Abstract
Wnt signalling drives a plethora of processes in development, homeostasis, and disease; however, the role and mechanism of individual ligand/receptor (Wnt/Frizzled, Fzd) interactions in specific biological processes remain poorly understood. Wnt9a is specifically required for the amplification of blood progenitor cells during development. Using genetic studies in zebrafish and human embryonic stem cells, paired with in vitro cell biology and biochemistry, we have determined that Wnt9a signals specifically through Fzd9b to elicit β-catenin-dependent Wnt signalling that regulates haematopoietic stem and progenitor cell emergence. We demonstrate that the epidermal growth factor receptor (EGFR) is required as a co-factor for Wnt9a/Fzd9b signalling. EGFR-mediated phosphorylation of one tyrosine residue on the Fzd9b intracellular tail in response to Wnt9a promotes internalization of the Wnt9a/Fzd9b/LRP signalosome and subsequent signal transduction. These findings provide mechanistic insights for specific Wnt/Fzd signals, which will be crucial for specific therapeutic targeting and regenerative medicine.
Keywords: haematopoietic stem cell (HSC), Wnt, Wnt9a, signalling specificity, Fzd, Fzd9b, FZD9, EGFR, APEX2
Wnt genes encode highly conserved, lipid-modified glycoproteins involved in the regulation of a plethora of developmental processes; yet, their specific functions are poorly understood. Although the mammalian genome encodes 19 Wnts and 10 Frizzled (Fzd) receptors, there is little evidence for signalling specificity through cognate Wnt-Fzd pairings1. There is an exquisitely specific requirement for Wnt9a in directing an early amplification of haematopoietic stem and progenitor cells (HSPCs), a surprising finding because Wnts have been thought to be functionally promiscuous2-5. Using HSPC development as a platform for validation, we demonstrate that the epidermal growth factor receptor (EGFR) is required as a co-factor to mediate the specificity of the Wnt9a-Fzd9b signalling interaction, a finding which may indicate a general paradigm for Wnt-Fzd signalling specificity.
Haematopoietic stem cells (HSCs) are the tissue-specific stem cells that provide blood and immune cells for the duration of an organism’s life. During development, these cells arise directly in major arterial vessels, from a specialized population of cells termed hemogenic endothelium (HE), specified from progenitors in the lateral plate mesoderm6,7. HE cells receive inductive cues from nearby tissues like the somite and neural crest cells, including fibroblast growth factors, Notch, and Wnt, to establish their fate and future function as HSPCs6-12. After their specification, HSPCs emerge directly from endothelium comprising the ventral floor of the dorsal aorta (hereafter aorta) in a process termed the endothelial-to-haematopoietic transition (EHT) 13,14. They then enter circulation and migrate to secondary haematopoietic organs, such as the fetal liver in mammals or the caudal haematopoietic tissue in teleosts, before seeding the final sites of residence in the bone marrow of mammals, or the kidney marrow of teleosts6,7.
Wnts are important to HSPC development and homeostasis15-26. We previously determined that an early Wnt9a cue is specific in driving a proliferative event in the aorta, after HSCs have emerged, but before they have seeded the secondary haematopoietic organs. We hypothesized that this specific function of Wnt9a may be mediated through specific interaction with one of the 14 zebrafish Fzd receptors3.
Here, we identify Fzd9b as the cognate signalling partner for Wnt9a in the process of haematopoietic stem and progenitor cell (HSPC) development, upstream of β-catenin, a process conserved in human haematopoiesis in vitro. Intracellular Fzd9b domains mediate the specificity of this Wnt-Fzd pairing, implicating a transmembrane spanning co-factor in establishing specificity. Using APEX2-mediated proximity labeling27, we identified the receptor tyrosine kinase EGFR as required for this specific signalling interaction. Altogether, these results demonstrate a conserved Wnt-Fzd pairing that mediates a precise Wnt cue required for haematopoiesis, shifting the paradigm of how specific Wnt-Fzd interactions are established and opening the field for discovery of other cofactors that mediate specific signalling through cognate Wnt-Fzd pairs.
Results
HSPCs require Fzd9b-Wnt9a interaction
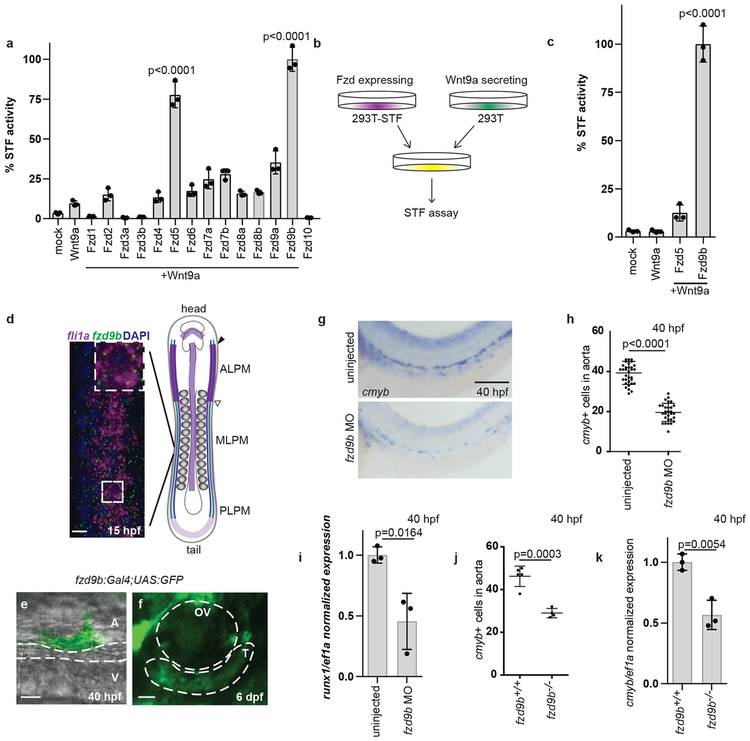
Specificity of Wnt signals likely involves Wnt-Fzd pairings; we sought to identify a cognate Fzd for the specific Wnt9a signal, which occurs when the somite signals to ingressing HE precursors prior to 20 hours post fertilization (hpf)3. A fzd expression screen in 16.5 hpf fli1a+ (endothelial) cells indicated that a majority of Fzds were expressed (Supplementary Fig. 1a-b). Since the Wnt9a signal involves β-catenin3, we employed an established β-catenin dependent Wnt reporter assay28, called Super-TOP-Flash (STF), to identify Fzd candidates. STF reporter activity indicated a synergistic interaction between Wnt9a and Fzd5, or Wnt9a and Fzd9b, but no other Fzd (Supplementary Fig. 1c-d, Fig. 1a). Wnt9a signals to its cognate receptor on neighboring cells, which we assessed using a co-culture approach (Fig. 1b). In this assay, Fzd9b, but not Fzd5, was able to transduce the Wnt9a signal and activate STF reporter activity (Fig. 1c), indicating that Fzd9b acts as a specific Wnt9a receptor. These observations were further supported by an absence of signal using the Super FOP:FLASH reporter (reporter lacking β-catenin activity, Supplementary Fig. 1e), and an absence of Fzd9b specific signal in response to the prototypical ligand, Wnt3a (Supplementary Fig. 1f). Altogether, these data provide evidence for a Wnt9a-Fzd9b interaction and suggest that Fzd9b is specifically involved in Wnt9a-mediated HSPC development.
Figure 1: Fzd9b is required for zebrafish haematopoietic stem cell development.
a. STF assay screen with zWnt9a and zFzds, n=3 biological replicates for each. b. zWnt9a cells were mixed with zFzd9b or zFzd5 STF cells and assayed for Wnt activity, quantified in c (n=3 biological replicates for each). d. FISH for fzd9b and fli1a in 16 hpf zebrafish embryos. MLPM-medial lateral plate mesoderm PLPM- posterior lateral plate mesoderm, ALPM- anterior lateral plate mesoderm. Scale bar=10um. Diagram shows haematopoietic precursors (green), vascular precursors (blue) and fzd9b (purple). e. Cell emerging from the aorta labeled by fzd9b:Gal4; UAS:GFP at 40 hpf. Scale bar=10um. A- aorta, V-vein. f. Thymus cells labeled by fzd9b:Gal4; UAS:GFP at 6 dpf. T-thymus, OV- otic vesicle; scale bar=25um. g. WISH for cmyb at 40 hpf in fzd9b morphants and controls. Scale bar=30um, quantified in h. Each dot represents a biological replicate; n=32 control, n=30 MO. In d-g, images are representative of 10 embryos examined in 3 independent experiments. i. qPCR for cmyb (black) and runx1 (white) in fzd9b morphants (n=3) and controls (n=3) at 40 hpf. j. Quantification of WISH for cmyb at 40 hpf in fzd9b mutants (n=4) and controls (n=5). Each dot represents a biological replicate. k. qPCR for cmyb in fzd9b mutants (n=3) and controls (n=3) at 40 hpf. In all graphs, dots represent biological replicates from a single experiment, bars represent the mean and error bars represent the standard deviation. All STF assays were repeated independently with a similar trend. All qPCR data was generated from biological replicates from dissected trunks and tails each represented as a dot. n.s. not significant. Statistical analyses by ANOVA compared to controls as indicated.
Consistent with a role in Wnt9a signal reception, we found by fluorescent in situ hybridization that at 15hpf, fzd9b mRNA is co-expressed with the endothelial marker fli1a in the lateral plate mesoderm, the tissue from which HE is derived6,7 (Fig. 1d). To test if haematopoietic cells were derived from cells expressing fzd9b, we performed two lineage tracing experiments using fzd9b promoter sequences driving expression of Gal4. First, Gal4 activates an upstream activating sequence (UAS)-driven green fluorescent protein (GFP) (fzd9b:Gal4;UAS:GFP); second, UAS:Cre is activated to excise a loxP-flanked sequence encoding blue fluorescent protein (BFP), ultimately leading to expression of dsRed (fzd9b:Gal4; UAS:Cre; loxP-BFP-loxP-dsRed). Using this strategy, we were able to observe GFP+ or dsRed+ (pseudo colored green in Supplementary Fig. 2a) cells in the floor of the dorsal aorta in the characteristic cup shape observed during the EHT (at 40 hpf), indicating that nascent HSPCs had expressed fzd9b prior to their emergence (Fig. 1e, Supplementary Fig. 2a, left). Thymocytes derived from HSCs reside in the thymus beginning around 4 days post-fertilization (dpf); these expressed GFP at 6 and 7 dpf (Fig. 1f, Supplementary Fig. 2a, right), consistent with a function for fzd9b in HSPC development.
To assess the function of Fzd9b in zebrafish HSPC development, we used an antisense morpholino (MO) oligonucleotide, which blocked translation of an ectopically provided Fzd9b-mKate fusion transcript in vivo (Supplementary Fig. 2b). Similar to wnt9a loss of function3, fzd9b morphants had normal specification, as measured by the early HSPC marker runx1 (Supplementary Fig. 2c-d); however, HSPC expansion markers (cmyb, gata2b and runx1) were significantly reduced in fzd9b morphants at 40 hpf (Fig. 1g-i, Supplementary Fig. 2e-f). This loss of HSPCs persisted throughout development, where rag1+ thymocytes were reduced in fzd9b morphants (Supplementary Fig. 2g). This effect was specific to HSPCs, as markers for aorta (dlc, dll4, notch1b), vasculature (kdrl) and pronephros (cdh17) were normal (Supplementary Fig. 2h). The effects of the MO were specific, as the fzd9b MO could be rescued with fzd9b mRNA (Supplementary Fig. 2i-j), and two germline mutants of fzd9b (predicted to produce severely truncated proteins of approximately 30 residues) had reduced cmyb+ cells at 40 hpf (Fig. 1j-k). Furthermore, MO-injected fzd9b mutants did not have a more severe phenotype than their siblings (Supplementary Fig. 2k). Finally, we used an established transgenic approach where a guide RNA to fzd9b is expressed ubiquitously and Cas9 expression is spatially regulated to conditionally inactivate fzd9b in early endothelial cells29, where the number of cmyb+ cells at 40 hpf was also reduced (Supplementary Fig. 2l-m). Taken together with our finding that the Wnt signal is received by HE3, these results indicate Fzd9b is required for HSPC development, downstream of fate specification, and specifically within HE.
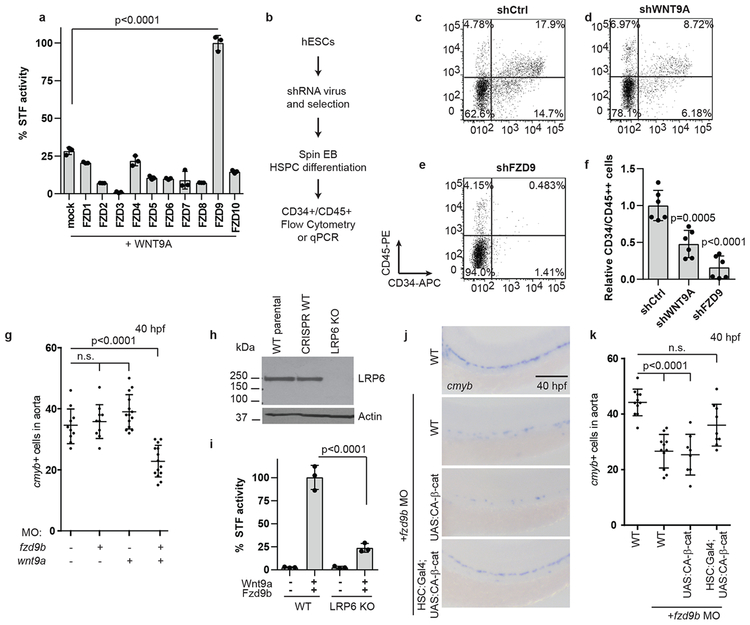
Having identified a specific zebrafish Wnt9a-Fzd9b signal in vivo, we used the STF reporter assay and determined that only human FZD9 coupled effectively with WNT9A (Fig. 2a). Furthermore, using an established differentiation protocol to drive human embryonic stem cells (hESCs) towards haematopoietic fates30 (Fig. 2b), we found that disrupting WNT9A or FZD9 expression using short hairpin RNAs (shRNAs, Supplementary Fig. 3a-b) significantly compromised the ability of hESCs to generate HSPCs, as assessed by flow cytometry for CD34 and CD45 (Fig. 2c-f) and by expression of endothelial (CD34) and haematopoietic markers (CD31, CD45, CMYB) (Supplementary Fig. 3c-f). These differences were not due to loss of pluripotency of the undifferentiated hESCs, as they still abundantly expressed the pluripotency markers TRA1–81 and SSEA431 (Supplementary Fig. 3g-i). Thus, Wnt9a/Fzd9b and WNT9A/FZD9 are required for both zebrafish and human HSPC development.
Figure 2: Fzd9b interacts genetically with Wnt9a.
a. Quantification of STF assay screen with human WNT9A and FZDs; n=3 biological replicates each. b. Schematic of experimental design for HSPC derivation. Representative flow cytometry plots of CD34 vs CD45 cells after 14 days of differentiation towards HSPCs in shControl (c), shWNT9A (d) and shFZD9 (e) transduced cells. Note the loss of double positive cells with the loss of WNT9A or FZD9, quantified in f, n=6 biological replicates each. g. Quantification of WISH for cmyb in suboptimal MO (0.1ng wnt9a, 0.5ng fzd9b) treated zebrafish at 40 hpf; n=10, 10, 14, 14 biological replicates from left to right. h. LRP6 immunoblot of lysates from WT HEK293T STF cells (WT parental), CRISPR-treated cells without disruption of LRP6 (CRISPR WT), and LRP6 null mutant line (LRP6 KO). Image representative of 4 experimental replicates. i. STF activity in WT and LRP6 KO HEK293T STF cells transfected with zWnt9a and zFzd9b. n=3 biological replicates for each. j. WISH for cmyb at 40 hpf in WT, UAS:CA-β-catenin and gata2b:Gal4;UAS:CA-β-catenin, injected with fzd9b MO. Scale bar=30um. k. Quantification of j; n=11, 11, 8 , 9 biological replicates from left to right. In all graphs, each dot represents a biological replicate from a single experiment, bars represent the mean, error bars represent the standard deviation. All STF assays were repeated independently with a similar trend. n.s. not significant. Statistical analyses by ANOVA compared to controls as indicated.
Fzd9b/Wnt9a operate upstream of β-catenin
To confirm that Wnt9a and Fzd9b function in the same pathway in vivo, we used genetic non-complementation with suboptimal MO dosages. A low dose of either wnt9a or fzd9b MO was not sufficient to affect HSPCs, while compound morphant animals had a reduction in cmyb+ cells similar to either wnt9a or fzd9b loss of function, supporting that these components operate in the same genetic pathway (Fig. 2g, Supplementary Fig. 4a).
The effect of Wnt9a on HSPCs required β-catenin (or the canonical Wnt pathway)3, which relies on the assembly of Fzd-Lrp5/6 heterodimers in response to a Wnt ligand1. Consistent with this, the Wnt9a-Fzd9b signal could be synergistically increased in vitro in cells co-transfected with Lrp6 (Supplementary Fig. 4b). To test the requirement for LRP6 in the Wnt9a-Fzd9b signal, we generated a HEK293T STF line deficient for LRP6 (Fig. 2h), which were compromised in their STF reporter activity upon Wnt3a addition (Supplementary Fig. 4c). Importantly, treatment with a GSK3 inhibitor (CHIR98014), which activates signalling independent of Wnt-Fzd-LRP interactions, stimulated STF activity, indicating that downstream signalling components were intact (Supplementary Fig. 4d). Wnt9a-Fzd9b signalling also required LRP6 (Fig. 2i), consistent with a role upstream of β-catenin. Accordingly, loss of fzd9b, the putative Wnt9a receptor, should be rescued by expression of constitutively-active (CA) β-catenin. Indeed, HE-specific regulatory sequences (gata2b promoter) driving expression of CA-β-catenin was sufficient to rescue loss of fzd9b (Fig. 2j-k), indicating that Fzd9b functions upstream of β-catenin. Altogether, these data indicate that Wnt9a and Fzd9b function in the same genetic pathway, upstream of β-catenin.
Intracellular Fzd9b domains mediate specificity
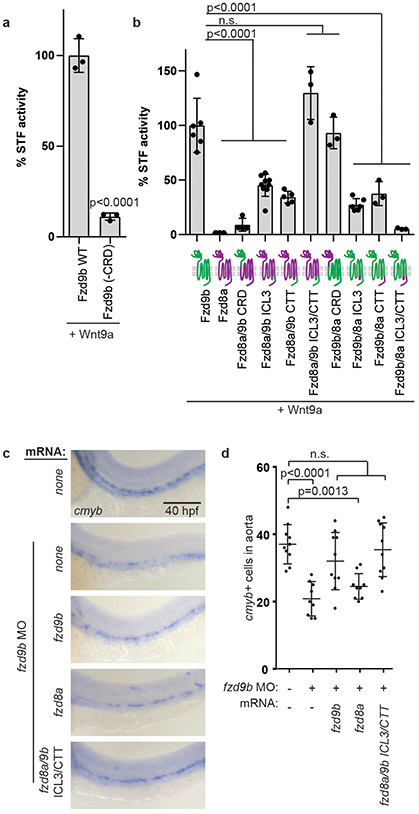
How the Wnt-Fzd signalling complex relays its signal and establishes specificity is poorly understood, but is thought to rely on varying affinities of Wnts for the extracellular cysteine rich domain (CRD) of Fzd1,32-34. Using the STF assay system and zebrafish cDNAs, we found that like other Wnts1, the Fzd9b CRD is required to mediate the Wnt9a signal in vitro (Fig. 3a). Signalling differences were not due to differences in protein expression (Supplementary Fig. 5a).
Figure 3: Wnt9a-Fzd9b specificity is mediated intracellularly.
a. STF assay with zWnt9a and zFzd9b with and without the cysteine rich domain (CRD); n=3 biological replicates each. b. STF assay with zWnt9a and zFzd9b/zFzd8a chimeras (schematics on x-axis; magenta=fzd8a; green=fzd9b); n=6, 3, 4, 9, 5, 3, 3, 6, 3, 3 biological replicates from left to right. c. WISH for cmyb at 40 hpf in fzd9b morphants injected with mRNAs for fzd9b, fzd8a and fzd8a with ICL3 and CTT from fzd9b and uninjected control. Scale bar=30um. d. Quantification of c; n=10, 9, 10, 9, 10 biological replicates from left to right. In all graphs, each dot represents a biological replicate; bars represent the mean and error bars represent the standard deviation. n.s. not significant. Panel b shows data from 2 experiments, while all other graphs represent biological replicates from the same experiment. All STF assays were repeated independently with a similar trend. Statistical analyses by ANOVA compared to controls as indicated.
We next sought to determine which domains are required for Wnt9a-Fzd9b signalling specificity by constructing a series of chimeric Fzd transgenes between Fzd9b and Fzd8a, a Fzd that did not activate STF reporter activity with Wnt9a (Fig. 1a). Surprisingly, a chimeric receptor in which the CRD of Fzd8a was replaced with that of Fzd9b did not signal (Fig. 3b), while the opposite chimeric Fzd protein produced wild-type Wnt9a signalling activity (Fig. 3b), suggesting that other Fzd domains other than the CRD are critical determinants in Wnt9a-Fzd9b signalling specificity.
The signalling events downstream of Wnt-Fzd-Lrp interaction are thought to involve interaction with intracellular mediator proteins such as Disheveled (Dsh/Dvl), which interacts with Fzd at the third intracellular loop (ICL3) and the C-terminal tail (CTT)1. Using further Fzd9b-Fzd8a chimeras, we found that substituting both the ICL3 and CTT from Fzd9b with those of Fzd8a was sufficient to completely ablate the signal, and the opposite was sufficient to produce wild-type signalling levels (Fig. 3b), whereas single substitutions resulted in partial levels of signalling (Fig. 3b), suggesting that signalling specificity for Wnt9a-Fzd9b lies entirely within the ICL3 and CTT domains. These chimeras were expressed to the same level (Supplementary Fig. 5b), indicating differences in signalling were not due to differences in expression. These findings were recapitulated using zebrafish and human cDNAs encoding chimeras for Fzd9b/FZD9 and Fzd4/FZD4 (Supplementary Fig. 5c-d), indicating that the ICL3 and CTT are required for zebrafish Fzd9b and human FZD9 signalling.
V5-tagged Fzd constructs had no change in the ability to mature or be modified post-translationally (Supplementary Fig. 5b), as assessed by their shift in size in immune-blots35-37. We also confirmed that the non-signalling chimeric Fzd9bs were transported to the cell surface using immunofluorescence and flow cytometry with a Fzd9b antibody directed to the extracellular region between the CRD and the first transmembrane domain of Fzd9b (Supplementary Fig. 5d-f). Therefore, differences in signalling were not due to differences in Fzd protein expression, maturation, or transport to the cell surface.
Finally, to determine if these domains were able to fulfill Fzd9b function in HSPC development, we co-injected mRNAs for fzd9b, fzd8a or fzd8a with the ICL3 and CTT from fzd9b (fzd8a/9b ICL3/CTT) in the context of fzd9b MO and found that only fzd9b and fzd8a/9b ICL3/CTT were able to rescue loss of fzd9b (Fig. 3c-d). Taken together, these data indicate that Wnt9a-Fzd9b specificity is regulated by the intracellular ICL3 and CTT domains of Fzd9b.
Wnt9a, Fzd9b and EGFR form a complex
Since Wnt9a-Fzd9b specificity is mediated intracellularly, we postulated the existence of another signalling component that spanned the membrane to contact both Wnt9a and the intracellular portion of Fzd9b. Our ability to analyze Wnt9a-Fzd9b signalling specificity using zebrafish cDNAs in human cells further suggested that this signalling component would be highly conserved. To identify proteins are in proximity to intracellular portions of Fzd9b, we generated a stable HEK293T cell line expressing Fzd9b fused via a Glycine-Serine linker to the peroxidase APEX227 (Fzd9b-5GS-APEX2). In the presence of hydrogen peroxide and biotin-phenol, endogenous proteins proximal to APEX2 (generally within 30 nm) are biotinylated within 1 minute, allowing for their enrichment with streptavidin beads, and subsequent identification by mass spectrometry (MS) 27. The Fzd9b-5GS-APEX2 cells labelled specifically in the presence of biotin-phenol and hydrogen peroxide induction (Supplementary Fig. 6a), had correct Fzd9b localization (Supplementary Fig. 6b-c), and were able to signal in response to Wnt9a (Supplementary Fig. 6d).
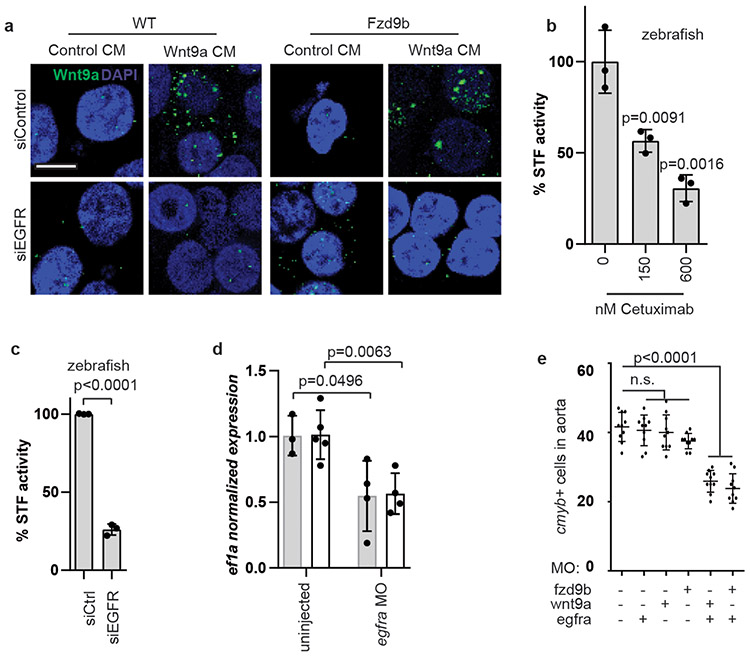
Gene ontology (GO) analysis of the APEX-MS data revealed that in response to Wnt9a, the most changed biological processes included ERBB signalling (Supplementary Fig. 6e). Genes from ERBB family encode single-pass transmembrane receptor tyrosine kinases that homo- and hetero-dimerize in response to multiple ligands to stimulate a number of signalling cascades38. The transmembrane protein most enriched by proximity labeling was EGFR (also known as ERBB1) (Supplementary Fig. 6f). We hypothesized that EGFR may play a role in Wnt9a recruitment to Fzd9b at the cell membrane. Indeed, disrupting EGFR expression in HEK293T cells stably expressing Fzd9b with short interfering RNA (siRNA) reduced cell surface binding of Wnt9a (Fig. 4a), suggesting that EGFR expression promotes Wnt9a binding to the cell surface. Interestingly, this also occurred in the absence of Fzd9b (Fig. 4a), suggesting that Wnt9a binds directly to EGFR. In addition, provision of Cetuximab39, which blocks the EGFR ligand binding, dampened the Wnt9a-Fzd9b signal (Fig. 4b), consistent with a model in which EGFR forms a complex with Wnt9a and Fzd9b to transmit the Wnt signal.
Figure 4: EGFR mediates Wnt9a-Fzd9b signalling.
a. Immunofluorescence for zWnt9a in cells treated as shown. Scale bar=15 um. Representative of 10 fields of view from 2 experiments. b. STF assay of zWnt9a and zFzd9b treated with Cetuximab; n=3 biological replicates each. c. STF assay of zWnt9a and zFzd9b with siRNAs; n=3 biological replicates each. d. qPCR for runx1 (grey) and gata2b (white) at 40 hpf generated from trunks and tails; n=3, 5, 4, 4 biological replicates from left to right. e. Quantification of WISH for cmyb at 40 hpf (suboptimal MO dosages 0.1ng wnt9a, 0.5ng fzd9b and 0.1ng egfra) n=11, 9, 10, 8, 10, 10 biological replicates from left to right. In all graphs, each dot represents a biological replicate from the same experiment; bars represent the mean and error bars represent the standard deviation. n.s. not significant. All STF assays were repeated independently with a similar trend. Statistical analyses by ANOVA compared to controls as indicated.
HEK293T cells transfected with siRNA to EGFR compromised the ability of both zebrafish and human Wnt9a/WNT9A and Fzd9b/FZD9 to stimulate STF reporter activity (Supplementary Fig. 6g-h, Fig. 4c). Using a previously validated MO to egfra40, the number of HSPCs at 40 hpf was decreased (Supplementary Fig. 6i-j), an effect that was validated with qPCR (Fig. 4d), similar to the phenotypes of fzd9b or wnt9a loss of function, and consistent with a role for Egfr in regulating the Wnt9a-Fzd9b signal. Suboptimal MO dosing indicated that both Fzd9b and Wnt9a synergize genetically with Egfr during HSPC development (Fig. 4e). Furthermore, treatment of cells or zebrafish with the selective EGFR tyrosine kinase inhibitor AG147841 significantly attenuated STF reporter activity or HSPC development, respectively (Supplementary Fig. 6k-m). Altogether, these data demonstrate that EGFR and its kinase activity are required for the Wnt9a-Fzd9b signal.
There are seven other known ligands for EGFR38, which led us to hypothesize that these may be required to assemble EGFR, Wnt9a and Fzd9b into a complex. Of these, only heparin bound EGF (HBEGF) is expressed in HEK293T cells42 (Supplementary Fig. 6n). To assess the requirement for HBEGF and other EGFR ligands in the Wnt9a/Fzd9b signal, we cultured HEK293T STF reporter cells in serum-free media lacking all known EGFR ligands, and added a neutralizing antibody targeting HBEGFR. STF reporter activity with Wnt9a/Fzd9b stimulation was maintained in these conditions (Supplementary Fig. 6o), indicating this interaction occurs in the independently of known EGFR ligands.
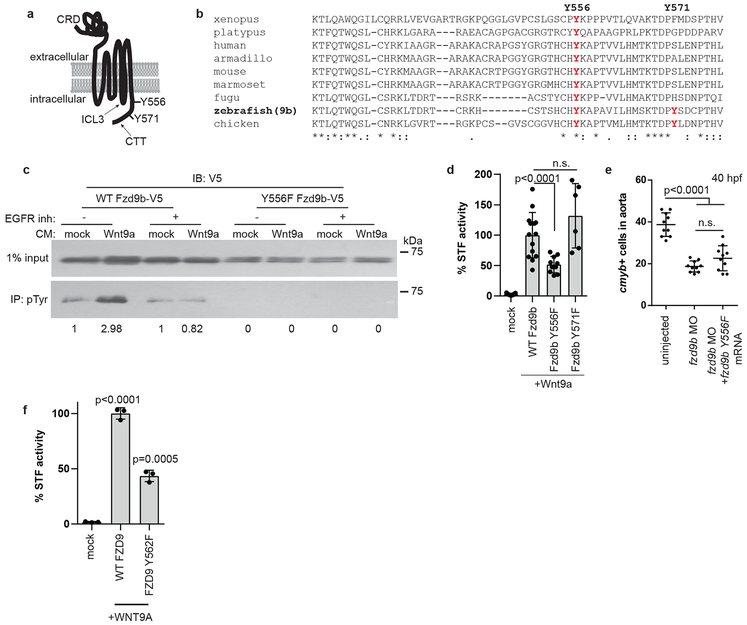
The C-terminal tail of Fzd9b contains two tyrosine (Y) residues, at 556 and 571 (Fig. 5a), which are predicted to be potential kinase substrates43. Additionally, the Y556 on Fzd9b is highly conserved among vertebrates, indicative of putative functional importance (Fig. 5b). Consistent with these predictions, treatment with Wnt9a increased Y-phosphorylation of Fzd9b, which was dependent on EGFR kinase activity as this increase was not observed in the presence of the EGFR tyrosine kinase inhibitor AG1478 (Fig. 5c). In addition, mutation of the Y556 (but not Y571) sites decreased the in vitro signalling capacity of Wnt9a (Fig. 5d), and was not able to rescue fzd9b morphants (Fig. 5e); we also found the corresponding residue in human FZD9, Y562F is required (Fig. 5f). Together, these data indicate that Fzd9b is phosphorylated on tyrosine residue 556 in response to Wnt9a, which is required for its downstream signal.
Figure 5: EGFR is required to phosphorylate the Fzd9b C-terminal tail in response to Wnt9a.
a. zFzd9b protein with putative EGFR tyrosine phosphorylation sites. CRD: cysteine rich domain, ICL3: intracellular loop 3, CTT: C-terminal tail. b. Protein alignment of Fzd9 C-terminal tails from species shown, using ClustalOmega. c. (See also Supplementary Fig. 7) Immunoblot for V5 from phosphotyrosine immunoprecipitation with increase in pY-zFzd9b (2.98 vs 1, arbitrary units). Trend observed in 4 independent experiments. d. STF assay of zFzd9b point mutants; n=9, 14, 10, 6 from left to right. e. Quantification of WISH for cmyb+ cells in uninjected, fzd9b MO injected and fzd9b MO+fzd9bY556F mRNA injected fish at 40 hpf; n=10 zebrafish each. f. Quantification of HEK293T cell STF assay with human WNT9A and FZD9 WT and Y562F mutant; n=3 biological replicates each. In all graphs, dots represent biological replicates from a single experiment; bars represent the mean and error bars represent the standard deviation. n.s. not significant. Statistical analyses by ANOVA compared to control as indicated. All STF assays were repeated independently with a similar trend.
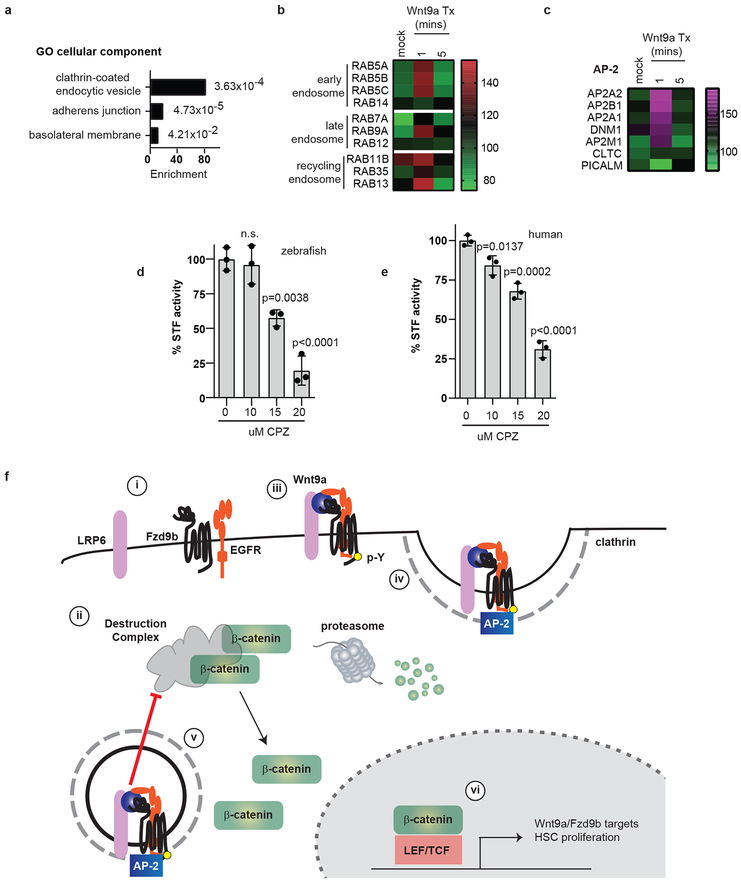
EGFR is known to have effects on signal transduction, as well as receptor internalization and trafficking44-46. Consistent with these functions, GO analysis of the APEX-MS data indicated that the most enriched cellular component was “clathrin-coated endocytic vesicle” (Fig. 6a). The APEX data also showed enrichment for proteins associated with early endosomes (RAB5A, RAB5B, RAB5C and RAB14), late endosomes (RAB7A, RAB9A and RAB12), and recycling endosomes (RAB11B, RAB35 and RAB13) (Fig. 6b), consistent with Fzd9b internalization in response to Wnt9a.
Figure 6: Clathrin-mediated endocytosis is required for the Wnt9a-Fzd9b signal.
a. Fold enrichment of GO terms for cellular components identified from the top 5% of changed proteins in Fzd9b-APEX2 HEK293T cells treated with Wnt9a. p-values are listed next to each bar. n=3 biological replicates, see methods for analysis and statistics. b. Heatmap of Fzd9b-APEX2 proximity labeled normalized intensities of RAB members in the early endosome, late endosome and recycling endosome over time from n=3 biological replicates. c. zFzd9b-APEX2 proximity labeled normalized intensity averages of 3 biological replicates. d. STF assay with zWnt9a and zFzd9b cultured with chlorpromazine (CPZ); n=3 experiments each. e. STF assay of hWNT9A CHO/hFZD9 STF co-culture treated with chlorpromazine (CPZ); n=3 experiments each. In all graphs, dots represent biological replicates from a single experiment; bars represent the mean and error bars represent the standard deviation. n.s. not significant. Statistical analyses by ANOVA compared to control as indicated. All STF assays were repeated independently with a similar trend. f. i. Fzd9b, LRP6 and EGFR are resident in proximity at the cell surface; f. ii. the destruction complex targets β-catenin for degradation in the absence of a ligand; f. iii. in the presence of Wnt9a, EGFR phosphorylates the Fzd9b tail at Y556. f. iv. AP-2 and clathrin are recruited. f. v. Fzd9b-LRP6 oligomerization leads to dissociation of the destruction complex and release of β-catenin into the cytosol, allowing for nuclear translocation. f. vi. Nuclear β-catenin transactivates a program for HSC proliferation.
Internalization of transmembrane proteins can be mediated by clathrin- or caveolin-mediated endocytosis, as can Fzd-Wnt complexes47,48. The APEX-MS data indicated that the AP-2 complex and clathrin-mediated endocytosis machinery were recruited to Fzd9b in response to Wnt9a, suggesting that internalization was mediated by clathrin (Fig. 6c). Indeed, clathrin was required for both the zebrafish Wnt9a/Fzd9b and human WNT9A/FZD signals, as STF activity was reduced in the presence of the clathrin inhibitor chlorpromazine (Fig. 6d-e). These results indicate that Fzd9b is internalized and sorted through the endosome-lysosome in response to Wnt9a.
Taken together, our data suggest a mechanism for the specificity of Wnt9a-Fzd9b signalling, and a role for EGFR in directly regulating this Wnt signal. Fzd9b, LRP6 and EGFR are resident in proximity at the cell surface (Fig. 6f.i); in the absence of a ligand, β-catenin is targeted for proteasomal degradation by the destruction complex (Fig. 6f.ii); in the presence of Wnt9a, these are bridged, allowing EGFR-mediated phosphorylation of the Fzd9b tail at Y556 (Fig. 6f.iii), leading to the recruitment of AP-2 and clathrin (Fig. 6f.iv). Once inside the cell, Fzd9b-LRP6 oligomerization leads to dissociation of the destruction complex and release of β-catenin (Fig. 6f.v). Finally, nuclear β-catenin transactivates a program for HSC proliferation (Fig. 6f.vi).
Discussion
One longstanding puzzle in the Wnt field has centered around the requirement for genetically encoding such a diverse set of ligands and receptors if Wnt ligands are as promiscuous as reported4,33,49. The specificity dogma has been that Wnt-Fzd interactions are regulated by (1) physical binding affinities between Wnt and the Fzd CRD32, and (2) spatio-temporal localization, driven by the findings that different Wnt-Fzd combinations have different affinities, and that many Wnts can physically interact with multiple (if not all) Fzd CRDs4,33,49,50. In addition, (3) the ligand availability in the extracellular space is regulated by co-receptors, such as Reck and Gpr12451-54, in a mechanism distinct from our findings. Our data indicate that signalling specificity is regulated at an additional level involving the activation of co-receptors. To this model, we propose the addition of (4) co-factors enzymatically activating Wnt-Fzd complexes for signalling through internalization. We hypothesize that different co-factors are recruited extracellularly by specific Wnt-Fzd pairs to target the receptor complex for signalling. These results support the notion that specificity of Wnt-Fzd pairs relies on co-receptor complexes; however, the concept that Wnt stimulation recruits a co-receptor (EGFR), leading to its enzymatic regulation of the Fzd intracellular domains (Y-phosphorylation), and subsequent internalization for signalling is unprecedented.
These discoveries have important implications in our understanding of development and disease, where overlapping functions of receptor tyrosine kinase families and Wnt, for example, may represent co-operative functions in signalling. Understanding Wnt-Fzd and co-factor specific interactions will be critical to the advancement of regenerative medicine, such as in the development of protocols to derive different tissues in vitro from pluripotent precursor cells, where in large part, the requirement for β-catenin dependent Wnt signalling in these protocols has been substituted for with the prototypical ligand, Wnt3a, or with GSK3 inhibitors, and not a specific ligand/receptor. Finally, determining how individual WNTs and FZDs are coupled will have important therapeutic implications, where pan-WNT inhibitory therapies cause toxic side effects. Furthermore, our observation that Cetuximab, which blocks ligand binding to EGFR, disrupts Wnt9a/Fzd9b (WNT9A/FZD9 in humans) signalling suggests potential alternative mechanisms of action for this chemotherapeutic agent. Therapies targeting specific WNT-FZD pairs will allow for more precise targeting of these cancer cells.
Methods:
Cell culture and luciferase reporter assays
HEK293T cells or HEK293T cells lacking FZD1, FZD2 and FZD7 (kindly provided by Professor Michael Boutros)5 with a stably integrated Super-TOP-Flash reporter (STF)55 and Chinese hamster ovary (CHO) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) under standard conditions. HEK293 cells are known to have potential cross contamination with HeLa cells (ICLAC Register of Misidentified Cell Lines (iclac.org/databases/cross-contaminations/)), which is irrelevant to this study.
For most assays, cells were seeded into six-well plates and transfected using polyethyleneimine (PEI), 50ng of renilla reporter vector, 200ng of cDNA expression vector, with a total of 1 ug of DNA/well. For co-culture experiments, cells were passaged 24 hours after transfection and plated together for analysis. For siRNA experiments, a well of a six-well plate was also treated with 10pmol of siRNA using RNAiMax transfection reagent (Invitrogen). All human WNT9A cell culture experiments except for the initial screen were performed by co-culturing stably expressing WNT9A CHO cells with stably expressing FZD9 HEK293T STF cells. Assays with Wnt3a were conducted in HEK293T cells lacking FZD1, FZD2 and FZD75.
Serum-free conditions were established by gradually adapting our HEK293T STF reporter cells to SFM II (Invitrogen), which lacks all known EGFR ligands. Serum-free transfections were performed according to the manufacturer recommendations using Lipofectamine 3000. 500ng/mL HBEGF neutralizing antibody (R&D systems, AF-259-SP) or normal Goat Serum (R&D systems, AB-108-C) were added 6 hours after transfection.
All transfected cells were harvested 48-hours post-transfection and all conditioned medium or co-cultured cells were harvested 24 hours post-treatment; the lysates processed and analyzed using the Promega Dual Luciferase Assay System according to the manufacturer’s instructions. Each experiment was performed with at minimum biological triplicate samples and reproduced at least one time with a similar trend. Wnt activity was calculated by normalizing Firefly Luciferase output to Renilla Luciferase; maximum fold induction was set to 100%. All STF reporter assays were conducted with a minimum of 3 biological replicates; the assays were always reproduced with a similar trend at least once.
10 cm plates of 293T cells were transfected with 10ug of constructs encoding chimeric Fzd cDNAs with a C-terminal V5 tag. For immunofluorescence, cells were plated onto glass coverslips after 24 hours, and stained with our Fzd9b antibody, generated to a region between the CRD and the first transmembrane domain, under non-permeabilized conditions, and according to standard protocols. For flow cytometry, cells were harvested with Accutase, pelleted, resuspended in PBS with 1% BSA and 1mM EDTA, filtered through an 80um filter and sorted using a BD Fortessa flow cytometer. For immune-blots, cells were harvested 48 hours after transfection in TNT buffer (1% Triton X-100, 150mM NaCl, 50mM Tris, pH8.0), with protease inhibitors. Immunoblotting was performed according to standard procedures, using antibodies for V5 (1:5,000, GeneTex, GTX628529) and β-actin (1:20,000, Sigma, A2228–100UL).
For immunoprecipitation, cells were washed 3 times in PBS and lysed in radio immunoprecipitation assay (RIPA) buffer (10mM Tris-HCl, pH8, 10mM EDTA, 0.5mM EGTA, 1%Tx-100, 0.1% deoxycholate, 0.1% SDS, 140mM NaCl), supplemented with protease and phosphatase inhibitor tablets (Pierce) and 20mM N-ethylmaleimide, for 30 minutes at 4°C with rocking. Resultant lysates were cleared of cell debris by centrifugation at 15,000g for 10 minutes at 4°C and quantified by Bradford Assay. A minimum of 200ug of protein was diluted into 400uL total volume with RIPA buffer; 2ug of antibody was added and incubated at room temperature for 30–60 minutes. Antibody-protein complexes were precipitated using Protein A Dynabeads (Invitrogen) for 30 minutes at room temperature, washed 3 times with 1mL of RIPA buffer and eluted using Laemmli buffer at room temperature. Precipitates were analyzed by Fzd9b immunoblot. Immunoblot intensities from pulldowns were quantified by densitometry using ImageJ, normalized to input controls.
EGFR inhibition was performed using a 5mM stock of AG1478 in 50:50 Ethanol: DMSO. The final concentration used was 2.5uM. Clathrin-mediated endocytosis was inhibited using chlorpromazine from a 1M stock in 50:50 Ethanol: DMSO. Final concentrations used are as indicated in the figure.
Wnt9a surface binding assay
Conditioned medium was collected from stably expressing Wnt9a or parental CHO cells and concentrated 10X using a 30kDa molecular weight cutoff ultra-filtration device (Millipore). HEK293T cells stably expressing Fzd9b were transfected with siControl or siEGFR and plated on 0.1% gelatin coated glass coverslips after 24 hours; after a further 24 hours, cells were treated with cold conditioned medium for 3 hours at 4°C, rinsed with PBS and fixed with 4% PFA at 4°C for 20 minutes and at room temperature for 10 minutes. Immunofluorescence was performed using standard non-permeabilizing methods with a rabbit polyclonal antibody generated to zebrafish Wnt9a.
Generation of LRP6 knockout HEK293T STF line
A confluent 10cm plate of HEK293T STF cells was transfected with 3μg each of Cas9 and two guide RNAs under regulatory control of a U6 promoter. The guide RNAs (GGGCTTGGAGGATGCAGCTG and GGATCTAAGGCAATAGCTCT) targeted the second exon of LRP6. Single cell clones were validated for loss of LRP6 by sequencing the genomic locus, immunoblotting using a rabbit monoclonal antibody (1:1000, C47E12, Cell Signalling, 2560S) and STF activity with mouse Wnt3a, which requires LRP6 for signalling35. Cell lines are available upon request.
Fzd9b and Wnt9a antibody generation
GST fusion proteins for immunogens of Fzd9b (residues 115–226) and Wnt9a (residues 233–295) were purified by standard methods. Rabbits were immunized with GST proteins, boosted and bled for serum according to standard methods (Lampire Biologicals). Antibodies for Wnt9a or Fzd9b were affinity purified against the same antigens fused to maltose binding protein (MBP), according to the manufacturer’s recommendations (Fisher), and stored in PBS/50% glycerol at −80°C. Antibodies are available upon request.
Animals
Zebrafish were maintained and propagated according to University of California and local Institutional Animal Care and Use Committee (IACUC) policies. AB*, Tg(kdrl:Cherry-CAAX)y171, Tg(fli1a:eGFP)zf544, Tg(cdh5:Gal4)mu101, Tg(UAS:CA-β-catenin)sd47Tg Tg(gata2b:KalTA4sd32; UAS:Lifeact:eGFPmu271), wnt9a Δ28/Δ28,sd49, Tg(UAS:Cre)t32240Tg Tg(bactin2:loxP-BFP-loxP-DsRed)sd27Tg lines have been previously described3,10,11,14,36,37. For simplicity in the text, these lines are referred to with short forms listed in square brackets: Tg(kdrl:Cherry-CAAX) [kdrl:mCherry], Tg(fli1a:eGFP) [fli1a:eGFP], Tg(UAS:CA-β-catenin)sd47Tg [UAS:CA-β-catenin], Tg(gata2b:KalTA4)[HSC:Gal4] and (UAS:Lifeact:eGFP) [UAS:eGFP], Tg(UAS:Cre) [UAS:Cre], Tg(bactin2:loxP-BFP-loxP-DsRed) [loxP BFP loxP dsRed].
MO for fzd9b was targeted to block the ATG start codon with sequence 5’- AGGTGAGCTTCCCATTCTGGATTTT −3’ from GeneTools. 1-cell stage zygotes were injected with 2ng of fzd9b MO and disruption of protein expression was confirmed with a fluorescently tagged mRNA. Suboptimal MO dosage was 0.5ng. The wnt9a MO has been previously described3 and was used at 0.1ng. The egfra MO has been previously described40 and was used at 2.5ng or 1ng. Rescue experiments were performed using 20 pg fzd chimeric mRNA synthesized using the SP6 mMessage machine kit (Ambion) according to the manufacturer’s recommendations.
CRISPR/Cas9 was used to generate germline mutants for fzd9b; sgRNAs were chosen according to their ability to cleave DNA in vitro as previously described57. Mutation of the fzd9b locus at the N-terminal was achieved 100ng of cas9 mRNA (Trilink) and 100ng of sgRNA (GGCTCTTATGACCTGGAGAG) and generated mutants with either a 2bp insertion (fzd9b2bp) fb203, or a 7bp deletion (fzd9b7bp)fb204. Similarly, mutation at the C-terminal tail was achieved by injecting the sgRNA (GGACTCTTCAGTGCCCACAG). For simplicity, in the text, these are referred to as fzd9b−/− and fzd9bΔCTT/ΔCTT, respectively. Mutations were confirmed by sequencing individuals. Zebrafish lines are available upon request.
Tg(fzd9b:Gal4) founders were established by injecting 25pg of a Tol2 kit58 generated plasmid with 100pg of transposase mRNA at the 1-cell stage. The transgenic plasmid encoded a 4.3 kb fzd9b promoter region amplified using the primers: 5’ CTCCCATGAGGCAGAACGTGTGT 3’ and 5’ AGTCCGCGAGCAGCTTGTCTGTT 3’; this was cloned into a p5E MCS Tol2 entry vector using XhoI and SacII restriction sites and then combined with a Gal4 middle entry and polyA 3 prime entry vector by Gateway Assembly to make a full transgene construct with cmlc2:gfp in the backbone. The resultant animals were crossed to Tg(UAS:YFP), and expression compared to in situ hybridization for fzd9b to identify founders that recapitulated endogenous fzd9b expression.
Lineage tracing experiments were visualized on a Zeiss LSM 880 with Airyscan. Representative images were produced by combining 3–4 Z-slices per scan.
Whole-mount in situ hybridization (WISH) and Fluorescent WISH (FISH)
RNA probe synthesis was carried out according to the manufacturer’s recommendations using the DIG-RNA labeling kit, or the fluorescein labeling kit for FISH (Roche). Probes for fli1a, rag1, dll4, dlc, notch1b, kdrl, cdh17, cmyb and runx1 and WISH protocols have been previously described11. The probe for fzd9b was generated from the full-length cDNA. FISH signal was developed as previously described.
Fluorescence activated cell sorting and quantitative PCR
Zebrafish were dissociated using Liberase TM (Roche) and filtered through an 80um filter. Cells were sorted on a BD Influx cell sorter according to standard procedures. RNA and cDNA were synthesized by standard means and qPCR was performed using FastStart Universal SYBR Green Master Mix (Roche) according to the manufacturer’s recommendations and analyzed using the 2-ΔΔCt method, as previously3. Sequences of primers are shown in the Supplementary table.
Quantifying HSPCs
HSPCs were quantified by counting the number of kdrl:mCherry; gata2b:GFP double positive cells in floor of the DA in the region above the yolk extension in a 625μm confocal Z-stack encompassing the entire mediolateral segment of the aorta. The number of HSPCs per mm was calculated from this data. Confocal images were generated by stacking 1–4 individual Z-slices. When quantifying WISH data, the number of cmyb+ or runx1+ cells were counted above the yolk extension.
Human embryonic stem cell culture and HSPC differentiation
All experiments described in this study were approved by a research oversight committee (IRB/ESCRO Protocol #100210, PI Willert). Human embryonic stem (hES) cell H9 (WA09, NIH Registration number 062) cells were obtained from WiCell. Cells were maintained in Essential 8 (E8) media, with minor modifications, as previously described59. Plasmids encoding pools of shRNAs for FZD9 and WNT9A were obtains from ABM. The cell lines harboring shRNAs for control, WNT9A and FZD9 were generated by lentiviral transduction, as previously described59,60. Virally-infected cells were selected with puromycin (4 ug/mL), and differentiated to HSPCs as previously described30. Cells were dissociated and HSPCs were quantified by flow cytometry as previously described2.
APEX2-mediated proximity labeling
Chinese hamster ovary (CHO) cells were stably integrated with a CMV:Wnt9a construct. Conditioned medium from CHO cells with or without this construct was collected for two weeks, pooled together, filtered through a 0.22um filter and tested for Wnt9a activity by luciferase STF assay with Fzd9b. HEK293T cells were stably integrated with a CMV:Fzd9b-5GS-APEX2 construct. APEX2-mediated proximity labeling was carried out as described27. Briefly, confluent 150mm plates of CMV:Fzd9b-5GS-APEX2 cells were treated with biotin-phenol for a total of 30 minutes each, ending at the time of hydrogen peroxide treatment. Cells were treated with Wnt9a conditioned medium for 1 or 5 minutes, or with WT CHO conditioned medium for 5 minutes. Cells were treated with hydrogen peroxide for 1 minute, quenched, lysed, and biotinylated proteins enriched by streptavidin pulldown, as previously described.
Protein digestion
To denature the eluted proteins, an equal volume of 8 M Urea in 50 mM HEPES, pH 8.5 was added to each sample. Protein disulfide bonds were reduced by Dithiothreitol (DTT, Sigma) and alkylated with Iodoacetamide (Sigma) as previously described61. Proteins were precipitated using trichloroacetic acid and resuspended in 300 μl of buffer (1 M urea (Fisher), 50 mM HEPES, pH 8.5) for proteolytic digestion.
Proteins were serially digested with 30 μg of LysC overnight at room temperature, then with 3 μg of trypsin for 6 hours at 37 °C62, quenched by the addition of trifluoracetic acid (TFA, Pierce), and peptides were desalted with C18-StageTips extraction columns (Waters) as previously described62. Peptides were dried in a speed vac, then re-suspended in 50% Acetonitrile/5% formic acid and quantified by the Pierce Quantitative Colorimetric Peptide Assay (Thermo); an equal amount of each sample to be run on a pooled bridge channel63. Aliquots were dried under vacuum and stored at −80°C until they were labeled with Tandem Mass Tag (TMT) reagents.
TMT labeling
Peptides were labeled with 10-plex TMT reagents (Thermo) as previously described62. Briefly, TMT reagents were reconstituted in dry acetonitrile (Sigma) at 20 μg/mL. Dried peptides were re-suspended in 30% dry acetonitrile in 200 mM HEPES, pH 8.5, and 8 μL of the appropriate TMT reagent was added to the peptides. Reagent 126 (Thermo) was used as a bridge between mass spectrometry runs. Remaining reagents were used to label samples in a random order. Labeling was carried out for 1 hour at room temperature and was quenched by adding 9 μL of 5% hydroxylamine (Sigma) which was allowed to react for 15 mins at room temperature. Labeled samples were acidified by adding 50 μL of 1%TFA, pooled into appropriate 10-plex TMT samples and desalted with C18 Sep-Paks.
LC-MS2/MS3 Analysis
All LC-MS2/MS3 experiments were performed on an Orbitrap Fusion mass spectrometer (Thermo) with an in-line Easy-nLC 1000 (Thermo). Home-pulled, home-packed columns (100 mm ID x 30 cm, 360 mm OD) were used for analysis. Analytical columns were triple-packed with 5 μm C4 resin, 3 μm C18 resin, and 1.8 μm C18 resin (Sepax) to lengths of 0.5 cm, 0.5 cm, and 30 cm, respectively. Peptides were eluted with a linear gradient from 11 to 30% acetonitrile in 0.125% formic acid over 165 min at a flow rate of 300 nL/minute and heating the column to 60°C. Nano-electrospray ionization was performed by applying 2000 V through a stainless-steel T-junction at the inlet of the microcapillary column.
The mass spectrometer was operated in a data-dependent mode, with a survey scan performed over a mass to charge (m/z) range of 500–1200 at a resolution of 1.2 × 105 in the Orbitrap. The target automatic gain control (AGC) was set to 2 × 105 with a maximum inject time of 100 ms and an s-lens RF of 60. Top Speed mode was used to select the most abundant ions for tandem MS analysis. All data collected was centroided.
Ions above an intensity threshold of 5 × 105 were isolated in the quadrupole and fragmented using collision-induced dissociation (normalized energy: 30%) for MS2 analysis. MS2 fragments were detected in the ion trap using the rapid scan rate setting with an AGC of 1 × 104 and a maximum injection time of 35 ms.
For MS3 analysis, synchronous precursor selection was used to maximize quantitation sensitivity of the TMT reporter ions64. Up to 10 MS2 ions were simultaneously isolated and fragmented with high energy collision induced dissociation (normalized energy: 50%). MS3 fragment ions were analyzed in the Oribtrap with a resolution of 6 × 104. The AGC was set to 5 × 104 using a maximum injection time of 150 ms. MS2 ions 40 m/z below and 15 m/z above the MS1 precursor ion were excluded from MS3 selection.
Data Processing
Raw spectral data were processed using Proteome Discoverer 2.1.0.81 (Thermo). MS2 spectra were identified using the Sequest HT node65, searching against the Human Uniprot database (downloaded: 5/1½017) with the zebrafish Fzd9b sequence appended. False discovery rate (FDR) estimation was performed using a reverse decoy database66-68. Search parameters were as follows. Mass tolerances were set to 50 ppm and 0.6 Da for MS1 and MS2 scans, respectively. Full trypsin digestion was specified with a maximum of two missed cleavages per peptide. Modifications included static 10-plex TMT tags on peptide n-termini and lysine, static carbamidomethylation of cysteine and variable oxidation of methionine. Data were filtered to a 1% FDR at both the peptide and protein level.
The intensities of TMT reporter ions were extracted from the MS3 scans for quantitative analysis. Prior to quantitation, spectra were filtered to have an average signal to noise of 10 across all labels and an isolation interference less than 25%. Data were normalized in a two-step process as previously described63, by normalizing each protein the pooled bridge channel value and then normalizing to the median of each reporter ion channel and the entire dataset.
Statistics and Reproducibility
For APEX results, two-tailed student’s t-tests were used to determine significantly enriched proteins at each time point. If the variances between samples were determined to be unequal by an F-test, Welch’s correction was used. Significantly changing proteins were prioritized using pi score69, a metric that takes both p-value and fold-change into account. Gene ontology of the significant proteins was performed using the database for annotation, visualization and integrated discovery (DAVID) server70. For STF assays and qPCR or cell counting comparing more than two populations, one-way ANOVA, followed by post-test analysis were conducted. For qPCR or cell counting comparing only two populations, two-tailed student’s t-tests were used. All data analysis and statistical findings are available in the Supplementary Table 3.
Plasmids
Expression constructs were generated by standard means using PCR from cDNA libraries generated from zebrafish larvae at 24 hpf, or from hES cells; these were cloned into pCS2+, downstream of a CMV promoter, and upstream of IRES:mKate2. Addgene provided expression vectors for Cas9 (47929), guide RNAs (46759) and zebrafish ctnnb1 (17199).
Reagents
A list of reagents and catalogue numbers is available in Supplementary Table 2.
Data availability
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD010649 through MassIVE (MSV000082677). Summary data is seen in Extended data Figs. 6e, 6f, 7d, 7e. Source data for main and supplementary figures have been provided as Supplementary Table 3.
Previously published sequencing data that were re-analysed here are available from the European Nucleotide Archive under the accession number PRJEB4197 https://www.ebi.ac.uk/ena.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
C. Fine and J. Olvera for FACS assistance; R. Espin-Palazon, J. Nussbacher and I.J. Huggins for manuscript reading; I.J. Huggins for Cetuximab; N. Gohad for microscopy assistance; M. Boutros for cell lines. SG was supported by awards from the American Heart Association (14POST18340021) and the Leukemia and Lymphoma Society (5431–15). Research reported in this publication was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number K99HL133458 (awarded to SG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JR was supported by was supported in part by the UCSD Interdisciplinary Stem Cell Training Program (CIRM TG2–01154) and by the American Heart Association Predoctoral Fellowship 16PRE27340012. This work was supported in part by funding to KW from the UCSD Stem Cell Program and was made possible in part by the CIRM Major Facilities grant (FA1–00607) to the Sanford Consortium for Regenerative Medicine. DT was supported by Scholar Award (1657–13) from The Leukemia and Lymphoma Society and CIRM (RB4–06158). This project was additionally supported by NIH/NHLBI grant R01HL135205 (awarded to DT and KW) and by NIH/NIGMS grant R01GM110304 (awarded to KW). JMW was supported by Graduate Training in Cellular and Molecular Pharmacology Training Grant NIH T32 GM007752. Harvard Stem Cell Institute and NIDDK grant UH2/3DK107372 to IAD.
Footnotes
Competing Interests
The authors do not report any competing interests.
References
- 1.Nusse R & Clevers H Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Richter J et al. WNT9A Is a Conserved Regulator of Hematopoietic Stem and Progenitor Cell Development. Genes (Basel) 9(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grainger S et al. Wnt9a Is Required for the Aortic Amplification of Nascent Hematopoietic Stem Cells. Cell Rep 17, 1595–1606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ring L, Neth P, Weber C, Steffens S & Faussner A beta-Catenin-dependent pathway activation by both promiscuous “canonical” WNT3a-, and specific “noncanonical” WNT4- and WNT5a-FZD receptor combinations with strong differences in LRP5 and LRP6 dependency. Cell Signal 26, 260–267 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Voloshanenko O, Gmach P, Winter J, Kranz D & Boutros M Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. FASEB J (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlin JR, Robertson AL & Zon LI Efforts to enhance blood stem cell engraftment: Recent insights from zebrafish hematopoiesis. J Exp Med 214, 2817–2827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements WK & Traver D Signalling pathways that control vertebrate haematopoietic stem cell specification. Nature Reviews Immunology 13, 336–348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhen F, Lan Y, Yan B, Zhang W & Wen Z Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development (Cambridge, England) 140, 3977–3985 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Leung A et al. Uncoupling VEGFA functions in arteriogenesis and hematopoietic stem cell specification. Dev Cell 24, 144–158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butko E et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development 142, 1050–1061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi I et al. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512, 319–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damm EW & Clements WK Pdgf signalling guides neural crest contribution to the haematopoietic stem cell specification niche. Nat Cell Biol 19, 457–467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissa K & Herbomel P Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Bertrand JY et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luis TC et al. Wnt3a deficiency irreversibly impairs hematopoietic stem cell self-renewal and leads to defects in progenitor cell differentiation. Blood 113, 546–554 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Zhao C et al. Loss of β-Catenin Impairs the Renewal of Normal and CML Stem Cells In Vivo. Cancer Cell 12, 528–541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming HE et al. Wnt Signaling in the Niche Enforces Hematopoietic Stem Cell Quiescence and Is Necessary to Preserve Self-Renewal In Vivo. Cell Stem Cell 2, 274–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reya T, Duncan AW, Ailles L, Domen J & Scherer DC A role for Wnt signaling in self-renewal of haematopoietic stem cells. Nature 423, 409 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Willert K et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Malhotra S et al. Contrasting responses of lymphoid progenitors to canonical and noncanonical Wnt signals. J Immunol 181, 3955–3964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba Y et al. Constitutively active beta-catenin promotes expansion of multipotent hematopoietic progenitors in culture. Journal of Immunology (Baltimore, Md.: 1950) 177, 2294–2303 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Kirstetter P, Anderson K, Porse BT, Jacobsen SE & Nerlov C Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol 7, 1048–1056 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Scheller M, Huelsken J, Rosenbauer F, Taketo MM & Birchmeier W Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat. Immunol. 10, 1038 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Luis TC, Ichii M, Brugman MH, Kincade P & Staal FJ Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 26, 414–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luis TC et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell stem cell 9, 345–356 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Goessling W et al. Genetic Interaction of PGE2 and Wnt Signaling Regulates Developmental Specification of Stem Cells and Regeneration. Cell 136, 1136–1147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam SS et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veeman MT, Slusarski DC, Kaykas A, Louie SH & Moon RT Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Current biology: CB 13, 680–685 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Ablain J, Durand EM, Yang S, Zhou Y & Zon LI A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell 32, 756–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng ES, Davis R, Stanley EG & Elefanty AG A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nature Protocols 3, 768–776 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Janda CY, Waghray D, Levin AM, Thomas C & Garcia KC Structural Basis of Wnt Recognition by Frizzled. Science (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dijksterhuis JP et al. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J Biol Chem 290, 6789–6798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rulifson EJ, Wu CH & Nusse R Pathway specificity by the bifunctional receptor frizzled is determined by affinity for wingless. Mol Cell 6, 117–126 (2000). [PubMed] [Google Scholar]
- 35.Tamai K et al. LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Lawson ND & Weinstein BM In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Developmental Biology 248, 307–318 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Lewis JL et al. Reiterated Wnt signaling during zebrafish neural crest development. Development 131, 1299–1308 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Mishra R, Hanker AB & Garrett JT Genomic alterations of ERBB receptors in cancer: clinical implications. Oncotarget 8, 114371–114392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doody JF et al. Inhibitory activity of cetuximab on epidermal growth factor receptor mutations in non small cell lung cancers. Mol Cancer Ther 6, 2642–2651 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y & Lin S Essential role of SH3-domain GRB2-like 3 for vascular lumen maintenance in zebrafish. Arterioscler Thromb Vasc Biol 33, 1280–1286 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Osherov N & Levitzki A Epidermal-growth-factor-dependent activation of the src-family kinases. Eur J Biochem 225, 1047–1053 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Sultan M et al. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics 15, 675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blom N, Gammeltoft S & Brunak S Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294, 1351–1362 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Schafer B, Gschwind A & Ullrich A Multiple G-protein-coupled receptor signals converge on the epidermal growth factor receptor to promote migration and invasion. Oncogene 23, 991–999 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Prenzel N et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402, 884–888 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Tomlins SA, Bolllinger N, Creim J & Rodland KD Cross-talk between the calcium-sensing receptor and the epidermal growth factor receptor in Rat-1 fibroblasts. Exp Cell Res 308, 439–445 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto H, Komekado H & Kikuchi A Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev Cell 11, 213–223 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Blitzer JT & Nusse R A critical role for endocytosis in Wnt signaling. BMC Cell Biol 7, 28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Ye X, Guo N & Nathans J Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 139, 4383–4394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsieh JC, Rattner A, Smallwood PM & Nathans J Biochemical characterization of Wnt-frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc Natl Acad Sci U S A 96, 3546–3551 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho C, Smallwood PM & Nathans J Reck and Gpr124 Are Essential Receptor Cofactors for Wnt7a/Wnt7b-Specific Signaling in Mammalian CNS Angiogenesis and Blood-Brain Barrier Regulation. Neuron 95, 1221–1225 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y & Nathans J Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev Cell 31, 248–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eubelen M et al. A molecular mechanism for Wnt ligand-specific signaling. Science (2018). [DOI] [PubMed] [Google Scholar]
- 54.Lai MB et al. TSPAN12 Is a Norrin Co-receptor that Amplifies Frizzled4 Ligand Selectivity and Signaling. Cell Rep 19, 2809–2822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez A et al. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proceedings of the National Academy of Sciences 111, 1409–1414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jao LE, Wente SR & Chen W Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grainger S et al. CRISPR Guide RNA Validation In Vitro. Zebrafish (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwan KM et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Huggins IJ et al. The WNT target SP5 negatively regulates WNT transcriptional programs in human pluripotent stem cells. Nature Communications 8, 1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moya N, Cutts J, Gaasterland T, Willert K & Brafman David A., Endogenous WNT Signaling Regulates hPSC-Derived Neural Progenitor Cell Heterogeneity and Specifies Their Regional Identity. Stem Cell Reports 3, 1015–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haas JP Measurement of infection control department performance: state of the science. Am J Infect Control 34, 543–549 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Lapek JD Jr., Lewinski MK, Wozniak JM, Guatelli J & Gonzalez DJ Quantitative Temporal Viromics of an Inducible HIV-1 Model Yields Insight to Global Host Targets and Phospho-Dynamics Associated with Protein Vpr. Mol Cell Proteomics 16, 1447–1461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lapek JD Jr. et al. Defining Host Responses during Systemic Bacterial Infection through Construction of a Murine Organ Proteome Atlas. Cell Syst 6, 579–592 e574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McAlister GC et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eng JK, McCormack AL & Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 66.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Elias JE, Haas W, Faherty BK & Gygi SP Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2, 667–675 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Peng J, Elias JE, Thoreen CC, Licklider LJ & Gygi SP Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Xiao Y et al. A novel significance score for gene selection and ranking. Bioinformatics 30, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD010649 through MassIVE (MSV000082677). Summary data is seen in Extended data Figs. 6e, 6f, 7d, 7e. Source data for main and supplementary figures have been provided as Supplementary Table 3.
Previously published sequencing data that were re-analysed here are available from the European Nucleotide Archive under the accession number PRJEB4197 https://www.ebi.ac.uk/ena.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.