Abstract
The prognosis for patients with metastatic bladder cancer (BCa) is poor, and has not been improved by current treatment methods. Long noncoding RNAs (lncRNAs) are involved in the pathology of various tumors, including bladder cancer. However, the role of zinc finger E-box-binding homeobox 1-antisense 1 (ZEB1-AS1) in BCa progression and metastasis remains unclear. The present study determined the expression level of ZEB1-AS1 in BCa and additionally investigated the functional role of ZEB1-AS1 in BCa metastasis. Reverse transcription quantitative polymerase chain reaction analysis showed that ZEB1-AS1 was upregulated in BCa cells compared with normal epithelial cells. Functionally, knockdown of ZEB1-AS1 suppressed BCa cell migration and invasion in vitro, and metastasis in vivo. Mechanistic investigations revealed that ZEB1-AS1 bound to heterogenous nuclear ribonucleoprotein D0 (AUF1), thereby activating the translation of ZEB1 mRNA without affecting its mRNA level. In addition, ZEB1-AS1 was significantly upregulated in BCa tissues and muscle-invasive BCa cases. ROC curve analysis demonstrated that ZEB1-AS1 expression was associated with metastasis in patients with BCa. In conclusion, the data from the present study demonstrated that ZEB1-AS1 induced BCa metastasis via an AUF1-mediated translation activation of ZEB1 mRNA mechanism. ZEB1-AS1 may serve as a promising target for clinical intervention in advanced BCa.
Keywords: bladder cancer, heterogenous nuclear ribonucleoprotein D0, zinc finger E-box-binding homeobox 1, zinc finger E-box-binding homeobox 1-antisense 1, metastasis
Introduction
Bladder cancer (BCa) is the most common malignancy of the urinary system in China (1) and worldwide (2), and is the primary cause of mortality in patients with urinary tract disease (3). Muscle-invasive BCa (MIBC) represents 25-40% of all BCa, and may spread from the bladder to the pelvic lymph nodes and then to visceral organs (4). The 5-year mortality rate of MIBC patients with lymph node (LN) metastasis (77.6%) is higher compared with that of MIBC patients without LN metastasis (18.6%) even when treatment with radical cystectomy was available (5,6). Metastasis to LNs and distant organs is a complex multistep process that involves dissemination of cancer cells to lymphatic vessels, transport, settlement and colonization expansion of cancer cells (7,8). However, the biological characteristics and molecular mechanisms of BCa cell invasion and metastasis remains largely unknown.
Long non-coding RNAs (lncRNAs) are a class of poorly conserved endogenous RNAs >200 nucleotides that do not encode proteins but regulate gene expression (9). On a functional level, lncRNAs are involved in complex biological processes via diverse mechanisms. These comprise, among others, gene regulation by titration of transcription factors, alternative splicing, sponging of microRNAs (miRNAs) and recruitment of chromatin modifying enzymes (10-13). Accumulating evidence indicates that lncRNAs serve diverse roles in the initiation and progression of human cancer. For example, lncRNAs HOX transcript antisense RNA, upregulated in colorectal cancer liver metastasis and small nucleolar RNA host gene 14 participate in the metastatic cascade by regulating cell migration and invasion (14-16). lncRNAs urothelial cancer associated 1 and H19 have been demonstrated to serve critical roles in bladder cancer metastasis.
The lncRNA zinc finger E-box-binding homeobox 1 antisense 1 (ZEB1-AS1) derives from the promoter region of ZEB1, a transcriptional factor that serves important roles in physiology and tumorigenesis. As a well-known epithelial-mesenchymal transition (EMT) promoter, ZEB1 serves important roles in cancer metastasis, including BCa. Previously, Lin et al (17) demonstrated that lncRNA ZEB1-AS1 was associated with higher histopathological grade and promoted tumorigenesis in BCa, indicating its oncogenic role in cancer progression. However, the biological function and molecular mechanism of lncRNA ZEB1-AS1 in BCa metastasis remains unknown.
The present study identified that lncRNA ZEB1-AS1 was significantly upregulated in BCa and closely associated with poor prognosis. Through gain or loss of function, it was demonstrated that ZEB1-AS1 promoted migration and invasion of BCa cells in vitro, and enhanced tumor metastasis in vivo. Mechanistically, ZEB1-AS1 guided heterogenous nuclear ribonucleoprotein D0 (AUF1) to promote the translation of ZEB1 mRNA, contributing to an increase in ZEB1 protein expression. Therefore, targeting ZEB1-AS1 may be a potential therapeutic strategy leading to decreased rates of growth and metastasis in BCa.
Materials and methods
Ethics statement and tissue samples
A total of 60 snap-frozen fresh BCa tissues [30 MIBC and 30 non-MIBC (NMIBC)] and 60 normal adjacent tissues were obtained with the written consent of patients who underwent surgery at Peking Union Medical College Hospital (Beijing, China) between January 2014 and January 2016. The diagnosis of recruited patients was pathologically confirmed, and primary cancer tissues (no biopsy samples) were collected prior to radiotherapy or chemotherapy. The obtained tissue samples were immediately snap-frozen in liquid nitrogen upon resection and then stored at -80°C until further use. The present study was approved by Research Scientific Ethics Committee of Peking Union Medical College Hospital. All participants signed informed consent prior to use of the tissues for scientific purposes.
Cell culture and reagents
The human BCa T24 and UM-UC-3 cell lines and normal bladder epithelial SV-HUC-1 cell line were purchased from American Tissue Culture Collection. UM-UC-3 and T24 cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Normal bladder epithelial SV-HUC-1 cells were grown in F-12K medium (HyClone; GE Healthcare Life Sciences) containing 10% FBS and 1% antibiotics. The cultures were incubated at 37°C in 5% CO2. Cycloheximide (CHX) was purchased from Sigma-Aldrich; Merck KGaA and used at the concentration of 20 µg/ml.
Vector construction and cell transfection
Short interfering RNA (siRNA) oligonucleotides targeting ZEB1-AS1, AUF1, ZEB1 and negative control siRNAs were purchased from Shanghai GenePharma Co., Ltd. The siRNA sequences are listed in Table IA. siRNA transfections were performed using 75 nM siRNA and Lipofectamine RNAimax (Thermo Fisher Scientific, Inc.). To establish stable ZEB1-AS1-silencing cell lines, short hairpin RNA targeting ZEB1-AS1 (sh-ZEB1-AS1) were cloned into pCDH-CMV-MCS-EF1-Puro or pLKO.1-Puro vectors and further loaded into a lentiviral vector (Shanghai GeneChem Co., Ltd.). The infection concentration was 1×108 pfu/ml. The sequences of the siRNAs and shRNAs are listed in Table IA.
Table I.
qPCR primer and siRNA sequences.
| A, siRNA sequences
| |
|---|---|
| siRNAs | Sequence (5′-3′) |
| si-ZEB1-AS1#1 | GGACCAACTTTATGGAATA |
| si-ZEB1-AS1#2 | GCTGAAGTCTGATGATTTA |
| si-ZEB1-AS1#3 | GGAGCCATCTAGTGCATAA |
| si-AUF1 | UCGACUAUCUGCUCCAAG |
| si-ZEB1 | TGATCAGCCTCAATCTGCA |
| si-NC | GACCTACAACTACCTATCA |
| sh-ZEB1-AS1 | CUUCAAUGAGAUUGAACUUCA |
| sh-NC C | AACAAGATGAAGAGCACCAA |
|
B, qPCR primer sequences | |
| Primers | Sequence (5′-3′) |
|
| |
| ZEB1-AS1 | F: TCCCTGCTAAGCTTCCTTCAGTGT |
| ZEB1-AS1 | R: GACAGTGATCACTTTCATATCC |
| ZEB1 | F: CGCAGTCTGGGTGTAATCGTAA |
| ZEB1 | R: GACTGCCTGGTGATGCTGAAA |
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| GAPDH | R: ATGGTGGTGAAGACGCCAGT |
| U6 | F: CTCGCTTCGGCAGCACA |
| U6 | R: AACGCTTCACGAATTTGCGT |
| U1 | F: GGGAGATACCATGATCACGAAGGT |
| U1 | R: CCACAAATTATGCAGTCGAGTTTCCC |
qPCR, quantitative polymerase chain reaction; siRNA, small interfering RNA; ZEB1, zinc finger E-box-binding homeobox 1; ZEB1-AS1, ZEB1 antisense 1; AUF1, heterogenous nuclear ribonucleoprotein D0; NC, negative control; F, forward; R, reverse.
Reverse transcription quantitative polymerase chain reac- tion (RT-qPCR)
Total RNA was extracted from BCa tissues or cells using the Qiagen RNeasy Mini kit according to the manufacturer's protocol (Qiagen GbmH). RT and qPCR kits were used to evaluate the expression of target RNAs. RT reactions (20 µl) were performed using the PrimeScript® RT reagent kit (Takara Biotechnology Co., Ltd.) and incubated for 30 min at 37°C followed by 5 sec at 85°C. For qPCR, 2 µl diluted RT product was mixed with 23 µl reaction buffer (Takara Biotechnology Co., Ltd.) to a final volume of 25 µl. All reactions were performed using an Eppendorf Mastercycler EP Gradient S (Eppendorf) under the following conditions: Initial denaturation at 95°C for 30 sec; and 45 cycles of denaturation (95°C for 5 sec), and annealing and elongation (60°C for 30 sec). The transcript expression of GAPDH was used for the normalization of detected RNAs using the comparative 2-ΔΔCq method (18). The primer sequences for qPCR are presented in Table IB.
Cell migration and invasion assay
Cell migration ability was evaluated by performing wound-healing assay. Cells were seeded onto 6-well plates at a density of 5×105 cells/well. A total of 12 h after transfection with the respective vectors, the cell layer was scratched to form wounds using a sterile 20-µl pipette tip; the non-adherent cells were washed away with culture medium, then the cells were additionally incubated for 48 h and images were captured to identify the wound size. Cell invasion was evaluated using a Transwell invasion assay with Boyden chambers (BD Biosciences) and membranes with 8 µm pores coated with Matrigel (coated at 37°C for 1 h). Cells in serum-free media were placed in the upper chamber of the insert. Medium containing 10% FBS was added to the lower chamber. After 12 h of incubation, the cells that had invaded through the membrane were fixed with 4% paraformaldehyde at 4°C for 20 min, followed by staining with 0.1% crystal violet for 10 min at room temperature. The images were visualized using an inverted microscope (×20 magnification) (Leica Microsystems GbmH).
In vivo lung-metastasis mice model
A total of 12 male BALB/c nude mice (19-22 g; 6 weeks old) were obtained from the Shanghai Laboratory Animal Center, Chinese Academy of Science. They were randomly divided into two groups of 6 mice in each, and housed with 3 mice/cage in a suitable pathogen-free sterile environment at 28°C and 50% humidity with a 12-12 h light-dark cycle, and were fed ad libitum with sterile chow food and water. The experimental protocol was approved by the Committee on the Ethics of Animal Experiments of Peking Union Medical College Hospital. Experimental lung metastases were induced by injections of single-cell suspension (2×106 eGFP-luc2-marked UM-UC-3 cells in 100 µl) into the mouse lateral tail vein. Cells were stably transfected with sh-ZEB1-AS1 or control vectors, and all cell injections were administered in a total volume of 500 µl PBS containing 0.1% BSA over a duration of 60 sec, as described previously (19). A total of 5 weeks later, prior to in vivo imaging, the mice were I.P. anaesthetized with sodium phenobarbital (75 mg/kg). During anesthesia (duration 15 to 20 min) and while recovering, mice were kept warm under a red heat lamp. The established lung metastases images were observed using the LB983 NIGHTOWL II system (Berthold Technologies GmbH & Co. KG).
Immunohistochemical (IHC) staining and scoring analyses
IHC staining and score calculation were conducted as described previously (20). Anti-ZEB1 antibody (1:100; cat. no. ab203829; Abcam) was used to detect their expression levels in mouse tumors. Images were visualized using a Nikon ECLIPSE Ti (Nikon Corporation) inverted microscope system (x20 magnification) and processed using Nikon software (CaptureNX2, version 2.4.7).
Cytosolic/nuclear fractioning and RNA florescence in situ hybridization (RNA FISH)
The cellular fraction was isolated to locate the sublocation of ZEB1-AS1. Briefly, 1×107 cells were harvested, resuspended in 1 ml ice-cold RNase-free PBS, 1 ml C1 buffer (1.28 M Sucrose, 40 mM Tris-HCl, pH 7.5, 20 mM MgCl2 and 4% Triton X-100) and 3 ml RNase-free water, and incubated for 15 min on ice. The cells were then centrifuged for 15 min at 3,000 × g at 4°C, and the supernatant containing the cytoplasmic constituents and the nuclear pellet were retained for RNA extraction.
For RNA FISH, BCa cells were seeded into a 6-well plate (1x105 cells/well) and fixed with 4% paraformaldehyde for 15 min at 4°C and treated with 0.5% Triton in PBS, followed by pre-hybridization. They were then hybridized with the ZEB1-AS1 probe (5 µM) at 37°C overnight. The ZEB1-AS1 probes were synthesized by Sangon Biotech Co., Ltd. The cells were visualized under a confocal microscope (x100 magnification; Carl Zeiss Microscopy GmbH).
RNA pulldown, electrophoresis and mass spectrometry
The RNA pulldown assay was performed using a Magnetic RNA-Protein Pull-down kit (Thermo Fisher Scientific, Inc.) according to manufacturer's protocol. BCa cells (2x107) were cross-linked for each hybridization reaction. The cell lysates were hybridized with a mixture of biotinylated DNA probes for 4 h at 37°C. The binding proteins were separated by electrophoresis. A total of 25 µg proteins in each lane were loaded for SDS-PAGE (10% gel) at 4°C. The concentrating voltage was 80 V and separating voltage was 110 V. The binding proteins were also identified by mass spectrometry to identify the potential binding proteins (H. Wayen Biotechnology, Shanghai, China) as previously described (21).
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using the EZ-Magna RIP kit (EMD Millipore) according to the manufacturer's protocol. Briefly, 1×107 cells were lysed with RIP lysis buffer using 1 freeze-thaw cycle. Cell extracts were coimmunoprecipitated with anti-AUF1 (1:200; cat. no. ab61193; Abcam) antibody, and the retrieved RNA was subjected to the aforementioned RT-qPCR analysis. Normal IgG was used as a negative control. For RT-qPCR analysis, GAPDH was used as the non-specific control.
Western blot analysis
Radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck KGaA) was used to lyse the cells to obtain total protein lysates. Protein concentration was measured using the bicinchoninic acid assay (Sigma-Aldrich; Merck KGaA). The quantified protein (25 µg) was transferred onto polyvinylidene fluoride membranes (Sigma-Aldrich; Merck KGaA) following 10% SDS-PAGE gel electrophoresis. Then, the membranes were blocked with 5% non-fat dry milk in TBS + Tween buffer (0.1%) for 2 h at room temperature and incubated with anti-ZEB1 antibody (1:1,000; cat. no. ab203829; Abcam) or anti-GAPDH antibody (1:5,000; cat. no. PA1-987; Invitrogen; Thermo Fisher Scientific, Inc.) at 4°C overnight, followed by horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. ab7090; Abcam) at room temperature for 1 h. Protein bands were detected using ECL reagent (Amersham; GE Healthcare). Gray analysis by image analysis was performed using the software Gel-Pro Analyzer (version 4.0; United States Biochemical) after scanning.
Bioinformatics analysis
The correlation between ZEB1-AS1 and ZEB1 mRNA levels was analyzed by using The Cancer Genome Atlas BCa dataset with the online database StarBase (http://starbase.sysu.edu.cn/panCancer.php).
Statistical analysis
The Kolmogorov-Smirnov test was applied to analyze the distribution of data in each group. Data were presented as median (interquartile range). Mann-Whitney U tests were performed to compare the data between the two groups. The Kruskal-Wallis test followed by Bonferroni correction post-hoc test was used for evaluating the difference among multiple groups. Receiver operation characteristic (ROC) analysis was performed to evaluate the diagnostic performance of ZEB1-AS1. Spearman's correlation analysis was performed to determine the correlation between variables. A two-sided P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software, Inc.).
Results
Knockdown of ZEB1-AS1 inhibits migration of BCa cells in vitro and metastasis in vivo
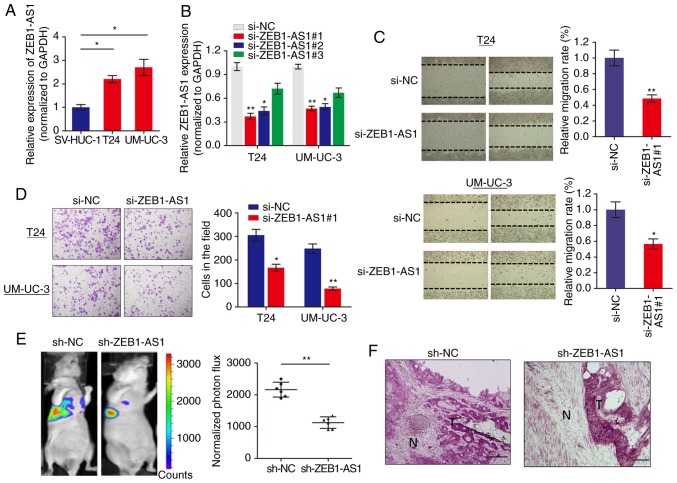
We measured the expression level of lncRNA ZEB1-AS1 in BCa cells via RT-qPCR and it was identified that ZEB1-AS1 was significantly increased in BCa cells when compared with normal SV-HUC-1 cells (Fig. 1A). To investigate the role of ZEB1-AS1 in BCa progression, ZEB1-AS1 expression was silenced in BCa cells using 3 different siRNAs. RT-qPCR analysis showed that ZEB1-AS1 was remarkably downregulated in T24 and UM-UC-3 cells (Fig. 1B), and si-ZEB1-AS1#1 was selected for use in functional experiments. The function of ZEB1-AS1 on cell motility and tumor metastasis was investigated using wound healing and invasion assays, respectively. Wound healing assays demonstrated that downregulation of ZEB1-AS1 significantly decreased the migratory ability of T24 and UM-UC-3 cells (Fig. 1C). In addition, ZEB1-AS1 knockdown inhibited the invasive ability of BCa cells (Fig. 1D).
Figure 1.
Knockdown of ZEB1-AS1 impairs migration and metastasis of BCa cells. (A) The expression levels of ZEB1-AS1 in BCa cells and normal epithelial bladder cells were measured via RT-qPCR. *P<0.05. (B) RT-qPCR determination of the silencing effect of ZEB1-AS1 following infection with respective silencing oligonucleotides. *P<0.05 and **P<0.01 vs. si-NC group. (C) A wound healing assay was performed to identify the effect of ZEB1-AS1 knockdown on BCa cell migration ability. *P<0.05 and **P<0.01 vs. si-NC group. (D) A Matrigel Transwell analysis was performed to determine the effect of ZEB1-AS1 on BCa cell invasion. *P<0.05 and **P<0.01. (E) Lung metastases were quantified using bioluminescence imaging 5 weeks after initial implantation. Representative in vivo bioluminescent images (left) and quantitative photon flux (right) are presented. **P<0.01. (F) Representative images showing that tumors had invaded into the surrounding muscle in the ZEB1-AS1 knockdown group and respective control. Scale bars, 100 µm. ZEB1-AS1, zinc finger E-box-binding homeobox 1-antisense 1; BCa, bladder cancer; RT-qPCR, reverse transcription quantitative polymerase chain reaction; si, small interfering; NC, negative control; N, non-tumor area; T, tumor tissue.
To additionally confirm the effects of ZEB1-AS1 in BCa metastasis, an experimental lung metastases model was induced by injections of single-cell suspension (2x106 UM-UC-3 cells in 100 µl) into the lateral tail vein of the mice. The luciferase flux count of lung metastases was significantly less in ZEB1-AS1-knockdown group compared to control group (Fig. 1E). Notably, it was observed that the tumors formed by the ZEB1-AS1-knockdown BCa cells grown in the nude mice exhibited sharp edges, while the control tumors exhibited spike-like structures that invaded the surrounding muscle tissues (Fig. 1F), which additionally supported the hypothesis that knockdown of ZEB1-AS1 suppressed BCa metastasis. Collectively, these results indicated that ZEB1-AS1 regulates BCa cells migration and invasion in vitro, and metastasis in vivo.
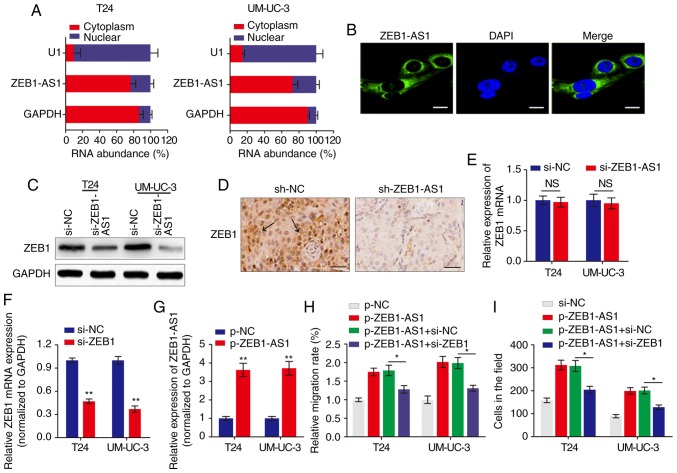
ZEB1-AS1 regulates BCa metastasis via the upregulation of ZEB1 protein
The subcellular localization of a lncRNA is associated closely with its biological mechanism. Cellular fractionation and RNA-FISH assays demonstrated that ZEB1-AS1 was primarily distributed in the cytoplasm in BCa cells (Fig. 2A and B). lncRNAs located in the cytoplasm are usually associated with post-transcriptional regulation (15,22). As ZEB1 is a critical transcription factor in cancer metastasis, we hypothesized that the antisense transcript ZEB1-AS1 may regulate the expression of ZEB1 and thereby induce the metastatic effect in BCa. Western blot analysis indicated that ZEB1 was downregulated by ZEB1-AS1 knockdown in BCa cells (Fig. 2C). IHC analysis of the tumor tissues grown in nude mice also suggested that ZEB1 expression was inhibited in the ZEB1-AS1-silencing group compared with the sh-NC group (Fig. 2D). However, when the expression of ZEB1 mRNA was analyzed using RT-qPCR, knockdown of ZEB1-AS1 demonstrated no effect on ZEB1 mRNA expression in BCa cells (Fig. 2E). Then, the functional role of ZEB1 in ZEB1-AS1-mediated BCa metastasis was examined by overexpression of ZEB1-AS1 and knockdown of ZEB1 (Fig. 2F and G). As indicated in Fig. 2H and I, enhanced expression of ZEB1-AS1 promoted BCa cell migration and invasion, however, this effect was significantly reversed by co-expression of siZEB1 in the ZEB1-AS1 overexpressing cells. These data demonstrated that ZEB1-AS1 regulates BCa metastasis via the upregulation of ZEB1 protein.
Figure 2.
ZEB1 is a functional target of ZEB1-AS1 in BCa. (A) The relative expression level of ZEB1-AS1 in the nucleus and cytoplasm of BCa cells was measured by RT-qPCR. U1 (retained in the nucleus) and GAPDH (exported to cytoplasm) were used as controls. (B) The subcellular distribution of ZEB1-AS1 was visualized by RNA fluorescence in situ hybridization in UM-UC-3 cells. Scale bars, 10 µm. (C) Western blot analysis of ZEB1 protein levels in BCa cells with ZEB1-AS1 knockdown. (D) Immunohistochemistry assay was used to measure the levels of ZEB1 expression in tumor tissues grown in mice. The arrows indicate the ZEB1-rich area. Scale bars, 50 µm. (E) RT-qPCR was performed to evaluate the effect of ZEB1-AS1 knockdown on ZEB1 mRNA level. (F and G) The effects of (F) ZEB1 knockdown and (G) ZEB1-AS1 overexpression were verified using RT-qPCR. **P<0.01. (H) Wound-healing assays and (I) Transwell invasion assays were performed to detect the effect of ZEB1 knockdown and ZEB1-AS1 overexpression on BCa cell migration and invasion, respectively. *P<0.05. ZEB1-AS1, zinc finger E-box-binding homeobox 1-antisense 1; BCa, bladder cancer; RT-qPCR, reverse transcription quantitative polymerase chain reaction; si, small interfering; sh, short hairpin; NC, negative control; NS, not significant.
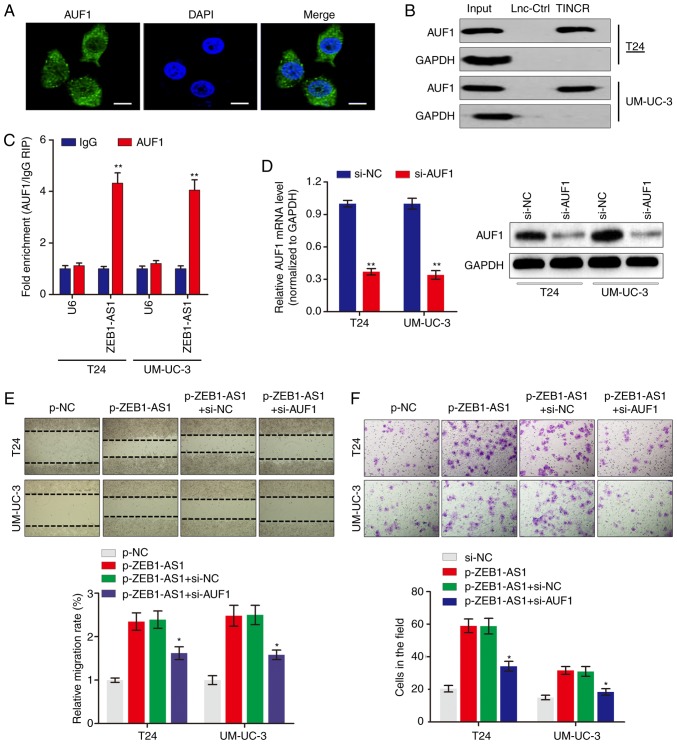
ZEB1-AS1 directly interacts with AUF1 in BCa
To examine the potential mechanism by which ZEB1-AS1 regulates the protein levels of ZEB1, RNA-pulldown experiments were performed followed by mass spectrometry to identify ZEB1-AS1-interacting proteins. The data identified a number of potential ZEB1-AS1-interacting proteins (Table II), among which AUF1 was identified. AUF1 is able to bind to (A + U)-rich elements within 3'-untranslated region (3'-UTR) of target mRNA and promote translation without affecting the mRNA level (23). Consistent with ZEB1-AS1 localization, AUF1 was also primarily distributed in the cytoplasm in BCa cells (Fig. 3A). The significance of the interaction between ZEB1-AS1 and AUF1 was confirmed using RNA pulldown (Fig. 3B) and RIP (Fig. 3C) assays. To explore the role of the ZEB1-AS1-AUF1 association in the function of ZEB1-AS1, ZEB1-AS1 was overexpressed, and then AUF1 expression was knocked down in BCa cells (Fig. 3D). Notably, AUF1 knockdown eliminated the ZEB1-AS1-mediated enhancement of the migration (Fig. 3E) and invasion (Fig. 3F) of BCa cells in vitro. These data suggested that the direct interaction between ZEB1-AS1 and AUF1 may be important for the ZEB1-AS1-induced BCa metastasis.
Table II.
Identification of ZEB1-AS1 binding proteins by mass spectrometry.
| Protein | Beads | ZEB1-AS1 | Ratio (ZEB1-AS1/Beads) |
|---|---|---|---|
| AUF1 | 0 | 3 | NA |
| LAS1L | 0 | 3 | NA |
| STT3B | 0 | 3 | NA |
| PCH2 | 1 | 3 | 3 |
| MRP1 | 0 | 3 | NA |
| ARF6 | 1 | 3 | 3 |
| AKAP8 | 0 | 3 | NA |
| GSTO1 | 0 | 3 | NA |
| AP1B1 | 0 | 3 | NA |
| DPM1 | 0 | 3 | NA |
| PSDE | 1 | 3 | 3 |
| PLST | 0 | 3 | NA |
| PSAL | 0 | 3 | NA |
| TTL12 | 0 | 3 | NA |
| ERLN1 | 0 | 3 | NA |
| NSF | 0 | 3 | NA |
| KTN1 | 1 | 3 | 3 |
ZEB1-AS1, zinc finger E-box-binding homeobox 1-antisense 1; NA, not available. The 'Beads' column represents the spectral counts of proteins in the beads only group. The 'ZEB1-AS1' column represents the spectral counts of proteins in the ZEB1-AS1 group. The 'Ratio (ZEB1-AS1/Beads)' column represents the comparison of the spectral count ratio of proteins the ZEB1-AS1 group with the beads only group.
Figure 3.
ZEB1-AS1 interacts with AUF1 to serve key roles in BCa. (A) Immunofluorescence analysis of AUF1 protein in UM-UC-3 cells. Scale bars, 10 µm. (B) The interaction between ZEB1-AS1 and AUF1 was confirmed by RNA pulldown assays and western blot analysis. GAPDH served as negative control. (C) RNA immunoprecipitation assays were performed using anti-AUF1 and control IgG antibodies, followed by RT-qPCR to examine the enrichment of ZEB1-AS1 and U6. U6 served as negative control. **P<0.01. (D) The silencing effect of si-AUF1 infection in BCa cells was determined by RT-qPCR (left panel) and western blot analysis (right panel). (E) Wound healing assays and (F) Transwell invasion assays were performed to detect the effect of AUF1 knockdown and ZEB1-AS1 overexpression on BCa cell migration and invasion, respectively. *P<0.05 vs. p-ZEB1-AS1+si-NC group. ZEB1-AS1, zinc finger E-box-binding homeobox 1-antisense 1; AUF1, heterogenous nuclear ribonucleoprotein D0; BCa, bladder cancer; RT-qPCR, reverse transcription quantitative polymerase chain reaction; si, small interfering; NC, negative control.
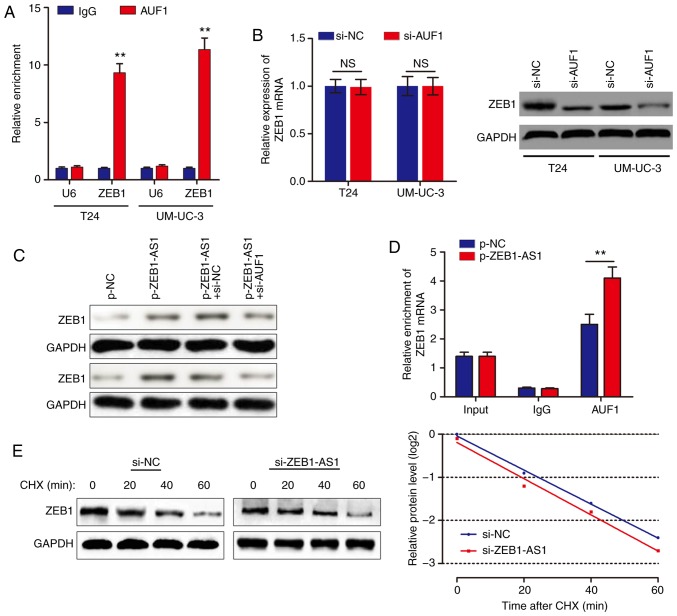
ZEB1-AS1 activates the translation of ZEB1 mRNA via recruiting AUF1
Based on the above results, we hypothesized that ZEB1-AS1 promoted ZEB1 mRNA translation by binding with AUF1. To examine this hypothesis, a RIP assay was performed, and it was identified that ZEB1 was enriched in AUF1 precipitates (Fig. 4A). AUF1 knockdown decreased ZEB1 protein level without affecting the ZEB1 mRNA level (Fig. 4B). In addition, knockdown of AUF1 eliminated the ZEB1-AS1-mediated increases in ZEB1 protein levels (Fig. 4C). As ZEB1-AS1 and AUF1 regulate ZEB1 expression synergistically, we hypothesized that ZEB1-AS1 may increase ZEB1 protein level through recruiting AUF1 to bind to the ZEB1 3'-UTR. In support of this hypothesis, ZEB1-AS1 overexpression promoted endogenous AUF1 binding to ZEB1 mRNA by RIP (Fig. 4D). In addition, UM-UC-3 cells were treated with CHX, which enabled the measurement of the degradation of pre-existing proteins. The data indicated that knockdown of ZEB1-AS1 or AUF1 had no effect on half-life of ZEB1 protein in BCa cells (Fig. 4E). Taken together, these results indicated that ZEB1-AS1 assists AUF1 in binding to ZEB1 mRNA, activating its translation without affecting the mRNA level.
Figure 4.
ZEB1-AS1 activates the translation of ZEB1 mRNA via binding with AUF1. (A) RNA immunoprecipitation assays were performed using anti-AUF1 and control IgG antibodies, followed by RT-qPCR to examine the enrichment of ZEB1-AS1 and U6. U6 served as a negative control. **P<0.01. (B) The effects of AUF1 knockdown on ZEB1 mRNA expression were determined via RT-qPCR. (C) Western blot analysis was performed to detect the expression level of ZEB1 protein in bladder cancer cells with ZEB1-AS1 overexpression and/or AUF1 silencing. (D) The endogenous binding of AUF1 to ZEB1 mRNA was modified by ZEB1-AS1 overexpression. (E) Control siRNA- or si-ZEB1-AS1-transfected UM-UC-3 cells were cultured with 20 µg/ml CHX for 0-60 min, then subjected to western blot analysis. ZEB1-AS1, zinc finger E-box-binding homeobox 1-antisense 1; AUF1, heterogenous nuclear ribonucleoprotein D0; RT-qPCR, reverse transcription quantitative polymerase chain reaction; si, small interfering; NC, negative control; NS, not significant; CHX, cycloheximide.
ZEB1-AS1 expression is associated with metastasis in patients with BCa
A preliminary study was performed to identify the clinical role of ZEB1-AS1 using 60 BCa tissues (30 MIBC and 30 NMIBC) and paired non-cancer tissues from patients with primary BCa. RT-qPCR analysis showed that ZEB1-AS1 was upregulated in BCa tissues in contrast to non-cancer tissues (Fig. 5A). In addition, the expression level of ZEB1-AS1 in MIBC tissues was significantly increased compared with that in NMIBC tissues (Fig. 5B). ROC curve analysis was then used to investigate the predictive value of ZEB1-AS1 in differentiating patients with cancer from the noncancerous population. As demonstrated in Fig. 5C, the area under the curve (AUC) was 0.860, with the diagnostic sensitivity and specificity measuring 71.7 and 85.0% with the cut-off value of 0.134, respectively. Notably, the ROC curve using the ZEB1-AS1 level to differentiate MIBC from NMIBC demonstrated the AUC, sensitivity and specificity values were 0.856; 76.7; and 80.0%, respectively (Fig. 5D). By analyzing The Cancer Genome Atlas BCa dataset with the online database StarBase, positive associations between the RNA expression of ZEB1-AS1 and ZEB1 mRNA were identified in BCa (Fig. 5E). This positive association was additionally verified in the 60 cancer tissues (Fig. 5F), which supports the regulatory mechanism of ZEB1-AS1 and ZEB1 in BCa metastasis.
Figure 5.
ZEB1-AS1 expression is associated with metastasis in patients with BCa. (A) ZEB1-AS1 expression level was detected by RT-qPCR in 60 BCa tissues and paired non-tumor tissues. (B) ZEB1-AS1 expression level was measured by RT-qPCR in 30 patients with MIBC and 30 patients with NMIBC. (C and D) ROC analysis was performed to investigate the diagnostic value of ZEB1-AS1 expression in differentiating between (C) patients with BCa and patients without cancer, and between (D) patients with MIBC from patients with NMIBC. (E) Correlation between RNA expression of ZEB1-AS1 and ZEB1 was analyzed from the Cancer Genome Atlas BLCA dataset using the online database StarBase. (F) The associations between the RNA expression of ZEB1-AS1 and ZEB1 in 60 BCa tissues were analyzed using Spearman's correlation analysis. ZEB1-AS1, zinc finger E-box-binding homeobox 1-anti-sense 1; BCa, bladder carcinoma; RT-qPCR, reverse transcription quantitative polymerase chain reaction; MIBC, muscle-invasive BCa; NMIBC, non-MIBC; AUC, area under the curve; BLCA, Bladder Urothelial Carcinoma.
Discussion
Numerous previous studies have assisted in gaining an improved understanding of the molecular mechanisms during cancer progression and chemoresistance. However, the specific regulatory model remains largely unknown in cancer, including BCa. Therefore, it is of importance to identify new molecular signatures which may be useful for cancer prevention and therapy. In the present study, it was demonstrated that over-expression of ZEB1-AS1 in BCa promoted the migration and metastasis of BCa cells in vitro and in vivo. Mechanistically, ZEB1-AS1 increased ZEB1 protein expression by guiding AUF1 to activate the translation of ZEB1 mRNA. In addition, ZEB1-AS1 expression was upregulated in BCa tissues and associated with metastasis in patients with BCa.
lncRNAs have been demonstrated to regulate biological functions via diverse mechanisms: Guider; decoy; scaffold effect on DNA, RNA or protein; and post-transcriptional effects (24). The expression of ZEB1-AS1 has been described, and it has been identified as an oncogene in several cancer types, including gastric (25), prostate (26) and colorectal cancer (27), and glioma (28). Lin et al (17) demonstrated that lncRNA ZEB1-AS1 serves an oncogenic role in BCa through the promotion of cell growth, migration and the inhibition of apoptosis in BCa 5637 and SW780 cell lines. Consistent with the data from Lin et al (17), the present study showed that ZEB1-AS1 enhanced metastasis and invasion in vitro, and promoted metastasis in vivo, confirming that ZEB1-AS1 may serve as a oncogene in BCa.
The subcellular localization of a lncRNA is closely associated with its biological mechanism. The results from the present study showed that ZEB1-AS1 was primarily distributed in the cytoplasm in BCa cells, indicating it may regulate cancer progression at the post-transcriptional level. ZEB1-AS1 is a noncoding antisense transcript generated from ZEB1 promoters and located in physical contiguity with ZEB1. It is well known that ZEB1 is a transcription factor that promotes tumor invasion and metastasis by inducing EMT in carcinoma cells. Emerging studies have indicated that overexpression of ZEB1-AS1 increased ZEB1 levels and promoted tumor progression in different kinds of malignancies (26,27,29-31). However, the regulatory interaction between lncRNA ZEB1-AS1 and ZEB1 has not been described in BCa. Notably, the present study verified that ZEB1 was positively regulated by ZEB1-AS1 at protein level but not at the mRNA level, which suggests that ZEB1-AS1 may regulate ZEB1 expression without affecting mRNA level.
As ZEB1-AS1 affects ZEB1 protein level but not mRNA level, we hypothesized that this regulatory model may occur at the post-transcriptional level. To uncover the underlying mechanism by which ZEB1-AS1 regulates ZEB1 protein level, the ZEB1-AS1-interacting proteins were verified. It was identified that AUF1 was associated with ZEB1-AS1 and may serve as an adaptor protein that cooperates with ZEB1-AS1 to bind to ZEB1 mRNA. AUF1 an RNA-binding protein that produces 4 transcript variants following alternative pre-messenger RNA (pre-mRNA) splicing, with canonical roles in controlling the stability or translation of mRNA targets based on recognition of AU-rich sequences within 3'-UTR of target mRNA (32). As ZEB1-AS1 affected ZEB1 protein level without affecting the mRNA, it was assumed that AUF1 regulated the translation of ZEB1 mRNA. The results of the RIP assay verified the direct interaction between AUF1 and ZEB1. In addition, ZEB1-AS1 showed no effect on the ZEB1 protein stability, which confirmed our hypothesis. Notably, Li et al (33) suggested that AUF1 could bind to the 3'-UTR of ZEB1 mRNA and affect the mRNA stability in thyroid cancer. Therefore, the regulatory mechanism of AUF1 for ZEB1 in cancer requires additional study. In addition, whether miRNAs are regulated by ZEB1-AS1 in BCa progression also requires verification, as there is a close association between lncRNAs and miRNAs.
Take a step further, the present study also investigated the clinical role of ZEB1-AS1 expression in patients with BCa. It was observed that ZEB1-AS1 was upregulated in BCa tissues, and expressed at significantly increased levels in MIBC tissues. ROC curve analysis clearly demonstrated a high predictive value of ZEB1-AS1 in differentiating patients with MIBC from patients with NMIBC, indicating a high predictive value for BCa metastasis. These data validated the experimental conclusion derived from the in vitro and in vivo studies.
In summary, the present study demonstrated that ZEB1-AS1 functionally and clinically participated in the metastasis and progression of BCa, based on an AUF1-mediated translation activation of ZEB1 mRNA. Identification of the precise role of ZEB1-AS1 in the progression of BCa will not only improve the understanding of lncRNA-induced tumorigenesis and metastasis, but also enable the development of novel therapeutic strategies to treat BCa.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and ZJ acquired the data and created a draft of the manuscript; YD and JZ collected clinical samples and performed the experimental assays; DW and GL analyzed and interpreted the data, and performed statistical analysis; XZ, DW and ZJ reviewed the manuscript, figures and tables. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Clinical Research Ethics Committee of Peking Union Medical College Hospital, and the experimental protocols for the animal model was approved by the Committee on the Ethics of Animal Experiments of Peking Union Medical College Hospital. Written informed consent was obtained from each participant prior to tissue collection.
Patient consent for publication
Written informed consent was obtained from each participant prior to tissue collection.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Van Batavia J, Yamany T, Molotkov A, Dan H, Mansukhani M, Batourina E, Schneider K, Oyon D, Dunlop M, Wu XR, et al. Bladder cancers arise from distinct urothelial sub-populations. Nat Cell Biol. 2014;16:982–991. 1–5. doi: 10.1038/ncb3038. [DOI] [PubMed] [Google Scholar]
- 4.Youssef RF, Raj GV. Lymphadenectomy in management of invasive bladder cancer. Int J Surg Oncol. 2011;2011:758189. doi: 10.1155/2011/758189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu XR. Urothelial tumorigenesis: A tale of divergent pathways. Nat Rev Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 6.Hautmann RE, de Petriconi RC, Pfeiffer C, Volkmer BG. Radical cystectomy for urothelial carcinoma of the bladder without neoadjuvant or adjuvant therapy: Long-term results in 1,100 patients. Eur Urol. 2012;61:1039–1047. doi: 10.1016/j.eururo.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y. Opinion: Emerging mechanisms of tumour lymphan-giogenesis and lymphatic metastasis. Nat Rev Cancer. 2005;5:735–743. doi: 10.1038/nrc1693. [DOI] [PubMed] [Google Scholar]
- 8.Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124:922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa S, Kageyama Y. Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochim Biophys Acta. 2014;1839:215–222. doi: 10.1016/j.bbagrm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, Chen K, Chen L, Ding Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquiti-nation of LATS1. J Hematol Oncol. 2017;10:91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Ye Z, Zhao X, Ji Z. SP1-induced up-regulation of lncRNA SNHG14 as a ceRNA promotes migration and invasion of clear cell renal cell carcinoma by regulating N-WASP. Am J Cancer Res. 2017;7:2515–2525. [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Zhan Y, Liu Y, Chen Z, Liang J, Li W, He A, Zhou L, Mei H, Wang F, Huang W. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8:24202–24212. doi: 10.18632/oncotarget.15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Gu P, Xie R, Han J, Liu H, Wang B, Xie W, Xie W, Zhong G, Chen C, et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. 2017;21:1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong X, Du X, Xu Y, Zheng W. LINC00037 inhibits proliferation of renal cell carcinoma cells in an epidermal growth factor receptor-dependent way. Cell Physiol Biochem. 2018;45:523–536. doi: 10.1159/000487030. [DOI] [PubMed] [Google Scholar]
- 22.Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med. 2018;22:1527–1537. doi: 10.1111/jcmm.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore AE, Chenette DM, Larkin LC, Schneider RJ. Physiological networks and disease functions of RNA-binding protein AUF1. Wiley Interdiscip Rev RNA. 2014;5:549–564. doi: 10.1002/wrna.1230. [DOI] [PubMed] [Google Scholar]
- 24.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu TP, Wang YF, Xiong WL, Ma P, Wang WY, Chen WM, Huang MD, Xia R, Wang R, Zhang EB, et al. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. doi: 10.1038/cddis.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong WC, Han N, Wu N, Zhao KL, Han C, Wang HX, Ping GF, Zheng PF, Feng H, Qin L, He P. Interplay between long noncoding RNA ZEB1-AS1 and miR-101/ZEB1 axis regulates proliferation and migration of colorectal cancer cells. Am J Transl Res. 2018;10:605–617. [PMC free article] [PubMed] [Google Scholar]
- 28.Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin CZ, Qu Q, Wang GH, He CJ, Zhou HH. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int J Mol Sci. 2016;17:E1431. doi: 10.3390/ijms17091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng R, Li N, Yang S, Liu L, Han S. Long non-coding RNA ZEB1-AS1 promotes cell invasion and epithelial to mesenchymal transition through inducing ZEB1 expression in cervical cancer. Onco Targets Ther. 2018;11:7245–7253. doi: 10.2147/OTT.S179937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Lin J. Long noncoding RNA-ZEB1 AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 31.Qu R, Chen X, Zhang C. LncRNA ZEB1-AS1/miR-409-3p/ZEB1 feedback loop is involved in the progression of non-small cell lung cancer. Biochem Biophys Res Commun. 2018;507:450–456. doi: 10.1016/j.bbrc.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 32.Choi YJ, Yoon JH, Chang JH. Crystal structure of the N-Terminal RNA recognition motif of mRNA decay regulator AUF1. Biomed Res Int. 2016;2016:3286191. doi: 10.1155/2016/3286191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Zhang HY, Du ZX, Li C, An MX, Zong ZH, Liu BQ, Wang HQ. Induction of epithelial-mesenchymal transition (EMT) by Beclin 1 knockdown via posttranscriptional upregulation of ZEB1 in thyroid cancer cells. Oncotarget. 2016;7:70364–70377. doi: 10.18632/oncotarget.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.