Abstract
MicroRNA (miRNA/miR) has been identified to be a promising tool in treating pharyngolaryngeal cancer. The present study aimed to investigate the role of miR-490-5p in the regulation of proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) of pharyngolaryngeal cancer cells. The data of miR-490-5p expression levels of 45 cases were obtained from the People's Hospital of Xinjiang Uygur Autonomous Region, and the prediction of the target of miR-490-5p was conducted by bioinformatics and verified using a luciferase assay. Cell viability was determined by cell counting kit-8. Migration and invasion rates were measured by wound healing test and Transwell apparatus, respectively. Colony formation rate was measured by plate colony formation assay. mRNA and protein levels were determined by quantitative polymerase chain reaction and western blotting, respectively. miR-490-5p expression was significantly depressed in primary pharyngolaryngeal cancer tissues and cell lines, leading to an unfavorable prognosis. Evidently, miR-490-5p overexpression decreased the cell viabilities of BICR 18 and FaDu cells. Mechanically, miR-490-5p could target mitogen-activated protein kinase kinasekinase 9 (MAP3K9). The overexpression of MAP3K9 could promote cell viability, migration and invasion rates, EMT process and ability of cloning, miR-490-5p could target MAP3K9 and further modulate the proliferation, migration, invasion and EMT of pharyngolaryngeal cancer cells. The results of the present study provide a novel entry point to the treatment of pharyngolaryngeal cancer.
Keywords: miR-490-5p, hallmarks of cancer, pharyngolaryngeal cancer, mitogen-activated protein kinase kinasekinase 9
Introduction
Pharyngolaryngeal cancer is one of the common malignant tumors that occurs in the head and neck, and its development is a highly complex progress during which plenty of mechanisms, for example, oncogene activation, tumor suppressor gene inactivation and abnormal expression of apoptosis-associated genes, are involved (1,2). Although comprehensive treatment for pharyngolaryngeal cancer has been developed recently, the overall survival rate has not yet improved to an acceptable level (3,4). Investigating the molecular mechanism of laryngeal cancer and searching for an effective therapeutic molecular target remains to be done (3).
MicroRNA (miRNA/miR) has been proved to be able to inhibit the synthesis of specific proteins by binding to specific 3′-untranslated region (UTR) of the target mRNAs, while they participate in the regulation of cell growth, proliferation, differentiation and apoptosis (5,6). Under the circumstance of mutation, deletion or abnormal expression, miRNAs can serve roles similar to oncogenes or tumor suppressor genes, participating in the process of proliferation, differentiation and apoptosis of tumor cells (7,8). Early in 2008, Chang et al (9) screened the differential expression profiles of miRNAs in pharyngolaryngeal cancer and normal tissues using gene chip techniques, and demonstrated that the levels of 13 miRNAs in pharyngolaryngeal cancer tissues were significantly different compared with in normal tissues. Furthermore, a previous study has revealed that miRNA was strongly associated with the development of laryngeal carcinoma (10). In addition, a comprehensive study revealed that smoking-specific miRNAs are altered in head and neck squamous cell carcinoma as well (11). Therefore, investigating the association between miRNA and pharyngolaryngeal cancer is remains necessary as it would be able to provide a comprehensive understanding of the mechanism of the development of pharyngolaryngeal cancer.
Furthermore, previous studies have demonstrated that miRNAs are a potential tumor biomarkers that could interact with a variety of signaling pathways, including mitogen-activated protein kinase (MAPK) and Wnt/Frizzled, therefore participating in the occurrence and development of tumors (12,13). MAPKs serve an important role in multiple biological processes including cell proliferation and differentiation. MAPK kinase kinase 9 (MAP3K9), alternatively known as mixed-lineage kinases1, has been classified as an oncoprotein (14). It has been reported that miRNAs could inhibit the proliferation, migration, invasion and EMT process of cancer cells by regulating MAP3K9 (15,16).
In this study the aim was to investigate whether miR-490-5p, the miRNA reported previously to act as a critical modulator in tumors (17), could affect the hallmarks of pharyngolaryngeal cancer including proliferation, migration, invasion and the epithelial-mesenchymal transition (EMT) process. The underlying mechanism of the modulation of miR-490-5p on these effects was also investigated.
Materials and methods
Tissue samples
Tumor and tumor-adjacent tissues samples of 45 patients with laryngeal carcinoma admitted to the People's Hospital of Xinjiang Uygur Autonomous Region (Xinjiang, China) from February 2009 to October 2013 were selected. These patients including 41 males and 4 females, the age of these patients ranged from 42-76 years old, with an average of 59.6±7.9 years. The data of overall survival (OS) rates of 45 cases were also collected from the hospital. The cut-off line of low and high expression of miR-490-5p were determined according to the median of its expression in these cases. The present study was approved by the Ethics Committee of the People's Hospital of Xinjiang Uygur Autonomous Region, and an informed consent was obtained from each patient.
Bioinformatics analysis
To investigate the target gene of miR-490-5p, the biological information online analysis software of Targetscan (http://www.targetscan.org/vert_72/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and miRDB (http://mirdb.org/miRDB/) were used.
Cell culturing, transfection and grouping
NP69, BICR 18, FaDu, HNE-3 and Detroit 562 cell lines were purchased from the Type Culture Collection of the Chinese Academy of Sciences. HaCaT cell lines (cat. no. 300493) were purchased from CLS Cell Lines Service GmbH. NP69 cell lines acted as the normal laryngeal cells, while HaCaT cell lines acted as normal tissue cells. The HNE-3, FaDu, NP69 cell lines were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc.), whereas Detroit 562, BICR 18 and HaCaT cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.). The medium was supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). Cells were cultured in an incubator filled with 5% CO2 at 37°C and subcultured every 2-3 days. The overexpression plasmid of miR-490-5p (mimics, 5′-CCAUGGAUACUCCCCAGGCGGGU-3′) and MAP3K9 as well as their corresponding negative control (NC) plasmids were synthesized by Shanghai Genepharma Company and loaded into the pcDNA 3.1 plasmids. The transfection of all plasmids (50 nM) was conducted using Lipofectamine™ 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Following transfection for 24 h, the efficiency of transfection was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). For observing the effect of upregulated miR-490-5p on cell viability and predicting the target of miR-490-5p, BICR 18 and FaDu cells were treated with medium alone, negative control (NC) plasmid of miR-490-5p or the overexpression plasmid of miR-490-5p (mimics) and grouped respectively as control, NC and miR-490-5p mimics groups. The expression vector of MAP3K9 cDNA was transfected into BICR 18 and FaDu cells to achieve the effect of MAP3K9 overexpression. For determining the effectiveness of the overexpression of MAP3K9 in cells, BICR 18 and FaDu cells were treated with medium alone, NC plasmid of MAP3K9 or overexpression plasmid of MAP3K9-cDNA, which were respectively grouped as control, NC and MAP3K9 groups. For the investigation of the role of MAP3K9 in the regulation of miR-490-5p on cell viability, migration, invasion, EMT and colony formation, BICR 18 and FaDu cells were treated with medium alone, NC plasmid of miR-490-5p, overexpression plasmid of miR-490-5p (mimics), overexpression plasmid of miR-490-5p (mimics) and MAP3K9 as well as the overexpression plasmid of MAP3K9, which were grouped as control, NC, mimics, mimics + MAP3K9 and MAP3K9 group, respectively.
RT-qPCR analysis
According to the manufacturer's protocol, total RNA of cells was extracted by TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNAs were synthesized by iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc.). The RT reaction was performed at 42°C for 30 min, followed by reverse transcriptase inactivation at 85°C for 15 sec. qPCR was performed using Fast Start Universal SYBR-Green Master kit (Roche Diagnostics) and the gene-specific primers. The reactions were run in 96-well plates using the following formula and program: 2X SYBR-Green master mix 10 μl, cDNA template 1 μl, forward primer (10 μM) 1 μl, reverse primer (10 μM) 1 μl, ddH2O 7 μl, 2 min of hot start at 95°C 40 cycles for 15 sec at 95°C for 60 sec at 60°C and for 2 min at 10°C. The data were analyzed by the 2-ΔΔCq method (18) with normalization to the control. Primers used are listed in Table I.
Table I.
The sequences of primers.
| Primer name | Direction | Sequence (5′-3′) |
|---|---|---|
| GAPDH | F | GAGTCAACGGATTTGGTCGT |
| GAPDH | R | CATGGGTGGAATCATATTGGA |
| E-cadherin | F | TACACTGCCCAGGAGCCAGA |
| E-cadherin | R | TGGCACCAGTGTCCGGATTA |
| Vimentin | F | AGGCAAAGCAGGAGTCCACTGA |
| Vimentin | R | ATCTGGCGTTCCAGGGACTCAT |
| MMP-9 | F | CTCTGGAGGTTCGACGTGAA |
| MMP-9 | R | TCAACTCACTCCGGGAACTC |
| TIMP-2 | F | CCAGAAGAAGAGCCTGAACCA |
| TIMP-2 | R | GTCCATCCAGAGGCACTCATC |
| miR-490-5p-RT | CTCAACTGGTGTCGTGGAGTC | |
| GGCAATTCAGTTGAGUCCACCCA | ||
| miR-490-5p | F | CATGGATCTCCAGGTGG |
| miR-490-5p | R | GAACATGTCTGCGTATCTC |
| TNKS2 | F | GTGAATGCCCAAGACAAAGGAGG |
| TNKS2 | R | GGTGTGAAAGCCCATTTGTCCG |
| MAP3K9 | F | AGGGTTCACCAGCCTTATGGAG |
| MAP3K9 | R | GGTGAATGCTGTAGGCGACTCT |
| NUFIP2 | F | AGCACCAGGAAACGCCGAAGAA |
| NUFIP2 | R | CTTGCTGGTTGCCATTGAGGAC |
| CLCC1 | F | TTCAGACTGGCAACAAGAGCCC |
| CLCC1 | R | TTCCACCTGTCTCCTTGGGCTT |
| ABCC2 | F | GCCAACTTGTGGCTGTGATAGG |
| ABCC2 | R | ATCCAGGACTGCTGTGGGACAT |
F, forward; R, reverse; RT, reverse transcription; miR, microRNA; MMP, matrix metalloproteinase; E, epithelial; TIMP-2, tissue inhibitor of metalloproteinase; MAPK, mitogen activated protein kinase.
The assessment of cell viability by Cell Counting Kit-8 (CCK-8)
The assessment of cell viability was conducted using CCK-8 (HY-K0301; MedChemExpress). Cells in each group described above were seeded in a 96-well plate at a density of 104-105 cells/well in 100 μl of culture medium and incubated in an incubator with CO2 at 37°C for 24 and 48 h. 10 μl of CCK-8 solution was added to each well of the plate, which was further incubated for another 1 h in the incubator. The absorbance of each sample was measured at 450 nm using a microplate reader (Multiskan; Thermo Fisher Scientific, Inc.) after the plate had been agitated on an orbital shaker for 1 min.
Luciferase assay
Cells were seeded in a 24-well plate at a cell density 6×104 cells/well. The point mutation of MAP3K9 in BICR 18 and FaDu cells was created by the Quick-Change Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.). Cells carrying wild or mutant type of MAP3K9 were grouped and treated as aforementioned. miR-490-5p was co-transfected with control vectors pRL-TK (Promega Corporation) coding for Renilla luciferase. Transfection was conducted using Lipofectamine™ 2000. Following transfection for 24 h, the luciferase activity was determined by GloMax 20/20 Luminometer (Promega Corporation). DNA sequences were confirmed by sequencing. Dual-Glo luciferase assay kit (Promega Corporation) was used to detect the luciferase activity after the cells had been gathered and lysed 48 h. Values of the firefly luciferase were normalized to Renilla.
The extraction of total protein
Cells were lysed in 1 ml of radioimmunoprecipitation assay buffer (cat. no. 89900; Thermo Fisher Scientific, Inc.) and centrifuged at 4°C 16,000 × g for 30 min, and the supernatant of cells was then gathered. The concentration of total protein was determined by the Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). According to the protocol given by the manufacturer, 20 μl of standard proteins was diluted and 25 μl of samples were added into a 96-well plate. A total of 200 μl of the Working Regent was added to each well and well mixed for 30 sec. Plate was placed in the dark at 37°C for 30 min. The OD at 562 nm was read on a microplate reader (Multiskan; Thermo Fisher Scientific, Inc.).
Western blotting
Protein samples (20 μg/lane) were separated on 10% SDS-PAGE for 60 min at 80 V and transferred to a polyvinylidene difluoride membrane for 60 min at 80 V. The membrane was washed once in PBS with 0.2% Tween 20 (PBST) and then blocked with 5% non-fat milk for 1 h at room temperature (RT). The membrane was further probed with anti-MAP3K9 antibody (cat. no. ab228752), anti-β actin antibody (cat. no. ab115777), anti-matrix metalloproteinase (MMP)-9 antibody (cat. no. ab76003), anti-tissue inhibitor of metalloproteinase 1 (TIMP2) antibody (cat. no. ab180630), anti-epithelial (E)-Cadherin antibody (cat.no. ab40772), anti-vimentin antibody (cat. no. ab92547; all 1:1,000; Abcam) in non-fat milk and incubated overnight at 4°C. The membranes were then washed 3 times in PBST for 5 min and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibodies (cat. no. ab205718; Abcam) were added at 1:2,000 dilution in non-fat milk for 1 h at RT. The membranes were then exposed to Pierce™ enhanced chemiluminescence (ECL) plus western blotting substrate (Thermo Fisher Scientific, Inc.) for 1 min in the dark. The resulting bands were scanned and the images were captured in the ChemiDoc MP (Bio-Rad Laboratories Inc.) and the gray level of the bands was analyzed by the ImageJ software (version 1.46; National Institutes of Health).
Wound healing test
Cells were seeded in the wells of a 6-well plate at a density of 5×104 cells/well until 90% confluence was reached. Each confluent cell monolayer was scratched by a sterile plastic tip in order to produce a straight gap. The debris and the edge of the gap were washed with PBS. The cells were cultured in complete medium for 24 h and the relative wound distance was analyzed based on the images under the inverted light microscope (Olympus IX71; Olympus Corporation) at the same observation site. The distance between the gaps was set as the mean of the distance between upper, middle and lower edges. The distance in the images was measured by ImageJ software.
The assessment of cell invasion rate by Transwell
A total of 3×104 cells with 100 μl serum-free medium were planted in the top chamber of a Transwell apparatus (8-μm) with a Matrigel-coated membrane (BD Biosciences) for Transwell invasion assay. The bottom chamber was filled with medium supplimented with 10% FBS. Following incubation of the Transwell for 12 h, the cells that invaded into the surface of the bottom chamber were fixed with 4% paraformaldehyde for 15 min at room temperature, stained with 0.5% crystal violet for 20 min at room temperature and counted under an Olympus IX71 microscope. The images of the bottom of each camber in the same site were imaged.
Plate colony formation assay
Cells were collected and seeded into the wells of a 6-well plate at a density of 200 cells/dish for 48 h. A total of 700 μg/ml of G418 (Abcam) was used to detect the positive cell clones for 3 weeks till the visible cell colony was visible. Medium was replaced every 3 days. Next, cell colony formations were gently washed with PBS and fixed with methanol for 15 min at room temperature. Colonies were stained with 0.5% crystal violet for 20 min at room temperature. The stain was then carefully washed off with running water and dried. Each colony in the wells was imaged and counted under an Olympus IX71 microscope.
Statistical analysis
All the data analysis was performed by GraphPad Prism v7.0 software (Graphpad Software, Inc.) and presented as mean ± standard deviation. The variantions between different groups were analyzed by one-way analysis of variance followed by Turkey's multiple comparison. The comparison of the OS rates was analyzed by log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-490-5p expression is depressed in primary pharyngolaryngeal cancer tissues and cell lines, leading to an unfavorable prognosis
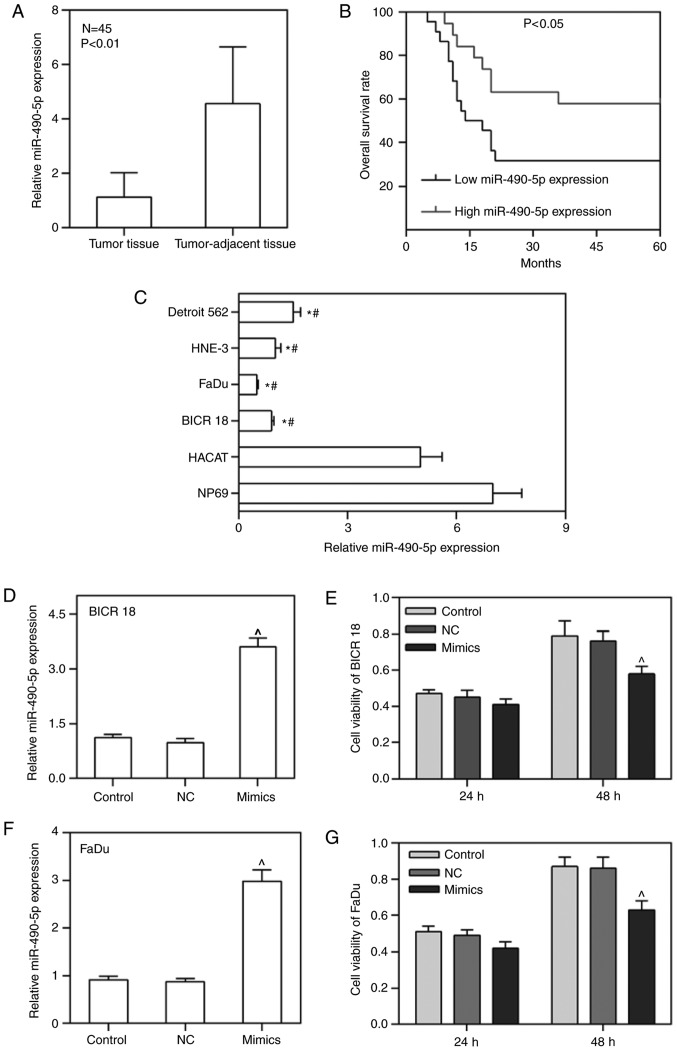
To confirm that the expression of miR-490-5p was downregulated in primary pharyngolaryngeal cancer tissues and cell lines as well as its effect on the prognosis, the mRNA level of miR-490-5p in tumor tissues, tumor-adjacent tissues, NP69, HaCaT, BICR 18, FaDu, HNE-3 and Detroit 562 cell lines was measured. As expected, the expression of miR-490-5p in tumor tissues was significantly decreased compared with in tumor-adjacent tissues (P<0.01; Fig. 1A). Similarly, the expression of miR-490-5p in BICR 18, FaDu, HNE-3 and Detroit 562 cells were decreased compared with HaCaT and NP69, particularly in BICR 18 and FaDu cells (P<0.05; Fig. 1C). Therefore, BICR 18 and FaDu cells were selected for later experiments. Furthermore, patients with low miR-490-5p expression demonstrated a significantly decreased OS rate compared with high miR-490-5p expression (P<0.05; Fig. 1B). The data from the present study suggested that the downregulation of miR-490-5p in pharyngolaryngeal cancer was a common condition, leading to an unfavorable prognosis.
Figure 1.
Fold-change in expression of miR-490-5p relative to the internal control gene (U6) in laryngeal tumor, tumor-adjacent tissues and tumor cell lines as well as the overall survival curves of pharyngolaryngeal cancer with different expression levels of miR-490-5p and the alteration of cell viabilities in control, NC and miR-490-5p mimics groups of BICR 18 and FaDu cells. (A) Relative miR-490-5p expression in laryngeal tumor and tumor-adjacent tissues. (B) The overall survival curves of pharyngolaryngeal cancer with low or high expression level of miR-490-5p. (C) Relative miR-490-5p expression in Detroit 562, HNE-3, FaDu, BICR 18, HaCaT and NP 69 cell lines. (D) Relative miR-490-5p expression in each group of BICR 18 cells. (E) Cell viability of BICR 18 cells in each group following 24 and 48 h of culturing since the transfection. (F) Relative miR-490-5p expression in each group of FaDu cells. (G) Cell viability of FaDu cells in each group following 24 and 48 h of culturing since the transfection. Bars indicated the mean ± standard deviation. *P<0.05 vs. NP69 cells; #P<0.05 vs. HaCaT cells; ^P<0.05 vs. NC groups. NC, negative control; miR, microRNA.
miR-490-5p overexpression decreases the cell viability of pharyngolaryngeal cancer cells
In order to determine whether the upregulation of miR-490-5p could affect the cell viability of pharyngolaryngeal cancer cells, the miR-490-5p overexpression plasmid (mimics) was transfected into BICR 18 and FaDu cells, and the cell viabilities of BICR 18 and FaDu cells 24 and 48 h following culturing were measured. miR-490-5p was successfully overexpressed in BICR 18 and FaDu cells. The cell viabilities in BICR 18 and FaDu cells were significantly decreased in the mimic group compared with the NC groups following 48 h of culturing (P<0.05; Fig. 1D-G). The results demonstrated that the upregulation of miR-490-5p was able to lower the proliferation of pharyngolaryngeal cancer cells.
miR-490-5p could target MAP3K9 in BICR 18 and FaDu cell lines
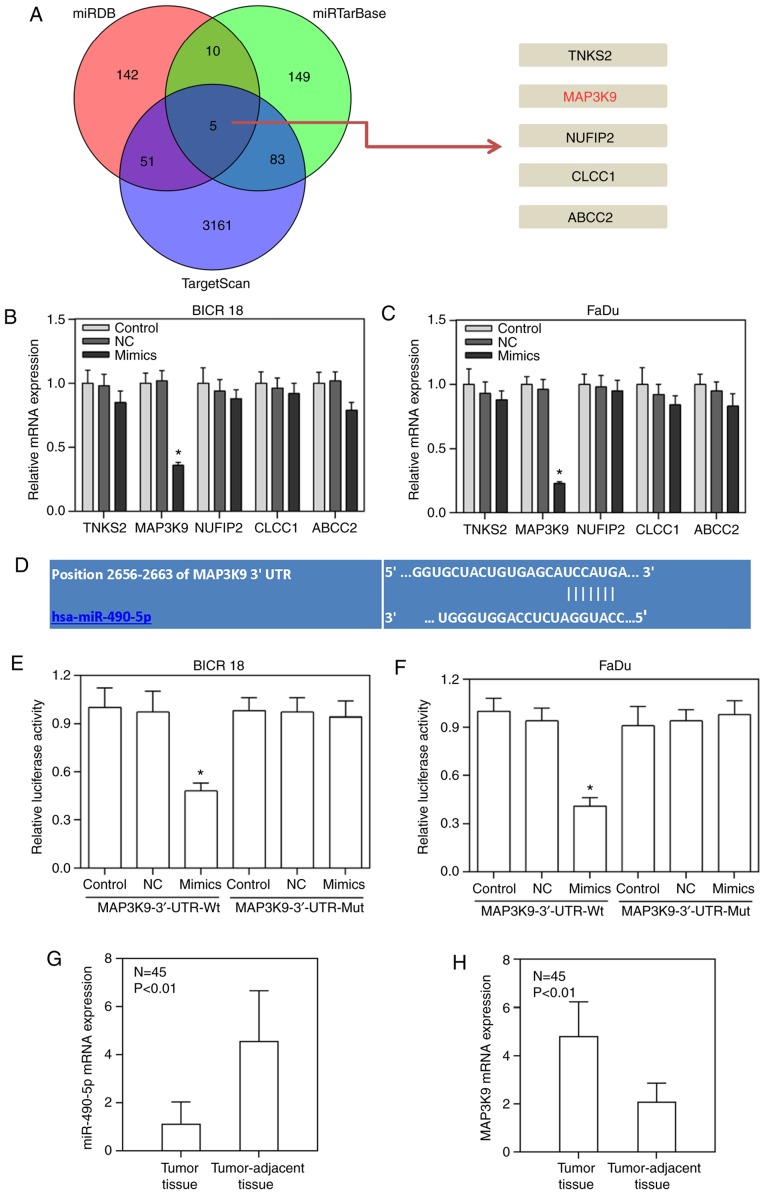
To understand how the upregulation of miR-490-5p led to decreased cell viability of BICR 18 and FaDu cells, the targets of miR-490-5p were predicted using targetscan, miRTarBase and miRDB databases and verified using a luciferase assay. The results of the Venn diagram demonstrated that TNKS2, MAP3K9, NUFIP2, CLCC1 and ABCC2 were target genes for miR-490-5p in results from targetscan, miRTarBase and miRDB databases (Fig. 2A). It was suggested that these five genes may be the target for miR-490-5p. Furthermore, the relative mRNA expression of MAP3K9 in miR-490-5p mimics group was significantly decreased compared with in NC groups in the BICR 18 and FaDu cells, while the other four genes exhibited no significant difference between the miR-490-5p mimics group and NC group (P<0.05; Fig. 2B and C). Moreover, the relative luciferase activities in the miR-490-5p mimics groups were significantly decreased compared with NC groups when the sequence of MAP3K9-3′-UTR was wild type, however, no significance difference was exhibited between NC and miR-490-5p mimics groups when the sequence of MAP3K9-3′-UTR was mutated in both BICR 18 and FaDu cells (P<0.05; Fig. 2D-F). Therefore, it was proved that MAP3K9 was the target gene for miR-490-5p. The expression of MAP3K9 in tumor tissues was significantly increased compared with in tumor-adjacent tissues (P<0.01; Fig. 2H), and the expression of MAP3K9 in tumor tissues and tumor-adjacent tissues was opposite to that of miR-490-5p (P<0.01; Fig. 2G and H).
Figure 2.
Prediction of the target of miR-490-5p. (A) Venn diagram exhibiting the most possible targets of miR-490-5p (TNKS2, MAP3K9, NUFIP2, CLCC1 and ABCC2) predicting from targetscan, miRTarBase and miRDB databases. (B) Relative mRNA expressions of TNKS2, MAP3K9, NUFIP2, CLCC1 and ABCC2 in control, NC and miR-490-5p mimics groups of BICR 18 cells. (C) Relative mRNA expressions of TNKS2, MAP3K9, NUFIP2, CLCC1 and ABCC2 in control, NC and miR-490-5p mimics groups of FaDu cells. (D) The possible complementary sequences of MAP3K9 3′-UTR and miR-490-5p. (E) Relative luciferase activities in control, NC, mimics groups of BICR 18 cells when the sequences of MAP3K9-3′-UTR was wild or mutant type. (F) Relative luciferase activities in control, NC, mimics groups of FaDu cells when the sequences of MAP3K9-3′-UTR was wild or mutant type. (G) Relative miR-490-5p expression in laryngeal tumor and tumor-adjacent tissues. (H) Relative MAP3K9 expression in laryngeal tumor and tumor-adjacent tissues. Bars indicated the mean ± standard deviation. *P<0.05 vs. NC groups. miR, microRNA; NC, negative control; UTR, untranslated region; MAPK, mitogen activated protein kinase.
MAP3K9 overexpression partially reverses the inhibitory effect of the miR-490-5p mimic on cell viability
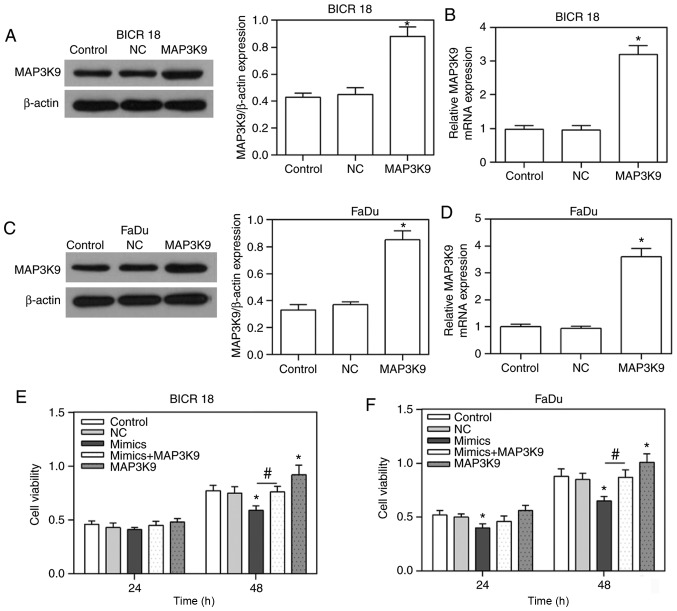
In order to confirm that MAP3K9 was involved in the regulation of miR-490-5p on the cell proliferation, the expression of MAP3K9 was overexpressed in BICR 18 and FaDu cells, and further observed whether it could affect the cell viability in miR-490-5p-upregulated BICR 18 and FaDu cells. As demonstrated in the results of the present study, MAP3K9 was significantly overexpressed in BICR 18 and FaDu cells (P<0.05; Fig. 3A-D). Furthermore, the cell viabilities in miR-490-5p mimics + MAP3K9 groups were increased compared with the miR-490-5p mimics groups in both BICR 18 and FaDu cells, the cell viabilities in miR-490-5p mimics groups were decreased compared with in the NC groups in both BICR 18 and FaDu cells, while the cell viabilities in MAP3K9 groups were increased compared with in NC groups (P<0.05; Fig. 3E and F). Therefore, it indicated that miR-490-5p could target and inhibit the expression of MAP3K9 and lead to the downregulation of proliferation of pharyngolaryngeal cancer cells.
Figure 3.
Overexpression of MAP3K9 in BICR 18 and FaDu cells as well as the alteration of cell viability in control, NC, miR-490-5p mimics, miR-490-5p mimics + MAP3K9 and MAP3K9 groups in BICR 18 and FaDu cells. (A) The protein expression of MAP3K9 in control, NC and MAP3K9 groups of BICR 18 cells. (B) Relative MAP3K9 mRNA expression in control, NC and MAP3K9 groups of BICR 18 cells. (C) The protein expression of MAP3K9 in control, NC and MAP3K9 groups of FaDu cells. (D) Relative MAP3K9 mRNA expression in control, NC and MAP3K9 groups of FaDu cells. (E) The cell viabilities in each group of BICR 18 cells following 24 and 48 h of culturing since the transfection. (F) The cell viabilities in each group of FaDu cells following 24 and 48 h of culturing since the transfection. Bars indicate the mean ± standard deviation. *P<0.05 vs. NC groups; #P<0.05 vs. miR-490-5p mimics groups. miR, microRNA; NC, negative control; MAPK, mitogen activated protein kinase.
MAP3K9 overexpression partially reverses the inhibitory effect of miR-490-5p mimic on cell migration
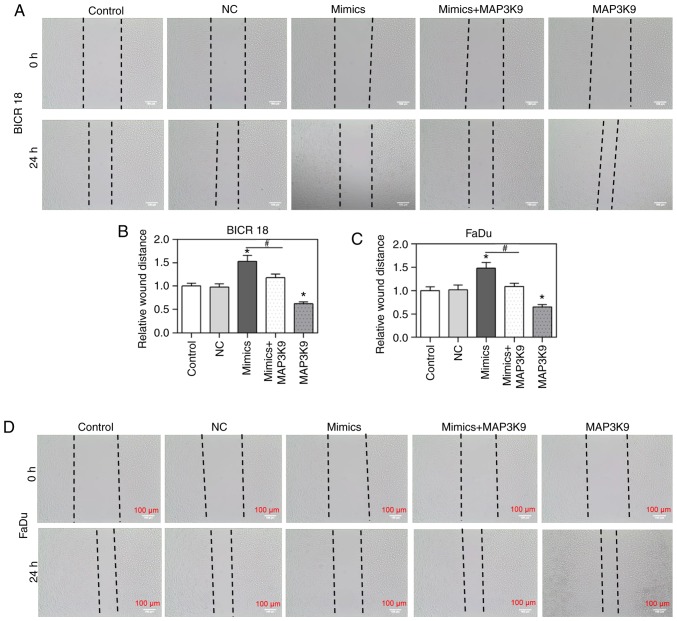
To confirm that MAP3K9 was involved in the regulation of miR-490-5p on the cell migration, the alteration of cell migration rate was measured by wound healing assay in BICR 18 and FaDu cells. The relative wound distances in miR-490-5p mimics groups were identified to be increased compared with in the NC groups but decreased in MAP3K9 groups compared with in NC groups in BICR 18 and FaDu cells (P<0.05; Fig. 4). Significantly, the cell viability in miR-490-5p mimics + MAP3K9 groups was decreased compared with in the miR-490-5p mimics groups in BICR 18 and FaDu cells (P<0.05; Fig. 4). The results of the present study suggested that miR-490-5p could target and inhibit the expression of MAP3K9, further leading to the downregulation of the migration of pharyngolaryngeal cancer cells.
Figure 4.
Relative wound distances in the control, NC, miR-490-5p mimics, miR-490-5p mimics + MAP3K9 and MAP3K9 groups of BICR 18 and FaDu cells. (A) The images demonstrated the distances between the edges of gaps of BICR 18 cell monolayer following 0 and 24 h of culturing in each group. (B) The statistical results of relative wound distance in each group of BICR 18 cells. (C) The statistical results of relative wound distance in each group of FaDu cells. (D) The images demonstrated the distances between the edges of gaps of FaDu cell monolayer following 0 and 24 h of culturing in each group. Bars indicate the mean ± standard deviation. *P<0.05 vs. NC groups; #P<0.05 vs. miR-490-5p mimics groups. miR, microRNA; NC, negative control; MAPK, mitogen activated protein kinase.
MAP3K9 overexpression partially reverses the inhibitory effect of miR-490-5p mimic on cell invasion
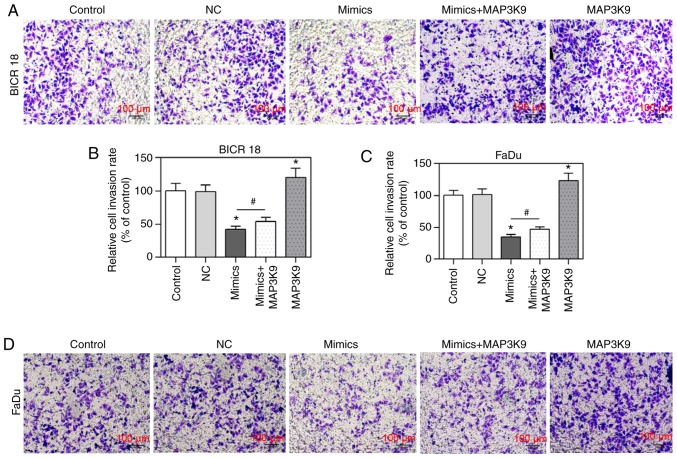
To confirm that MAP3K9 was involved in the regulation of miR-490-5p on cell invasion, the change of cell invasion rate was measured by Transwell invasion assay in BICR 18 and FaDu cells. In the present study, the relative cell invasion rates (of control) in miR-490-5p mimics group were observed to be significantly decreased compared with in the NC groups in BICR 18 and FaDu cells and the relative cell invasion rates (of control) in MAP3K9 group was significantly increased compared with in NC groups in BICR 18 and FaDu cells (P<0.05; Fig. 5). Moreover, the relative cell invasion rates (of control) in miR-490-5p mimics + MAP3K9 groups were significantly increased compared with in the miR-490-5p mimics groups in BICR 18 and FaDu cells (P<0.05; Fig. 5). It suggested that miR-490-5p could target and inhibit the expression of MAP3K9 and further lead to the downregulation of the invasion of pharyngolaryngeal cancer cells.
Figure 5.
Relative cell invasion rates in control, NC, miR-490-5p mimics, miR-490-5p mimics + MAP3K9 and MAP3K9 groups of BICR 18 and FaDu cells. (A) The images demonstrated the cells invaded into the bottom chamber of Transwell apparatus in each group of BICR 18 cells. (B) The statistical outcomes of relative cell invasion rates (of control) in each group of BICR 18 cells. (C) The statistical outcomes of relative cell invasion rates (of control) in each group of FaDu cells. (D) The images demonstrated the cells invaded into the bottom chamber of Transwell apparatus in each group of FaDu cells. Bars indicate the mean ± standard deviation. *P<0.05 vs. NC groups; #P<0.05 vs. miR-490-5p mimics groups. miR, microRNA; NC, negative control; MAPK, mitogen activated protein kinase.
MAP3K9 overexpression partially reverses the inhibitory effect of miR-490-5p mimic on EMT process
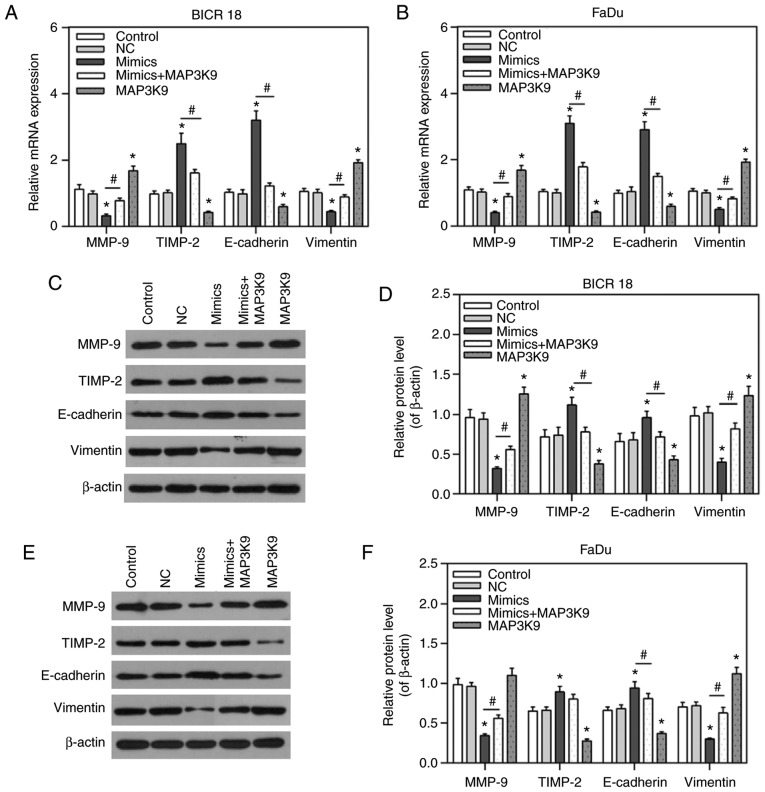
To confirm that MAP3K9 was involved in the regulation of miR-490-5p on the migration, invasion and EMT process, the expression of MMP-9, TIMP-2, E-cadherin and vimentin was measured in BICR 18 and FaDu cells. It was demonstrated that the expression of MMP-9 and vimentin were significantly decreased, while the expression of TIMP-2 and E-cadherin were significantly increased in the miR-490-5p mimics group (P<0.05; Fig. 6). Compared with BICR 18 and FaDu cells in the NC groups, the expression of MMP-9 and vimentin were significantly increased, while the expression of TIMP-2 and E-cadherin were significantly decreased in MAP3K9 groups, compared with BICR 18 and FaDu cells in the NC groups (P<0.05; Fig. 6). In addition, the expression of MMP-9 and vimentin in miR-490-5p mimics + MAP3K9 groups were significantly increased compared with in the miR-490-5p mimics groups, while the expression of TIMP-2 and E-cadherin in miR-490-5p mimics + MAP3K9 groups were significantly decreased in BICR 18 and FaDu cells (P<0.05; Fig. 6) in the miR-490-5p mimics groups. The results of mRNA and protein expression levels were consistent, apart from the relative protein level of TIMP-2 in FaDu cells. It indicated that miR-490-5p could target and inhibit the expression of MAP3K9 and lead to the further downregulation of migration, invasion and EMT process of pharyngolaryngeal cancer cells.
Figure 6.
Expression of MMP-9, TIMP-2, E-cadherin and vimentin in the control, NC, miR-490-5p mimics, miR-490-5p mimics + MAP3K9 and MAP3K9 groups of BICR 18 and FaDu cells. (A) Relative mRNA expressions of MMP-9, TIMP-2, E-cadherin and vimentin in each group of BICR 18 cells. (B) Relative mRNA expressions of MMP-9, TIMP-2, E-cadherin and vimentin in each group of FaDu cells. (C) The original outcomes of western blot of the expressions of MMP-9, TIMP-2, E-cadherin and vimentin in each group of BICR 18 cells. (D) Relative protein levels of MMP-9, TIMP-2, E-cadherin and vimentin (of β-actin) in each group of BICR 18 cells (E) The original outcomes of western blotting of the expression of MMP-9, TIMP-2, E-cadherin and vimentin in each group of FaDu cells. (F) Relative protein levels of MMP-9, TIMP-2, E-cadherin and vimentin (of β-actin) in each group of FaDu cells. Bars indicates the mean ± standard deviation. *P<0.05 vs. NC groups; #P<0.05 vs. miR-490-5p mimics groups. miR, microRNA; NC, negative control; MAPK, mitogen activated protein kinase; E, epithelial; TIMP-2, tissue inhibitor of metalloproteinase; MMP, matrix metalloproteinase.
MAP3K9 overexpression partially reverses the inhibitory effect of the miR-490-5p mimic on the cloning of ability of pharyngolaryngeal cancer cells
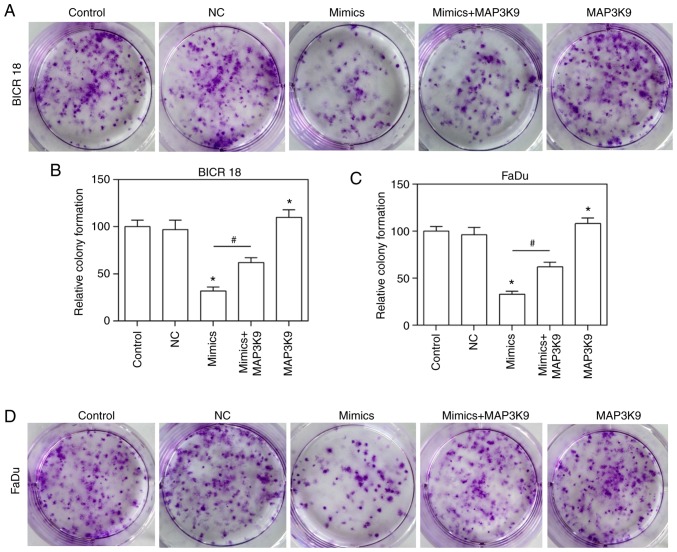
To confirm that MAP3K9 was involved in the regulation of miR-490-5p on the ability of growth, the growth ability of BICR 18 and FaDu cells was measured by plate clone formation assay. The relative colony formation in the miR-490-5p mimics and MAP3K9 groups was significantly decreased and increased, respectively compared with in NC groups in BICR 18 and FaDu cells (P<0.05; Fig. 7). Furthermore, the relative colony formation in miR-490-5p mimics + MAP3K9 groups was significantly increased compared with the miR-490-5p mimics groups in BICR 18 and FaDu cells (Fig. 7; P<0.05). It suggested that miR-490-5p could target and inhibit the expression of MAP3K9 and lead to the downregulation of the ability of cloning of pharyngolaryngeal cancer cells.
Figure 7.
Relative colony formation in the control, NC, miR-490-5p mimics, miR-490-5p mimics + MAP3K9 and MAP3K9 groups of BICR 18 and FaDu cells. (A) The images exhibited the stained colony in each group of BICR 18 cells. (B) Relative colony formation in each group of BICR 18 cells. (C) Relative colony formation in each group of FaDu cells. (D) The images demonstrated the stained colony in each group of FaDu cells. Bars indicates the mean ± standard deviation. *P<0.05 vs. NC groups; #P<0.05 vs. miR-490-5p mimics groups. miR, microRNA; NC, negative control; MAPK, mitogen activated protein kinase.
Discussion
In the present study, it was identified that miR-490-5p was downregulated in laryngeal tumor tissue, tumor-adjacent tissue and tumor cell lines, and that patients who demonstrated a low expression level of miR-490-5p exhibited a poor prognosis during the development of pharyngolaryngeal cancer. Furthermore, it was demonstrated that miR-490-5p could precisely target MAP3K9, which served a critical role in the modulation of miR-490-5p on the proliferation, migration, invasion, EMT and colony formation of BICR 18 and FaDu cells.
In the present study, it was identified that the expression of miR-490-5p was decreased in laryngeal tumor tissues and cell lines, leading to an unfavorable prognosis. According to the study of Xu et al (17) decreased expression of miR-490-5p was detected in hepatocellular carcinoma (HCC) tissues and cells. Furthermore, Fang et al (19) reported that the expression level of miR-490-5p was connected with survival rate of HCC patients. Therefore, it was hypothesized that miR-490-5p seemed to be downregulated in most pharyngolaryngeal cancer tissues and cell lines, leading to a poor prognosis of patients. miR-490-5p expression was also detected in NP69, HaCaT, BICR 18, FaDu, HNE-3 and Detroit 562 cells, and the result demonstrated that miR-490-5p expression in pharyngolaryngeal cancer cells was decreased, particularly in BICR 18 and FaDu cells, which further confirmed that the expression of miR-490-5p was downregulated in pharyngolaryngeal cancer cells. In addition, laryngeal carcinoma is one of the most common head and neck neoplasms, which is mainly represented by squamous cell carcinoma was predominant, BICR 18 and FaDu cells are squamous cells (20,21). Therefore, BICR 18 and FaDu cells were selected for later experiments.
Furthermore, it was identified that miR-490-5p could decrease the cell viabilities of BICR 18 and FaDu cells. Lan et al (22) revealed that the overexpression of miR-490-5p inhibited the proliferation of T24 cells (bladder cancer cells). Therefore, it was hypothesized that the overexpression of miR-490-5p could eventually decrease the proliferation of pharyngolaryngeal cancer cells.
The underlying mechanism of the effect of miR-490-5p on the proliferation of pharyngolaryngeal cancer cells attracted the authors' attention. Targetscan, miRTarBase and miRDB databases are widely used tools for predicting the targets of microRNAs. A total of five candidates were screened out as the targets for miR-490-5p and it was confirmed that MAP3K9 was the most accurate target for miR-490-5p by luciferase assay. To date, to the best of our knowledge, no study has demonstrated that MAP3K9 was the target of miR-490-5p. Therefore, it was the first time that MAP3K9 was proved to be the target for miR-490-5p in BICR 18 and FaDu cell lines.
Through elevating the expression of MAP3K9 in BICR 18 and FaDu cells, which overexpressed miR-490-5p (mimic), it was demonstrated that miR-490-5p may regulate the proliferation of these two cell lines via targeting MAP3K9. In the present study, it was identified that the overexpression of miR-490-5p could decrease cell viability or colony formation by the downregulation of MAP3K9 in cells. MAP3K9 is the upstream kinase in the MAPK signaling pathway, belonging to the MAPK kinase kinase (MAPKK) family (23,24). MAP3K9 is composed of 1,104 amino acids, receiving various stimulatory signals from receptors of cell membranes (23,24). Its phosphorylation can activate the downstream MAPKK and further activate MAPK, which ultimately activate the whole MAPK signaling pathway, which is the classic pathway that regulates plenty of cellular functions including cell proliferation, differentiation, migration and apoptosis (23,24). Nie et al (15) reported that miR-148b increased proliferation in human renal cancer cells by targeting MAP3K9. Xia et al (16) also revealed that MAP3K9 directly targets miR-7, which increased the proliferation of pancreatic cancer cells. This evidence indirectly supported the hypothesis that MAP3K9 participated in the regulation of miR-490-5p on the proliferation of pharyngolaryngeal cancer cells.
Migration and invasion represent the metastatic ability of cancer which is the most important hallmark of tumors (25). It was identified that the overexpressed miR-490-5p could decrease cell migration and invasion by inhibiting MAP3K9. Chen et al (26) reported that miR-490-5p inhibited the migration and invasion of HCC. Furthermore, the participation of MAP3K9 in regulating the migration and invasion of PC cells had been demonstrated (16). Therefore, miR-490-5p may regulate the migration and invasion of pharyngolaryngeal cancer cells by targeting MAP3K9.
MMP-9 belongs to the MMP family and degrades type IV collagen effectively, therefore destroying the integrity of the basal membrane, which is associated with infiltration and metastasis of tumors (27,28). MMP-9 is one of the invasion and migration marker proteins, and a number of studies have demonstrated that the upregulation of MMP-9 could enhance the migration and invasion of a variety of cancer cells including gastric cancer, esophageal squamous cell cancer and oral cancer (29-31). E-cadherin not only inhibits the communication between tumor cells, but also the movement and invasion of tumors (32). The downregulation of E-cadherin could significantly lower the risk of EMT of tumor cells (32,33). vimentin is one of the cytoskeletal proteins and is expressed mainly in cells derived from mesoderm, which is considered to be a key factor in EMT (34,35). TIMP-2 is a type of endogenous enzyme that inhibits the degradation of the extracellular matrix and maintains the stability of the environment within the tissues, therefore resisting the initiation of the EMT process (36,37). Notably, it was identified that miR-490-5p overexpression could suppress MMP-9, vimentin expression and increase E-cadherin, TIMP-2 expression in the BICR 18 and FaDu cells by downregulating MAP3K9 expression. This suggests that miR-490-5p could inhibit the migration, invasion and EMT process of pharyngolaryngeal cancer cells by targeting MAP3K9. Chen et al (38) revealed that miR-490-5p may be a tumor suppressor and was involved in clear cell renal cell carcinomas metastasis. Luo et al (39) demonstrated that the overexpression of miR-148a inhibited cutaneous squamous cell carcinoma cell metastasis by downregulating MAP3K9. Though this study was identified to directly support the present study's findings, it was hypothesized that miR-490-5p may regulate the EMT progress of pharyngolaryngeal cancer cells by targeting MAP3K9.
In the present study, the MAP3K9 expression cDNA vector was transfected into cells to achieve the effect of MAP3K9 overexpression, the cDNA contains the complete UTR region and miR-490-5p can bind to the target site of MAP3K9 to rescue the phenotype of miR-490-5p. It was identified that overexpression of miR-490-5p inhibited the proliferation, migration, invasion and EMT process of BICR 18 and FaDu cells by targeting MAP3K9, however, the effect of silencing miR-490-5p on the proliferation, migration, invasion and EMT process of laryngeal cancer cells remains unknown, and needs to be further investigated. In addition, to verify whether miR-490-5p/MAP3K9 serves a role in the apoptosis, migration, invasion and EMT of laryngeal cancer cells via participating in specific signaling pathways, also needs to be discussed in the future. This study has certain limitations, in the experiment to detect the expression of miR-490-5p in tumor and tumor-adjacent tissues, the results would have been more reliable if the experiment of the in-situ hybridization had been conducted. Furthermore, with reference to the research methods of other studies (40-42), the effect of cell proliferation on migration and invasion was not excluded when detecting the effect of miR-490-5p on cell migration and invasion.
In conclusion, miR-490-5p could target MAP3K9 and further modulate the proliferation, migration, invasion and EMT of pharyngolaryngeal cancer cells. The present study provides a novel entry point to the treatment of pharyngolaryngeal cancer.
Acknowledgments
Not applicable.
Funding
No funding received.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
Substantial contributions to conception and design: AA. Data acquisition, data analysis and interpretation: XC, ML and YT. Drafting the article or critically revising it for important intellectual content: XC and ML. Final approval of the version to be published: All authors, Agreement to be accountable for all aspects of the work in ensuring that questions associated with the accuracy or integrity of the work are appropriately investigated and resolved: AA and XC.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The present study was approved by the Ethics Committee of the People's Hospital of Xinjiang Uygur Autonomous Region, and an informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
References
- 1.de Miguel-Luken MJ, Chaves-Conde M, de Miguel-Luken V, Muñoz-Galván S, López-Guerra JL, Mateos JC, Pachón J, Chinchón D, Suarez V, Carnero A. MAP17 (PDZKIP1) as a novel prognostic biomarker for laryngeal cancer. Oncotarget. 2015;6:12625–12636. doi: 10.18632/oncotarget.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler A, Rigby MH, Scott J, Trites J, Hart R, Taylor SM. A retrospective review in the management of T3 laryngeal squamous cell carcinoma: An expanding indication for transoral laser microsurgery. J Otolaryngol Head Neck Surg. 2016;45:34. doi: 10.1186/s40463-016-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Liu Y, Hu G, Wang R, Zhao Y, Zhang M. The survival rate and larynx preservation in elderly cancer patients who received surgical operation: A retrospective cohort study. Int J Surg. 2016;36:342–346. doi: 10.1016/j.ijsu.2016.11.073. [DOI] [PubMed] [Google Scholar]
- 4.Henriques de Figueiredo B, Grégoire V. How to minimize morbidity in radiotherapy of pharyngolaryngeal tumors? Curr Opin Otolaryngol Head Neck Surg. 2016;24:163–169. doi: 10.1097/MOO.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 5.Cui N, Hao G, Zhao Z, Wang F, Cao J, Yang A. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J Cell Mol Med. 2016;20:1503–1512. doi: 10.1111/jcmm.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meza-Sosa KF, Valle-Garcia D, Pedraza-Alva G, Perez-Martinez L. Role of microRNAs in central nervous system development and pathology. J Neurosci Res. 2012;90:1–12. doi: 10.1002/jnr.22701. [DOI] [PubMed] [Google Scholar]
- 7.Leng R, Zha L, Tang L. miR-718 represses VEGF and inhibits ovarian cancer cell progression. FEBS Lett. 2014;588:2078–2086. doi: 10.1016/j.febslet.2014.04.040. [DOI] [PubMed] [Google Scholar]
- 8.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 9.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin C, Zhang Y, Li J. Upregulation of miR-196a promotes cell proliferation by downregulating p27kip1 in laryngeal cancer. Biol Res. 2016;49:40. doi: 10.1186/s40659-016-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan AR, Zheng H, Kwok JG, Qu Y, Zou AE, Korrapati A, Li PX, Califano JA, Hovell MF, Wang-Rodriguez J, Ongkeko WM. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017;72:56–64. doi: 10.1016/j.oraloncology.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan YH, Liu J, Qu XJ, Hou KZ, Wang KF, Liu YP, Wu B. β-Elemene induces apoptosis in human renal-cell carcinoma 7860 cells through inhibition of MAPK/ERK and PI3K/Akt/mTOR signalling pathways. Asian Pac J Cancer Prev. 2012;13:2739–2744. doi: 10.7314/APJCP.2012.13.6.2739. [DOI] [PubMed] [Google Scholar]
- 13.Du HF, Ou LP, Song XD, Fan YR, Yang X, Tan B, Quan Z, Luo CL, Wu XH. Nuclear factor-κB signaling pathway is involved in phospholipase Cε-regulated proliferation in human renal cell carcinoma cells. Mol Cell Biochem. 2014;389:265–275. doi: 10.1007/s11010-013-1948-4. [DOI] [PubMed] [Google Scholar]
- 14.Hannes S, Abhari BA, Fulda S. Smac mimetic triggers necroptosis in pancreatic carcinoma cells when caspase activation is blocked. Cancer Lett. 2016;380:31–38. doi: 10.1016/j.canlet.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 15.Nie F, Liu T, Zhong L, Yang X, Liu Y, Xia H, Liu X, Wang X, Liu Z, Zhou L, et al. MicroRNA-148b enhances proliferation and apoptosis in human renal cancer cells via directly targeting MAP3K9. Mol Med Rep. 2016;13:83–90. doi: 10.3892/mmr.2015.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia J, Cao T, Ma C, Shi Y, Sun Y, Wang ZP, Ma J. miR-7 suppresses tumor progression by directly targeting MAP3K9 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:121–132. doi: 10.1016/j.omtn.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu B, Xu T, Liu H, Min Q, Wang S, Song Q. miR-490-5p suppresses cell proliferation and invasion by targeting BUB1 in hepatocellular carcinoma cells. Pharmacology. 2017;100:269–282. doi: 10.1159/000477667. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using rea time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Fang ZQ, Li MC, Zhang YQ, Liu XG. miR-490-5p inhibits the metastasis of hepatocellular carcinoma by down-regulating E2F2 and ECT2. J Cell Biochem. 2018;119:8317–8324. doi: 10.1002/jcb.26876. [DOI] [PubMed] [Google Scholar]
- 20.Lu W, Feng L, Li P, Wang Y, Du Y, Chen X, Wu S, Zhao G, Lou W. Effects of HPV-16 infection on hypopharyngeal squamous cell carcinoma and FaDu cells. Oncol Rep. 2016;35:99–106. doi: 10.3892/or.2015.4340. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Wawrose JS, Gooding WE, Garraway LA, Lui VW, Peyser ND, Grandis JR. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: A rational approach to preclinical model selection. Mol Cancer Res. 2014;12:571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan G, Yang L, Xie X, Peng L, Wang Y. MicroRNA-490-5p is a novel tumor suppressor targeting c-FOS in human bladder cancer. Arch Med Sci. 2015;11:561–569. doi: 10.5114/aoms.2015.52359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durkin JT, Holskin BP, Kopec KK, Reed MS, Spais CM, Steffy BM, Gessner G, Angeles TS, Pohl J, Ator MA, Meyer SL. Phosphoregulation of mixed-lineage kinase 1 activity by multiple phosphorylation in the activation loop. Biochemistry. 2004;43:16348–16355. doi: 10.1021/bi049866y. [DOI] [PubMed] [Google Scholar]
- 24.Slattery ML, Hines LH, Lundgreen A, Baumgartner KB, Wolff RK, Stern MC, John EM. Diet and lifestyle factors interact with MAPK genes to influence survival: The Breast Cancer Health Disparities Study. Cancer Causes Control. 2014;25:1211–1225. doi: 10.1007/s10552-014-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Ye L, Wen D, Chen F. miR-490-5p inhibits hepatocellular carcinoma cell proliferation, migration and invasion by directly regulating ROBO1. Pathol Oncol Res. 2019;25:1–9. doi: 10.1007/s12253-017-0305-4. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Li K, Liang Q, Zheng G, Zhang S, Lao X, Liang Y, Liao G. Elevated hydrostatic pressure promotes ameloblastoma cell invasion through upregulation of MMP-2 and MMP-9 expression via Wnt/β-catenin signalling. J Oral Pathol Med. 2018;47:836–846. doi: 10.1111/jop.12761. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Tao Y, Li X. Expression of MMP-9/TIMP-2 in nasal polyps and its functional implications. Int J Clin Exp Pathol. 2015;8:14556–14561. [PMC free article] [PubMed] [Google Scholar]
- 29.Chang X, Xu X, Xue X, Ma J, Li Z, Deng P, Chen J, Zhang S, Zhi Y, Dai D. NDRG1 controls gastric cancer migration and invasion through regulating MMP-9. Pathol Oncol Res. 2016;22:789–796. doi: 10.1007/s12253-016-0071-8. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Song X, Zhu J, Li M, Ji Y, Wu F, Chen Y, Cui X, Hu J, Wang L, et al. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J Oncol. 2017;51:378–388. doi: 10.3892/ijo.2017.4015. [DOI] [PubMed] [Google Scholar]
- 31.Hsu LS, Huang RH, Lai HW, Hsu HT, Sung WW, Hsieh MJ, Wu CY, Lin YM, Chen MK, Lo YS, Chen CJ. KLF6 inhibited oral cancer migration and invasion via downregulation of mesenchymal markers and inhibition of MMP-9 activities. Int J Med Sci. 2017;14:530–535. doi: 10.7150/ijms.19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazdani J, Ghavimi MA, Jabbari Hagh E, Ahmadpour F. The role of E-cadherin as a prognostic biomarker in head and neck squamous carcinoma: A systematic review and meta-analysis. Mol Diagn Ther. 2018;22:523–535. doi: 10.1007/s40291-018-0351-y. [DOI] [PubMed] [Google Scholar]
- 33.Tiwari N, Gheldof A, Tatari M, Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012;22:194–207. doi: 10.1016/j.semcancer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Du Y, Beckford J, Alachkar H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J Transl Med. 2018;16:170. doi: 10.1186/s12967-018-1539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CY, Lin HH, Tang MJ, Wang YK. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6:15966–15983. doi: 10.18632/oncotarget.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Łukaszewicz-Zając M, Mroczko B, Guzińska-Ustymowicz K, Pryczynicz A, Gryko M, Kemona A, Kędra B, Szmitkowski M. Matrix metalloproteinase 2 (MMP-2) and their tissue inhibitor 2 (TIMP-2) in gastric cancer patients. Adv Med Sci. 2013;58:235–243. doi: 10.2478/ams-2013-0018. [DOI] [PubMed] [Google Scholar]
- 37.Urbaniak-Kujda D, Kapelko-Slowik K, Prajs I, Dybko J, Wolowiec D, Biernat M, Slowik M, Kuliczkowski K. Increased expression of metalloproteinase-2 and -9 (MMP-2, MMP-9), tissue inhibitor of metalloproteinase-1 and -2 (TIMP-1, TIMP-2), and EMMPRIN (CD147) in multiple myeloma. Hematology. 2016;21:26–33. doi: 10.1179/1607845415Y.0000000043. [DOI] [PubMed] [Google Scholar]
- 38.Chen K, Zeng J, Tang K, Xiao H, Hu J, Huang C, Yao W, Yu G, Xiao W, Guan W, et al. miR-490-5p suppresses tumour growth in renal cell carcinoma through targeting PIK3CA. Biol Cell. 2016;108:41–50. doi: 10.1111/boc.201500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Q, Li W, Zhao T, Tian X, Liu Y, Zhang X. Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch Biochem Biophys. 2015;583:47–54. doi: 10.1016/j.abb.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Mao D, Qiao L, Lu H, Feng Y. B-cell translocation gene 3 overexpression inhibits proliferation and invasion of colorectal cancer SW480 cells via Wnt/β-catenin signaling pathway. Neoplasma. 2016;63:705–716. doi: 10.4149/neo_2016_507. [DOI] [PubMed] [Google Scholar]
- 41.Jia S, Qu T, Wang X, Feng M, Yang Y, Feng X, Ma R, Li W, Hu Y, Feng Y, et al. KIAA1199 promotes migration and invasion by Wnt/β-catenin pathway and MMPs mediated EMT progression and serves as a poor prognosis marker in gastric cancer. PLoS One. 2017;12:e0175058. doi: 10.1371/journal.pone.0175058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang D, Du G, Xu A, Xi X, Li D. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res. 2017;7:2209–2219. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.