Abstract
Accumulating evidence indicates that the health impact of dietary phenolic compounds, including the principal grape-derived polyphenols, ( + )-catechin and ( − )-epicatechin, is exerted by not only the parent compounds but also their phenolic metabolites generated by the gut microbiota. In this work, a new high-throughput, sensitive and reproducible analytical method was developed employing ultra-high performance liquid chromatography coupled with triple quadrupole tandem mass spectrometry (UHPLC-QqQ-MS/MS) for the simultaneous analysis of 16 microbial-generated phenolic acid metabolites (PAMs) along with their precursors, catechin and epicatechin. Following optimizing the solvent system, LC conditions and MS parameters, method validation was carried out to evaluate the sensitivity, selectivity, accuracy and precision of the proposed method, and to ensure promising recovery of all analytes extracted from the matrix prior to bioanalysis. Results showed that the optimized analytical method allowed successful confirmation and quantitation of all analytes under dynamic multiple reaction monitoring mode using trans-cinnamic acid-d7 as an internal standard (I.S.). Excellent sensitivity and linearity were obtained for all analytes, with lower limits of detection (LLODs) and lower limits of quantification (LLOQs) in the ranges of 0.225–2.053 ng/mL and 0.698–8.116 ng/mL, respectively. By examining blank matrix spiked with standard mixture at different concentration levels, promising recoveries at two spiking levels (low level, 91.2–115%; high level 90.2–121%), and excellent precision (RSD < 10%) were obtained. This method was then successfully applied to an in vitro study where catechin/epicatechin-enriched broth samples were anaerobically fermented with gut microbes procured from healthy human donors. All sources of bacteria employed showed remarkable activity in metabolizing grape polyphenols and distinct variations in the production of PAMs. The successful application of this method in the in vitro fermentation assays demonstrates its suitability for high-throughput analysis of polyphenol metabolites, particularly catechin/epicatechin-derived PAMs, in biological studies.
Keywords: UHPLC-QqQ-MS/MS, Method validation, Grape polyphenols, Phenolic acid metabolites, Gut microbiota, In vitro fermentation
1. Introduction
Research involving the health benefits of dietary polyphenols continues to increase, echoing public awareness and consumer interest in foods and nutraceuticals rich in polyphenols. However, the actual in vivo bioefficacy of phenolic compounds, especially the polyphenolic constituents, remains unclear [1,2]. Among the most popular polyphenol-rich foods, grape-derived products encompass an abundant array of polyphenols, with proanthocyanidins (PACs), including monomeric flavan-3-ols, anthocyanidins, flavonols, stilbenes and phenolic acids, being the primary contributors [3]. Following oral ingestion, phenolic compounds are metabolized by phase I/II enzymes and gut microbiota, and as such the bioavailability of grape polyphenols can vary from very low to moderate levels depending on their chemical and physiological properties [4–6]. Considering that parent phenolic compounds are present at much lower abundances in biological fluids or tissues than their metabolites [7], their wide spectrum of bioactivities should be largely attributed to their metabolites rather than the native compounds. The extensive efforts devoted to elucidating the metabolic fates of dietary polyphenols [8] also propel the evolution of metabolomics research including the related bio-analytical techniques [9–12].
The human gastrointestinal (GI) microbiota represents a unique and versatile bio-reactor responsible for metabolism of the non-absorbed phenolic proportions, generating a wealth of microbial phenolic metabolites [8,13]. The combination of major monomeric PAC microbial metabolites has been recognized as an excellent biomarker for indicting profound colonic microbial activity towards grape polyphenols and for partially evaluating their bioefficacy in vivo [14]. Microbial-derived phenolic metabolites are generally comprised of derivatives of carboxylic acid, particularly hydroxybenzoic and hydroxycinnamic acids, representing a cluster of low molecular weight, highly polar and chemically distinct compounds [5,6]. Challenges in the analysis of these small polar organic acids reside in the difficulties in resolving chromatographic peaks, e.g. serious coelution and peak distortion problem [15], and problems with mass spectrometry (MS) detection, e.g. in-source fragmentation, ionization suppression and ion cluster formation [16]. The best metabolomics analytical method today appears to be based on tandem mass spectrometry (MS/MS), coupled with a gas or liquid chromatography system. More specifically, for quantitative analysis of known compounds, triple quadrupole (QqQ) and Q-trap MS, normally operated under the multiple reaction monitoring (MRM) or selective reaction monitoring (SRM) mode, are two of the preferred techniques [13,17]. The QqQ-MS/MS technique enables simultaneous profiling of multiple analytes with high selectivity and sensitivity and, as such, allows for large-scale screening of potential gut bacteria candidates with high capacity in bioconverting polyphenols into bioactive phenolic metabolites [18, 19].
To better understand the metabolic fates of grape polyphenols in human intestine following oral administration, this study aimed to develop and validate an efficient and reliable analytical method for the precise measuring of the most characteristic microbial PAMs derived from two major grape polyphenols, ( + )-catechin (( + )-C) and ( − )-epicatechin (( − )-EC), in biosamples.
2. Materials and methods
2.1. Standards and reagents
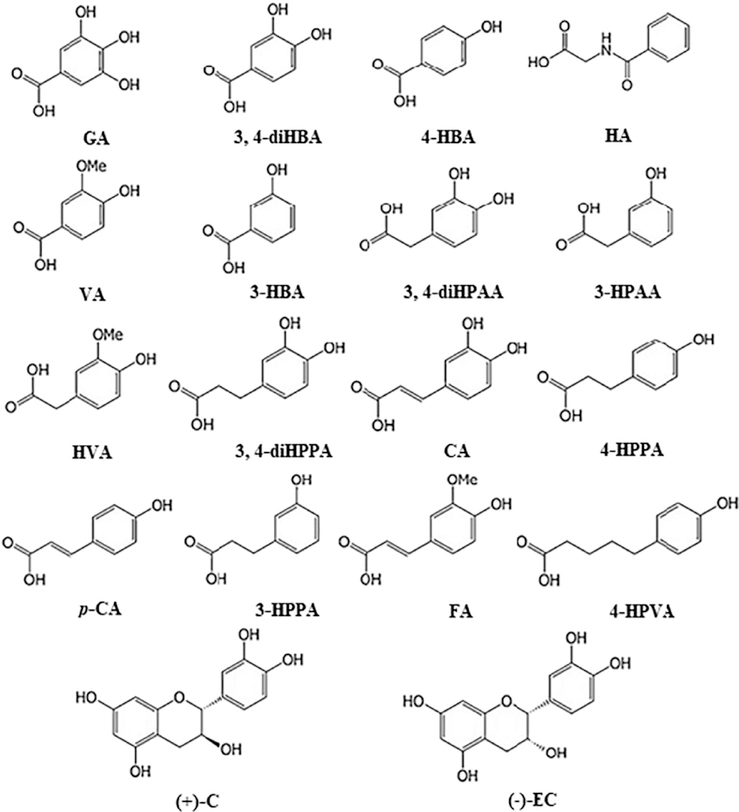
Phenolic standards including gallic acid (GA), caffeic acid (CA), trans-p-coumaric acid (p-CA), catechin (( + )-C), epicatechin (( − )-EC), dihydrocoumaric acid (diHCA), 3-(3,4-dihydroxyphenyl)propionic acid (3,4-diHPPA), 3,4-dihydroxybenzoic acid (3,4-diHBA), hippuric acid (HA), homovanillic acid (HVA), 3-hydroxybenzoic acid (3-HBA), 4-hydroxybenzoic acid (4-HBA), 3-hydroxyphenylacetic acid (3-HPAA), 3-(3-hydroxyphenyl)propionic acid (3-HPPA), vanillic acid (VA) and trans-cinnamic acid-d7 as well as ascorbic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA); 3,4-dihydroxyphenylacetic acid (3,4-diHPAA) and ferulic acid (FA) were from ChromaDex Inc. (Irvine CA); 5-(4-hydroxyphenyl)valeric acid (4-HPVA) was from Alfa Aesar (Lancashire, UK). The structures of two grape polyphenol precursors and phenolic acid metabolites investigated in this study are presented in Fig. 1. Acids and solvents (all HPLC Grade) including glacial acetic acid (AA), formic acid, acetonitrile (ACN), methanol (MeOH) and ethyl acetate were obtained from Fisher Scientific (Pittsburgh, PA, USA), and Pierce™ LC-MS water from Thermo Fisher Scientific (Waltham, MA, USA).
Fig. 1.
Chemical structures of grape polyphenol precursors (catechin and epicatechin) and phenolic acid metabolites included in the study. Abbreviations are presented in Table 1.
2.2. Standard solution preparation
Stock solution (ca. 0.5 mg/mL) of 18 phenolic compounds and the internal standard were prepared in 70% MeOH in water containing 0.1% formic acid. Stock solutions of phenolic compounds were aliquoted into 1.0 mL Eppendorf tubes and stored at −20 °C in dark. Working solutions were prepared by diluting stock solutions (after conditioned to room temperature) in 45% aqueous MeOH containing 0.1% formic acid. For preparing the calibration dilution series, all 18 standards were pipetted into one container and constituted to form a single standard mixture with each analyte at 20 μg/mL and the mixture was then serially diluted to 15 concentration levels (ca. 1.25 to 6000 ng/mL) in 45% aqueous MeOH containing 0.1% formic acid. An internal standard (I.S.) (ca. 20 μg/mL), trans-cinnamic acid-d7, was spiked into each dilution to a final concentration of 100 ng/mL. All standard solutions were filtered through a 0.45-μm Millex-FH membrane (Millipore, Bedford, MA, USA) before loaded onto a UHPLC column.
2.3. UHPLC QqQ MS/MS method optimization
2.3.1. Apparatus
Analysis of ( + )-catechin, ( − )-epicatechin and phenolic acids was performed on an Agilent 1290 Infinity II UHPLC (Agilent Technology, Palo Alto, CA, USA) system interfaced with an Agilent 6470 Triple Quadrupole Mass Spectrometer with an electrospray ionization (ESI) source. Chromatographic separation of compounds was achieved with a Waters Acquity UHPLC BEH C8 column (2 × 150 mm, 1.7 μm) (Milford, Massachusetts, USA) equipped with a Waters VanGuard Acquity C8 guard column (2.1 × 5 mm, 1.7 μm).
2.3.2. Optimization of LC-MS conditions
To ensure the best performance of the LC-MS system for individual analytes, optimization was carried out in regard to dilution solvent, LC conditions and MS-related parameters.
The methanol percentages of the diluent for preparing standard working solutions from stock solutions (Section 2.2) and for re-constituting phenolic compounds after sample extraction (Section 2.4.2) were examined. Water:MeOH (v/v) combinations of 4/96, 20/80, 40/60, 45/55, 50/50, 60/40 and 80/20 were tested and 45/55 was finally selected as the best combination. The subsequent optimization experiments were carried out using standard working solutions diluted with water:MeOH:formic acid (45/55/0.1, v/v/v).
To optimize LC-dependent conditions, we tested multiple mobile phase compositions focusing on the type and the concentration of mobile phase organic modifiers added to the aqueous and the organic phases. The following aqueous phase modifiers were tested: ammonium acetate (5 mM, at pH 3, 4 and 6.5), ammonium hydroxide (5 mM, pH 4), formic acid (0.1%, 0.2%, 1.0%, 2.0%) and AA (0.1%, 0.2%, 0.25%, 0.5%, 1.0%, 2.0%). AA (0.1%, 0.2% and 1.0%), and formic acid (0.1% and 1.0%) were also tested as additives to the organic phase. Following selection of the optimal mobile phase, elution gradient, column temperature and solvent flow rate were also adjusted.
The third part of optimization focused on source-dependent and compound-dependent MS operation parameters. The source-dependent parameters applied to all target compounds included sheath gas temperature, capillary voltage, multiplier voltage (delta EMV) and cell accelerator voltage (CAV). For individual analytes, parameters of precursor-product ion pair transition operated under multiple reaction monitoring (MRM) mode was tuned by the Agilent MassHunter Optimizer (software version B.07.00) using authentic standards for reference. Two of the most intense precursor-product ion transitions were identified. For the two transitions of each analyte, the one with higher response was selected to be the quantifier ion, while the other one was the qualifier ion.
2.3.3. Optimized LC-MS conditions and compound identification
After optimizing the LC-MS system, a binary mobile phase system consisting of phase A (0.2% AA in water) and phase B (0.1 % AA in ACN) and a flow rate at 0.3 mL/min were used for qualitative and quantitative analysis of targeted phenolic compounds. Thermostats of the column and the autosampler were set at 40 °C and 4 °C, respectively. Injection volume was 5 μL for all standards and biosamples studied. The LC gradient program for each run started at 4% (B%), held for 1.5 min before increasing to 12% in 12.5 min, to 90% in 1 min and held for another 2 min, and then returned to initial conditions in 1 min. The column was equilibrated for another 6 min before the next injection.
Mass spectral data acquisition was achieved under negative polarity (ESI −) and dynamic multiple reaction monitoring (dMRM) mode. Two specific transitions for each analyte were monitored over a 1-min delta retention time frame with a dwell time of 150 ms. Integral MS parameters were set as follows: ESI capillary voltage at −2.5kV, nozzle voltage at −1.0 kV, nebulizer gas (N2) pressure at 30 psi, dry gas temperature at 350 °C with a flow rate of 12.0 L/min, sheath gas temperature at 200 °C with a flow rate of 12.0L/min.
Identification of PAMs, ( + )-C and ( − )-EC was accomplished by comparing their MRM precursor-product ion pair transitions (both quantifier and qualifier) and retention times with those of the authentic standards. Quantitation was achieved with calibration curves established using the analyte-to-I.S. peak area of quantifier ions.
2.3.4. Method validation
To ensure analytical consistency, in most cases the analytical sequence consisted of a calibration standard set, quality control (QC) samples and biological samples. All QC samples and biological samples were processed with the same procedures each time. Four analysis sequences were completed within one month with each preparation injected twice.
2.3.5. Selectivity
Selectivity was evaluated by comparing the MS chromatograms of blank broth, broth spiked with 18 analytes and an I.S. With the final chromatographic conditions and MRM transition used, all the analytes should be resolved without interference from the matrix at the retention time and both mass transitions of the analytes, also as compared to the standards analyzed in solvent.
2.3.6. Calibration, linearity and sensitivity
The standard mixtures were analyzed using the optimized LC-MS method under dMRM mode. The calibration standard mixtures were prepared at 15 concentration levels ranging from ca. 1.25 to 6000 ng/mL. The analyte-to-I.S. peak area ratios were plotted against the exact concentration of individual analyte spiked in blank broth to establish calibration curves with four replicates at each concentration. Due to the diverse concentration levels of individual analytes in different biosamples, two calibration curves were constructed for each analyte, corresponding to the lower (ca. 1.25–1000 ng/mL) and the higher (ca. 1000–6000 ng/mL) concentration ranges, using linear regression and the origins were not forced through zero.
Sensitivity of the method were evaluated by investigating the response of target analytes in consecutive dilutions of a concentrated working solution (in solvent) until the signal-to-noise (S/N) ratio of individual analyte reached > 3 for lower limit of detection (LLOD), and > 10 for lower limit of quantification (LLOQ).
2.3.7. Accuracy and precision
For quantitative analyses of diverse phenolic compounds in biological samples, method accuracy and precision were evaluated by analyzing six replicates of blank broth samples and spiked QC samples at two concentration levels, LQC and HQC. Specifically, 10 μL of 20 μg/mL or 200 μg/mL standard mixture were spiked into 1 mL of blank broth to achieve a final concentration at ca. 200 ng/mL for LQC, or a final concentration at ca. 2000 ng/mL for HQC, with the addition of 5 μL of LS. (final concentration at ca. 100 ng/mL). Precision was expressed as relative standard deviation (RSD, %) of the measured concentrations in spiked replicates. Accuracy was reported as relative error (RE, %) between the measured and the actual concentrations spiked, calculated using the following equation:
2.3.8. Recovery and matrix effect
Recoveries of analytes from bacterial broth samples were also determined at two QC levels as aforementioned. Matrix effect was evaluated by comparing the measured peak area from analytes extracted from spiked broth samples and the theoretical value obtained with standards prepared in solvent. Six independent replicates were prepared and injected into the LC-MS system in duplicate. Recovery was calculated using the following formula:
where “Peak area (spiked matrix/blank matrix/solvent)” refers to the peak area of target quantifier ion observed in standard-spiked broth/non-spiked blank broth/standards dissolved in 45% aqueous MeOH containing 0.1% formic acid.
2.4. Method application to analysis of microbial phenolic acid metabolites
2.4.1. In vitro anaerobic fermentation of catechins by human gut microbiota
A nutrient-rich broth (see Supplementary material 1 for the detailed composition) with or without the supplementation of ( + )-C and (−)-EC was inoculated with gut microbiota from a healthy human donor and incubated under anaerobic conditions at 37 °C for 24 h. Blank nutrient broth sample, and broth samples incubated in the absence of bacteria with or without ( + )-C and ( − )-EC served as controls. After incubation, all samples were centrifuged at 4000 ×g for 5 min. The clarified bacterial broth samples were then recovered and immediately acidified with formic acid to a final concentration of 0.2%.
2.4.2. Preparation of phenolic extracts
For extracting phenolic compounds from bacterial broth, 500 μL of bacterial broth was acidified with 100 μL of 4 M HCl solution and spiked with 5 μL of trans-cinnamic acid (d7) solution (20 μg/mL in 70% MeOH containing 0.1% formic acid), and the cocktail was then mixed well. The mixture was then extracted with ethyl acetate (500 μL), followed by vortexing vigorously for 1 min, and centrifuged at 3000 ×g for 5 min using a micro-centrifuge. The upper organic phase (450 μL) was transferred to a 1-dram glass vial. The aqueous phase was extracted twice more with ethyl acetate (500 μL). All the recovered organic supernatants were combined, mixed with 10 μL of 2% ascorbic acid and dried under a gentle stream of nitrogen. The residue was reconstituted in 1000 μL of 45% methanol containing 0.1% formic acid and centrifuged at 16,000 ×g for 10 min. For each sample extract, 5 μL was injected into the LC-MS system for analysis. Each extract was injected twice to prevent any instrumental error that may affect identification and quantitation of analytes. The final measured concentrations were multiplied by two to compensate for the two-fold dilution made during extraction.
3. Results and discussion
3.1. UHPLC-QqQ-MS/MS method development and optimization
To meet the ever-growing demands for large-scale identification of target metabolites in biological samples and to better understand the actual metabolites responsible for nutraceutical and medicinal importance of grape-derived products, a sensitive and reproducible UHPLC-QqQ-MS/MS method was developed for the analysis of major phenolic acid metabolites derived from grape polyphenols. Optimization of the method was achieved by fine-tuning multiple key parameters closely related to chromatographic and mass spectrometric behaviors. Structural information of 18 phenolic compounds and an I.S., corresponding retention time, optimized MS parameters, and precursor-product ion pair transitions specific to individual analytes are summarized in Table 1.
Table 1.
Structural information, retention time and the optimized LC-QqQ/MS parameters for analyzing target phenolic compounds.
| Compound (abbreviation) | Formula | Retention time (min) | MS/MS transition (dMRM) |
Fragmentor voltage (V) | Collision energy (V) | |
|---|---|---|---|---|---|---|
| Precursor ion (m/z) | Product ion (m/z) (quantifier/qualifier) | |||||
| C6C1 | ||||||
| Gallic acid (GA) | C7H6O5 | 2.24 | 169.0 | 125.0/79.1 | 86 | 12/24 |
| 3,4-Dihydroxybenzoic acid (3,4-diHBA) | C7H6O4 | 4.24 | 153.0 | 109.1/108.1 | 86 | 12/28 |
| 4-Hydroxybenzoic acid (4-HBA) | C7H6O3 | 7.11 | 137.0 | 93.1/80.1 | 74 | 16/20 |
| Hippuric acid (HA) | C9H9NO3 | 8.65 | 178.1 | 134.1/77.1 | 80 | 8/16 |
| Vanillic acid (VA) | C8H8O4 | 9.16 | 167.0 | 152.0/108.1 | 80 | 12/16 |
| 3-Hydroxybenzoic acid (3-HBA) | C7H6O3 | 9.91 | 137.0 | 93.1/N.D. | 74 | 16/8 |
| C6C2 | ||||||
| 3, 4-Dihydroxyphenylacetic acid (3,4-diHPAA) | C8H8O4 | 5.87 | 167.0 | 123.1/N.D. | 54 | 4/12 |
| 3-Hydroxyphenylaeetie acid (3-HPAA) | C8H8O3 | 10.75 | 151.0 | 107.1/105.0 | 48 | 4/0 |
| Homovanillic acid (HVA) | C9H10O4 | 11.22 | 181.1 | 137.1/122.1 | 58 | 4/12 |
| C6C3 | ||||||
| 3-(3,4-Dihydroxyphenyl)propionic acid (3,4-diHPPA) | C9H10O4 | 8.93 | 181.1 | 137.1/59.1 | 72 | 8/16 |
| Caffeic acid (GA) | C9H8O4 | 9.51 | 179.0 | 135.1/97.0 | 88 | 4/16 |
| Dihydrocoumaric acid (diHCA) | C9H10O3 | 13.87 | 165.1 | 119.1/121.1 | 88 | 12/8 |
| trons-p-Coumaric acid (p-CA) | C9H8O3 | 14.18 | 163.0 | 119.1/93.1 | 80 | 12/16 |
| 3-(3-Hydroxyphenyl)propionic acid (3-HPPA) | C9H10O3 | 15.56 | 165.1 | 121.1/106.1 | 88 | 16/20 |
| Ferulic acid (FA) | C10H10O4 | 15.96 | 193.1 | 178.1/134.1 | 88 | 8/24 |
| trans-Cinnamic acid-d7 (I.S.) | C9D8O2 | 16.22 | 154.2 | 110.1/82.2 | 74 | 8/20 |
| C6C5 | ||||||
| 5-(4-Hydroxyphenyl)valeric acid (4-HPVA) | C11H14O3 | 16.15 | 193.1 | 175.2/149.0 | 86 | 12/12 |
| Flavanol | ||||||
| Catechin (( + )-C) | C15H14O6 | 8.03 | 289.1 | 245.2/123.1 | 134 | 12/32 |
| Epicatechin (( − )-EC) | C15H14O6 | 11.88 | 289.1 | 245.2/203.1 | 134 | 12/20 |
I.S., internal standard.
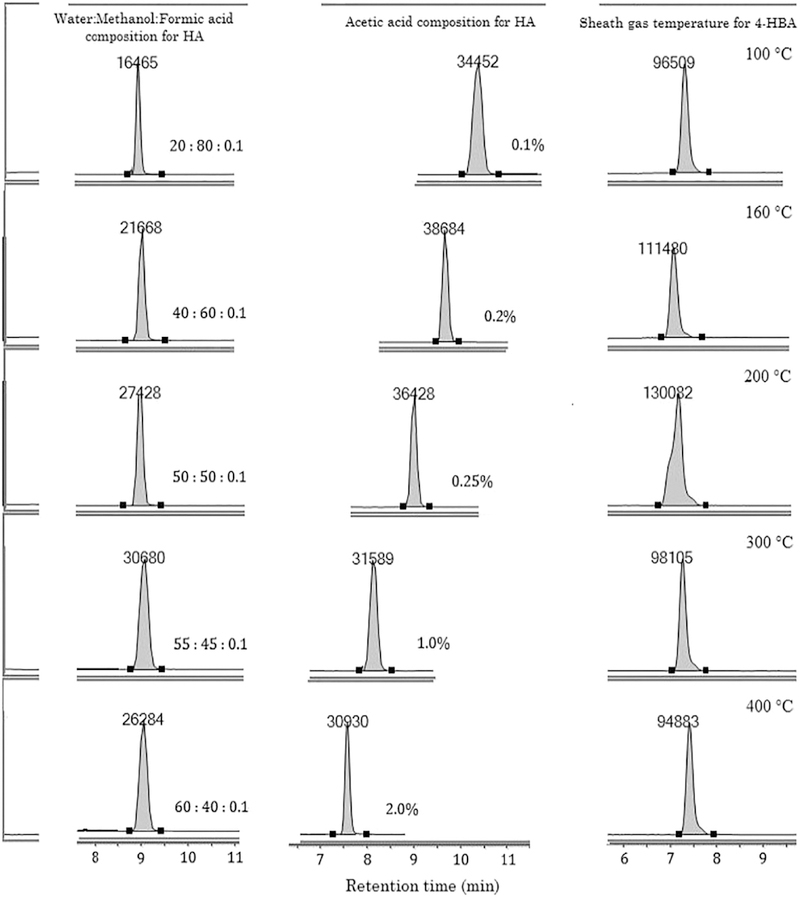
In addition to LC-MS methodologies, we also optimized the composition of the dilution solvent, which can greatly affect the injection solution composition, compound solubility in matrix and in mobile phase, peak shape and MS detector response [20,21]. Preliminary tests in SIM mode indicated that the QqQ analyzer was unable to acquire strong precursor ion signals for some of the analytes, particularly 3, 4-diHPAA, 3-HBA and diHCA. In addition, peak shape of 4-HPVA, diHCA and ( − )-EC was distorted when using high-water-content diluent (H20% > 60%). Thus, the effects of varying MeOH percentages in the dilution solvent for preparing standard working solutions and for reconstituting phenolic compounds extracted from biosamples were investigated. As illustrated in Fig. 2 (left column), a higher organic content supported much better ionization and MS response and improved peak shape compared to the using of high-water-content solvents. The optimum water:MeOH composition was found to be 45/55 (v/v), with the addition of 0.1% formic acid as modifier that helps sharpen peaks and prevents degradation of phenolic compounds during sample preparation.
Fig. 2.
Representative SIM chromatograms of selected phenolic compounds (4-hydroxybenzoic acid (4-HBA) and hippuric acid (HA) obtained under different optimization conditions showing changes in instrument response (arbitrary unit), peak shape and retention time (min). Left column: dilution solvent composition for preparing standard working solutions and for reconstituting phenolic extract; Middle column: acetic acid concentration in aqueous mobile phase; Right column: sheath gas temperature of the QqQ analyzer.
Another influential factor on LC-MS performance is mobile phase composition, especially the modifier, which can dramatically affect MS ionization efficiency. The mobile phase ionic strength and pH are known to influence chromatographic behaviors (analyte solubility, retention, resolution and peak shape), and MS performance (selectivity, reproducibility and sensitivity) [22,23]. To date, the preferred modifiers have mainly been volatile organic acids, such as acetic acid and formic acid [22,24], although neutral or alkaline additives have also been reported [23,25]. Following the investigation of a series of mobile phase compositions, the results indicated that the addition of low concentrations of acid (AA and formic acid) to aqueous phase greatly improved peak shape, detector signal intensity and S/N ratio of the precursor ion detected under SIM mode. In terms of peak shape and sensitivity, AA exhibited advantages over formic acid at concentrations of 0.1 % and 0.2%. As shown in Fig. 2 (middle column), at lower AA percentages (< 1.0%), the response to HA (m/z at 178.1), for example, was much stronger than that obtained at 1.0% or 2.0%, and retention time of HA was delayed. Theoretically, the target compounds are all Lewis acids and should better ionize under alkaline conditions. To test this hypothesis, we also tested the influence of two weak bases (ammonium acetate and ammonium hydroxide) on the ionization and retention of target analytes. However, results showed distorted peaks, poor ionization and serious coelution of the target compounds. Considering all these factors and results, AA was selected as the modifier, and the analytical performance of organic phase (ACN) with the addition of varying amounts of AA was also similarly optimized. The optimal mobile phase system was 0.2% AA in water and 0.1% AA in ACN. Afterwards, the gradient elution program was also tuned to facilitate the separation of analytes and to attain better resolution and MS response.
3.2. Triple quadrupole MS conditions
One great challenge in detecting and quantifying PAMs in complicated matrix originates from their high polarity, low retention in reversed phase LC column and the tendency of becoming clustered during ionization [15,24]. In this investigation, the operating parameters of the QqQ analyzer, including source-dependent and compound-dependent ones, were fully optimized to obtain the best MS performance for the analysis of PAMs and their precursors (Table 1).
Source-dependent parameters affect the integral MS performance for all target compounds. Both positive and negative polarities were tested to ensure efficient ionization of target analytes and acquisition of adequate MS responses. Negative mode was finally selected since it facilitated ionization of the majority of analytes although 3, 4-diHPAA, p-CA and ( + )-C gave higher responses in positive mode (data not shown). One possible explanation for the polarity preference is that polyphenols and their metabolites do not contain any nitrogen atom (except for HA), and thus ionization (protonation) is more difficult in the positive mode than in the negative mode [26]. By tuning delta EMV, the additional voltage applied to the MS detector, from 0 to 350 V, an 8-fold increase in S/N was observed. In excess of 350 V, there was compromise in S/N ratios despite of minor augmentation in the response to certain analytes. By adjusting sheath gas temperatures, the MS performance was further improved. Taking 4-HBA as an example (Fig. 2, right column), the peak area obtained at the default temperature 400 °C was only 72% of that obtained at 200 °C. Moreover, the precursor ion of 3, 4-diHPAA (m/z at 167.0), diHCA (m/z at 165.1) and GA (m/z at 160.0) were not detected in high abundance at 400 °C under SIM mode, while when the temperature was tuned to 200 °C, the signal increased by more than three times. This could be attributed to the reduced in-source fragmentation at a lower temperature. As Ostrowski et al. pointed out, registration the low molecular mass compounds with an ESI method, as employed in our study, can easily trigger bond cleavage of parent compounds, reducing abundances of these ions, and consequently lowering detector sensitivity [16]. Taking these factors into consideration, the generic MS parameters were optimized to produce high peak intensity at high S/N ratios for most analytes, as described in Section 2.3.3.
To finely adjust the compound-dependent parameters, i.e. the MRM transitions specific to each analyte, the MassHuntcr Optimizer was employed. It first selected the most abundant [M – H]− ion under MS2 scan mode as the precursor ion, after which the fragmentor voltage and collision energy (CE) were tuned stepwisely by a difference of 2 V in order to acquire two strongest precursor-product ion transitions. For the two transitions of each analyte, the one with higher response was selected as the quantifier for quantification purpose and the other one as the qualifier for confirmation. It should be noted that fragmentation of 3-HBA and 3, 4-diHPAA to form a second product ion with a strong signal was not successful even when the upper limit of CE was set to 60 V. This was not surprising, given that no qualifier ion for either of these compounds has been reported under negative mode in other related studies [26,27]. Since MS response to the respective MRM transition for 3-HBA and 3, 4-diHPAA was very strong and both were resolved without interference from the broth matrix at their respective retention time, 3-HBA and 3, 4-diHPAA were confirmed and quantified using a single ion pair transition. In addition, the efficiency and reproducibility of the developed method can also be attributed to the advanced dynamic MRM mode. In contrast to traditional MRM which continuously performs ion pair transition scans throughout the entire run, dMRM allows peak clusters to be better resolved, and MRM transitions to be more accurate and sensitive due to the longer dwell time around the expected retention time of the analyte [28].
3.3. Method validation
Following method optimization (parameters summarized in Section 2.3.3 and Table 1), the proposed LC-MS method was validated in terms of selectivity, linearity, low limit of detection/quantification (LLOD/LLOQ), accuracy, precision and recovery. Method validation was carried out following method optimization and complied with FDA guidelines [29], Validation results are presented in Table 2 with detailed calibration curve parameters of target compounds included in Supplementary material 2.
Table 2.
Calibration parameters, sensitivity, recovery, accuracy and precision of the optimized LC-QqQ/MS method.
| Compound | Calibration range | LLOD (ng/ mL) | LLOQ (ng/ mL) | Linear dynamic range | R2 | Spiked amount (ng/ mL) | Accuracy (%) | Precision (%) | Recovery (%) |
|---|---|---|---|---|---|---|---|---|---|
| GA | Low | 0.35 | 1.02 | 5.100–2040 | 0.998 | 200 | −3.82 | 4.38 | 115 (8.82) |
| High | 2040–6120 | 0.995 | 2000 | −17.1 | 6.53 | 121 (10.2) | |||
| 3,4-diHBA | Low | 0.62 | 2.55 | 2.560–2040 | 0.998 | 200 | 3.31 | 3.21 | 92.0 (7.41) |
| High | 2040–6120 | 0.999 | 2000 | 7.49 | 3.65 | 102 (5.32) | |||
| 3,4-diHPAA | Low | 1.23 | 5.01 | 2.652–2080 | 0.999 | 200 | −15.5 | 8.96 | 108 (12.0) |
| High | 2080–6240 | 0.996 | 2000 | −17.7 | 4.13 | 116 (6.34) | |||
| 4-HBA | Low | 0.29 | 0.97 | 1.325–1060 | 0.999 | 200 | – | – | – |
| High | 1060–6360 | 0.998 | 2000 | −9.23 | 4.55 | 102 (1.87) | |||
| ( + )-c | Low | 0.41 | 1.32 | 1.325–1060 | 0.999 | 200 | 4.57 | 9.69 | 91.2 (2.78) |
| High | 1060–6360 | 0.996 | 2000 | 2.89 | 4.54 | 95.0 (6.44) | |||
| 3,4-diHPPA | Low | 0.51 | 2.60 | 5.204–1040 | 0.998 | 200 | −8.75 | 12.2 | 110 (9.43) |
| High | 1040–6240 | 0.997 | 2000 | −20.8 | 3.16 | 117 (6.56) | |||
| HA | Low | 0.42 | 1.29 | 2.505–1002 | 0.999 | 200 | – | – | – |
| High | 1002–6012 | 0.998 | 2000 | −3.57 | 3.77 | 114 (9.02) | |||
| VA | Low | 0.75 | 2.36 | 2.602–1040 | 0.999 | 200 | 5.82 | 0.98 | 104 (2.84) |
| High | 1040–6240 | 0.999 | 2000 | 2.21 | 4.05 | 102 (8.04) | |||
| CA | Low | 0.25 | 1.08 | 2.464–980.1 | 0.999 | 200 | −11.0 | 9.55 | 102 (2.90) |
| High | 980.1–6020 | 0.997 | 2000 | −7.81 | 4.44 | 108 (4.15) | |||
| ( − )-EC | Low | 0.51 | 1.60 | 1.275–1020 | 0.999 | 200 | 12.6 | 5.81 | 93.6 (4.59) |
| High | 1020–6120 | 0.998 | 2000 | 14.3 | 5.35 | 91.1 (7.18) | |||
| HVA | Low | 0.63 | 1.62 | 2.700–2160 | 0.999 | 200 | −3.28 | 5.90 | 100 (5.83) |
| High | 2160–6480 | 0.999 | 2000 | −6.48 | 1.58 | 106 (2.62) | |||
| 3-HBA | Low | 1.25 | 3.75 | 2.452–1020 | 0.999 | 200 | −0.67 | 5.12 | 108 (5.42) |
| High | 1020–6120 | 0.998 | 2000 | −1.96 | 2.81 | 102 (2.25) | |||
| 3-HPAA | Low | 0.74 | 2.07 | 2.718–1040 | 0.999 | 200 | −3.10 | 4.33 | 106 (6.78) |
| High | 1040–6240 | 0.999 | 2000 | −1.85 | 1.76 | 94.6 (4.10) | |||
| diHCA | Low | 2.05 | 8.11 | 10.80–1080 | 0.997 | 200 | −15.9 | 4.70 | 112 (5.45) |
| High | 1080–6480 | 0.995 | 2000 | −19.9 | 2.19 | 116 (4.73) | |||
| p-CA | Low | 0.22 | 0.69 | 1.245–996 | 0.999 | 200 | −1.24 | 1.89 | 101 (5.14) |
| High | 996–5976 | 0.996 | 2000 | −5.74 | 4.22 | 100 (3.45) | |||
| FA | Low | 0.50 | 1.51 | 1.374–1080 | 0.999 | 200 | 4.96 | 4.67 | 96.6 (5.80) |
| High | 1080–6480 | 0.999 | 2000 | 6.04 | 7.84 | 93.6 (5.15) | |||
| 3-HPPA | Low | 0.31 | 2.65 | 5.306–1060 | 0.999 | 200 | – | – | – |
| High | 1060–5360 | 0.998 | 2000 | 4.27 | 3.19 | 90.2 (4.94) | |||
| 4-HPVA | Low | 0.76 | 2.61 | 2.607–1040 | 0.999 | 200 | 6.20 | 3.40 | 94.4 (4.13) |
| High | 1040–5240 | 0.996 | 2000 | 8.25 | 2.93 | 96.8 (2.20) | |||
| trans-Cinnamic acid-d7 (I.S.) | 0.23 | 0.71 | 1.272–1018 | 0.999 | – | – | – | – |
Data are expressed as means of six independent replicates. Standard deviations for Recoveries (%) were shown in parentheses.
Abbreviations are the same as in Table 1. LLOD, lower limit of detection; LLOQ, lower limit of quantification.
Calibration concentrations were divided into two ranges (low, ca. 1.25–1000 ng/mL and high, ca. 1000–6000 ng/mL), with optimum fit-to-line linear response. Some values for the 200 ng/mL spiked samples were null (−) due to the high response from the amount of the compounds existing endogenously.
3.3.1. Selectivity
Selectivity of the method was evaluated by comparing the SIM and MRM chromatograms of blank broth samples with those obtained from the spiked ones. No interference was observed at the retention time of individual analytes and I.S. from substances existing endogenously, i.e. there were no peaks overlapping within the 1-min delta retention time frame operated under dMRM mode, and complete separation of all analytes was achieved using the optimized method described. These characteristics indicate promising specificity and selectivity with our method.
3.3.2. Calibration, linearity and sensitivity
Calibration curve parameters, coefficient of determination (R2), LLODs and LLOQs of all target compounds were obtained with authentic standard solutions analyzed under the optimized LC-MS conditions (Table 2). Great calibration linearity was achieved over two concentration ranges for each compound present at low (ca. 1.25 to 1000 ng/mL) and high (ca. 1000 to 6000 ng/mL) levels, with coefficients of determination above 0.994 in all instances. The linear dynamic ranges were separated into two to obtain better fitted lines, particularly at higher concentrations. The measured LLODs and LLOQs were in the ranges of 0.22–2.05 ng/mL and 0.69–8.11 ng/mL, respectively, evincing the great sensitivity achieved with this analytical method. The lowest LODs/LOQs were obtained for three compounds t-cinnamic acid, CA and p-CA, possibly attributed to the same skeletal structure. We also tried to perform the matrix-matching calibration test, but the endogenous presence of high level of a few phenolic acids, namely 4-HBA, HA, VA, diHCA and 3-HPPA, greatly complicated the study. Consequently, calibration series were only prepared in the dilution solvent. An efficient and well-validated method for profiling polyphenol microbial metabolites developed by Gasperotti et al. included 19 PAMs [27], while our method was able to reach much lower LODs and LOQs, considering the different injection volumes in their study (10 μL) and ours (5 μL).
3.3.3. Accuracy and precision
For GA, 3, 4-diHBA, 3, 4-diHPAA, 4-HBA, HA, VA, diHCA, p-CA, FA and 3-HPPA, which existed endogenously in blank broth, the determination of accuracy and precision was done by subtracting content in blank broth from that in spiked broth samples. Thus, the corrected concentrations corresponded to the net changes occurred before and after in vitro fermentation. The values of accuracy, measured at two QC concentrations and expressed as relative error (RE, %), were within ± 12% in most cases, except for the slightly higher values determined for GA, 3, 4-diHPAA, 3, 4-diHPPA and diHCA (Table 2). For almost all compounds, the precision results, expressed as RSD (%), fell below 10%. These results met the criteria set forth in FDA guidelines [29], indicating that phenolic contents measured in samples using the proposed method should be consistent and exact at both low and high concentrations.
3.3.4. Recovery and matrix effect
Consistent and satisfactory results for recovery evaluation at different concentration levels are regarded as one prerequisite for a well-established analytical method. The recovery of spiked samples was similarly measured as for accuracy. By using the proposed method, we obtained excellent recoveries ranging from 91.2% to 115% at LQC, and 90.2% to 121% at HQC (Table 2). The mean recovery values for all analytes were 103%. Complex matrices can exert serious interfering effects on the stability of target compounds during processing, and the extraction efficiencies, which may account for the slightly over-ranged values obtained for some compounds [27,30]. Prior research using the traditional LC-MS/MS instrumentations has demonstrated the importance of matrix effects [31,32]. As such, we carefully considered potential matrix effects during method development to ensure minimal ion suppression and to reduce chances that matrix components and our analytes share the same MRM transitions or similar retention times. In the recovery test, matrix effects were also taken into account by comparing the analyte response obtained from spiked blank broth with those obtained from authentic standard prepared in pure solvent. There was no significant signal suppression or augmentation observed for all analytes studied. This is in agreement with the minimal matrix effects in various complex bio-matrices as reported by Hurtado-Gaitán et al., who focused on grapevine stilbcncs while employing a similar state-of-the-art analytical instrument as ours [33]. Ion suppression of a few phenolic compounds was however observed by Caprioli et al. also using the advanced analytical instrumentation but this was likely due to the high protein and carbohydrate content found in their pulse extracts [34]. Overall, the holistic performance of the extraction process and LC-MS method should be considered satisfactory, and analyses of biological samples containing the broth matrix were not affected by the matrix effect.
3.4. Method application
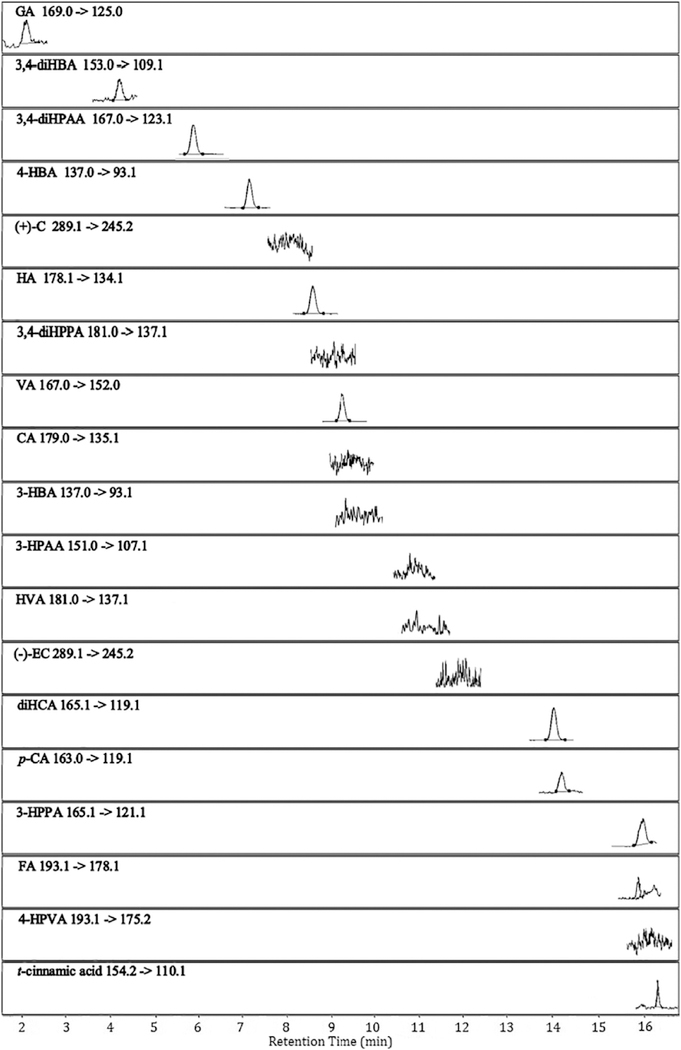
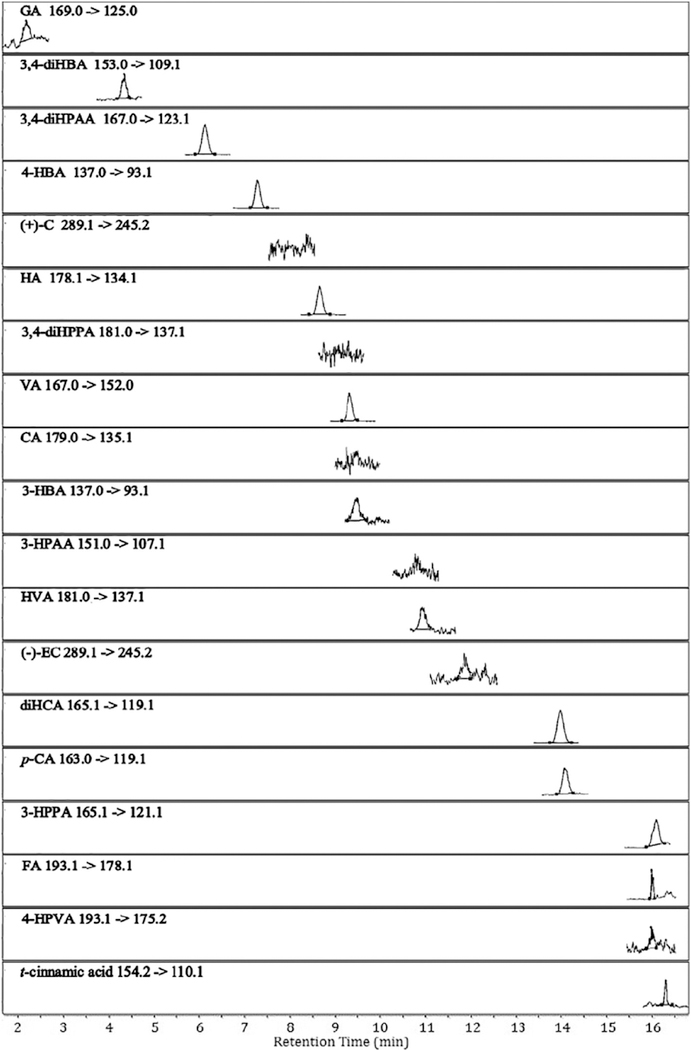
Using the proposed UHPLC-QqQ-MS/MS method, two precursor polyphenols, ( + )-C and ( − )-EC, and their characteristic PAMs (structures shown in Fig. 1) were successfully identified and quantified in ( + )-C/ ( − )-EC-enriched nutrient broth anaerobically fermented with gut microbiota from health human donors. This allows us to acquire a comprehensive metabolic profile of the two major monomeric PACs in grape-related products. The developed analytical method proved to be efficient, sensitive and reproducible for the quantitative analysis of ( + )-C, ( − )-EC and their phenolic acid catabolites in bacterial broth. All analyte peaks were well resolved within 16 min without interference from endogenous components in the matrix. The representative MRM chromatograms showing the precursor-product ion pair transitions of 18 target phenolic compounds, and an I.S. in a blank and a fermented bacterial sample are presented in Figs. 3 and 4, respectively. Detailed quantitation results are presented in Table 3.
Fig. 3.
Representative LC-MS chromatograms obtained under dynamic MRM mode showing precursor/product ion transitions of target phenolic compounds and the internal standard (trans-cinnamic acid) detected in a blank broth sample. Abbreviations are presented in Table 1.
Fig. 4.
Representative LC-MS chromatograms obtained under dynamic MRM mode showing precursor/product ion transitions of target phenolic compounds and the internal standard (trans-cinnamic acid) in a bacterial broth sample. The broth was enriched with catechin and epicatechin followed by fermentation with gut microbiota from a healthy human donor. Abbreviations are presented in Table 1.
Table 3.
Concentrations of phenolic acid metabolites and precursor polyphenols (catechin/epicatechin) detected in blank broth and bacterial fermented broth samples.
| Sample ID | GA | 3,4-diHBA | 3,4-diHPAA | ( + )-C | 4-HBA | 3,4-diHPPA | HA | VA | CA | ( − )-EC | HVA | 3-HBA | 3-HPAA | diHCA | p-CA | FA | 3-HPPA | 4-HPVA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blank | 407 | 151 | 619 | N.D. | 909 | N.D. | 1249 | 1152 | N.D. | N.D. | N.D. | N.D. | N.D. | 1067 | 96.9 | 59.4 | 1213 | N.D. |
| (2.44) | (4.24) | (11.4) | (−) | (25.4) | (−) | (24.8) | (31.3) | (−) | (−) | (−) | (−) | (−) | (11.8) | (0.83) | (0.96) | (2.87) | (−) | |
| Ctl ( − ) | 719 | 138 | 631 | N.D. | 972 | N.D. | 1289 | 1129 | N.D. | N.D. | N.D. | N.D. | N.D. | 1184 | 97.4 | 63.1 | 1281 | N.D. |
| (10.7) | (6.35) | (24.0) | (−) | (47.8) | (−) | (17.0) | (50.6) | (−) | (−) | (−) | (−) | (−) | (14.1) | (2.99) | (1.40) | (14.8) | (−) | |
| Ctl ( + ) | 963 | 128 | 631 | 42389 | 912 | N.D. | 1276 | 1059 | N.D. | 42710 | N.D. | N.D. | N.D. | 1164 | 93.5 | 61.2 | 1282 | N.D. |
| (30.6) | (6.65) | (6.66) | (271) | (17.2) | (−) | (20.6) | (11.0) | (−) | (389) | (−) | (−) | (−) | (3.71) | (0.88) | (0.73) | (7.67) | (−) | |
| Donor 1 ( − ) | 772 | 570 | 637 | N.D. | 1017 | N.D. | 13.4 | 1012 | N.D. | N.D. | 238 | N.D. | N.D. | 1203 | 57.0 | 121 | 1353 | 274 |
| (16.9) | (3.81) | (4.55) | (−) | (10.8) | (−) | (1.73) | (20.8) | (−) | (−) | (10.0) | (−) | (−) | (11.9) | (0.40) | (1.34) | (3.78) | (6.91) | |
| Donor 1 ( + ) | 990 | 648 | 617 | 55.1 | 1097 | N.D. | 19.2 | 986 | N.D. | 45.8 | 267 | Trace | N.D. | 1260 | 59.7 | 120 | 1316 | 322 |
| (19.3) | (15.3) | (12.9) | (1.46) | (28.6) | (−) | (0.45) | (25.9) | (−) | (1.38) | (1.29) | (−) | (−) | (9.95) | (0.19) | (1.09) | (5.73) | (3.43) | |
| Donor 2 ( − ) | 1912 | 708 | 643 | N.D. | 1005 | N.D. | 91.4 | 751 | N.D. | N.D. | 222 | N.D. | N.D. | 1352 | 29.2 | 95.4 | 1278 | N.D. |
| (52.9) | (30.7) | (8.23) | (−) | (42.2) | (−) | (2.16) | (19.7) | (−) | (0.00) | (8.12) | (−) | (−) | (5.85) | (0.67) | (3.34) | (4.28) | (−) | |
| Donor 2 ( + ) | 2012 | 611 | 646 | 54.0 | 927 | N.D. | 106 | 748 | N.D. | Trace | 219 | Trace | N.D. | 1260 | 28.7 | 54.2 | 1283 | N.D. |
| (110) | (21.6) | (15.0) | (1.59) | (27.4) | (−) | (1.79) | (21.1) | (−) | (−) | (9.02) | (−) | (−) | (11.3) | (0.08) | (0.40) | (5.39) | (−) | |
| Bact 1 ( − ) | 378 | 135 | 677 | N.D. | 1004 | N.D. | 1323 | 1034 | N.D. | N.D. | Trace | N.D. | N.D. | 1177 | 85.9 | 41.0 | 1258 | 774 |
| (15.3) | (11.0) | (13.9) | (−) | (67.7) | (−) | (19.3) | (18.1) | (−) | (−) | (−) | (−) | (−) | (11.3) | (0.94) | (0.85) | (10.5) | (34.2) | |
| Bact 1 ( + ) | 1063 | 120 | 648 | 15645 | 939 | N.D. | 1351 | 1051 | N.D. | 6735 | Trace | Trace | N.D. | 1182 | 92.4 | 109 | 1360 | Trace |
| (26.7) | (1.76) | (12.1) | (359) | (4.62) | (−) | (15.5) | (10.1) | (−) | (219) | (−) | (−) | (−) | (6.10) | (1.41) | (4.07) | (3.30) | (−) |
Data are expressed as ng/mL (standard deviation shown in parentheses). Abbreviations are the same as in Table 1.
The “Blank” sample refers to non-incubated nutrient broth in the absence of ( + )-C/( − )-EC or bacteria. Six extractions were carried out to find out the endogenous levels of targeted phenolic acids. Ctl samples refers to negative control “Ctl ( − )” or positive control “Ctl ( + )”, containing pure nutrient broth incubated at 37 °C for 24 h; Bact 1 refers to a bacteria strain isolated from human feces. A plus sign ( + ) refers to the supplementation of grape polyphenols, ( + )-C and ( − )-EC.
N.D., not detected. Trace, trace amount, below the limit of quantification.
As shown in Table 3, gut bacteria from different human donors were able to degrade ( + )-C/( − )-EC, but showed varying capacities to generate PAMs. With respect to the precursor polyphenols, it is clear that microbiota from both healthy donors showed greater capacity in metabolizing ( + )-C/( − )-EC compared with the isolated bacteria culture. Almost all ( + )-C and ( − )-EC were metabolized by bacteria from two donors following 24 h fermentation, while 63% of ( + )-C and 84% of ( − )-EC remained in the broths treated with the bacterial isolate. Regarding the PAMs, the presence of endogenous phenolic acids was confirmed by examining the phenolic extract of blank nutrient broth (Fig. 3). Both the blank sample (Blank) and negative control sample (Ctl ( − )) were found to contain moderate levels of GA, 3, 4-diHBA, 3, 4-diHPAA, 4-HBA, HA, VA, 3-HBA, HVA, diHCA, p-CA, 3-HPPA and FA. Following incubation of broths containing ( + )-C and ( − )-EC with the complete microbiota collection or the isolated strain, there were significant production of GA, 3, 4-diHBA, HVA and slight increases in 4-HPVA and 3-HBA. By contrast, varying degrees of reduction in HA, VA and p-CA (Donor 1 only) were observed. Concentrations of 3, 4-diHPAA, diHCA, 3-HPPA and 4-HBA remained relatively constant in all samples regardless of the treatment applied. There was none or only trace amount of 3, 4-diHPPA, 3-HPAA and CA detected in samples. Considering the changes in the abundance of precursors and PAMs in bacterial broth before and after fermentation (Table 3), it appears that the GI microbiota from different donors possessed similar metabolic activity towards polyphenols but distinct capacity in generating PAMs. This also highlights the well-recognized inter-personal differences in gut microbiota that are responsible for the varied bioefficacies of orally ingested polyphenols in human subjects [35]. In addition, single bacterial isolate also demonstrated remarkable ability to metabolize ( + )-C and ( − )-EC into multiple PAMs, showing potential to be developed into “next-generation probiotics”. This new functional culture may assist in improving bioefficacy of polyphenols with low absorption in the upper GI tract, and also in generating bioavailable and bioactive phenolic metabolites with extended half-life and/or increased tissue deposition.
4. Conclusion
In this study, a high-throughput, sensitive and reproducible UHPLC-QqQ-MS/MS method was developed and validated for the identification and quantification of 16 microbial PAMs derived from ( + )-C and ( − )-EC. Optimization of the method was conducted by adjusting parameters in relation to dilution solvent, LC conditions and MS behaviors. Results obtained from optimization and validation studies indicated that the developed LC-MS method was highly selective, sensitive, accurate and reproducible for the detection and quantitation of target phenolic compounds even at trace levels in complex biological matrices. The analytical method developed was then applied to in vitro fermentation studies incorporating grape polyphenols and human gut microbiota.
To the best of our knowledge, this is the most sensitive and comprehensive LC-MS/MS-based method for targeted analysis of GI microbial PAMs derived from grape polyphenols. The accurate measuring of major microbial phenolic metabolites can lead to a comprehensive elucidation of the metabolic pathway of polyphenols in human intestine and a better interpretation of the beneficial effects of consuming grape-derived products. In addition, this new analytical method can be of great value to polyphenol-associated metabolomics investigations for the discovery of reliable biomarkers in animals and human subjects exposed to polyphenol-rich botanical preparations.
Supplementary Material
Acknowledgements
This work was done as part of the Core B research program in NIH Botanical Center with funds provided by NIH ODS and NCCAM IP50AT008661–01 to Mt. Sinai and with Supplemental Funding by the NIH to Mt. Sinai (Grant number 0254–3831-4609). We thank Agilent Instruments for providing the state-of-art analytical instrumentation as part of a scientific collaborative agreement to Rutgers that was used to conduct this study. Funds were also provided by the New Jersey Agricultural Experiment Station Hatch Project Number NJ12158. The authors are very thankful to Dr. Mario Ferruzzi in North Carolina State University for his constructive suggestions. We thank Daniel Giurleo for preparing phenolic extracts from the biological samples.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jchromb.2018.09.014.
References
- [1].Yang CS, et al. , Bioavailability issues in studying the health effects of plant polyphenolic compounds, Mol. Nutr. Food Res 52 (S1) (2008) S139–S151. [DOI] [PubMed] [Google Scholar]
- [2].Halliwell B, Dietary polyphenols: good, bad, or indifferent for your health? Cardiovasc. Res 73 (2) (2007) 341–347. [DOI] [PubMed] [Google Scholar]
- [3].Pasinetti GM, et al. , Roles of resveratrol and other grape-derived polyphenols in Alzheimer’s disease prevention and treatment, Biochim. Biophys. Acta (BBA) - Mol. Basis Dis 1852 (6) (2015) 1202–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Del Rio D, et al. , Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases, Antioxid. Redox Signal 18 (14) (2012) 1818–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Heleno SA, et al. , Bioactivity of phenolic acids: metabolites versus parent compounds: a review, Food Chem. 173 (2015) 501–513. [DOI] [PubMed] [Google Scholar]
- [6].Sanchez-Patán F, et al. , Gut microbial catabolism of grape seed flavan-3-ols by human faecal microbiota. Targetted analysis of precursor compounds, intermediate metabolites and end-products, Food Chem. 131 (1) (2012) 337–347. [Google Scholar]
- [7].Crazier A, Jaganath IB, Clifford MN, Dietary phenolics: chemistry, bioavailability and effects on health, Nat. Prod. Rep 26 (8) (2009) 1001–1043. [DOI] [PubMed] [Google Scholar]
- [8].van Duynhoven J, et al. , Metabolic fate of polyphenols in the human superorganism, Proc. Natl. Acad. Sci 108 (Supplement 1) (2011) 4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scalbert A, et al. , The food metabolome: a window over dietary exposure, Am. J. Clin. Nutr 99 (6) (2014) 1286–1308. [DOI] [PubMed] [Google Scholar]
- [10].van Dorsten FA, et al. , The metabolic fate of red wine and grape juice polyphenols in humans assessed by metabolomics, Mol. Nutr. Food Res 54 (7) (2010) 897–908. [DOI] [PubMed] [Google Scholar]
- [11].Zamora-Ros R, et al. , Measuring exposure to the polyphenol metabolome in observational epidemiologic studies: current tools and applications and their limits, Am. J. Clin. Nutr 100 (1) (2014) 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wishart DS, Emerging applications of metabolomics in drug discovery and precision medicine, 15 (2016) 473. [DOI] [PubMed] [Google Scholar]
- [13].Margalef M, et al. , A rapid method to determine colonic microbial metabolites derived from grape flavanols in rat plasma by liquid chromatography-tandem mass spectrometry, J. Agric. Food Chem 62 (31) (2014) 7698–7706. [DOI] [PubMed] [Google Scholar]
- [14].Ou K, Gu L, Absorption and metabolism of proanthocyanidins, J. Funct. Foods 7 (2014) 43–53. [Google Scholar]
- [15].Kalili KM, de Villiers A, Recent developments in the HPLC separation of phenolic compounds, J. Sep. Sci 34 (8) (2011) 854–876. [DOI] [PubMed] [Google Scholar]
- [16].Ostrowski W, et al. , Mass spectrometric behavior of phenolic acids standards and their analysis in the plant samples with LC/ESI/MS system, J. Chromatogr. B 967 (2014) 21–27. [DOI] [PubMed] [Google Scholar]
- [17].González RR, et al. , Development and validation of an ultra-high performance liquid chromatography-tandem mass-spectrometry (UHPLC-MS/MS) method for the simultaneous determination of neurotransmitters in rat brain samples, J. Neurosci. Methods 198 (2) (2011) 187–194. [DOI] [PubMed] [Google Scholar]
- [18].Tabasco R, et al. , Effect of grape polyphenols on lactic acid bacteria and bifido-bacteria growth: resistance and metabolism, Food Microbiol 28 (7) (2011) 1345–1352. [DOI] [PubMed] [Google Scholar]
- [19].Urpi-Sarda M, et al. , Epicatechin, procyanidins, and phenolic microbial metabolites after cocoa intake in humans and rats, Anal. Bioanal. Chem 394 (6) (2009) 1545–1556. [DOI] [PubMed] [Google Scholar]
- [20].VanMiddlesworth BJ, Dorsey JG, Quantifying injection solvent effects in reversed-phase liquid chromatography, J. Chromatogr. A 1236 (2012) 77–89. [DOI] [PubMed] [Google Scholar]
- [21].Keunchkarian S, et al. , Effect of sample solvent on the chromatographic peak shape of analytes eluted under reversed-phase liquid chromatogaphic conditions, J. Chromatogr. A 1119 (1–2) (2006) 20–28. [DOI] [PubMed] [Google Scholar]
- [22].Inbaraj BS, et al. , Simultaneous determination of phenolic acids and flavonoids in Lycium barbarum Linnaeus by HPLC-DAD-ESI-MS, J. Pharm. Biomed. Anal 51 (3) (2010) 549–556. [DOI] [PubMed] [Google Scholar]
- [23].Dai J, Mumper RJ, Plant phenolics: extraction, analysis and their antioxidant and anticancer properties, Molecules 15 (10) (2010) 7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stalikas CD, Extraction, separation, and detection methods for phenolic acids and flavonoids, J. Sep. Sci 30 (18) (2007) 3268–3295. [DOI] [PubMed] [Google Scholar]
- [25].Irakli MN, et al. , Development and validation of an HPLC-method for determination of free and bound phenolic acids in cereals after solid-phase extraction, Food Chem. 134 (3) (2012) 1624–1632. [DOI] [PubMed] [Google Scholar]
- [26].Magiera S, Baranowska I, Kusa J, Development and validation of UHPLC-ESI-MS/MS method for the determination of selected cardiovascular drugs, polyphenols and their metabolites in human urine, Talanta 89 (2012) 47–56. [DOI] [PubMed] [Google Scholar]
- [27].Gasperotti M, et al. , Development of a targeted method for twenty-three metabolites related to polyphenol gut microbial metabolism in biological samples, using SPE and UHPLC-ESI-MS/MS, Talanta 128 (2014) 221–230. [DOI] [PubMed] [Google Scholar]
- [28].Chen S, et al. , Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry, Anal. Chem 85 (17) (2013) 8326–8333. [DOI] [PubMed] [Google Scholar]
- [29].FDA, US Department of Health and Human Services, Draft Guidance for Industry: Bioanalytical Method Validation (Revised), (2013).
- [30].Jiang H, et al. , Systematic evaluation of supported liquid extraction in reducing matrix effect and improving extraction efficiency in LC-MS/MS based bioanalysis for 10 model pharmaceutical compounds, J. Chromatogr. B 891–892 (2012) 71–80. [DOI] [PubMed] [Google Scholar]
- [31].Matuszewski BK, Constanzer ML, Chavez-Eng CM, Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC – MS/MS, Anal. Chem 75 (13) (2003) 3019–3030. [DOI] [PubMed] [Google Scholar]
- [32].Little JL, Wempe MF, Buchanan CM, Liquid chromatography-mass spectrometry/mass spectrometry method development for drug metabolism studies: examining lipid matrix ionization effects in plasma, J. Chromatogr. B 833 (2) (2006) 219–230. [DOI] [PubMed] [Google Scholar]
- [33].Hurtado-Gaitán E, et al. , A focused multiple reaction monitoring (MRM) quantitative method for bioactive grapevine stilbenes by ultra-high-performance liquid chromatography coupled to triple-quadrupole mass spectrometry (UHPLC-QqQ), Molecules 22 (3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Caprioli G, et al. , Optimization of an extraction method for the simultaneous quantification of sixteen polyphenols in thirty-one pulse samples by using HPLC-MS/MS dynamic-MRM triple quadrupole, Food Chem. 266 (2018) 490–497. [DOI] [PubMed] [Google Scholar]
- [35].Manach C, et al. , Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies, Am. J. Clin. Nutr 81 (1) (2005) 230S–242S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.