Abstract
In contrast to other teleosts, Atlantic cod (Gadus morhua) has an expanded repertoire of MHC-I and TLR components, but lacks the MHC-II, the invariant chain/CD74, and CD4+ T cell response, essential for production of antibodies and prevention of bacterial infectious diseases. The mechanisms by which G. morhua fight bacterial infections are not well understood. Aeromonas salmonicida subsp. salmonicida is a recurrent pathogen in cultured and wild fish, and has been reported in Atlantic cod. Macrophages are some of the first responders to bacterial infection and the link between innate and adaptive immune response. Here, we evaluated the viability, reactive oxygen species (ROS) production, cell morphology, and gene expression of cod primary macrophages in response to A. salmonicida infection. We found that A. salmonicida infects cod primary macrophages without killing the cod cells. Likewise, infected Atlantic cod macrophages up-regulated key genes involved in the inflammatory response (e.g., IL-1β and IL-8) and bacterial recognition (e.g., BPI/LBP). Nevertheless, our results showed a down-regulation of genes related to antimicrobial peptide and ROS production, suggesting that A. salmonicida utilizes its virulence mechanisms to control and prevent macrophage anti-bacterial activity. Our results also indicate that Atlantic cod has a basal ROS production in non-infected cells, and this was not increased after contact with A. salmonicida. Transmission electron microscopy results showed that A. salmonicida was able to infect the macrophages in a high number, and release outer membrane vesicles (OMV) during intracellular infection. These results suggest that Atlantic cod macrophage innate immunity is able to detect A. salmonicida and trigger an anti-inflammatory response, however A. salmonicida controls the cell immune response to prevent bacterial clearance, during early infection.
Keywords: Atlantic cod, Gadiform, innate immunity, primary macrophages, Aeromonas salmonicida, Gram-negative
Introduction
Atlantic cod (Gadus morhua), one of the most important commercial fish species in the North Atlantic fisheries, has unusual modifications of the immune gene repertoire that set it apart from other teleosts (1). This Gadiform fish lacks the genes for the major histocompatibility complex class II (MHC-II), the invariant chain/CD74 (Ii), and CD4+ T cell response, representing an important evolutionary diversification of the adaptive immune system of vertebrates (2). The MHC-II binds antigens from extracellular pathogens, and the MHC-II-antigen complex activates helper CD4+ T cells, which play an essential role fighting bacterial infectious diseases (3).
The Atlantic cod appears to have compensated for the lack of the MHC-II pathway by expanding the number of MHC-I genes (4). This expanded MHC-I gene family has been divided into two clades, one maintaining the classical MHC-I functionality, and the other showing a MHC-II-like function (2). Indeed, around 80–100 copies of the MHC-I loci are found in the Atlantic cod genome, in contrast to other gadiformes that present only 40 copies (1, 2, 5), or to humans that harbor only ~10 copies (6). In addition to the MHC-I diversification, the Atlantic cod has expanded some Toll-like receptor (TLR) families, which have an important role in the innate immune response and pathogen detection (2, 7, 8). The Atlantic cod lacks TLR1, TLR2, and TLR5 that recognize bacterial surface antigens, however, this seems to be compensated by an expansion of the TLR7, TLR8, TLR9, and TLR22 families related to nucleic acids recognition (2, 6, 9, 10).
The Atlantic cod is a very successful teleost species and not particularly susceptible to infectious diseases (11), even though some of the prevalent marine bacterial pathogens such as Vibrio anguillarum, Francisella noatunensis, and Aeromonas salmonicida have been reported in wild and cultured cod (12–14).
A. salmonicida, is worldwide found in aquatic environments and have been implicated in the etiology of a large variety of fish diseases (15). A. salmonicida has five subspecies, salmonicida, achromogenes, smithia, masoucida, and pectinolytica (15). Immune response of Atlantic cod to subsp. achromogenes infection has been described (13, 16). In contrast, immune response of Atlantic cod to subsp. salmonicida has not been studied.
Aeromonas salmonicida subsp. salmonicida (hereafter A. salmonicida), a causative agent of the furunculosis, is a recurrent health problem for several marine fish species (17). This Gram negative, facultative anaerobic, non-motile, and bacillus shaped bacterium (18, 19), contains among others, a type-three secretion system that translocates to the eukaryotic cell several effector proteins, which influence immune response, including inflammation (20, 21).
Inflammation is a protective reaction of the host in response to bacterial infection, involving the migration of leukocytes, including macrophages, to the site of infection (22).
Macrophages, in addition to neutrophils, are the first defense line of vertebrates, including in Atlantic cod. Upon infection, macrophages are activated, secreting antimicrobial peptides (AMPs), cytokines, and chemokines, among others immune modulatory molecules (11, 23, 24). Activated macrophages have an increased phagocytic activity, correlated with an increased production of reactive oxygen species (ROS), and up-regulation of anti-bacterial gene transcription (25–27).
How Atlantic cod primary macrophages respond to A. salmonicida infection is unknown. Therefore, the aim of this study was to investigate the immune response of Atlantic cod head kidney primary macrophages to A. salmonicida infection.
We determined that Atlantic cod primary macrophages are able to mount an immune response and survive during the first 6 h of A. salmonicida infection. However, A. salmonicida invade the macrophages in a short period of time, remain in intracellular A. salmonicida containing vesicles, and modify macrophage immunity, likely preventing the action of AMPs and apoptosis.
Materials and Methods
Aeromonas salmonicida Growth Conditions
A single colony of A. salmonicida J223 (17) was grown routinely in 3 ml of Trypticase Soy Broth (TSB, Difco, Franklin Lakes, NJ) at 15°C in a 16 mm diameter glass tube and placed in a roller for 24 h. After growth, 300 μl of the overnight culture were added in 30 ml of TSB media using a 250 ml flask and incubated for 24 h at 15°C with aeration (180 rpm). The bacterial growth was monitored spectrophotometrically until O.D. 600 nm ~0.7 (1 × 108 CFU ml−1) using the Genesys 10 UV spectrophotometer (Thermo Spectronic, Thermo Fischer Scientific Inc., Waltham, MA, USA). Then the bacterial culture was centrifuged at 6,000 rpm at room temperature for 10 min. The pellet was washed twice with PBS and centrifuged at 6,000 rpm at room temperature for 5 min, and finally resuspended in 300 μl of PBS (~5 × 1010 CFU ml−1). The concentrated bacterial inoculum was serial diluted and quantified by plating onto TSA supplemented with Congo red (50 μg ml−1).
Formalin-Killed A. salmonicida
A. salmonicida J223 strain was grown in TSB media supplemented with 100 μM 2, 2′-dypyridyl at 15°C with aeration (180 rpm) up to an optical density of O.D. 600 nm ~0.7 (~1 × 108 CFU ml−1). The bacterial cells were washed three times with PBS and then fixed with 6% formalin for 3 days at room temperature with gentile agitation. Formalin killed cells were dialyzed (Molecular Weight cut off 3.5 kDa; Spectra/Por, Laguna Hills, CA) in PBS three times and stored at 4°C at the concentration of 6 × 1010 CFU ml−1 until utilization.
Fish Holding
Adult specimens of Atlantic cod 1.5 ± 0.2 kg (mean ± SE) were obtained from the Dr. Joe Brown Aquatic Research Building (JBARB) at the Department of Ocean Sciences, Memorial University of Newfoundland, Canada. The animals were kept in 21 m3 tanks, with flow-through (75.l × min−1) of sea water (6°C) and ambient photoperiod. The individuals were fed with commercial dry pellets (Skretting: 50% protein, 18% fat, 1.5% carbohydrate, 3% calcium, 1.4% phosphorus) with a ratio of 1% of body weight 3 days per week. The experiment was performed in accordance with the guidelines of the Canadian Council on Animal Care and approved by Memorial University of Newfoundland's Institutional Animal Care Committee (protocols #17-01-JS; #17-02-JS).
Macrophage Isolation
Primary macrophages were isolated from Atlantic cod head kidney in accordance to the protocol established by Eslamloo et al. (28) with modifications. Briefly, head kidney tissues from six fish were aseptically removed and individually minced through 100 μm nylon sterile cell strainers (Fisher Scientific, Thermo Fisher Scientific, Waltham, MA, USA) in isolation media [(Leibovitz-15 (Gibco®, Gran Island, NY, USA) supplemented with 2 mM L-glutamine, 4.2 mM NaHCO3, 25 mM HEPES, 1.8 mM glucose, 20 U ml−1 heparin, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, and 1% Fetal Bovine Serum (FBS)]. After this period, 3 ml of cell suspension were centrifuged (400 × g at 4°C) for 40 min in a 25/51% Percoll gradient (GE Healthcare, Uppsala, Sweden). Macrophages collected from the macrophage-enriched interface were washed with phosphate buffered saline [PBS; 136 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.5 mM KH2PO4 (pH 7.2)] (29) twice and the number and viable cells were determined using the Countness™ cell counter (Invitrogen), and trypan blue stain (Invitrogen). After determining the numbers of cells from each sample, the primary macrophages were seeded in 22 mm 12-well or 35 mm 6-well cell-culture multidishes (Thermo Scientific, Roskilde, Denmark) at a concentration of 1 × 107 cells ml−1. The plates were incubated at 15°C for 24 h in isolation media. After this period the cells were washed with PBS and incubated at 15°C for additional 24 h in 1 ml of culture media [Leibovitz-15 (Gibco®), supplemented with 2 mM L-glutamine, 4.2 mM NaHCO3, 25 mM HEPES, 1.8 mM glucose, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, and 1% FBS] to allow cell attachment until the infection assay.
Gentamicin Exclusion Assay
Infections of primary macrophages with A. salmonicida were performed according to a previously established protocol (17). To remove the antibiotic present in the culture media, the isolated primary macrophages were washed once with 1 ml of PBS, and then inoculated with 1 ml of cultured media without antibiotics. After this, the primary macrophage monolayers were infected with 10 μL of bacterial suspension [~1 × 106 cells ml−1; Multiplicity of Infection (MOI) 1:1 (bacteria:macrophage)] and incubated at 15°C. After 1 h post infection, the A. salmonicida attached to the Atlantic cod macrophages were quantified. The infected primary macrophage monolayers were washed 3 times with PBS, and lysed with 400 μl of Triton X100 (0.01%; Sigma) during 10 min (30) and then 600 μl of PBS were added to complete 1 ml of lysed macrophage suspension. Then the lysed macrophage suspensions were serially diluted (1:10) and plate/counted on TSA plates supplemented with Congo Red to determine the number of viable A. salmonicida per monolayer. The plates were incubated at 15°C for 5 days to determine the CFU per well. In addition, samples were taken for cell viability, RNA extraction, and transmission electron microscopy (see below for details).
For the invasion assay, cell monolayers were infected for 1 h, washed 3 times with PBS, followed by the addition of 1 ml of fresh culture media supplemented with gentamicin (10 μg ml−1, a higher concentration than the minimal inhibitory concentration for A. salmonicida) (31), and incubated at 15°C. Samples were taken at 2, 3, and 6 h post infection for bacterial count, cell viability, RNA extraction, and transmission electron microscopy. All the macrophages were isolated from three individual fish and triplicates were utilized for each treatment in the assays.
Primary Macrophage Viability Determination
To determine the viability of infected primary macrophages, the cells were seeded in 12 wells plates, infected with A. salmonicida, and processed as described in the gentamicin exclusion assay. For each time point post A. salmonicida infection, the cells were washed with 1 ml of PBS and then treated with 500 μl of trypsin-EDTA (0.5%; Gibco) for 10 to 15 min. After this period, the trypsin was inactivated with 500 μl of culture media. The cells were stained with trypan blue (0.4%; Invitrogen) in a ratio of 1:1 (10 μl: 10 μl) and quantified using Countess™ Cell Counting Chamber Slides (Invitrogen) and Countess® Automated Cell Counter (Invitrogen) according to the manufacturer's instructions. The numbers of alive and dead cells were determined at each time point post- infection. All the macrophages were isolated from three individual fish and technical triplicates were utilized in the assays.
RNA Extraction and qPCR
To determine the effect of A. salmonicida on the innate immune response of Atlantic cod primary macrophages, samples of RNA were isolated from infected cells at 1, 2, and 6 h post A. salmonicida infection, using the previously described gentamycin exclusion methodology. Primary macrophages that were either mock infected with PBS or inoculated with 1 × 106 CFU of formalin killed A. salmonicida were utilized as controls. Total RNA was extracted using TRIzol (Invitrogen), and purified using RNeasy (QIAGEN) following manufacturers' instruction (32). RNA samples were treated with TURBO DNA-free™ Kit (Invitrogen) for complete digestion of DNA and removal of remaining DNase and divalent cations, such as magnesium and calcium. Purified RNA samples were quantified and evaluated for purity using a Nano-quant spectrophotometer (Genway, UK), and evaluated for integrity by 1% agarose gel electrophoresis (29). cDNA was synthetized with the SuperScript™ III First-Strand Synthesis System (Invitrogen) using 500 ng of RNA per reaction and random hexamers according to the manufacturer's instructions.
Primer pair efficiencies were analyzed using a 20 ng μl−1 pooled cDNA from each set of samples, which was serially diluted (dilutions starting with 1 (20 ng μl−1), 1:3 (6.67 ng μl−1), 1:9 (2.22 ng μl −1), 1:27 (0.74 ng μl−1), 1:81 (0.25 ng μl−1), 1:243 (0.08 ng μl−1), 1:729 (0.03 ng μl−1)). Primer pair efficiencies were calculated using the formula E = 10(−1/slope) (33).
All qPCR reactions were done in a final volume of 20 μL, containing 10 μL of 1 × PowerUp-SYBR Master Mix (Applied BioSystems, Foster City, CA, USA), 1 μL (10 μM) of each primer, 6 μL of nuclease free water (Ambion), and 2 μL of cDNA. All samples were amplified and detected in a QuantStudio 3 (Applied BioSystems). The reaction mixtures were incubated for 2 min at 95°C, followed by 40 cycles of 1 s at 95°C, 30 s at 60°C, and finally 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. Initially, a total of five Atlantic cod genes were tested as reference gene candidates (EF-1α, ß-actin, Eif3, 18S, 60S). cDNA from a sub-set of samples (Supplementary Table 1) were utilized for evaluation of reference gene stability. The most stable gene for this set of individual samples was determined by using geNorm (M value 0.102) and BestKeeper (Value 0.101) (Supplementary Table 1). After this determination, the individual samples were analyzed. The mRNA gene expression was normalized to the Atlantic cod elongation factor 1 alpha (EF-1a) due to its stability across different treatments. Gene expression was determined using the comparative −ΔΔCt method (34).
The primers used in this study are listed in Table 1. In all cases, each qPCR was performed with triplicate samples and repeated with six independent fish.
Table 1.
Primer sequences used in this experiment.
| Gene | Forward (5′to 3′) | Reverse (5′to 3′) | Tm°C | Efficiency (%) | References |
|---|---|---|---|---|---|
| IL-1b | TGAGGACCTGCTCAACCTCT | TCTTCTGGTGGTCCCTCAAC | 55.6 | 103.7 | (35) |
| IL-8 | GGTTTGTTCAATGATGGGCTGTT | GACCTTGCCTCCTCATGGTAATACT | 56.5 | 98.4 | (36) |
| IL-10 | CCTATAAAGCCATCGGCGAGTTA | TGAAGTCGTCGTTTTGAACCAAG | 56.6 | 100.1 | (36) |
| MHC-I | CTAGCGTGGGACCTGAAGAC | CAGAGTGCTCTTCCCGTAGG | 56.5 | 108.4 | (35) |
| g-type lysozyme | CATTGACCAAGCCACTGGAATCCT | ATTCGACTCTACCGTCTCCAGTGT | 59.3 | 102.3 | (35) |
| BPI/LBP | GACCGTCAACGTGATGGCCCCGGT | CTTTGTTGGCCTCTATGCTGGAGAG | 59.4 | 96.8 | (37) |
| Cathelicidin (CAMP) | ATTGCAATTTCACCCTGAGC | CCAGACCTGCTCCTTCTCAC | 56.4 | 108.1 | (38) |
| Transferrin | GAGCTCCCATCGACAGCTAC | CAAACCCAGCAGAGGAGAAG | 56.7 | 108.9 | (39) |
| Hepcidin (HAMP) | CCACAGGCTCCTCTCAAGTC | CTGCAACTGCAATGCTGAAT | 56.4 | 105.1 | (38) |
| nrf2 | TCGCAGTAGGAGCTGGATGA | CTCCGGTCTGTCCTTGGAAA | 57.0 | 98.1 | (40) |
| nox1 | GCCTATATGATTGGCCTGATGAC | GCTGTGCTGAGTGGGTCGTA | 55.3 | 108.6 | (40) |
| Mn-Sod | ATGTGGCCTCCTCCATTGAA | GCATCACGCCACCTATGTCA | 55.1 | 109.2 | (40) |
| Cu/Zn-Sod | CATGGCTTCCACGTCCATG | CGTTTCCCAGGTCTCCAACAT | 56.8 | 98.0 | (40) |
| cat | GCCAAGTTGTTTGAGCACGTT | CTGGGATCACGCACCGTATC | 57.3 | 101.0 | (40) |
| EF-1a | GATGCACCACGAGTCTCTGA | GGGTGGTTCAGGATGATGAC | 56.2 | 98.3 | (35) |
Respiratory Burst Assay
Respiratory burst of primary cod macrophages infected with A. salmonicida was determined according to the protocol established by Smith et al. (41, 42) with some modifications. Briefly, isolated primary macrophages were infected with 10 μl of bacterial suspension (~1 × 106 cells ml−1; MOI 1:1) and incubated at 15°C for 48 h. Primary macrophages inoculated with PBS were utilized as negative control, and phorbol myristate acetate (PMA 1 mM; Sigma) dissolved in dimethyl sulfoxide (DMSO) was utilized as positive control.
After 1 h post infection, cells were washed and the culture media was replaced with respiratory burst assay buffer (Leibovitz L-15 media supplemented with 1% BSA and 1 mM CaCl2). Then, 1 μl of dihydrorhodamine 123 (DHR, 5 mg/ml; Sigma) was diluted in 1 ml of PBS, and 50 μl of the dilution added to the macrophages for 15 min. Subsequently, 125 μl of PBS for negative control, or 125 μl of PMA (1 mM, final concentration 0.185 μM PMA) were added to the macrophages monolayers for 45 min to stimulate ROS production (43, 44). Finally, the macrophages were detached using 1 ml of trypsin-EDTA (0.5%; Gibco), washed with PBS, centrifuged for 5 min (500 × g at 4°C), and resuspended in fluorescence-activated cell sorting (FACS) buffer (1% FBS in PBS). Fluorescence was detected from 10,000 cells using a BD FACS Aria II flow cytometer (Becton Dickinson™) and analyzed using BD FACS Diva v7.0 software (BD Biosciences, San Jose, CA, USA). The PBS control cells were used to define the region of ROS negative cells, and based on this gating the FITC positive cells were identified. The mean fluorescence intensity and percentage of FITC-positive cells were determined for each condition. The experiments were conducted in macrophages isolated from six independent fish, and 10,000 events were measured for each sample.
Transmission Electron Microscopy (TEM)
Primary macrophages were fixed in anhydrous paraformaldehyde (4%; Electron Microscope Sciences, Hatfield, PA, USA) at 4°C until the samples were processed at the Electron Microscopy/Flow Cytometry Unit at Memorial University of Newfoundland. The cells were pelleted and resuspended in Karnovsky fixative for 20 min (45), washed in 0.1 M sodium cacodylate buffer pH 7.4 for 5 min, and post-fixed in 1% Osmium tetroxide during 15 min. After this, the fixed cells were dehydrated in increasing concentrations of ethanol and acetone followed by infiltration with EPON resin (Sigma). Cells were pelletized between incubation steps. Resin blocks were polymerized in BEEM capsules (Electron Microscope Sciences) overnight at 70°C and ultra-thin sections were cut with a diamond knife (Diatome, Hatfield, PA, USA). The ultra-thin sections were mounted on 300 copper mesh grids, stained with uranyl acetate and lead citrate, and examined in a Tecnai™ Spirit TMA with an accelerating voltage of 80 kV. Cells incubated for 3 h with PBS (control), J223 strain, and formalin-killed A. salmonicida were observed.
Statistical Analysis
All data are shown as the mean ± standard error (SE). Assumptions of normality and homogeneity were tested for the detected variances. A Kruskal-Wallis nonparametric test was performed for gentamicin exclusion assay results. Gene expression and ROS data were analyzed using a repeated measures two-way ANOVA test, followed by Sidak multiple comparisons post hoc test to identify significant differences of each treatment in different times and between treatments in the same time point. Differences were considered significant at P < 0.05. All statistical analyses were performed using GraphPad Prism (GraphPad Software, La Jolla California USA, www.graphpad.com).
Results
Macrophage Viability and Aeromonas salmonicida Infection
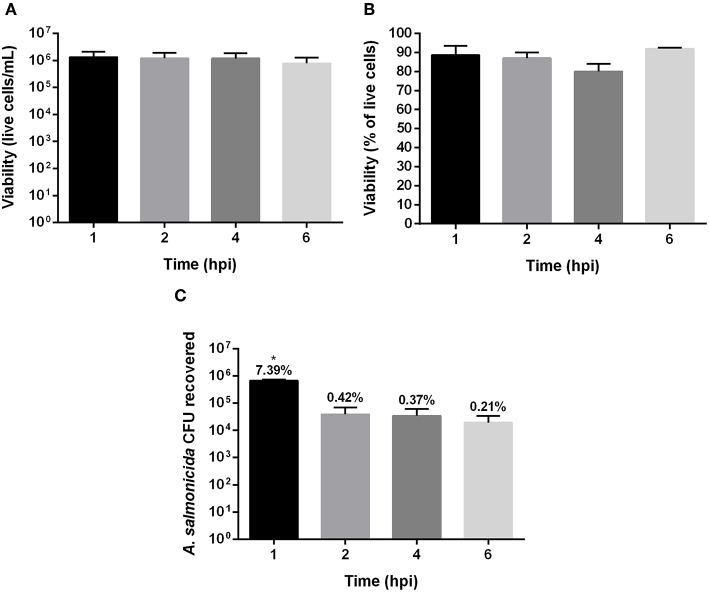
The viability of Atlantic cod primary macrophages infected with A. salmonicida was determined at 1, 2, 4, and 6 h post-infection. The results did not show significant differences between the time points post-infection in the number of live cells and percentage of viability. For instance, after 1, 2, 4, and 6 h post-infection 1.32 × 106 ± 8.02 × 105, 1.18 × 106 ± 7.13 × 105, 1.17 × 106 ± 6.64 × 105, and 7.77 × 105 ± 4.77 × 105 cells were quantified, respectively (Figures 1A,B). The percentage of viability during the infection process also did not show significant differences between time points post-infection. After 1, 2, 4, and 6 h the infected cells showed a viability of 89% ± 4.8, 87% ± 3.1, 80% ± 4.2%, and 92% ± 6%, respectively (Figure 1B).
Figure 1.
Gentamicin exclusion assay in Atlantic cod macrophages infected with Aeromonas salmonicida subsp. salmonicida. (A) Live cells (A) and (B) percentage of viability, after 1, 2, 4, and 6 h post infection. (C) Colony forming unit (CFU) recovered after 1, 2, 4, and 6 h post infection with A. salmonicida. The percentage showed above bars indicate the total % of attachment (1 h post infection) and invasion (2, 4, and 6 h post invasion) of A. salmonicida in Atlantic cod macrophages, p < 0.005. Each value represents the ± S.E.M (n = 3). Symbol (*) indicate statistical differences between each time post infection.
Although the primary cod macrophage cells seemed to survive the A. salmonicida infection, the bacteria infected and invaded the cell monolayers. The Atlantic cod macrophages were infected with a MOI of 1:1 (bacteria: macrophage) with an initial inoculum of 9.6 × 106 CFU. After 1 h post-infection, 7.39% (6.8 × 105 CFU) was attached to the macrophage monolayer, and after 2, 4, and 6 h post-infection, 0.42% (3.87 × 104 CFU), 0.37% (3.42 × 104 CFU), and 0.21% (1.94 × 104 CFU) of A. salmonicida were intracellularly located, respectively (Figure 1C).
Gene Expression Response of Atlantic Cod Macrophages to A. salmonicida Infection
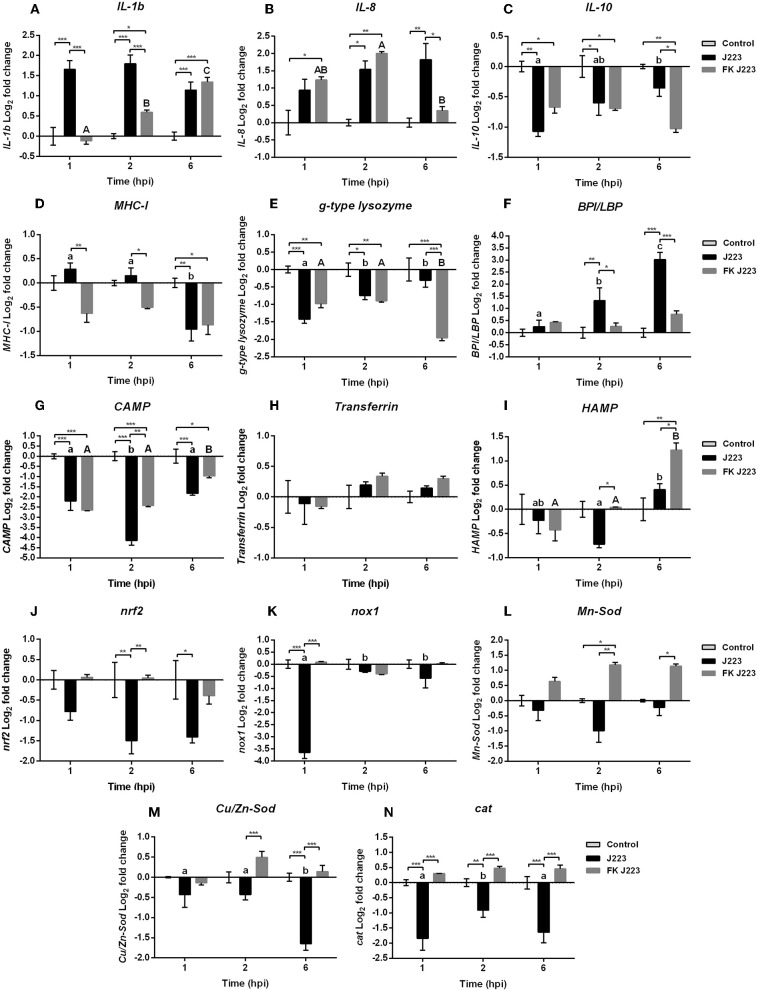
Quantitative real-time expression of selected genes related to Atlantic cod macrophage immunity was evaluated during A. salmonicida infection, and compared with macrophages inoculated with inactivated A. salmonicida (formalin-killed), and PBS inoculated controls. Significant increases in the expression of the pro-inflammatory cytokine interleukin 1β (IL-1β) gene were observed 1, 2, and 6 h post A. salmonicida infection compared to the time-matched PBS controls (Figure 2A). An up-regulation of the pro-inflammatory cytokine interleukin 8 (IL-8) gene, was also observed, nonetheless, this up-regulation occurred at 2 h, and 6 h post A. salmonicida infection compared to the PBS controls (Figure 2B). In contrast, the macrophages inoculated with inactivated A. salmonicida showed a higher expression of IL-1β at 2 and 6 h post-inoculation compared to their respective PBS controls (Figure 2A). IL-8 was up-regulated in cells treated with formalin-killed bacteria after 1 and 2 h post-inoculation (Figure 2B). At 6 h post-inoculation with the inactivated A. salmonicida, the expression of IL-8 in cod macrophages did not show differences compared to the PBS inoculated cells (Figure 2B). The expression of IL-1b was significantly up-regulated at 1 and 2 h after A. salmonicida infection compared with bacterin-exposed macrophages, whereas for IL-8 significant up-regulation in infected vs. bacterin-exposed macrophages was only seen at the 6 h time point (Figures 2A,B).
Figure 2.
Gene expression of (A) Interleukin 1b (IL-1b), (B) Interleukin 8 (IL-8), (C) Interleukin 10 (IL-10), (D) Major histocompatibility complex class 1 (MHC-I), (E) Goose type lysozyme (g-type lysozyme), (F) Bactericidal permeability increasing protein / lipopolysaccharide-binding protein (BPL/LBP), (G) Cathelicidin (CAMP), (H) Transferrin, (I) Hepcidin (HAMP), (J) Nuclear factor erythroid 2-related factor 2 (nrf2), (K) NADPH oxidase 1 (nox1), (L) Mn superoxide dismutase (Mn-Sod), (M) CuZn superoxide dismutase (Cu/Zn-Sod), and (N) Catalase (cat) in Atlantic cod primary macrophages isolated from head kidney and infected with live (J223) and formalin-killed A. salmonicida (FK 223) at different times post infection (1, 2, and 6 h). Relative expression was calculated using the 2(−ΔΔCt) method and Log2 converted using EF-1a as internal reference gene. Different letters represent significant differences between primary macrophages infected with J223 strain (lower case) or inoculated with FK J223 (upper case) at different times-points. Asterisks (*) represent significant differences between treatments on each time-point (*p < 0.05, **p < 0.01, ***p < 0.001). Each value is the mean ± S.E.M (n = 6).
In contrast, the relative expression of the anti-inflammatory cytokine Interleukin 10 (IL-10) gene, was significantly down-regulated after 1 and 2 h post A. salmonicida infection compared to the non-infected control macrophages (Figure 2C). Cod macrophages inoculated with the inactivated pathogen also showed a significant down-regulation at 1, 2, and 6 h compared with PBS controls (Figure 2C).
Genes involved in antigen recognition and host defense showed different patterns of expression after macrophage exposure to the live or inactivated A. salmonicida. In the case of the major histocompatibility complex class I (MHC-I) gene, a significant down-regulation was observed after 6 h post live A. salmonicida infection and post-inoculation with the formalin-killed bacteria (Figure 2D).
A down-regulation of the relative expression of the Goose-type lysozyme (g-type lysozyme) gene was observed at 1 and 2 h post A. salmonicida infection, and at 1, 2, and 6 h post inoculation with the formalin-killed pathogen, compared to their respective controls (Figure 2E). At 6 h post-treatment, g-type lysozyme expression was significantly different in infected vs. inactivated pathogen exposed macrophages (Figure 2E).
In contrast, the bactericidal permeability-increasing protein/lipopolysaccharide-binding protein (BPI/LBP) gene, involved in the antimicrobial defense against Gram negative bacteria, showed a significant up-regulation at 2 and 6 h post-infection only in cells inoculated with the live bacteria compared with the control and formalin-killed inoculated treatments (Figure 2F).
The relative expression of AMPs encoding genes showed different expression patterns. Cathelicidin (CAMP) gene was down-regulated in macrophages infected with A. salmonicida and those inoculated with formalin-killed A. salmonicida at 1, 2, and 6 h post-infection compared with time-matched PBS controls (Figure 2G). Macrophages infected with A. salmonicida showed a significant down-regulation of CAMP 2 h post-infection compared to the cells inoculated with inactivated bacteria (Figure 2G).
Transferrin did not show variation in the level of expression of both treatments compared with the time-PBS matched controls during the assays, as well as, between macrophages inoculated with live and bacterin A. salmonicida (Figure 2H). Hepcidin (HAMP) expression was significantly up-regulated only at 6 h post-exposure to the formalin-killed A. salmonicida (Figure 2I). However, HAMP was significantly down-regulated at 2 h post-infection with A. salmonicida (Figure 2I).
Expression of genes involved in the synthesis of ROS was also evaluated (Figures 2J–N). Nuclear factor erythroid 2-related factor 2 (nrf2), a transcriptional factor that is translocated into the nucleus under oxidative stress and initiates transcription of antioxidative genes (40), did not show transcriptional variation in cells inoculated with formalin-killed A. salmonicida compared to their respective controls (Figure 2J). In contrast, macrophages infected with A. salmonicida showed a significant down-regulation in the expression of nrf2 at 2 and 6 h post-infection (Figure 2J). After 1 h of infection, nrf2 transcript was significantly lower expressed in pathogen-infected macrophages compared to bacterin-exposed macrophages.
The expression of NADPH oxidase 1 (nox1) gene, which encodes a membrane-bound pro-oxidant enzyme that catalyzes superoxide synthesis, was significantly down-regulated 1 h post-infection with A. salmonicida compared to the non-treated cells and the bacterin-exposed macrophages (Figure 2K). Furthermore, this gene was significantly lower expressed at 1 h compared with 2 and 6 h post-infection (Figure 2K).
The relative expression of the Mn superoxide dismutase (Mn-Sod) and the CuZn superoxide dismutase (Cu/Zn-Sod) genes, involved in the transformation of superoxide into H2O2 in the mitochondria and the cytosol, respectively, also was evaluated. The Mn-Sod was significantly up-regulated at 2 h post-inoculation with inactivated A. salmonicida, compared to the controls (Figure 2L). In contrast, macrophages infected with A. salmonicida showed a Mn-Sod down-regulation tendency, with a significant down-regulation at 2 and 6 h post-infection compared to the bacterin-exposed macrophages (Figure 2L).
Similar patterns were observed in Cu/Zn-Sod and catalase (cat) expression, where a higher expression was observed post-inoculation with the formalin-killed A. salmonicida and a down-regulation was observed in cells infected with A. salmonicida (Figures 2M,N). For instance, Cu/Zn-Sod show a significant down-regulation at 6 h post A. salmonicida infection compared to the PBS control inoculated cells, and 2 and 6 h post-infection compared to cells treated with the inactivated pathogen (Figure 2M).
The relative expression of cat, which encodes for the catalase enzyme that plays an important role in H2O2 detoxification (40), showed a down-regulation at 1, 2, and 6 h in macrophages infected with A. salmonicida compared to the control and the cells treated with the inactivated pathogen (Figure 2N). In contrast, macrophages exposed to inactivated A. salmonicida did not showed significant differential expression of cat, compared to their respective controls (Figure 2N).
Reactive Oxygen Species (ROS) Production
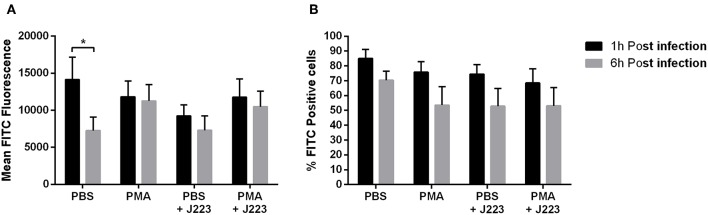
ROS production was determined in Atlantic cod primary macrophages infected with A. salmonicida. Macrophages inoculated with PBS or PMA were utilized as negative and positive controls, respectively. ROS production was analyzed at 1 and 6 h post A. salmonicida infection. We did not observe significant differences in ROS production between treatments. Macrophages treated with PBS showed that 85.1% ± 6.1 and 70.4% ± 6.2 were producing ROS after 1 and 6 h, respectively. Macrophages treated with PMA showed that 75.8% ± 7.0 and 53.6% ± 12.4 were producing ROS after 1 and 6 h, respectively. Similarly, macrophages treated with PBS and infected with A. salmonicida showed that 74.5% ± 6.4 and 52.9% ± 11.9 of the cells were producing ROS after 1 and 6 h post infection, respectively. Additionally, macrophages treated with PMA and infected with A. salmonicida showed that 68.5% ± 9.5 and 53.1% ± 12.4 of the cells were producing ROS after 1 and 6 h post treatment, respectively (Figures 3A,B).
Figure 3.
Reactive oxygen species (ROS) production in Atlantic cod macrophages after 1 and 6 h post-infection with A. salmonicida. (A) Mean FITC fluorescence and (B) percentage of FITC positive cells were obtained by flow cytometry. PBS inoculated cells (PBS) and PMA inoculated cells were utilized as negative and positive controls, respectively. Each value is the mean ± S.E.M (n = 6). Symbol (*) indicate differences on each group at different times of infection, p < 0.005.
Transmission Electron Microscopy (TEM)
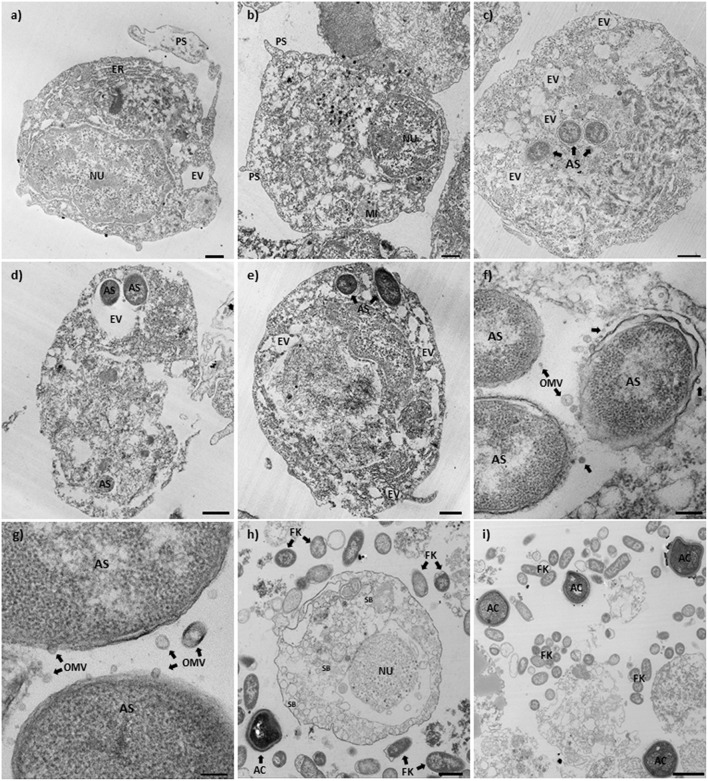
Cod primary macrophages infected with A. salmonicida were visualized 3 h post-infection using TEM. A group of non-infected cells were utilized as reference control (Figures 4a,b). These cells showed a rounded cell morphology, large nucleus, and evident presence of cell organelles (e.g., mitochondria, endoplasmic reticulum, endocytic vesicles) and pseudopodias (Figures 4a,b). In contrast, Atlantic cod macrophages infected with A. salmonicida showed poorly defined nuclei, membrane ruffling, and large vesicles containing A. salmonicida cells (average of 2–3 bacterial cells per macrophage, a maximum of 8 bacterial cells per macrophages, and 70–80% macrophages infected) (Figures 4c–e). Furthermore, secretion of A. salmonicida outer membrane vesicles (OMVs) was observed in intracellular bacterial cells (Figures 4f,g).
Figure 4.
Transmission electron microscopy of Atlantic cod macrophages infected with A. salmonicida. (a,b) Mock infected Atlantic cod head kidney macrophages (control). (× 2,100, scale bar 1 μm). (c–e) Atlantic cod head kidney macrophages infected with A. salmonicida. (c and d: × 2700, scale bar 1 μm; e: × 2,100, scale bar 1 μm). (f,g) Intracellular A. salmonicida. (f: × 15,000, scale bar 200 nm; g: × 30,000, scale bar 100 nm). (h,i) Atlantic cod head kidney macrophages infected with the formalin killed A. salmonicida. (h: × 2,700, scale bar 1 μm; i: × 1,650, scale bar 2 μm). NU, Nucleus; PS, Pseudopodia; ER, Endoplasmic reticulum; MI, Mitochondria; AS, A. salmonicida; EV, Endoplasmic vesicles; OMV, Outer membrane vesicles (arrows); FK, Formalin killed A. salmonicida; SB, Secretion bodies; and AC, Apoptotic bodies.
Macrophages inoculated with the formalin killed A. salmonicida showed a defined nucleus and a large number of secretion bodies within the cytoplasm (Figure 4h). Apoptotic-like bodies were observed in high numbers in the presence of extracellular A. salmonicida bacterin (Figure 4i).
Discussion
Atlantic cod lacks the genes for MHC-II, the invariant chain/CD74 (Ii), and CD4+ T cell response, representing a paradigm in the context of adaptive immunity against bacterial infectious diseases (2, 3). Additionally, Atlantic cod lacks TLR1, TLR2, and TLR5 that recognize bacterial surface antigens (2, 6, 9, 10). However, this absence seems to be compensated for by an expansion of the MHC-I (1, 4, 5) and the TLR7, TLR8, TLR9, and TLR22 families (2, 8). Nevertheless, how Atlantic cod fight bacterial infections is unknown. Here, we evaluated the early response of Atlantic cod primary macrophages to A. salmonicida infection.
To evaluate the early response of Atlantic cod primary macrophages to A. salmonicida infection, a gentamicin exclusion assay was conducted. The macrophage viability results obtained 1 h post infection (attachment) and 2, 4, and 6 h post-infection (invasion) showed similar viability, with 89% ± 4.8, 87% ± 3.1, 80% ± 4.2%, and 92% ± 6%, respectively (Figure 1B). In contrast, Atlantic salmon primary macrophages isolated and infected under similar conditions, showed 3 h post A. salmonicida infection a viability around ~40% (Supplementary Data and Supplementary Figures 1A,B). Similar reduction in viability was reported in non-phagocytic Chinook salmon embryo cell line (CHSE-214) infected with the A. salmonicida J223 strain, showing a viability of ~40% at 2 and 4 h post-infection (17). These results reveal that Atlantic cod macrophages are more resistant to A. salmonicida infection compared with Atlantic salmon macrophages and CHSE-214 cells.
The attachment of A. salmonicida in cod primary macrophages was 7.39%, and only 0.42, 0.37, and 0.21% was able to invade 2, 4, and 6 h post-infection, respectively (Figure 1C). The study conducted in CHSE-214 embryo cell line infected with A. salmonicida J223 strain showed an attachment of ~60% at 1 h post-infection, and an invasion of 0.47% and 0.29% after 2 and 4 h post-infection, respectively (17). Moreover, A. hydrophila isolated from ornamental fish, showed an attachment between 75 and 80% in the mammalian cell line CaCo-2 (cells from human colon adenocarcinoma) at 1 h post-infection (46). In contrast, the attachment of A. salmonicida J223 in Atlantic salmon primary macrophages was 10.7% at 1 h post-infection, and the invasion was 0.42, 0.26, and 0.28% after 2, 3, and 4 h post-infection, respectively (Supplementary Figure 1C). These results suggest that primary macrophages are less susceptible to be infected by A. salmonicida than non-phagocytic cells, even when the intracellular A. salmonicida recovered in non-phagocytic and phagocytic cells have shown to be relatively similar.
The transcriptional profiles of cytokine genes, antibacterial genes, antimicrobial peptide genes, and ROS related genes were determined using qPCR. The observed up-regulation of IL-1β and IL-8 after infection with A. salmonicida or inoculation with formalin killed A. salmonicida (Figures 2A,B) indicates a canonical macrophage innate immune response (24, 47). Coincident with the up-regulation of the pro-inflammatory IL-1β and IL-8 genes (Figures 2A,B), the anti-inflammatory IL-10 gene was down-regulated in macrophages infected with A. salmonicida or inoculated with the formalin killed pathogen (Figure 2C).
Similar up-regulation of the pro-inflammatory cytokine IL-1β was observed in Atlantic cod intramuscular injected with A. salmonicida subsp. achromogenes, meanwhile an up-regulation in both, IL-1β and IL-8, has been reported in Atlantic cod gill epithelial cells infected with Vibrio anguillarum and A. salmonicida (48), and in Atlantic cod macrophages infected with Francisella noatunensis (49), reinforcing the importance of these canonical interleukins against Gram-negative pathogens during the first hours of infection.
Antigen recognition and host defense genes, like MHC-I, g-type lysozyme, and BPI/LBP were also evaluated. As mentioned previously, BPI/LBP participates in the recognition of lipopolysaccharide (LPS) (50, 51), the major component of Gram negative bacterial outer membrane (52), MHC-I participate in the recognition of intracellular pathogens, like viruses or cytoplasmic invader bacteria (3), and g-type lysozyme is related to both Gram positive and Gram negative antibacterial activity (53). Interestingly, BPI/LBP was up-regulated only after infection with live A. salmonicida but not in presence of formalin-killed A. salmonicida (Figure 2F). BPI/LBP up-regulation has been observed in Atlantic cod vaccinated with inactivated V. anguillarum and A. salmonicida (11, 51), and in Atlantic cod intestinal epithelial cells after exposure with the bacterial probiotics GP21 (Pseudomonas spp.) and GP12 (Psychrobacter spp.) isolated from the intestinal tract of an adult Atlantic cod (54, 55).
The AMP-encoding genes CAMP and transferrin, both involved in the iron homeostasis, showed unexpected transcriptional profiles (Figures 2G,H), meanwhile HAMP, also related to the iron ion homeostasis, was the only AMP-encoding gene that showed an up-regulation after inoculation with the inactivated bacteria (Figure 2I). CAMP was down-regulated after infection with the live bacteria or inoculation with the formalin-killed pathogen (Figure 2G), meanwhile transferrin did not show variations compared to the controls (Figures 2H,I). In the case of HAMP, up-regulation was observed only at 6 h post-inoculation with the inactivated pathogen (Figure 2I). As previously mentioned, it has been reported that CAMP, transferrin, and HAMP AMPs are key during bacterial infection, and in general these genes are expressed in several immune tissues of Atlantic cod after bacterial (e.g., Mycobacterium chelonei, Aeromonas salmonicida subsp. salmonicida, and Aeromonas salmonicida subsp. achromogenes) and viral (infectious pancreatic necrosis virus) infection, or viral mimic [poly (I:C)] stimulation (11, 36, 56). However, according to our results, these genes were either lower expressed (CAMP and HAMP) or not affected (transferrin) in Atlantic cod macrophages compared with formalin-killed A. salmonicida stimulation (Figures 2G–I). A study in Atlantic cod intramuscular infected with A. salmonicida subsp. achromogenes showed an up-regulation in the expression of transferrin and HAMP (16). In contrast, we found that in Atlantic cod primary macrophages, the infection with A. salmonicida subsp. salmonicida down-regulated the expression of CAMP and HAMP. This can suggest a different mechanism of infection between A. salmonicida subsps.
The Atlantic cod CAMP has a potent antimicrobial activity against Gram-negative bacteria and fungi (57). Nonetheless, some bacterial pathogens, like V. anguillarum, A. salmonicida subsp. achromogenes and A. hydrophila are able to evade the action of CAMP (57), and this can be the case for A. salmonicida J223 in Atlantic cod macrophages.
Atlantic cod injected with turpentine oil, an inducer of acute immune response that involves inflammation and other biological processes (e.g., hemostasis), showed an increase in the relative expression of these AMP genes after 24 h of injection (39), and similar results were observed in intestinal epithelial cells after probiotic exposure to the probiotics GP21 and GP12 (55). Moreover, a study conducted in Atlantic cod stimulated with formalin-killed A. salmonicida showed lower levels of expression in the transcripts encoding CAMP and HAMP in head kidney and spleen, compare with PBS control samples (38). In this study, only a pick was observed after 24 h of stimulation with the inactivated pathogen (38). Therefore, our results suggest that likely more time is required for Atlantic cod macrophages to up-regulate the CAMP, transferrin, and HAMP genes after inactivated A. salmonicida exposition.
A study conducted in the Gram-negative bacteria, Pseudomonas syringae, showed that outer membrane vesicles (OMVs) can potentially suppress the action of AMPs (58). OMVs bind and sequester AMPs to prevent bacterial cell damage, and also induce the release of peptidases, proteases, and other lytic enzymes to degrade the host AMPs (58, 59). Combining this reported evidence with our results, where a high number of OMVs were released by A. salmonicida during intracellular infection (Figures 4f,g), we hypothesize that the presence of OMVs during intracellular infection might be related to a mechanism by which the bacterium tolerates the action of host AMPs or translocate virulence factors to the host cell. This is in addition to unknown A. salmonicida mechanism involving the down-regulation of AMP-related genes.
Typically, macrophages phagocytize the invading bacteria, assemble the lysosome, and eliminate them through the action of several enzymes and production of ROS (40). Here, we evaluated the expression of nrf2, nox1, Mn-Sod, Cu/Zn-Sod, and cat genes that are part of the redox system and antioxidant enzymes related to ROS synthesis (40, 60). Relative expression levels of nrf2, nox1, Mn-Sod, Cu/Zn-Sod, and cat genes in cod primary macrophages were down-regulated after infection with A. salmonicida (Figures 2J–N). In contrast, macrophages inoculated with the formalin-killed A. salmonicida, showed an up-regulation in the expression of the Mn-Sod and Cu/Zn-Sod genes (Figures 2L–N).
The macrophage gene expression of nrf2, nox1, Mn-Sod, Cu/Zn-Sod, and cat after A. salmonicida infection, together with the ROS flow cytometry results, shows that Atlantic cod macrophages do not increase ROS levels after exposure to A. salmonicida (Figures 3A,B).
High basal levels of ROS production have been observed in non-induced Atlantic cod blood phagocytes (43), suggesting that ROS synthesis in Atlantic cod cells is related to the in vitro culture conditions (43). However, we hypothesize that high basal production of ROS could be normal in G. morhua, and the lack of ROS production, above the basal levels after A. salmonicida infection, could be associated to mechanisms used by A. salmonicida to control the macrophage immune response, as described previously in other pathogens such as Mycobacterium leprae, R. salmoninarum, Salmonella spp, and Edwardsiella tarda (61–64).
Macrophages are usually highly efficient killers of bacteria, however pathogenic bacteria, like A. salmonicida, have evolved multiple strategies to infect, avoid enzymatic digestion, and trigger immunosuppression of the host (49). Our TEM results showed that A. salmonicida was localized intracellularly in bacteria containing vesicles (Figures 4c,e). Also, we observed that the cytoskeleton of the infected cells was rearranged, and several structures, like the nuclei, pseudopodia, and endoplasmic reticulum, were not observed in infected macrophages, in contrast to non-infected cells (Figures 4a–e). Additionally, the TEM images of infected macrophages showed a significant number of intracellular bacteria at 3 h post-infection (Figures 4c–e). Not all Gram-negative pathogens gain access to macrophages in high numbers. For instance, F. noatunensis and F. tularensis invade in lower numbers, even 24 h post-infection with high infectious doses. Thus, few F. tularensis cells are required to cause fatal diseases (49, 65–68). In contrast to these previous studies, our results indicate that A. salmonicida required a higher number of infecting bacterial cells, compared to other bacterial pathogens, to have a productive infection (Figures 4c–e), Additionally, a single macrophage is infected with a significant number of bacterial cells (maximum of 8 bacterial cells per macrophage was founded, 2–3 average) suggesting that these are the target cells of A. salmonicida.
An interesting finding of our study was the presence of several OMVs produced by A. salmonicida during intracellular infection (Figures 4f,g). The OMVs are virulence factors released by mammalian pathogens like Neisseria meningitides, Escherichia coli, Vibrio spp., Brucella spp., among others (69). These OMVs play an important role during pathogenesis, delivering toxins and immunomodulatory proteins to the host cell (69). Also, OMVs have been observed and described previously in marine pathogens such as F. noatunensis subsp. orientalis, Edwardsiella anguillarum, and Piscirickettsia salmonis (70–72). Our results suggest that A. salmonicida release the OMVs during intracellular infection in order to control the immune response of the Atlantic cod primary macrophages, and perhaps neutralizing antimicrobial peptides to avoid lysis.
Phagocytosis is a highly efficient mechanism for bacterial elimination used by macrophages. However, Atlantic cod macrophages exposed to formalin-killed A. salmonicida for 3 h showed that most of the formalin-killed bacteria were localized in the extracellular milieu, with a reduced number of bacterial cells in phagocytic vesicles (Figures 4h,i). This results suggest that A. salmonicida promote macrophage phagocytosis, in contrast to inactivated bacteria.
Also, we observed that Atlantic cod macrophages exposed to formalin-killed A. salmonicida produced a large amount of apoptotic-like bodies (Figures 4h,i). Programmed cell death (i.e., apoptosis), is a highly regulated process and an important mechanism used by the host to prevent infectious diseases (73). Cells undergoing apoptosis maintain membrane integrity until very late in the process, unlike cells undergoing necrosis, but produce several morphological and biochemical changes inside the cells, including chromatin condensation, nuclear segmentation, internucleosomal DNA fragmentation, and cytoplasmic vacuolization (74, 75). Similar morphological changes were observed in Atlantic cod macrophages inoculated with formalin-killed A. salmonicida (Figures 4h,i). This suggest that A. salmonicida is displaying pathogenesis mechanisms to avoid not only phagocytosis but also to prevent apoptosis.
Conclusion
In this study we evaluated the infection of Atlantic cod macrophages with A. salmonicida J223 strain. A. salmonicida infects and invades Atlantic cod primary macrophages. We found between 2 and 8 A. salmonicida cells per infected macrophage. The infected Atlantic cod macrophages survived during the first 6 h of A. salmonicida infection. Nevertheless, TEM observations showed that A. salmonicida remained in A. salmonicida-containing vesicles. Gene expression results from infected macrophages suggest that A. salmonicida modulate the expression of several genes involved in the innate immune response. For instance, relative expression of HAMP, nrf2, nox1, Mn-Sod, Cu/Zn-Sod, and cat genes were down-regulated and BPI/LBP was up-regulated after A. salmonicida infection. Additionally, we observed that A. salmonicida secrete OMVs during intracellular infection. These results suggest that A. salmonicida has immune suppressive mechanisms to control cod macrophage immune response, where OMVs could play an essential role.
In contrast, macrophages inoculated with formalin-killed A. salmonicida, showed a canonical innate immune response, where most of the evaluated genes were up-regulated or not induced, like the relative expression of HAMP, nrf2, nox1, Mn-Sod, Cu/Zn-Sod, and cat genes. Additionally, we observed that macrophages inoculated with inactivated A. salmonicida did not phagocytize the formalin-killed pathogen, and post-exposure to the bacterin, several apoptotic-like bodies were presented. These results suggest that inactivate A. salmonicida trigger a potent innate immunity modulated by the macrophage.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Ethics Statement
The animal experiments were performed in accordance with the guidelines of the Canadian Council on Animal Care and approved by Memorial University of Newfoundland's Institutional Animal Care Committee (protocols #17-01-JS; #17-02-JS).
Author Contributions
MS-D and JS: conception and design of research. MS-D and AH: macrophage isolation. MS-D and SC: live and formalin-filled A. salmonicida preparation. MS-D, AH, and SC: performed experiments. MS-D, MR, and JS: interpreted results of experiments, edited and revised manuscript. MS-D and JS: prepared figures, drafted manuscript, and funding support. MS-D, AH, SC, MR, and JS: approved final version of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the support provided by Vitamin Initiative–Ocean Frontier Institute; Canada First—Ocean Frontier Institute (Module J.3); MUN Seed, Bridge and Multidisciplinary Funds; and NSERC-Discovery. The author also thanks to the Dr. Joe Brown Aquatic Research Building (JBARB) staff for their assistance with the fish. We also thanks to Khalil Eslamloo and Nicole Smith (Memorial University, Department of Ocean Sciences) for their suggestions with the macrophage isolation protocol and respiratory burst assay technical support, respectively. We would like to thanks Stephanie Tucker (Memorial University, Faculty of Medicine) for her technical support with the TEM samples. Finally, we thanks to Dr. Wilfred Templeman Memorial Scholarship for the funding provided to the study of groundfish of Newfoundland and Labrador.
Footnotes
Funding. The funding provided for this research came from the Vitamin Initiative-Ocean Frontier Institute; Canada First-Ocean Frontier Institute (Module J); MUN Seed, Bridge and Multidisciplinary Funds, Memorial University; and NSERC-Discovery (RGPIN-2018-05942).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01237/full#supplementary-material
References
- 1.Malmstrøm M, Matschiner M, Torresen OK, Star B, Snipen LG, Hansen TF, et al. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. (2016) 48:1204–10. 10.1038/ng.3645 [DOI] [PubMed] [Google Scholar]
- 2.Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, Gregers TF, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. (2011) 477:207–10. 10.1038/nature10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham P. How the codfish changed its immune system. Nat Genet. (2016) 48:1103–4. 10.1038/ng.3684 [DOI] [PubMed] [Google Scholar]
- 4.Malmstrøm M, Jentoft S, Gregers TF, Jakobsen KS. Unraveling the evolution of the Atlantic cod's (Gadus morhua L.) alternative immune strategy. PLoS ONE. (2013) 8:e74004. 10.1371/annotation/18b70612-fd3d-46ce-a04b-652d18c82d5b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solbakken MH, Voje KL, Jakobsen KS, Jentoft S. Linking species habitat and past palaeoclimatic events to evolution of the teleost innate immune system. Proc Biol Sci. (2017) 284:2810. 10.1098/rspb.2016.2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buonocore F, Gerdol M. Alternative adaptive immunity strategies: coelacanth, cod and shark immunity. Mol Immunol. (2016) 69:157–69. 10.1016/j.molimm.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 7.Solbakken MH, Rise ML, Jakobsen KS, Jentoft S. Successive losses of central immune genes characterize the Gadiformes' alternate immunity. Genome Biol Evol. (2016) 8:3508–15. 10.1093/gbe/evw250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solbakken MH, Torresen OK, Nederbragt AJ, Seppola M, Gregers TF, Jakobsen KS, et al. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci Rep. (2016) 6:25211. 10.1038/srep25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Star B, Jentoft S. Why does the immune system of Atlantic cod lack MHC II? BioEssays. (2012) 34:648–51. 10.1002/bies.201200005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundaram AY, Kiron V, Dopazo J, Fernandes JM. Diversification of the expanded teleost-specific toll-like receptor family in Atlantic cod, Gadus morhua. BMC Evol Biol. (2012) 12:256. 10.1186/1471-2148-12-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnadottir B. The immune response of Atlantic cod, Gadus morhua L. Icel Agric Sci. (2014) 27:41–61. [Google Scholar]
- 12.Bakkemo KR, Mikkelsen H, Johansen A, Robertsen B, Seppola M. Francisella noatunensis subsp. noatunensis invades, survives and replicates in Atlantic cod cells. Dis Aquat Organ. (2016) 121:149–59. 10.3354/dao03043 [DOI] [PubMed] [Google Scholar]
- 13.Magnadóttir B, Bambir SH, Gudmundsdóttir BK, Pilström L, Helgason S. Atypical Aeromonas salmonicida infection in naturally and experimentally infected cod, Gadus morhua L. J Fish Dis. (2002) 25:583–97. 10.1046/j.1365-2761.2002.00407.x [DOI] [Google Scholar]
- 14.Samuelsen OB, Nerland AH, Jorgensen T, Schroder MB, Svasand T, Bergh O. Viral and bacterial diseases of Atlantic cod Gadus morhua, their prophylaxis and treatment: a review. Dis Aquat Org. (2006) 71:239–54. 10.3354/dao071239 [DOI] [PubMed] [Google Scholar]
- 15.Graf J. Aeromonas. Norfolk: Caister Academic Press; (2015). [Google Scholar]
- 16.Fazio A, Bragason BT, Magnadottir B, Faggio C, Gudmundsdottir S. The acute phase response of Atlantic cod (Gadus morhua L.): expression of immune response genes after infection with Aeromonas salmonicida subsp. achromogenes. J Biol Res. (2015) 88:12–3. [Google Scholar]
- 17.Valderrama K, Saravia M, Santander J. Phenotype of Aeromonas salmonicida sp. salmonicida cyclic adenosine 3',5'-monophosphate receptor protein (Crp) mutants and its virulence in rainbow trout (Oncorhynchus mykiss). J Fish Dis. (2017) 40:1849–56. 10.1111/jfd.12658 [DOI] [PubMed] [Google Scholar]
- 18.Janda JM, Abbott SL. The Genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. (2010) 23:35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dallaire-Dufresne S, Tanaka KH, Trudel MV, Lafaille A, Charette SJ. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet Microbiol. (2014) 169:1–7. 10.1016/j.vetmic.2013.06.025 [DOI] [PubMed] [Google Scholar]
- 20.Ebanks RO, Knickle LC, Goguen M, Boyd JM, Pinto DM, Reith M, et al. Expression of and secretion through the Aeromonas salmonicida type III secretion system. Microbiology. (2006) 152:1275–86. 10.1099/mic.0.28485-0 [DOI] [PubMed] [Google Scholar]
- 21.Frey J, Origgi FC. Type III secretion system of Aeromonas salmonicida undermining the host's immune response. Front Mar Sci. (2016) 3:130 10.3389/fmars.2016.00130 [DOI] [Google Scholar]
- 22.Suzuky Y, Lida T. Fish granulocytes in the process of inflammation. Annu Rev Fish Dis. (1992) 2:149–60. 10.1016/0959-8030(92)90061-2 [DOI] [Google Scholar]
- 23.Whyte SK. The innate immune response of finfish - a review of current knowledge. Fish Shellfish Immunol. (2007) 23:1127–51. 10.1016/j.fsi.2007.06.005 [DOI] [PubMed] [Google Scholar]
- 24.Tort L, Balasch JC, Mackenzie S. Fish immune system. A crossroads between innate and adaptive responses. Inmunología. (2003) 22:277–86. [Google Scholar]
- 25.Esteban MA, Cuesta A, Chaves-Pozo E, Meseguer J. Phagocytosis in teleosts. Implications of the new cells involved. Biology. (2015) 4:907–22. 10.3390/biology4040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stafford J, Belosevic M. Transferrin and the innate immune response of fish: identification of a novel mechanism of macrophage activation. Dev Comp Immunol. (2003) 27:539–54. 10.1016/S0145-305X(02)00138-6 [DOI] [PubMed] [Google Scholar]
- 27.Secombes CJ, Fletcher TC. The role of phagocytes in the protective mechanisms of fish. AnnuRev Fish Dis. (1992) 2:53–71. 10.1016/0959-8030(92)90056-4 [DOI] [Google Scholar]
- 28.Eslamloo K, Xue X, Booman M, Smith NC, Rise ML. Transcriptome profiling of the antiviral immune response in Atlantic cod macrophages. Dev Comp Immunol. (2016) 63:187–205. 10.1016/j.dci.2016.05.021 [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; (2001). [Google Scholar]
- 30.Sung K, Khan SA, Nawaz MS, Khan AA. A simple and efficient Triton X-100 boiling and chloroform extraction method of RNA isolation from Gram-positive and Gram-negative bacteria. FEMS Microbiol. Lett. (2003) 229:97–101. 10.1016/S0378-1097(03)00791-2 [DOI] [PubMed] [Google Scholar]
- 31.Aravena-Román M, Inglis TJJ, Henderson B, Riley TV, Changa BJ. Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob Agents Chemother. (2012) 56:1110–2. 10.1128/AAC.05387-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santander J, Kilbourne J, Park JY, Martin T, Loh A, Diaz I, et al. Inflammatory effects of Edwardsiella ictaluri lipopolysaccharide modifications in catfish gut. Infect Immun. (2014) 82:3394–404. 10.1128/IAI.01697-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. (2001) 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Anal method. Methods. (2001) 25:402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Perez-Casanova JC, Hamoutene D, Samuelson S, Burt K, King TL, Lee K. The immune response of juvenile Atlantic cod (Gadus morhua L.) to chronic exposure to produced water. Mar Environ Res. (2010) 70:26–34. 10.1016/j.marenvres.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 36.Seppola M, Larsen AN, Steiro K, Robertsen B, Jensen I. Characterisation and expression analysis of the interleukin genes, IL-1beta, IL-8 and IL-10, in Atlantic cod (Gadus morhua L.). Mol Immunol. (2008) 45:887–97. 10.1016/j.molimm.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 37.Caipang CM, Hynes N, Puangkaew J, Brinchmann MF, Kiron V. Intraperitoneal vaccination of Atlantic cod, Gadus morhua with heat-killed Listonella anguillarum enhances serum antibacterial activity and expression of immune response genes. Fish Shellfish Immunol. (2008) 24:314–22. 10.1016/j.fsi.2007.11.018 [DOI] [PubMed] [Google Scholar]
- 38.Feng CY, Johnson SC, Hori TS, Rise M, Hall JR, Gamperl AK, et al. Identification and analysis of differentially expressed genes in immune tissues of Atlantic cod stimulated with formalin-killed, atypical Aeromonas salmonicida. Physiol Genomics. (2009) 37:149–63. 10.1152/physiolgenomics.90373.2008 [DOI] [PubMed] [Google Scholar]
- 39.Audunsdottir SS, Magnadottir B, Gisladottir B, Jonsson ZO, Bragason BT. The acute phase response of cod (Gadus morhua L.): expression of immune response genes. Fish Shellfish Immunol. (2012) 32:360–7. 10.1016/j.fsi.2011.11.034 [DOI] [PubMed] [Google Scholar]
- 40.Skjærven KH, Penglase S, Olsvik PA, Hamre K. Redox regulation in Atlantic cod (Gadus morhua) embryos developing under normal and heat-stressed conditions. Free Radic Biol Med. (2013) 57:29–38. 10.1016/j.freeradbiomed.2012.11.022 [DOI] [PubMed] [Google Scholar]
- 41.Smith NC, Christian SL, Taylor RG, Santander J, Rise ML. Immune modulatory properties of 6-gingerol and resveratrol in Atlantic salmon macrophages. Mol Immunol. (2018) 95:10–9. 10.1016/j.molimm.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 42.Smith NC, Christian SL, Taylor RG, Santander J, Rise ML. Corrigendum to “Immune modulatory properties of 6-gingerol and resveratrol in Atlantic salmon macrophages” [Mol. Immunol. 95 (2018) 10–19]. Mol Immunol. (2018) 104:139 10.1016/j.molimm.2018.10.018 [DOI] [PubMed] [Google Scholar]
- 43.Nikoskelainen S, Kjellsen O, Lilius EM, Schroder MB. Respiratory burst activity of Atlantic cod (Gadus morhua L.) blood phagocytes differs markedly from that of rainbow trout. Fish Shellfish Immunol. (2006) 21:199–208. 10.1016/j.fsi.2005.11.008 [DOI] [PubMed] [Google Scholar]
- 44.Kalgraff CA, Wergeland HI, Pettersen EF. Flow cytometry assays of respiratory burst in Atlantic salmon (Salmo salar L.) and in Atlantic cod (Gadus morhua L.) leucocytes. Fish Shellfish Immunol. (2011) 31:381–8. 10.1016/j.fsi.2011.05.028 [DOI] [PubMed] [Google Scholar]
- 45.Karnovsky MJ. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. (2003) 27:137A. [Google Scholar]
- 46.Saidi N, Snoussi M, Usai D, Zanetti S, Bakhrouf A. Adhesive properties of Aeromonas hydrophila strains isolated from Tunisian aquatic biotopes. Afr J Microbiol Res. (2011) 5:5644–55. 10.5897/AJMR11.803 [DOI] [Google Scholar]
- 47.Secombes CJ, Wang T, Hong S, Peddie S, Crampe M, Laing KJ, et al. Cytokines and innate immunity of fish. Dev Comp Immunol. (2001) 25:713–23. 10.1016/S0145-305X(01)00032-5 [DOI] [PubMed] [Google Scholar]
- 48.Caipang CM, Lazado CC, Brinchmann MF, Kiron V. Infection-induced changes in expression of antibacterial and cytokine genes in the gill epithelial cells of Atlantic cod, Gadus morhua during incubation with bacterial pathogens. Comp Biochem Physiol B Biochem Mol Biol. (2010) 156:319–25. 10.1016/j.cbpb.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 49.Bakkemo KR, Mikkelsen H, Bordevik M, Torgersen J, Winther-Larsen HC, Vanberg C, et al. Intracellular localisation and innate immune responses following Francisella noatunensis infection of Atlantic cod (Gadus morhua) macrophages. Fish Shellfish Immunol. (2011) 31:993–1004. 10.1016/j.fsi.2011.08.020 [DOI] [PubMed] [Google Scholar]
- 50.Kono T, Sakai M. Molecular cloning of a novel bactericidal permeability-increasing protein/lipopolysaccharide-binding protein (BPI/LBP) from common carp Cyprinus carpio L. and its expression. Mol Immunol. (2003) 40:269–78. 10.1016/S0161-5890(03)00103-2 [DOI] [PubMed] [Google Scholar]
- 51.Stenvik J, Solstad T, Strand C, Leiros I, Jørgensen TØ. Cloning and analyses of a BPI/LBP cDNA of the Atlantic cod (Gadus morhua L.). Dev Comp Immunol. (2004) 28:307–23. 10.1016/j.dci.2003.09.002 [DOI] [PubMed] [Google Scholar]
- 52.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. (2007) 76:295–329. 10.1146/annurev.biochem.76.010307.145803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsen AN, Solstad T, Svineng G, Seppola M, Jorgensen TO. Molecular characterisation of a goose-type lysozyme gene in Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. (2009) 26:122–32. 10.1016/j.fsi.2008.03.021 [DOI] [PubMed] [Google Scholar]
- 54.Lazado CC, Caipang CM, Gallage S, Brinchmann MF, Kiron V. Expression profiles of genes associated with immune response and oxidative stress in Atlantic cod, Gadus morhua head kidney leukocytes modulated by live and heat-inactivated intestinal bacteria. Comp Biochem Physiol B Biochem Mol Biol. (2010) 155:249–55. 10.1016/j.cbpb.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 55.Lazado CC, Caipang CM. Bacterial viability differentially influences the immunomodulatory capabilities of potential host-derived probiotics in the intestinal epithelial cells of Atlantic cod Gadus morhua. J Appl Microbiol. (2014) 116:990–8. 10.1111/jam.12414 [DOI] [PubMed] [Google Scholar]
- 56.Solstad T, Larsen AN, Seppola M, Jørgensen TØ. Identification, cloning and expression analysis of a hepcidin cDNA of the Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. (2008) 25:298–310. 10.1016/j.fsi.2008.05.013 [DOI] [PubMed] [Google Scholar]
- 57.Broekman D, Zenz A, Gudmundsdottir B, Lohner K, Maier BH, Gudmundsson G. Functional characterization of codCath, the mature cathelicidin antimicrobial peptide from Atlantic cod (Gadus morhua). Peptides. (2011) 32:2044–51. 10.1016/j.peptides.2011.09.012 [DOI] [PubMed] [Google Scholar]
- 58.Kulkarni HM, Swamy Ch .V, Jagannadham MV. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res. (2014) 13:1345–58. 10.1021/pr4009223 [DOI] [PubMed] [Google Scholar]
- 59.Chua SL, Tan SY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, et al. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. (2013) 57:2066–75. 10.1128/AAC.02499-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kansanen E, Jyrkkänen H-K, Levonen A-L. Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic Biol Med. (2012) 52:973–82. 10.1016/j.freeradbiomed.2011.11.038 [DOI] [PubMed] [Google Scholar]
- 61.Holzer TJ, Nelson KE, Crispen RG, Andersen BR. Mycobacterium leprae fails to stimulate phagocytic cell superoxide anion generation. Infect Immun. (1986) 51:514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bandin I, Ellis AE, Barja JL, Secombes CJ. Interaction between rainbow trout macrophages and Renibacterium salmoninarum in vitro. Fish Shellfish Immunol. (1993) 3:25–33. 10.1006/fsim.1993.1003 [DOI] [Google Scholar]
- 63.Foster JW, Spector MP. How Salmonella survive against the odds. Annu Rev Microbiol. (1995) 49:145–74. 10.1146/annurev.mi.49.100195.001045 [DOI] [PubMed] [Google Scholar]
- 64.Rao PSS, Lim TM, Leung KY. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect Immun. (2001) 69:5689–97. 10.1128/IAI.69.9.5689-5697.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anthony LD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect Immun. (1991) 59:3291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fortier AH, Leiby DA, Narayanan RB, Asafoadjei E, Crawford RM, Nacy CA, et al. Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect Immun. (1995) 63:1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. (2003) 71:5940–50. 10.1128/IAI.71.10.5940-5950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirimanjeswara GS, Olmos S, Bakshi CS, Metzger DW. Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol Rev. (2008) 225:244–55. 10.1111/j.1600-065X.2008.00689.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avila-Calderón ED, Araiza-Villanueva MG, Cancino-Diaz JC, López-Villegas EO, Sriranganathan N, Boyle SM, et al. Roles of bacterial membrane vesicles. Arch Microbiol. (2015) 197:1–10. 10.1007/s00203-014-1042-7 [DOI] [PubMed] [Google Scholar]
- 70.Shahin K, Thompson KD, Inglis NF, McLean K, Ramirez-Paredes JG, Monaghan SJ, et al. Characterization of the outer membrane proteome of Francisella noatunensis subsp. orientalis J Appl Microbiol. (2018) 125:686–99. 10.1111/jam.13918 [DOI] [PubMed] [Google Scholar]
- 71.LiHua D, JianJun F, Peng L, SongLin G, Le H, YiQun X. Evaluation of an outer membrane protein as a vaccine against Edwardsiella anguillarum in Japanese eels (Anguilla japonica). Aquaculture. (2019) 498:143–50. 10.1016/j.aquaculture.2018.08.012 [DOI] [Google Scholar]
- 72.Oliver C, Valenzuela K, Hernández M, Sandoval R, Haro RE, Avendaño-Herrera R, et al. Characterization and pathogenic role of outer membrane vesicles produced by the fish pathogen Piscirickettsia salmonis under in vitro conditions. Vet Microbiol. (2016) 184:94–101. 10.1016/j.vetmic.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 73.Jacobson MD, Weil M, Raff MC. Programmed Cell death in animal development. Cell. (1997) 88:347–54. 10.1016/S0092-8674(00)81873-5 [DOI] [PubMed] [Google Scholar]
- 74.Häcker G. The morphology of apoptosis. Cell Tissue Res. (2000) 301:5–17. 10.1007/s004410000193 [DOI] [PubMed] [Google Scholar]
- 75.do Vale A, Marques F, Silva MT. Apoptosis of sea bass (Dicentrarchus labrax L.) neutrophils and macrophages induced by experimental infection with Photobacterium damselae subsp. piscicida. Fish Shellfish Immunol. (2003) 15:129–44. 10.1016/S1050-4648(02)00144-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.