Abstract
Aim
The aim of this study was to resolve the phylogenetic placement of island taxa, reconstruct ancestral origins and resolve competing hypotheses of dispersal patterns and biogeographical histories for oceanic island endemic taxa within subgenus Plantago (Plantaginaceae).
Location
Juan Fernández Islands, the Auckland Islands, Lord Howe Island, New Amsterdam Island, New Zealand, Tasmania, Falkland Islands, Rapa Iti and the Hawaiian Islands.
Taxon
Island endemics within Plantago (Plantaginaceae), a globally distributed taxonomic group comprising approximately 250 species.
Methods
We use Bayesian phylogenetic and divergence time analyses and historical biogeographical analysis of molecular sequence data to infer the ancestral origins of the oceanic island species in Plantago.
Results
Taxa within subgenus Plantago form clades based on geographic proximities and challenge previous phylogenetic relationships and classification based on morphology. We infer that biogeographic histories of oceanic island taxa from multiple islands were shaped by dispersal at different scales and possibly by different types of birds. The highly remote Hawaiian Islands and Rapa Iti were colonized from North American taxa in a pattern corresponding to known migration routes of large marine birds, rather than from New Zealand as previously hypothesized. The island endemics of Juan Fernández, the Falkland Islands, Lord Howe, Auckland Islands and New Zealand are found to have sources in the nearest continental areas. The analyses confirm recent speciation within subgenus Plantago – which is particularly heightened in island lineages in Hawaii and Rapa Iti – but show slightly older divergence times than previous molecular dating studies.
Main conclusions
Using molecular data to infer ancestral ranges for plants with uncertain taxonomic relationships can greatly improve our understanding of biogeographical histories and help elucidate origins, dispersal modes and routes in widespread lineages with complex distribution patterns such as Plantago. We improve understanding of important floristic exchange areas between continents and islands as a result of long‐distance dispersal. We infer that a combination of both stepping stone dispersal and extreme long‐distance dispersal can shape insular floras, and that multiple floristic areas can be the sources of closely related island taxa. However, despite the successful dispersal of Plantago, radiation in island archipelagos is generally limited suggesting specific traits may limit diversification.
Keywords: biogeographical range reconstruction, bird dispersal, island taxa, long‐distance dispersal, oceanic islands, Plantago
1. INTRODUCTION
For over a century, evolutionary biologists have investigated island taxa regarding their ancestral origins and evolutionary histories. The isolation and often young age of oceanic islands are considered excellent systems in which to study dispersal and speciation (Emerson, 2002; Losos & Ricklefs, 2009). In recent decades, owing to the advancement of molecular phylogenetic and divergence analyses, evidence has been increasing that long‐distance dispersal (LDD) events by wind, oceanic drift and animal migrations play a larger role than originally thought in explaining widespread species distributions (de Queiroz, 2005; Gillespie et al., 2012; le Roux et al., 2014; Nathan, 2006; Nathan et al., 2008; Raxworthy, Forstner, & Nussbaum, 2002; Vences et al., 2003; Viana, Gangoso, Bouten, & Figuerola, 2016; Winkworth, Wagstaff, Glenny, & Lockhart, 2002). Long‐distance dispersal, rather than geological vicariance, regardless of the geological histories and ages of island systems, appears to be the principal driver of range evolution and subsequent speciation (Christenhusz & Chase, 2013; de Queiroz, 2005; Dupin et al., 2017; Gallaher, Callmander, Buerki, & Keeley, 2015; Givnish et al., 2004; Mitchell et al., 2016).
In the case of plants, the movement of propagules either internally or externally by birds is recognized as the most common type of LDD responsible for the distribution of disjunct plant lineages on remote oceanic islands, though dispersal by oceanic drift and weather events also play roles (Carlquist, 1966, 1967; Gillespie et al., 2012; Kistler et al., 2014; Nathan et al., 2008; Pole, 1994; Sanmartin & Ronquist, 2004). Birds are more often implicated as the main dispersal vectors for island plants based on an over‐representation of plant traits that favour bird dispersal in island taxa (Baldwin & Wagner, 2010; Carlquist, 1967; Gillespie et al., 2012). The nearest landmasses are often implicated as the primary sources for island taxa, yet, as a result of bird‐mediated LDD across extreme distances such as via the Pacific or Asia‐Australian bird flyways, thousands of kilometres can separate some of the most closely related taxa (Baldwin & Wagner, 2010; Gillespie et al., 2012). Thus, LDD events mediated by bird vectors can result in multiple theories of biogeographic origins for island plant lineages (Birch & Keeley, 2013; de Queiroz, 2005; Kainulainen, Razafimandimbison, Wikstrom, & Bremer, 2017; le Roux et al., 2014). Applying biogeographical models and using molecular phylogenetic relationships to reconstruct source areas for island taxa can help resolve complex biogeographical histories, identify important areas of floristic exchange and elucidate dispersal routes that shape island floras (Bacon, Simmons, Archer, Zhao, & Andriantiana, 2016; Bouckaert et al., 2014; Ho et al., 2015; Johnson, Clark, Wagner, & McDade, 2017; Matzke, 2014).
Plantago L., a genus of approximately 250 species, is a model group to study LDD processes and compare different hypotheses regarding dispersal modes and routes, due to its worldwide distribution, high dispersal capabilities and high number of single island endemic taxa (Dunbar‐Co, Wieczorek, & Morden, 2008; Hassemer, De Giovanni, & Trevisan, 2016; Rahn, 1996; Tay, Meudt, Garnock‐Jones, & Ritchie, 2010a). The genus is largely temperate in distribution, but occurs at high altitudes in tropical areas and on oceanic islands (van der Aart & Vulto, 1992). Previous phylogenetic and molecular divergence analyses have estimated that Plantago s.l. (including Littorella P.J.Bergius) diverged from its closest known relative, Aragoa Kunth, in the late Miocene to Pliocene, 7.1 million years ago (Ma) (Bello, Chase, Olmstead, Rønsted, & Albach, 2002; Rønsted, Chase, Albach, & Bello, 2002) or 2.8 Ma (Tay et al., 2010a). This finding of recent diversification of the genus rules out earlier hypotheses of vicariance in explaining the global distribution of the genus (i.e. as proposed by Rahn, 1996), and thus, long‐distance dispersal – presumably by birds – is the accepted scenario for Plantago (Dunbar‐Co et al., 2008; Rønsted et al., 2002; Tay et al., 2010a). Not only are the seeds of Plantago species known to be eaten by birds and other animals (Buse & Filser, 2014; Czarnecka & Kitowski, 2013; Panter & Dolman, 2012) but also Plantago species often grow alongside graminoids and other plants that are known to be typically eaten as fodder by birds prior to long migratory flights (Carlquist, 1966; Meudt, 2012; Rahn, 1996). Furthermore, dispersal in the genus is thought to be facilitated by the mucilaginous properties of wetted seeds, a specialized adaptation that assists with seed dispersal by increasing chance of seeds adhering to animals (Fischer et al., 2004; Rønsted et al., 2002; Tay et al., 2010a) or by attracting animals and facilitating dispersal via ingestion (Buse & Filser, 2014; Western, 2012). The mucilage in P. lanceolata L. seeds for example has been shown to be strongly adhesive to feathers and fur, especially when it has dried, which increases the potential of being transported externally over vast distances (Kreitschitz, Kovalev, & Gorb, 2016), while mucilage in P. major L. has been shown to attract invertebrates and facilitate endozoochory (Buse & Filser, 2014).
In addition to its adaptations to long‐distance dispersal, the majority of Plantago species have traits that likely assist in the successful colonization of new locations, including remote oceanic islands. For example, species are wind pollinated and many are self‐compatible – two important traits that are common in other island endemics (Carlquist, 1967; Rahn, 1996; Stuessy, Crawford, & Ruiz, 2018). Additionally, many Plantago island endemics grow on cliffs and other inaccessible habitats that may serve as refuges from natural threats, thereby facilitating successful establishment and survival in new areas (Dunbar‐Co, Sporck, & Sack, 2009; Stuessy et al., 2018).
The genus Plantago has been classified by Rahn (1996) into six subgenera, that is, Plantago, Coronopus (Lam. & DC.) Rahn, Littorella (P.J.Bergius) Rahn, Bougueria (Decne.) Rahn, Psyllium (Mill.) Harms & Reiche and Albicans Rahn; however, recent molecular phylogenetic studies have challenged classical taxonomic treatment. Subgenus Albicans, for example, has subsequently been included in subgenus Psyllium based on molecular phylogenetic studies (Rønsted et al., 2002), and subgenus Littorella was found to be sister to the remainder of Plantago and is most often considered a separate genus (Hassemer, Moroni, & O'Leary, 2018; Hoggard, Kores, Molvray, Hoggard, & Broughton, 2003; Kolář, 2014).
Subgenus Plantago, sensu Rahn (1996), is the largest of the subgenera with about 130 species currently recognized (Rahn, 1996; Rønsted et al., 2002). The subgenus is monophyletic (Hoggard et al., 2003; Rønsted et al., 2002; Tay et al., 2010a) and is distributed on all continents. This group is the focus of this work, as the subgenus includes the highest number of native Plantago species on oceanic islands, yet taxonomic, phylogenetic and biogeographic relationships between taxa in the subgenus remain some of the most poorly resolved, and morphological variation between many of the species is low (Meudt, 2012; Rahn, 1996; Rønsted et al., 2002). Previous molecular analyses have found Rahn's (1996) taxonomic sections within subgenus Plantago to be paraphyletic (Hoggard et al., 2003; Ishikawa, Yokoyama, Ikeda, Takabe, & Tsukaya, 2009; Tay et al., 2010a) though additional sampling is necessary to confirm this and improve our understanding of biogeographic histories.
Forty‐five taxa in subgenus Plantago have been described from 18 oceanic islands or island systems (Appendix I, Supporting Information). Several species are single island endemics to South Pacific Ocean islands or island archipelagos, Juan Fernández Islands off the coast of Chile, Galápagos Islands, Tonga Islands and Lord Howe Island and the Auckland Islands in Australasia. A single species is endemic to Madagascar, nine to mainland New Zealand, six to Tasmania, one species is native to both the Auckland Islands and Tasmania. Two species are endemic to St. Paul and New Amsterdam Islands, seven to New Guinea, two to Java, one species is recognized from Japan, and three species are recognized in Jeju. Three endemic species are currently recognized in the Hawaiian Islands, two species are endemic to Rapa Iti. In the South Atlantic Ocean, single endemic island species are known from Saint Helena, from Trindade Island and from the Falkland Islands.
The remoteness of the oceanic islands, the extreme distance between the nearest landmasses as well as uncertainty in the taxonomic relationships have resulted in numerous and often competing hypotheses as to the biogeographical histories of the island taxa in Plantago (Dunbar‐Co et al., 2008; Hoggard et al., 2003; Meudt, 2012; Rahn, 1996; Rønsted et al., 2002; Tay et al., 2010a, 2010b). Given adaptations of Plantago plants to LDD by birds, dispersal routes could be many, and relatedness between taxa may not reflect geographic proximity. For example, some of the island taxa (P. rupicola, P. fernandezia, P. princeps s.l. and P. robusta) exhibit typical island traits such as woody stems (Carlquist, 1970; Rahn, 1996; Stuessy et al., 2018), and based on such morphological similarities, P. fernandezia is thought to be more closely related to P. princeps s.l. in the Hawaiian Islands rather than to taxa in the Americas (Pilger, 1937; Rahn, 1996). However, the question remains whether plant propagules arrived in the Juan Fernández Islands from the western Pacific or from South America – the closest continental landmass where extant Plantago species are known (Rahn, 1996; Stuessy et al., 2018). Along similar lines, at least two competing hypotheses exist for the species endemic to the Hawaiian Islands, either arising from a LDD colonization event from a North American ancestor or from an ancestor in Australasia via Rapa Iti (Dunbar‐Co et al., 2008).
Until now, inferences of dispersal patterns within subgenus Plantago based on molecular data have been hindered by poorly resolved phylogenetic relationships due to too few of the 130 taxa being available for previous analyses [i.e. Rønsted et al. (2002) included 19 species; Hoggard et al. (2003) included 14 species, Ishikawa et al. (2009) included 24 species; and Tay et al. (2010a) included 20 species] as well as insufficient resolution obtained by the few genetic sequences used. Additionally, only 19 of the 45 island endemics in subgenus Plantago were available for previous phylogenetic analyses covering seven islands or island systems with known Plantago species (most of these were restricted to Australasia [Meudt, 2012; Tay et al., 2010a], see Appendix I, Supporting Information). This severely limits the testing of biogeographic histories for island taxa in the globally distributed genus Plantago.
In this study, molecular data from 14 of the island endemics were obtained – covering 11 of the 18 different oceanic islands or island systems where Plantago species are known – and the number of taxa represented from within subgenus Plantago was substantially increased (Appendix I, Supporting Information). We include for the first‐time endemic species from Juan Fernandez Islands, Lord Howe Island, Java and Jeju. This sampling increase not only results in improved phylogenetic and biogeographical inference but also allows for a greater number of island calibration points to investigate divergence times within this challenging taxonomic group. We use phylogenetic analyses of nuclear internal transcribed spacer (ITS) sequence data along with sequence data of four plastid regions (ndhF‐rpl32, rpl32‐trnL, rps16 and trnLF) to produce a well‐supported phylogenetic tree and further aim to (1) resolve the placement of island taxa within subgenus Plantago, (2) infer source areas for island taxa by reconstructing the most probable ancestral ranges and resolve competing hypotheses regarding the biogeographic histories of taxa and (3) infer the degree to which biogeographical proximity (nearest landmass) versus extreme long‐distance dispersal events is responsible for the distribution of the taxa in a genus that is likely to be dispersed by birds across extreme long distances.
2. MATERIALS AND METHODS
2.1. Taxon sampling
The present study follows the most recent and comprehensive circumscription of the genus published by Rahn (1996), except for the Hawaiian taxa, in which case we follow Wagner, Herbst, and Sohmer (1990). To ascertain biogeographic histories of island taxa within subgenus Plantago, we sampled as many oceanic island taxa as possible, plus the representation of species from across the global distribution for the subgenus. In total, 11 of the 18 islands or island systems for which species from subgenus Plantago are known are represented (Appendix I). In total, 54 taxa were available for our analyses (Table 1), including 48 species belonging to subgenus Plantago (representing 37% of the subgenus) of which 14 species (and three varieties) are endemic to oceanic islands. An additional six species from other taxonomic subgenera in Plantago was included to confirm monophyly of subgenus Plantago. Initially, multiple specimens were obtained for several critical or rarely collected species, including P. aucklandica, P. canescens Adams, P. fernandezia, P. hawaiensis, P. macrocarpa Cham. & Schltdl., P. palmata Hook.f., P. princeps, P. rapensis and P. rupicola, to test DNA amplification on degraded tissue samples. In all of the above cases, the multiple specimens were monophyletic (data not shown), and therefore, only one specimen per species was used in the final analyses, except for the four varieties of the Hawaiian P. princeps s.l. (Dunbar‐Co et al., 2008; Wagner et al., 1990). Material from herbarium specimens of P. robusta from St. Helena was also obtained and would have provided an additional calibration point, but could not be included due to lack of amplification of the highly degraded DNA.
Table 1.
List of Plantago taxa and outgroups, including specimen information, and DNA regions used in the present study
| Rahn No. | Species | Distribution | Voucher information | DNA Bank ID | ITS | rps16 | trnLF | ndhF–rpl32 | rpl32–trnL |
|---|---|---|---|---|---|---|---|---|---|
| Genus Plantago L. | |||||||||
| Subgenus Plantago | |||||||||
| Section Plantago | |||||||||
| 1 | P. canescens Adams | Northern Asia, northwestern North America | R. Ernst, Alberta, CA, CSU | 30434, K | MK487840 | MK487941 | MK487990 | MK487866 | MK487899 |
| 2 | P. macrocarpa Cham. & Schltdl. | Northwestern North America* | M. Woodbridge, 181046, OSC, Lincoln, Oregon. | 20556, K | EU602337b | EU580432b | MK487919 | ||
| 3 | P. rupicola Pilg. | Rapa Iti | T. J. Motley 2678, NY/K, Rapa Iti, Polynesia. | 20558, K | EU602338b | MK487939 | MK487988 | EU580427b | EU594462b |
| 4 | P. princeps var. anomala Rock | Hawaii | Dunbar, 216, PTBG, Hawaii | 2297, HPDL | MK487831 | MK487983 | MK487880 | MK487906 | |
| 4 | P. princeps var. laxifolia A.Gray | Hawaii | Dunbar, 111, PTBG, Maui, Hawaii. | 2288, HPDL | EU602322b | MK487984 | EU580412b | EU594447b | |
| 4 | P. princeps var. longibracteata H.Mann | Hawaii | Dunbar, 287, PTBG, Hawaii | 2308, HPDL | MK487832 | MK487985 | MK487881 | MK487909 | |
| 4 | P. princeps var. princeps Cham. & Schltdl. | Hawaii | Dunbar, 256, PTBG, Hawaii | 2303, HPDL | MK487833 | MK487986 | MK487877 | MK487908 | |
| 5 | P. fernandezia Bertero ex Barnéoud | Juan Fernández Islands | H. Valdebenito & A. Landero 6595, OS | 20546, K | MK487825 | MK487938 | MK487994 | MK487861 | MK487930 |
| 6 | P. hawaiensis (A.Gray) Pilg. | Eastern Hawaii | Dunbar, 10, PTBG, Hawaii | 1993, HPDL | MK487830 | MK487981 | MK487876 | MK487910 | |
| 7 | P. pachyphylla A.Gray | Eastern Hawaii | Dunbar, 59, PTBG, Hawaii | 2046, HPDL | MK487829 | MK487982 | MK487879 | MK487907 | |
| 14 | P. rapensis Pilg. | Rapa Iti | T. J. Motley 2740, K | 20557, K | MK487837 | MK487940 | MK487987 | MK487850 | MK487911 |
| 15 | P. aucklandica Hook.f. | Auckland Island | B. D. Rance, No Data | 20547, K | MK487823 | MK487943 | MK487991 | MK487854 | MK487932 |
| 16 | P. fischeri Engl. | Eastern Africa | Grimshaw, J.M. 93799 K | 31944, K | MK487836 | MK487900 | |||
| 18 | P. longissima Decne. | Southern Africa | H. F. Glen 4054, PRE/K, South Africa | 20555, K | MK487835 | MK487944 | MK487968 | MK487855 | MK487917 |
| 20 | P. sparsiflora Michx. | Southeastern USA | R. LeBlond, 5305, CSU | 30433, K | AJ548979c | MK487945 | MK487971 | MK487883 | |
| 21 | P. gentianoides subsp. griffithii (Decne.) Rech.f. | Southern Asia | Koelz, W. 2076 | 31948, K | MK487839 | MK487996 | |||
| 22 | P. reniformis Beck | Southeastern Europe | Rønsted 42, C, Cultivated | 9446, K | AY101858a | AY101914a | MK487856 | MK487905 | |
| 23 | P. cornutii Gouan | Southern Europe | Rønsted 31, C, Cultivated | 11180, K | AY101859a | AY101915a | MK487859 | MK487903 | |
| 24 | P. palmata Hook.f. | Tropical Africa | Rahn 670, C, Rwanda | 9396, K | AY101860a | MK487946 | AY101916a | MK487853 | MK487902 |
| 25 | P. africana Verdc. | Eastern Africa | Thulin, M. 1636 K | 31942, K | MK487834 | MK487970 | MK487852 | MK487901 | |
| 26 | P. major L. | Cosmopolite, Europe and western Asia | Rønsted 41, C, Cultivated | 11185, K | AY101861a | MK487947 | AY101917a | MK487860 | MK487918 |
| 29 | P. asiatica L. | Southern and eastern Asia | Rønsted 42, C, China | 9585, K | AY101862a | MK487948 | AY101918a | MK487874 | |
| 30 | P. taquetii H.Lév. | Korea | H‐K. Choi, 21088, | 20551, K | MK487822 | MK487949 | MK487972 | MK487872 | MK487923 |
| 31 | P. himalaica Pilg. | Himalaya | Townsend, C.C. 87/159 K | 31935, K | MK487827 | MK487989 | MK487888 | MK487926 | |
| 33 | P. erosa Wall. | India | Luo, Lin‐bo 0148, K | 31932, K | MK487821 | MK487980 | MK487873 | ||
| 34 | P. incisa Hassk. | Java | J.L. Filip, H578184‐52, K | 11191, K | MK487828 | ||||
| 35 | P. rugelii Decne. | Eastern North America | Rønsted 37, C, Ontario | 9447, K | AY101863a | MK487950 | AY101919a | MK487882 | MK487928 |
| 36 | P. eriopoda Torr. | Western North America | P.J. Cotterill, Edmonton, AB | 20541, K | MK487824 | MK487951 | MK487974 | MK487889 | MK487913 |
| 37 | P. tweedyi A.Gray | USA | Hoggard R, 518, CSU | 30436, K | MK487838 | MK487952 | MK487973 | MK487868 | MK487912 |
| 38 | P. cordata Lam. | Eastern North America | Allison JR, 12478, CSU | 31947, K | MK487841 | MK487904 | |||
| 39 | P. hedleyi Maiden | Lord Howe Island | Crawford 3833, KANU | 20549, K | MK487826 | MK487995 | MK487887 | MK487922 | |
| 40 | P. maxima Juss. ex. Jacq. | Eastern Europe | Rønsted 28, C, Cutivated | 11181, K | AY101864a | MK487969 | MK487867 | MK487897 | |
| 41 | P. media L. | Europe | Rønsted 50, C, Cultivated | 9441, K | AY101865a | MK487942 | AY101920a | MK487890 | MK487898 |
| Section Micropsyllium Decne. | |||||||||
| 43 | P. tenuiflora Waldst. & Kit. | Eastern Europe, Central Asia | Rønsted 30, C, Hungary | 11186, K | AY101866a | MK487953 | AY101921a | MK487871 | MK487895 |
| 48 | P. bigelovii A.Gray | Eastern USA | Sivinski R 5274, CSU | 30445, K | MK487819 | MK487954 | MK487975 | MK487870 | MK487896 |
| Section Mesembrynia Decne. | |||||||||
| 52 | P. camtschatica Link | Eastern Asia | Rahn 684, C | 9402, K | MK487842 | MK487955 | MK487976 | MK487875 | MK487925 |
| 54 | P. depressa Willd. | Central and eastern Asia | Zhen Yu Li 11339, PE | 20544, K | MK487843 | MK487956 | MK487977 | MK487869 | MK487929 |
| 60 | P. debilis R.Br. | Australia, Tasmania | Rønsted 45, C, Cultivated | 9443, K | AY101868a | MK487957 | AY101922a | MK487885 | MK487915 |
| 68 | P. raoulii Decne. | New Zealand | Rahn 692, C, Cultivated | 9428, K | AY101867a | MK487958 | AY101923a | MK487884 | MK487921 |
| 78 | P. stauntonii Reichardt | New Amsterdam & St. Paul Islands | Rahn 706, C, New Amsterdam & St. Paul Islands | 9586, K | AY101870a | MK487959 | AY101925a | MK487886 | MK487916 |
| Section Virginica Decne. & Steinh. ex Barnéoud | |||||||||
| 84 | P. tomentosa Lam. | South America | Rønsted 29, C, Argentina | 11182, K | AY101872a | MK487960 | AY101927a | MK487865 | |
| 91 | P. myosuros Lam. | South America | Rønsted 47, C, Cultivated | 9405, K | AY101873a | MK487961 | AY101928a | MK487863 | MK487933 |
| 93 | P. virginica L. | Eastern USA | Hoggard R 250, CSU | 30428, K | MK487844 | MK487962 | MK487864 | ||
| 108 | P. australis Lam. | North and South America | DTU‐6/94, C, Cultivated | 9425, K | AY101874a | MK487963 | AY101929a | ||
| Section Oliganthos Barnéoud | |||||||||
| 110 | P. barbata G.Forst. | South America | Hoggard R 528, CSU | 30429, K | MK487845 | MK487964 | MK487992 | MK487857 | MK487920 |
| 111 | P. moorei Rahn | Falkland Islands | Hoggard R 528, CSU | 30431, K | MK487846 | MK487965 | MK487993 | MK487858 | MK487931 |
| 121 | P. paradoxa Hook.f. | Tasmania | D. Burns, 2136, 9011116, CBG | 30449, K | AJ548969c | MK487966 | MK487978 | MK487862 | MK487914 |
| 131 | P. muelleri Pilg. | Australia, Tasmania | Craven 10162, QRS, CSIRO | 30437, K | MK487847 | MK487967 | MK487979 | ||
| Subgenus Bougueria (Decne.) Rahn | |||||||||
| 162 | P. nubicola (Decne.) Rahn | South America | HHCH 5079, C, Peru. | 9639, K | MK487820 | AY101948a | |||
| Subgenus Psyllium (Mill.) Harms & Reiche | |||||||||
| Section Lanceifolia Barnéoud | |||||||||
| 170 | P. lanceolata L. | Cosmopolite, Europe and western Asia | Rønsted 33, C, cultivated | 9391, K | AY101898a | MK487935 | AY101952a | MK487849 | MK487893 |
| 205 | P. tandilensis (Pilg.) Rahn | Eastern Argentina | Rønsted 51, C, Argentina | 9488, K | AY101908a | AY101961a | |||
| Section Psyllium (Tourn. ex Juss.) Lam. & DC. | |||||||||
| 149 | P. sempervirens Crantz | Southwestern Europe | Rønsted 27, C, France | 9430, K | AY101889a | MK487936 | AY101942a | MK487848 | MK487892 |
| Subgenus Coronopus (Lam. & DC.) Rahn | |||||||||
| 140 | P. coronopus L. | Mediterranean basin | Rønsted 8, C, Denmark | 9439, K | AY101882a | MK487937 | AY101937a | MK487851 | MK487894 |
| Genus Littorella P.J.Bergius | |||||||||
| 145 | L. uniflora (L.) Asch. | South America | Chase 2798, K, England | 2798, K | AY101885a | MK487934 | AY101940a | MK487891 | |
Notes: Herbaria: (C) Natural History Museum of Denmark, University of Copenhagen, Denmark; (CBG) Australian National Botanic Gardens, Canberra, Australia; (CSU) University of Central Oklahoma, USA; (K) Royal Botanic Gardens, Kew, UK; (KANU) University of Kansas, USA; (OS) Ohio State University, USA; (OSC) Oregon State University, USA; (PE) Institute of Botany, Chinese Academy of Sciences, China; (PTBG) National Tropical Botanical Garden, Hawaii, USA; (QRS) SCIRO, Australian National Herbarium, Queensland.
DNA banks: (HPDL) Hawaiian Plant DNA Library; (K) Royal Botanic Gardens, Kew DNA bank.
Rønsted et al. (2002).
Dunbar‐Co et al. (2008).
Hoggard et al. (2003).
Plantago macrocarpa has been reported in northeastern Russia (Hultén, 1930), but no vouchers could be located to confirm this.
Dried leaf material was obtained from the Botanic Gardens of the University of Copenhagen (C), from our in‐house herbarium collection or from collaborators. Genomic DNA samples were obtained from the Hawaiian Plant DNA Library (HPDL) and from new extractions of herbarium collections (K) provided by the DNA bank of the Royal Botanic Gardens, Kew. Origin and voucher information of materials are listed in Table 1. Additionally, 42 sequences from the previous study of Rønsted et al. (2002), 3 from Dunbar‐Co et al. (2008) and 2 from Hoggard et al. (2003) were downloaded from GenBank as listed in Table 1. In total, 17 species were sequenced for the first time in this study, most of the remaining included species were supplemented with additional sequence data, and 176 new sequences were submitted to GenBank.
2.2. DNA extractions, amplification and sequencing
Total genomic DNA was extracted from 15 to 30 mg of dried leaf fragments or herbarium material following Rønsted et al. (2002). Amplification of ITS and the trnLF intron was performed following Rønsted et al. (2002), while amplification of the intergenic spacers ndhF–rpl32 and rpl32–trnL followed Dunbar‐Co et al. (2008). The rps16 intron was amplified following Oxelman, Lidén, and Berglund (1997). Primers used are listed in Appendix II. Amplified products were purified with the Qiagen PCR purification kit (Qiagen, Germany) following the manufacturer's protocols. Cycle sequencing reactions were carried out using the BigDye™ Terminator Mix (Applied Biosystems, USA). Products were run on an ABI 3730 DNA Analyzer according to the manufacturer's protocols (Applied Biosystems, USA) at the Jodrell Laboratory in Kew Gardens, at the National Sequencing Centre, Natural History Museum of Denmark, or by Macrogen Inc. (Europe). Both strands were sequenced for each region for all taxa.
2.3. Phylogenetic analysis
Sequences were assembled, edited and subsequently aligned with MAFFT 7.2 (Katoh & Standley, 2013) using the bioinformatics software platform geneious 9.1.8 (www.geneious.com, Kearse et al., 2012). Gaps were coded for all regions following a simple gap coding scheme (Simmons and Ochoterena (2000). The best‐fit nucleotide substitution model for each marker was chosen based on the corrected Akaike information criterion (AICc) as calculated using jmodeltest 2.1.10 (Darriba, Taboada, Doallo, & Posada, 2012). The best‐fit models are listed in Appendix II (Supporting Information). The partitioned data set was analysed with mrbayes 3.2.6 (Ronquist & Huelsenbeck, 2003), running for 5 million generations and sampling every 200 generations. Littorella uniflora was set as the outgroup based on the results of previous phylogenetic analyses for Plantago (see Hoggard et al., 2003; Rønsted et al., 2002). Chain convergence and ESS parameters were inspected with Tracer 1.6 (Rambaut, Suchard, Xie, & Drummond, 2014) and the first 25% of the trees sampled from the posterior were discarded as burn‐in. A 50% majority rule consensus tree was calculated and visualized together with the posterior probabilities using figtree 1.4.3 (Rambaut, 2012). Maximum likelihood analyses were performed in RaxML (Stamatakis, 2014), defining L. uniflora as outgroup and setting the number of bootstrap iterations to 1000.
2.4. Divergence time analysis
beast 2.4.7 was used to compute divergence times (Bouckaert et al., 2014). The nucleotide substitution models used in the beast analysis were identical to the ones used in the mrbayes analysis (Appendix II). Littorella uniflora was defined as the outgroup. The appropriate molecular clock model was determined by using pathsampler 1.3.4, which is integrated in beast 2.4.7 (Bouckaert et al., 2014). The chain length for this path sampling analysis was set at 1 million generations and the number of steps at 100. The marginal likelihood estimates for a strict, a lognormal and an exponential clock were obtained, and an uncorrelated relaxed lognormal clock was determined to be the most likely model and thus selected for the analyses.
Assuming the timing of dispersal to a datable oceanic island occurred soon after emergence of the island from the ocean, the age of the island can be used as an approximate maximum date for the occurrence of endemic species to that island (Ho et al., 2015; Richardson et al., 2001; Rønsted et al., 2002). In the absence of reliable fossil data for Plantago, previous studies of Plantago have used the endemicity of P. stauntonii on New Amsterdam Island as calibration point (Plantago section Mesembrynia Decne.; Rønsted et al., 2002; Tay et al., 2010a). In the present study, the ages of five oceanic islands were used as calibration points for the occurrence of endemic species to those islands and a normal distribution was used for the estimation of those priors (Table 2). Using this type of data may cause overestimation of divergence times (Hipsley & Müller, 2014). We therefore applied a large confidence interval for each calibration point, allowing for the lower limit to be zero (i.e. present day). Three independent MCMC runs were performed with an uncorrelated relaxed lognormal clock prior on molecular rates and a restricted calibrated Yule speciation process prior on tree shapes. Chain length for each run was set at 100 million generations and sampling was conducted every 5,000 generations. Chain convergence and ESS parameters were inspected with tracer 1.6 (Rambaut et al., 2014). The burn‐in of each run was removed and the outputs of the three independent runs were pooled using logcombiner 2.4.7 (Bouckaert et al., 2014). A maximum clade credibility tree was produced from the combined file composed of the three replicate runs using treeannotator 2.4.7 (Bouckaert et al., 2014) with mean heights and a posterior probability limit of 0.5 and visualized using figtree 1.4.3 (Rambaut, 2012).
Table 2.
Oceanic island calibration points used for the divergence time analysis for subgenus Plantago
| Island | Mean age | Reference | Endemic taxa |
|---|---|---|---|
| Lord Howe Island | 6.9 Ma | Zielske, Ponder, & Haase, 2017; McDougall, Embleton, & Stone, 1981 | P. hedleyi |
| New Amsterdam Island | 0.3 Ma | Doucet, Weis, Scoates, Debaille, & Giret, 2004 | P. stauntonii |
| Juan Fernández Islands | 5.8 Ma | Anderson, Bernardello, Stuessy, & Crawford, 2001; Stuessy, Foland, Sutter, & Silva, 1984 | P. fernandezia |
| Hawaiian Islands | 5.1 Ma | Ree & Smith, 2008; Fleischer, McIntosh, & Tarr, 1998 |
P. hawaiensis
P. pachyphylla P. princeps s.l. |
| Rapa Iti | 5.1 Ma | Zielske et al., 2017; Krummenacher & Noetzlin, 1966 |
P. rupicola
P. rapensis |
2.5. Biogeographical analysis and ancestral range estimation
The R package ‘biogeoBEARS’ (Matzke, 2013) was used to compare biogeographical models and estimate the ancestral ranges of island taxa in subgenus Plantago. The Bayesian maximum clade credibility tree obtained in the beast analysis was used as input. biogeoBEARS uses a maximum likelihood framework (ML) for the three most commonly used models in historical biogeography, the dispersal–extinction–cladogenesis (DEC; Ree & Smith, 2008), dispersal–vicariance analysis (DIVA, Ronquist, 1997) and Bayesian Inference of Historical Biogeography for Discrete Areas (BayArea; Landis, Matzke, Moore, & Huelsenbeck, 2013). The DIVA model and the BayArea model are implemented in ML, rather than the parsimony or Bayesian frameworks originally developed for these models; therefore, the models are called DIVALIKE and BAYAREALIKE in biogeoBEARS (Matzke, 2013). biogeoBEARS also allows for the modelling of founder‐event speciation, with the implementation of the + j parameter. We applied seven biogeographical regions in our analyses, following the works of others for plants with global distributions (i.e. Dupin et al., 2017): Europe and western Asia; eastern Asia and Southeast Asia; Africa; Pacific Islands; Australasia; North America; South America. We assigned the area of each species to the seven areas according to distributions listed in Rahn (1996), Flora of China (Missouri Botanical Garden & Harvard University Herbaria, eFloras, 2017) and Flora Iranica (Patzak & Rechinger, 1965). For the cosmopolitan weeds, P. major L. and P. lanceolata, putative centres of origin were used to code their distribution being from Europe and western Asia (based on the floras listed above). Maximum range areas in biogeoBEARS was set to two areas, based on there being a maximum of two geographic areas occupied by the extant taxa included in our analyses.
All six possible models (DEC, DEC + j, DIVALIKE, DIVALIKE + j, BAYAREALIKE and BAYAREALIKE + j) were fitted to the data, and the selection of the best‐fit model was based on comparing the log‐likelihood values and corrected Akaike Information Criterion (Table 3). The results of the ancestral range analysis are visualized as pie charts on each supported node of the tree, signifying the probabilities for all estimated ranges of all possible biogeographic histories.
Table 3.
Summary statistics for the six models in BioGeoBEARS for ancestral state reconstruction in subgenus Plantago. Abbreviations: log‐likelihood (LnL), dispersal parameter (d), extinction parameter (e), founder effect parameter (j) and corrected Akaike information criterion (AICc). Bold text indicates that the data are best explained by a DEC + j model
| Model | LnL | No. Parameters | d | e | j | AICc |
|---|---|---|---|---|---|---|
| DEC | −100.4 | 2 | 0.011 | 8.27E‐3 | 0 | 204.7 |
| DEC+J | −77.6 | 3 | 0.0017 | 1.00E‐12 | 0.032 | 161.3 |
| DIVALIKE | −99.3 | 2 | 0.014 | 5.4E‐3 | 0 | 202.6 |
| DIVALIKE+J | −78.8 | 3 | 0.0021 | 1.00E‐12 | 0.033 | 163.5 |
| BAYAREALIKE | −120.3 | 2 | 0.019 | 0.11 | 0 | 244.6 |
| BAYAREALIKE+J | −79.5 | 3 | 0.0015 | 1.00E‐07 | 0.037 | 164.9 |
3. RESULTS
3.1. Phylogenetic relationships and divergence times for section Plantago
The resulting alignment from concatenating all sequences from the sampled taxa is 4639 base pairs long; however, not all five regions could be amplified for all taxa (see Table 1). No hard incongruences between the ITS and the plastid topologies were obtained and only the combined tree is presented herein. Separate trees for ITS and plastid regions are presented as supplementary files (Figures S1 and S2), along with a tree from the RAxML analysis (see Supporting Information, Figure S3). Bayesian phylogenetic analysis of the five DNA regions resulted in a robust phylogenetic tree with nodes with high posterior probability (<0.95) in all but a few cases. In the backbone of the topology, the sister relationship of a clade containing P. canescens, P. maxima and P. media to the remainder of subgenus Plantago was only supported by PP = 0.53, and a trichotomy was found consisting of a European clade (3), a Hawaiian clade (13) and the remainder of subgenus Plantago. The consensus tree and posterior probabilities (PP) from the mrbayes analysis are presented in Figure 1, and the results of the beast divergence analyses are presented in Figure 2. Island endemics are indicated with an asterisk (“*”) in both Figures 1 and 2. Current geographic distribution for all taxa is listed in Table 1. Maximum likelihood analysis resulted in the same topology, though support was lower at many nodes (Supplementary Information, Figure S3).
Figure 1.
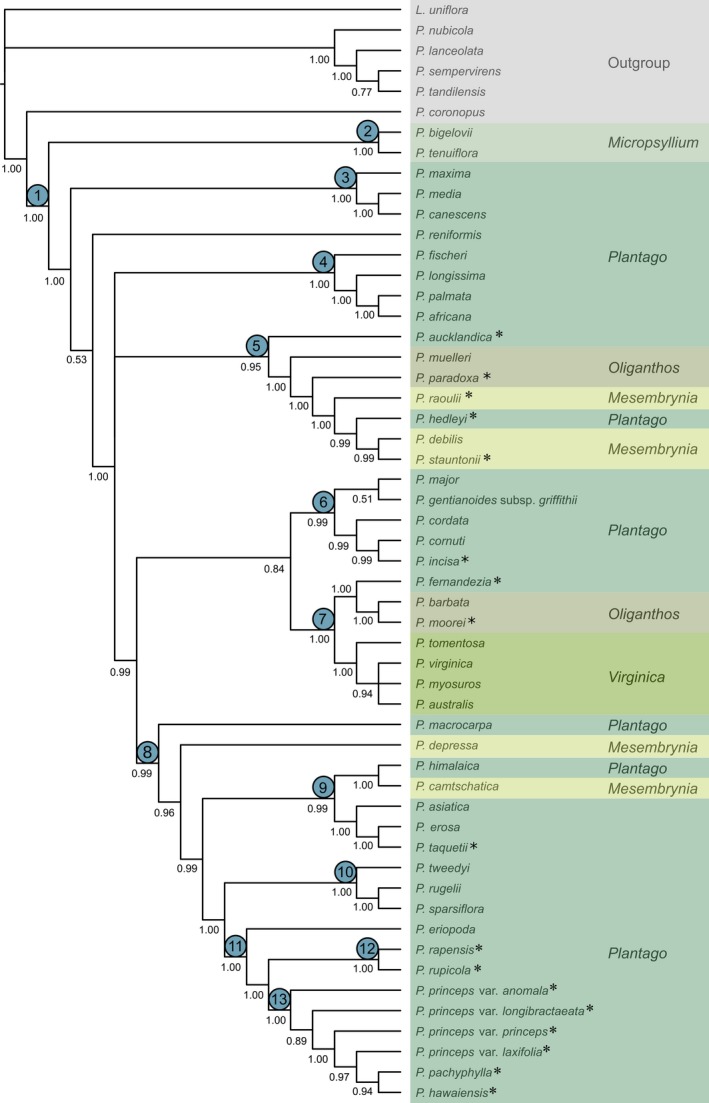
The 50% majority rule consensus cladogram from the mrbayes analysis for members from subgenus Plantago. The posterior probabilities are listed below the branches. Taxonomic sections of Plantago are shown in different colours. Numbers listed above the nodes are the numbers for clades discussed in the text. Island taxa are denoted by an asterisk (*) following the taxon name [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.
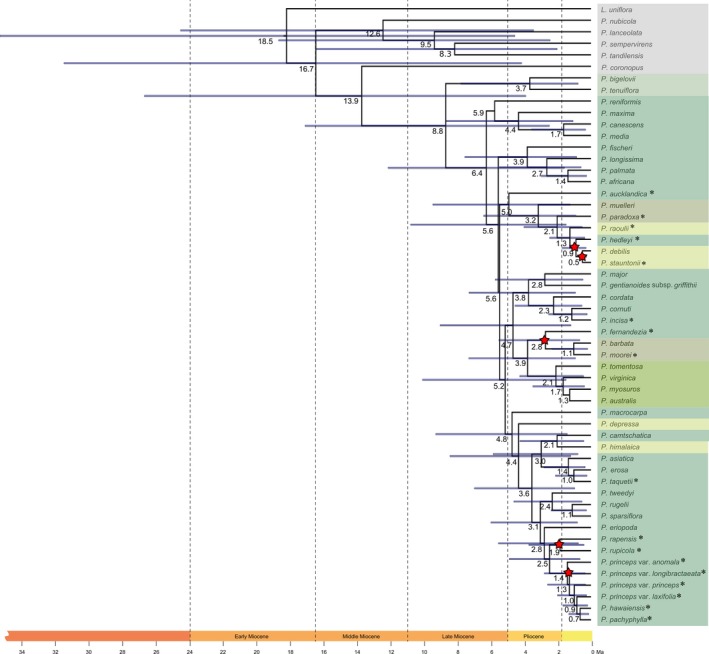
Divergence time tree of subgenus Plantago from the beast analysis. The five calibration points are indicated with red stars, and represent (from top to bottom) Lord Howe Island, New Amsterdam Island, the Juan Fernández Islands, Rapa Iti and the Hawaiian Islands. Island taxa from subgenus Plantago are denoted by an asterisk (*) following the taxon name [Colour figure can be viewed at wileyonlinelibrary.com]
Subgenus Plantago is found to be monophyletic (Clade 1; PP = 1.00; Figure 1) and started diverging 8.8 Ma (with an error range of 17.4–2.5 Ma) (Figure 2). The 14 sampled island taxa were resolved in six different clades (Clades 5, 6, 7, 9, 12, 13) and each is described below. All divergence time estimates presented in the text are followed by error estimates in square parentheses based on the HPD 95% confidence intervals (as shown in Figure 2).
Two taxa classed into section Micropsyllium Decne. form a monophyletic clade (Clade 2; PP = 1.00; 3.9 Ma (7.9–0.8 Ma) which is sister to the rest of the subgenus. The other three taxonomic sections (as defined by Rahn, 1996) within the subgenus are polyphyletic. There are up to 12 different clades that can be recognized within subgenus Plantago, all of which have high support. These clades are mainly formed by species that share common geographic distributions rather than taxonomic sections as previously described (Rahn, 1996). Of particular note are the oceanic island species P. aucklandica, P. hedleyi and P. fernandezia, which are taxonomically classed as part of the section Plantago but are phylogenetically clustered with species from other sections in clades that constitute coherent geographic groups.
A clade of European species is found to be the next diverging lineage (Clade 3; PP = 1.00; 6.0 Ma [8.8–1.1 Ma]) and consists of the species P. media L., P. maxima Juss. ex. Jacq. and P. canescens, though the last species also occurs in North America (Table 1). A minor incongruence between the mrbayes and beast trees is the positioning of the European species P. reniformis (compare Figures 2 and 3). In the beast analysis (Figure 2), P. reniformis is placed closest to the European clade, but the relationship between them is uncertain due to low node support. However, in the mrbayes analyses (Figure 1), P. reniformis is closer to the rest of the subgenus than to the three species in Clade 3, though also with low support (PP = 0.53).
Figure 3.
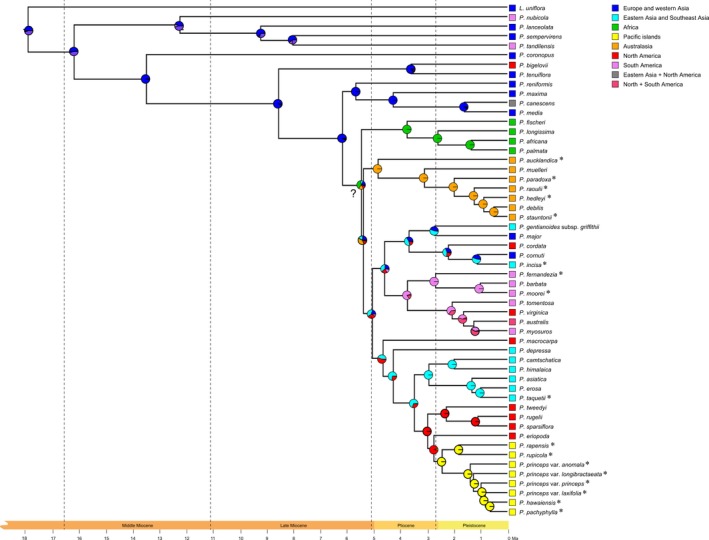
Ancestral range reconstruction of subgenus Plantago (−LnL = 76.98) under the best‐fitted model, DEC + j, based on the divergence time tree from the beast analysis and the distribution ranges of the extant species. The relative probabilities of the estimated ranges are represented in pie charts at each supported node. Colours correspond to the coded geographic states (single and two area ranges) listed in the legend. The node labelled with a question mark “?” denotes doubtful results from the biogeographical analyses based on poor node support in the mrbayes analyses. Island taxa from subgenus Plantago are denoted by an asterisk (*) following the taxon name [Colour figure can be viewed at wileyonlinelibrary.com]
A clade of African species (all previously classed in section Plantago) is also found (Clade 4; PP = 1.00; 4.0 Ma (7.7–0.88 Ma); P. africana Verdc., P. fischeri Engl., P. longissima Decne. and P. palmata).
Clade 5 is a highly supported group of Australasian taxa (PP = 0.94; 5.1 Ma) that consists of a mix of members of sections Oliganthos Barnéoud and Mesembrynia, and includes the largest number of island endemics including P. aucklandica from the Auckland Islands, P. paradoxa Hook.f. from Tasmania, P. raoulii from New Zealand, P. stauntonii from Amsterdam Island and P. hedleyi from Lord Howe Island. Within this Australasian clade, P. aucklandica is found to diverge the earliest of all island taxa sampled in the subgenus (5.1 Ma [9.6–1.3 Ma]), whereas P. stauntonii from New Amsterdam Island is found to be the most recently diverged oceanic island endemic in this clade (0.5 Ma [0.8–0.3 Ma]).
Clade 6 consists of a mix species that occur in various geographic areas (PP = 0.99; 3.8 Ma [7.4–0.9 Ma]): the cosmopolitan P. major, P. gentianoides subsp. griffithii (Decne.) Rech.f. from western Asia, P. cordata Lam. from North America, P. cornutii Gouan from Europe and western Asia and the island species P. incisa Hassk. from Java.
Clade 7 (PP = 1.00; 3.9 Ma [7.4–0.9 Ma]) is an entirely South American clade comprised of taxa classed in section Virginica Decne. & Steinh. ex Barnéoud (P. australis Lam., P. myosuros Lam., P. tomentosa Lam. and P. virginica L.), and South American taxa from section Oliganthos, including the single island endemics P. fernandezia, which diverged 2.8 Ma [5.6–0.7 Ma] on the Juan Fernández Islands, and P. moorei Rahn from the Falkland Islands which diverged 1.1 Ma [2.4–0.2 Ma]).
A large and more recently diverged clade (Clade 8; PP = 0.99; 4.8 Ma [9.4–1.4]) includes a mixture of taxa from North America, eastern Asia and species from Pacific Island systems, Rapa Iti and Hawaii.
Clade 9 (PP = 0.99; 3.0 Ma [5.9–0.8 Ma]) consists of the majority of the eastern Asian species from sections Plantago (P. asiatica, P. erosa Wall. and P. himalaica Pilg.) and Mesembrynia (P. camtschatica Link), including P. taquetii H.Lév., from Jeju which diverged (1.0 Ma [2.2–0–2 Ma].
Clade 10 (PP = 1.00; 2.4 Ma) is a North American group containing the species P. rugelii Decne., P. sparsiflora Michx. and P. tweedyi A.Gray.
In Clade 11 (P = 1.00; 2.8 Ma [5.6–0.8 Ma]), P. eriopoda Torr. from western North America is sister to the two Rapa Iti taxa (Clade 12; PP = 1.00; 1.9 Ma [3.8–0.4 Ma]; P. rapensis and P. rupicola) and the Hawaiian Islands taxa (Clade 13; PP = 1.00; 1.5 Ma [2.8–0–4]; P. princeps s.l., P. hawaiensis and P. pachyphylla).
3.2. Ancestral range reconstruction
Model selection for the ancestral range reconstruction indicated that the data are best explained by a DEC + j model (LnL = −76.98 and AICc = 160.4; Table 3). The results of the ancestral range reconstruction are depicted in Figure 3, where the probabilities for possible ancestral areas are shown in pie charts at each supported node. The selection of the DEC + j model indicates that founder‐event speciation (i.e. the parameter j) is a major contributor to the currently observed biogeographical patterns within section Plantago.
Interpreting the results of the ancestral range reconstruction at the deeper nodes should be done carefully as incomplete taxon sampling has a major influence on the outcome, and in particular, the presence of phylogenetic uncertainty at some nodes may hinder a correct interpretation. For example, the result presented at the node directly above an unsupported node (indicated with a question mark “?” in Figure 3) is doubtful. The most recent common ancestor (MRCA) of the African clade (Clade 4), the Australasian clade (Clade 5) and the remainder of the tree cannot be confidently inferred, as long as the phylogenetic relationships between these groups are not completely resolved. However, the ancestral ranges at other nodes that are well supported can be estimated with higher confidence.
Based on our sampling, ancestral range reconstruction shows that subgenus Plantago, as well as the first diverging clades, likely has geographic origins in Europe. The MRCA of the African species in section Plantago is African and the MRCA of the Australasian species, including the island endemics from Lord Howe Island, the Auckland Islands, New Zealand, Tasmania and Amsterdam Island originated in Australasia. The ancestral range of the MRCA of the remaining species is inferred to be in the northern hemisphere, likely in eastern Asia or North America. From there, some taxa could have recolonized Europe or may be descendants of early European ancestors (P. major and P. cornutii), while other taxa moved into South America, finally also giving rise to the island endemics from the Juan Fernández Islands (P. fernandezia) and Falkland Islands (P. moorei). The MRCA of P. macrocarpa and the remainder of the species are indicated as North American because P. macrocarpa was coded as North American in our analysis. However, if unconfirmed reports of P. macrocarpa being present in Asia are correct (Hultén, 1930), there would be a greater likelihood that the ancestor is Asian. The ancestral ranges reconstructed at the more recent nodes are more straightforward and the analysis shows that the island endemics from Rapa Iti and the Hawaiian Islands have their most likely ancestral origins in North America, meanwhile P. taquetii from Jeju is inferred to have origins in Asia.
4. DISCUSSION
4.1. Phylogenetic relationships and divergence times
The current work is the most comprehensive phylogenetic and biogeographical study for subgenus Plantago to date – and the first to focus on estimating ancestral origins of island endemics. The most important phylogenetic result of this study was that taxa within the subgenus, particularly from sections Mesembrynia, Oliganthos and Plantago, form clades based on their geographic proximities, rather than on previous taxonomic placement. Phylogenetic relatedness, based on our molecular data, is thus found to be incongruent with morphological phylogenetic analyses and previous taxonomic classifications (i.e. Rahn, 1996). Thus, morphological traits are not always useful in inferring evolutionary relationships for this group. Rahn (1996) admitted that taxonomic sections such as section Plantago that are geographically inconsistent were evidently paraphyletic in relation to the rest of subgenus Plantago, and he urged for further studies to be done to clarify the taxonomy and phylogeny of the species included in this group. With our expanded molecular data set, we confirm the earlier findings of Rønsted et al. (2002), Hoggard et al. (2003), Ishikawa et al. (2009) and Tay et al. (2010a) that sections within subgenus Plantago are polyphyletic, and echo that the concepts of taxonomic sections in subgenus Plantago need to be revisited.
Using five islands as calibration points, we obtained a date of 8.8 Ma [17.4–2.5 Ma] for the diversification of subgenus Plantago. As a consequence, our estimate of the origin and diversification of the entire genus is older than previously estimated, however within the error margins since earlier works dated the split of Plantago‐Littorella from Aragoa at 7.1 or 2.8 Ma (Rønsted et al., 2002; Tay et al., 2010a). The dating approach used by Tay et al. (2010a) was similar to the approach used herein (using a Bayesian modelling in beast), but only a single calibration point was used (i.e. the young New Amsterdam Island) and therefore recovered a much earlier date of 2.8 Ma (Tay et al., 2010a). Known records for fossil pollen for the genus only extend to the Late Miocene at approximately 6 Ma (Mueller, 1981; Rahn, 1996; Rønsted et al., 2002), though it is possible that the genus is older because fossil ages define only minimum ages. Our divergence analyses were limited to using beast, which in comparative dating analyses have previously been shown to give older estimates of dates compared to other methods (Goodall‐Copestake, Harris, & Hollingsworth, 2009). Regardless of the timing of the origin of subgenus Plantago, our analyses are still congruent with earlier hypotheses of recent speciation (Rønsted et al., 2002; Tay et al., 2010a), especially in lineages with island endemics, such as the Hawaiian Islands, New Amsterdam Island and Lord Howe that are inferred to have diverged most recently in the group, sometime in the Pleistocene.
Three of the island taxa were resolved within clades different from their classification in taxonomic sections by Rahn (1996; P. aucklandica and P. hedleyi were resolved within Australasian sections Oliganthos and Mesembrynia; and P. fernandezia in a clade with South American sections Oliganthos and Virginica). Interestingly, P. aucklandica was found to diverge much earlier than any of the other island taxa in our study, which is in keeping with the findings of Tay et al. (2010a) that P. aucklandica is part of an early diverging clade, which was sister to all other Australasian taxa in subgenus Plantago. Considering that less than half of all known species described in subgenus Plantago were included in the current work, further investigation should focus on a more comprehensive sampling of species from the subgenus to resolve these relationships, taxonomy and divergence times with higher certainty.
4.2. Historical biogeography of island endemics
Our sampling in subgenus Plantago was extensive enough to infer a first approximation of the biogeographical history for subgenus Plantago and suggest ancestral ranges of the 14 island taxa we sampled. The ancestor to all subgenus Plantago likely originated in Europe. However, the island endemics were found to come from six different lineages each with different ancestral ranges. This suggests that several lineages within subgenus Plantago were successful in dispersing to, and speciating on, oceanic islands, which is not surprising for a group with a global distribution and adaptations to bird dispersal (Birch & Keeley, 2013; de Queiroz, 2005). It confirms earlier theories that the genus Plantago is particularly efficient at dispersing, especially over long ranges (Rønsted et al., 2002; Tay et al., 2010a).
Our analyses further show that different patterns of dispersal are inferred for island taxa in Plantago. Geographic proximity is found to be a key factor in determining relatedness and defining the biogeographic histories of some of the island taxa within subgenus Plantago, such that the nearest continents and landmasses were inferred to be source areas for the taxa endemic to Lord Howe Island, Auckland Islands, New Zealand, Tasmania, Juan Fernández Islands and the Falkland Islands. Only in the case of the most remote islands in our study, that is, the Hawaiian Islands and Rapa Iti, and New Amsterdam Island, do we find evidence of extreme long‐distance events that defy the rules of proximity, which is in keeping with previous findings on the origins of island floras such as Hawaii (Baldwin & Wagner, 2010). Interestingly, it is only on these remote islands that multiple Plantago species are known; the remaining islands host single island endemics. The specific biogeographical findings for each island taxon included in this study are discussed below (and illustrated in Figures 1, 2, 3.
Plantago fernandezia, from the Juan Fernández Islands off the coast of Chile, and P. moorei from the Falkland Islands were inferred to have been dispersed from ancestors of close geographic proximity in South America. Plantago fernandezia was previously thought to be more closely related to other island taxa from Hawaii and Rapa Iti because it shares more morphological features (such as woody stems typical of insular taxa) in common with them (Pilger, 1937; Rahn, 1996). Our results are, however, in line with the biogeographical histories of other plant lineages from the Juan Fernández Islands having their sources in neighbouring South America (Stuessy et al., 2018). This result also supports the idea that, at least in the case of island taxa, growth forms such as woodiness and other morphological traits (i.e. the presence of only two ovules) can be derived rather than of relict origin and reflect convergent evolution (Carlquist, 1970; Emerson, 2002) rather than phylogenetic relationships.
Plantago hedleyi from Lord Howe Island, P. aucklandica from the Auckland Islands, the mainland New Zealand taxon (P raoulii) and Tasmanian species (P. paradoxa) have their ancestral ranges in Australasia (Figure 3). Plantago stauntonii from New Amsterdam Island was also found to have ancestral ranges in Australasia, despite the island being situated midway between Africa and Australia, and that representatives of extant species in subgenus Plantago are known from both continents. Molecular phylogenetic studies aimed at reconstructing origins of New Amsterdam flora are scarce; however, Bauret et al. (2017) hypothesized that a likely dispersal scenario for a fern species endemic to New Amsterdam was from Madagascar via the Neotropics. We demonstrate that Australasia is an important floristic source for Plantago on Amsterdam Island.
Plantago incisa from Java has its ancestral range within Europe or Asia though it is phylogenetically most closely related to European species, and is geographically closer to the Australasian species. Sampling other Plantago species from New Guinea and mainland Asia would be needed to improve confidence in our ancestral range estimations.
Island taxa from the Hawaiian Islands and Rapa Iti were found to have a common ancestor in western North America, thousands of kilometres further away from their closest landmasses. Plantago species from the Hawaiian Islands have previously been proposed to have arisen from a single LDD colonization event either from a North American ancestor or from an ancestor in New Zealand via Rapa Iti (Dunbar‐Co et al., 2008). Our findings rule out the possibility of Hawaiian Plantago originating from New Zealand and dispersing via a stepping stone pattern, despite evidence of dispersal moving in that direction for some Hawaiian plant groups (Dunbar‐Co et al., 2008; and references therein; Birch & Keeley, 2013). North America is increasingly recognized as the source for the majority of Hawaiian lineages due the Pacific flyway (Baldwin & Wagner, 2010). Despite the seemingly higher possibility of stepping stone dispersal or island hoping driving the dispersal of island taxa in the Pacific, extreme long‐distance dispersal events are now considered equally as likely and explain the biogeographies of many globally distributed plant groups (Birch & Keeley, 2013; Dupin et al., 2017; Gillespie et al., 2012; Givnish et al., 2009; Nathan, 2006). Our findings therefore further demonstrate the importance of the Pacific flyway and the occurrence of extreme LDD events for the movement of flora from North America across thousands of kilometres to the Hawaiian Islands and also Rapa Iti (Baldwin & Wagner, 2010). Given that the Hawaiian Islands and Rapa Iti have multiple endemic Plantago species present on them and are among the most extreme with regard to island remoteness in our study group, these island areas may represent the most extreme cases of recent speciation in island Plantago lineages. Further genetic and ecological study of these species may assist in determining what traits are important in not only successful dispersal and colonization to islands but also the subsequent diversification (Baker, 1955, 1955; Carvajal‐Endara, Hendry, Emery, & Davies, 2017).
Although our analyses were sufficient to provide a first approximation of ancestral ranges of island taxa, ancestral range reconstruction at one of the deeper nodes in the tree (shown with a question mark “?” in Figure 3) is highly uncertain due to poorly supported phylogenetic relationships, and increased sampling from subgenus Plantago would be necessary to improve node support and confidence in the ancestral range reconstruction, and clarify the biogeographic histories and relationships between extant Plantago taxa from Africa, Asia and Australasia. For example, our sampling and/or the molecular data produced herein was insufficient to test whether dispersal to Australasia came via the west (Africa) or north (Asia). A more comprehensive sampling of subgenus Plantago would be needed in order to further emphasize that, for biogeographic studies such as this, it is the phylogenetic sampling that is critical, not the taxonomy as proposed by Rahn (1996). Additionally, the definition of biogeographic regions used in our analyses limits the testing of ancestry of insular taxa being in temperate locations (van der Aart & Vulto, 1992). Future analyses could investigate the degree of climate niche matching between island taxa and their ancestral ranges.
4.3. Long‐distance dispersal by birds
Our findings support the notion that extant plant species in subgenus Plantago are well adapted to dispersal (Rønsted et al., 2002; Tay et al., 2010a), and this was also likely the case for common ancestors. However, the distance that the propagules can disperse may be a factor of the type of birds involved rather than differences in propagule traits. Due to the closer proximity between some oceanic islands to their nearest landmasses (i.e. Lord Howe 600 km from Australia, Auckland Island 500 km south of New Zealand, Juan Fernández 600 km off the coast of Chile and the Falkland Islands 480 km off the coast of Argentina), the dispersal patterns we inferred may be due to movement by smaller birds (Nogales, Heleno, Traveset, & Vargas, 2012; and references therein). Meanwhile, the extreme long‐distance dispersal patterns inferred for taxa on the Hawaiian Islands, as well as on Rapa Iti, coincide with the migration of large marine birds, such as the Pacific Plover, that fly thousands of kilometres from the coasts of Alaska to the Pacific Islands over the Pacific flyway (Gillespie et al., 2012; Henshaw, 1910; Jenni & Jenni‐Eiermann, 1998; Nogales et al., 2012). Similarly, Plantago may have found its way to New Amsterdam Island (midway between Australia and Africa in the southern Indian Ocean) with the help of larger sea birds or accidental birds capable of flying thousands of kilometres. However, the rarity of LDD events and limitations in studying historical bird movements and behaviour makes it difficult to conclude which birds may have been responsible (Nogales et al., 2012).
Our findings show that, for a globally distributed plant genus that is well suited to LDD by birds, differing scales and modes of dispersal may be equally important in explaining the biogeographical histories. Similarly, differing dispersal modes have also been inferred in explaining the historical biogeographies of other bird‐dispersed plant families with unique taxa on multiple oceanic islands (i.e. Asteliaceae [Birch & Keeley, 2013], Rubiaceae [Kainulainen et al., 2017]), and thus, evidence is building that there is not a single LDD model that fits all, but rather that a combination of stepping stone dispersal and extreme LDD can both shape insular floras within closely related plant groups, and that multiple floristic areas can be the sources of closely related island taxa.
4.4. Limitations to diversification
Plantago is an example of a plant group, which is particularly well adapted to long‐distance dispersal, and possesses traits such as wind pollination and self‐compatibility as well as ability to grow in harsh environments that are conducive to establishment and speciation in insular settings (Baker, 1955). However, the majority of Plantago taxa are single island endemics and thus not as successful in radiation and speciation in the insular setting compared to, for example, ferns that are generally over‐represented on islands (Hennequin, Kessler, Lindsay, & Schneider, 2014). As plants with adaptations to LDD such as Plantago are not always over‐represented in island floras, it is increasingly being accepted that factors other than dispersal limitations are important for the successful colonization and speciation of insular species (Baker, 1955; Carvajal‐Endara et al., 2017; Cheptou, 2012; Heleno & Vargas, 2015). For example, the flora of the Galápagos Islands – of which, 30% are single island endemics – was found to be shaped by habitat filtering rather than by dispersal limitations. Consequently, the match between the species continental climate niche and the island climate was the single best predictor of colonization success (Carvajal‐Endara et al., 2017). In the case of orchids, specific traits such as their pollination biology and association with mycorrhizal fungi are considered factors limiting successful colonization (McCormick & Jacquemyn, 2014). The genus Plantago thus remains an interesting case not only to study long‐distance dispersal patterns but also to test hypothesis for limitations to successful establishment and radiation in insular habitats.
5. CONCLUSIONS
Our study provides further insights into the importance of geographic proximity as sources of island flora, even for a group of plants that is arguably well adapted to long‐distance dispersal by birds, and that differing scales and modes of dispersal may be equally important in explaining the biogeographical histories of insular species. This further suggests that factors other than dispersal success are important for the establishment and subsequent speciation of insular taxa. This work further emphasizes that classical cladistics approaches to infer closely related species (i.e. using morphological traits) can often mislead the reconstruction of accurate biogeographical histories of island species (de Queiroz, 2005); however, using molecular data to infer ancestral ranges can greatly improve our understanding of biogeographical histories and help elucidate origins, dispersal routes and means in widespread lineages with complex distribution patterns such as Plantago. The genus Plantago is an excellent case to study to further improve our understanding of organismal traits and ecological factors involved in the successful colonization of insular habitats.
BIOSKETCH
Natalie Iwanycki Ahlstrand's research interests are in using multidisciplinary techniques to retrace migration and dispersal patterns of useful plants, in evolutionary time‐scales and also in the Anthropocene. This work was a part of her PhD research on the dispersal, migration and local adaptation of species with global distributions in the genus Plantago (see http://snm.ku.dk/da/plant_evolutionary_interactions/).
Author contributions: N.R., N.I.A. and R.H. conceptualized the study and assembled the plant material. N.R., R.H. and S.D.C. performed DNA extractions, and NR and RH conducted DNA sequencing. N.I.A. and B.V. processed and analysed sequence data. N.I.A. wrote the manuscript with B.V. and N.R. All authors read and commented on the manuscript and approved the final version. Authors declare no conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
This work received funding from the Marie Curie Actions of the 7th European Community Framework Programme: FP7/2007‐2013/, REA grant agreement n° 606895‐MedPlant to NR, and grant agreement n° 656853 to NR and BV. The authors thank Charlotte Hansen for DNA sequencing. Thanks to KEW DNA Bank manager, L. Cziba and the Hawaiian Plant DNA Library manager C. Morden for DNA extracts. Thanks to the following individuals J.R. Allison, D.A. Broughton, R. Burns, H.‐K. Choi, L. Craven, R. Ernst, R. LeBlond, Z.‐Y. Li, T.J. Motley, R. Sivinski and T. Stuessy for supplying leaf material. R. Olmstead and two independent reviewers are thanked for insightful improvement of the manuscript.
Iwanycki Ahlstrand N, Verstraete B, Hassemer G, et al. Ancestral range reconstruction of remote oceanic island species of Plantago (Plantaginaceae) reveals differing scales and modes of dispersal. J Biogeogr. 2019;46:706–722. 10.1111/jbi.13525
Editor: Hans‐Peter Comes
DATA AVAILABILITY
All newly generated sequence data for this study have been submitted to GenBank.
REFERENCES
- Anderson, G. J. , Bernardello, G. , Stuessy, T. F. , & Crawford, D. J. (2001). Breeding system and pollination of selected plants endemic to Juan Fernández Islands. American Journal of Botany, 88, 220–233. 10.2307/2657013 [DOI] [PubMed] [Google Scholar]
- Bacon, C. D. , Simmons, M. P. , Archer, R. H. , Zhao, L. C. , & Andriantiana, J. (2016). Biogeography of the Malagasy Celastraceae: Multiple independent origins followed by widespread dispersal of genera from Madagascar. Molecular Phylogenetics and Evolution, 94, 365–382. 10.1016/j.ympev.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Baker, H. G. (1955). Self‐compatibility and establishment after ‘long distance’ dispersal. Evolution, 9, 347–348. [Google Scholar]
- Baldwin, B. G. , & Wagner, W. L. (2010). Hawaiian angiosperm radiations of North American origin. Annals of Botany, 105, 849–879. 10.1093/aob/mcq052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauret, L. , Rouhan, G. , Hirai, R. Y. , Perrie, L. , Prado, J. , Salino, A. , & Gaudeul, M. (2017). Molecular data, based on an exhaustive species sampling of the fern genus Rumohra (Dryopteridaceae), reveal a biogeographical history mostly shaped by dispersal and several cryptic species in the widely distributed Rumohra adiantiformis. Botanical Journal of the Linnean Society, 185, 463–481. [Google Scholar]
- Bello, M. A. , Chase, M. W. , Olmstead, R. G. , Rønsted, N. , & Albach, D. (2002). The páramo endemic Aragoa is the sister genus of Plantago (Plantaginaceae; Lamiales): Evidence from plastid rbcL and nuclear ribosomal ITS sequence data. Kew Bulletin, 57, 585–597. 10.2307/4110987 [DOI] [Google Scholar]
- Birch, J. L. , & Keeley, S. C. (2013). Dispersal pathways across the Pacific: The historical biogeography of Astelia s.l. (Asteliaceae, Asparagales). Journal of Biogeography, 40, 1914–1927. [Google Scholar]
- Bouckaert, R. , Heled, J. , Kuhnert, D. , Vaughan, T. , Wu, C. H. , Xie, D. , … Drummond, A. J. (2014). beast 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10, e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buse, T. , & Filser, J. (2014). Mucilaginous seeds and algal diets attract soil Collembola in preference tests. European Journal of Soil Biology, 65, 706–6. 10.1016/j.ejsobi.2014.08.005 [DOI] [Google Scholar]
- Carlquist, S. (1966). The biota of long‐distance dispersal. I. Principles of dispersal and evolution. The Quarterly Review of Biology, 41, 247–270. 10.1086/405054 [DOI] [PubMed] [Google Scholar]
- Carlquist, S. (1967). The biota of long‐distance dispersal. V. Plant dispersal to Pacific Islands. Bulletin of the Torrey Botanical Club, 94, 129–162. 10.2307/2484044 [DOI] [Google Scholar]
- Carlquist, S. (1970). Wood anatomy of insular species of Plantago and the problem of raylessness. Bulletin of the Torrey Botanical Club, 97, 353–361. 10.2307/2483855 [DOI] [Google Scholar]
- Carvajal‐Endara, S. , Hendry, A. P. , Emery, N. C. , & Davies, J. T. (2017). Habitat filtering not dispersal limitation shapes oceanic island floras: Species assembly of the Galápagos archipelago. Ecology Letters, 20, 495–504. 10.1111/ele.12753 [DOI] [PubMed] [Google Scholar]
- Cheptou, P.‐O. (2012). Clarifying Baker's Law. Annals of Botany, 109, 633–641. 10.1093/aob/mcr127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz, M. J. M. , & Chase, M. W. (2013). Biogeographical patterns of plants in the Neotropics—Dispersal rather than plate tectonics is most explanatory. Botanical Journal of the Linnean Society, 171, 277–286. 10.1111/j.1095-8339.2012.01301.x [DOI] [Google Scholar]
- Czarnecka, J. , & Kitowski, I. (2013). The white stork as an engineering species and seed dispersal vector when nesting in Poland. Annales Botanici Fennici, 50, 706–12. 10.5735/085.050.0101 [DOI] [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. , & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz, A. (2005). The resurrection of oceanic dispersal in historical biogeography. Trends in Ecology & Evolution, 20, 68–73. 10.1016/j.tree.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Doucet, S. , Weis, D. , Scoates, J. S. , Debaille, V. , & Giret, A. (2004). Geochemical and Hf‐Pb‐Sr‐Nd isotopic constraints on the origin of the Amsterdam‐St. Paul (Indian Ocean) hotspot basalts. Earth and Planetary Science Letters, 218, 179–195. 10.1016/S0012-821X(03)00636-8 [DOI] [Google Scholar]
- Dunbar‐Co, S. , Sporck, M. J. , & Sack, L. (2009). Leaf trait diversification and design in seven rare taxa of the Hawaiian Plantago radiation. International Journal of Plant Science, 170, 61–75. 10.1086/593111 [DOI] [Google Scholar]
- Dunbar‐Co, S. , Wieczorek, A. M. , & Morden, C. W. (2008). Molecular phylogeny and adaptive radiation of the endemic Hawaiian Plantago species (Plantaginaceae). American Journal of Botany, 95, 1177–1188. 10.3732/ajb.0800132 [DOI] [PubMed] [Google Scholar]
- Dupin, J. , Matzke, N. J. , Sarkinen, T. , Knapp, S. , Olmstead, R. G. , Bohs, L. , & Smith, S. D. (2017). Bayesian estimation of the global biogeographical history of the Solanaceae. Journal of Biogeography, 44, 887–899. 10.1111/jbi.12898 [DOI] [Google Scholar]
- Emerson, B. C. (2002). Evolution on oceanic islands: Molecular phylogenetic approaches to understanding pattern and process. Molecular Ecology, 11, 951–966. 10.1046/j.1365-294X.2002.01507.x [DOI] [PubMed] [Google Scholar]
- Fischer, M. H. , Yu, N. , Gray, G. R. , Ralph, J. , Anderson, L. , & Marlett, J. A. (2004). The gel‐forming polysaccharide of psyllium husk (Plantago ovata Forsk). Carbohydrate Research, 339, 2009–2017. 10.1016/j.carres.2004.05.023 [DOI] [PubMed] [Google Scholar]
- Fleischer, R. C. , McIntosh, C. E. , & Tarr, C. L. (1998). Evolution on a volcanic conveyor belt: Using phylogeographic reconstructions and K‐Ar‐based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Molecular Ecology, 7, 533–545. 10.1046/j.1365-294x.1998.00364.x [DOI] [PubMed] [Google Scholar]
- Gallaher, T. , Callmander, M. W. , Buerki, S. , & Keeley, S. C. (2015). A long‐distance dispersal hypothesis for the Pandanaceae and the origins of the Pandanus tectorius complex. Molecular Phylogenetics and Evolution, 83, 20–32. 10.1016/j.ympev.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Gillespie, R. G. , Baldwin, B. G. , Waters, J. M. , Fraser, C. I. , Nikula, R. , & Roderick, G. K. (2012). Long‐distance dispersal: A framework for hypothesis testing. Trends in Ecology and Evolution, 27, 47–56. 10.1016/j.tree.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Givnish, T. J. , Millam, K. C. , Evans, T. M. , Hall, J. C. , Pires, J. C. , Berry, P. E. , & Sytsma, K. J. (2004). Ancient vicariance or recent long‐distance dispersal? Inferences about phylogeny and South American‐African disjunctions in Rapateaceae and Bromeliaceae based on ndhF sequence data. International Journal of Plant Sciences, 165, S35–S54. 10.1086/421067 [DOI] [Google Scholar]
- Givnish, T. J. , Millam, K. C. , Mast, A. R. , Paterson, T. B. , Theim, T. J. , Hipp, A. L. , … Sytsma, K. J. (2009). Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proceedings of the Royal Society B‐Biological Sciences, 276, 407–416. 10.1098/rspb.2008.1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall‐Copestake, W. P. , Harris, D. J. , & Hollingsworth, P. M. (2009). The origin of a mega‐diverse genus: Dating Begonia (Begoniaceae) using alternative datasets, calibrations and relaxed clock methods. Botanical Journal of the Linnean Society, 159, 363–380. 10.1111/j.1095-8339.2009.00948.x [DOI] [Google Scholar]
- Hassemer, G. , De Giovanni, R. , & Trevisan, R. (2016). The use of potential distribution models in the study of the distribution and conservation status of plants: The case of Plantago L. (Plantaginaceae) in Brazil. Journal of the Torrey Botanical Society, 143, 38–49. 10.3159/TORREY-D-14-00070 [DOI] [Google Scholar]
- Hassemer, G. , Moroni, P. , & O'Leary, N. (2018). A nomenclatural revision of Littorella (Plantaginaceae, Plantagineae). Taxon, 67, 1024–1028. 10.12705/675.14 [DOI] [Google Scholar]
- Heleno, R. , & Vargas, P. (2015). How do islands become green? Global Ecology and Biogeography, 24, 518–526. 10.1111/geb.12273 [DOI] [Google Scholar]
- Hennequin, S. , Kessler, M. , Lindsay, S. , & Schneider, H. (2014). Evolutionary patterns in the assembly of fern diversity on the oceanic Mascarene Islands. Journal of Biogeography, 41, 1651–1663. 10.1111/jbi.12339 [DOI] [Google Scholar]
- Henshaw, H. W. (1910). Migration of the Pacific Plover to and from the Hawaiian Islands. The Auk, 27, 245–262. 10.2307/4071308 [DOI] [Google Scholar]
- Hipsley, C. A. , & Müller, J. (2014). Beyond fossil calibrations: Realities of molecular clock practices in evolutionary biology. Frontiers in Genetics, 5, 706–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S. Y. W. , Tong, K. J. , Foster, C. S. P. , Ritchie, A. M. , Lo, N. , & Crisp, M. D. (2015). Biogeographic calibrations for the molecular clock. Biology Letters, 11, 20150194 10.1098/rsbl.2015.0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard, R. K. , Kores, P. J. , Molvray, M. , Hoggard, G. D. , & Broughton, D. A. (2003). Molecular systematics and biogeography of the amphibious genus Littorella (Plantaginaceae). American Journal of Botany, 90, 429–435. 10.3732/ajb.90.3.429 [DOI] [PubMed] [Google Scholar]
- Hultén, E. (1930). Flora of Kamtchatka and the adjacent islands (Vol. 4). Stockholm: Almqvist & Wiksell. [Google Scholar]
- Ishikawa, N. , Yokoyama, J. , Ikeda, H. , Takabe, E. , & Tsukaya, H. (2009). Evaluation of morphological and molecular variation in Plantago asiatica var. densiuscula, with special reference to the systematic treatment of Plantago asiatica var. yakusimensis . Journal of Plant Research, 119, 385–395. 10.1007/s10265-006-0286-y [DOI] [PubMed] [Google Scholar]
- Jenni, L. , & Jenni‐Eiermann, S. (1998). Fuel supply and metabolic constraints in migrating birds. Journal of Avian Biology, 29, 521–528. 10.2307/3677171 [DOI] [Google Scholar]
- Johnson, M. A. , Clark, J. R. , Wagner, W. L. , & McDade, L. A. (2017). A molecular phylogeny of the Pacific Glade of Cyrtandra (Gesneriaceae) reveals a Fijian origin, recent diversification, and the importance of founder events. Molecular Phylogenetics and Evolution, 116, 30–48. 10.1016/j.ympev.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Kainulainen, K. , Razafimandimbison, S. G. , Wikstrom, N. , & Bremer, B. (2017). Island hopping, long‐distance dispersal and species radiation in the Western Indian Ocean: Historical biogeography of the Coffeeae alliance (Rubiaceae). Journal of Biogeography, 44, 1966–1979. 10.1111/jbi.12981 [DOI] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , … Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, L. , Montenegro, A. , Smith, B. D. , Gifford, J. A. , Green, R. E. , Newsom, L. A. , & Shapiro, B. (2014). Transoceanic drift and the domestication of African bottle gourds in the Americas. Proceedings of the National Academy of Sciences of the United States of America, 111, 2937–2941. 10.1073/pnas.1318678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolář, J. (2014). Littorella uniflora (L.) Ascherson: A review. Scientia Agriculturae Bohemica, 45, 147–154. [Google Scholar]
- Kreitschitz, A. , Kovalev, A. , & Gorb, S. N. (2016). “Sticky invasion”—The physical properties of Plantago lanceolata L. seed mucilage. Beilstein Journal of Nanotechnology, 7, 1918–1927. 10.3762/bjnano.7.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis, M. J. , Matzke, N. J. , Moore, B. R. , & Huelsenbeck, J. P. (2013). Bayesian analysis of biogeography when the number of areas is large. Systematic Biology, 62, 789–804. 10.1093/sysbio/syt040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Roux, J. J. , Strasberg, D. , Rouget, M. , Morden, C. W. , Koordom, M. , & Richardson, D. M. (2014). Relatedness defies biogeography: The tale of two island endemics (Acacia heterophylla and A. koa). New Phytologist, 204, 230–242. 10.1111/nph.12900 [DOI] [PubMed] [Google Scholar]
- Losos, J. B. , & Ricklefs, R. E. (2009). Adaptation and diversification on islands. Nature, 457, 830–836. 10.1038/nature07893 [DOI] [PubMed] [Google Scholar]
- Matzke, N.J. (2013). BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts, CRAN: The Comprehensive R Archive Network, Vienna, Austria. http://cran.r-project.org/package=BioGeoBEARS.
- Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder‐event speciation is a crucial process in island clades. Systematic Biology, 63, 951–970. 10.1093/sysbio/syu056 [DOI] [PubMed] [Google Scholar]
- McCormick, M. K. , & Jacquemyn, H. (2014). What constrains the distribution of orchid populations? New Phytologist, 202, 392–400. 10.1111/nph.12639 [DOI] [Google Scholar]
- McDougall, I. , Embleton, B. J. J. , & Stone, D. B. (1981). Origin and evolution of Lord‐Howe Island, southwest Pacific Ocean. Journal of the Geological Society of Australia, 28, 155–176. 10.1080/00167618108729154 [DOI] [Google Scholar]
- Meudt, H. M. (2011). Amplified fragment length polymorphism data reveal a history of auto and allopolyploidy in New Zealand endemic species of Plantago (Plantaginaceae): New perspectives on a taxonomically challenging group. International Journal of Plant Sciences, 172(220), 237. [Google Scholar]
- Meudt, H. M. (2012). A taxonomic revision of native New Zealand Plantago (Plantaginaceae). New Zealand Journal of Botany, 50, 101–178. 10.1080/0028825X.2012.671179 [DOI] [Google Scholar]
- Missouri Botanical Garden and Harvard University Herbaria (2017). eFloras. Retrieved from http://www.efloras.org (Accessed: 10th February 2017)
- Mitchell, T. C. , Williams, B. R. M. , Wood, J. R. I. , Harris, D. J. , Scotland, R. W. , & Carine, M. A. (2016). How the temperate world was colonised by bindweeds: Biogeography of the Convolvuleae (Convolvulaceae). BMC Evolutionary Biology, 16, 16 10.1186/s12862-016-0591-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. (1981). Fossil pollen records of extant angiosperms. Botanical Review, 47, 706–142. [Google Scholar]
- Nathan, R. (2006). Long‐distance dispersal of plants. Science, 313, 786–788. 10.1126/science.1124975 [DOI] [PubMed] [Google Scholar]
- Nathan, R. , Schurr, F. M. , Spiegel, O. , Steinitz, O. , Trakhtenbrot, A. , & Tsoar, A. (2008). Mechanisms of long‐distance seed dispersal. Trends in Ecology & Evolution, 23, 638–647. 10.1016/j.tree.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Nogales, M. , Heleno, R. , Traveset, A. , & Vargas, P. (2012). Evidence for overlooked mechanisms of long‐distance seed dispersal to and between oceanic islands. New Phytologist, 194, 313–317. 10.1111/j.1469-8137.2011.04051.x [DOI] [PubMed] [Google Scholar]
- Oxelman, B. , Lidén, M. , & Berglund, D. (1997). Chloroplast rps16 intron phylogeny of tribe Sileneae (Caryophyllaceae). Plant Systematics and Evolution, 206, 393–410. 10.1007/BF00987959 [DOI] [Google Scholar]
- Panter, C. J. , & Dolman, P. M. (2012). Mammalian herbivores as potential seed dispersal vectors in ancient woodland fragments. Wildlife Biology, 18, 292–303. 10.2981/11-112 [DOI] [Google Scholar]
- Patzak, A. , & Rechinger, K. H. (1965). Plantaginaceae In Rechinger K. H. (Ed.), Flora Iranica (23 pp). Vienna, Austria: Naturhistorisches Museum. [Google Scholar]
- Pilger, R. (1937). Plantaginaceae In Engler A. (Ed.), Das Pflanzenreich IV 269 (102, Heft). Leipzig: Wilhelm Engelmann. [Google Scholar]
- Pole, M. (1994). The New‐Zealand Flora—Entirely long‐distance dispersal. Journal of Biogeography, 21, 625–635. 10.2307/2846036 [DOI] [Google Scholar]
- Rahn, K. (1996). A phylogenetic study of the Plantaginaceae. Botanical Journal of the Linnean Society, 120, 145–198. [Google Scholar]
- Rambaut, A. (2012). FigTree v1.4. Retrieved from http://tree.bio.ed.ac.uk/software/figtree.
- Rambaut, A. , Suchard, M.A. , Xie, D. , & Drummond, A. (2014). Tracer v1.6. Retrieved from http://beast.bio.ed.ac.uk/Tracer.
- Raxworthy, C. J. , Forstner, M. R. J. , & Nussbaum, R. A. (2002). Chameleon radiation by oceanic dispersal. Nature, 415, 784–787. 10.1038/415784a [DOI] [PubMed] [Google Scholar]
- Ree, R. H. , & Smith, S. A. (2008). Maximum Likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology, 57, 4–14. 10.1080/10635150701883881 [DOI] [PubMed] [Google Scholar]
- Richardson, J. E. , Weitz, F. M. , Fay, M. F. , Cronk, Q. C. B. , Linder, H. P. , Reeves, G. , & Chase, M. W. (2001). Phylogenetic analysis of Phylica L. with an emphasis on island species: Evidence from plastid trnL‐F DNA and nuclear internal transcribed spacer (ribosomal DNA) sequences. Taxon, 50, 405–427. 10.2307/1223889 [DOI] [Google Scholar]
- Ronquist, F. (1997). Dispersal‐Vicariance Analysis: A new approach to the quantification of historical biogeography. Systematic Biology, 46, 195–203. 10.1093/sysbio/46.1.195 [DOI] [Google Scholar]
- Ronquist, F. , & Huelsenbeck, J. P. (2003). mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rønsted, N. , Chase, M. W. , Albach, D. C. , & Bello, M. A. (2002). Phylogenetic relationships within Plantago (Plantaginaceae): Evidence from nuclear ribosomal ITS and plastid trnL‐F sequence data. Botanical Journal of the Linnean Society, 139, 323–338. 10.1046/j.1095-8339.2002.00070.x [DOI] [Google Scholar]
- Sanmartin, I. , & Ronquist, F. (2004). Southern hemisphere biogeography inferred by event‐based models: Plant versus animal patters. Systematic Biology, 53, 216–243. 10.1080/10635150490423430 [DOI] [PubMed] [Google Scholar]
- Shaw, J. , Lickey, E. B. , Schilling, E. E. , & Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. American Journal of Botany, 94, 275–288. 10.3732/ajb.94.3.275 [DOI] [PubMed] [Google Scholar]
- Simmons, M. P. , & Ochoterena, H. (2000). Gaps as characters in sequence‐based phylogenetic analyses. Systematic Biology, 49, 369–381. 10.1093/sysbio/49.2.369 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuessy, T. F. , Crawford, D. J. , & Ruiz, E. A. (2018). Patterns of phylogeny In Stuessy T. F., Crawford D. J., Loppez‐Sepulveda P., Baeza C. M., & Ruiz E. A. (Eds.), Plants of Oceanic Islands: Evolution, biogeography, and conservation of the flora or the Juan Fernández (Robinson Crusoe) Archipelago. Cambridge, United Kingdom: Cambridge University Press. [Google Scholar]
- Stuessy, T. F. , Foland, K. A. , Sutter, J. F. , & Silva, M. (1984). Botanical and geological significance of potassium‐argon dates from the Juan Fernández islands. Science, 225, 49–51. 10.1126/science.225.4657.49 [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Skinner, D. Z. , Liang, G. H. , & Hulbert, S. H. (1994). Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics, 89, 26–32. 10.1007/BF00226978 [DOI] [PubMed] [Google Scholar]
- Taberlet, P. , Gielly, L. , Pautou, G. , & Bouvet, J. (1991). Universal primers for amplification of three non‐coding regions of chloroplast DNA. Plant Molecular Biology, 17, 1105–1109. 10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Tay, M. L. , Meudt, H. M. , Garnock‐Jones, P. J. , & Ritchie, P. A. (2010a). DNA sequences from three genomes reveal multiple long‐distance dispersals and non‐monophyly of sections in Australasian Plantago (Plantaginaceae). Australian Systematic Botany, 23, 47–68. 10.1071/SB09040 [DOI] [Google Scholar]
- Tay, M. L. , Meudt, H. M. , Garnock‐Jones, P. J. , & Ritchie, P. A. (2010b). Testing species limits of New Zealand Plantago (Plantaginaceae) using internal transcribed spacer (ITS) DNA sequences. New Zealand Journal of Botany, 48(205), 224. [Google Scholar]
- van der Aart, P. J. M. , & Vulto, J. C. (1992). General biology of Plantago. Biogeography and human effects In Kuiper P. J. C., & Bos M. (Eds.), Plantago: A multidisciplinary study (pp. 5–6)., Ecological Studies, 89 Paris: Springer‐Verlag. [Google Scholar]
- Vences, M. , Vieites, D. R. , Glaw, F. , Brinkmann, H. , Kosuch, J. , Veith, M. , & Meyer, A. (2003). Multiple overseas dispersal in amphibians. Proceedings of the Royal Society B‐Biological Sciences, 270, 2435–2442. 10.1098/rspb.2003.2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, D. S. , Gangoso, L. , Bouten, W. , & Figuerola, J. (2016). Overseas seed dispersal by migratory birds. Proceedings of the Royal Society B‐Biological Sciences, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, W. L. , Herbst, D. R. , & Sohmer, S. H. (1990). Manual of flowering plants of Hawaii. Honolulu, Hawaii, USA: University of Hawaii Press and Bishop Museum Press. [Google Scholar]
- Western, T. L. (2012). The sticky tale of seat coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Science Research, 22, 706–25. 10.1017/S0960258511000249 [DOI] [Google Scholar]
- Winkworth, R. C. , Wagstaff, S. J. , Glenny, D. , & Lockhart, P. J. (2002). Plant dispersal N.E.W.S from New Zealand. Trends in Ecology and Evolution, 17, 514–520. 10.1016/S0169-5347(02)02590-9 [DOI] [Google Scholar]
- Zielske, S. , Ponder, W. F. , & Haase, M. (2017). The enigmatic pattern of long‐distance dispersal of minute freshwater gastropods (Caenogastropoda, Truncatelloidea, Tateidae) across the South Pacific. Journal of Biogeography, 44, 195–206. 10.1111/jbi.12800 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly generated sequence data for this study have been submitted to GenBank.


