements
Human pancreatic islets were obtained from The Nordic Network for Clinical Islet Transplantation. We would like to thank Manju Nidagodu Jayakumar for FCAS flow analysis. This work is supported by the Swedish National Strategic Research Initiative EXODIAB (Excellence of Diabetes Research in Sweden) and the Juvenile Diabetes Research Foundation. L.H. and P.A. are funded by National Institute of Health grant DK48280. J.T. is funded by Al Jalila Foundation (AJF201723), University of Sharjah and Boehringer Ingelheim (1701090119-P).
Footnotes
Conflicts of interest
The authors declare that there is no duality of interest associated with this manuscript.
References
- Alkan I, Altunkaynak BZ, Altun G, Erener E The investigation of the effects of topiramate on the hypothalamic levels of fat mass/obesity-associated protein and neuropeptide Y in obese female rats Nutr. Neurosci. (2017), pp. 1–10 [DOI] [PubMed] [Google Scholar]
- Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB Establishment of2-mercaptoethanol-dependent differentiated insulin-secreting cell lines Endocrinology, 130 (1992), pp. 167–178 [DOI] [PubMed] [Google Scholar]
- Blodgett DM, Nowosielska A, Afik S, et al. Novel observations from next generation RNA sequencing of highly purified human adult and fetal islet cell subsets Diabetes (2015), Article db150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns SM, Vetere A, Walpita D, et al. High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic Beta-cell function Cell Metabol, 21 (2015), pp. 126–137 [DOI] [PubMed] [Google Scholar]
- Church C, Lee S, Bagg EA, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene PLoS Genet, 5 (2009), Article e1000599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayeh T, Volkov P, Salö S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion PLoS Genet., 10 (2014), Article e1004160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity Nat. Genet, 39 (2007), pp. 724–726 [DOI] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study Diabetes, 57 (2008), pp. 1147–1150 [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL ARF family G proteins and their regulators: roles in membrane transport, development and disease Nat. Rev. Mol. Cell Biol, 12 (2011), p. 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Vikman P, Laakso EO, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism Proc. Natl. Acad. Sci. U. S. A, 111 (2014), pp. 13924–13929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Vikman P, Laakso EO, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism Proc. Natl. Acad. Sci. Unit. States Am, 111 (2014), pp. 13924–13929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HQ, He W, Xu KF, Wang ZX, Xu XY, Chen H FTO inhibits insulin secretion and promotes NF-kappaB activation through positively regulating ROS production in pancreatic beta cells PLoS One, 10 (2015), Article e0127705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, et al. 963201. Inactivation of the Fto gene protects from obesity Nature, 458 (2009), pp. 894–898 [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity Science, 316 (2007), pp. 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Hägglund M, Olszewski PK, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain Endocrinology, 149 (2008), pp. 2062–2071 [DOI] [PubMed] [Google Scholar]
- Gao X, Shin Y-H, Li M, Wang F, Tong Q, Zhang P The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice PLoS One, 5 (2010), Article e14005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung Y-CL, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase Science, 318 (2007), pp. 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grarup N, Overvad M, Sparsø T, et al. The diabetogenic VPS13C/C2CD4A/C2CD4B rs7172432 variant impairs glucose-stimulated insulin response in 5,722 non-diabetic Danish individuals Diabetologia, 54 (2011), pp. 789–794 [DOI] [PubMed] [Google Scholar]
- Guillam M-T, Hümmler E, Schaerer E, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2 Nat. Genet, 17 (1997), pp. 327–330 [DOI] [PubMed] [Google Scholar]
- Haataja L, Snapp E, Wright J, et al. Proinsulin intermolecular interactions during secretory trafficking in pancreatic β cells J. Biol. Chem, 288 (2013), pp. 1896–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Marchetti P, Purrello F, et al. Type 2 diabetes susceptibility gene expression in normal or diabetic sorted human alpha and beta cells: correlations with age or BMI of islet donors PLoS One, 5 (2010), Article e11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkestein M, McTaggart JS, Lee S, et al. Changes in gene expression associated with FTO overexpression in mice PLoS One, 9 (2014), Article e97162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meur G, Simon A, Harun N, et al. Insulin gene mutations resulting in early-onset diabetes: marked differences in clinical presentation, metabolic status, and pathogenic effect through endoplasmic reticulum retention Diabetes, 59 (2010), pp. 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters A, Beckers S, Verrijken A, et al. Variants in the FTO gene are associated with common obesity in the Belgian population Mol. Genet. Metabol, 93 (2008), pp. 481–484 [DOI] [PubMed] [Google Scholar]
- Peters T, Ausmeier K, Rüther U Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation Mamm. Genome, 10 (1999), pp. 983–986 [DOI] [PubMed] [Google Scholar]
- Poritsanos N, Lew P, Fischer J, et al. Impaired hypothalamic Fto expression in response to fasting and glucose in obese mice Nutr. Diabetes, 1 (2011) e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouli AE, Kennedy HJ, Schofield JG, Rutter GA Insulin targeting to the regulated secretory pathway after fusion with green fluorescent protein and firefly luciferase Biochem. J, 331 (1998), pp. 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound LD, Oeser JK, O’Brien TP, et al. G6PC2: a negative regulator of basal glucose-stimulated insulin secretion Diabetes, 62 (2013), pp. 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravassard P, Hazhouz Y, Pechberty S, et al. A genetically engineered human pancreatic β cell liqne exhibiting glucose-inducible insulin secretion J. Clin. Invest, 121 (2011), p. 3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Waterworth D, O’Rahilly S, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels PLoS Genet, 5 (2009), Article e1000768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Payne F, Huang-Doran I, et al. The metabolic syndrome-associated small G protein ARL15 plays a role in adipocyte differentiation and adiponectin secretion Sci. Rep, 7 (2017), p. 17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MA, Morgan NG Conditional expression of the FTO gene product in rat INS-1 cells reveals its rapid turnover and a role in the profile of glucose-induced insulin secretion Clin. Sci, 120 (2011), pp. 403–413 [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants Science, 316 (2007), pp. 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways Nat. Genet, 44 (2012), p. 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen W-M, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits PLoS Genet, 3 (2007)e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia Am. J. Hum. Genet, 68 (2001), pp. 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence Nat. Genet, 15 (1997), pp. 106–110 [DOI] [PubMed] [Google Scholar]
- Stratigopoulos G, Padilla SL, LeDuc CA, et al. Regulation of Fto/Ftm gene expression in mice and humans Am. J. Physiol. Regul. Integr. Comp. Physiol, 294 (2008), pp. R1185–R1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera J, Lang S, Sharma A, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets Cell Metabol., 16 (2012), pp. 122–134 [DOI] [PubMed] [Google Scholar]
- Taneera J, Fadista J, Ahlqvist E, et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia Hum. Mol. Genet, 24 (2015), pp. 1945–1955 [DOI] [PubMed] [Google Scholar]
- Thomsen SK, Ceroni A, van de Bunt M, et al. Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants Diabetes, 65 (12) (2016. December), pp. 3805–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesekara N, Dai F, Hardy A, et al. Beta cell-specific Znt8 deletion in mice causes marked defects in insulin processing, crystallisation and secretion Diabetologia, 53 (2010), pp. 1656–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajnik CS, Janipalli CS, Bhaskar S, et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians Diabetologia, 52 (2009), pp. 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang M, Deng W, et al. ADP-ribosylation factor-like GTPase 15 enhances insulin-induced AKT phosphorylation in the IR/IRS1/AKT pathway by interacting with ASAP2 and regulating PDPK1 activity Biochem. Biophys. Res. Commun, 486 (2017), pp. 865–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.